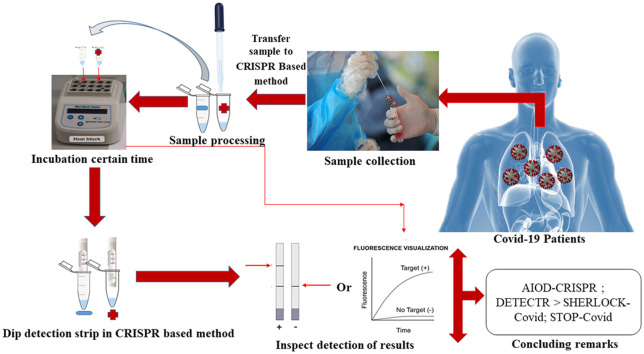
Abstract
The recent pandemic of novel coronavirus disease (COVID-19) has spread globally and infected millions of people. The quick and specific detection of the nucleic acid of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) remains a challenge within healthcare providers. Currently, quantitative reverse transcription-polymerase chain reaction (RT-qPCR) is the widely used method to detect the SARS-CoV-2 from the human clinical samples. RT-qPCR is expensive equipment and needs skilled personnel as well as lengthy detection time. RT-qPCR limitation needed an alternative healthcare technique to overcome with a fast and cheaper detection method. By applying the principles of CRISPR technology, several promising detection methods giving hope to the healthcare community. CRISPR-based detection methods include SHERLOCK-Covid, STOP-Covid, AIOD-CRISPR, and DETECTR platform. These methods have comparative advantages and drawbacks. Among these methods, AIOD-CRISPR and DETECTR are reasonably better diagnostic methods than the others if we compare the time taken for the test, the cost associated with each test, and their capability of detecting SARS-CoV-2 in the clinical samples. It may expect that the promising CRISPR-based methods would facilitate point-of-care (POC) applications in the CRISPR-built next-generation novel coronavirus diagnostics.
Keywords: Nucleic acid, SARS-CoV-2, COVID-19, CRISPR, Diagnostics
Graphical Abstract
1. Introduction
The first case of SARS-CoV-2 infection was found in December 2019 in Wuhan, China. The coronavirus spread to different parts of the world, and then the WHO declared this alarming situation as a COVID-19 pandemic disease [1], [2]. Many countries were overwhelmed by the sudden increase of an unknown infectious disease. The healthcare providers were trying to understand this disease by collecting information from different sources [3], [4]. The symptoms of SARS-CoV-2 infections are being varied widely with pneumonia and life-threatening complications such as multisystem organ failure, acute respiratory distress syndrome, and so on [5], [6], [7]. People with a pre-existing condition, especially aged people with respiratory or cardiovascular problems, diabetes, are the most vulnerable to SARS-CoV-2 infection [6], [7]. People with severe illness can develop acute respiratory distress syndrome (ARDS) may lead to death.
Furthermore, the infected individual with no symptoms caused the transmission of SARS-CoV-2 to others [8], [9]. The treatment of Covid-19 patients composed of supportive care, such as invasive and non-invasive oxygen support and treatment with antibiotics, due to the unavailability of an effective vaccine in the community [10], [11]. To provide immediate treatment, a quick diagnosis of SARS-CoV-2 has surged a demand. For this purpose, several laboratories, including the US CDC, developed reverse transcription-polymerase chain reaction (RT-qPCR) assays to detect the novel coronavirus within 4–6 h [12]. RT-qPCR has been considered a gold standard for viral nucleic acid diagnosis because of its high sensitivity and specificity [13]. However, RT-qPCR has some limitations, such as it requires expensive equipment and well-trained personnel. Contrary, serology tests are rapid and need minimal equipment. However, the utility is limited for diagnosing acute SARS-CoV-2 infection, including taking days to weeks to develop an antibody response [14]. The necessity for instrument-free nucleic acid detection technologies has led to the evolution of multiple isothermal amplification methods, including Recombinase Polymerase Amplification (RPA) [15] and Loop-mediated isothermal amplification (LAMP) [16]. These methods are advantageous compared to conventional PCR methods because of their simplicity, quickness, and low cost. Recently, the CRISPR system was adapted for the precise and quick sensing of nucleic acids [17], [18], [19]. For instance, a crRNA-target binding activated Cas nucleases (including Cas12a, Cas12b, and Cas13a) have distinctive enzymatic properties so that they can cleave surrounding single-stranded DNA for detection of nucleic acids [20], [21], [22], [23], [24]. In different circumstances, specific high-sensitivity enzymatic reporter unlocking (SHERLOCK) [25] and All-In-One dual CRISPR (AIOD-CRISPR) [26] systems have been developed by combining RPA pre-amplification with Cas13a and Cas12a respectively for the ultra-sensitive detection of nucleic acid. On the other hand, CRISPR-Cas12a combining with RT-LAMP and LAMP have been developed DNA Endonuclease-Targeted CRISPR Trans Reporter (DETECTR) [16] and STOP test for COVID-19 (STOP-Covid) [27] platforms, aside from the RPA method, respectively. In this review, we discussed CRISPR-mediated nucleic acid detection for COVID-19 in clinical samples.
2. Methods
Different publication databases have been searched. The top searched engine was Google Scholar, PubMed, and Science direct. We explored some keywords including, novel coronavirus, SARS-CoV-2, Nucleic acid, and COVID-19 detection. The emphasis gave to the recently published articles.
3. Discussion
3.1. Overview of SARS-CoV-2 detection
To mitigate the spreading of SARS-CoV-2 is detection. Without proper detection, the spreading of viral infections will continuously grow at its rate. Recently asymptomatic infected person number is increasing. Different countries follow different tests, such as PCR testing, antigen, and antibody testing, to diagnose SARS-CoV-2. Even incorrect detection enabling spread aided by the false-negative test result.
