Abstract
Targeting inhibitory immune checkpoint molecules has significantly altered cancer treatment regimens. T cell immunoglobulin and mucin domain 3 (TIM3) is one of the major inhibitory immune checkpoints expressed on T cells. Blocking the engagement of TIM3 and its inhibitory ligand galectin-9 may potentiate the effects of immunotherapy or overcome the adaptive resistance to the therapeutic blockade of programmed cell death protein 1, cytotoxic T-lymphocyte-associated protein 4, B- and T-lymphocyte attenuator and lymphocyte-activation gene 3, amongst others, as each of these immune checkpoints harbors unique properties that set it apart from the rest. Heavy chain variable fragment (VH)-derived single-domain antibodies (sdAbs) represent a class of expanding drug candidates. These sdAbs have unique advantages, including their minimal size in the antibody class, ease of expression, broad scope for modular structure design and re-engineering, and excellent tumor penetration. In the present study, two sdAbs, TIM3-R23 and TIM3-R53, were generated by immunizing rabbits with the recombinant extracellular domain of TIM3 and applying phage display technology. These sdAbs were easily expressed in mammalian cells. The purified sdAbs were able to bind to both recombinant and cell surface TIM3, and blocked it from binding to the ligand galectin-9. In vivo studies demonstrated that TIM3-R53 was able to potentiate the antitumor activity of chimeric antigen receptor T cells that targeted mesothelin. In conclusion, the results of the present study suggested that TIM3-R53 may be a novel and attractive immune checkpoint inhibitor against TIM3, which is worthy of further investigation.
Keywords: T cell immunoglobulin and mucin domain 3, galectin-9, single-domain antibody, chimeric antigen receptor T cells
Introduction
Immunotherapy has become the mainstay treatment for cancer. One of the most successful, and still rapidly progressing areas in immunotherapy is the blockade of inhibitory immune checkpoint molecules expressed on T cells, such as programmed cell death protein 1 (PD-1) and cytotoxic T-lymphocyte-associated protein 4 (CTLA-4). T cells are the primary regulators of immune cells that participate in immune surveillance and cancer immunity. Due to the potency of T cells and the cascade amplification effect in immune responses initiated by T cells, the activation state of T cells has to be intricately regulated. Inhibitory immune checkpoints, such as PD-1 and CTLA-4, negatively regulate the function and dampen the antitumor immunity of activated T cells. Therefore, unleashing a pre-existing immune response against tumors through neutralization antibodies targeting inhibitory immune checkpoints can enhance the antitumor activity of T cells. Currently, anti-PD-1 antibodies have been approved by the U.S. Food and Drugs Administration and are used in the treatment of malignant melanoma (1), lung cancer (2), renal cancer (3) and lymphoma (4).
T cell immunoglobulin and mucin domain 3 (TIM3) represents a new target beyond the first generation of immune checkpoints, CTLA-4 and PD-1. TIM3 is one of the major negative regulators on the surface of activated T cells. The TIM family has eight members in mice, but only three members in humans (TIM1, TIM3 and TIM4) located on chromosome 5 (5). TIM3 was identified as a molecule expressed by interferon γ-producing CD4+ and CD8+ T cells (6) and numerous other cell types, including regulatory T cells, myeloid cells (7), natural killer cells (8) and mast cells (9). Previous data have revealed that TIM3 served a critical role in regulating the activities of different innate immune cells, and it was associated with autoimmune diseases, chronic viral infections and cancer (10). Several ligands of TIM3 have been identified, including galectin-9, phosphatidylserine (PtdSer), carcinoembryonic antigen-related cell adhesion molecule 1 (CEACAM1), and high-mobility group box 1 protein (11,12). TIM3 and its ligand galectin-9 may constitute one of the negative regulatory pathways that can inhibit T cell immunity (6). Therefore, the blockade of TIM3 is currently being investigated in clinical trials for the treatment of cancer (13).
Single-domain antibodies (sdAbs) are antibody fragments, typically derived from the heavy chain variable fragment (VH) domain, with a molecular weight of ~12 kD. The best studied sdAbs include camelid VHH, cartilaginous fish VNAR and human VH. Due to the small size, excellent thermal stability, reversible refolding capacity and other outstanding properties, sdAbs have demonstrated significant potential in medical applications or as research tools (14,15). Numerous sdAbs are being actively evaluated in clinical trials, and one sdAb drug, caplacizumab (a humanized camelid VHH), has been recently approved for the treatment of acquired thrombotic thrombocytopenic purpura and thrombosis (16,17). Rabbits have been identified as a convenient model to generate VH domain antibodies via immunization and phage display (18). In the present study, a similar strategy was adopted to generate TIM3 inhibitory sdAbs that could be combined with chimeric antigen receptor (CAR) T cell therapy to boost CAR T cell efficacy in cancer treatment.
Materials and methods
Cell lines
The human mesothelioma cell line H226 (ATCC® CRL-5826™; Genetimes ExCell Technology, Inc.) was maintained as an adherent monolayer culture in RPMI-1640 medium (Invitrogen; Thermo Fisher Scientific, Inc.) supplemented with 10% FBS (HyClone; Cytiva), 2 mM L-glutamine and 100 IU/ml penicillin-streptomycin (Invitrogen; Thermo Fisher Scientific, Inc.). H9 (Feng Lab, Huazhong Agricultural University, Wuhan, China) cells were also used, which were an engineered human epithelial carcinoma A431 (ATCC® CRL-1555; Genetimes ExCell Technology, Inc.) cell line (mesothelin negative) that overexpressed cell surface mesothelin. A431 and all A431-derived cells were maintained in DMEM (Invitrogen; Thermo Fisher Scientific, Inc.), and incubated in 5% CO2 at 37°C. These cells were harvested, and the media were refreshed twice a week. Human 293F cells (cat. no. 11625019, Invitrogen; Thermo Fisher Scientific, Inc.) were maintained in SMM 293-TII Expression medium (cat. no. M293TII; SinoBiological, Inc.) in 5% CO2 at 37°C. Cells were passaged every 4 days at the seeding density of 3×105 cells/ml.
Preparation of recombinant proteins
The extracellular domain (ECD) of TIM3 (NP_033665.1, a.a. 22–202) was fused with human IgG1-Fc and cloned into mammalian expression vector pFUSE-hIgG1-Fc2 (pfuse-hg1fc2; InvivoGen) to generate TIM3-hFc. Galectin-9 (NP_033665.1, a.a. 2-355) was fused with rabbit IgG-Fc and cloned into mammalian expression vector pFUSE-rIgG-Fc2 (pfuse-rfc2; InvivoGen) to generate Gal9-rFc. The mesothelin-coding sequence (NP_005814.2, a.a. 296-580) was fused with either human IgG1-Fc or rabbit IgG-Fc and cloned into the same expression vector. The expression vectors were then introduced into 293F cells using polyethylenimine (PEI) (cat. no. 23966-1, Polysciences, Inc.) transfection. The PEI was dissolved at 1 µg/µl in endotoxin-free deionized water that had been heated to ~65°C. For transfection, 300 µg expression plasmid and 750 µl PEI was pre-diluted in 5 ml Opti-MEM medium (Invitrogen; Thermo Fisher Scientific, Inc.) separately and then mixed for 20 min at room temperature before being added to 3×108 293F cells that were passaged in 100 ml of SMM 293-TII Expression medium. Following 24 h of transfection, 200 ml fresh cell culture medium was added and cells were cultured at 37°C and 5% CO2, with gently shaking at 110–175 rotation per minute. The cell density was maintained at 6×106 cells/ml by daily supplementation with fresh medium. The cell culture medium was collected 1 week after transfection, centrifuged at 4°C and 10,000 × g for 1 h and then filtered through 0.2-µm Nalgene Bottle Top filters (cat. no. 291-4520; Thermo Fisher Scientific, Inc.) and subsequently loaded onto the Protein A affinity chromatography column (cat. no. C600952-0501; Sangon Biotech Co., Ltd.). After being washed with PBS buffer, the recombinant protein was eluted with pH 2.0 buffer and immediately neutralized with pH 8.0 Tris-HCl buffer. Protein concentration was measured by bicinchoninic acid (BCA) method. The purity of the preparations was analyzed by 10% SDS-PAGE, and 5 µg protein was loaded per lane. The protein band was visualized with Coomassie Blue staining solution at room temperature for 4 h.
