Abstract
Periodontitis is an inflammation of tooth-supporting tissues, which is caused by bacteria in the subgingival plaque (biofilm) and the host immune response. Traditionally, subgingival pathogens have been investigated using methods such as culturing, DNA probes, or PCR. The development of next-generation sequencing made it possible to investigate the whole microbiome in the subgingival plaque. Previous studies have implicated dysbiosis of the subgingival microbiome in the etiology of periodontitis. However, details are still lacking. In this study, we conducted a metagenomic analysis of subgingival plaque samples from a group of Japanese individuals with and without periodontitis. In the taxonomic composition analysis, genus Bacteroides and Mycobacterium demonstrated significantly different compositions between healthy sites and sites with periodontal pockets. The results from the relative abundance of functional gene categories, carbohydrate metabolism, glycan biosynthesis and metabolism, amino acid metabolism, replication and repair showed significant differences between healthy sites and sites with periodontal pockets. These results provide important insights into the shift in the taxonomic and functional gene category abundance caused by dysbiosis, which occurs during the progression of periodontal disease.
Keywords: periodontal disease, periodontitis, subgingival plaque biofilm, periodontal pathogens, metagenomics, human oral microbiome
1. Introduction
Periodontitis is a highly prevalent inflammatory and infectious disease of the tooth-supporting tissues. It is caused by bacteria present in the subgingival plaque (biofilm) and the host immune responses [1]. Imbalance in inflammatory and immune responses leads to the destruction of periodontal tissues, including alveolar bone loss, ultimately causing loss of teeth. The percentage of adults with periodontitis is very high. Approximately 42% of dentate adults between 30 and 79 years in the USA suffer from periodontitis [2], and the global age-standardized prevalence of severe periodontitis is approximately 10% [3]. Cumulative evidence has indicated that periodontal disease can underlie or exacerbate systemic diseases such as atherosclerosis, diabetes, rheumatoid arthritis [4], and Alzheimer’s disease [5]. These reports indicated that the eradication of periodontitis could greatly contribute to public health. However, to date, the etiologic agents of periodontitis are yet to be fully identified.
The results of the traditional investigations of the pathogens in the subgingival plaque, using methods such as culturing [6], DNA probes [7], and polymerase chain reaction (PCR) [8], have implicated gram-negative anaerobes such as Porphyromonas gingivalis and Tannerella forsythia, in the pathogenesis and progression of periodontitis. However, these conventional methods cannot provide a full picture of periodontal pathogens in subgingival plaque samples. Therefore, further comprehensive analysis of the subgingival microbiome is necessary. In addition, the results of animal experiments have shown increases in the bacterial species and their numbers following the infection by P. gingivalis, and a synergistic increase in the virulence of mixed infections by P. gingivalis and Streptococcus gordonii [9,10]. Based on these reports, dysbiosis of the subgingival microbiome is thought to be a major cause of periodontitis [11]. A comprehensive investigation of the microbiome shifts is essential to understand the mechanism underlying dysbiosis. Recently, several approaches for investigating the microbiome have been developed. One of these is next-generation sequencing, such as 16S rRNA sequencing and metagenome analysis, and another is a culture-based method, such as culturomics, which involves isolation of bacteria by culturing under different conditions and identification by using matrix-assisted laser desorption/ionization-time of flight mass spectrometry [12]. Among these, methods using next-generation sequencing have made comprehensive analysis of the whole microbiome of multiple samples possible. Recently, analyses using 16S rRNA sequencing have clarified the relative abundance of pathogens at the sites of periodontitis (periodontal pockets) [13,14].
During dysbiosis, a number of polymicrobial synergies occur [15]. In the subgingival plaques, more than 700 bacterial taxa have been detected [16]. Multiple species and genes may be involved in these synergies. Endo et al. [17] reported that essential genes involved in butyric acid metabolism within the fatty acid biosynthesis pathway are complemented in P. gingivalis, T. forsythia, and Treponema denticola. The composition of the microbiomes of several body areas, including supragingival plaque, buccal mucosa and tongue dorsum, showed diversity among subjects at the phylum level; however, the functional genes in this metabolic pathway were similar among all subjects [18]. These reports indicate the necessity of functional gene analysis. In addition, differences in the virulence within each species have been reported. The virulence of P. gingivalis with type II fimbriae was reportedly higher than that of the same species that produce other types of fimbriae [19]. A variant of Aggregatibacter actinomycetemcomitans, another major pathogen implicated in periodontitis, with mutations in its leukotoxin promoter region, produces significantly higher levels of leukotoxin than the original strain does [20]. These reports indicate that metagenomic analyses of subgingival plaque are essential to clarify the mechanisms of interspecies synergy inducing dysbiosis. Here, we present a metagenomic analysis of subgingival plaque samples from a group of Japanese individuals with or without periodontitis, aiming to clarify the taxonomic and functional genomic abundance in periodontitis.
2. Results
2.1. Sequence Overview of Plaque Samples
We obtained 64 subgingival plaque samples from 33 participants (age range 25–79 years old) (Table S1). After quality control of the reads, removal of the human genome, and extraction of the samples satisfying the threshold of the read number for metagenomic analysis, 42 samples were subjected to further data analysis (Table 1.).
Table 1.
