Abstract
N6-methyladenosine (m6A) is the most prevalent, conserved, and abundant RNA modification of the mRNAs of most eukaryotes, including mammals. Similar to epigenetic DNA modifications, m6A has been proposed to function as a critical regulator for gene expression. This modification is installed by m6A methylation “writers” (Mettl3/Mettl14 methyltransferase complex), and it can be reversed by demethylase “erasers” (Fto and Alkbh5). Furthermore, m6A can be recognized by “readers” (Ythdf and Ythdc families), which may be interpreted to affect mRNA splicing, stability, translation or localization. Levels of m6A methylation appear to be highest in the brain, where it plays important functions during embryonic stem cell differentiation, brain development, and neurodevelopmental disorders. Depletion of the m6A methylation writer Mettl14 from mouse embryonic nervous systems prolongs cell cycle progression of radial glia and extends cortical neurogenesis into postnatal stages. Recent studies further imply that dysregulated m6A methylation may be significantly correlated with neurodegenerative diseases. In this review, we give an overview of m6A modifications during neural development and associated disorders, and provide perspectives for studying m6A methylation.
Keywords: m6A, Epitranscriptome, RNA, Neural development, Neurodegeneration
Introduction
Gene expression and cell division are controlled by genetic and epigenetic regulation. Abnormal genetic changes (such as gene mutation, deletion, or amplification, as well as chromosomal translocation or epigenetic abnormalities such as DNA methylation or histone modification) may result in developmental defects or diseases. In recent years, RNA modifications have gained increasing attention for their largely unexplored roles in gene regulation (i.e., RNA epitranscriptomics) [1]. Since the 1950s, over 100 types of RNA modification have been identified. With the power of new high-throughput sequencing methods, a diversity of mRNA modifications—including N6-methyladenosine (m6A), N1-methyladenosine (m1A), 5-methylcytosine (m5C), 5-hydroxymethylcytosine (hm5C), and pseudouridine (ψ)—have been revealed in various organisms [2]. Among them, dynamic and reversible m6A mRNA modification, discovered in the 1970s [3], arguably represents the most widely distributed form of mRNA modification in mammals. m6A is deposited by the m6A methyltransferase complex (termed a “writer”) that comprises Mettl3 (methyltransferase-like 3), Mettl14 (methyltransferase-like 14), Wtap (Wilms tumor 1-associated protein), Virma (VIR-like m6A methyltransferase associated), RBM15 (RNA-binding motif protein 15) and its paralogue (RBM15B) [4, 5]. Conversely, m6A can be removed by m6A demethylases (termed “erasers”), such as Fto (fat mass and obesity-associated protein) and Alkbh5 (alkB homolog 5). m6A tagging has multiple functions, including in mRNA splicing, stability, nuclear export, localization, translational efficiency activation, and decay of target mRNA stability (Fig. 1) [6–8]. Notably, m6A deposition manifests at the highest levels in the central nervous system (CNS), where it plays major roles in embryonic stem cell differentiation, brain development, and neurodevelopmental disorders [9, 10]. Recent studies have shown that constitutive knockout of Mettl14—a key element of the m6A methyltransferase complex—is embryonically lethal, whereas conditional knockout (cKO) of Mettl14 in neural progenitor cells disrupts cortical development and leads to premature death in mice [11, 12]. Interestingly, levels of m6A are relatively low in mouse brain tissue during embryogenesis, but drastically increase by adulthood [10], suggesting that m6A modification plays a unique role in the adult brain. This scenario also raises the possibility that m6A might play an important role in adult RNA homeostasis and that associated imbalances might lead to onset or progression of neurodegeneration. This notion is supported by studies showing that the m6A demethylase Fto plays an important role in learning and behavior [13–15]. In addition to m6A’s functions in the CNS, Mettl3, another component of the m6A methyltransferase complex, has been shown to affect plant growth, yeast meiosis, mammalian metabolism, synaptic signaling, stem cell self-renewal, and differentiation [16]. Furthermore, deletion of Mettl3 from cardiomyocytes reduces m6A levels, resulting in long-term loss of normal cardiac homeostasis and function in adult mouse heart [17]. Thus, overall, m6A modification plays versatile roles during embryonic development, as well as functioning in adult homeostasis, but its most prominent roles lie in the CNS.
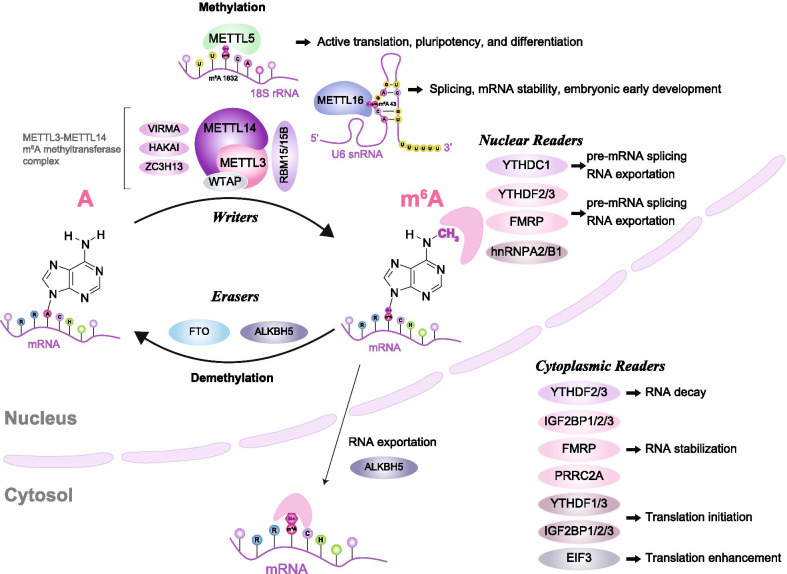
Fig. 1.
m6A-associated proteins and molecular pathways. N6-methyladenosine (m6A) modifications have important roles in a series of RNA-centric regulatory mechanisms. The illustration depicts the roles of m6A “writer”, “eraser” and “reader” complexes in regulating mRNAs. m6A biogenesis in mammalian cells (nucleus) is catalysed by a core methyltransferase complex comprising METTL3, METTL14 and WTAP, and it can be reversed by m6A demethylases (FTO or ALKBH5). METTL5 and METTL16 have recently been reported as additional writers to catalyze the addition of m6A on the 3′UTR of 18S rRNA and U6 snRNA, respectively. m6A can be recognized by reader proteins in both the nucleus and cytoplasm. For example, YTHDF2/3 can participate in post-transcriptional regulation by recruiting different complexes to m6A sites. m6A-modification of mRNAs can impact RNA nuclear splicing, export, stability, trafficking, and translation efficiency. METTL, methyltransferase-like; WTAP, WT1-associated protein; RBM15, RNA-binding motif protein 15 or its paralog RBM15B; VIRMA, vir-like m6A methyltransferase-associated; HAKAI, E3 ubiquitin-protein ligase Hakai (also known as CBLL1, cbl proto-oncogene-like 1); ZC3H13, zinc finger CCCH-type containing 13 proteins; FTO, fat-mass and obesity-associated protein; ALKBH5, alkB homolog 5; YTHDF, YT521-B homology domain family; FMRP, fragile X mental retardation protein; hnRNPA2/B1, heterogeneous nuclear ribonucleoprotein A2/B1; PRRC2A, proline rich coiled-coil 2A; IGF2BP1/2/3, insulin like growth factor 2 mRNA binding protein 1/2/3; elF3, E74-like factor 3
As many previous reviews have illustrated the importance of m6A modification of mRNAs during early embryonic development [12, 18, 19], here we provide an overview of current progress on m6A epitranscriptomic regulation of neural development and its biological implications. In addition, we review the roles of m6A during adult neurogenesis and potential consequences for neurological disorders (Fig. 2 and Table 1). Finally, we present our perspectives for future research directions of m6A in neural development and degeneration.
