Abstract
Background
Early identification of glioma molecular phenotypes can lead to understanding of patient prognosis and treatment guidance. We aimed to develop a multiparametric MRI texture analysis model using a combination of conventional and diffusion MRI to predict a wide range of biomarkers in patients with glioma.
Methods
In this retrospective study, patients were included if they (1) had diagnosis of gliomas with known IDH1, EGFR, MGMT, ATRX, TP53, and PTEN status from surgical pathology and (2) had preoperative MRI including FLAIR, T1c+ and diffusion for radiomic texture analysis. Statistical analysis included logistic regression and receiver-operating characteristic (ROC) curve analysis to determine the optimal model for predicting glioma biomarkers. A comparative analysis between ROCs (conventional only vs conventional + diffusion) was performed.
Results
From a total of 111 patients included, 91 (82%) were categorized to training and 20 (18%) to test datasets. Constructed cross-validated model using a combination of texture features from conventional and diffusion MRI resulted in overall AUC/accuracy of 1/79% for IDH1, 0.99/80% for ATRX, 0.79/67% for MGMT, and 0.77/66% for EGFR. The addition of diffusion data to conventional MRI features significantly (P < .05) increased predictive performance for IDH1, MGMT, and ATRX. The overall accuracy of the final model in predicting biomarkers in the test group was 80% (IDH1), 70% (ATRX), 70% (MGMT), and 75% (EGFR).
Conclusion
Addition of MR diffusion to conventional MRI features provides added diagnostic value in preoperative determination of IDH1, MGMT, and ATRX in patients with glioma.
Keywords: glioma, MR diffusion, multiparametric MRI, radiogenomics, texture analysis
Key Points.
Combination of T1c+, FLAIR and diffusion texture features can predict several glioma biomarkers.
Addition of diffusion MRI features significantly improved prediction for IDH1, MGMT and ATRX.
No texture features were identified to predict PTEN and TP53.
Importance of the Study.
In patients with glioma, our knowledge about the status of tumoral biomarkers has changed our approach in terms of histopathological classification with provided prognostic and therapeutic implications. Prior studies have assessed glioma biomarkers statuses using texture data from either conventional or diffusion MR imaging with some success. This study aims to construct a multiparametric model combining radiomic data from T1c+, FLAIR, and diffusion MR features to predict individual glioma biomarker statuses. Addition of MR diffusion to conventional MRI features provides added diagnostic value in preoperative determination of IDH1, MGMT, and ATRX in patients with glioma. Early and noninvasive recognition of these biomarkers would help neuro-oncologists to construct a more specific prognostic and treatment plan for patients with glioma.
Gliomas are the most common primary brain neoplasms accounting for roughly 40%–50% of all malignant primary central nervous system tumors.1 Despite advances in diagnosis and treatment, the prognosis of patients with high-grade gliomas or glioblastomas (GBM) remain dismal with 5-year survival rate of 5%–10%.2 As molecular phenotypes have been found to successfully predict prognosis and guide treatment options, there is increasing emphasis on identification of these biomarkers to better understand the pathophysiology of gliomas and to explore more specific targeted treatment options.
The earliest biomarkers to make up the genetic hallmarks of GBMs were upregulation of EGFR,3 mutations in TP53,4 and mutations in PTEN.5 Since then, methylation of the MGMT gene promoter was found to be a predictor of treatment outcome of temozolomide and radiotherapy.6 More recently, IDH1 mutation was found to be an independent positive prognostic biomarker with significantly longer progression-free survival and better treatment outcome for chemotherapy plus radiation compared to IDH1 wildtype.7 The WHO Classification of 2016 now further categorizes GBMs by IDH status and encourages routine testing for mutational status. Radiogenomic mapping has emerged as a promising noninvasive tool for predicting these biomarkers. To date, using conventional MRI sequences (T1c+/FLAIR), several investigators have found radiomic associations with IDH1 mutation,8,9MGMT methylation status,10,11EGFR amplification,12,13ATRX mutation,14,15PTEN deletion,16 and TP53 mutation17 with varying success. The ability to predict biomarker status via radiomics noninvasively is invaluable as large tissue specimens are often needed for accurate histopathological diagnosis and there are limited laboratories that can perform these tests. Radiomics may have the potential to provide complimentary information to overcome some of the limitations of histologic assessment as related to insufficient sampling or tumor heterogeneity.18 Furthermore, presurgical identification of these biomarkers can facilitate patient counseling and contribute to surgical planning and optimal patient management when complete molecular characterization is not possible.
In this study, we aimed to develop a multiparametric MRI texture analysis model using a combination of conventional (T1c+/FLAIR) and diffusion MRI to predict individual glioma biomarkers. We specifically investigated the potential added value of combining MR diffusion radiomics with conventional MRI in order to improve the accuracy of our predictive model.