Construction of phage-displayed sdAb library
In total, 2 rabbits (3–4 months old, 1.5–2.0 kg, provided by the Animal Facility of Huazhong Agricultural University, Wuhan, China) were immunized with the recombinant TIM3-hFc. Each rabbit was primed with a subcutaneous injection of 100 µg protein mixed with 100 µl Complete Freund's adjuvant (Sigma-Aldrich; Merck KGaA). Then, 14 days later, rabbits were boosted with half the amount of the protein mixed with Incomplete Freund's adjuvant. Seven days later after boosting, rabbits were euthanized by ear vein injection of sodium pentobarbital at 100 mg/kg according to the previous protocol (19), and the immunized spleens were harvested. The total splenic RNA was isolated using TRIzol® (cat. no. 15596018; Invitrogen; Thermo Fisher Scientific, Inc.) and reverse transcribed into cDNA at 37°C for 15 min using a PrimeScript™ kit (Takara Bio, Inc.). PCR amplification of the VH domains was performed as previously described (18). The VH fragments were digested with the restriction enzyme SfiI and cloned into the phage display vector pComb3× (cat. no. 3498425, Biovector NTCC, Inc.). Then, 10 µg ligation product was introduced into 0.5 ml competent E.coli TG1 cells (cat. no. DE1055; Shanghai Weidi Biotechnology Co., Ltd.) by electroporation using the following parameters: 25 µF, 200 Ω and 1.8 kV. The resultant library contained 1×108 original transformants.
Isolation of TIM3 sdAbs
Briefly, TIM3-hFc protein was immobilized on a 96-well Nunc MaxiSorp™ plate (Thermo Fisher Scientific, Inc.) at a concentration of 100 µg/ml in PBS for 30 min. After being washed with PBS-0.1% Tween-20 (PBST) buffer, 50 µl phage solution was added to the plate and incubated for 1 h at room temperature. After being washed with PBST four times, the bound phage was eluted with pH 2.0 buffer (0.2 M glycine-hydrochloric acid) and re-amplified in TG1 cells for the next round of panning. After three rounds of panning, single colonies were randomly selected and monoclonal phage enzyme-linked immunosorbent assays (ELISAs) were performed to identify the TIM3-specific binders. The detailed phage panning and phage ELISA methods have been previously described (20).
Production of TIM3 sdAb
Rabbit VH domain antibodies were fused with His-FLAG tag, inserted into the expression vector pFUSE-hIgG1-Fc2 (pfuse-hg1fc2; InvivoGen), and expressed as a VH-His-FLAG-hFc format in 293F cells according to the aforementioned method. The 6× His tag was used for Nickel column affinity purification and the hFc tag was used in the cell binding assay by flow cytometry that used goat anti-human polyclonal antibody (cat. no. 109-135-170, Jackson ImmunoResearch Laboratories, Inc.) as secondary antibody.
ELISAs
A 96-well MaxiSorp plate was coated with 5 µg/ml TIM3-hFc protein and incubated at 37°C for 30 min. After being washed twice with PBST, the plate was blocked for 30 min with blocking buffer (2% BSA in PBS buffer). After removing the blocking buffer, biotinylated TIM3 sdAbs were incubated with the plate at variable concentrations starting from 200 µg/ml and followed by 1:2 serial dilutions. TIM3 sdAbs binding was detected using HRP-conjugated streptavidin (Invitrogen; Thermo Fisher Scientific, Inc.; 1:4,000).
Neutralization assay of TIM3 sdAbs
The neutralization assay was performed using a competitive ELISA to determine the ability of the TIM3 sdAbs to block TIM3 from binding to its ligand galectin-9. Briefly, a 96-well MaxiSorp plate was coated with 5 µg/ml TIM3-hFc at 37°C for 30 min. After blocking, variable amounts of TIM3 sdAbs were added to the plate and incubated at 37°C for 30 min to allow the sdAbs to bind to TIM3-hFc. Following the incubation, 5 µg/ml biotinylated galectin-9-rFc was directly added to the wells without removing the TIM3 sdAbs solution and galetin-9-rFc binding was performed at 37°C for 30 min. After the plate was washed twice with PBST, galectin-9-rFc binding was detected by HRP-conjugated streptavidin (1:4,000; cat. no. D111054-0001, Sangon Biotech Co. Ltd.). Subsequently, the plate was washed four times and the binding was visualized using the HRP substrate 3,3′,5,5′-tetramethylbenzidine (Beyotime Institute of Biotechnology), and quantified using a spectrometer at 450 nm.
Packaging of lentiviral virus
Lentivirus was generated from 293T cells (ATCC® CRL-3216; Genetimes ExCell Technology, Inc., China) that were co-transfected with a 3rd generation lentiviral expression plasmid pLenti-LG or pLenti-MesoCAR-IRES-mScarletI (both were constructed in Feng Lab, Huazhong Agricultural University, Wuhan, China) carrying the gene of interest, together with packaging plasmid psPAX2 and enveloping plasmid pMD2.G. A total of 10 µg of the plasmids were mixed at 1:3:1 ratio in 0.5 ml Opti-MEM medium, associated with 25 µg PEI that was diluted in 0.5 ml Opti-MEM medium. The complex of DNA-PEI was formed by incubating at room temperature for 20 min, and then transferred to 1×107 293T cells seeded on T-75 flask. The lentivirus was collected at 72 h post transfection, then filtered with 0.45-µm filter unit (EMD Millipore), immediately used or stored at −80°C before the transduction experiment. For the transductions, the lentiviral particles were directly added to the target cell culture at MOI of 10, followed by 24 h of co-incubation. After changing the medium, transduced cells were cultured for 3 days before subsequent experimentation.
Production of CAR T cells
The CAR expression cassette was based on the second generation format, and was constructed by de novo gene synthesis, which consisted of the CD8a leader sequence (NP_001139345, a.a. 1–21), followed by anti-mesothelin scFv YP158 (21), a CD8α hinge (NP_001139345, a.a. 138–182), a CD8α transmembrane region (NP_001139345, a.a. 183–206), a 4-1BB intracellular domain (NP_001552.2, a.a. 214–255), a CD3ζ intracellular domain (NP_932170, a.a. 52–164), an internal ribosome entry site (from encephalomyocarditis virus) and a red fluorescent protein mScarlet-I. The amino acid sequence of the CAR-YP158 cassette was as follows: MAL PVT ALL LPL ALL LHA ARP DIQSLEESGGDLVKPGASLTLTCTASGFSFSGDYYMCWVRQAPGKGLEWIACIGGGSNTATYYATWAKGRFTISKTSSTTVTLQMTSLTAADTATYFCARDLGFVDYALELWGPGTLVTVSSGGGGSGGGGSGGGGSDVVMTQTPASVEVAVGGTVTIKCQASENMYNSLAWYQQKPGQPPKLLIYRASTLESGVPSRFKGSGSGTEYTLTISDLECADAATYYCQCTFYSHNNNYGGAFGGGTEVVVKSGT TTP APR PPT PAP TIA SQP LSL RPE ACR PAA GGA VHTRGL DFA CDI YIW APL AGT CGV LLL SLV ITL YCK RGRKKL LYI FKQ PFM RPV QTT QEE DGC SCR FPE EEE GGCELR VKF SRS ADA PAY QQG QNQ LYN ELN LGR REE YDV LDK RRG RDP EMG GKP QRR KNP QEGL YNE LQK DKM AEA YSE IGM KG ERR RGK GHD GLY QGL STA TKD TYD ALH MQA LPP R (underlined sections correspond to the cloning enzyme sites EcoRV and BspEI).