Sequence result overview.
| Sample Name | Indiv. No. |
Status of Individual | Sampling Site and Status | Age | Sex 1 | Raw Reads | Reads for Metagenomic Analysis |
|---|---|---|---|---|---|---|---|
| HH1 | 1 | Healthy | 46, Healthy | 29 | M | 5,775,202 | 1,402,887 |
| HH2 | 2 | Healthy | 46, Healthy | 43 | M | 3,831,266 | 204,308 |
| HH3 | 3 | Healthy | 46, Healthy | 37 | M | 5,422,140 | 995,685 |
| HH4 | 4 | Healthy | 46, Healthy | 41 | F | 4,866,418 | 1,100,826 |
| HH5 | 5 | Healthy | 46, Healthy | 31 | F | 4,247,496 | 1,089,164 |
| HH6 | 6 | Healthy | 46, Healthy | 27 | M | 4,265,064 | 663,850 |
| PH11_2 | 11 | Periodontitis | 43, Healthy | 48 | M | 2,624,566 | 548,873 |
| PH12_1 | 12 | Periodontitis | 23, Healthy | 69 | M | 1,205,826 | 695,215 |
| PH12_2 | 12 | Periodontitis | 25, Healthy | 69 | M | 2,697,604 | 1,750,204 |
| PH14 | 14 | Periodontitis | 43, Healthy | 48 | F | 8,165,844 | 239,717 |
| PH15 | 15 | Periodontitis | 12, Healthy | 79 | M | 7,358,454 | 366,238 |
| PH17 | 17 | Periodontitis | 24, Healthy | 56 | F | 2,125,774 | 278,004 |
| PH23_1 | 23 | Periodontitis | 23, Healthy | 42 | M | 2,340,430 | 463,933 |
| PH25_1 | 25 | Periodontitis | 41, Healthy | 43 | M | 5,952,692 | 1,716,829 |
| PH25_2 | 25 | Periodontitis | 41, Healthy | 43 | M | 4,593,914 | 1,556,673 |
| PH26_2 | 26 | Periodontitis | 31, Healthy | 60 | M | 4,678,568 | 225,769 |
| PH29 | 29 | Periodontitis | 27, Healthy | 58 | F | 5,253,782 | 516,845 |
| PH30 | 30 | Periodontitis | 13, Healthy | 76 | M | 6,012,650 | 390,445 |
| PH31 | 31 | Periodontitis | 23, Healthy | 46 | F | 4,890,982 | 213,660 |
| PP12_2 | 12 | Periodontitis | 25, Periodontal pocket | 69 | M | 2,398,604 | 1,894,262 |
| PP13_1 | 13 | Periodontitis | 26, Periodontal pocket | 47 | F | 327,016 | 245,066 |
| PP13_2 | 13 | Periodontitis | 25, Periodontal pocket | 47 | F | 309,300 | 306,024 |
| PP14_1 | 14 | Periodontitis | 16, Periodontal pocket | 48 | F | 7,440,288 | 535,904 |
| PP14_2 | 14 | Periodontitis | 27, Periodontal pocket | 48 | F | 7,213,688 | 630,156 |
| PP15_1 | 15 | Periodontitis | 11, Periodontal pocket | 79 | M | 7,346,724 | 414,834 |
| PP15_2 | 15 | Periodontitis | 47, Periodontal pocket | 79 | M | 6,530,194 | 350,097 |
| PP16_1 | 16 | Periodontitis | 24, Periodontal pocket | 55 | F | 1,904,568 | 452,740 |
| PP16_2 | 16 | Periodontitis | 26, Periodontal pocket | 55 | F | 3,093,452 | 226,275 |
| PP17 | 17 | Periodontitis | 47, Periodontal pocket | 56 | F | 3,646,976 | 297,839 |
| PP18_1 | 18 | Periodontitis | 16, Periodontal pocket | 36 | F | 1,604,552 | 439,199 |
| PP18_2 | 18 | Periodontitis | 25, Periodontal pocket | 36 | F | 3,920,256 | 912,649 |
| PP21_1 | 21 | Periodontitis | 27, Periodontal pocket | 50 | M | 1,099,140 | 998,367 |
| PP21_2 | 21 | Periodontitis | 13, Periodontal pocket | 50 | M | 2,426,890 | 987,197 |
| PP22 | 22 | Periodontitis | 37, Periodontal pocket | 48 | F | 7,794,000 | 1,322,677 |
| PP23 | 23 | Periodontitis | 35, Periodontal pocket | 42 | M | 8,557,728 | 1,484,973 |
| PP24_1 | 24 | Periodontitis | 13, Periodontal pocket | 59 | F | 5,485,608 | 262,343 |
| PP24_2 | 24 | Periodontitis | 16, Periodontal pocket | 59 | F | 8,658,802 | 768,780 |
| PP25_1 | 25 | Periodontitis | 27, Periodontal pocket | 43 | M | 9,030,826 | 2,508,972 |
| PP25_2 | 25 | Periodontitis | 47, Periodontal pocket | 43 | M | 6,575,378 | 817,066 |
| PP26_1 | 26 | Periodontitis | 18, Periodontal pocket | 60 | M | 1,427,528 | 1,008,404 |
| PP26_2 | 26 | Periodontitis | 37, Periodontal pocket | 60 | M | 11,622,702 | 440,551 |
| PP27 | 27 | Periodontitis | 24, Periodontal pocket | 56 | F | 8,580,870 | 3,233,325 |