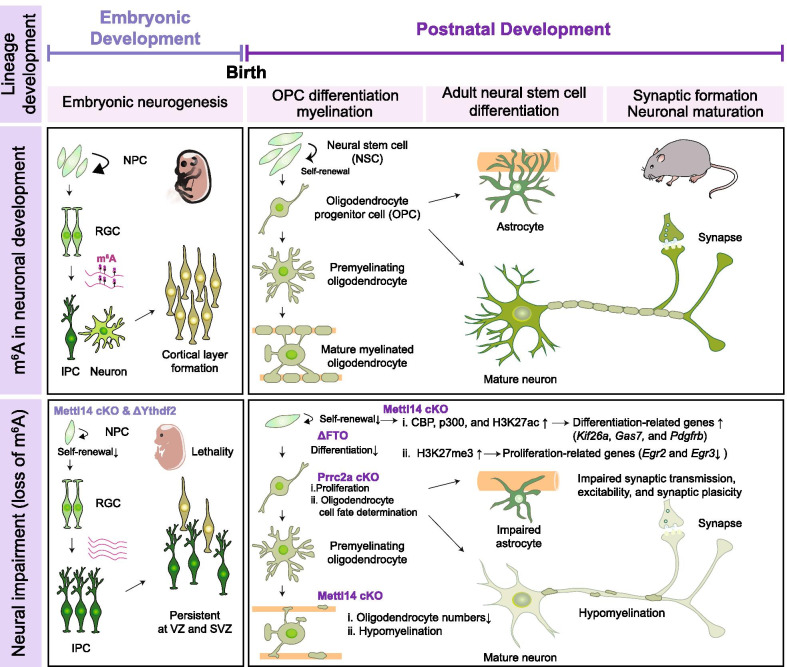
Fig. 2.
The roles of m6A in the developing central nervous system. m6A RNA modifications play important roles in regulating neural development, including embryonic neurogenesis, OPC differentiation, myelination, adult NSC differentiation, synaptic formation and neuronal maturation. The panels summarize the physiological functions of m6A during neural development, as revealed by loss-of-function studies on m6A-associated proteins. NPC, neural progenitor cell; RGC, radial glial cell; IPC, intermediate progenitor cell; NSC, neural stem cell; OPC, oligodendrocyte progenitor cell; CBP, CREB binding protein; H3K27ac, histone H3 lysine 27 acetylation; H3K27me3, histone H3 lysine 27 trimethylation
Table 1.
Roles of m6A methylation in the developing CNS
| Roles of m6A methylation in developing CNS | ||||||
|---|---|---|---|---|---|---|
| Lineage development | m6A methyltransferase component involved | Type | m6A-methylated target mRNAs | Mouse models | Neural development function affected | Refs. |
| Cortical neurogenesis | Mettl14 | Writer | Neuronal differentiation-related transcription factors (Pax6, Sox1, Sox2, Emx2, Neurog2, and Neurogenin 2), and proneural genes (Neurod1/2) | Nestin-cre;Mettl14f/f |
1. Temporal specification and cell-cycle progression of NPCs 2. Transcriptional regulation in NSCs 3. Cell-cycle progression of cortical neural progenitors |
[12] |
| Histone acetyltransferase (CBP/p300) | Nestin-cre;Mettl14f/f | 1. Self-renewal of NSCs | [19] | |||
| Ythdf2 | Reader |
Positive regulation of cell differentiation and GTPase activity Negative regulation of JAK-STAT cascade genes (Flrt2/3, Ptprd, and Lrrtm1/4) |
Ythdf2 cKO | 1. Clearance of negative regulators of neurogenesis | [18] | |
| Neural differentiation | Fmrp | Reader | Nuclear retention of m6A-tagged Fmrp target mRNAs | Fmrp KO | 1. Promotion of mRNA nuclear export during neural differentiation | [74] |
| Oligodendrocyte development and myelination | Mettl14 | Writer | Oligodendrocyte lineage genes (Ptprz1, Gsn, and Map2) |
Olig2-cre;Mettl14f/f and CNP-Cre;Mettl14f/f |
1. Differentiation and myelination of OPCs | [57] |
| Prrc2a | Reader | Oligodendroglial transcriptional factor (Olig2) | Nestin-Cre;Prrc2af/f |
1. Proliferation and differentiation of OPCs 2. Stabilization of Olig2 mRNA for oligodendrocyte maturation |
[56] | |
| Cerebellar development | Alkbh5 | Eraser |
Loss of methylated RNAs has been linked to altered cell division (Cenpe), cell cycle (Cdca2), and cell projection organization (Erbb4) genes Hypermethylated RNAs are related to metabolic processes (Ccr5), ion transport (Camk2g), and and axon guidance (Rora) |
Alkbh5 KO |
1. Proliferation and differentiation in the cerebellum 2. Disrupted RNA metabolism of a subset of cell fate determination genes 3. Defected cerebellar development under hypobaric hypoxia |
[60] |
| Neurogenesis in the brain | Fto | Eraser | Loss of Fto led to altered expression of several brain-related neurotrophic factors | Fto KO |
1. Decreased brain size and body weight 2. Reduced pool of NSCs in the SGZ region 3. Reduced proliferation and neuronal differentiation in NSCs 4. Impairment of learning and memory |
[52] |
| Hippocampal neurogenesis | Ythdf1 | Reader | Synaptic plasticity transcripts (Gria1, Grin1, and Camk2a) | Ythdf1 KO |
1. Learning and memory 2. Basal transmission and long-term potentiation at synapses in the hippocampus |
[61] |
| Axon guidance | Ythdf1 | Reader | Axon guidance receptor Robo3.1 | Ythdf1 KO | 1. Axon guidance in spinal commissural axons | [75] |
| Axon regeneration | Ythdf1 | Reader | Axon regeneration-related genes (Tet3 and Gadd45a) | Ythdf1 KO |
1. Axon regeneration in the peripheral nervous system 2. Translation of injury-induced protein 3. Axon regeneration in adult mouse dorsal root ganglions |
[11] |
N6-methyladenosine (m6A)
m6A is known to be chemically stable, but the methyl modification is dynamic due to low cellular abundance of mRNAs (representing 2–5% of total RNA), with 0.2–0.5% of total mRNA adenines being m6A-modified [7, 9, 31, 32]. Although few genes are m6A-modified, this modification appears to play a significant regulatory role in gene expression [33]. Over the past decade, multiple studies have shown that Mettl3/Mettl14 together with other components of the m6A methyltransferase complex can dynamically exert m6A mRNA modifications on mRNAs or long non-coding RNAs to regulate mRNA stability, translation efficiency, localization, and splicing (Fig. 1). An early study provided the first evidence that reversible post-transcriptional RNA modifications enact regulatory functions to fine-tune the structure and function of RNAs [1]. In that study and a subsequent one, a m6A demethylase (Fto) was shown to catalyze oxidative m6A demethylation of nuclear RNA [1, 34]. Later, another m6A demethylase, Alkbh5, was shown to affect mouse fertility and spermatogenesis [35]. Following these breakthrough findings, dynamic m6A methylation and gene regulation have gained increasing attention. Several insightful reviews have illustrated details of the molecular pathways underlying m6A modification [36–41]. In this review, we focus on providing a brief overview of recent exciting discoveries on how the m6A epigenome regulates neural development and degeneration.
