Abstract
The most abundant cells in the tumor microenvironment are cancer-associated fibroblasts (CAFs). They play an important role in oral squamous cell carcinoma (OSCC) angiogenesis, invasion and metastasis. Platelet-derived growth factor (PDGF)-BB has an obvious regulating effect on the formation of CAFs through binding to PDGF receptor (PDGFR)-β, but the role of long non-coding (lnc)RNA in PDGF-BB-induced transformation of fibroblasts into CAFs remains poorly understood. Using an lncRNA ChIP, 370 lncRNA transcripts were identified to be significantly and differentially expressed between fibroblasts and PDGF-BB-induced fibroblasts, including 240 upregulated lncRNAs and 130 downregulated lncRNAs, indicating that lncRNAs are involved in the regulation of the transformation of CAFs. Previous studies have shown that the nuclear factor (NF)-κB signaling pathway plays an important role in the activation of CAFs. Dual-luciferase reporter assay and co-immunoprecipitation were conducted to confirm that the leucine-rich adaptor protein 1-like (LURAP1L), which is the target of lncRNA LURAP1L antisense RNA 1 (LURAP1L-AS1) had a positive regulatory effect on I-κB kinase (IKK)/NF-κB signaling. Therefore, LURAP1L-AS1 was selected and PDGF-BB was demonstrated to upregulate the expression of LURAP1L-AS1 and LURAP1L, which was reversed by a PDGFR-β inhibitor. Subsequently, knocking down LURAP1L-AS1 suppressed the expression of PDGF-BB-induced fibroblast activation marker protein α-smooth muscle actin, fibroblast activation protein-α, PDGFR-β and phosphorylated (p)-PDGFR-β. IKKα, p-IĸB and p-NF-κB were downregulated by the knockdown of LURAP1L-AS1 and upregulated by overexpression of LURAP1L-AS1. The present study indicates that LURAP1L-AS1/LURAP1L/IKK/IĸB/NF-κB plays an important regulatory role in PDGF-BB-induced fibroblast activation and may become a potential target for the treatment of OSCC.
Keywords: oral squamous cell carcinoma, nuclear factor-κB, long non-coding RNA, fibroblasts, cancer-associated fibroblasts, platelet-derived growth factor
Introduction
Oral squamous cell carcinoma (OSCC) is a type of malignant tumor derived from the oral epithelium (1,2). It is the sixth most common malignant tumor in the world (3). Resection combined with radiotherapy and chemotherapy is currently the predominantly used treatment for OSCC, but the prognosis is poor and serious side effects are common (1,2,4). Therefore, investigating the underlying molecular mechanisms of the development of OSCC is required for identifying novel therapeutic targets (5).
The tumor microenvironment (TME) plays an important role in regulating the development of OSCC (6). The most abundant cells in the TME are cancer-associated fibroblasts (CAFs), which have similar characteristics to myofibroblasts (7). CAFs are primarily derived from stromal fibroblasts (8). Indeed, CAFs can be activated by signals, such as paracrine transforming growth factor (TGF)-β, epidermal growth factor (EGF) and platelet-derived growth factor (PDGF) from tumor cells (9–12), and specifically express α-smooth muscle actin (α-SMA), fibroblast activation protein-α (FAP-α), PDGF receptor (PDGFR)-β and fibroblast specific protein (13,14). Conversely, CAFs release a large number of cytokines to regulate the biological behavior of tumors (15) and promote tumors by promoting microvessel formation, interstitial remodeling, drug resistance, immunosuppression, tumor cell proliferation, invasion and metastasis (16).
The PDGF family plays key roles in normal embryonic development, cell growth, cell differentiation and response to tissue damage (17,18). PDGF-BB is one of the important members of the PDGF family and its specific receptor is PDGFR-β (19). The binding of PDGF-BB to its receptor PDGFR-β triggers the formation of dimeric complexes of the receptor, thereby activating downstream signaling molecules to produce a series of biological effects and promote the occurrence and development of tumors (19). Zhang et al (20) demonstrated that PDGF-BB induces human normal fibroblasts (NFs) to transform into CAFs.
Long non-coding (lnc)RNA are RNA molecules 0.2–100 kb in length that lack protein-coding ability. They are key regulators of cell growth, apoptosis, differentiation, invasion and stem cell diversity (21). Studies have shown that metastasis-associated lung adenocarcinoma transcript 1, maternally expressed gene 3 and lncRNA AC132217.4 are closely related to the development, invasion and metastasis of OSCC (22–24). Ding et al (25) identified that lnc-CAF/interleukin-33 transforms NFs into CAFs, promoting the development of OSCC. In addition, Zhao et al (26) found that long intergenic non-protein coding RNA 92 in ovarian cancer cells could maintain CAF activation through glycolysis. Therefore, lncRNAs are important modulators between cancer cells and CAFs, and demonstrate potential application prospects in cancer treatment.
Nuclear factor (NF)-κB is an inducible transcriptional regulator that manipulates the expression of multiple inflammation and immune genes (27). It is a key signaling molecule connecting inflammation and tumors, and has important effects on the TME (28). Phosphorylated (p) NF-κB enters the nucleus to induce the reorganization of the fibroblast structure, as characterized by the increased expression of cytoskeletal protein α-SMA and activation of fibroblasts into CAFs (29,30). Leucine-rich adaptor protein 1-like (LURAP1L) is a protein of the LURAP1 family that activates the canonical NF-κB pathway (31). The present study hypothesized that LURAP1L was the target of lncRNA LURAP1L antisense RNA 1 (LURAP1L-AS1). Notably, the preliminary lncRNA ChIP results indicated that expression of lncRNA LURAP1L-AS1 was upregulated during the activation of fibroblasts induced by PDGF-BB. Therefore, it was hypothesized that PDGF-BB might upregulate LURAP1L by promoting lncRNA LURAP1L-AS1 expression and subsequently activating the canonical NF-κB pathway, thereby inducing the transformation of NFs into CAFs.
Materials and methods
Cell culture and treatment
Normal human oral mucosa (p3) 500K fibroblast (hOMF) cells were purchased from CellResearch Corporation and grown in complete DMEM, high glucose (Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% fetal bovine serum (cat. no 16000-04; Gibco; Thermo Fisher Scientific, Inc.; fetal bovine serum:DMEM=1:9) and Gibco penicillin-streptomycin solution (0.1 U/ml penicillin and 0.1 µg/ml streptomycin; Thermo Fisher Scientific, Inc.) at 37°C in humidified air with 5% CO2.
For activation, recombinant human PDGF-BB (32) was used to stimulate the conventional hOMF cells for 72 h before passage. After three consecutive passages, the cells were collected for subsequent experiments.
Protein extraction and western blotting
Total protein was extracted from cell cultures using Cell Extraction Buffer (Thermo Fisher Scientific, Inc.) and the Mini Protease Inhibitor Cocktail (Roche Diagnostics). The protein concentration was determined using a BCA Protein assay kit (Beyotime Institute of Biotechnology). Equal quantities (20 µg) of proteins were separated using 10% SDS-PAGE and then transferred to a PVDF membrane (EMD Millipore). The membrane was blocked in 5% skimmed milk in TBS containing 0.1% Tween-20 (TBST, pH 7.2) for 1 h at room temperature, incubated with an appropriate quantity of primary antibody at 4°C overnight and washed three times with TBST for 15 min each time and then incubated with secondary antibodies. Immunoreactivity was revealed by chemiluminescence using a western blot imaging system (ImageQuant LAS 4000; GE Healthcare) and the gray value was analyzed using ImageJ software 1.41 (National Institutes of Health). β-tubulin, β-actin and GAPDH served as internal references. All of the samples were run in triplicate as a minimum. The following antibodies were used in the present study: α-SMA (cat. no. ab5694; 1:1,000), FAP-α (cat. no. ab53066; 1:500), IKKα (cat. no. ab32041; 1:1,000), IκBα (cat. no. ab32518; 1:1,000), NF-κB p65 (cat. no. ab16502; 1:1,000), p-NF-κBp65 (p-S536; cat. no. ab86299; 1:1,000), GAPDH (cat. no. ab181602; 1:10,000), β-tubulin (cat. no. ab179511; 1:1,000) and β-actin (cat. no. ab8227; 1:1,000) were obtained from Abcam; LURAP1L (cat. no. PA5-55072; 1:1,000) was obtained from Thermo Fisher Scientific, Inc., and PDGFR-β (cat. no. bs-0232R; 1:1,000) and p-PDGFR-β (Tyr740; cat. no. bs-3323R; 1:1,000) were obtained from BIOSS. Goat Anti-Rabbit IgG H&L (HRP) (cat. no. zs-2301; 1:5,000) was obtained from ZSBIO.