Material and Methods
This retrospective study was approved by an institutional review board and informed consent was waived. Patients with initial diagnosis of glioma between January 2016 to September 2018 were reviewed. A total of 151 patients were reviewed (Supplementary Figure 1). Patients were included if they (1) had diagnosis of gliomas with known IDH1, EGFR, MGMT, ATRX, PTEN, and TP53 status from surgical pathology and (2) had preoperative MRI including FLAIR, T1c+, and diffusion within 30 days of biopsy or surgical resection. Patients were excluded if they had insufficient MR image quality (motion artifact, n = 8), prior surgeries involving the tumoral bed (n = 8) or treated with radiotherapy previously (n = 4). In addition, 20 patients were excluded due to lack of preoperative diffusion imaging. This yielded a final cohort of 111 patients. A total of 91 patients (82%) were categorized as training dataset for model development and 20 patients (18%) as testing dataset for assessment of predictive accuracy.
Histopathological Data
Tissue samples were obtained from patients undergoing targeted tissue biopsy or resection, as part of routine clinical care and diagnostic neuropathology and molecular evaluation. Immunohistochemistry was used to detect mutant status of IDH1 (specifically IDH1R132H immunoreactivity) and ATRX (loss of nuclear staining). Chromogenic in situ hybridization was used to assess EGFR amplification signal. Targeted next-generation sequencing was used to detect PTEN and TP53 mutational status. Pyrosequencing of bisulfite-treated genomic DNA (CpG sites 74–78, QIAGEN) was used to detect MGMT promoter methylation status.
Image Acquisition
MR imaging was obtained using 7 MRI scanners (2 Skyra 3T and 2 Aera 1.5T from Siemens Healthineers, Erlangen Germany; 2 Signa 1.5T and one Discovery 3T from GE Healthcare) within our Radiology Department. Image acquisition was performed using a standardized preoperative brain tumor MRI protocol within our radiology department including: FLAIR (TR/TE/TI, 8000–12,000/98–130/2400–2700 ms, voxel size: 0.5 × 0.5 × 1 mm3), DWI (TR/TE: 4025–4600/65–82 ms, with b values of 0 and 1000 s/mm2, voxel size: 0.9 × 0.9 × 5.0 mm3) and post-contrast T1W imaging (TR/TE, 600–1800/9–19 ms, voxel size: 0.5 × 0.5 × 1 mm3). A total volume of 0.1 mmol/kg of gadobenate dimeglumine was injected intravenously for post-contrast T1W imaging.
Image Analysis
Using our training dataset (n = 91 patients), image analysis was performed by a commercially available FDA-approved software (Olea Sphere software, Olea Medical SAS). Automatic preprocessing was standardized for each case involving intensity normalization, resampling, and discretization. Since MR images were obtained using different MRI scanners from 2 vendors and with different magnetic fields, a normalization step was implemented to normalize images by centering at the mean with standard deviation using all gray values in the image. The resampling grid was aligned to the input origin enabling in-plane resampling. Size and number of bins was set to 25 and 64, respectively, for every case standardizing the process of making histogram and discretion of the image gray level. T1c+, FLAIR, and diffusion images (ADC/b1000) were coregistered on each examination using a 6-df transformation and a mutual information cost function.
Tumor segmentation was performed manually on every slice that the tumor was visualized using FLAIR images. This was performed by a trained radiologist and under supervision of a board certified neuroradiologist. Subsequently, a VOI was generated encompassing the entire region of FLAIR hyperintensity and overlaid onto coregistered T1c+ and diffusion datasets for radiomic texture analysis (Supplementary Figure 2).
A total of 92 radiomic features were assessed. These included 19 first-order metrics, such as the mean, standard deviation, skewness, and kurtosis, and second-order metrics including 23 gray level run length matrix (GLCM), 16 gray level run length matrix (GLRLM), 15 gray level size zone matrix (GLSZM), 5 neighboring gray tone difference matrix (NGTDM), and 14 gray level dependence matrix (GLDM). Details of the definitions and calculations of these features have previously been reported.19–23 Texture feature extraction through Olea sphere software was in compliance with the image biomarker standardization initiative with the above 92 features categorized into (1) histogram features, which included grey intensity or brightness information of the lesion, (2) form factor features, which describe the shape and compactness of the lesions, and (3) texture features, which includes the remainder of the second-order metrics as above, and has been cited in prior studies.24,25
Statistical Analysis
Statistical analysis was performed using Matlab R2019b and Statistics and Machine Learning Toolbox (The MathWorks, Inc.) and SAS 9.4M6 (TS1M6) 2020 (SAS Institute Inc.). Ninety-two texture features were obtained from each imaging sequence (T1c+, FLAIR, ADC, and b1000) resulting in a total of 368 features for each patient in our training cohort. One-way analysis of variance for each imaging parameter (n = 368) was performed with each biomarker (IDH1 [wildtype vs mutated], MGMT [methylated vs unmethylated], EGFR [amplified vs nonamplified], ATRX [wildtype vs mutated], TP53 [wildtype vs mutated], and PTEN [wildtype vs mutated] as the independent variable. From the texture feature means that differed with statistical significance (P < .05) between biomarker positivity, Least Absolute Shrinkage and Selection Operator (LASSO)26 regularization was employed to select contributing variables to the models, thereby reducing potential risk of overfitting and increased interpretation. The significant contributing variables were then entered into a stepdown logistic regression analysis. A stepwise method was used to avoid collinearity because redundant variables were omitted. A 10-fold cross-validation scheme was used for evaluation of the training cohort, where 90% of the data was randomly assigned into the training cohort and 10% used for validation. This process was repeated 10 times.