To understand the fundamental functions of m6A in mRNAs, it is necessary to determine the positions of m6A sites in gene transcripts. In 2012, two groups independently reported on antibody-based m6A RNA immunoprecipitation sequencing (RIP-Seq/MeRIP-Seq) techniques and subsequent mapping of the m6A transcriptome [10, 31]. Comprehensive analyses of those sequencing data revealed that primary m6A sites are enriched in the regions of translational stop codons and in 3′ untranslated regions (3′UTR), suggesting m6A may serve a role in mRNA translation [10, 31, 42]. Moreover, the consensus motif RRACH (in which R represents A or G, and H represents A, C or U) mediates most m6A depositions. Although m6A does not alter Watson–Crick base pairing, the m6A modification promotes destabilization of A/U pairings and alters RNA secondary structure [43]. As the frequency of this consensus motif is much higher in the genome than m6A occurrence, additional sequences or RNA structures may also play as yet unidentified roles in determining methylation sites. For instance, recent reports have demonstrated that m6A reshapes protein and RNA binding, thereby affecting mRNA secondary structure [44, 45].
By revealing m6A deposition on mRNAs via MeRIP-Seq, m6A abundance has been demonstrated as both tissue-specific and species-specific [46]. Brain tissue displays the highest levels of m6A deposition, with > 30% of its transcripts being m6A-modified [24]. Detailed characterizations have further highlighted that m6A occurs within various types of RNA other than mRNA, such as tRNA, rRNA, non-coding RNA (ncRNA), and snRNAs [5, 22, 27, 30, 45, 47–49], but the consensus motifs in those RNAs differ from those in mRNA. For example, m6A uses the U6 snRNA as substrate to form 3′-stem loop structures in vitro [30, 50]. Therefore, m6A may serve some unique functions in mRNAs that are different from those in other RNA types, perhaps being cell-type-dependent and tissue-specific. Collectively, current evidence indicates a functional significance for m6A modifications of RNA. However, the exact molecular mechanisms underlying m6A methylation sites in consensus motifs of mRNAs remain to be elucidated.
m6A epitranscriptomics in neural development
m6A has been reported to be the most abundant epitranscriptomic mark for mRNA modification in eukaryotes. Emerging studies of the CNS have shown that most mRNAs present low levels of m6A modification in the brain from the embryonic to postnatal stages, but levels dramatically increase in adulthood [10, 24]. The importance of m6A in the CNS seems to be conserved from flies to mammals. In Drosophila, the m6A epitranscriptome is dynamic and remarkably enriched in early embryogenesis, but it declines dramatically two hours after fertilization. Then, m6A remains at a low level throughout the rest of embryogenesis and during early larval stages. During the third larval instar, m6A levels increase again, reaching a peak in pupal phases. Although overall levels of m6A decrease in adult Drosophila, it remains substantially elevated in head and ovary tissues. When the Mettl3 orthologue, Ime4, is knocked out, the mutant flies manifest reduced lifespan, severe behavioral defects, and altered neural gene expression. Recent works have also shown that m6A regulates neural development and brain function in mouse models [12, 39]. In human, two studies have reported that children with homozygous missense mutations in FTO (m6A demethylase) presented severe neurodevelopmental disorders, including microcephaly, developmental delay, behavioral abnormalities, dysmorphic facial features, hypotonia, and various other phenotypic abnormalities, suggesting an essential, yet unexplored, role for m6A RNA modification in brain development [21, 25]. Thus, neural development represents one of the best paradigms for elucidating the functional significance of m6A modification.
Role of m6A in regulating neurogenesis
Embryonic neurogenesis is coordinated between neural progenitor cell (NPC) proliferation and cell fate specification. NPCs differentiate into various neural and glial cell subtypes before migrating to their final destinations in the CNS [20, 29]. In addition, long non-coding RNA represses progenitor genes and maintains neural cell fate identity representing a highly organized topographic migratory process from embryonic to postnatal stages [51]. Notably, Yoon et al. showed that Mettl3 is highly enriched during the early stage of neurogenesis [12], whereas Fto is expressed more prominently during the later stage of neurogenesis [52]. Conditional Mettl14 knockout (cKO) in mouse NPCs using Nestin-Cre impairs NPC differentiation, prolongs cell cycle progression of radial glia, and extends cortical neurogenesis into postnatal stages [12]. Similar phenomena have also been observed upon knockdown of another m6A writer, Mettl3 [12]. Prolongation of cell cycle progression in neural stem cells (NSCs) upon loss of either Mettl3 or Mettl14 delays production of upper-layer neurons in postnatal mouse cortex. Human induced pluripotent stem cell (iPSC)-derived brain organoids were also used to confirm that m6A regulates NPC cell cycle progression in a human context [12]. Further comparison of the m6A-seq data from human forebrain organoids and E13.5 mouse forebrains revealed that m6A-modified transcripts are conserved and have distinct m6A epitranscriptomic landscape features. Interestingly, many transcripts encoding transcription factors are m6A-tagged, such as Pax6, Sox1, Sox2, Emx2, and Neurog2/Neurogenin 2 [12]. Gene ontology (GO) analysis of the m6A-modified transcripts in both mouse and human revealed enrichment of genes related to neurogenesis, neuronal differentiation, and development. Furthermore, disease ontology analysis of those transcripts uniquely m6A-tagged in human highlighted enrichment for neurodevelopmental diseases such as schizophrenia and autism, implying that m6A may contribute to human psychiatric or neural disorders. Thus, Yoon et al. have provided the first proof-of-principle of a m6A epitranscriptomic mechanism contributing to conserved transcriptional coordination during mammalian cortical neurogenesis.
Similarly, Wang et al. used a mouse genetic model to conditionally inactivate Mettl14 in embryonic NPCs, which also revealed that Mettl14 is required for NPC proliferation, with consequent loss of m6A in the CNS slowing NPC cell cycle progression so that the NPCs remained in an undifferentiated state [19]. To further examine the consequences of m6A loss, they systematically characterized Mettl14 cKO NPCs in vitro and observed that loss of m6A reduced NSC proliferation and resulted in precocious NPC differentiation in vitro. Furthermore, cortical radial glial cells (RGCs) in the brain were found to be smaller and numbers of late-born neurons were reduced in the Mettl14-cKO mutant mice. Profiling of histone modifications upon m6A loss via Mettl14 knockout revealed increased histone H3 acetylation at lysine 27 (H3K27ac), histone H3 trimethylation at lysine 4 (H3K4me3), and histone H3 trimethylation at lysine 27 (H3K27me3) in cell-proliferation related genes. Most of the changes in histone levels could be rescued by treating cells with H3K27ac or H3K27me3 inhibitors, indicating that m6A RNA methylation serves an essential function in regulating NSC self-renewal via m6A-mediated histone modification during NSC-related gene expression. These changes in histone modification were also partially attributable to m6A-mediated destabilization of transcripts of the histone acetyltransferase CBP (CREB binding protein) and p300, both of which were stabilized upon loss of m6A. This study by Wang et al. has provided new insights into crosstalk between RNAs and histone modification. Importantly, their study also indicates that different m6A-regulated histone marks coordinate active/repressive gene expression, implying that m6A-regulated active and repressive histone modifications work synergistically to ensure an NSC differentiation program.