Immunofluorescence (IF)
For IF, cells were plated on coverslips, washed three times with PBS, fixed in 4% paraformaldehyde for 10 min at room temperature, permeabilized with 0.5% Triton X-100 for 15 min and incubated with primary antibodies, anti-α-SMA, anti-FAP-α and anti-PDGFR-β at 4°C overnight, followed by a 1-h incubation with rhodamine-conjugated goat anti-rabbit IgG (DyLight 649; cat. no. A23620; Abbkine Scientific Co., Ltd. and Alexa Fluor®488, cat. no. ab150081; Abcam) at 37°C. The nuclei were counterstained with 4′,6-diamidino-2-phenylindole (Invitrogen; Thermo Fisher Scientific, Inc, 0.5 µg/ml) for 5 min in the dark at room temperature. The coverslips were evaluated by fluorescence microscopy using an 80i Eclipse microscope (Nikon Corporation).
RNA extraction
Total RNA was extracted using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's instructions. RNA quantity and quality were measured using a NanoDrop® ND-1000 (NanoDrop Technologies; Thermo Fisher Scientific, Inc.). RNA integrity was assessed by standard denaturing agarose gel electrophoresis.
Microarray
An Agilent array platform (Agilent Technologies, Inc.) was used for microarray analysis. The sample preparation and microarray hybridization were performed according to the manufacturer's instructions with minor modifications. Briefly, mRNA was purified from total RNA after removal of rRNA with an mRNA-ONLY™ Eukaryotic mRNA Isolation kit (Epicentre; Illumina, Inc.). Then, each sample was amplified and transcribed into fluorescent cRNA along the entire length of the transcripts without 3′bias using a random priming method (Flash RNA Labeling kit; Arraystar, Inc.). The labeled cRNAs were hybridized onto a Human LncRNA Microarray V4.0 (8×60k; Arraystar, Inc.) designed for 40,173 lncRNAs and 20,730 coding transcripts. The lncRNAs were carefully constructed using public transcriptome databases, including RefSeq (http://www.ncbi.nlm.nih.gov/refseq/), UCSC Known Genes (http://www.biomedsearch.com/nih/UCSC-Known-Genes/16500937.html) and GENCODE (http://www.gencodegenes.org/) as well as landmark publications (33–35). Each transcript was accurately identified by a specific exon or splice junction probe. Positive probes for housekeeping genes and negative probes were printed onto the array for hybridization quality control. After washing the slides with Gene Expression Wash Buffer 1 (p/n 5188–5325; Agilent Technologies, Inc.), the arrays were scanned using a G2505C scanner (Agilent Technologies, Inc.) and the acquired array images were analyzed with the Feature Extraction software Version 11.0.1.1 (Agilent Technologies, Inc.). Quantile normalization and subsequent data processing were performed using the GeneSpring GX V12 1 software package (Agilent Technologies, Inc.). Microarray was performed by Kangchen Bio-tech Inc.
Reverse transcription-quantitative PCR (RT-qPCR) validation of lncRNAs
Total RNA (2 µg) was reverse transcribed into cDNA using SuperScriptTM III Reverse Transcriptase (cat. no. 18080093; Invitrogen; Thermo Fisher Scientific, Inc.). LncRNA expression was measured by qPCR using the SYBR Premix Ex Taq (Thermo Fisher Scientific, Inc.) with an ABI PRISM® 7000 Sequence Detection System (Thermo Fisher Scientific, Inc.). The total reaction volume was 10 µl, including 5 µl Master Mix (2X), 2 µl cDNA template, 0.5 µl forward primer, 0.5 µl reverse primer (10 µM) and 2 µl double-distilled water. The qPCR reaction was performed with an initial denaturation step of 10 min at 95°C, then 95°C (5 sec) and 60°C (60 sec) for a total of 40 cycles, with a final extension step at 72°C for 5 min. All experiments were performed in triplicate and all samples were normalized to GAPDH. The median for each triplicate was used to calculate the relative lncRNA concentrations (∆Cq=Cq median lncRNAs-Cq median GAPDH) (36). The primer sequences used were as follows: GAPDH forward, 5′-GGGAAACTGTGGCGTGAT-3′ and reverse, 5′-GAGTGGGTGTCGCTGTTGA-3′; LURAP1L forward, 5′-CCTCCTCAGGCAAGAGATGGT-3′ and reverse, 5′-TGCTGCCTCTGCTGGTAATG-3′; and LURAP1L-AS1 forward, 5′-GAGCGGTCAAATAGAGGATAT-3′ and reverse, 5′-ATATCCTCTATTTGACCGCTC-3′.
Bioinformatics prediction and dual-luciferase reporter assays
The LURAP1L-AS1 potential mRNA binding sites were predicted using network analysis (http://atlasgeneticsoncology.org). The luciferase reporter plasmid encoding both Renilla luciferase (hRluc) pmiR-RB-REPORT™ the control firefly luciferase (hluc+) (both Guangzhou RiboBio Co., Ltd.) were used for all assays. The dual-luciferase reporter assays were performed by Shanghai GeneChem Co., Ltd. according to the method described by Fish et al (37). The 3′-untranslated region (UTR) of the target gene or the relevant negative control was constructed to the back of luciferase. The lncRNA LURAP1L-AS1 was provided and constructed into the reporter plasmid pmir-GLO (Shanghai GeneChem Co., Ltd.; LURAP1L-AS1 mimic). The empty pmir-GLO plasmid served as the negative control for lncRNA LURAP1L-AS1 (mimic control). Constructed vectors (or constructed and negative vectors) were used to co-transfect HEK293T cells (Shanghai GeneChem Co., Ltd.) into four experimental groups: 3′UTR-NC+LURAP1L-AS1-NC; 3′UTR-NC+LURAP1L-AS1; 3′UTR-LURAP1L+LURAP1L-AS1-NC; and 3′UTR- LURAP1L+LURAP1L-AS1. HEK293T cells were seeded into 96-well plates and transfection solution (100 µl) containing 0.5 µl Lipofectamine 2000® (Invitrogen; Thermo Fisher Scientific, Inc.) and 25 ng reporter plasmids with LURAP1L-AS1 mimic (50 nM) or mimic control (50 nM) was added. The cells were washed with phosphate buffer saline and then fully lysed with Passive Lysis Buffer 1× (Shanghai GeneChem Co., Ltd.) for 20 min at 4°C 24 h after transfection.
Luciferase activities were measured using the Dual-luciferase Reporter assay kit (Promega Corporation) and Centro XS (Titertek-Berthold). The ratio of firefly luciferase to Renilla luciferase in the same sample pore represented the relative expression of luciferase. For the two groups transfected with the same luciferase plasmid, the relative expression of luciferase in the LURAP1L-AS1-NC group was normalized to 1 and the relative expression of luciferase in the LURAP1L-AS1 group was normalized to that in the LURAP1L-AS1-NC group. The normalized data of the 3′UTR-NC+LURAP1L-AS1 group and relative expression of luciferase in 3′UTR-NC+LURAP1L and 3′UTR-LURAP1L+LURAP1L-AS1 were compared.