The whole blood samples were collected with informed consent from healthy donors using a protocol approved by the Wuhan Blood Center. Ethics of using human blood samples was reviewed and approved by Biomedical Research Ethical Committee, Huazhong Agriculture University and ethics committee-approved oral consent was obtained from donors. Human peripheral blood mononuclear cells (PBMCs) were isolated by Ficoll gradient centrifugation (Stemcell Technologies, Inc.). The PBMCs were activated with Dynabeads™ CD3/CD28 Human T-Activator (cat. no. 11131D; Thermo Fisher Scientific, Inc.) and cultured in RPMI-1640 medium supplemented with 200 IU/ml human recombinant interleukin (IL)-2 (cat. no. GMP-TL104, T&L Biological Technology) for 3 days, according to the manufacturer's protocol. The activated T cells were subsequently transduced with the lentivirus carrying the CAR cassette by co-incubation for 24 h. After transduction, fresh medium containing 200 IU/ml IL-2 was supplemented to maintain the growth of CAR T cells for an additional 3 days before the functional studies. The expression of CAR genes was indicated by the expression of mScarlet-I protein, which can be detected by flow cytometry.
In vitro cytotoxicity assay of CAR T cells
The cancer cell lines, A431, H9 and H226, were stably transduced with a 3rd generation lentiviral vector pLenti-LG (Feng Lab, Huazhong Agricultural University, Wuhan, China; details mentioned previously) to express firefly luciferase (ffLuc2)-green fluorescent protein (GFP) fusion protein. The ffLuc2-GFP gene was synthesized by GenScript and the pLenti-LG plasmid was constructed by the Feng Lab, Huazhong Agricultural University, Wuhan, China. Subsequently, 200 µl cancer cells were plated into a 96-well plate (1×104 cells/well), followed by the addition of CAR T cells at a 5:1 ratio (CAR T: Cancer cells). Following an 18 h co-incubation, the surviving cells were collected by centrifuging the plate at 400 × g at 4°C for 10 min. After removing the medium, the cell pellet was resuspended in 100 µl PBS buffer and lysed by two rounds of freezing-thawing. The activity of the released luciferase was measured with 10 µl cell lysate mixed with 100 µl substrate D-Luciferin buffer (Pierce Firefly Luc One-Step Glow Assay Kit, cat. no. 16196, Thermo Fisher Scientific Inc.) according to the manufacturer's instruction. The enzymatic activity was used to quantify the cell viability. The interval between CAR T cell transduction and activity measurement was 3 days. The viability percentage (%) was, therefore, equal to the experimental signal/maximal signal.
Flow cytometry
Unstimulated PBMCs and CAR T cells were co-incubated with 10 µg/ml TIM3 sdAbs that had the fused hFc tag for 1 h on ice. Cell binding of TIM3 sdAbs was detected with APC-conjugated goat anti-human polyclonal antibody (1:1,000; cat. no. 109-135-170, Jackson ImmunoResearch Laboratories, Inc.). The fluorescence intensity was measured on a FACS Calibur flow cytometer (BD Biosciences) and used to analyze sdAbs binding. The expression level of CAR in CAR T cells was quantified by measuring the fluorescence intensity of the co-expressed mScarlet-I.
In vivo animal studies
The therapeutic potential of sdAb TIM3-R53 was tested using H9 ×enograft model mice. Four to six-week old female athymic nude mice (average ~22 g, provided by Animal Facility of Huazhong Agricultural University, Wuhan, China) were assigned to 4 groups, 5 mice per group. Mice were housed at 25°C, 45% humidity, 12 h light/dark cycle and had free access to food and water. Briefly, nude mice were injected subcutaneously on the right flank with 2×106 H9 cells. After the tumor had formed and reached a size of 100 mm3, treatment was started by intraperitoneal injection of 5×106 mesothelin-targeted CAR T cells or control PBMCs. TIM3 sdAb was intravenously delivered every two days at a concentration of 10 mg/kg. Tumor growth was measured every two days with a caliper; the tumor volume (mm3) was calculated using the following formula: (a × b2) ×0.5, where a is the tumor length and b is the tumor width in mm. Five mice per group were assigned and mice were considered at the end point when tumor size exceeded 1,000 mm3. Mice were euthanized with inhaled CO2 when the tumors grew to >1,000 mm3. The flow rate for CO2 was 10% as the volume displacement per min, and death was verified by lack of cardiac pulse and fixed and dilated pupils.
Statistical analysis
For the quantitative experiments, ≥3 experimental repeats were performed. All statistical analyses were conducted using GraphPad Prism 5 software (GraphPad Software, Inc.). The data are expressed as the mean ± SEM. Comparisons between two groups were performed using an unpaired Student's t test (two-tailed). Comparisons among ≥3 groups were performed using a one-way ANOVA followed by Tukey's multiple comparison test. P<0.05 was considered to indicate a statically significant difference.
Results
Preparation of TIM3-hFc and Gal9-rFc proteins
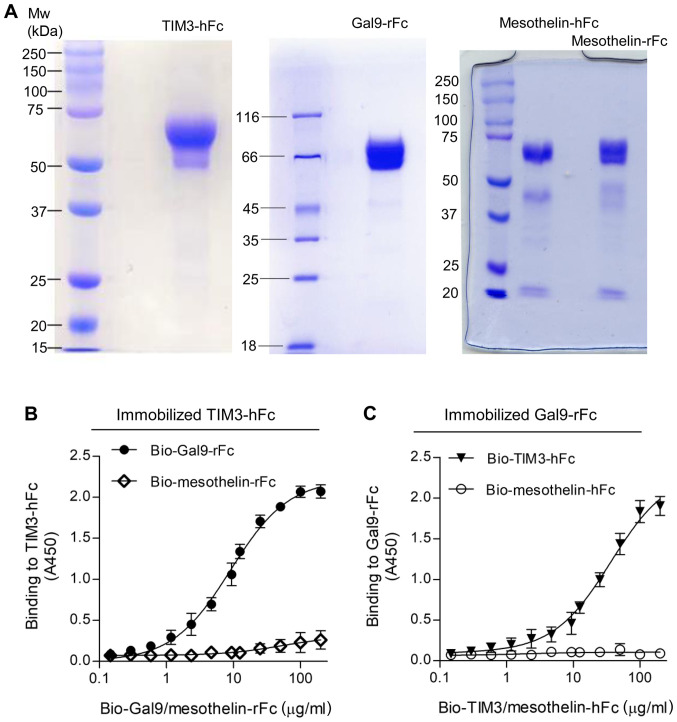
The ECD of TIM3 was fused with human IgG1 Fc (TIM3-hFc) and galectin-9 was fused with rabbit Fc (Gal9-rFc). Recombinant mesothelin-hFc and mesothelin-rFc were used as the Fc isotype control. Mesothelin is a glycosylphosphatidylinositol-linked cell surface protein that has been discovered to be highly expressed in numerous types of malignancy, such as mesothelioma, ovarian cancer, pancreatic cancer and non-small cell lung cancer (22). All the recombinant proteins were expressed in 293F cells and purified via protein A chromatography. The yield of the recombinant proteins was 1–3 mg/l of the cell culture medium, depending on the protein. As shown in Fig. 1, SDS-PAGE analysis revealed that the purity of the recombinant proteins was estimated to be >90%. The reduced monomers of both TIM3-hFc and Gal9-rFc migrated at ~60 kD, and mesothelin Fc fusions migrated at ~70 kD (Fig. 1A). The binding of Gal9-rFc to the immobilized TIM3-hFc was confirmed by ELISA, and the half maximal effective concentration (EC50) value was calculated to be 138 nM (Fig. 1B). A reverse binding assay was also performed, which tested the binding of TIM3-hFc to immobilized Gal9-rFc (Fig. 1C). In both cases, the rFc and hFc isotype control did not reveal any non-specific binding to TIM3 or galectin-9.
Figure 1.
Preparation of TIM3-hFc and Gal9-rFc. (A) SDS-PAGE analysis of the purified TIM3-hFc, Gal9-rFc, mesothelin-hFc and mesothelin-rFc; 5 µg protein was loaded per lane. (B) ELISA analysis of biotinylated Gal9-rFc binding to immobilized TIM3-hFc. Biotinylated mesothelin-rFc was used as the isotype control. (C) ELISA analysis of biotinylated TIM3-hFc binding to immobilized Gal9-rFc. Biotinylated mesothelin-hFc was used as the isotype control. TIM3, T cell immunoglobulin and mucin domain 3; ELISA, enzyme-linked immunosorbent assay; Gal9, galectin-9.