1 M, male; F, female.
2.2. Taxonomic Composition Analysis
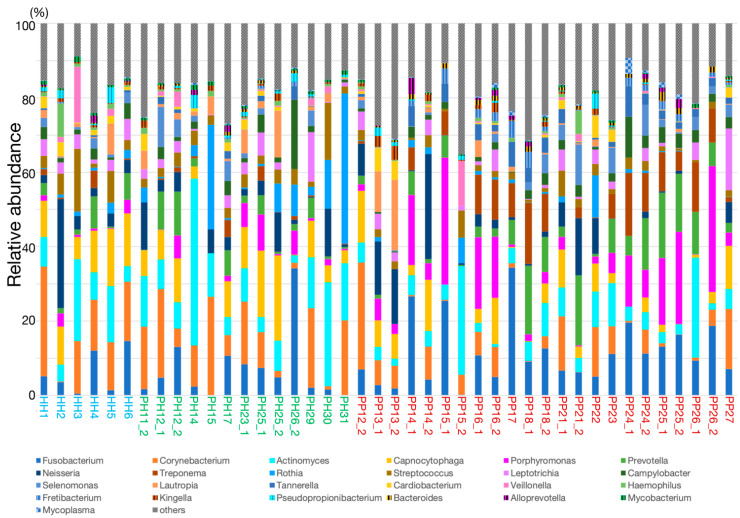
Taxonomic compositions were estimated from fragments of 16S rDNA in metagenomic reads. Less than 1% relative abundance in all samples was categorized as “others”, and we identified 60 genera (Figure S1). We have summarized the top 25 genera for average of all samples in Figure 1.
Figure 1.
The genus level taxonomic composition in the top 25 genera, representing the mean of all samples.
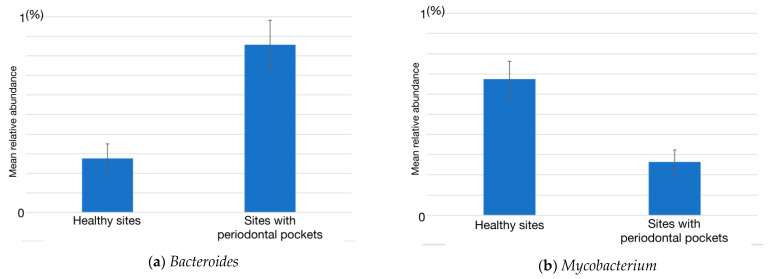
Taxonomic compositions varied among individuals and the disease status of the sites. At the healthy sites, Corynebacterium, Actinomyces, Capnocytophaga, Fusobacterium, Rothia and Neisseria were predominant (Table S2). At the sites with periodontal pockets, Fusobacterium, Porphyromonas, Prevotella Actinomyces, Treponema, Corynebacterium, and Neisseria were predominant. Bacteroides, Porphyromonas, Tannerella, Burkholderia, Moraxella, Treponema, Fretibacterium, Mogibacterium, Shuttleworthia, Filifactor, and Megasphaera were increased with >3-fold higher abundance in the sites with periodontal pockets than the healthy sites (Table S3, Figure S2). In addition, Genera including Abiotrophia, Dialister, Filifactor and Fretibacterium were increased with >3-fold abundance in the samples from healthy sites in periodontitis patient compared with that from the healthy sites in the healthy individuals (Table S4). We further compared the abundances of 60 genera between healthy sites and sites with periodontal pockets using Mann-Whitney U test. The results from the taxonomic comparison indicated the change of abundance in the following two genera: Bacteroides (p = 0.047, power = 0.359) and Mycobacterium (p = 0.021, power = 0.397). The comparison of the mean abundance of these genera is shown in Figure 2.
Figure 2.
The relative abundances of the genera Bacteroides (a) and Mycbacterium (b). The relative abundance of Bacteroides and Mycobacterium were significantly different between the healthy sites and sites with periodontal pockets (p < 0.05). Data are shown as the mean ± standard error of the mean.