Co-immunoprecipitation (co-IP)
Co-IP analysis was performed as described previously (38). Briefly, the cultured cells were lysed using RIPA buffer (Beijing Solarbio Science & Technology Co., Ltd.) containing 1% protease inhibitors. The lysates were immunoprecipitated with anti-LURAP1L or anti-IKKα for 1 h at 37°C, followed by incubation with 100 µl Protein A Agarose (cat. no. #9863; Cell Signaling Technology, Inc.) overnight at 4°C. The next day, the Protein A Agarose-antigen-antibody complexes were collected by centrifugation at 12,000 × g for 2 min at 4°C and the immunoprecipitation-HAT buffer (cat. no. 10009330-1; Aimeijie Technology Co., Ltd.) was used to wash the complexes five times. Western blot was used to detect bound proteins.
Knockdown and overexpression of LURAP1L-AS1
To observe the effects of LURAP1L-AS1 knockdown on fibroblasts activated by PDGF-BB, three different small interfering (si)RNAs that targeted LURAP1L-AS1 RNA and a scrambled siRNA control were provided by Shanghai GenePharma Co. Ltd. The three siRNAs were transfected into hOMF cells using Lipofectamine 2000 according to the manufacturer's instructions. Twenty-four hours after transfection, the LURAP1L-AS1 expression levels were measured through RT-qPCR and it was observed that siRNA-LURAP1L-AS1 (forward, 5′-GAGCGGTCAAATAGAGGATAT-3′ and reverse, 5′-ATATCCTCTATTTGACCGCTC-3′) yielded the highest degree of LURAP1L-AS1 silencing. Subsequently, the LURAP1L-AS1-targeting sequence was designed, synthesized and inserted into a SuperSilencing shRNA Expression Vector System (Shanghai GenePharma Co. Ltd.). An unrelated sequence lentiviral vector GV248 (Shanghai GenePharma Co. Ltd.) served as a negative control. hOMF cells were then plated into 6-well plates and allowed to adhere for 24 h. The lentivirus was transfected according to the manufacturer's instructions. Stably transfected cells were selected with puromycin (MilliporeSigma) and confirmed through fluorescence microscopy and RT-qPCR.
The overexpression of LURAP1L-AS1 in hOMF was achieved using a transient transfection method. The primers for LURAP1L-AS1-targeting sequence cloning were designed and synthesized in the present study (LURAP1L-AS1-p1 forward, TACCGGACTCAGATCTCGAGTTAGAATGATCTAATGAAAAC and reverse, TACCGTGACTGCAGAATTCACAAATTGAAGAATATTTATTTTAGGTTAAAATATTTTTAAG; Shanghai Genechem Co., Ltd.). The cloned LURAP1L-AS1 was verified by sequencing (sequencing primer forward, CGCAAATGGGCGGTAGGCGTG and reverse, AACGCACACCGGCCTTATTC) and subsequently cloned into a GV146 overexpression vector (Shanghai Genechem Co., Ltd.). The GV146 empty vector served as a negative control, while PDGF-BB served as a positive control. hOMF cells were transfected using Lipofectamine 2000 according to the manufacturer's instructions. After culturing for 3–4 days, RT-qPCR was performed to confirm cell transfection.
Statistical analysis
All experiments were repeated at least three times and data are presented as means ± standard errors of the mean. Graphpad Prism 5.0 (GraphPad Software Inc.) was used to perform statistical analysis. Differences between two groups were compared with an independent samples t-test. Differences among three or more groups were compared with one-way analysis of variance and the Dunnett's post hoc test. P<0.05 was considered to indicate a statistically significant difference.
Results
Effects of PDGF-BB on fibroblast activation
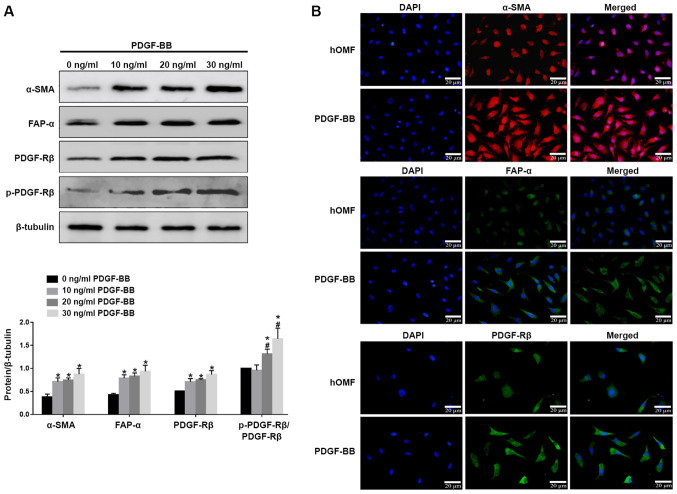
PDGF-BB (10, 20 and 30 ng/ml) was used to stimulate conventionally cultured fibroblasts for 72 h. The cells from the third passage were used. Western blotting showed that all the three concentrations of PDGF-BB could upregulate the expression of α-SMA, FAP-α, PDGFR-β and p-PDGFR-β, while the effect of PDGF-BB was in a dose-dependent manner (Fig. 1A). IF confirmed that PDGF-BB upregulated the expression levels of α-SMA, FAP-α and PDGFR-β, as shown by an increased fluorescence signal (Fig. 1B). These results indicate that PDGF-BB could induce fibroblast activation.
Figure 1.
PDGF-BB induced the formation of the CAF phenotype in hOMF. (A) The cells were treated with PDGF-BB in a dose-dependent manner (10, 20, 30 ng/ml) in 10% fetal bovine serum for 72 h for three passages. The cells were subjected to western blot analysis with antibodies against CAF markers α-SMA, FAP-α, PDGFR-β and p-PDGFR-β. β-tubulin served as a loading control and sample loading was 20 µg. (B) Fluorescence microscopy of hOMF stained with α-SMA, FAP-α and PDGFR-β (probed with a primary and a secondary antibody). Cells were counterstained with DAPI. Data were expressed as means ± SEM (n=3). *P<0.05 vs. hOMF, #P<0.05 vs. 10 ng/ml PDGF-BB. PDGF, platelet-derived growth factor; CAF, cancer-associated fibroblast; hOMF, human oral mucosa (p3) 500K fibroblast; α-SMA, α-smooth muscle actin; FAP-α, fibroblast activation protein-α; PDGFR-β, platelet-derived growth factor receptor-β; p, phosphorylated.