3.2. Current SARS-CoV-2 diagnostics
Xu et al. [28] reviewed currently available SARS-CoV-2 diagnostics. Some common diagnostics like, in viral culture test, respiratory samples used as specimen type where testing time requires 3–7 days. The main problem in this detection is time-consuming and cannot be used widely in clinical settings. In real-time PCR, respiratory samples, stool, and blood use as specimen types. This method is widely used for laboratory confirmation. By this method, testing time is only 1.5–3 h. But in this detection, there is a possibility of getting a false-negative result. As real-time PCR NAAT, isothermal amplification test also has some probability of getting false-negative results. However, it is faster than the real-time PCR, only 0.5–2 h. In serological testing, serum, plasma, and blood use as specimen types, and its most important characteristic is its less time, within 15–45 min detection process is done. But it has a drawback as sometimes its cross-reaction with other subtypes of coronavirus. Last, the point-of-care test is the fastest testing procedure; it takes only 5–30 min. In this detection, the respiratory sample is used as a specimen, provides rapid actionable information with good sensitivity.
3.3. Background on CRISPR-cas9 technology
CRISPR-Cas9 is a unique technology that enables geneticists and medical researchers to edit parts of the genome by removing, adding, or altering sections of the DNA sequence. It is a rapidly evolving technology that has revolutionized biological research. It represents a family of DNA repeats in most archaeal (~ 90%) and bacterial (~40%) genomes. Regulation of endogenous gene expression, epigenome editing, live-cell labeling of chromosomal loci, edition of single-stranded RNA, and high-throughput gene screening performed using CRISPR-cas9 technology [29]. In our review, we described this technology for nucleic acid detection of SARS-CoV-2.
3.4. The mechanism of CRISPR-based nucleic acid detection
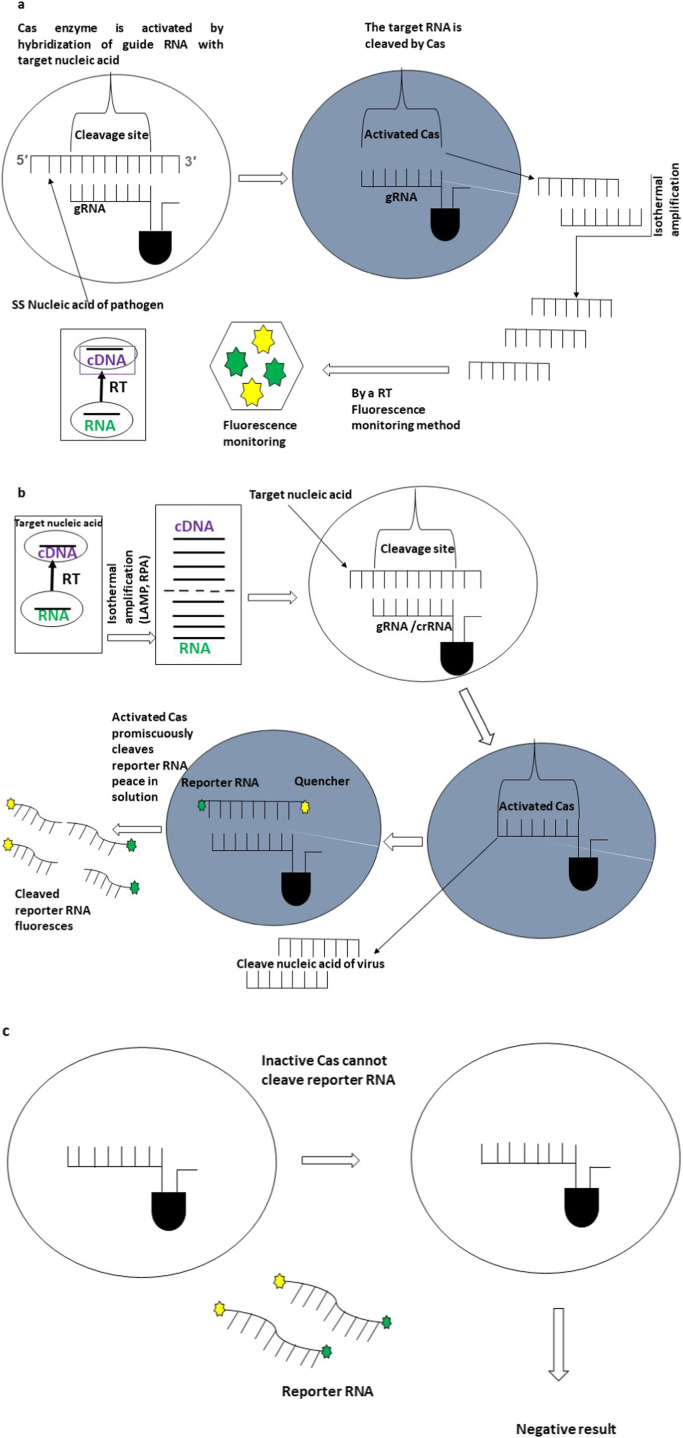
The CRISPR system detects desired DNA or RNA sequences with CRISPR-Cas enzymology and amplification process in disease diagnosis platforms. A Cas endonuclease and a single-stranded guide RNA (sgRNA) form the CRISPR-Cas system. The three stages which can describe the CRISPR-based disease diagnosis are the adaptation stage, amplification processes such as RPA or LAMP/RT-LAMP, and target detection ( Fig. 1) [30].
Fig. 1.
Mechanism of CRISPR-based nucleotide detection. a Fluorescent tracking detection of nucleotide-based on specific binding and cutting activity of CRISPR/Cas9. Firstly, crRNA identifies the target nucleic acid for the spacer of the CRISPR Cas system. After identifying and detecting the sequence, specific Cas nucleases cut the target nucleic acid. After that, the vast amount of target fragments of nucleic acids was synthesized with the assistance of an isothermal amplification process. At last, fluorescent tracking finds out the target nucleotides in detection platforms. b Collateral cleavage based on CRISPR/Cas effectors. In the first instance, the vast amount of target nucleic acids synthesized with an isothermal amplification process, including LAMP, RPA, RT-LAMP. After that, crRNA identifies the target nucleic acid for the spacer of the CRISPR Cas system. After identifying and detecting the target sequence, specific Cas nucleases (Cas12a, AapCas12b, or Cas13) cut the target nucleic acid. Finally, the reporters cut by the activated Cas effectors to liberate visual fluorescent signals within detection platforms. c Cleavage of the reporter RNA and fluorescence generation do not occur without the Cas enzyme activation.