2.3. Functional Gene Category Analysis
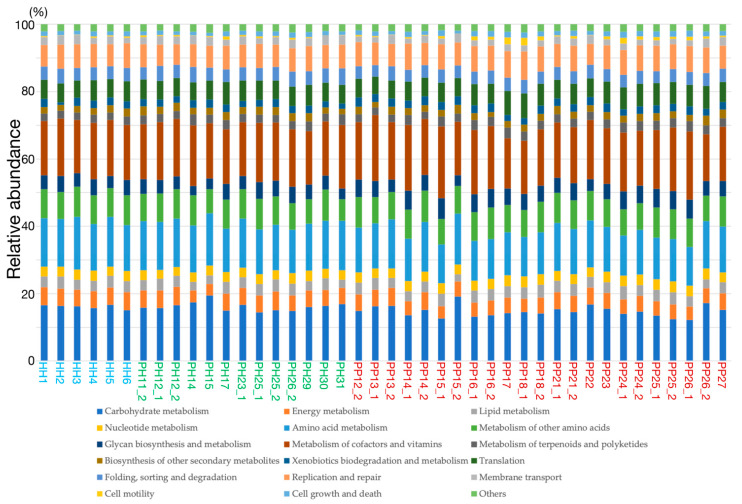
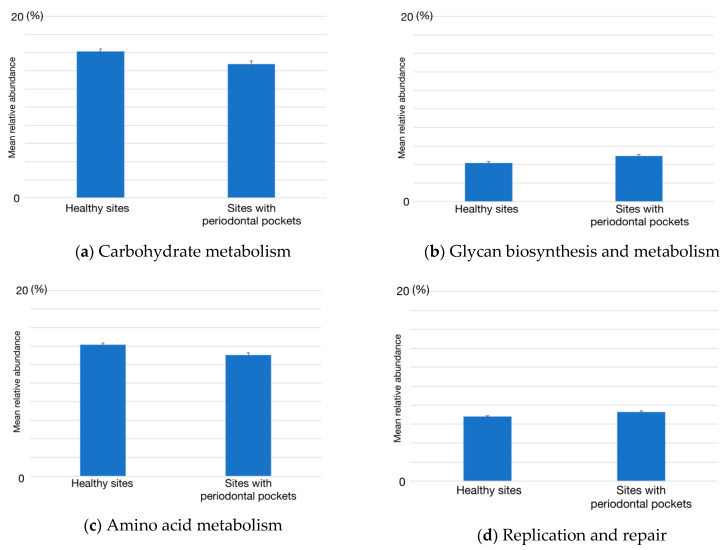
The relative abundances of functional gene categories were estimated from metagenomic reads using the KEGG database (Figure 3). Compared to the taxonomic composition, the relative abundances of functional gene categories seemed to be uniform between individuals, especially at healthy sites. For a more detailed exploration, we again performed the Mann–Whitney U test for each of the 19 functional gene categories in healthy or periodontitis sampling sites. The results from this analysis indicated differences in the abundance of genes functioning in the following five categories: carbohydrate metabolism (p = 0.032, power = 0.328), amino acid metabolism (p = 0.035, power = 0.324), glycan biosynthesis and metabolism (p = 0.023, power = 0.347), and replication and repair (p = 0.035, power = 0.324). Comparisons of the mean abundances of these gene categories in healthy and periodontitis sites are shown in Figure 4.
Figure 3.
The relative abundances of functional gene categories.
Figure 4.
Relative abundances of four functional gene categories in the healthy sites and sites with periodontal pockets. (a) Carbohydrate metabolism, (b) glycan biosynthesis and metabolism, (c) amino acid metabolism, and (d) replication and repair. The four functional gene categories were significantly different between healthy and periodontitis sites (p < 0.05). Data are shown as the mean ± standard error of the mean.
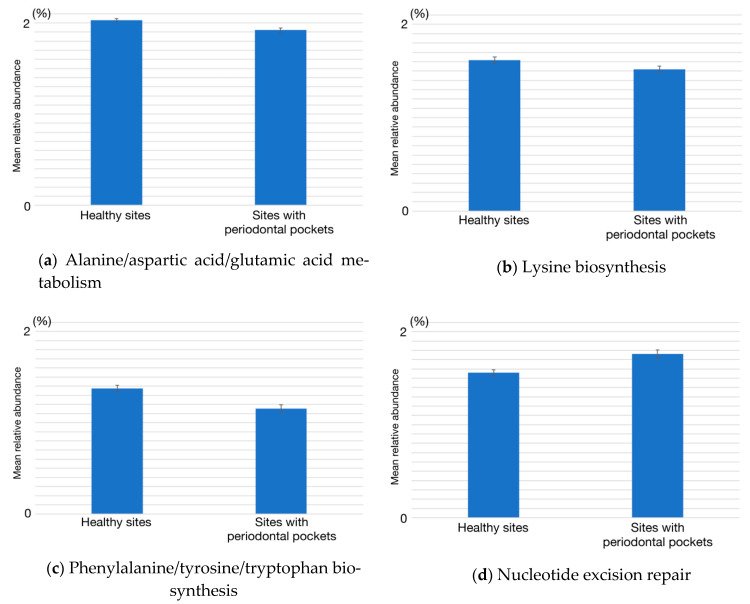
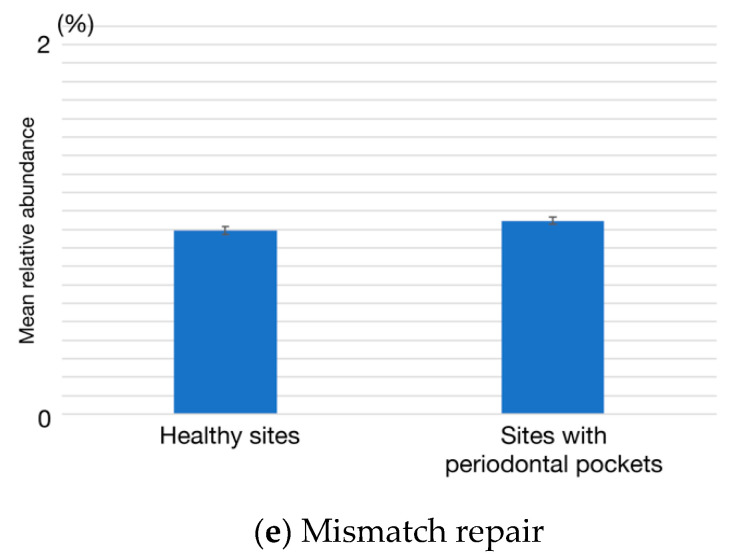
Currently, little is known about the decrease in amino acid metabolism, and the increase in the replication and repair in periodontitis sites. For a more detailed exploration of these three categories, we conducted the Mann-Whitney U test (p < 0.05) on one lower-level category. From lower levels of amino acid metabolism, a decrease in phenylalanine/tyrosine/tryptophan biosynthesis (p = 0.032, power = 0.314) and lysine biosynthesis (p = 0.040, power = 0.160) at the sites with periodontal pockets were indicated (Figure 5). We also found that alanine/aspartic acid/glutamic acid metabolism decreased (p = 0.004, power = 0.439) and high abundances of the genes belonging to replication and repair, nucleotide excision repair (p = 0.024, power = 0.177), and mismatch repair (p = 0.005, power = 0.378) at the sites with periodontal pockets.