Generation of anti-TIM3 sdAbs
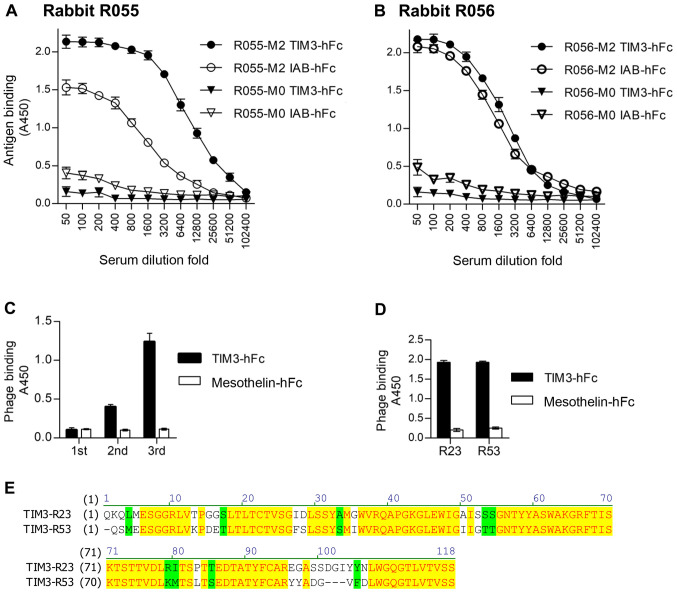
After immunization of the two rabbits with TIM3-hFc, the titer and specificity were analyzed using an ELISA, with IAB-hFc as the hFc control (Fig. 2A and B). IAB was a fragment of mesothelin (Q13421, amino acid 296–359) (22). The serum of the immunized rabbit R055 (R055-M2) showed increased binding to TIM3-hFc compared with the IAB-hFc control. Therefore, this rabbit was chosen to make the phage-displayed sdAb library. After three rounds of panning, the results of polyclonal phage ELISAs from each round of panning revealed the gradual increased binding to TIM3-hFc (Fig. 2C). A total of 96 colonies from the third panning were randomly picked, and a monoclonal phage ELISA was performed to determine the TIM3-specific binders. Sequencing of the resultant 89 ELISA-positive clones identified that only two representative sequences, named TIM3-R23 and TIM3-R53, were enriched. The monoclonal phage ELISA data and the amino acid sequences of these two clones are presented in Fig. 2D and E.
Figure 2.
Generation of TIM3 sdAbs. (A) Serum titer of rabbit R055 immunized with the recombinant TIM3-hFc. The control antigen, IAB-hFc, was a recombinant fragment of mesothelin (Q13421, amino acid 296–359). M0, pre-immunization serum; M2, post-immunization serum. (B) Serum titer of rabbit R056. The same experimental settings were followed. (C) Polyclonal phage ELISA. Eluted phage from the first, second and third round of panning was tested for its binding to TIM3-hFc. (D) Monoclonal phage ELISA. Monoclonal phage clones R23 and R53 were tested for their binding to TIM3-hFc. (E) Amino acid sequences of sdAbs R23 and R53. TIM3, T cell immunoglobulin and mucin domain 3; ELISA, enzyme-linked immunosorbent assay; sdAbs, single-domain antibodies.
Binding and neutralization activity of TIM3 sdAbs
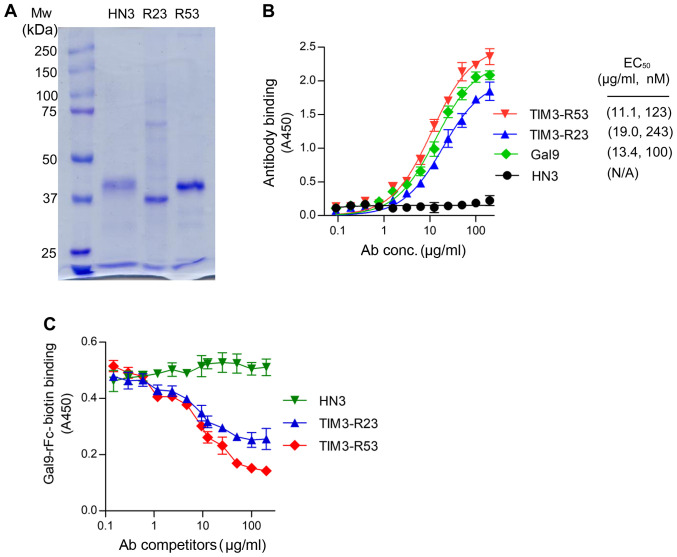
The sdAbs TIM3-R23 and TIM3-R53 were expressed in 293F cells as a VH-His-FLAG-hFc format and purified using a HiTrap Nickel column. SDS-PAGE analysis revealed that the purified sdAbs migrated at ~40 kD as a monomer under reducing conditions (Fig. 3A). The control sdAb HN3 is a VH domain antibody that targets glypican-3 (20). The binding affinity of the sdAbs to TIM3 was analyzed using ELISAs and the EC50 values for R53 and R23 were calculated to be 123 and 243 nM, respectively (Fig. 3B), which were similar to that of the galectin-9 binding (100 nM). The HN3 sdAb control did not reveal any background binding in the assay.
Figure 3.
Binding and neutralization properties of TIM3 sdAbs. (A) SDS-PAGE analysis of purified TIM3 sdAbs; 1 µg protein per lane was loaded. HN3, a sdAb to glypican-3 (20), was used as an irrelevant sdAb-hFc control. (B) ELISA analysis of sdAbs binding to TIM3. The plate was coated with 5 µg TIM3-hFc and incubated with increasing amounts of biotinylated sdAbs. Binding of the sdAbs was detected using HRP-conjugated streptavidin. Biotinylated Gal9-rFc was also included for comparison. (C) Neutralization assay of the sdAbs. The ability of TIM3 sdAbs to block galectin-9 binding to TIM3 was analyzed using a competitive ELISA. The MaxiSorp™ plate was coated with 5 µg TIM3-hFc and preincubated with increasing amount of sdAbs, followed by the addition of 5 µg/ml biotinylated Gal9-rFc. The binding was detected using HRP-conjugated streptavidin. TIM3, T cell immunoglobulin and mucin domain 3; ELISA, enzyme-linked immunosorbent assay; sdAbs, single-domain antibodies; Gal9, galectin-9.
Subsequently, the neutralization activity of TIM3 sdAbs was analyzed by measuring its ability to inhibit TIM3 binding to galectin-9 (Fig. 3C). The results of the competitive ELISA illustrated that both sdAbs were able to inhibit TIM3-galectin-9 binding (Fig. 3C); TIM3-R53 exhibited stronger blocking activity compared with TIM3-R23, and therefore the ability of this sdAb to boost the function of CAR T cells was further investigated in vivo.
Cellular binding activity of TIM3 sdAbs
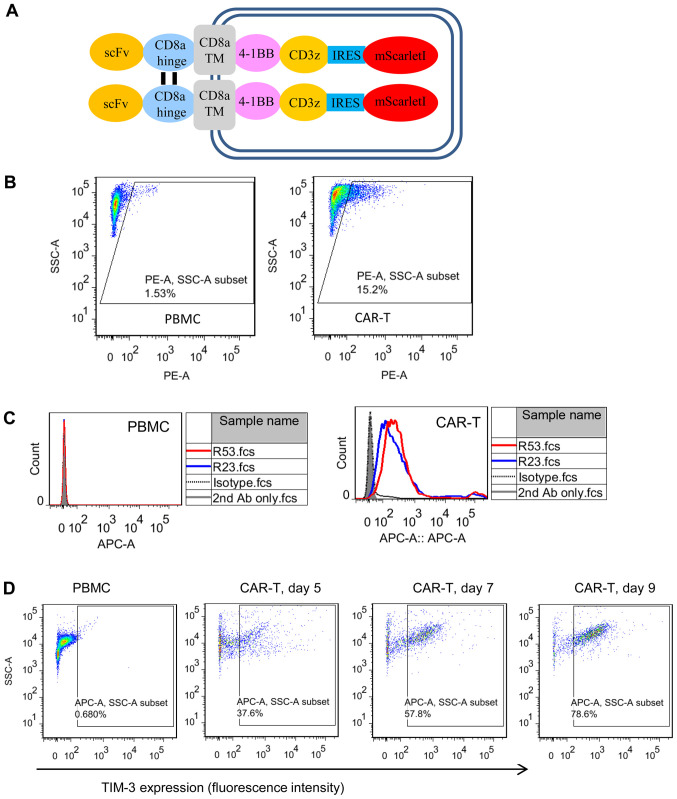
Cell surface TIM3 binding was analyzed using mesothelin-targeted CAR T cells. Unstimulated PBMCs were used as the TIM3-negative counterpart. CAR T cells were generated by lentiviral transduction of activated PBMCs, which produced an ~15% transduction efficiency (Fig. 4A). Flow cytometric analysis demonstrated that the two TIM3 sdAbs selectively bound to CAR T cells, but not to PBMCs (Fig. 4B and C). To further confirm the cell binding and TIM3 expression kinetics on activated CAR T cells, the CAR T cells were cultured for different days and sdAbs binding was analyzed (Fig. 4D). The results clearly demonstrated that the percentage of TIM3-positive CAR T cells was increased as the culturing time was prolonged from day 5 to 9.