3.5. Techniques of CRISPR for nucleic acid detection
The CRISPR technology and the variety of CRISPR-associated Cas enzymes provide genomes editing and nucleic acid detection. The identification of the nucleic acid of SARS-CoV-2 is crucial for confirming the COVID-19 disease. CRISPR-based methods such as SHERLOCK-Covid, STOP-Covid, AIOD-CRISPR, and DETECTR lateral flow establish to detect the nucleic acid in the clinical samples.
3.6. SHERLOCK-Covid assay
Kellner et al. (2019) [31] established a CRISPR-based diagnostic system that uses recombinase polymerase amplification (RPA), CRISPR-Cas13 complex fluorescence reporter to detect RNA or DNA from clinical samples. The pre-amplification step of SHERLOCK introduces the T7 RNA polymerase promoter. During detection reaction, T7 RNA polymerase generates nucleic acid for Cas13, which induces single-stranded nucleic acid trans-cleavage as a collateral effect. The fluorescence reporter signal was released after RNA cleavage and subsequently detected through the detector during post-incubation. SHERLOCK can provide results in < 1 h.
3.7. STOP-Covid assay
Joung et al. (2020) [27] developed the STOP-Covid method to detect SARS-CoV-2 requires a SHERLOCK pair AapCas12b-crRNA complex generated gRNA to bind the related sites that are imminent to the recognition sites of LAMP amplification primers in the target sequence. This complex load into the solution containing LAMP amplification primers, lateral flow reporter, taurine, isothermal amplification buffer, target sequence, etc. During incubation of the STOP-Covid system in one pot, the sample lysed to extract RNA by Quick Extract at 95 °C for 5 min or 60 °C for 10 min (Inhibition of proteinase K) and followed by LAMP with CRISPR- mediated detection step. The activation of AapCas12b in the virus's presence within two minutes in the lateral flow strip confirmed the collateral activity. This Assay only takes 40 (fluorescence readout) or 70 min (lateral flow readout) to detect the N gene (of the virus) of about 100 copies of the viral genome in a nasopharyngeal swab or saliva sample.
3.8. AIOD-CRISPR assay
Recently, Ding et al. (2020) [26] conducted an All-In-One dual CRISPR-Cas12a (termed AIOD-CRISPR) assay method to identify SARS-CoV-2. Two individual crRNAs bind two related sites of Cas12a to come up with a pair of Cas12a-crRNA complexes that are individually prepared before being loaded into the answer containing different components, including two RPA primers, ssDNA-FQ reporters, recombinase, ssDNA binding protein, strand-displacement DNA polymerase, and target sequence. RPA amplification is first initiated and exposes the binding sites of the Cas12a-crRNA complexes thanks to the strand displacement when incubating the AIOD-CRISPR system in one pot at 37 °C. After binding Cas12a-crRNA complexes in the target sites, Cas12a endonuclease is activated and subsequently cleaves the adjacent ssDNA-FQ reporters to inflict fluorescence incessantly trigger CRISPR-Cas12a based collateral cleavage activity. Therefore, AIOD-CRISPR- based detection could detect SARS‐CoV‐2 N RNA and the human RPP30 gene in 40 min incubation (activated after reaction temperature arrives at 37 °C).
3.9. DETECTR lateral flow assay
Recently Broughton, James et al. (2020) [32] developed a CRISPR-Cas12a based assay called DETECTR to detect SARS-CoV-2 from extracted patient sample RNA. Nasopharyngeal or oropharyngeal swabs in universal transport medium used as a sample. This Assay depends on RT-LAMP for reverse transcription and isothermal amplification using loop-mediated amplification for extracted sample RNA at 62˚ C for 20–30 min and subsequently generate nucleic acid for Cas12a, which induces robust single-stranded DNA trans-cleavage as a collateral effect. The virus's presence is confirmed by the collateral activity of Cas12a, which cleaves reporter molecules at 37˚C for 10 min, visualized by a fluorescent reader or lateral flow strip. This method only takes 30–40 min and detects the virus's E gene and N gene and the human RNase P gene to control about ten copies/µL viral nucleic acids.
3.10. Comparison of variant CRISPR system-mediated detection of SARS-Cov-2
Rapid detection of nucleic acids is integral to applications in clinical diagnostics and biotechnology [31]. For the quick and sensitive detection of desired cDNA or RNA sequences of SARS-CoV-2, CRISPR-based diagnosed platforms such as AIOD-CRISPR, STOP-Covid, SHERLOCK-Covid, DETECTR have been established recently (Table 1). These assays have the potential to be comparable with RT-qPCR because the later consistently count on costly equipment and compliant personnel why it is not satisfactory for the point-of-care diagnostic application. But when it comes to the AIOD-CRISPR assay, the detection results can be visually judged based on the fluorescence and color change of reaction solutions, making it more suitable for the point-of-care application. AIOD-CRISPR has some peculiar advantages compare with previously reported CRISPR-based nucleic acid detection [23], [25], [33], [34], [35], [36]. Hence AIOD-CRISPR employing an entire single reaction system may well be a quick, powerful, ultra-specific, versatile, and nearly single-molecule sensorial isothermal nucleic acid detection method [26].
Table 1.