Both the Yoon et al. and Wang et al. studies utilized conditional knockout of the m6A writer Mettl14 in the developing forebrain as an experimental paradigm. Although the mechanisms revealed by these two studies as underlying m6A function differ, both studies revealed similar phenotypes, such as delayed NPC cell cycle progression. Another study has provided evidence that the m6A “reader” protein Ythdf2 also participates in cortical neurogenesis [18]. Ythdf2 knockout mice die at late embryonic developmental stages. Furthermore, Ythdf2-deficient NSCs display diminished proliferation and differentiation, and neurons derived from Ythdf2-deficient NSCs have shorter neurites and are vulnerable to oxidative stress. When Li et al. examined the proliferative and differentiation capabilities of neural stem/progenitor cells (NSPCs), they observed a dramatically reduced overall cortical thickness of Ythdf2-deficient embryonic forebrains [18]. Furthermore, NPSC self-renewal and spatiotemporal generation of neurons and other cell types were severely negatively impacted in embryonic neocortex upon loss of Ythdf2. Since neurite outgrowth is critical for neuronal development and maturation, as well as synapse formation, the abnormal neurite branching and extension presented by Ythdf2-deficient neurons might contribute to defective neurogenesis during neural development. Consistent with the findings of Yoon et al. [12], deletion of Mettl14 and Ythdf2 led to enlarged ventricles and decreased cortical thickness, respectively [18]. As Ythdf2 is known to bind m6A-methylated mRNAs and promotes mRNA decay [8, 53], Li et al. further demonstrated increased expression of m6A-tagged gene transcripts associated with neural development and cortical neuron differentiation upon loss of Ythdf2-mediated RNA degradation. This scenario adds an additional layer of m6A-dependent control of the neural development-related mRNA targets recognized by Ythdf2 and that modulate neural development. Thus, fundamentally, the Ythdf2-mediated functions of m6A epitranscriptomic regulation are not only essential for post-transcriptional regulation of the maternal transcriptome and oocyte competence [54], but are also crucial for regulating cortical neurogenesis during embryonic neural development. However, how the m6A reader Ythdf2 interprets m6A epitranscriptomic regulation of cortical neurogenesis for neural development and differentiation is not entirely deciphered. Since a battery of m6A “reader” proteins have already been identified in various cell types [23, 28, 46, 55], further neuron-specific activities could multiply the highly variable functions of m6A during neural development.
Role of m6A in oligodendrocytes
Differential m6A methylation may also play important roles in oligodendrocyte development and CNS myelination [56, 57]. Xu et al. characterized the pathological consequences of Mettl14 ablation for oligodendrocyte lineage progression [56, 57]. In that study, numbers of mature oligodendrocytes were reduced in the corpus callosum of both Olig2-Cre;Mettl14f/f and CNP-Cre;Mettl14f/f mutant mice. Although Olig2-Cre;Mettl14f/f mutant mice exhibited a relatively normal postnatal phenotype, they began displaying occasional hindlimb flexion, slight ataxia, and mild tremors after 6 months. In addition, CNP-Cre;Mettl14f/f mutant mice started to display tremors and hindlimb clenching at ~ 4 months of age, with a gradual worsening of the ataxic phenotype after onset. Both the corpus callosum and optic nerve were hypomyelinated in Olig2-Cre;Mettl14f/f and CNP-Cre;Mettl14f/f mutant mice, as revealed by electron microscopy at postnatal day 18 (P18). This observation indicates that m6A methylation is important for oligodendrocyte development and CNS myelination. The severe developmental phenotypes observed in vitro and in vivo are consistent with oligodendrocyte lineage progression being controlled by dynamic changes in m6A modification of numerous transcripts. Mechanistically, Mettl14 deletion was shown to differentially alter Nfasc155 alternative splicing and expression in Olig2-Cre;Mettl14f/f mutant mice. Thus, m6A might act in generating functional isoforms of myelin proteins via alternative splicing to ensure precise oligodendrocyte lineage progression.
Consistent with the study by Xu et al. [56, 57], Wu et al. reported a novel m6A “reader” protein, proline rich coiled-coil 2 A (Prrc2a), which modulates oligodendrocyte progenitor cell (OPC) specification, proliferation, fate determination and CNS myelination, affirming the importance of m6A modification in the glial lineage [56]. Genetic deletion of Prrc2a from brain of Nestin-Cre;Prrc2af/f and Olig2-Cre;Prrc2af/f mutant mice led to hypomyelination in the corpus callosum. Moreover, the mutant mice displayed locomotive and cognitive disabilities, as well as decreased lifespan, though neurogenesis was not affected. When Wu et al. compared Prrc2a binding targets that contain m6A peaks with the downregulated differentially-expressed genes (DEG) of Olig2-Cre;Prrc2af/f mutant mice, they found that Prrc2a binds and stabilizes m6A-methylated transcripts of oligodendrocyte transcription factor 2 (Olig2), a key oligodendroglial lineage-determining transcription factor. Prrc2a-knockdown reduced both mRNA and protein levels of Olig2, whereas Prrc2a overexpression enhanced them, indicating that Prrc2a post-transcriptionally regulates Olig2 expression. Additionally, knockout of the m6A demethylase Fto recapitulated the phenotype of enhanced Olig2 expression displayed upon Prrc2a overexpression. That study has revealed yet another function for m6A RNA modification associated with the myelination process of oligodendrocytes. Thus, apart from its essential roles in neurons, m6A also participates in myelination by regulating oligodendrocyte development.
Role of m6A in adult brain development
In addition to the roles of Mettl14 and m6A in embryonic neural development, m6A also functions in adult mouse brain. One prominent activity is its regulation of synaptic function and stress-induced responses. In this scenario, most cortical genes expressed in adult mouse are m6A-methylated, with m6A also being enriched in synaptic transcripts [24]. Moreover, Mettl14, Fto, and Ythdf1/2/3 are enriched during dendritic development of cortical neurons [58]. In hippocampal neurons, more than 1000 transcripts are m6A-methylated, which related to synaptic organization, assembly, maturation, and transmission modulation [58]. Mettl14 deletion reduced m6A methylation levels in synaptic plasticity-related transcripts that are correlated with impaired neuronal excitability levels, without altering cell numbers or morphology [59]. It also increased neuronal excitability, reduced spike frequency adaptation, and profoundly impaired striatal-mediated behaviors, suggesting that m6A is important for maintaining normal striatal function in adult mice [60]. The functions of m6A in the adult CNS have been further revealed by two additional sets of studies. First, increased m6A methylation of synaptic transmission-related mRNAs has been observed in the midbrain of Fto-deficient mice [13]. Second, m6A mRNA methylation was shown to facilitate learning and memory formation in mouse hippocampus at postnatal days 30 and 120. Upon loss of Ythdf1, learning and memory defects, as well as functional deficits in hippocampal excitatory synaptic transmission, were manifested by promoting the translation of m6A-modified transcripts [58, 61]. These observations highlight critical functions for m6A in neural circuit formation, especially for synaptic plasticity, and highlight a new aspect of m6A mRNA methylation-dependent translational regulation. Overall then, m6A is an important epitranscriptomic RNA modification that is highly expressed during neurogenesis in the brain, from embryonic to adult stages. m6A methylation broadly affects the CNS, acting in NSC self-renewal, glioma cell proliferation, brain development, synaptic growth, learning and memory. Thus, intuitively, it is not surprising that m6A might play a prominent role in neurological disorders and diseases.