Target gene prediction and dual-luciferase reporter assay
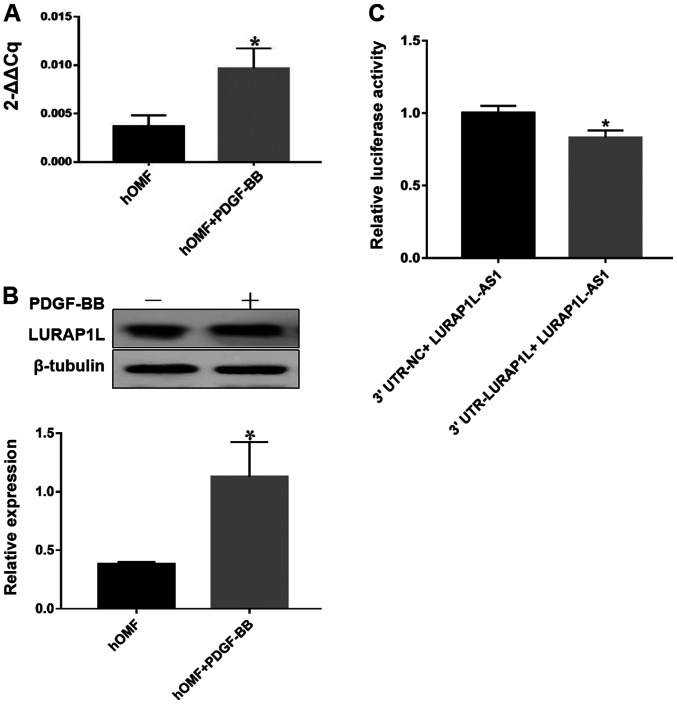
As the aim of the present study was to identify the lncRNA interaction with LURAP1L, a key regulator activating the canonical NF-κB pathway, western blotting and RT-qPCR were used to evaluate the expression of LURAP1L. The results indicated that LURAP1L expression was upregulated by PDGF-BB (Fig. 2A and B). Next, we predicted LURAP1L-AS1 may have an impact on the expression levels of LURAP1L, as well as the binding sites between LURAP1L and LURAP1L-AS1 using network analysis. The predicted results demonstrated that there was a significant correlation between LURAP1L-AS1 and LURAP1L (Fig. S1). Furthermore, the dual-luciferase reporter assay indicated that LURAP1L-AS1 significantly repressed the activity of luciferase derived from RNAs containing the 3′UTR of LURAP1L (Fig. 2C). This implies that LURAP1L is the target gene of LURAP1L-AS1 and that PDGF-BB induces the upregulation of LURAP1L via LURAP1L-AS1.
Figure 2.
Result of the target gene prediction and dual-luciferase reporter assay. The cells were treated with 30 ng/ml PDGF-BB in 10% fetal bovine serum for 72 h for three passages. (A) Reverse transcription-quantitative PCR analysis was performed to evaluate the expression of LURAP1L. Data are presented as the mean ± SEM of triplicate experiments. (B) Western blot analysis of LURAP1L. Densitometry was used to determine the LURAP1L/β-actin ratios. Data are presented as means ± SEM of three independent experiments. (C) Dual-luciferase reporter assay was performed to verify the regulatory effect of LURAP1L-AS1 on LURAP1L. *P<0.05. LURAP1L-AS1-NC, LURAP1L-AS1 empty plasmid as the negative control; LURAP1L-AS1, vector plasmid expressing target LURAP1L-AS1; 3′UTR-NC: Empty plasmid without 3′UTR-LURAP1L negative control; 3′UTR-LURAP1L, plasmid harboring LURAP1L at the 3′UTR. PDGF, platelet-derived growth factor; LURAP1L, leucine-rich adaptor protein 1-like; LURAP1L-AS1, LURAP1L antisense RNA 1; NC, negative control; UTR, untranslated region; hOMF, human oral mucosa (p3) 500K fibroblast.
PDGF-BB regulates lncRNA expression in fibroblasts, as confirmed by RT-PCR
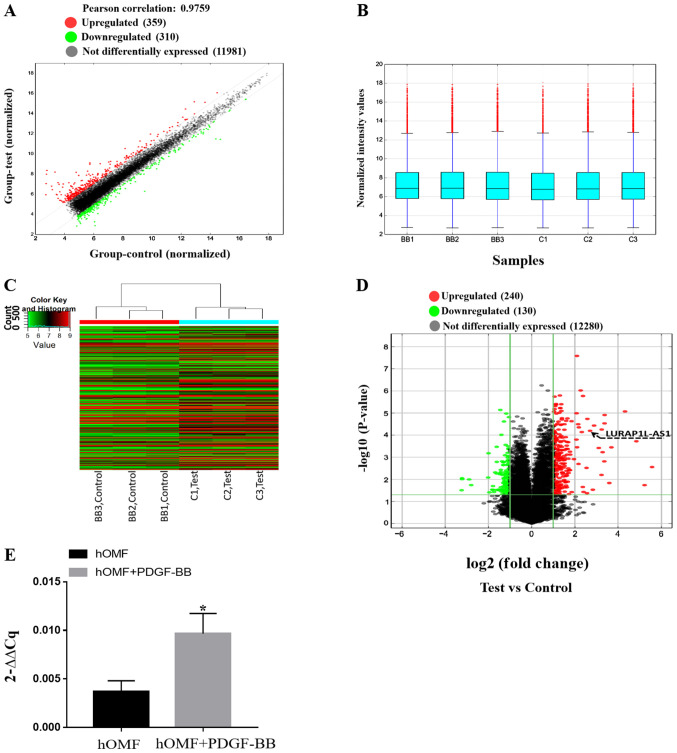
In order to confirm the results above and the mechanisms of activation of fibroblasts induced by PDGF-BB, an lncRNA microarray was used to screen the samples from fibroblasts and PDGF-BB-activated fibroblasts. After normalization and data filtering, 12,650 lncRNAs were identified. The heat map showed a distinguishable lncRNA expression profile between the two groups (Fig. 3A-C). Compared with the control group, 240 lncRNAs were upregulated, and 130 were downregulated (fold-change ≥2) as shown in Fig. 3D (P<0.05). Table I presents the lncRNAs that were up- or downregulated by PDGF-BB (fold-change ≥5; P<0.05). Among these, it was confirmed that LURAP1L-AS1 was upregulated, as demonstrated by the arrow in Fig. 3D. RT-qPCR for LURAP1L-AS1 was also performed and the results were consistent with the microarray results (P<0.05; Fig. 3E). These results indicated significantly upregulated expression of LURAP1L-AS1 in fibroblasts following PDGF-BB treatment and supported the prediction of the present study that LURAP1L-AS1 activates LURAP1L in fibroblasts.
Figure 3.
Expression profiles of lncRNAs between PDGF-BB-hOMF and hOMF by microarray analysis to confirm the expression of LURAP1L-AS1 in activated hOMF cells. (A) The scatterplot is a visualization method used for assessing the lncRNA expression variation between PDGF-BB-hOMF and hOMF cells. The gray lines are the fold-change lines (the default fold-change value given is 2.0). (B) The box plot compares the distributions of the intensities from all samples. Following normalization, the distributions of the log2 ratios among the three pairs of samples were almost the same. (C) The differentially expressed lncRNAs were analyzed using hierarchical clustering. Red indicates high relative expression and blue indicates low relative expression. (D) Differentially expressed lncRNAs with statistical significance between two groups were identified through volcano plot filtering. The vertical lines correspond to two-fold up- and downregulation, respectively and the horizontal line represents P=0.05. The red points in the plot represent significantly upregulated lncRNAs where LURAP1L-AS1 is identified by the black arrow. The green points in the plot represent significantly downregulated lncRNAs. (E) Reverse transcription-quantitative PCR verification was performed for LURAP1L-AS1 expression in hOMF cells. *P<0.05. lncRNA, long non-coding RNA; PDGF, platelet-derived growth factor; hOMF, human oral mucosa (p3) 500K fibroblast; LURAP1L, leucine-rich adaptor protein 1-like; LURAP1L-AS1, LURAP1L antisense RNA 1.
Table I.
Differentially expressed long non-coding RNAs.
| Probe name | P-value | Fold-change | LncRNA expression | Gene symbol |
|---|---|---|---|---|
| ASHGV40002366 | 1.54×10−3 | 10.2773694 | Upregulated | LOC102724467 |
| ASHGV40016892 | 2.78×10−3 | 47.6213517 | Upregulated | RORA-AS1 |
| ASHGV40003735 | 1.69×10−6 | 5.186901 | Upregulated | SNHG8 |
| ASHGV40024576 | 2.78×10−4 | 28.7604505 | Upregulated | XLOC_013370 |
| ASHGV40035139 | 3.04×10−3 | 5.8165393 | Upregulated | LINC00886 |
| ASHGV40048333 | 6.30×10−3 | 9.386177 | Upregulated | G077644 |
| ASHGV40059227 | 5.59×10−5 | 9.4526926 | Upregulated | G090807 |
| ASHGV40010343 | 5.91×10−4 | 9.7206085 | Upregulated | G019348 |
| ASHGV40005652 | 3.70×10−5 | 7.4065884 | Upregulated | T042114 |
| ASHGV40035740 | 1.84×10−5 | 5.9734672 | Upregulated | T239636 |
| ASHGV40055486 | 8.45×10−6 | 19.9812999 | Upregulated | G090385 |
| ASHGV40033198 | 1.23×10−5 | 10.3854729 | Upregulated | RP4-756G23.5 |
| ASHGV40007819 | 3.57×10−4 | 12.9009956 | Upregulated | RP11-264E20.2 |
| ASHGV40051519 | 6.35×10−5 | 6.5042034 | Upregulated | LURAP1L-AS1 |
| ASHGV40041072 | 8.92×10−3 | 9.313185 | Downregulated | CTC-436K13.5 |
Fold-change ≥5. P<0.05.