Figure 5.
The relative abundances of five lower functional gene categories in the healthy sites and sites with periodontal pockets. (a) Alanine/aspartic acid/glutamic acid metabolism, (b) lysine biosynthesis, (c) phenylalanine/tyrosine/tryptophan biosynthesis, (d) nucleotide excision repair, and (e) mismatch repair. The five functional gene categories were significantly different between healthy and periodontitis sites (p < 0.05). Data are shown as the mean ± standard error of the mean.
3. Discussion
In the present study, Fusobacterium, Porphyromonas, Prevotella, Actinomyces, Treponema, Corynebacterium, and Neisseria were predominant at the sites with periodontal pockets (Table S2). Among 11 increased genera, Bacteroides, Porphyromonas, Tannerella, Treponema, and Filifactor have been reported as the core bacterial genera in the sites with periodontal pockets [13]. Moreover, the association between Fretibacterium and periodontitis has also been reported [21]. The observed shift of genera in the subgingival microbiome at the sites with periodontal pockets was consistent with the previous results. At the healthy sites of the patients with periodontitis, Dialister, Filifactor, Fretibacterium, and Parvimonas were highly abundant compared with the healthy sites of the periodontally healthy subjects (Table S4). Increases in these species have been reported at the sites with periodontal pockets [13,22,23]. A previous report indicated that the disease-associated bacteria were detected not only in deep periodontal pockets but also in shallow pockets in individuals with periodontitis [14]. The results suggested that the shift of the microbiome precedes the appearance of symptoms.
Among the genera with increased abundances at the sites with periodontal pockets, a significant increase in the relative abundance of Bacteroides and decrease in that of Mycobacterium at the sites with periodontal pockets were noted compared with their abundances in the healthy sites. Previous studies have also reported a significantly high abundance of Bacteroides in the sites with periodontal pockets [13,14,24]. Bacteroides detected from the sites with periodontal pockets in these studies and the present study have yet to be isolated and characterized. Because the species in Bacteroides are implicated in dysbiosis, the isolation and detailed characterization of these species is required. Mycobacterium tuberculosis, a causative bacterium of tuberculosis, is a well-known representative of this genus. However, a recent study revealed that a member of the genus Mycobacterium, which does not cause tuberculosis, is present in the oral cavity of healthy humans [25]. Bacteria that belong to the genus Mycobacterium are generally aerobic. Thus, it is conceivable that their population was reduced in the subgingival milieu, which is mostly anerobic. This genus was not predominant in the microbiome at the healthy sites, although significantly decreased in the sites with periodontal pockets. The difference in the abundance of Mycobacterium between the healthy sites and the sites with periodontal pockets has not reported previously. Further analysis of the prevalence of this genus concerning geographical or ethnical difference is required to clarify the association with the dysbiosis.
In the analysis of functional genes, the abundance of genes related to carbohydrate metabolism was decreased in the sites with periodontal pockets. Species in most of the increased genera listed above were asaccharolytic or weak saccharolytic [26,27,28]. These bacteria gain their nutrients from the broken cells and gingival crevicular fluid (GCF). A decrease in the abundance of functional genes for glycolysis_gluconeogenesis and galactose metabolism at the sites with periodontal pockets has been reported [29]. This decrease suggested that the major energy source of the microbiome in the sites with periodontal pockets shifts from carbohydrate to amino acid or intermediate metabolites from microorganisms in the consortia.
Glycan biosynthesis and metabolism were increased at the sites with periodontal pockets. A previous study reported that the clinical isolates of Prevotella intermedia strain possessing a strong ability to produce exopolysaccharide (EPS) showed enhanced abscess-forming activity compared with low EPS producing strains [30]. In P. gingivalis, inactivation of sinR, which negatively regulates lipopolysaccharide and capsular polysaccharide synthesis, increased the biofilm formation [31]. The capsular polysaccharide of P. gingivalis plays an important role in evasion from the host defense [32]. These reports indicate that polysaccharide synthesis is important for this microorganism in the biofilm formation and evasion from host defense. T. forsythia is highly associated with the severe form of periodontitis. It has a specific glycan core on the cell surface. This glycan core plays an important role in the suppression of human Th17 cells [33]. The abundance of these three species was significantly increased at the sites with periodontal pockets. The observed increase in the glycan biosynthesis and metabolism suggested the importance of polysaccharides in periodontopathic biofilms.