Neurological disorders
Although several studies have shown that the m6A epitranscriptome is important for neural development, how m6A mRNA methylation contributes to neurological disorders is largely unexplored. Alzheimer’s disease (AD) is one of the most prevalent human neurodegenerative diseases in the elderly, and synaptic changes are widely regarded as disease-causative. The salient clinical feature of AD is progressive decline of memory function, leading to impaired cognitive function [26]. However, the pathogenesis of AD remains unclear. Recent studies have shown that m6A methylation deficiency impairs hippocampal excitatory synaptic transmission [58, 61]. Studies of an AD mouse model (APP/PS1 transgenic mice) have revealed that the mice display increased m6A methylation in the cortex and hippocampus, and that expression of Mettl3 is increased whereas Fto expression is reduced in the AD mice [62, 63]. These studies support that dysregulation of the Mettl3/Fto axis may alter global patterns of m6A methylation of mRNAs, further impacting dendritic development, synaptic growth, synaptic assembly, axon guidance, and long-term potentiation, thereby potentially linking m6A epitranscriptomic regulation and neurological disorders.
Apart from AD, some genetic variants of FTO have been linked to major depressive disorder (MDD) [64, 65]. Moreover, YTHDC2 has been reported as a potential risk factor for autism spectrum disorder (ASD) in east Asian populations [65, 66]. Integrative analysis of genome-wide association studies (GWAS) for m6A single nucleotide polymorphisms (SNPs) has also highlighted potential causal genes important in neurodegenerative disease [67, 68]. For example, ALKBH5 is associated with various clinical features of MDD, including anxiety, retardation, and cognitive dysfunction [67, 68]. Those studies also suggest that m6A may contribute to various other neurodevelopmental and neurodegenerative diseases.
Concluding remarks and future perspectives
The field of epitranscriptomics has emerged rapidly in recent years, and increasing numbers of researchers are tackling its implications for human disease. In this review, we have summarized m6A-mediated epitranscriptomic gene regulation and the mechanisms involved in neural development and neurodegenerative disease. Dynamic m6A-dependent transcriptomic regulation has been demonstrated to be involved in embryonic and postnatal neural development, with activities ranging from NSC establishment to maintenance of adult neuronal function. However, we still lack a complete map of the global m6A transcriptome during neural development and degeneration in different parts of the CNS. In particular, how m6A contributes to neural disorders remains unclear. Fortunately, site-specific m6A incorporation in distinct cellular compartments is now feasible since nucleus-localized dCas13 can be fused with a truncated METTL3 methyltransferase domain or cytoplasm-localized Cas can be fused with a modified METTL3/14 methyltransferase complex [69]. We envision these approaches will provide researchers with a powerful arsenal for dissecting cause-and-effect relationships that will reveal the importance of m6A in neurological disorders, including AD, Parkinson's disease, and amyotrophic lateral sclerosis (ALS).
How a complete protein-RNA regulome participants are involved in m6A mRNA modification remain to be uncovered, and whether its dynamic regulation is cell context-dependent remains to be characterized in detail. Currently, epitranscriptome mapping technologies primarily rely on the sensitivity and specificity of m6A modification-specific antibodies. However, it remains challenging to detect a specific RNA modification at single-nucleotide resolution with low background signal. Improvements in detection techniques enabling characterization (quantitatively and at the genome-wide scale) of the distribution of RNA modifications at nucleotide resolution and with greater sensitivity and accuracy will greatly help us understand this critical RNA modification. Antibody-independent approaches have recently been reported [70–72], which will greatly help establish the m6A epitranscriptomic landscape in various cellular and developmental contexts with single base resolution. These novel cutting-edge techniques will ultimately facilitate experiments that can characterize m6A epitranscriptomic marks in vivo and confirm their regulatory functions at the single cell level.
Finally, the crosstalk between epitranscriptomics and epigenetics is emerging as a new regulatory axis [19]. Some studies have suggested that m6A could directly or indirectly regulate chromatin-mediated transcription and accessibility by controlling chromatin regulatory complexes and long noncoding RNAs [5, 19, 73]. Whether or not m6A regulates chromatin states is an interesting research avenue that may reveal mechanisms by which epigenetic regulation can contribute to gene regulation during neural development. Such investigations may also help us to better understand if m6A modification may control the protein levels of genes involved in neurodegenerative diseases and aging [63]. Our review provides a brief overview of the roles of m6A in neural development and degeneration. We believe we are only beginning to understand the mysterious roles of RNA modification in the CNS.
Acknowledgements
We apologize to researchers not cited in the manuscript due to limited space. Special thanks to Dr. John O’Brien for further editing the manuscript.
Authors' contributions
YPY and JAC drafted, revised and approved the manuscript.
Funding
Work in the Jun-An Chen lab is supported by grants from the NHRI (NHRI-EX110-10831NI), Academia Sinica (CDA-107-L05; AS-GC-109-03), and MOST (109-2326-B-001-017-; 109-2314-B-001-010-MY3). Y.P.Y. is funded by Academia Sinica Postdoctoral Research Fellowships.
Availability of data and materials
Not applicable.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Ya-Ping Yen, Email: yapingyen@gate.sinica.edu.tw.
Jun-An Chen, Email: jachen@imb.sinica.edu.tw.
References
- 1.He C. Grand challenge commentary: RNA epigenetics? Nat Chem Biol. 2010;6:863. doi: 10.1038/nchembio.482. [DOI] [PubMed] [Google Scholar]
- 2.Li X, Xiong X, Yi C. Epitranscriptome sequencing technologies: decoding RNA modifications. Nat Methods. 2016;14:23. doi: 10.1038/nmeth.4110. [DOI] [PubMed] [Google Scholar]