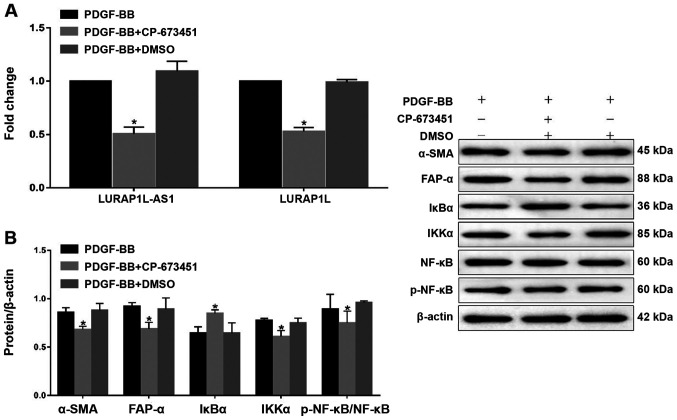
Inhibition of PDGFR-β affects LURAP1L-AS1- LURAP1L/IKK/IκB/NF-κB signaling and CAF programming. To examine the PDGF-BB-induced signaling pathway via PDGFR-β, a PDGFR-β tyrosine kinase inhibitor, CP-673451 (Selleck Chemicals) was used prior to constructing a fibroblast activation model using PDGF-BB (32). The results indicated that CP-673451 downregulated LURAP1L-AS1 and LURAP1L (Fig. 4A). In addition, western blotting showed that CP-673451 downregulated α-SMA, FAP-α, IKKα and p-p65, but upregulated IκBα (Fig. 4B). These results demonstrate that PDGFR-β is involved in LURAP1L-AS1-LURAP1L/IKK/IκB/NF-κB signaling and CAF programming.
Figure 4.
Effects of the PDGFR-β inhibitor, CP-673451 on LURAP1L-AS1-LURAP1L/IKK/IκB/NF-κB signaling and CAF programming. (A) Reverse transcription-quantitative PCR analysis of LURAP1L and LURAP1L-AS1 expression levels. (B) Western blot analysis of FAP-α, α-SMA, IKKα, IκBα, NF-κB p65 and p-p65 expression following overexpression of LURAP1L-AS1 and treatment with PDGF-BB. *P<0.05 vs. PDGF-BB. PDGF, platelet-derived growth factor; LURAP1L, leucine-rich adaptor protein 1-like; LURAP1L-AS1, LURAP1L antisense RNA 1; IKKα, IκB kinase α; IKK, I-κB kinase; IκBα, nuclear factor of κ light polypeptide gene enhancer in B-cells inhibitor α; NF-κB, nuclear factor-κB; CAF, cancer-associated fibroblasts; p, phosphorylated; α-SMA, α-smooth muscle actin; FAP-α, fibroblast activation protein-α.
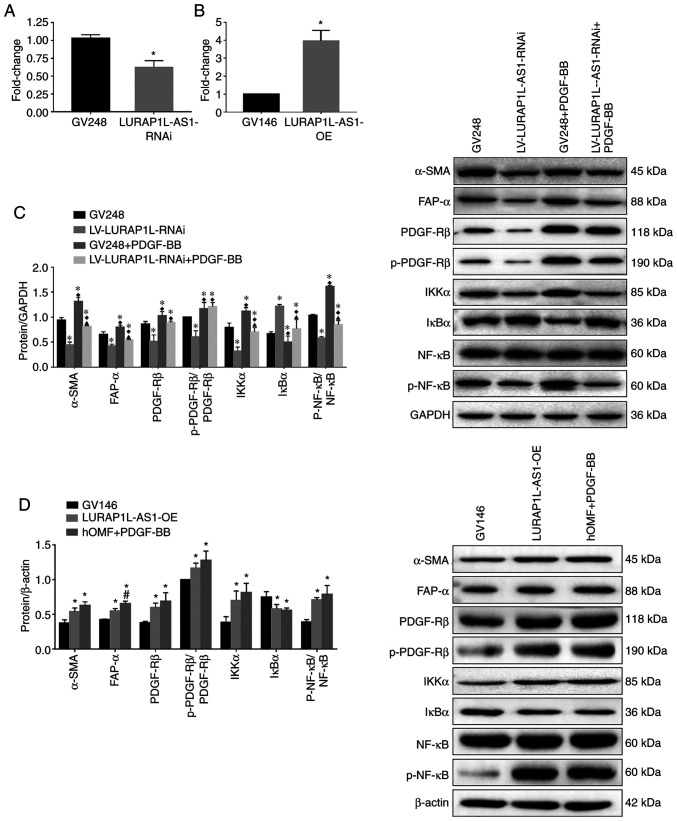
Knockdown and overexpression of lncRNA LURAP1L-AS1 alter the effects of PDGF-BB on fibroblast activation and IKKα/IκBα/p-NF-κB expression
To verify the role of LURAP1L-AS1 in PDGF-BB-induced fibroblast activation, LURAP1L-AS1 was knocked down or overexpressed in fibroblast cells, as confirmed by RT-qPCR (Fig. 5A and B). The fibroblast activation model was then established using these cells. The results indicated that levels of the activation marker proteins α-SMA, FAP-α, PDGFR-β and p-PDGFR-β were significantly lower after knocking down LURAP1L-AS1 (Fig. 5C). Furthermore, IKKα and p-p65 were downregulated, while IκBα was upregulated by knocking down LURAP1L-AS1 (Fig. 5C; P<0.05). Contrasting results were obtained by overexpressing LURAP1L-AS1 (Fig. 5D). The results indicated that the LURAP1L-AS1/IKKα/IκBα/NF-κB axis might be involved in PDGF-BB-induced fibroblast activation into CAFs. Furthermore, LURAP1L-AS1 associated with PDGF-BB-induced fibroblast activation and affected the NF-κB signaling pathway.
Figure 5.
Knockdown and overexpression of lncRNA LURAP1L-AS1 alters the effects of PDGF-BB on fibroblast activation and interaction between LURAP1L and IKKα. hOMF cells were constructed for knocking down and overexpressing LURAP1L-AS1. The fibroblast activation model was established using transfected cells. Expression of lncRNA LURAP1L-AS1 in hOMF cells with LURAP1L-AS1 (A) knocked down and (B) overexpressed. (C) Western blot analysis of the expression of FAP-α, α-SMA, IKKα, IκBα, NF-κB p65, and p-p65 after knocking down LURAP1L-AS1 and treatment with PDGF-BB. *P<0.05 vs. GV248; ♦P<0.05 vs. LURAP1L-AS1-RNAi; ▲P<0.05 vs. GV248+PDGF-BB. (D) Western blot analysis of the expression of FAP-α, α-SMA, IKKα, IκBα, NF-κB p65 and p-p65 after overexpressing LURAP1L-AS1 and treatment with PDGF-BB. *P<0.05 vs. GV146; #P<0.05 vs. LURAP1L-AS1-OE. lncRNA, long non-coding RNA; OE, overexpression; GV248, negative control; GV146, negative control; PDGF, platelet-derived growth factor; hOMF, human oral mucosa (p3) 500K fibroblast; LURAP1L, leucine-rich adaptor protein 1-like; LURAP1L-AS1, LURAP1L antisense RNA 1; p, phosphorylated; α-SMA, α-smooth muscle actin; FAP-α, fibroblast activation protein-α; IKKα, IκB kinase α; IκBα, nuclear factor of κ light polypeptide gene enhancer in B-cells inhibitor α; RNAi, RNA interference.