Receiver-operating characteristic (ROC) curves were generated and area under the curve (AUC) was estimated for cross-validated models utilizing conventional MRI (T1c+, FLAIR) features first and then by addition of MRI-diffusion features. A comparative analysis between ROCs (conventional vs conventional + diffusion) was performed using nonparametric methods described by DeLong et al.27 Optimal thresholds were determined to maximize sensitivity and specificity for each biomarker utilizing the Youden’s index. The final constructed model was applied to a testing dataset to calculate the accuracy of biomarker prediction.
Results
Clinical Characteristics of Patient Population
Our final patient cohort consisted of a total of 111 patients (Table 1). The mean ± standard deviation of age (years) was 57 ± 15 with median of 59. Sixty-four patients were male and 47 were female. There were a total of 92 patients with GBM and 19 patients with lower-grade glioma including grade II glioma (n = 7) and grade III glioma (n = 12). There were 19 nonenhancing tumors and 92 enhancing tumors (Table 1). The demographic data including age, sex, tumor grade, and enhancement status are grouped based on biomarker mutation status and summarized in Table 1.
Table 1.
Demographic Data for Each Glioma Biomarker
| Status | N (%) | Age (mean ± SD) | Sex (M/F) | Enhancement (%) | WHO Grade (II/III/IV) | |
|---|---|---|---|---|---|---|
| Total | 111 (100) | 57 ± 15 | 64/47 | 92 (83) | 7/12/92 | |
| IDH1 | ||||||
| Wild-type | 88 (79) | 61 ± 11* | 48/40 | 86 (98) | 0/0/88 | |
| Mutant | 23 (21) | 40 ± 11 | 16/7 | 6 (26) | 7/12/4 | |
| MGMT | ||||||
| Nonmethylated | 59 (47) | 58 ± 12 | 30/29 | 53 (90) | 4/2/53 | |
| Methylated | 52 (53) | 55 ± 15 | 34/18 | 39 (75) | 3/10/39 | |
| EGFR | ||||||
| Wild-type | 76 (68) | 56 ± 14 | 39/37 | 59 (78) | 7/12/57 | |
| Amplified | 35 (32) | 59 ± 12 | 25/10 | 33 (94) | 0/0/35 | |
| ATRX | ||||||
| Wild-type | 92 (83) | 60 ± 11* | 50/42 | 86 (93) | 3/2/87 | |
| Mutant | 19 (17) | 39 ± 9 | 14/5 | 6 (32) | 4/10/5 | |
| TP53 | ||||||
| Wild-type | 84 (76) | 59 ± 13 | 44/40 | 74 (88) | 4/6/74 | |
| Mutant | 27 (24) | 47 ± 15 | 20/7 | 18 (67) | 3/6/18 | |
| PTEN | ||||||
| Wild-type | 100 (90) | 56 ± 14 | 60/40 | 81 (81) | 7/12/81 | |
| Mutant | 11 (10) | 63 ± 10 | 4/7 | 11 (100) | 0/0/11 |
*Statistically significant (P < .05).
Model Development in Training Cohort (n = 91)
IDH1.—
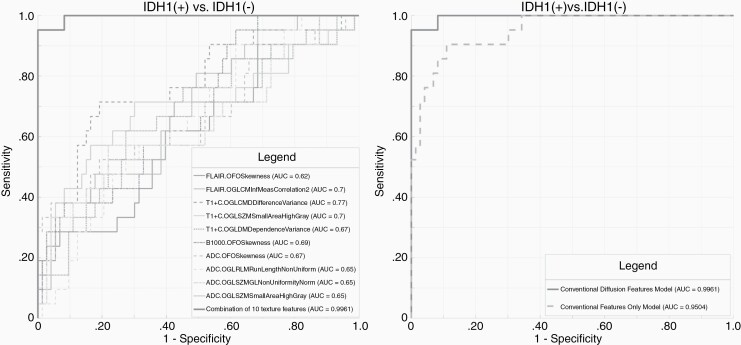
Following LASSO regularization and logistic regression analysis, a total of 10 texture features from conventional MR imaging remained as significant contributors in our predictive model with resultant AUC of 0.95 (Supplementary Table 1). After addition of diffusion data, a combination of 5 conventional and 5 diffusion MR features remained as significant contributors (Supplementary Table 2), improving the AUC to 1.0 (Figure 1). The addition of diffusion data significantly (P = .03) increased the predictive performance for IDH1 (Table 2). The AUC, sensitivity, specificity, and threshold for the conventional versus conventional + diffusion model are summarized in Table 2.
Figure 1.
Receiver-operating characteristic (ROC) curves for prediction of IDH1 mutational status. (A) Combined model constructed with a combination of 5 conventional and 5 diffusion MR texture features with AUC: 1.0. (B) Comparative analysis of ROCs shows significant (P = .03) improvement in prediction of IDH1 status in the combined model (conventional + diffusion MR) in comparison to the conventional only model. Parameters (n = 10) included in the final model are GLCM informal measure correlation 2 (T1c+), first-order skewness (T1c+), GLCM difference variance (T1c+), GLSZM small area high gray (T1c+), GLDM dependence variance (T1c+), first-order skewness (b1000), first-order skewness (ADC), GLRLM run length non-uniformity (ADC), GLSZMGL non-uniformity normalized (ADC), and GLSZM small area high gray (ADC).