Potential strategies for variant CRISPR system-mediated detection of Nucleic acid of SARS-CoV-2.
| CRISPR variants | Reaction Assay | Features | Target | Advantages | Disadvantages | Reference |
|---|---|---|---|---|---|---|
| AIOD- CRISPR | One-pot reaction assay | A pair of Cas12a-crRNA complexes, ssDNA-FQ reporters, SSB protein, recombinase, strand-displacement DNA polymerase, and target sequences | SARS- CoV- 2 N RNA target as well as human RPP30 gene | Rapid, visual, Ultrasensitive, and specific detection to SARS-CoV-2 | [25] | |
| STOP-Covid | One-step reaction assay | AapCas12b-crRNA, LAMP amplification primers, Lateral flow reporter, Taurine, Isothermal amplification buffer and target sequence | N gene of the virus | Rapid, point-of-care diagnosis of SARS-CoV-2 | Moderate cost and no-modular heater capable | [26] |
| SHERLOCK-Covid | Two-step reaction assays | RPA, CRISPR–Cas13 complex, a fluorescence reporter, and T7 RNA polymerase | Detection of DNA or RNA | Highly sensitive, specific, and rapid | Time-consuming, increase complexity and cross-contamination, quantification difficulties | [26], [30] |
| DETECTR | One-pot reaction | RT-LAMP, Cas12-crRNA, Lateral flow strip and fluorescence reporters | E gene and N gene of virus and the human RNase P gene | Visual and faster, low-cost, and accurate | [31], [50] |
Additionally, AIOD-CRISPR without pre-amplification can detect as low as 4.6 copies of RNA targets and 1.2 copies of DNA targets in 40 min incubation. Likewise, SHERLOCK is rapid, ultra-sensitive, and precise, low-cost platform detection over other detection like TaqMan qPCR [31]. The multi-step nucleic acid amplification process is the potential limitation of SHERLOCK, including fluid handling steps, complicating their deployment outside clinical labs, affecting accurate target quantification [30]. Alongside STOP-Covid, which has an additional feature (such as united reaction) over SHERLOCK. Notwithstanding STOP-Covid provides sensitivity and does not a require sample extraction process and may be performed at a single temperature with one fluid handling step, it should further develop into a low-cost assay, and a modular heater should be installed to streamline the workflow and simplify the test to be used [27]. CRISPR-Cas12a based Assay [25], [33], [37], [38] for identifying SARS-CoV-2 called SARS-CoV-2 DETECTR, which performs simultaneous, reverse transcription RT-LAMP [16]. Therefore, DETECTR technology develops a quick (~ 30 min), low-cost test that contains a limit of detection of 10 copies/µL of viral nucleic acid in the sample, which does not need expensive lab instrumentation, and availability currently restricted to public health laboratories [32]. Thus, all CRISPR system-mediated detection of SARS-CoV-2 represents a promising platform for developing point-of-care COVID-19 diagnostics and has the potential to play a crucial role in effective test-trace-isolate measures to contain the COVID-19 pandemic and reopen society. These straightforward CRISPR-nucleic acid sensing systems encompass excellent potential in enabling CRISPR-based next-generation molecular diagnostics towards quantitation, point-of-care, and digital analysis.
3.11. Real-time problems in the clinical setting for detecting SARS-CoV-2
RT-PCR is a widely used method for detecting the infection of SARS-CoV-2 based on the nasopharyngeal specimen [39]. Several studies have shown that this method has excessive rates of false-negative results and the conversion of nucleic acid is additionally prolonged here [40], [41], [42]. The sensitivity of PCR-based SARS-CoV-2 was reported by Miller et al., 2020 as 70–90%, which become agitated with the appeared date of symptoms [43]. The sensitivity was shown to be decreased by 30.9% in an alternative study on VIASURE RT-PCR Kit in clinical samples [44]. Extensive distribution of RT-PCR-based SARS-CoV-2 diagnosis worldwide would face difficulties regarding the high-cost instruments, mass peoples with sufficient skills for performing the test and analyzing the results, and the required lengthy turnaround times. Due to the high worldwide demands of SARS-CoV-2 diagnosis, it would be a significant issue to supply the necessary instruments and reagents of RT-PCR with proper validation for manufacturers and buyers (especially for developing countries). The complications mentioned above related to the sensitivity and clinical setting of RT-PCR facilities indicate a disturbing situation in checking COVID-19. Therefore, other possibilities should be developed that can replace RT-PCR and overcome the complications as well. Nowadays isothermal amplification-based methods (LAMP and RT-LAMP) are used as an alternative to RT-PCR diagnostics as it utilizes basic instruments and inexpensive reagents [45], [46]. However, these methods possess a few limitations, including high rates of false-positive results, lacking specificity, and the disadvantage of adjusting these methods for well-organized diagnostics as POC [47], [48]. As a result, following CRISPR based method such as SHERLOCK [49], AIOD-CRISPR [26], DETECTR [50], STOP-COVID [27], and iSCAN [51] have been developed to get over the complications comprising false results, low sensitivity & poor specificity, lengthy turnaround times as well as clinical settings. Table 1 presents the comprehensive features and merits of CRISPR-based techniques. Generally, the SARS-CoV-2 viral load in the early start point of symptoms was > 10^3/µL in the clinical specimen [52]. SherlockTM CRISPR SARS-CoV-2 kit relied on RT-LAMP and CRISPR complex to detect SARS-CoV-2 at 6.75 copies/µL VTM as the limit of detection (LoD) within 1 h, and both sensitivity and specificity were found to be 100% by applying 5x LoD, 3x LoD, and 2x LoD from nasopharyngeal specimens [49]. SARS-CoV-2 with 95% and 100% positive and negative predictive agreement can be visually diagnosed by DETECTR assay, respectively. Needful LoD and time of DETECTR assay is 10 copies/µL and close to 45 min, respectively, whereas CDC developed qRT-PCR required 1–3.2 copies/µL and 4 h [50]. The results of one pot (37 °C) AIOD-CRISPR assay can be visually judged, showing high sensitivity and specificity, detecting 1.2 copies and 4.6 copies of target DNA and RNA, respectively, within 40 min of incubation time [26]. Based on the viral load (> 10^3/µL) in clinical samples, the LoD of CRISPR-based diagnostics is far enough to diagnose SARS-CoV-2 effectively. Hence, CRISPR-based assays could be employed as a successful substitute to remaining established methods of SARS-CoV-2 diagnosis due to their demand for basic instrumentation, little turnaround times, inexpensive reagents, very low viral load, high specificity, and sensitivity. Finally, Sherlock Biosciences, Inc Company commercialized the SherlockTM CRISPR SARS-CoV-2 kit approved under FDA Emergency Use Authorization (EUA) [49].