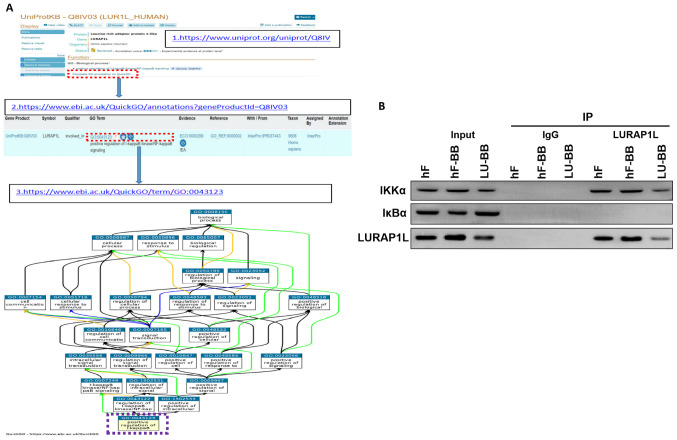
Co-IP indicates an interaction between LURAP1L and IKKα
The results demonstrated that LURAP1L-AS1 could affect the changes in the NF-κB signaling pathway and that LURAP1L was a target gene of LURAP1L-AS1, but the regulatory effect of LURAP1L on the NF-κB signaling pathway had to be verified. Through protein-protein interaction analysis, an interaction was identified between LURAP1L and IKKα in the NF-κB signaling pathway (Fig. 6A). Therefore, co-IP was performed for verification. The results demonstrated that the IKKα protein was detected in the interacting protein of LURAP1L, indicating that LURAP1L interacts with IKKα and promotes the activation of the NF-κB signaling pathway to affect the activation of fibroblasts (Fig. 6B). It was also observed that lncRNA LURAP1L-AS1 knockdown weakened the interaction between LURAP1L and IKKα (Fig. 6B).
Figure 6.
Demonstration of interaction between LURAP1L and IKKα in the NF-κB signaling pathway. (A) Protein-protein interaction analysis in database (www.uniprot.org) (49). (B) Co-immunoprecipitation was used to analyze the interaction between LURAP1L and IKKα. hF, hOMF; hF-BB, hOMF+PDGF-BB; LU-BB, LURAP1L-AS1-RNAi+PDGF-BB; LURAP1L, leucine-rich adaptor protein 1-like; LURAP1L-AS1, LURAP1L antisense RNA 1; IKKα, IκB kinase α.
Discussion
CAFs are matrix fibroblasts that were first isolated from prostate cancer tissues by Olumi et al (39) in 1999. In the present study, the results showed that compared with the normal group, α-SMA, FAP-α, PDGFR-β and p-PDGFR-β were significantly increased by PDGF-BB, indicating fibroblast activation. The lncRNA microarray showed that the expression of LURAP1L-AS1 was upregulated by PDGF-BB. Furthermore, the present study showed that the target gene of LURAP1L-AS1 is LURAP1L. The LURAP1L protein interacts with IKKα to activate NF-κB, thereby promoting the activation of NFs.
CAFs are derived from the host's fibroblasts and are induced by a variety of cytokines secreted by tumor cells, such as TGF-β, PDGF and fibroblast growth factor (40). CAFs specifically express α-SMA, FAP-α and PDGFR-β (13). The present study showed that the expression of α-SMA, FAP-α, PDGFR-β and p-PDGFR-β were upregulated in fibroblasts induced by PDGF-BB, indicating that PDGF-BB could activate NFs into CAFs. This is consistent with the results by Zhang et al (20). Indeed, CAFs are mainly derived from resting fibroblasts (NFs) in the matrix. When stimulated by external factors, NFs will activate into CAFs with very different shapes and functions. Another previous study showed that the overexpression of PDGF-BB and its receptor could promote the occurrence and development of tumors and was closely related to CAFs (41). Aoto et al (42) and Rizvi et al (43) suggested the blockade of PDGFRβ as a potential strategy to prevent the formation of CAFs, supporting the role of PDGF-BB in the formation of CAFs in the TME, which is consistent with the present study.
CAF activation can be regulated by many factors, such as growth factors, inflammatory factors, transcription factors, hypoxia, reactive oxygen species and non-coding RNA (44,45). The biological functions of lncRNA are complex and they regulate transcription and post-transcription. Tumorigenesis and development are closely related to tumor cell proliferation, invasion, metastasis and recurrence. lnc003875 also has a significant regulatory effect on CAF activation (46). The present study found that lncRNA LURAP1L-AS1 expression was significantly upregulated during PDGF-BB-induced fibroblast activation. Following RNA interference, lncRNA LURAP1L-AS1 could inhibit the expression of the activation marker proteins α-SMA, FAP-α, PDGFR-β and p-PDGFR-β, indicating that the lncRNA LURAP1L-AS1 is involved in the process by which PDGF-BB activates fibroblasts.
The present study showed that the LURAP1L- AS1/LURAP1L/IKKα/IκBα/NF-κB axis might be involved in PDGF-BB-induced fibroblast activation into CAFs. NF-κB primarily exists in eukaryotic cells. It is a multidirectional and multifunctional nuclear transcription factor that plays a pivotal role in mediating intracellular signal transduction to promote tumor cell proliferation, inhibit apoptosis, promote migration and stimulate angiogenesis. NF-κB in tumor cells and the TME is continuously activated, and the activation of NF-κB depends on the decrease in the expression of IκB, which is the inhibitor of NF-κB. The phosphorylation of IKK is closely associated with this process. Zheng et al (47) found that downregulation of the cell membrane protein CD146 in pancreatic cancer stroma could also stimulate NF-κB signaling and promote CAF activation. Liu et al (29) showed that TNF-α could activate CAFs by enhancing the transcriptional activity of NF-κB, thereby promoting tumorigenesis and cancer development. Reactive oxygen species produced by tumors induce the expression of the chloride intracellular channel 4 and C-C motif chemokine ligand 2 in CAFs, stimulate the expression of TGF-β1 and NF-κB and induce CAF activation (30,48). The abovementioned studies showed that the NF-κB signaling pathway plays an important role in the activation of CAFs.
LURAP1L is a key regulator that activates the canonical NF-κB pathway (31). In the present study, potential direct mRNA targets of LURAP1L-AS1 were identified through bioinformatics and LURAP1L was identified as the target gene of LURAP1L-AS1. Additionally, its encoding protein LURAP1L interacts with IKKα, which was demonstrated by co-IP. Knockdown of LURAP1L-AS1 downregulated the expression levels of IKKα and p-NF-κB and increased the expression of IκBα, indicating that LURAP1L-AS1 may have a regulatory effect on the NF-κB signaling pathway. Therefore, the present study showed that the LURAP1L-AS1/LURAP1L/IKK/IκB/NF-κB axis plays an important role in the regulation of PDGF-BB-induced fibroblast activation. However, additional studies are required to determine the exact mechanisms involved in this process. Furthermore, the present study focused on LURAP1L-AS1, but other lncRNAs are also involved in CAF activation and require investigation in the future.
Thus, the present study identified that the LURAP1L- AS1/LURAP1L/IKK/IĸB/NF-κB axis plays an important regulatory role in PDGF-BB-induced fibroblast activation into CAFs, indicating that this axis might represent a potential target for the treatment of OSCC.
Supplementary Material
Acknowledgements
Not applicable.