Table 2.
Prediction Performance for Glioma Biomarkers Through Optimal Combination of Conventional and Diffusion MR Features
| T1c+/FLAIR | T1c+/FLAIR + Diffusion | ROC Comparison | |
|---|---|---|---|
| (AUC/Sens/Spec/Acc/threshold) | (AUC/Sens/Spec/Acc/threshold) | P value | |
| IDH1 | 0.95/67/85/76/0.33 | 1/87/71/79/0.76 | .03 |
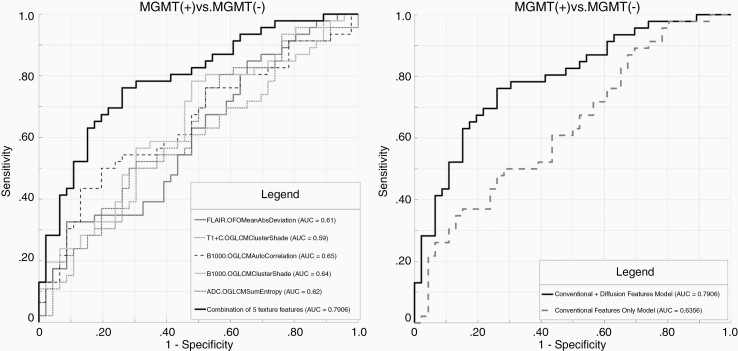
| MGMT | 0.64/41/61/51/0.56 | 0.79/70/65/67/0.51 | .006 |
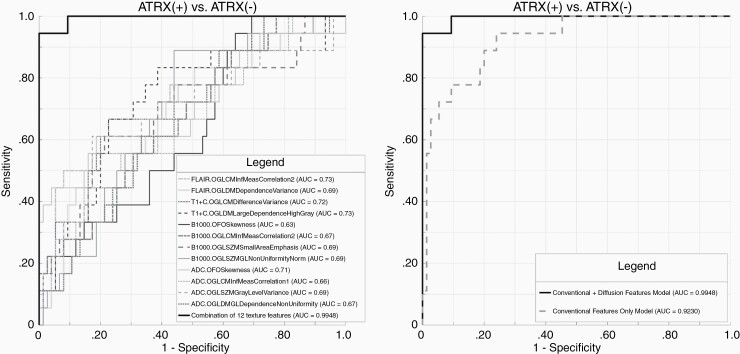
| ATRX | 0.92/78/76/77/0.1 | 0.99/72/88/80/0.64 | .01 |
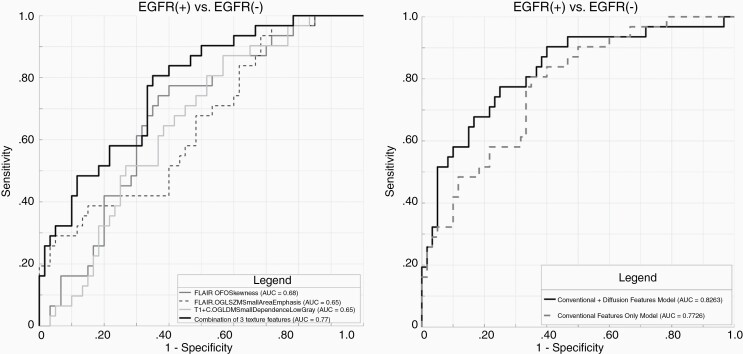
| EGFR | 0.77/74/62/68/0.3 | 0.83/65/68/66/0.38 | .17 |
| TP53 | * | Noncontributory | NA |
| PTEN | * | Noncontributory | NA |
*No variables remained as significant contributors following logistic regression analysis.
MGMT.—
Following LASSO regularization and logistic regression analysis, only a total of 2 conventional imaging features remained as significant contributors with overall AUC of 0.64 (Supplementary Table 1). With the addition of diffusion features, a combination of 2 conventional and 3 diffusion MR features (Supplementary Table 2), resulted in significant (P = .006) improvement in predicting model, increasing the AUC to 0.79 (Figure 2). The AUC, sensitivity, specificity, and threshold for the conventional versus conventional + diffusion model are summarized in Table 2.
Figure 2.
Receiver-operating characteristic (ROC) curves for prediction of MGMT methylation status. (A) Combined model constructed with a combination of 2 conventional and 3 diffusion MR texture features with AUC: 0.79. (B) Comparative analysis of ROCs shows significant (P = .006) improvement in prediction of MGMT methylation status in combined model (conventional + diffusion MR) in comparison to conventional only model. Parameters (n = 5) included in the final model are first-order mean absolute deviation (FLAIR), GLCM cluster shade (T1c+), GLCM autocorrelation (b1000), GLCM cluster shade (b1000), and GLCM sum entropy (ADC).