3.12. Rapid and precise CRISPR based detection approach of SARS-CoV-2
The existing methods for nucleic acid detection are precise. However, the sample manipulation and expensive machinery with duration slow down the viral detection. Therefore, a rapid, inexpensive, and sensitive diagnostic test urgently develops viral detection for disease monitoring [53]. Although CRISPR is a robust technique well-known for its use in gene editing, it has also use for viral nucleic acid detection. This recent development in the field of molecular diagnostics is a benediction for COVID-19 outbreaks. CRISPR/Cas-based diagnostic is a better option to develop rapid non-laboratory-based detection for SARS-CoV-2 and other pathogens [54]. Some Cas enzymes target DNA, single-effector RNA-guided RNases, such as Cas13a, which can be reprogrammed with CRISPR RNAs (crRNAs) to provide a vast platform for specific RNA sensing. Upon recognition of its RNA target, activated Cas13a engages in 'Collateral' cleavage of nearby non-targeted RNAs, RNAs, which allows Cas13a to detect the presence of a specific RNA in vivo by triggered programmed cell death or in vitro by nonspecific degradation of labeled RNA [55]. The SHERLOCK based on nucleic acid amplification and Cas13a-mediated collateral cleavage of a reporter RNA allows for real-time, rapid, and specific target detection with attomole sensitivity [56]. A recent study reported that CRISPR-Cas13a also targeting SARS-CoV-2 spike protein and provides a fast design pipeline for a CRISPR-Cas13a-based antiviral tool through cleavage effect on S against SARS-CoV-2 [57].
The development of an amplification-free CRISPR-Cas13a assay enables a mobile phone microscope to detect SARS-CoV-2 from nasal swab RNA. The Assay achieved ˜100 copies/µL sensitivity within 30 min of measurement time and accurately detected pre-extracted RNA from a set of positive clinical samples under 5 min [58]. However, this study only demonstrates a proof-of-concept for sensitive and rapid SARS-CoV-2 RNA detection by a mobile phone-based device. A light-up transcription method of CRISPR-Cas13a enables the detection of SARS-CoV-2 and its mutated variants. It accommodated to sense as low as 82 copies of SARS-CoV-2. Specifically, it allowed to strictly discriminate key mutation of the SARS-CoV-2 variant, D614G, which may induce higher epidemic and pathogenic risk [59]. CRISPR-based approaches have some advantages over the RT-PCR-based method: Isothermal signal amplification obviating the need for thermocycling, rapid turnaround time, single nucleotide target specificity, integration with accessible and easy to use reporting formats such as lateral flow strips, and no requirements for complex laboratory infrastructure [50]. Hence, CRISPR-based approaches expect to use for the rapid, sensitive, and visual detection of SARS-CoV-2.
3.13. Drawbacks of using CRISPR based detection
The CRISPR-based detection is slightly less sensitive than the most used RT-PCR test [60]. For achieving a similar sensitivity, the CRISPR-based test must go through the technology's future advancement. In the case of point-of-care use at home, this diagnostic may face ethical challenges [61]. The regulatory agency must give point-of-care approval to overcome this limitation.
4. Conclusion
DETECTR method is 90% sensitive and 100% specific for identifying the novel coronavirus in respiratory swab and economical because it does not require expensive lab instrumentation. It only takes 30–40 min and detects the virus's E gene and N gene (and the human RNase P gene as a control) of about ten copies/ µL viral nucleic acid [31]. Likewise, the AIOD-CRISPR assay possesses high specificity detecting N gene from SARS-CoV-2 (and human RPP30 gene for control) while avoiding cross-reactions for non-SARS-CoV-2 targets. It detects as low as 1.2 copies of DNA targets and 4.6 copies of RNA targets in 40 min incubation without cDNA preparation [25]. STOP-Covid Assay takes 40 (fluorescence readout) or 70 min (lateral flow readout) minutes. It detects the N gene of about 100 copies of the viral genome in saliva or nasopharyngeal swabs samples. While the STOP-Covid Assay does not require sample extraction and can perform at a single temperature with one fluid handling step, it should be developed into a low-cost assay. A modular heater should be installed to streamline the workflow and simplify the test to be used [26]. The above discussion can say that AIOD-CRISPR, and DETECTR are comparatively better diagnostic tests than the other two regarding the time taken for the test, cost associated, and their capability to detect SARS-CoV-2 in clinical samples.
Ethics approval and consent to participate
Not applicable.
Funding
This work did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Consent for publication
All authors have given consent to publish the work.
Availability of data and materials
All data generated or analyzed during this study were included in this article.
CRediT authorship contribution statement
MRR: Conceptualization, designed, and coordinated the work; MAH, MM, FAM, and MSAM: have written the manuscript sections; MSAM, MAIA, and AF: analyzed and checked the manuscript thoroughly. All authors read and approved the manuscript.
Competing interest
The authors declare no conflict of interest. The authors are solely responsible for the writing and content of this article.
Acknowledgments
Not applicable.