- 3.Desrosiers R, Friderici K, Rottman F. Identification of methylated nucleosides in messenger RNA from Novikoff hepatoma cells. Proc Natl Acad Sci USA. 1974;71(10):3971–3975. doi: 10.1073/pnas.71.10.3971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mazloomian A, Meyer IM. Genome-wide identification and characterization of tissue-specific RNA editing events in D. melanogaster and their potential role in regulating alternative splicing. RNA Biol. 2015;12(12):1391–1401. doi: 10.1080/15476286.2015.1107703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Patil DP, Chen CK, Pickering BF, Chow A, Jackson C, Guttman M, Jaffrey SR. m(6)A RNA methylation promotes XIST-mediated transcriptional repression. Nature. 2016;537(7620):369–373. doi: 10.1038/nature19342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Deng X, Su R, Weng H, Huang H, Li Z, Chen J. RNA N6-methyladenosine modification in cancers: current status and perspectives. Cell Res. 2018;28(5):507–517. doi: 10.1038/s41422-018-0034-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu J, Yue Y, Han D, Wang X, Fu Y, Zhang L, Jia G, Yu M, Lu Z, Deng X, Dai Q, Chen W, He C. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat Chem Biol. 2014;10(2):93–95. doi: 10.1038/nchembio.1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang X, Lu Z, Gomez A, Hon GC, Yue Y, Han D, Fu Y, Parisien M, Dai Q, Jia G, Ren B, Pan T, He C. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature. 2014;505(7481):117–120. doi: 10.1038/nature12730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Geula S, Moshitch-Moshkovitz S, Dominissini D, Mansour AA, Kol N, Salmon-Divon M, Hershkovitz V, Peer E, Mor N, Manor YS, Ben-Haim MS, Eyal E, Yunger S, Pinto Y, Jaitin DA, Viukov S, Rais Y, Krupalnik V, Chomsky E, Zerbib M, Maza I, Rechavi Y, Massarwa R, Hanna S, Amit I, Levanon EY, Amariglio N, Stern-Ginossar N, Novershtern N, Rechavi G, Hanna JH. Stem cells. m6A mRNA methylation facilitates resolution of naive pluripotency toward differentiation. Science. 2015;347(6225):1002–1006. doi: 10.1126/science.1261417. [DOI] [PubMed] [Google Scholar]
- 10.Meyer KD, Saletore Y, Zumbo P, Elemento O, Mason CE, Jaffrey SR. Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell. 2012;149(7):1635–1646. doi: 10.1016/j.cell.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weng Y-L, Wang X, An R, Cassin J, Vissers C, Liu Y, Liu Y, Xu T, Wang X, Wong SZH, Joseph J, Dore LC, Dong Q, Zheng W, Jin P, Wu H, Shen B, Zhuang X, He C, Liu K, Song H, Ming G-L. Epitranscriptomic m6A regulation of axon regeneration in the adult mammalian nervous system. Neuron. 2018;97(2):313–325.e316. doi: 10.1016/j.neuron.2017.12.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yoon KJ, Ringeling FR, Vissers C, Jacob F, Pokrass M, Jimenez-Cyrus D, Su Y, Kim NS, Zhu Y, Zheng L, Kim S, Wang X, Dore LC, Jin P, Regot S, Zhuang X, Canzar S, He C, Ming GL, Song H. Temporal control of mammalian cortical neurogenesis by m(6)A methylation. Cell. 2017;171(4):877–889 e817. doi: 10.1016/j.cell.2017.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hess ME, Hess S, Meyer KD, Verhagen LAW, Koch L, Brönneke HS, Dietrich MO, Jordan SD, Saletore Y, Elemento O, Belgardt BF, Franz T, Horvath TL, Rüther U, Jaffrey SR, Kloppenburg P, Brüning JC. The fat mass and obesity associated gene (Fto) regulates activity of the dopaminergic midbrain circuitry. Nat Neurosci. 2013;16:1042. doi: 10.1038/nn.3449. [DOI] [PubMed] [Google Scholar]
- 14.Walters BJ, Mercaldo V, Gillon CJ, Yip M, Neve RL, Boyce FM, Frankland PW, Josselyn SA. The role of the RNA demethylase FTO (Fat Mass and Obesity-Associated) and mRNA methylation in hippocampal memory formation. Neuropsychopharmacology. 2017;42:1502. doi: 10.1038/npp.2017.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Widagdo J, Zhao QY, Kempen MJ, Tan MC, Ratnu VS, Wei W, Leighton L, Spadaro PA, Edson J, Anggono V, Bredy TW. Experience-dependent accumulation of N6-methyladenosine in the prefrontal cortex is associated with memory processes in mice. J Neurosci. 2016;36(25):6771–6777. doi: 10.1523/JNEUROSCI.4053-15.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin S, Choe J, Du P, Triboulet R, Gregory RI. The m6A methyltransferase METTL3 promotes translation in human cancer cells. Mol Cell. 2016;62(3):335–345. doi: 10.1016/j.molcel.2016.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dorn LE, Lasman L, Chen J, Xu X, Hund TJ, Medvedovic M, Hanna JH, van Berlo JH, Accornero F. The N(6)-methyladenosine mRNA methylase METTL3 controls cardiac homeostasis and hypertrophy. Circulation. 2019;139(4):533–545. doi: 10.1161/CIRCULATIONAHA.118.036146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li M, Zhao X, Wang W, Shi H, Pan Q, Lu Z, Perez SP, Suganthan R, He C, Bjoras M, Klungland A. Ythdf2-mediated m(6)A mRNA clearance modulates neural development in mice. Genome Biol. 2018;19(1):69. doi: 10.1186/s13059-018-1436-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Y, Li Y, Yue M, Wang J, Kumar S, Wechsler-Reya RJ, Zhang Z, Ogawa Y, Kellis M, Duester G, Zhao JC. N(6)-methyladenosine RNA modification regulates embryonic neural stem cell self-renewal through histone modifications. Nat Neurosci. 2018;21(2):195–206. doi: 10.1038/s41593-017-0057-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen KW, Chen JA. Functional roles of long non-coding RNAs in motor neuron development and disease. J Biomed Sci. 2020;27(1):38. doi: 10.1186/s12929-020-00628-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boissel S, Reish O, Proulx K, Kawagoe-Takaki H, Sedgwick B, Yeo GS, Meyre D, Golzio C, Molinari F, Kadhom N, Etchevers HC, Saudek V, Farooqi IS, Froguel P, Lindahl T, O'Rahilly S, Munnich A, Colleaux L. Loss-of-function mutation in the dioxygenase-encoding FTO gene causes severe growth retardation and multiple malformations. Am J Hum Genet. 2009;85(1):106–111. doi: 10.1016/j.ajhg.2009.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Alarcon CR, Lee H, Goodarzi H, Halberg N, Tavazoie SF. N6-methyladenosine marks primary microRNAs for processing. Nature. 2015;519(7544):482–485. doi: 10.1038/nature14281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aas A, Isakson P, Bindesboll C, Alemu EA, Klungland A, Simonsen A. Nucleocytoplasmic shuttling of FTO does not affect starvation-induced autophagy. PLoS ONE. 2017;12(3):e0168182. doi: 10.1371/journal.pone.0168182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chang M, Lv H, Zhang W, Ma C, He X, Zhao S, Zhang ZW, Zeng YX, Song S, Niu Y, Tong WM. Region-specific RNA m(6)A methylation represents a new layer of control in the gene regulatory network in the mouse brain. Open Biol. 2017;7(9):170166. doi: 10.1098/rsob.170166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Caglayan AO, Tuysuz B, Coskun S, Quon J, Harmanci AS, Baranoski JF, Baran B, Erson-Omay EZ, Henegariu O, Mane SM, Bilguvar K, Yasuno K, Gunel M. A patient with a novel homozygous missense mutation in FTO and concomitant nonsense mutation in CETP. J Hum Genet. 2016;61(5):395–403. doi: 10.1038/jhg.2015.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bateman RJ, Xiong C, Benzinger TL, Fagan AM, Goate A, Fox NC, Marcus DS, Cairns NJ, Xie X, Blazey TM, Holtzman DM, Santacruz A, Buckles V, Oliver A, Moulder K, Aisen PS, Ghetti B, Klunk WE, McDade E, Martins RN, Masters CL, Mayeux R, Ringman JM, Rossor MN, Schofield PR, Sperling RA, Salloway S, Morris JC, Dominantly Inherited Alzheimer N. Clinical and biomarker changes in dominantly inherited Alzheimer's disease. N Engl J Med. 2012; 367(9):795–804. [DOI] [PMC free article] [PubMed]