ATRX.—
A total of 6 conventional imaging features remained as significant contributors in our predictive model with resultant AUC of 0.92 (Supplementary Table 1). After incorporating diffusion features, the final model consisted of a combination of 4 conventional and 8 diffusion features (Supplementary Table 2). Addition of diffusion features resulted in significant (P = 0.01) improvement in predictive performance with an AUC of 0.99 (Figure 3). The AUC, sensitivity, specificity, and threshold for the conventional versus conventional + diffusion model are summarized in Table 2.
Figure 3.
Receiver-operating characteristic (ROC) curves for prediction of ATRX mutational status. (A) Combined model constructed with a combination of 4 conventional and 8 diffusion MR texture features with AUC: 0.99. (B) Comparative analysis of ROCs shows significant (P = 0.01) improvement in prediction of ATRX mutational status in combined model (conventional + diffusion MR) in comparison to conventional only model. Parameters (n = 12) included in the final model are GLCM informal measure correlation 2 (FLAIR), GLDM dependence variance (FLAIR), GLCM difference variance (T1c+), GLDM large dependence high gray (T1c+), first-order skewness (b1000), GLCM informal measure correlation 2 (b1000), GLSZM small area emphasis (b1000), GLSZMGL non-uniformity normalized (b1000), first-order skewness (ADC), GLCM informal measure correlation 1 (ADC), GLSZM gray level variance (ADC), and GLDMGL dependence non-uniformity (ADC).
EGFR.—
A total of 3 conventional imaging features remained as significant contributors in our predictive model with resultant AUC of 0.77 (Supplementary Table 1). After integrating diffusion features, there was no statistically significant (P = .17) improvement in diagnostic performance (Figure 4). The AUC, sensitivity, specificity, and threshold for the model constructed from conventional MRI features are summarized in Table 2.
Figure 4.
Receiver-operating characteristic (ROC) curves for prediction of EGFR amplification. (A) Final model was constructed with 3 conventional imaging features with AUC: 0.77. (B) Addition of diffusion features did not result in significant (P = .17) improvement in prediction of EGFR amplification. Parameters (n = 3) included in the final model are first-order skewness (FLAIR), GLSZM small area emphasis (FLAIR), and GLDM small dependence low gray (T1c+).
TP53.—
Following LASSO regularization and logistic regression analysis, no texture features remained as significant contributors in our predictive model.
PTEN.—
Following LASSO regularization and logistic regression analysis, no texture features from conventional MRI remained as significant contributors in our predictive model.
Independent Testing Cohort (n = 20)
The predictive accuracy of the validated models was tested in a total of 20 patients whose imaging data were not used for model development. The breakdown of biomarkers for this group were the following: IDH1 wildtype/mutant (14/6), ATRX wildtype/mutant (16/4), MGMT nonmethylated/methylated (10/10), and EGFR amplification nondetected/detected (16/4). The overall accuracy of the final (combined conventional-diffusion) models in predicting biomarkers in the test group was 80% for IDH1 (16/20 were correctly identified), 70% for ATRX (14/20 were correctly identified), 70% for MGMT (14/20 were correctly identified), and 75% for EGFR (15/20 were correctly identified).
Discussion
Our results showed that constructed multiparametric model from MRI radiomics features can identify IDH1, ATRX, MGMT, and EGFR in preoperative MRI scans of patients with glioma.
We specifically demonstrated that addition of diffusion data to FLAIR and T1c+ can significantly improve predictive performance for IDH1, MGMT, and ATRX, with the highest incremental value achieved for prediction of MGMT. MR feature analysis in our study did not contribute in determination of TP53 or PTEN mutational status.
The AUC/diagnostic accuracy of our constructed cross-validated model in prediction of IDH1 status was 0.95/75.8% from conventional MRI features, and was significantly improved to 1.0/79.3% after addition of MR diffusion features. Prior multimodal radiomic studies have predicted IDH1 status with AUCs ranging from 0.86 to 0.90.8,9,17,28 Within LGGs, Eichinger et al.29 demonstrated IDH1 status prediction with AUC of 0.92 using DWI features. IDH1 wildtype status has been associated with poor survival outcome regardless of WHO grade.30 Furthermore, aggressive surgical tumor resection has not been shown to provide survival benefit, specifically in IDH1 wildtype gliomas.31 Therefore, preoperative identification of IDH1 status can play an important role with prognostic and treatment implications.
Within anaplastic astrocytomas, ATRX mutation is a favorable prognostic biomarker associating with longer survival outcome and ATRX wildtype has been found to associate with recurrence.32 To date, only one prior study has assessed radiomic prediction of ATRX status.14 They demonstrated predictability with AUC of 0.94 in LGGs using T2-weighted images. Interestingly, a recent study by Ren et al.15 showed radiomic prediction of co-occurrence of mutations in IDH1 and ATRX with AUC of 0.93 by combining texture features from FLAIR and DWI images in LGGs. The AUC/diagnostic accuracy of our constructed model in prediction of ATRX status was 0.92/76.9% from conventional MRI features, and was significantly improved to 0.99/80.1% after addition of MR diffusion features. A strong predictive performance is promising as ATRX mutation is becoming increasingly recognized as an important prognostic biomarker and is now incorporated in the decision-making algorithm for differentiating oligodendroglial and astrocytic gliomas in the 2016 WHO classification.