In the amino acid metabolism category, abundance of genes involved in phenylalanine/tyrosine/tryptophan biosynthesis was decreased. In the subgingival pocket, microorganisms were bathed in GCF, which included a mixture of proteins such as albumin and globulin. The protein concentration of GCF at the sites with periodontal pockets was similar to that in the serum [34]. The predominant species, such as P. gingivalis and T. denticola, possess arginine- and lysin-specific proteases and prolyl-phenylalanine-specific protease, respectively [35,36]. In the presence of these proteolytic enzymes, lysin, phenylalanine, tyrosine and tryptophan may be accessible without synthesis. Although it was not statistically significant, a reduction in phenylalanine/tyrosine/tryptophan biosynthesis was also previously reported [23]. In addition, the amount of phenylalanine/tyrosine/tryptophan reportedly increased in the saliva of the patients with periodontitis [37]. The reduction in the genes belonging to the biosynthesis of lysine, phenylalanine, tyrosine, and tryptophan may depend on the supply of a large quantity of proteins from the host tissue. In the previous metagenomic analysis, phenylalanine-specific permease was significantly decreased at the sites with periodontal pockets, although phenylalanine/tyrosine/tryptophan biosynthesis did not change [38]. It suggested some difference in the metabolism of the phenylalanine at the sites with periodontal pockets. Interestingly, a decrease in phenylalanine, tyrosine and tryptophan biosynthesis was reported in the gut microbiome of mice, after they were administrated P. gingivalis [39]. The decrease in the abundance of functional genes associated with phenylalanine/tyrosine/tryptophan biosynthesis might be a characteristic of the P. gingivalis-induced microbiome shift. A significant reduction in phenylalanine in saliva has been observed by debridement of supragingival plaque from individuals with periodontitis [40], suggestive of the phenylalanine supply from the supragingival plaque. Further analysis of the proteome of the subgingival microbiome in periodontitis is required.
Abundance of genes associated with alanine/aspartic acid/glutamic acid metabolism also decreased in the sites with periodontal pockets. In the microbiome, complicated synergy in metabolism was organized among the organisms in the microbiome. Previous reports indicated the abundance of P. gingivalis, T. denticola and T. forsythia increase in periodontitis [7], and synergy among the species was reported [41,42]. The genes involved in the butyric acid metabolism were complemented in the three species, although, individually, they did not have the genes for the complete pathway [17]. A synergistic effect on biofilm formation between P. gingivalis and Fusobacterium nucleatum has been reported [43]. The decrease in alanine/aspartic acid/glutamic acid metabolism may be associated with the synergy among multiple species of bacteria. The evidence is limited for the metabolism of the entire microbiome in the oral cavity. Clarification of the change in the abundance of the functional genes requires further transcriptome and proteome analyses of the total microbiome.
High abundances of the genes belonging to replication and repair, nucleotide excision repair, and mismatch repair at the sites with periodontal pockets were observed. In the subgingival microbiome, bacteria are exposed to many types of stress, including oxygen stress [44]. Oxygen stress is induced by oxygen and H2O2 produced from microorganisms and neutrophils in the subgingival milieu [45,46]. Without enzyme neutralizing the H2O2, such as catalase, reactive oxygen species damage the bacterial DNA. In the present study, most genera with an increased relative abundance in the sites with periodontal pockets were anaerobes, and they do not contain catalase. In P. gingivalis, the gene involved in the repair of oxidative stress-induced DNA mismatching was reported [47]. It is possible that the genes involved in response to oxygen stress may partially reflect the increase in the abundance of genes associated with the nucleotide excision repair.
The increase in nucleotide excision repair suggested the involvement of the response to host defense response to functional genes of subgingival bacteria. Crosstalk between periodontopathic bacteria and immune response has been investigated. Macrophage isolated from the patients with periodontitis showed the less ability to polarize to M1 or M2 phenotype compared with that from the healthy subjects [48]. Polymorphisms of IL-10 have been associated with those of tumor necrosis factor α IL-1α, IL-1β and IL-1RA in periodontitis [49]. In patients with periodontitis, the response of neutrophil and monocytes against P. gingivalis-derived lipopolysaccharide was exaggerated and the immune alterations associated with periodontitis were lost after treatment of periodontitis [50]. The attenuation of the immune response was suggested to play an important role in dysbiosis [11]. Recent analysis using mice periodontitis model indicated the decrease in the amount of dental plaque and change of the microflora composition by the inactivation of IL-17 using a mice periodontitis model [51]. Investigation of the immune response with the functional analysis requires the further analysis of the meaning of the shift of the functional genes.
In the present study, the abundance of genes associated with the virulence of bacteria did not increase. Previous studies have indicated an increase in the abundance of genes associated with chemotaxis and motility, and two component system in the sites with periodontal pockets [23,52]. Here, part of the healthy site samples were collected from the patients diagnosed with periodontitis. Previous studies indicated that the relative abundances of functional genes and taxa are different between the healthy sites in healthy subjects and the gingival crevice or deep periodontal pocket in patients with periodontitis, but are not different between the gingival crevice and deep periodontal pocket in patients with periodontitis. In the microbiome composition found in the present study, abundances of genera, such as Bacteroides, Tannerella, Fretibacterium, and Filifactor, frequently increased in periodontitis. It is possible that a subliminal increase in periodontopathic bacterial abundance attenuated the detection of virulence-associated genes.