References
- 1.Cucinotta D., Vanelli M. WHO declares COVID-19 a pandemic. Acta Biomed. 2020;91:157–160. doi: 10.23750/abm.v91i1.9397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Spinelli A., Pellino G. COVID-19 pandemic: perspectives on an unfolding crisis. Br. J. Surg. 2020;107:785–787. doi: 10.1002/bjs.11627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fauci A.S., Lane H.C., Redfield R.R. Covid-19-navigating the uncharted. N. Engl. J. Med. 2020;382:1268–1269. doi: 10.1056/NEJMe2002387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mahase E., Kmietowicz Z. Covid-19: doctors are told not to perform CPR on patients in cardiac arrest. BMJ. 2020;368:1282. doi: 10.1136/bmj.m1282. [DOI] [PubMed] [Google Scholar]
- 5.Rodriguez-Morales A.J., Cardona-Ospina J.A., Gutiérrez-Ocampo E., Villamizar-Peña R., Holguin-Rivera Y., Escalera-Antezana J.P., Alvarado-Arnez L.E., Bonilla-Aldana D.K., Franco-Paredes C., Henao-Martinez A.F., Paniz-Mondolfi A., Lagos-Grisales G.J., Ramírez-Vallejo E., Suárez J.A., Zambrano L.I., Villamil-Gómez W.E., Balbin-Ramon G.J., Rabaan A.A., Harapan H., Dhama K., Nishiura H., Kataoka H., Ahmad T., Sah R., Latin American Network of Coronavirus Disease -COVID- Research (LANCOVID-). Electronic address h. Clinical, laboratory and imaging features of COVID-19: a systematic review and meta-analysis. Travel. Med. Infect. Dis. 2020;34 doi: 10.1016/j.tmaid.2020.101623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weiss P., Murdoch D.R. Clinical course, and mortality risk of severe COVID-19. Lancet. 2020;395:1014–1015. doi: 10.1016/S0140-6736(20)30633-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu C., Chen X., Cai Y., Xia J., Zhou X., Xu S., Huang H., Zhang L., Zhou X., Du C., Zhang Y., Song J., Wang S., Chao Y., Yang Z., Xu J., Zhou X., Chen D., Xiong W., Xu L., Zhou F., Jiang J., Bai C., Zheng J., Song Y. Risk factors associated with acute respiratory distress syndrome and death in patients with coronavirus disease 2019 pneumonia in Wuhan, China. JAMA Intern. Med. 2020;180:934–943. doi: 10.1001/jamainternmed.2020.0994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bai Y., Yao L., Wei T., Tian F., Jin D.Y., Chen L., Wang M. Presumed asymptomatic carrier transmission of COVID-19. JAMA. 2020;323:1406–1407. doi: 10.1001/jama.2020.2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rothe C., Schunk M., Sothmann P., Bretzel G., Froeschl G., Wallrauch C., Zimmer T., Thiel V., Janke C., Guggemos W., Seilmaier M., Drosten C., Vollmar P., Zwirglmaier K., Zange S., Wölfel R., Hoelscher M. Transmission of 2019-nCoV infection from an asymptomatic contact in Germany. N. Engl. J. Med. 2020;382:970–971. doi: 10.1056/NEJMc2001468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Onder G., Rezza G., Brusaferro S. Case fatality rate and characteristics of patients dying in relation to COVID-19 in Italy. JAMA. 2020;323:1775–1776. doi: 10.1001/jama.2020.4683. [DOI] [PubMed] [Google Scholar]
- 11.Poston J.T., Patel B.K., Davis A.M. Management of critically ill adults with COVID-19. JAMA. 2020;323:1839–1841. doi: 10.1001/jama.2020.4914. [DOI] [PubMed] [Google Scholar]
- 12.Centers for Disease Control and Prevention. Real-time RT-PCR Panel for Detection 2019- nCoV (US Centers for Disease Control and Prevention, 2020), 〈https://www.cdc.gov/coronavirus/2019-ncov/lab/rt-PCR-detection-instructions.html〉.
- 13.Tahamtan A., Abdollah A. Real-time RT-PCR in COVID-19 detection: issues affecting the results. Expert Rev. Mol. Diagn. 2020;20:453–454. doi: 10.1080/14737159.2020.1757437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang W., Du R.H., Li B., Zheng X.S., Yang X.L., Hu B., Wang Y.Y., Xiao G.F., Yan B., Shi Z.L., Zhou P. Molecular and serological investigation of 2019-nCoV infected patients: implications of multiple shedding routes. Emerg. Microbes Infect. 2020;9:386–389. doi: 10.1080/22221751.2020.1729071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Piepenburg O., Williams C.H., Stemple D.L., Armes N.A. DNA detection using recombination proteins. PLoS. Biol. 2006;4:204. doi: 10.1371/journal.pbio.0040204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Notomi T., Okayama H., Masubuchi H., Yonekawa T., Watanabe K., Amino N., Hase T. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000;28:63. doi: 10.1093/nar/28.12.e63. e63-e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bhattacharyya R.P., Thakku S.G., Hung D.T. Harnessing CRISPR effectors for infectious disease diagnostics. ACS Infect. Dis. 2018;4:1278–1282. doi: 10.1021/acsinfecdis.8b00170. [DOI] [PubMed] [Google Scholar]
- 18.Batista A.C., Pacheco L.G.C. Detecting pathogens with Zinc-Finger, TALE, and CRISPR-based programmable nucleic acid-binding proteins. J. Microbiol. Methods. 2018;152:98–104. doi: 10.1016/j.mimet.2018.07.024. [DOI] [PubMed] [Google Scholar]
- 19.Li Y., Li S., Wang J., Liu G. CRISPR/Cas systems towards next-generation biosensing. Trends Biotechnol. 2019;37:730–743. doi: 10.1016/j.tibtech.2018.12.005. [DOI] [PubMed] [Google Scholar]