For prediction of MGMT methylation status, we showed a modest predictive performance for the conventional MRI model with AUC of 0.64 and diagnostic accuracy of only 51.1%. The predictive performance was significantly improved in the combined model after addition of diffusion MRI features with resultant AUC of 0.79 and overall diagnostic accuracy of 67.4%. Prior studies have shown prediction of MGMT status with AUC as high as 0.85.10,11 Methylation of MGMT gene has been associated with longer overall survival and favorable prognostic indicator of response to temozolomide and radiotherapy.6 However, subsequent studies have reported conflicting results of the prognostic implication of MGMT methylation independent of therapy.33MGMT promoter methylation has been associated with mitotic counts and Phospho-histone-H3 values as measures of cellular proliferation.34 Diffusion MRI provides information about extracellular‐space tortuosity, tissue cellularity, and the integrity of cellular membranes and, therefore, has been used to draw association with MGMT status with some success.35 It is therefore plausible that addition of diffusion texture features likely exploited the existing differences between MGMT-methylated versus nonmethylated groups to explain the highest incremental added value in prediction accuracy obtained by adding diffusion to conventional features in our study.
The AUC/diagnostic accuracy of our constructed model in prediction of EGFR was 0.77/68% from conventional MRI features with no significant incremental improvement after addition of diffusion data. EGFR signal amplification has been shown to be a common feature of GBMs36 and is associated with aggressive phenotype and poor prognosis.3 Li et al.12 showed excellent radiomic prediction of EGFR amplification with AUC of 0.95 in LGGs. However, there is scarcity of data regarding radiomic prediction of EGFR amplification status in GBMs.
Finally, we showed no radiomic prediction of TP53 or PTEN status in either the conventional or combined model. TP53 is one of the most commonly deregulated genes in cancers and its pathway is deregulated in up to 85% of gliomas.37 To date, only Zhang et al.17 have assessed radiomic association with TP53 mutation reporting AUC of 0.95 in LGGs through multimodal combination of features from T1, T2, and FLAIR images. PTEN is a tumor suppressor gene significantly altered in 30%–40% of GBM and strongly associated with poor survival.16 The loss of PTEN function has been mechanistically linked to metastasis and a lack of response to radiotherapy and chemotherapy.2 Only one study to date, Li et al.,16 has assessed radiomic prediction of PTEN status showing excellent AUC of 0.93 by combining texture features from T1 and T2 images.
Establishing an accurate glioma biomarker prediction through radiogenomic approach enables noninvasive prediction of prognosis and contributes to treatment planning. Several studies have assessed individual glioma biomarker status using texture data from either conventional or diffusion imaging, however, only a few studies9,38 have combined conventional and diffusion texture data to predict for biomarker status. Qin et al.39 combined these features but only in relation to glioma grade without assessing individual glioma biomarkers. Understanding the mutation status of individual glioma markers is critical to guiding treatment.
Our study has several limitations. First, it was a retrospective study with potential for unknown bias. We acknowledge that our patient cohort had a skewed distribution consisting predominantly of GBMs. A prospective study would also allow us to recruit relatively balanced distribution of biomarker status although not all biomarkers statutes can be evenly distributed in the same cohort. Another challenge is the heterogeneous nature of gliomas, especially in GBMs, which vary across individual patients and spatially within each tumor. Thus, biomarker profiles may vary depending on the site of biopsy even within the same tumor, and comprehensive biomarker landscape may not be captured by biopsy alone. As biopsy results were used as gold standard in this study, the innate heterogeneity can introduce inconsistency and potentially inaccuracy which may underestimate the accuracy of our prediction algorithm. Our inability to develop models to predict TP53 or PTEN should therefore be interpreted in the context of this limitation. Techniques assessing tumor purity and multiple gene expression profile40 have been used to remedy this limitation, although not available for every patient in our study. Another limitation similar to other radiomic studies is the risk of overfitting considering a large number of variables included. We tried to mitigate this by adopting LASSO regularization followed by logistic regression to select contributing variables while minimizing the potential risk of overfitting and collinearity.41 Finally, although preprocessing steps such as signal normalization and resampling were performed to mitigate the effect of image variability related to different MR scanners (n = 7) with magnetic fields and vendors, our study was a single center study, external testing of the developed model through multi-institutional collaboration will improve the generalizability and clinical utility of our model.
It should be noted that tumor segmentation was performed by using the entire volume of T2-FLAIR to increase inclusivity. Segmentation on enhancing tumor only as being reported in prior studies,10,17 would have resulted in exclusion of 19 out of 111 patients in our study. Furthermore, similar to prior reports,8,11,39,42 we included the entire tumor volume on T2 FLAIR images including the cystic and necrotic components to make our approach more generalizable and limit manual segmentation variability. However, texture data may differ when compared with studies excluding these components.8,10,24 Further investigation is needed to assess whether inclusion or exclusion of these components may yield significant difference in biomarker prediction.
In conclusion, the described multiparametric MR texture model from combining conventional and diffusion features can predict individual glioma biomarker status in preoperative gliomas. In particular, addition of MR diffusion to conventional MRI features provided significant added diagnostic value in determination of IDH1, MGMT, and ATRX status.