Nonetheless, there are some limitations to this study. The metagenomic analysis is limited owing to its biases [53]: bias from the extraction of the gene sequence from the sample and methodology used for bioinformatics analysis of taxonomy and sequence depth (the method cannot detect < 105 per gram of sample). In this study, we aimed to obtain basic information on the potential difference between healthy sites and sites with periodontal pockets, and sample size calculation was not performed a priori. Therefore, it is possible that we could not find some of the features to be significant because of a type II error. Although this method is advantageous in investigating a large number of microbiome compositions in a short time, the determination of the genera and functional gene abundances was performed without the isolation of bacteria. To complement the metagenomic data, a culture-based approach, such as culturomics, is required [12]. Despite these limitations, our study provided important implications in the shift of functional gene abundances occurring during the progression of periodontal disease.
In conclusion, the taxonomic abundances were higher in sites with periodontal pockets than those in healthy sites. In the functional gene categories, carbohydrate metabolism, glycan biosynthesis and metabolism, amino acid metabolism, replication and repair showed significantly different abundances between healthy sites and sites with periodontal pockets. Among them, a decrease in phenylalanine/tyrosine/tryptophan biosynthesis was suggested as a candidate marker of dysbiotic shift in periodontitis. These differences might be useful for clarifying the mechanisms of the microbiome shift occurring in periodontitis. Future metagenomic research in larger Japanese cohorts combined with transcriptome and metabolome analyses should provide us with more information about the changes in taxonomic and functional gene category abundances caused by periodontitis.
4. Materials and Methods
4.1. Sample Collection
Systemically healthy individuals (aged 20 to 80 years) were recruited from the Tokyo Dental College hospitals. Subjects who had a history of antimicrobial agent use for the past 3 months, smoking, or serious underlying disease were excluded. We followed the description in the literatures for the diagnosis of periodontitis [54,55]. Periodontally healthy subjects showed no probing depth > 3 mm, and no attachment loss or detectable inflammation. Subjects with periodontitis had at least six sites with a probing depth >5 mm and two sites exhibiting bleeding upon probing. Subgingival plaque samples were collected from volunteers with healthy periodontium and patients with periodontitis. From patients with periodontitis, samples were obtained from healthy and diseased sites. After removal of supragingival plaque, the site was isolated with cotton rolls, gently air-dried, and subgingival plaque were collected with scaler. The obtained samples were suspended in PowerBead tubes of Power Soil DNA Isolation kit (MoBio, Carlsbad, CA, USA) and stored at −20 °C until DNA isolation. All participants provided written informed consent. Ethical approval for this study was obtained from the Tokyo Dental College Ethics Committee (No. 622).
4.2. DNA Extraction, Library Preparation and Sequencing
DNA was extracted from each sample using a Power Soil DNA Isolation kit (MoBio) according to the manufacturer’s protocol. The extracted DNA samples were quantified using a Quantus Fluorometer (Promega, Madison, WI, USA). One nanogram of input DNA was enzymatically fragmented and tagged with sequencing adapters using the Nextera XT DNA Library Preparation Kit (Illumina, San Diego, CA, USA) and then normalized.
Sequencing was conducted using the Illumina MiSeq with the MiSeq Reagent Kit v2 (300 cycles).
4.3. Sequencing Data Analysis
The obtained reads were subjected to quality control using PRINSEQ v0.20.4 [56] to obtain a quality score of more than 20 in all parts of the reads. The sequencing tags were trimmed using cutadapt v1.14 [57]. The reads derived from the human genome were excluded by mapping them to the human genome (GRCh38) using bowtie2 v2.3 [58]. Following the literature [59], samples retaining more than 200,000 reads were subjected to further analysis.
For taxonomic analysis at the genus level, the 16S ribosomal RNA database distributed by NCBI was downloaded in February 2019 and used. We aligned the reads to the database using BLASTN [60] in default settings. The relative taxonomic abundance was calculated using MEGAN6 [61].
For functional gene category analysis, the KEGG gene database [62] distributed by KEGG was downloaded in December 2019 and used. We aligned reads to the database using GHOSTZ-GPU [63] on TSUBAME3.0 in default settings. The relative abundance of functional gene categories was calculated using HUMAnN [64].
The obtained relative abundance data were subjected to a two-sided Mann–Whitney U test with Bonferroni correction. We set the significance level at p < 0.05 and calculations were conducted using the stats module implemented by SciPy v1.5. For convenience, the p-values indicated in the text were p-values from a single test multiplied by the number of tests. The statistical powers were calculated using G*power software [65].