Funding Statement
The present study was supported by the National Natural Science Foundation of China (grant no. 81660448 and 81360401), the Special Health Technical Personnel Training program of Yunnan, China (grant no. L-201612) and the Natural Science Foundation of Yunnan, China (grant number 2017FE468-006).
Funding
The present study was supported by the National Natural Science Foundation of China (grant no. 81660448 and 81360401), the Special Health Technical Personnel Training program of Yunnan, China (grant no. L-201612) and the Natural Science Foundation of Yunnan, China (grant number 2017FE468-006).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
XR and LL performed the experiments, participated in collecting data and drafted the manuscript. YH and LB designed the current study and confirm the authenticity of all the raw data. KL and JW analyzed the data and performed the experiments. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Argiris A, Karamouzis MV, Raben D, Ferris RL. Head and neck cancer. Lancet. 2008;371:1695–1709. doi: 10.1016/S0140-6736(08)60728-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wei WI, Sham JS. Nasopharyngeal carcinoma. Lancet. 2005;365:2041–2054. doi: 10.1016/S0140-6736(05)66698-6. [DOI] [PubMed] [Google Scholar]
- 3.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 4.Head and Neck Cancers. National Comprehensive Cancer Network; Fort Washington: 2020. NCCN Clinical Practice Guidelines in Oncology (NCCN Guidelines) Version 1.2020. [DOI] [PubMed] [Google Scholar]
- 5.Li CC, Shen Z, Bavarian R, Yang F, Bhattacharya A. Oral cancer: Genetics and the role of precision medicine. Surg Oncol Clin N Am. 2020;29:127–144. doi: 10.1016/j.soc.2019.08.010. [DOI] [PubMed] [Google Scholar]
- 6.Reina-Campos M, Moscat J, Diaz-Meco M. Metabolism shapes the tumor microenvironment. Curr Opin Cell Biol. 2017;48:47–53. doi: 10.1016/j.ceb.2017.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brouty-Boyé D. Developmental biology of fibroblasts and neoplastic disease. Prog Mol Subcell Biol. 2005;40:55–77. doi: 10.1007/3-540-27671-8_3. [DOI] [PubMed] [Google Scholar]
- 8.Kalluri R. The biology and function of fibroblasts in cancer. Nat Rev Cancer. 2016;16:582–598. doi: 10.1038/nrc.2016.73. [DOI] [PubMed] [Google Scholar]
- 9.Wu W, Zaal EA, Berkers CR, Lemeer S, Heck AJR. CTGF/VEGFA-activated fibroblasts promote tumor migration through Micro-environmental modulation. Mol Cell Proteomics. 2018;17:1502–1514. doi: 10.1074/mcp.RA118.000708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Abdul-Wahid A, Cydzik M, Fischer NW, Prodeus A, Shively JE, Martel A, Alminawi S, Ghorab Z, Berinstein NL, Gariépy J. Serum-derived carcinoembryonic antigen (CEA) activates fibroblasts to induce a local re-modeling of the extracellular matrix that favors the engraftment of CEA-expressing tumor cells. Int J Cancer. 2018;143:1963–1977. doi: 10.1002/ijc.31586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhu Y, Zhang L, Zha H, Yang F, Hu C, Chen L, Guo B, Zhu B. Stroma-derived Fibrinogen-like protein 2 activates cancer-associated fibroblasts to promote tumor growth in lung cancer. Int J Biol Sci. 2017;13:804–814. doi: 10.7150/ijbs.19398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tao L, Huang G, Song H, Chen Y, Chen L. Cancer associated fibroblasts: An essential role in the tumor microenvironment. Oncol Lett. 2017;14:2611–2620. doi: 10.3892/ol.2017.6497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sharon Y, Alon L, Glanz S, Servais C, Erez N. Isolation of normal and cancer-associated fibroblasts from fresh tissues by Fluorescence Activated Cell Sorting (FACS) J Vis Exp. 2013:e4425. doi: 10.3791/4425. doi: 10.3791/4425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ishii G, Ochiai A, Neri S. Phenotypic and functional heterogeneity of cancer-associated fibroblast within the tumor microenvironment. Adv Drug Deliv Rev. 2016;99:186–196. doi: 10.1016/j.addr.2015.07.007. [DOI] [PubMed] [Google Scholar]
- 15.Ziani L, Chouaib S, Thiery J. Alteration of the antitumor immune response by cancer-associated fibroblasts. Front Immunol. 2018;9:414. doi: 10.3389/fimmu.2018.00414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kashima H, Noma K, Ohara T, Kato T, Katsura Y, Komoto S, Sato H, Katsube R, Ninomiya T, Tazawa H, et al. Cancer-associated fibroblasts (CAFs) promote the lymph node metastasis of esophageal squamous cell carcinoma. Int J Cancer. 2019;144:828–840. doi: 10.1002/ijc.31953. [DOI] [PubMed] [Google Scholar]
- 17.Huber L, Birk R, Rotter N, Aderhold C, Lammert A, Jungbauer F, Kramer B. Effect of small-molecule tyrosine kinase inhibitors on PDGF-AA/BB and PDGFRα/β expression in SCC according to HPV16 Status. Anticancer Res. 2020;40:825–835. doi: 10.21873/anticanres.14575. [DOI] [PubMed] [Google Scholar]
- 18.Ouyang L, Zhang K, Chen J, Wang J, Huang H. Roles of platelet-derived growth factor in vascular calcification. J Cell Physiol. 2018;233:2804–2814. doi: 10.1002/jcp.25985. [DOI] [PubMed] [Google Scholar]
- 19.Heldin CH, Lennartsson J, Westermark B. Involvement of platelet-derived growth factor ligands and receptors in tumorigenesis. J Intern Med. 2018;283:16–44. doi: 10.1111/joim.12690. [DOI] [PubMed] [Google Scholar]
- 20.Zhang D, Wang Y, Shi Z, Liu J, Sun P, Hou X, Zhang J, Zhao S, Zhou BP, Mi J. Metabolic reprogramming of cancer-associated fibroblasts by IDH3α downregulation. Cell Rep. 2015;10:1335–1348. doi: 10.1016/j.celrep.2015.02.006. [DOI] [PubMed] [Google Scholar]
- 21.Maass PG, Luft FC, Bahring S. Long non-coding RNA in health and disease. J Mol Med (Berl) 2014;92:337–346. doi: 10.1007/s00109-014-1131-8. [DOI] [PubMed] [Google Scholar]
- 22.Chang L, Wang G, Jia T, Zhang L, Li Y, Han Y, Zhang K, Lin G, Zhang R, Li J, Wang L. Armored long non-coding RNA MEG3 targeting EGFR based on recombinant MS2 bacteriophage virus-like particles against hepatocellular carcinoma. Oncotarget. 2016;7:23988–24004. doi: 10.18632/oncotarget.8115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhu M, Zhang C, Chen D, Chen S, Zheng H. lncRNA MALAT1 potentiates the progression of tongue squamous cell carcinoma through regulating miR-140-5p-PAK1 pathway. Onco Targets Ther. 2019;12:1365–1377. doi: 10.2147/OTT.S192069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kolenda T, Guglas K, Ryś M, Bogaczynska M, Teresiak A, Bliźniak R, Łasińska I, Mackiewicz J, Lamperska KM. Biological role of long non-coding RNA in head and neck cancers. Rep Pract Oncol Radiother. 2017;22:378–388. doi: 10.1016/j.rpor.2017.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ding L, Ren J, Zhang D, Li Y, Huang X, Hu Q, Wang H, Song Y, Ni Y, Hou Y. A novel stromal lncRNA signature reprograms fibroblasts to promote the growth of oral squamous cell carcinoma via LncRNA-CAF/interleukin-33. Carcinogenesis. 2018;39:397–406. doi: 10.1093/carcin/bgy006. [DOI] [PubMed] [Google Scholar]