Figure 4.
Cell binding properties of TIM3 sdAbs. (A) Schematic illustration of the mesothelin-targeted CAR structure. Anti-mesothelin scFv YP158 (21) was fused with the following domains: CD8α hinge, CD8α transmembrane region, 4-1BB intracellular domain, CD3ζ intracellular domain, internal ribosome entry site and a red fluorescent protein mScarlet-I. (B) Transduction efficiency analysis of CAR T cells. Following the transduction of PBMCs with a CAR-bearing lentiviral vector, CAR expression was indicated by the fluorescence of mScarletI. (C) Flow cytometric analysis of TIM3 sdAbs binding to the unstimulated PBMCs and CAR T cells. Briefly, 10 µg sdAbs were co-incubated with 1×106 cells. Antibody binding was detected by APC-conjugated goat-anti-human IgG. HN3, a human sdAb antibody that targeted the glypican-3, was used as the isotype control. Shaded area, secondary antibody staining; dashed lines, isotype control; red and blue solid line, sdAbs R53 and R23 staining, respectively. (D) Expression of TIM3 on CAR T cells cultured for different lengths of time. TIM3, T cell immunoglobulin and mucin domain 3; sdAbs, single-domain antibodies; CAR, chimeric antigen receptor; PBMCs, peripheral blood mononuclear cells.
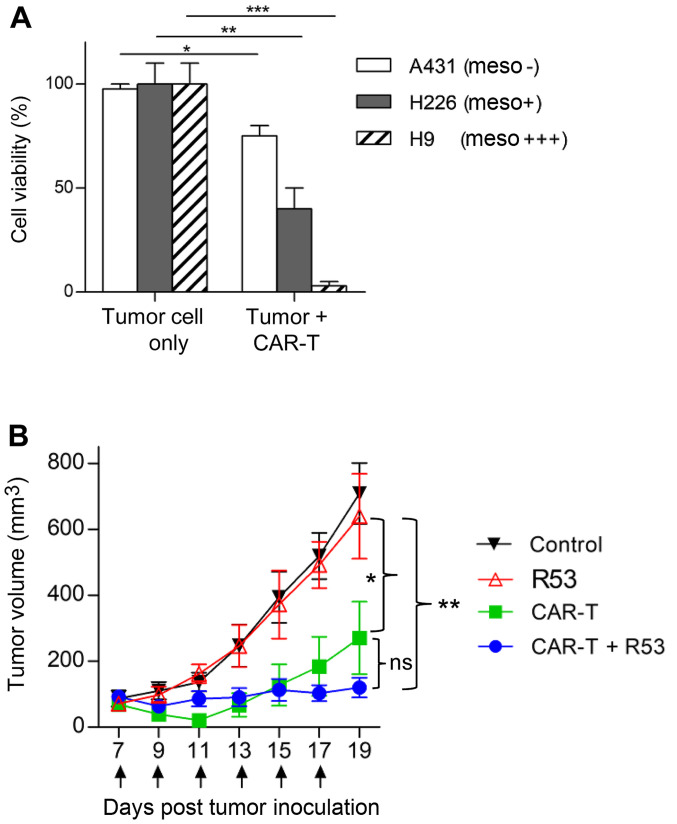
In vivo antitumor activity of CAR T cells is boosted by TIM3-R53
As TIM3-R53 revealed stronger in vitro neutralization activity, it was chosen for the in vivo study. Firstly, the in vitro cytotoxicity of the mesothelin-targeted CAR T cells was confirmed by the co-incubation with mesothelin-positive H226 and H9 cells and mesothelin-negative A431 cells. The aforementioned cells were genetically labeled with luciferase and the cell viability after CAR T cell killing was determined by measuring the remaining luciferase activity. As shown in Fig. 5A, the CAR T cells efficiently killed H226 and H9 cells, with only some background killing apparent on antigen-negative A431 cells. The in vivo activity testing of CAR T cells was performed on H9 nude model mice. As shown in Fig. 5B, mesothelin-targeted CAR T cells significantly suppressed H9 tumor growth compared with the PBS control group, and the largest tumor size was ~14×11 mm (Table I). In addition, the combination of TIM3-R53 and CAR T cells could moderately boost the activity of the CAR T cells, suggesting that the sdAb had the potential to be further explored as a TIM3 inhibitory drug candidate, but further optimization was required to increase its potency.
Figure 5.
In vivo antitumor activity of mesothelin-targeted CAR T cells combined with TIM3-R53 single-domain antibody. (A) Cytotoxicity measurement of mesothelin CAR T cells. All the cancer cell lines were stably transduced to express a firefly luciferase reporter gene (ffLuc2). Subsequently, 1×104 cancer cells were co-incubated with CAR T cells for 48 h at a ratio of 5:1 (CAR T: Cancer cells). The cell viability was quantified by measuring the intracellular luciferase activity. *P<0.05; **P<0.01; ***P<0.001. The experiment was performed in triplicate for each sample. Statistical significances were analyzed using unpaired Student's t-test. (B) In vivo efficacy of mesothelin-targeted CAR T cells combined with TIM3-R53. Briefly, 2×106 H9 cells were subcutaneously inoculated into female athymic nude mice. When the tumor reached a size of 100 mm3, treatment was initiated by the intraperitoneal injection of 5×106 CAR T cells or peripheral blood mononuclear cells. The arrows indicate the time point of 10 mg/kg TIM3-R53 delivery by tail vein injection; n=5/group. Tumor volume was measured every 2 days. *P<0.05; **P<0.01. Statistical significances were analyzed using a one-way ANOVA followed by Tukey's multiple comparison test. TIM3, T cell immunoglobulin and mucin domain 3; CAR, chimeric antigen receptor.
Table I.
Measurement of tumor growth. (a) × width (b) in mm; volume was calculated as (axb2)/2 in mm3.