- 27.Alarcon CR, Goodarzi H, Lee H, Liu X, Tavazoie S, Tavazoie SF. HNRNPA2B1 is a mediator of m(6)A-dependent nuclear RNA processing events. Cell. 2015;162(6):1299–1308. doi: 10.1016/j.cell.2015.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Batista PJ, Molinie B, Wang J, Qu K, Zhang J, Li L, Bouley DM, Lujan E, Haddad B, Daneshvar K, Carter AC, Flynn RA, Zhou C, Lim KS, Dedon P, Wernig M, Mullen AC, Xing Y, Giallourakis CC, Chang HY. m(6)A RNA modification controls cell fate transition in mammalian embryonic stem cells. Cell Stem Cell. 2014;15(6):707–719. doi: 10.1016/j.stem.2014.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen TH, Chen JA. Multifaceted roles of microRNAs: from motor neuron generation in embryos to degeneration in spinal muscular atrophy. Elife. 2019 doi: 10.7554/eLife.50848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bringmann P, Luhrmann R. Antibodies specific for N6-methyladenosine react with intact snRNPs U2 and U4/U6. FEBS Lett. 1987;213(2):309–315. doi: 10.1016/0014-5793(87)81512-0. [DOI] [PubMed] [Google Scholar]
- 31.Dominissini D, Moshitch-Moshkovitz S, Schwartz S, Salmon-Divon M, Ungar L, Osenberg S, Cesarkas K, Jacob-Hirsch J, Amariglio N, Kupiec M, Sorek R, Rechavi G. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012;485(7397):201–206. doi: 10.1038/nature11112. [DOI] [PubMed] [Google Scholar]
- 32.Ke S, Alemu EA, Mertens C, Gantman EC, Fak JJ, Mele A, Haripal B, Zucker-Scharff I, Moore MJ, Park CY, Vagbo CB, Kussnierczyk A, Klungland A, Darnell JE, Jr, Darnell RB. A majority of m6A residues are in the last exons, allowing the potential for 3′ UTR regulation. Genes Dev. 2015;29(19):2037–2053. doi: 10.1101/gad.269415.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yue Y, Liu J, He C. RNA N6-methyladenosine methylation in post-transcriptional gene expression regulation. Genes Dev. 2015;29(13):1343–1355. doi: 10.1101/gad.262766.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jia G, Fu Y, Zhao X, Dai Q, Zheng G, Yang Y, Yi C, Lindahl T, Pan T, Yang YG, He C. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat Chem Biol. 2011;7(12):885–887. doi: 10.1038/nchembio.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zheng G, Dahl JA, Niu Y, Fedorcsak P, Huang CM, Li CJ, Vagbo CB, Shi Y, Wang WL, Song SH, Lu Z, Bosmans RP, Dai Q, Hao YJ, Yang X, Zhao WM, Tong WM, Wang XJ, Bogdan F, Furu K, Fu Y, Jia G, Zhao X, Liu J, Krokan HE, Klungland A, Yang YG, He C. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol Cell. 2013;49(1):18–29. doi: 10.1016/j.molcel.2012.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chokkalla AK, Mehta SL, Vemuganti R. Epitranscriptomic regulation by m(6)A RNA methylation in brain development and diseases. J Cereb Blood Flow Metab. 2020;40(12):2331–2349. doi: 10.1177/0271678X20960033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Frye M, Harada BT, Behm M, He C. RNA modifications modulate gene expression during development. Science. 2018;361(6409):1346–1349. doi: 10.1126/science.aau1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang H, Weng H, Chen J. The biogenesis and precise control of RNA m(6)A methylation. Trends Genet. 2020;36(1):44–52. doi: 10.1016/j.tig.2019.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Livneh I, Moshitch-Moshkovitz S, Amariglio N, Rechavi G, Dominissini D. The m(6)A epitranscriptome: transcriptome plasticity in brain development and function. Nat Rev Neurosci. 2020;21(1):36–51. doi: 10.1038/s41583-019-0244-z. [DOI] [PubMed] [Google Scholar]
- 40.Shi H, Wei J, He C. Where, when, and how: context-dependent functions of RNA methylation writers, readers, and erasers. Mol Cell. 2019;74(4):640–650. doi: 10.1016/j.molcel.2019.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vissers C, Sinha A, Ming GL, Song H. The epitranscriptome in stem cell biology and neural development. Neurobiol Dis. 2020;146:105139. doi: 10.1016/j.nbd.2020.105139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Linder B, Grozhik AV, Olarerin-George AO, Meydan C, Mason CE, Jaffrey SR. Single-nucleotide-resolution mapping of m6A and m6Am throughout the transcriptome. Nat Methods. 2015;12(8):767–772. doi: 10.1038/nmeth.3453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Roost C, Lynch SR, Batista PJ, Qu K, Chang HY, Kool ET. Structure and thermodynamics of N6-methyladenosine in RNA: a spring-loaded base modification. J Am Chem Soc. 2015;137(5):2107–2115. doi: 10.1021/ja513080v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu N, Dai Q, Zheng G, He C, Parisien M, Pan T. N(6)-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature. 2015;518(7540):560–564. doi: 10.1038/nature14234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu N, Zhou KI, Parisien M, Dai Q, Diatchenko L, Pan T. N6-methyladenosine alters RNA structure to regulate binding of a low-complexity protein. Nucleic Acids Res. 2017;45(10):6051–6063. doi: 10.1093/nar/gkx141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu J, Li K, Cai J, Zhang M, Zhang X, Xiong X, Meng H, Xu X, Huang Z, Peng J, Fan J, Yi C. Landscape and regulation of m(6)A and m(6)Am methylome across human and mouse tissues. Mol Cell. 2020;77(2):426–440 e426. doi: 10.1016/j.molcel.2019.09.032. [DOI] [PubMed] [Google Scholar]
- 47.Mauer J, Sindelar M, Despic V, Guez T, Hawley BR, Vasseur JJ, Rentmeister A, Gross SS, Pellizzoni L, Debart F, Goodarzi H, Jaffrey SR. FTO controls reversible m(6)Am RNA methylation during snRNA biogenesis. Nat Chem Biol. 2019;15(4):340–347. doi: 10.1038/s41589-019-0231-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yang Y, Fan X, Mao M, Song X, Wu P, Zhang Y, Jin Y, Yang Y, Chen LL, Wang Y, Wong CC, Xiao X, Wang Z. Extensive translation of circular RNAs driven by N(6)-methyladenosine. Cell Res. 2017;27(5):626–641. doi: 10.1038/cr.2017.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhou C, Molinie B, Daneshvar K, Pondick JV, Wang J, Van Wittenberghe N, Xing Y, Giallourakis CC, Mullen AC. Genome-wide maps of m6A circRNAs identify widespread and cell-type-specific methylation patterns that are distinct from mRNAs. Cell Rep. 2017;20(9):2262–2276. doi: 10.1016/j.celrep.2017.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Warda AS, Kretschmer J, Hackert P, Lenz C, Urlaub H, Hobartner C, Sloan KE, Bohnsack MT. Human METTL16 is a N(6)-methyladenosine (m(6)A) methyltransferase that targets pre-mRNAs and various non-coding RNAs. EMBO Rep. 2017;18(11):2004–2014. doi: 10.15252/embr.201744940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yen YP, Hsieh WF, Tsai YY, Lu YL, Liau ES, Hsu HC, Chen YC, Liu TC, Chang M, Li J, Lin SP, Hung JH, Chen JA. Dlk1-Dio3 locus-derived lncRNAs perpetuate postmitotic motor neuron cell fate and subtype identity. Elife. 2018 doi: 10.7554/eLife.38080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li L, Zang L, Zhang F, Chen J, Shen H, Shu L, Liang F, Feng C, Chen D, Tao H, Xu T, Li Z, Kang Y, Wu H, Tang L, Zhang P, Jin P, Shu Q, Li X. Fat mass and obesity-associated (FTO) protein regulates adult neurogenesis. Hum Mol Genet. 