- 20.Li S.-Y., Cheng Q.X., Liu J.K., Nie X.Q., Zhao G.P., Wang J. CRISPR-Cas12a has both cis and trans-cleavage activities on single-stranded DNA. Cell Res. 2018;28:491–493. doi: 10.1038/s41422-018-0022-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen J.S., Ma E., Harrington L.B., Da Costa M., Tian X., Palefsky J.M., Doudna J.A. CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science. 2018;360:436–439. doi: 10.1126/science.aar6245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Abudayyeh O.O., Gootenberg J.S., Konermann S., Joung J., Slaymaker I.M., Cox D.B., Shmakov S., Makarova K.S., Semenova E., Minakhin L., Severinov K., Regev A., Lander E.S., Koonin E.V., Zhang F. C2c2 is a single-component programmable RNA-guided RNA targeting CRISPR effector. Science. 2016;353:5573. doi: 10.1126/science.aaf5573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li L., Li S., Wu N., Wu J., Wang G., Zhao G., Wang J. HOLMESv2: a CRISPR-Cas12b-assisted platform for nucleic acid detection and DNA methylation quantitation. ACS Synth. Biol. 2019;8:2228–2237. doi: 10.1021/acssynbio.9b00209. [DOI] [PubMed] [Google Scholar]
- 24.Jeon Y., Choi Y.H., Jang Y., Yu J., Goo J., Lee G., Jeong Y.K., Lee S.H., Kim I.S., Kim J.S., Jeong C., Lee S., Bae S. Direct observation of DNA target searching and cleavage by CRISPRCas12a. Nat. Commun. 2018;9:1–11. doi: 10.1038/s41467-018-05245-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gootenberg J.S., Abudayyeh O.O., Lee J.W., Essletzbichler P., Dy A.J., Joung J., Verdine V., Donghia N., Daringer N.M., Freije C.A., Myhrvold C., Bhattacharyya R.P., Livny J., Regev A., Koonin E.V., Hung D.T., Sabeti P.C., Collins J.J., Zhang F. Nucleic acid detection with CRISPR-Cas13a/C2c2. Science. 2017;356:438–442. doi: 10.1126/science.aam9321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ding X., Yin K., Li Z., Liu C. All-in-one dual CRISPR-Cas12a (AIOD-CRISPR) assay: a case for rapid, ultrasensitive and visual detection of novel coronavirus SARS-CoV-2 and HIV virus. bioRxiv. 2020 doi: 10.1038/s41467-020-18575-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Joung J., Ladha A., Saito M., Segel M., Bruneau R., Huang M.W., Kim N.G., Yu X., Li J., Walker B.D., Greninger A.L., Jerome K.R., Gootenberg J.S., Abudayyeh O.O., Zhang F. Point-of-care testing for COVID-19 using SHERLOCK diagnostics. medRxiv. 2020 [Google Scholar]
- 28.Xu Yuzhong, Cheng Minggang, Chen Xinchun, Zhu Jialou. Current approaches in laboratory testing for SARS-CoV-2. Int. J. Infect. Dis. 2020;100:7–9. doi: 10.1016/j.ijid.2020.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Torres-Ruiz Raul, Rodriguez-Perales Sandra. CRISPR-Cas9 technology: applications and human disease modeling. Brief. Funct. Genom. 2017;16:4–12. doi: 10.1093/bfgp/elw025. (Oxford University Press) [DOI] [PubMed] [Google Scholar]
- 30.Jia F., Li X., Zhang C., Tang X. The expanded development and application of CRISPR system for sensitive nucleotide detection. Protein Cell. 2020;11:624–629. doi: 10.1007/s13238-020-00708-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kellner M.J., Koob J.G., Gootenberg J.S., Abudayyeh O.O., Zhang F. SHERLOCK: nucleic acid detection with CRISPR nucleases. Nat. Protoc. 2019;14:2986–3012. doi: 10.1038/s41596-019-0210-2. 2986-2912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Broughton J.P., Deng X., Yu G., Fasching C.L., Singh J., Streithorst J., Granados A., Sotomayor-Gonzalez A., Zorn K., Gopez A., Hsu E., Gu W., Miller S., Pan C.Y., Guevara H., Wadford D.A., Chen J.S., Chiu C.Y. Rapid detection of 2019 novel coronavirus SARS-CoV-2 using a CRISPR-based DETECTR lateral flow assay. medRxiv. 2020 [Google Scholar]
- 33.Chen J.S., Ma E., Harrington L.B., Da Costa M., Tian X., Palefsky J.M., Doudna J.A. CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science. 2018;360:436–439. doi: 10.1126/science.aar6245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yeh E.-C., Fu C.C., Hu L., Thakur R., Feng J., Lee L.P. Self-powered integrated microfluidic point-of-care low-cost enabling (SIMPLE) chip. Sci. Adv. 2017;3 doi: 10.1126/sciadv.1501645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhou W., Hu L., Ying L., Zhao Z., Chu P.K., Yu X.F. A CRISPR-Cas9-triggered strand displacement amplification method for ultrasensitive DNA detection. Nat. Commun. 2018;9:5012. doi: 10.1038/s41467-018-07324-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang B., Wang R., Wang D., Wu J., Li J., Wang J., Liu H., Wang Y. Cas12aVDet: a CRISPR/Cas12a-based platform for rapid and visual nucleic acid detection. Anal. Chem. 2019;91:12156–12161. doi: 10.1021/acs.analchem.9b01526. [DOI] [PubMed] [Google Scholar]
- 37.Chiu C. Cutting-Edge Infectious Disease Diagnostics with CRISPR. Cell Host Microbe. 2018;23:702–704. doi: 10.1016/j.chom.2018.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Myhrvold C., Freije C.A., Gootenberg J.S., Abudayyeh O.O., Metsky H.C., Durbin A.F., Kellner M.J., Tan A.L., Paul L.M., Parham L.A., Garcia K.F., Barnes K.G., Chak B., Mondini A., Nogueira M.L., Isern S., Michael S.F., Lorenzana I., Yozwiak N.L., MacInnis B.L., Bosch I., Gehrke L., Zhang F., Sabeti P.C. Field-deployable viral diagnostics using CRISPR-Cas13. Science. 2018;360:444–448. doi: 10.1126/science. aas8836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mathuria J.P., Yadav R. Laboratory diagnosis of SARS-CoV-2-a review of current methods. J. Infect. Public Health. 2020;13:901–905. doi: 10.1016/j.jiph.2020.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Woloshin S., Patel N., Kesselheim A.S. False negative tests for SARS-CoV-2 infection-challenges and implications. N. Engl. J. Med. 2020;383:38. doi: 10.1056/NEJMp2015897. [DOI] [PubMed] [Google Scholar]