Supplementary Material
Acknowledgments
We thank the Pathology Department at Mount Sinai Hospital for collaboration through sharing histopathological data. We thank the Institute for Health Care Delivery Sciences, specifically Nicole Zubizarreta, for statistical assistance and guidance.
Funding
This study was supported by NCI Cancer Center Support Grant P30 CA196521-01 and partially supported by R01NS106229 (N.M.T).
Authorship Statement. Study design: S.K., N.M.T., J.H., F.K., A.H., D.H., and K.N. Data collection: S.K., N.M.T., Y.S., J.H., F.K., and A.H. Statistical analysis: A.B., N.Z., and K.N. Manuscript writing: S.K., K.N., and N.S.
Conflicts of interest statement. K.N.: Olea Medical, Consultant, none for others.
References
- 1. Wrensch M, Minn Y, Chew T, Bondy M, Berger MS. Epidemiology of primary brain tumors: current concepts and review of the literature. Neuro Oncol. 2002;4(4):278–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Buckner JC. Factors influencing survival in high-grade gliomas. In Brandes AA, ed.: Seminars in Oncology. Vol 30. W.B. Saunders; 2003:10–14. [DOI] [PubMed] [Google Scholar]
- 3. Perry A, Aldape KD, George DH, Burger PC. Small cell astrocytoma: an aggressive variant that is clinicopathologically and genetically distinct from anaplastic oligodendroglioma. Cancer. 2004;101(10):2318–2326. [DOI] [PubMed] [Google Scholar]
- 4. Zhang X, Tian Q, Wang L, et al. Radiomics strategy for molecular subtype stratification of lower-grade glioma: detecting IDH and TP53 mutations based on multimodal MRI. J Magn Reson Imaging. 2018;48(4):916–926. [DOI] [PubMed] [Google Scholar]
- 5. Verhaak RG, Hoadley KA, Purdom E, et al. ; Cancer Genome Atlas Research Network . Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17(1):98–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Stupp R, Mason WP, van den Bent MJ, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352(10):987–996. [DOI] [PubMed] [Google Scholar]
- 7. Wick W, Roth P, Hartmann C, et al. Long-term analysis of the NOA-04 randomized phase III trial of sequential radiochemotherapy of anaplastic glioma with PCV or temozolomide. Neuro Oncol. 2016;18(11):1529–1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Li ZC, Bai H, Sun Q, et al. Multiregional radiomics profiling from multiparametric MRI: Identifying an imaging predictor of IDH1 mutation status in glioblastoma. Cancer Med. 2018;7(12):5999–6009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lee MH, Kim J, Kim ST, et al. Prediction of IDH1 mutation status in glioblastoma using machine learning technique based on quantitative radiomic data. World Neurosurg. 2019;125:e688–e696. [DOI] [PubMed] [Google Scholar]
- 10. Korfiatis P, Kline TL, Coufalova L, et al. MRI texture features as biomarkers to predict MGMT methylation status in glioblastomas. Med Phys. 2016;43(6):2835–2844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Drabycz S, Roldán G, de Robles P, et al. An analysis of image texture, tumor location, and MGMT promoter methylation in glioblastoma using magnetic resonance imaging. Neuroimage. 2010;49(2):1398–1405. [DOI] [PubMed] [Google Scholar]
- 12. Li Y, Liu X, Xu K, et al. MRI features can predict EGFR expression in lower grade gliomas: a voxel-based radiomic analysis. Eur Radiol. 2018;28(1):356–362. [DOI] [PubMed] [Google Scholar]
- 13. Kickingereder P, Bonekamp D, Nowosielski M, et al. Radiogenomics of glioblastoma: machine learning-based classification of molecular characteristics by using multiparametric and multiregional MR imaging features. Radiology. 2016;281(3):907–918. [DOI] [PubMed] [Google Scholar]
- 14. Li Y, Liu X, Qian Z, et al. Genotype prediction of ATRX mutation in lower-grade gliomas using an MRI radiomics signature. Eur Radiol. 2018;28(7):2960–2968. [DOI] [PubMed] [Google Scholar]
- 15. Ren Y, Zhang X, Rui W, et al. Noninvasive prediction of IDH1 mutation and ATRX expression loss in low-grade gliomas using multiparametric MR radiomic features. J Magn Reson Imaging. 2019;49(3):808–817. [DOI] [PubMed] [Google Scholar]
- 16. Li Y, Liang Y, Sun Z, et al. Radiogenomic analysis of PTEN mutation in glioblastoma using preoperative multi-parametric magnetic resonance imaging. Neuroradiology. 2019;61(11):1229–1237. [DOI] [PubMed] [Google Scholar]
- 17. Zhang B, Chang K, Ramkissoon S, et al. Multimodal MRI features predict isocitrate dehydrogenase genotype in high-grade gliomas. Neuro Oncol. 2017;19(1):109–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Preusser M, Wöhrer A, Stary S, Höftberger R, Streubel B, Hainfellner JA. Value and limitations of immunohistochemistry and gene sequencing for detection of the IDH1-R132H mutation in diffuse glioma biopsy specimens. J Neuropathol Exp Neurol. 2011;70(8):715–723. [DOI] [PubMed] [Google Scholar]
- 19. Amadasun M, King R. Texural features corresponding to texural properties. Ieee Trans Syst Man Cybern. 1989;19(5):1264–1274. [Google Scholar]