2017;26(13):2398–2411. doi: 10.1093/hmg/ddx128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Roundtree IA, Evans ME, Pan T, He C. Dynamic RNA modifications in gene expression regulation. Cell. 2017;169(7):1187–1200. doi: 10.1016/j.cell.2017.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ivanova I, Much C, Di Giacomo M, Azzi C, Morgan M, Moreira PN, Monahan J, Carrieri C, Enright AJ, O'Carroll D. The RNA m(6)A reader YTHDF2 is essential for the post-transcriptional regulation of the maternal transcriptome and oocyte competence. Mol Cell. 2017;67(6):1059–1067 e1054. doi: 10.1016/j.molcel.2017.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhao BS, Roundtree IA, He C. Post-transcriptional gene regulation by mRNA modifications. Nat Rev Mol Cell Biol. 2017;18(1):31–42. doi: 10.1038/nrm.2016.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wu R, Li A, Sun B, Sun JG, Zhang J, Zhang T, Chen Y, Xiao Y, Gao Y, Zhang Q, Ma J, Yang X, Liao Y, Lai WY, Qi X, Wang S, Shu Y, Wang HL, Wang F, Yang YG, Yuan Z. A novel m(6)A reader Prrc2a controls oligodendroglial specification and myelination. Cell Res. 2019;29(1):23–41. doi: 10.1038/s41422-018-0113-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xu H, Dzhashiashvili Y, Shah A, Kunjamma RB, Weng YL, Elbaz B, Fei Q, Jones JS, Li YI, Zhuang X, Ming GL, He C, Popko B. m(6)A mRNA methylation is essential for oligodendrocyte maturation and CNS myelination. Neuron. 2020;105(2):293–309 e295. doi: 10.1016/j.neuron.2019.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Merkurjev D, Hong WT, Iida K, Oomoto I, Goldie BJ, Yamaguti H, Ohara T, Kawaguchi SY, Hirano T, Martin KC, Pellegrini M, Wang DO. Synaptic N(6)-methyladenosine (m(6)A) epitranscriptome reveals functional partitioning of localized transcripts. Nat Neurosci. 2018;21(7):1004–1014. doi: 10.1038/s41593-018-0173-6. [DOI] [PubMed] [Google Scholar]
- 59.Koranda JL, Dore L, Shi H, Patel MJ, Vaasjo LO, Rao MN, Chen K, Lu Z, Yi Y, Chi W, He C, Zhuang X. Mettl14 is essential for epitranscriptomic regulation of striatal function and learning. Neuron. 2018;99(2):283–292.e285. doi: 10.1016/j.neuron.2018.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ma C, Chang M, Lv H, Zhang ZW, Zhang W, He X, Wu G, Zhao S, Zhang Y, Wang D, Teng X, Liu C, Li Q, Klungland A, Niu Y, Song S, Tong WM. RNA m(6)A methylation participates in regulation of postnatal development of the mouse cerebellum. Genome Biol. 2018;19(1):68. doi: 10.1186/s13059-018-1435-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shi H, Zhang X, Weng YL, Lu Z, Liu Y, Lu Z, Li J, Hao P, Zhang Y, Zhang F, Wu Y, Delgado JY, Su Y, Patel MJ, Cao X, Shen B, Huang X, Ming GL, Zhuang X, Song H, He C, Zhou T. m(6)A facilitates hippocampus-dependent learning and memory through YTHDF1. Nature. 2018;563(7730):249–253. doi: 10.1038/s41586-018-0666-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Han M, Liu Z, Xu Y, Liu X, Wang D, Li F, Wang Y, Bi J. Abnormality of m6A mRNA methylation is involved in Alzheimer's disease. Front Neurosci. 2020;14:98. doi: 10.3389/fnins.2020.00098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shafik AM, Zhang F, Guo Z, Dai Q, Pajdzik K, Li Y, Kang Y, Yao B, Wu H, He C, Allen EG, Duan R, Jin P. N6-methyladenosine dynamics in neurodevelopment and aging, and its potential role in Alzheimer's disease. Genome Biol. 2021;22(1):17. doi: 10.1186/s13059-020-02249-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Choudhry Z, Sengupta SM, Grizenko N, Thakur GA, Fortier ME, Schmitz N, Joober R. Association between obesity-related gene FTO and ADHD. Obesity (Silver Spring) 2013;21(12):E738–744. doi: 10.1002/oby.20444. [DOI] [PubMed] [Google Scholar]
- 65.Milaneschi Y, Lamers F, Mbarek H, Hottenga JJ, Boomsma DI, Penninx BW. The effect of FTO rs9939609 on major depression differs across MDD subtypes. Mol Psychiatry. 2014;19(9):960–962. doi: 10.1038/mp.2014.4. [DOI] [PubMed] [Google Scholar]
- 66.Liu X, Shimada T, Otowa T, Wu YY, Kawamura Y, Tochigi M, Iwata Y, Umekage T, Toyota T, Maekawa M, Iwayama Y, Suzuki K, Kakiuchi C, Kuwabara H, Kano Y, Nishida H, Sugiyama T, Kato N, Chen CH, Mori N, Yamada K, Yoshikawa T, Kasai K, Tokunaga K, Sasaki T, Gau SS. Genome-wide association study of autism spectrum disorder in the east Asian populations. Autism Res. 2016;9(3):340–349. doi: 10.1002/aur.1536. [DOI] [PubMed] [Google Scholar]
- 67.Du T, Rao S, Wu L, Ye N, Liu Z, Hu H, Xiu J, Shen Y, Xu Q. An association study of the m6A genes with major depressive disorder in Chinese Han population. J Affect Disord. 2015;183:279–286. doi: 10.1016/j.jad.2015.05.025. [DOI] [PubMed] [Google Scholar]
- 68.Mo XB, Lei SF, Qian QY, Guo YF, Zhang YH, Zhang H. Integrative analysis revealed potential causal genetic and epigenetic factors for multiple sclerosis. J Neurol. 2019;266(11):2699–2709. doi: 10.1007/s00415-019-09476-w. [DOI] [PubMed] [Google Scholar]
- 69.Wilson C, Chen PJ, Miao Z, Liu DR. Programmable m(6)A modification of cellular RNAs with a Cas13-directed methyltransferase. Nat Biotechnol. 2020;38(12):1431–1440. doi: 10.1038/s41587-020-0572-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Garcia-Campos MA, Edelheit S, Toth U, Safra M, Shachar R, Viukov S, Winkler R, Nir R, Lasman L, Brandis A, Hanna JH, Rossmanith W, Schwartz S. Deciphering the "m(6)A Code" via antibody-independent quantitative profiling. Cell. 2019;178(3):731–747 e716. doi: 10.1016/j.cell.2019.06.013. [DOI] [PubMed] [Google Scholar]
- 71.Liu H, Begik O, Lucas MC, Ramirez JM, Mason CE, Wiener D, Schwartz S, Mattick JS, Smith MA, Novoa EM. Accurate detection of m(6)A RNA modifications in native RNA sequences. Nat Commun. 2019;10(1):4079. doi: 10.1038/s41467-019-11713-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sas-Chen A, Thomas JM, Matzov D, Taoka M, Nance KD, Nir R, Bryson KM, Shachar R, Liman GLS, Burkhart BW, Gamage ST, Nobe Y, Briney CA, Levy MJ, Fuchs RT, Robb GB, Hartmann J, Sharma S, Lin Q, Florens L, Washburn MP, Isobe T, Santangelo TJ, Shalev-Benami M, Meier JL, Schwartz S. Dynamic RNA acetylation revealed by quantitative cross-evolutionary mapping. Nature. 2020;583(7817):638–643. doi: 10.1038/s41586-020-2418-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Liu J, Dou X, Chen C, Chen C, Liu C, Xu MM, Zhao S, Shen B, Gao Y, Han D, He C. N (6)-methyladenosine of chromosome-associated regulatory RNA regulates chromatin state and transcription. Science. 2020;367(6477):580–586. doi: 10.1126/science.aay6018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Edens BM, Vissers C, Su J, Arumugam S, Xu Z, Shi H, Miller N, Rojas Ringeling F, Ming G, He C, Song H, Ma YC. FMRP modulates neural differentiation through m6A-dependent mRNA nuclear export. Cell Reports. 2019;28(4):845–854.e5. doi: 10.1016/j.celrep.2019.06.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhuang M, Li X, Zhu J, Zhang J, Niu F, Liang F, Chen M, Li D, Han P, Ji S-J. The m6A reader YTHDF1 regulates axon guidance through translational control of Robo3.1 expression. Nucleic Acids Res. 2019;47(9):4765–4777. doi: 10.1093/nar/gkz157. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.