| Group | ID# | Day 7 | Day 9 | Day 11 | Day 13 | Day 15 | Day 17 | Day 19 | |
|---|---|---|---|---|---|---|---|---|---|
| Control | 790 | Measurement, axb mm | 7.32×6.41 | 7.75×6.70 | 8.48×6.93 | 9.60×8.18 | 11.02×8.47 | 12.70×9.38 | 13.48×10.81 |
| Volume, mm3 | 150.38 | 173.95 | 203.63 | 321.18 | 395.29 | 558.70 | 787.61 | ||
| 564 | Measurement, axb mm | 7.38×6.18 | 7.52×6.80 | 8.19×7.02 | 9.61×9.61 | 10.92×10.88 | 11.97×10.92 | 12.57×12.06 | |
| Volume, mm3 | 140.93 | 173.95 | 201.80 | 443.75 | 646.32 | 713.69 | 913.69 | ||
| 609 | Measurement, axb mm | 5.74×4.42 | 6.28×5.26 | 6.93×5.67 | 7.84×7.47 | 9.46×8.57 | 10.52×9.75 | 11.47×10.38 | |
| Volume, mm3 | 56.07 | 86.88 | 111.40 | 218.74 | 347.39 | 500.03 | 617.83 | ||
| 566 | Measurement, axb mm | 5.58×4.47 | 6.14×5.04 | 6.87×5.53 | 8.74×6.47 | 12.28×8.25 | 13.27×9.06 | 14.47×10.69 | |
| Volume, mm3 | 55.75 | 77.98 | 105.05 | 182.93 | 417.90 | 544.62 | 826.90 | ||
| 565 | Measurement, axb mm | 4.91×3.66 | 5.08×4.01 | 5.57×4.71 | 6.60×4.56 | 8.99×6.02 | 9.37×7.98 | 10.53×8.75 | |
| Volume, mm3 | 32.89 | 40.84 | 61.78 | 68.62 | 162.90 | 298.48 | 402.72 | ||
| R53 | 661 | Measurement, axb mm | 7.21×5.73 | 7.8×6.45 | 8.21×6.86 | 9.53×9.21 | 11.62×10.98 | 12.01×10.88 | 13.41×12.17 |
| Volume, mm3 | 118.51 | 162.19 | 193.22 | 404.20 | 699.93 | 711.35 | 992.81 | ||
| 611 | Measurement, axb mm | 6.92×5.65 | 7.65×6.16 | 8.58×7.17 | 9.32×8.58 | 10.91×8.94 | 11.56×9.38 | 12.82×10.23 | |
| Volume, mm3 | 110.36 | 145.05 | 220.47 | 342.68 | 435.82 | 508.84 | 670.53 | ||
| 638 | Measurement, axb mm | 5.67×4.65 | 6.12×5.15 | 7.89×6.94 | 8.63×7.86 | 9.33×8.09 | 10.64×9.50 | 11.14×10.02 | |
| Volume, mm3 | 61.21 | 81.23 | 189.76 | 266.63 | 305.06 | 480.37 | 559.74 | ||
| 791 | Measurement, axb mm | 5.12×3.92 | 5.67×5.06 | 6.77×6.38 | 7.41×6.94 | 9.21×8.66 | 11.02×9.70 | 11.98×11.25 | |
| Volume, mm3 | 39.24 | 72.49 | 137.91 | 178.35 | 345.64 | 518.30 | 757.56 | ||
| 792 | Measurement, axb mm | 4.24×3.86 | 4.98×3.67 | 5.83×4.58 | 5.59×3.89 | 6.71×4.92 | 9.21×7.78 | 8.87×7.34 | |
| Volume, mm3 | 31.60 | 33.52 | 61.24 | 42.38 | 81.13 | 278.94 | 239.25 | ||
| CAR-T | 947 | Measurement, axb mm | 7.32×5.64 | 6.20×4.58 | 4.90×3.38 | 7.63×5.75 | 9.35×6.83 | 9.62×8.12 | 11.51×8.86 |
| Volume, mm3 | 116.42 | 65.03 | 27.99 | 126.05 | 218.08 | 317.40 | 451.46 | ||
| 568 | Measurement, axb mm | 7.17×4.59 | 6.70×3.98 | 6.58×3.43 | 8.34×6.56 | 9.96×8.02 | 10.28×9.43 | 10.95×10.23 | |
| Volume, mm3 | 75.53 | 52.94 | 38.71 | 179.62 | 320.33 | 456.99 | 612.79 | ||
| 610 | Measurement, axb mm | 6.23×4.63 | 5.31×3.98 | 3.74×3.31 | 3.87×3.67 | 5.89×5.13 | 6.35×5.45 | 7.03×6.32 | |
| Volume, mm3 | 66.65 | 41.99 | 20.44 | 26.00 | 77.43 | 94.24 | 140.36 | ||
| 563 | Measurement, axb mm | 5.45×4.59 | 4.13×3.84 | 3.66×3.30 | 4.23×1.66 | 4.89×2.95 | 5.20×4.00 | 6.79×5.30 | |
| Volume, mm3 | 57.42 | 30.45 | 19.97 | 5.82 | 21.30 | 41.56 | 95.22 | ||
| 578 | Measurement, axb mm | 4.07×3.66 | 2.57×2.07 | 1.00×1.00 | 0.00×0.00 | 0.00×0.00 | 2.91×2.75 | 4.96×4.92 | |
| Volume, mm3 | 27.26 | 5.51 | 0.50 | 0.00 | 0.00 | 11.00 | 60.00 | ||
| CAR-T+R53 | 960 | Measurement, axb mm | 7.02×5.50 | 6.42×5.10 | 7.55×6.14 | 7.63×6.60 | 7.88×7.32 | 7.56×7.08 | 8.56×7.16 |
| Volume, mm3 | 106.18 | 83.49 | 142.24 | 165.94 | 211.11 | 189.65 | 219.65 | ||
| 794 | Measurement, axb mm | 6.78×5.46 | 5.98×5.02 | 6.53×5.49 | 6.87×5.80 | 7.38×6.28 | 6.72×5.73 | 7.58×6.19 | |
| Volume, mm3 | 101.00 | 75.30 | 98.50 | 115.40 | 145.70 | 110.30 | 145.20 | ||
| 585 | Measurement, axb mm | 6.55×5.20 | 5.43×4.77 | 7.08×5.78 | 7.47×5.69 | 7.24×5.79 | 6.82×5.20 | 7.28×5.34 | |
| Volume, mm3 | 88.56 | 61.77 | 118.41 | 120.81 | 121.51 | 92.21 | 103.76 | ||
| 665 | Measurement, axb mm | 6.21×5.18 | 5.34×4.39 | 6.10×4.02 | 6.53×3.55 | 6.98×4.49 | 6.54×4.79 | 6.74×5.03 | |
| Volume, mm3 | 83.20 | 51.50 | 49.30 | 41.20 | 70.30 | 75.10 | 85.20 | ||
| 636 | Measurement, axb mm | 6.50×5.01 | 6.23×3.80 | 5.03×3.07 | 4.18×2.28 | 4.42×2.74 | 6.19×3.88 | 6.36×3.86 | |
| Volume, mm3 | 81.58 | 44.98 | 23.64 | 10.89 | 16.61 | 46.49 | 47.44 |
Volume was calculated as (axb2)/2. a, length; b, width; CAR, chimeric antigen receptor. ID#, ear ring tagging number of mice.
Discussion
TIM3 is generally considered as an inhibitory immune checkpoint that is preferentially expressed on activated T cells and other immune cells. TIM3 interacts with multiple ligands including galectin-9, CEACAM1, PtdSer, HMGB1, and causes exhaustion and apoptosis of antigen-specific Th1 cells and CTLs, which correlates with impaired antitumor immune response (23). The blockade of TIM3 has exhibited beneficial effects in T cell-based immunotherapies in some preclinical studies (24,25). However, more studies showed that antibodies to TIM3 could enhance T cell function, but the efficacy looks moderate or may work best in synergy with PD1/PDL1 blockade (26–28). A recently completed phase Ia/b study on non-small cell lung cancer indicated that TIM3 blocking antibody LY3321367 had acceptable safety profile with favorable pharmacokinetics/pharmacodynamics but only modest antitumor activity (27). In such a complicating and disappointing scenario, it was worthwhile to develop and test more antibodies with different epitopes and binding properties more rigorously in the preclinical studies.
To date, most TIM3-blockade antibodies are conventional IgG types. Plenty of preclinical studies heavily relied on one rat monoclonal antibody, RMT3-23, as a research tool to block TIM3 function (11,29,30). RMT3-23 is commercially available as research reagent and recognizes mouse TIM3 but not human TIM3. However, detailed characterization showed that RMT3-23 weakly blocks CEACMA1 and PtdSer from binding to TIM3, but not galectin-9 (11). Very recently, a fully human anti-human TIM3 monoclonal antibody M6903 was developed from human immunoglobulin (Ig) transgenic rats (OmniRat®) immunized with His-tagged recombinant human TIM3 extracellular domain (31). M6903 binds human TIM3 with strong affinity (Kd, 2.3 nM), but moderately blocks the engagement of three TIM3 ligands, PtdSer (IC90, ~1 µg/ml), CEACAM1 (IC90, ~0.5 µg/ml), and galectin-9 (IC90, ~1.5 µg/ml). However the in vivo antitumor activity by M6903 monotherapy was still moderate (31), probably due to the insufficient blocking activity. Apart from the aforementioned IgGs, one camel sdAb to human TIM3 was previously generated (32), however this sdAb has not been rigorously tested for the binding and blocking activity in vitro and in vivo. Taken together, good monoclonal antibodies that could potently block human TIM3 from binding to any of its ligands are still lacking. The present study aimed to develop new sdAbs that could block the binding of galectin-9 to human TIM3.