- 20. Chicklore S, Goh V, Siddique M, Roy A, Marsden PK, Cook GJ. Quantifying tumour heterogeneity in 18F-FDG PET/CT imaging by texture analysis. Eur J Nucl Med Mol Imaging. 2013;40(1):133–140. [DOI] [PubMed] [Google Scholar]
- 21. Haralick RM, Dinstein I, Shanmugam K. Textural Features for Image Classification. Ieee Trans Syst Man Cybern. 1973;SMC-3(6):610–621. [Google Scholar]
- 22. Liao X, Cai B, Tian B, Luo Y, Song W, Li Y. Machine-learning based radiogenomics analysis of MRI features and metagenes in glioblastoma multiforme patients with different survival time. J Cell Mol Med. 2019;23(6):4375–4385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Soni N, Priya S, Bathla G. Texture analysis in cerebral gliomas: a review of the literature. AJNR Am J Neuroradiol. 2019;40(6):928–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kickingereder P, Burth S, Wick A, et al. Radiomic profiling of glioblastoma: identifying an imaging predictor of patient survival with improved performance over established clinical and radiologic risk models. Radiology. 2016;280(3):880–889. [DOI] [PubMed] [Google Scholar]
- 25. Sakai Y, Yang C, Kihira S, et al. MRI radiomic features to predict idh1 mutation status in gliomas: a machine learning approach using gradient tree boosting. Int J Mol Sci. 2020;21(21):1–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tibshirani R. Regression shriknage and selectino via the lasso. J R Stat Soc Ser B. 1996;58(1):267–288. [Google Scholar]
- 27. DeLong ER, DeLong DM, Clarke-Pearson DL. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics. 1988;44(3):837–845. [PubMed] [Google Scholar]
- 28. Lewis MA, Ganeshan B, Barnes A, et al. Filtration-histogram based magnetic resonance texture analysis (MRTA) for glioma IDH and 1p19q genotyping. Eur J Radiol. 2019;113:116–123. [DOI] [PubMed] [Google Scholar]
- 29. Eichinger P, Alberts E, Delbridge C, et al. Diffusion tensor image features predict IDH genotype in newly diagnosed WHO grade II/III gliomas. Sci Rep. 2017;7(1):13396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Brat DJ, Verhaak RGW, Aldape KD, et al. Comprehensive, integrative genomic analysis of diffuse lower-grade gliomas. N Engl J Med. 2015;372(26):2481–2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Beiko J, Suki D, Hess KR, et al. IDH1 mutant malignant astrocytomas are more amenable to surgical resection and have a survival benefit associated with maximal surgical resection. Neuro Oncol. 2014;16(1):81–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Johnson BE, Mazor T, Hong C, et al. Mutational analysis reveals the origin and therapy-driven evolution of recurrent glioma. Science. 2014;343(6167):189–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Martinez R, Esteller M. The DNA methylome of glioblastoma multiforme. Neurobiol Dis. 2010;39(1):40–46. [DOI] [PubMed] [Google Scholar]
- 34. Duregon E, Bertero L, Pittaro A, et al. Ki-67 proliferation index but not mitotic thresholds integrates the molecular prognostic stratification of lower grade gliomas. Oncotarget. 2016;7(16):21190–21198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sunwoo L, Choi SH, Park CK, et al. Correlation of apparent diffusion coefficient values measured by diffusion MRI and MGMT promoter methylation semiquantitatively analyzed with MS-MLPA in patients with glioblastoma multiforme. J Magn Reson Imaging. 2013;37(2):351–358. [DOI] [PubMed] [Google Scholar]
- 36. Huang PH, Xu AM, White FM. Oncogenic EGFR signaling networks in glioma. Sci Signal. 2009;2(87):re6. [DOI] [PubMed] [Google Scholar]
- 37. Brennan CW, Verhaak RG, McKenna A, et al. ; TCGA Research Network . The somatic genomic landscape of glioblastoma. Cell. 2013;155(2):462–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Villanueva-Meyer JE, Wood MD, Choi BS, et al. MRI features and IDH mutational status of grade II diffuse gliomas: impact on diagnosis and prognosis. AJR Am J Roentgenol. 2018;210(3):621–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Qin JB, Liu Z, Zhang H, et al. Grading of gliomas by using radiomic features on multiple magnetic resonance imaging (MRI) sequences. Med Sci Monit. 2017;23:2168–2178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hu X, Martinez-Ledesma E, Zheng S, et al. Multigene signature for predicting prognosis of patients with 1p19q co-deletion diffuse glioma. Neuro Oncol. 2017;19(6):786–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Witten IH, Frank E, Hall MA. . Data Mining: Practical Machine Learning Tools and Techniques, 3rd ed. Cambridge, MA: Elsevier; 2011. [Google Scholar]
- 42. Li J, Liu S, Qin Y, Zhang Y, Wang N, Liu H. High-order radiomics features based on T2 FLAIR MRI predict multiple glioma immunohistochemical features: a more precise and personalized gliomas management. PLoS One. 2020;15(1):e0227703. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.