- 41.Long Q.X., Liu B.Z., Deng H.J., Wu G.C., Deng K., Chen Y.K., Liao P., Qiu J.F., Lin Y., Cai X.F., Wang D.Q. Antibody responses to SARS-CoV-2 in patients with COVID-19. Nat. Med. 2020;6:845–848. doi: 10.1038/s41591-020-0897-1. [DOI] [PubMed] [Google Scholar]
- 42.Xiao A.T., Tong Y.X., Zhang S. False negative of RT‐PCR and prolonged nucleic acid conversion in COVID‐19: Rather than recurrence. J. Med. Virol. 2020;92:1755–1756. doi: 10.1002/jmv.25855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Miller T.E., Garcia Beltran W.F., Bard A.Z., Gogakos T., Anahtar M.N., Astudillo M.G., Yang D., Thierauf J., Fisch A.S., Mahowald G.K., Fitzpatrick M.J. Clinical sensitivity and interpretation of PCR and serological COVID‐19 diagnostics for patients presenting to the hospital. FASEB J. 2020;34:13877–13884. doi: 10.1096/fj.202001700RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Matzkies L.M., Leitner E., Stelzl E., Assig K., Bozic M., Siebenhofer D., Mustafa M.E., Steinmetz I., Kessler H.H. Lack of sensitivity of an IVD/CE-labelled kit targeting the S gene for detection of SARS-CoV-2. Clin. Microbiol. Infect. 2020;26:1417–e1. doi: 10.1016/j.cmi.2020.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Niemz A., Ferguson T.M., Boyle D.S. Point-of-care nucleic acid testing for infectious diseases. Trends Biotechnol. 2011;29:240–250. doi: 10.1016/j.tibtech.2011.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mori Y., Notomi T. Loop-mediated isothermal amplification (LAMP): a rapid, accurate, and cost-effective diagnostic method for infectious diseases. J. Infect. Chemother. 2009;15:62–69. doi: 10.1007/s10156-009-0669-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chou P.H., Lin Y.C., Teng P.H., Chen C.L., Lee P.Y. Real-time target-specific detection of loop-mediated isothermal amplification for white spot syndrome virus using fluorescence energy transfer-based probes. J. Virol. Methods. 2011;173:67–74. doi: 10.1016/j.jviromet.2011.01.009. [DOI] [PubMed] [Google Scholar]
- 48.Nagai K., Horita N., Yamamoto M., Tsukahara T., Nagakura H., Tashiro K., Shibata Y., Watanabe H., Nakashima K., Ushio R., Ikeda M. Diagnostic test accuracy of loop-mediated isothermal amplification assay for Mycobacterium tuberculosis: systematic review and meta-analysis. Sci. Rep. 2016;6:1–10. doi: 10.1038/srep39090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sherlock BioSciences, SherlockTM CRISPR SARS-CoV-2 Kit, 2020. 〈https://www.fda.gov/media/137746/download〉, 1st November 2020.
- 50.Broughton J.P., Deng X., Yu G., Fasching C.L., Servellita V., Singh J., Miao X., Streithorst J.A., Granados A., Sotomayor-Gonzalez A., Zorn K. CRISPR–Cas12-based detection of SARS-CoV-2. Nat. Biotechnol. 2020;38:870–874. doi: 10.1038/s41587-020-0513-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ali Z., Aman R., Mahas A., Rao G.S., Tehseen M., Marsic T., Salunke R., Subudhi A.K., Hala S.M., Hamdan S.M., Pain A. iSCAN: An RT-LAMP-coupled CRISPR-Cas12 module for rapid, sensitive detection of SARS-CoV-2. Virus Res. 2020;288 doi: 10.1016/j.virusres.2020.198129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pan Y., Zhang D., Yang P., Poon L.L., Wang Q. Viral load of SARS-CoV-2 in clinical samples. Lancet Infect. Dis. 2020;20:411–412. doi: 10.1016/S1473-3099(20)30113-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Srivastava S., Upadhyay D.J., Srivastava A. Next-generation molecular diagnostics development by CRISPR/Cas tool: rapid detection and surveillance of viral disease outbreaks. Front. Mol. Biosci. 2020;7 doi: 10.3389/fmolb.2020.582499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xiong D., Dai W., Gong J., Li G., Liu N., Wu W., Pan J., Chen C., Jiao Y., Deng H. Rapid detection of SARS-CoV-2 with CRISPR-Cas12a. PLoS Biol. 2020;18 doi: 10.1371/journal.pbio.3000978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang W., Liu K., Zhang P., Cheng W., Li L., Zhang F., Yu Z., Li L., Zhang X. CRISPR-Based Approaches for Efficient and Accurate Detection of SARS-CoV-2. Lab. Med. 2021;52:116–121. doi: 10.1093/labmed/lmaa101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Joung J., Ladha A., Saito M., Kim N.-G., Woolley A.E., Segel M., Barretto R.P., Ranu A., Macrae R.K., Faure G. Detection of SARS-CoV-2 with SHERLOCK one-pot testing. N. Engl. J. Med. 2020;383:1492–1494. doi: 10.1056/NEJMc2026172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang L., Zhou J., Wang Q., Wang Y., Kang C. Rapid design and development of CRISPR-Cas13a targeting SARS-CoV-2 spike protein. Theranostics. 2021;11:649–664. doi: 10.7150/thno.51479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fozouni P., Son S., de León Derby M.D., Knott G.J., Gray C.N., D’Ambrosio M.V., Zhao C., Switz N.A., Kumar G.R., Stephens S.I. Amplification-free detection of SARS-CoV-2 with CRISPR-Cas13a and mobile phone microscopy. Cell. 2021;184:323–333. doi: 10.1016/j.cell.2020.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang Y., Zhang Y., Chen J., Wang M., Zhang T., Luo W., Li Y., Wu Y., Zeng B., Zhang K. Detection of SARS-CoV-2 and its mutated variants via CRISPR-Cas13-based transcription amplification. Anal. Chem. 2021;93:3393–3402. doi: 10.1021/acs.analchem.0c04303. [DOI] [PubMed] [Google Scholar]
- 60.〈https://www.scientificamerican.com/article/crispr-gene-editing-may-help-scale-up-coronavirus-testing/〉 (Accessed 4/2/2021).
- 61.〈https://www.clinicallabmanager.com/q-and-a/the-promise-of-crispr-based-diagnostics-274〉 (Accessed 4/2/2021).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study were included in this article.