By using protein immunization and phage display, the current study successfully isolated two rabbit TIM3 sdAbs, R53 and R23. The binding affinity of these two sdAbs to TIM3 was similar and close to that of the ligand galectin-9, all of which were within the 100 nM range. As lead molecules, these sdAbs demonstrated weak but acceptable affinity and it is still possible to further improve their affinity through antibody engineering. The neutralization assay revealed that sdAb R53 inhibited galectin-9-TIM3 binding to a greater extent compared with R23. Thus, the present study focused on R53 for the following functional studies.
It was reported that CAR T cells could overexpress TIM3 for ~10 days after transduction (33). The present study confirmed the cell binding specificity of the sdAbs through their ability to selectively bind to mesothelin-targeted CAR T cells, but not to PBMCs. In addition, the expression of TIM3 in the CAR T cells was discovered to gradually increase with the extended culture time. As a proof-of-concept study on the in vivo antitumor activity of the sdAb, mesothelin-targeted CAR T cells were constructed and the efficacy of CAR T cells in combination with TIM3 sdAbs were analyzed. The combination of R53 could boost the CAR T cell activity moderately (Fig. 5B), probably due to its low affinity. The affinity of R53 was about 50-fold lower than M6903 (31), however their in vivo efficacy seems quite similar, if disregarding the difference of the experimental conditions, such as tumor cell lines, immune effector cells, and the immune competency of the mice. To the best of our knowledge, R53 and M6903 are the only reported monoclonal antibodies against human TIM3 that have been well characterized and functionally tested in vitro and in vivo; further antibody engineering may improve their efficacy.
It has been reported that PD-1 blockade therapy could produce adaptive resistance in numerous patients, and that the combined therapy of anti-PD-1 and anti-TIM3 significantly inhibited the growth of tumors in preclinical studies (34,35). Therefore, it is worthwhile to further evaluate the efficacy of the sdAb isolated in the present study in more diverse and translational settings.
Acknowledgements
Not applicable.
Funding Statement
The present study was supported by the National Natural Science Foundation of China (grant. no. 31670943), the Fundamental Research Funds for the Central Universities (grant nos. 2662016PY113, 2662017PY111 and 2662019YJ013) and the Applied Basic Research Program of Wuhan Science and Technology Bureau (grant nos. 2017060201010195 and 2019020701011438).
Funding
The present study was supported by the National Natural Science Foundation of China (grant. no. 31670943), the Fundamental Research Funds for the Central Universities (grant nos. 2662016PY113, 2662017PY111 and 2662019YJ013) and the Applied Basic Research Program of Wuhan Science and Technology Bureau (grant nos. 2017060201010195 and 2019020701011438).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
LY performed the experiments, analyzed the data, prepared the figures and drafted the manuscript. XC, QW and XM performed the flow cytometry experiments. CW, DZ and YZ measured the CAR T cell killing ability both in vitro and in vivo. HH, LH, JL and CX provided assistance with the experiments and data analysis. SH analyzed the data and revised the manuscript. XY and MF designed the study and finalized the manuscript. LY and XC confirmed the authenticity of all the raw data. All authors read and approved the final manuscript.
Ethics approval and consent to participate
All the procedures used in the animal studies were approved by the Animal Care and Use Committee of Huazhong Agricultural University (approval no. HZAUMO-2017-051). Peripheral blood materials used in this study were obtained from healthy donors with informed consent. The use of human blood samples was reviewed and approved by Biomedical Research Ethical Committee, Huazhong Agriculture University (approval no. HZAUHU-2017-002).
Patient consent for publication
Not applicable.
Competing interests
A patent (patent no. CN110938146A, submitted on Nov. 20th, 2019) has been filed based on the domain antibodies and their applications. The authors have declared no other competing interest.
References
- 1.Hamid O, Robert C, Ribas A, Hodi FS, Walpole E, Daud A, Arance AS, Brown E, Hoeller C, Mortier L, et al. Antitumour activity of pembrolizumab in advanced mucosal melanoma: A post-hoc analysis of KEYNOTE-001, 002, 006. Br J Cancer. 2018;119:670–674. doi: 10.1038/s41416-018-0207-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Garon EB, Rizvi NA, Hui R, Leighl N, Balmanoukian AS, Eder JP, Patnaik A, Aggarwal C, Gubens M, Horn L, et al. Pembrolizumab for the treatment of non-small-cell lung cancer. N Engl J Med. 2015;372:2018–2028. doi: 10.1056/NEJMoa1501824. [DOI] [PubMed] [Google Scholar]
- 3.Motzer RJ, Escudier B, McDermott DF, George S, Hammers HJ, Srinivas S, Tykodi SS, Sosman JA, Procopio G, Plimack ER, et al. Nivolumab versus everolimus in advanced renal-cell carcinoma. N Engl J Med. 2015;373:1803–1813. doi: 10.1056/NEJMoa1510665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Langer CJ, Gadgeel SM, Borghaei H, Papadimitrakopoulou VA, Patnaik A, Powell SF, Gentzler RD, Martins RG, Stevenson JP, Jalal SI, et al. Carboplatin and pemetrexed with or without pembrolizumab for advanced, non-squamous non-small-cell lung cancer: A randomised, phase 2 cohort of the open-label KEYNOTE-021 study. Lancet Oncol. 2016;17:1497–1508. doi: 10.1016/S1470-2045(16)30498-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mcintire JJ, Umetsu SE, Akbari O, Potter M, Kuchroo VK, Barsh GS, Freeman GJ, Umetsu DT, Dekruyff RH. Identification of Tapr (an airway hyperreactivity regulatory locus) and the linked Tim gene family. Nat Immunol. 2001;2:1109–1116. doi: 10.1038/ni739. [DOI] [PubMed] [Google Scholar]
- 6.Zhu C, Anderson AC, Schubart A, Xiong H, Imitola J, Khoury SJ, Zheng XX, Strom TB, Kuchroo VK. The Tim-3 ligand galectin-9 negatively regulates T helper type 1 immunity. Nat Immunol. 2005;6:1245–1252. doi: 10.1038/ni1271. [DOI] [PubMed] [Google Scholar]
- 7.Anderson AC, Anderson DE, Bregoli L, Hastings WD, Kassam N, Lei C, Chandwaskar R, Karman J, Su EW, Hirashima M, et al. Promotion of tissue inflammation by the immune receptor Tim-3 expressed on innate immune cells. Science. 2007;318:1141–1143. doi: 10.1126/science.1148536. [DOI] [PubMed] [Google Scholar]