- 26.Zhao L, Ji G, Le X, Wang C, Xu L, Feng M, Zhang Y, Yang H, Xuan Y, Yang Y, et al. Long noncoding RNA LINC00092 acts in cancer-associated fibroblasts to drive glycolysis and progression of ovarian cancer. Cancer Res. 2017;77:1369–1382. doi: 10.1158/0008-5472.CAN-16-1615. [DOI] [PubMed] [Google Scholar]
- 27.Freitas R, Fraga CAM. NF-κB-IKKβ pathway as a target for drug development: Realities, challenges and perspectives. Curr Drug Targets. 2018;19:1933–1942. doi: 10.2174/1389450119666180219120534. [DOI] [PubMed] [Google Scholar]
- 28.Park MH, Hong JT. Roles of NF-kappaB in cancer and inflammatory diseases and their therapeutic approaches. Cells. 2016;5:15. doi: 10.3390/cells5020015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu J, Chen S, Wang W, Ning BF, Chen F, Shen W, Ding J, Chen W, Xie WF, Zhang X. Cancer-associated fibroblasts promote hepatocellular carcinoma metastasis through chemokine-activated hedgehog and TGF-β pathways. Cancer Lett. 2016;379:49–59. doi: 10.1016/j.canlet.2016.05.022. [DOI] [PubMed] [Google Scholar]
- 30.Li X, Xu Q, Wu Y, Li J, Tang D, Han L, Fan Q. A CCL2/ROS autoregulation loop is critical for cancer-associated fibroblasts-enhanced tumor growth of oral squamous cell carcinoma. Carcinogenesis. 2014;35:1362–1370. doi: 10.1093/carcin/bgu046. [DOI] [PubMed] [Google Scholar]
- 31.Jing Z, Yuan X, Zhang J, Huang X, Zhang Z, Liu J, Zhang M, Oyang J, Zhang Y, Zhang Z, Yang R. Chromosome 1 open reading frame 190 promotes activation of NF-κB canonical pathway and resistance of dendritic cells to tumor-associated inhibition in vitro. J Immunol. 2010;185:6719–6727. doi: 10.4049/jimmunol.0903869. [DOI] [PubMed] [Google Scholar]
- 32.Saini H, Rahmani Eliato K, Veldhuizen J, Zare A, Allam M, Silva C, Kratz A, Truong D, Mouneimne G, LaBaer J, et al. The role of tumor-stroma interactions on desmoplasia and tumorigenicity within a microengineered 3D platform. Biomaterials. 2020;247:119975. doi: 10.1016/j.biomaterials.2020.119975. [DOI] [PubMed] [Google Scholar]
- 33.Faghihi MA, Wahlestedt C. Regulatory roles of natural antisense transcripts. Nat Rev Mol Cell Biol. 2009;10:637–643. doi: 10.1038/nrm2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ørom UA, Derrien T, Beringer M, Gumireddy K, Gardini A, Bussotti G, Lai F, Zytnicki M, Notredame C, Huang Q, et al. Long noncoding RNAs with enhancer-like function in human cells. Cell. 2010;1:46–58. doi: 10.1016/j.cell.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cabili MN, Trapnell C, Goff L, Koziol M, Tazon-Vega B, Regev A, Rinn JL. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011;25:1915–1927. doi: 10.1101/gad.17446611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 37.Fish JE, Santoro MM, Morton SU, Yu S, Yeh RF, Wythe JD, Ivey KN, Bruneau BG, Stainier DY, Srivastava D. miR-126 regulates angiogenic signaling and vascular integrity. Dev Cell. 2008;15:272–284. doi: 10.1016/j.devcel.2008.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang S, Zhu M, Wang Q, Hou Y, Li L, Weng H, Zhao Y, Chen D, Ding H, Guo J, Li M. Alpha-fetoprotein inhibits autophagy to promote malignant behaviour in hepatocellular carcinoma cells by activating PI3K/AKT/mTOR signalling. Cell Death Dis. 2018;9:1027. doi: 10.1038/s41419-018-1036-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Olumi AF, Grossfeld GD, Hayward SW, Carroll PR, Tlsty TD, Cunha GR. Carcinoma-associated fibroblasts direct tumor progression of initiated human prostatic epithelium. Cancer Res. 1999;59:5002–5011. doi: 10.1186/bcr138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lu Y, Chen R, Ma JY, Wang LP, Qiu LL, Wang CP, Yan JC, Liu PJ. Platelet derived growth factor-BB regulates phenotype transformation of pulmonary artery smooth muscle cells via SIRT3 affecting glycolytic pathway. Zhonghua Xin Xue Guan Bing Za Zhi. 2019;47:993–999. doi: 10.3760/cma.j.issn.0253-3758.2019.12.009. (In Chinese) [DOI] [PubMed] [Google Scholar]
- 41.Gialeli C, Nikitovic D, Kletsas D, Theocharis AD, Tzanakakis GN, Karamanos NK. PDGF/PDGFR signaling and targeting in cancer growth and progression: Focus on tumor microenvironment and cancer-associated fibroblasts. Curr Pharm Des. 2014;20:2843–2848. doi: 10.2174/13816128113199990592. [DOI] [PubMed] [Google Scholar]
- 42.Aoto K, Ito K, Aoki S. Complex formation between platelet-derived growth factor receptor beta and transforming growth factor beta receptor regulates the differentiation of mesenchymal stem cells into cancer-associated fibroblasts. Oncotarget. 2018;9:34090–34102. doi: 10.18632/oncotarget.26124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rizvi S, Mertens JC, Bronk SF, Hirsova P, Dai H, Roberts LR, Kaufmann SH, Gores GJ. Platelet-derived growth factor primes cancer-associated fibroblasts for apoptosis. J Biol Chem. 2014;289:22835–22849. doi: 10.1074/jbc.M114.563064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shiga K, Hara M, Nagasaki T, Sato T, Takahashi H, Takeyama H. Cancer-associated fibroblasts: Their characteristics and their roles in tumor growth. Cancers (Basel) 2015;7:2443–2458. doi: 10.3390/cancers7040902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Primac I, Maquoi E, Blacher S, Heljasvaara R, Van Deun J, Smeland HY, Canale A, Louis T, Stuhr L, Sounni NE, et al. Stromal integrin α11 regulates PDGFR-β signaling and promotes breast cancer progression. J Clin Invest. 2019;129:4609–4628. doi: 10.1172/JCI125890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang S, Tao X, Cao Q, Feng X, Wu J, Yu H, Yu Y, Xu C, Zhao H. lnc003875/miR-363/EGR1 regulatory network in the carcinoma -associated fibroblasts controls the angiogenesis of human placental site trophoblastic tumor (PSTT) Exp Cell Res. 2020;387:111783. doi: 10.1016/j.yexcr.2019.111783. [DOI] [PubMed] [Google Scholar]
- 47.Zheng B, Ohuchida K, Chijiiwa Y, Zhao M, Mizuuchi Y, Cui L, Horioka K, Ohtsuka T, Mizumoto K, Oda Y, et al. CD146 attenuation in cancer-associated fibroblasts promotes pancreatic cancer progression. Mol Carcinog. 2016;55:1560–1572. doi: 10.1002/mc.22409. [DOI] [PubMed] [Google Scholar]
- 48.Yao Q, Qu X, Yang Q, Wei M, Kong B. CLIC4 mediates TGF-beta1-induced fibroblast-to-myofibroblast transdifferentiation in ovarian cancer. Oncol Rep. 2009;22:541–548. doi: 10.3892/or_00000469. [DOI] [PubMed] [Google Scholar]
- 49.Binns D, Dimmer E, Huntley R, Barrell D, O'Donovan C, Apweiler R. QuickGO: a web-based tool for Gene Ontology searching. Bioinformatics. 2009;25:3045–3046. doi: 10.1093/bioinformatics/btp536. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.