- 8.Ndhlovu LC, Lopez-Vergès S, Barbour JD, Jones RB, Jha AR, Long BR, Schoeffler EC, Fujita T, Nixon DF, Lanier LL. Tim-3 marks human natural killer cell maturation and suppresses cell-mediated cytotoxicity. Blood. 2012;119:3734–3743. doi: 10.1182/blood-2011-11-392951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Phong BL, Avery L, Sumpter TL, Gorman JV, Watkins SC, Colgan JD, Kane LP. Tim-3 enhances FcεRI-proximal signaling to modulate mast cell activation. J Exp Med. 2015;212:2289–2304. doi: 10.1084/jem.20150388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Han G, Chen G, Shen B, Li Y. Tim-3: An activation marker and activation limiter of innate immune cells. Front Immunol. 2013;4:449. doi: 10.3389/fimmu.2013.00449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sabatos-Peyton CA, Nevin J, Brock A, Venable JD, Tan DJ, Kassam N, Xu F, Taraszka J, Wesemann L, Pertel T, et al. Blockade of Tim-3 binding to phosphatidylserine and CEACAM1 is a shared feature of anti-Tim-3 antibodies that have functional efficacy. Oncoimmunology. 2017;7:e1385690. doi: 10.1080/2162402X.2017.1385690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dolina JS, Braciale TJ, Hahn YS. Liver-primed CD8+ T cells suppress antiviral adaptive immunity through galectin-9-independent T-cell immunoglobulin and mucin 3 engagement of high-mobility group box 1 in mice. Hepatology. 2014;59:1351–1365. doi: 10.1002/hep.26938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Markwick LJ, Riva A, Ryan JM, Cooksley H, Palma E, Tranah TH, Manakkat Vijay GK, Vergis N, Thursz M, Evans A, et al. Blockade of PD1 and TIM3 restores innate and adaptive immunity in patients with acute alcoholic hepatitis. Gastroenterology. 2015;148:590–602.e10. doi: 10.1053/j.gastro.2014.11.041. [DOI] [PubMed] [Google Scholar]
- 14.Khodabakhsh F, Behdani M, Rami A, Kazemi-Lomedasht F. Single-domain antibodies or nanobodies: A class of next-generation antibodies. Int Rev Immunol. 2018;37:316–322. doi: 10.1080/08830185.2018.1526932. [DOI] [PubMed] [Google Scholar]
- 15.Wesolowski J, Alzogaray V, Reyelt J, Unger M, Juarez K, Urrutia M, Cauerhff A, Danquah W, Rissiek B, Scheuplein F, et al. Single domain antibodies: Promising experimental and therapeutic tools in infection and immunity. Med Microbiol Immunol. 2009;198:157–174. doi: 10.1007/s00430-009-0116-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Elverdi T, Eskazan AE. Caplacizumab as an emerging treatment option for acquired thrombotic thrombocytopenic purpura. Drug Des Devel Ther. 2019;13:1251–1258. doi: 10.2147/DDDT.S134470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Scully M, Cataland SR, Peyvandi F, Coppo P, Knöbl P, Kremer Hovinga JA, Metjian A, de la Rubia J, Pavenski K, Callewaert F, et al. Caplacizumab treatment for acquired thrombotic thrombocytopenic purpura. N Engl J Med. 2019;380:335–346. doi: 10.1056/NEJMoa1806311. [DOI] [PubMed] [Google Scholar]
- 18.Shinozaki N, Hashimoto R, Fukui K, Uchiyama S. Efficient generation of single domain antibodies with high affinities and enhanced thermal stabilities. Sci Rep. 2017;7:5794. doi: 10.1038/s41598-017-06277-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.AVMA Panel on Euthanasia. American Veterinary Medical Association, corp-author. 2000 Report of the AVMA panel on euthanasia. J Am Vet Med Assoc. 2001;218:669–696. doi: 10.2460/javma.2001.218.669. [DOI] [PubMed] [Google Scholar]
- 20.Feng M, Gao W, Wang R, Chen W, Man YG, Figg WD, Wang XW, Dimitrov DS, Ho M. Therapeutically targeting glypican-3 via a conformation-specific single-domain antibody in hepatocellular carcinoma. Proc Natl Acad Sci USA. 2013;110:E1083–E1091. doi: 10.1073/pnas.1217868110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang YF, Ho M. Humanization of rabbit monoclonal antibodies via grafting combined Kabat/IMGT/Paratome complementarity-determining regions: Rationale and examples. MAbs. 2017;9:419–429. doi: 10.1080/19420862.2017.1289302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaneko O, Gong L, Zhang J, Hansen JK, Hassan R, Lee B, Ho M. A binding domain on mesothelin for CA125/MUC16. J Biol Chem. 2009;284:3739–3749. doi: 10.1074/jbc.M806776200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baghdadi M, Takeuchi S, Wada H, Seino K. Blocking monoclonal antibodies of TIM proteins as orchestrators of anti-tumor immune response. MAbs. 2014;6:1124–1132. doi: 10.4161/mabs.32107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Herrera-Camacho I, Anaya-Ruiz M, Perez-Santos M, Millán- Pérez Peña L, Bandala C, Landeta G. Cancer immunotherapy using anti-TIM3/PD-1 bispecific antibody: A patent evaluation of EP3356411A1. Expert Opin Ther Pat. 2019;29:587–593. doi: 10.1080/13543776.2019.1637422. [DOI] [PubMed] [Google Scholar]
- 25.Liu JF, Wu L, Yang LL, Deng WW, Mao L, Wu H, Zhang WF, Sun ZJ. Blockade of TIM3 relieves immunosuppression through reducing regulatory T cells in head and neck cancer. J Exp Clin Cancer Res. 2018;37:44. doi: 10.1186/s13046-018-0713-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou G, Sprengers D, Boor PPC, Doukas M, Schutz H, Mancham S, Pedroza-Gonzalez A, Polak WG, de Jonge J, Gaspersz M, et al. Antibodies against immune checkpoint molecules restore functions of tumor-infiltrating T cells in hepatocellular carcinomas. Gastroenterology. 2017;153:1107–1119.e10. doi: 10.1053/j.gastro.2017.06.017. [DOI] [PubMed] [Google Scholar]
- 27.Harding JJ, Moreno V, Bang YJ, Hong MH, Patnaik A, Trigo J, Szpurka AM, Yamamoto N, Doi T, Fu S, et al. Blocking TIM-3 in treatment-refractory advanced solid tumors: A phase Ia/b study of LY3321367 with or without an anti-PD-L1 antibody. Clin Cancer Res. 2021;27:2168–2178. doi: 10.1158/1078-0432.CCR-20-4405. [DOI] [PubMed] [Google Scholar]
- 28.Kim JE, Patel MA, Mangraviti A, Kim ES, Theodros D, Velarde E, Liu A, Sankey EW, Tam A, Xu H, et al. Combination therapy with Anti-PD-1, anti-TIM-3, and focal radiation results in regression of murine gliomas. Clin Cancer Res. 2017;23:124–136. doi: 10.1158/1078-0432.CCR-15-1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ngiow SF, von Scheidt B, Akiba H, Yagita H, Teng MW, Smyth MJ. Anti-TIM3 antibody promotes T cell IFN-γ-mediated antitumor immunity and suppresses established tumors. Cancer Res. 2011;71:3540–3551. doi: 10.1158/0008-5472.CAN-11-0096. [DOI] [PubMed] [Google Scholar]
- 30.Kurtulus S, Sakuishi K, Ngiow SF, Joller N, Tan DJ, Teng MW, Smyth MJ, Kuchroo VK, Anderson AC. TIGIT predominantly regulates the immune response via regulatory T cells. J Clin Invest. 2015;125:4053–4062. doi: 10.1172/JCI81187. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 31.Zhang D, Jiang F, Zaynagetdinov R, Huang H, Sood VD, Wang H, Zhao X, Jenkins MH, Ji Q, Wang Y, et al. Identification and characterization of M6903, an antagonistic anti-TIM-3 monoclonal antibody. Oncoimmunology. 2020;9:1744921. doi: 10.1080/2162402X.2020.1744921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Homayouni V, Ganjalikhani-Hakemi M, Rezaei A, Khanahmad H, Behdani M, Lomedasht FK. Preparation and characterization of a novel nanobody against T-cell immunoglobulin and mucin-3 (TIM-3) Iran J Basic Med Sci. 2016;19:1201–1208. [PMC free article] [PubMed] [Google Scholar]
- 33.Jiang Z, Jiang X, Chen S, Lai Y, Wei X, Li B, Lin S, Wang S, Wu Q, Liang Q, et al. Anti-GPC3-CAR T cells suppress the growth of tumor cells in patient-derived xenografts of hepatocellular carcinoma. Front Immunol. 2017;7:690. doi: 10.3389/fimmu.2016.00690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sakuishi K, Apetoh L, Sullivan JM, Blazar BR, Kuchroo VK, Anderson AC. Targeting Tim-3 and PD-1 pathways to reverse T cell exhaustion and restore anti-tumor immunity. J Exp Med. 2010;207:2187–2194. doi: 10.1084/jem.20100643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Koyama S, Akbay EA, Li YY, Herter-Sprie GS, Buczkowski KA, Richards WG, Gandhi L, Redig AJ, Rodig SJ, Asahina H, et al. Adaptive resistance to therapeutic PD-1 blockade is associated with upregulation of alternative immune checkpoints. Nat Commun. 2016;7:10501. doi: 10.1038/ncomms10501. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.