Supplementary Materials
Supplementary materials can be found at https://www.mdpi.com/article/10.3390/ijms22105298/s1: Figure S1. The genus-level taxonomic composition of all samples; Figure S2. The relative abundance of (a) Tannerella, (b) Porphyromonas, (c) Treponema and (d) Prevotella; Table S1. A summary of the sequence analysis of the samples; Table S2. Dominant genera at the healthy sites and sites with periodontal pocket; Table S3. Genera showed high abundance at sites with periodontal pockets compared with healthy sites; Table S4. Genera showed high abundance at healthy sites in subjects with periodontitis compared with that in healthy subjects
Author Contributions
Methodology, K.I. (Kazuki Izawa) and K.O.-S.; Investigation of DNA sequence, K.O.-S.; Resources, K.O.-S. and K.I. (Kazuyuki Ishihara); Sample collection and diagnosis of the participants, D.K. S.T. and A.S.; Data curation, K.I. (Kazuki Izawa) and K.O.-S.; Writing—original draft preparation, K.I. (Kazuki Izawa); Writing-review and editing, K.I. (Kazuki Izawa), K.O.-S. T.I. M.O. Y.A. A.S. and K.I. (Kazuyuki Ishihara); Visualization, K.I. (Kazuki Izawa); Supervision, Y.A. K.I. (Kazuyuki Ishihara) and A.S.; Project administration, K.O.-S. and K.I. (Kazuyuki Ishihara); Funding acquisition, K.I. (Kazuyuki Ishihara). All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by KAKENHI Grant number 18K09559 from the Japan Society for Promotion of Science and a grant from the Multidisciplinary Research Center for Jaw Disease (MRCJD), Tokyo Dental College, Tokyo, Japan (a MEXT Private University Research Branding Project).
Institutional Review Board Statement
The study was conducted according to the guidelines of the Declaration of Helsinki and approved by Tokyo Dental College Ethics Review Committee (No. 622, date of approval: 16 December 2015).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
The read data analyzed in this study were deposited in the DNA Data Bank of Japan under the accession numbers DRR285642-DRR285683.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Kinane D.F., Stathopoulou P.G., Papapanou P.N. Periodontal diseases. Nat. Rev. Dis. Primers. 2017;3:17038. doi: 10.1038/nrdp.2017.38. [DOI] [PubMed] [Google Scholar]
- 2.Eke P.I., Borgnakke W.S., Genco R.J. Recent epidemiologic trends in periodontitis in the USA. Periodontol. 2000. 2020;82:257–267. doi: 10.1111/prd.12323. [DOI] [PubMed] [Google Scholar]
- 3.Kassebaum N.J., Bernabe E., Dahiya M., Bhandari B., Murray C.J., Marcenes W. Global burden of severe periodontitis in 1990–2010: A systematic review and meta-regression. J. Dent. Res. 2014;93:1045–1053. doi: 10.1177/0022034514552491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Falcao A., Bullon P. A review of the influence of periodontal treatment in systemic diseases. Periodontol. 2000. 2019;79:117–128. doi: 10.1111/prd.12249. [DOI] [PubMed] [Google Scholar]
- 5.Dominy S.S., Lynch C., Ermini F., Benedyk M., Marczyk A., Konradi A., Nguyen M., Haditsch U., Raha D., Griffin C., et al. Porphyromonas gingivalis in Alzheimer’s disease brains: Evidence for disease causation and treatment with small-molecule inhibitors. Sci. Adv. 2019;5:1. doi: 10.1126/sciadv.aau3333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moore W.E., Holdeman L.V., Cato E.P., Smibert R.M., Burmeister J.A., Ranney R.R. Bacteriology of moderate (chronic) periodontitis in mature adult humans. Infect. Immun. 1983;42:510–515. doi: 10.1128/IAI.42.2.510-515.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Socransky S.S., Haffajee A.D., Cugini M.A., Smith C., Kent R.L., Jr. Microbial complexes in subgingival plaque. J. Clin. Periodontol. 1998;25:134–144. doi: 10.1111/j.1600-051X.1998.tb02419.x. [DOI] [PubMed] [Google Scholar]
- 8.Kuboniwa M., Amano A., Kimura K.R., Sekine S., Kato S., Yamamoto Y., Okahashi N., Iida T., Shizukuishi S. Quantitative detection of periodontal pathogens using real-time polymerase chain reaction with TaqMan probes. Oral Microbiol. Immunol. 2004;19:168–176. doi: 10.1111/j.0902-0055.2004.00135.x. [DOI] [PubMed] [Google Scholar]
- 9.Daep C.A., Novak E.A., Lamont R.J., Demuth D.R. Structural dissection and in vivo effectiveness of a peptide inhibitor of Porphyromonas gingivalis adherence to Streptococcus gordonii. Infect. Immun. 2011;79:67–74. doi: 10.1128/IAI.00361-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hajishengallis G., Liang S., Payne M.A., Hashim A., Jotwani R., Eskan M.A., McIntosh M.L., Alsam A., Kirkwood K.L., Lambris J.D., et al. Low-abundance biofilm species orchestrates inflammatory periodontal disease through the commensal microbiota and complement. Cell Host Microbe. 2011;10:497–506. doi: 10.1016/j.chom.2011.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hajishengallis G., Lamont R.J. Dancing with the stars: How choreographed bacterial interactions dictate nososymbiocity and give rise to keystone pathogens, accessory pathogens, and pathobionts. Trends Microbiol. 2016;24:477–489. doi: 10.1016/j.tim.2016.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lagier J.C., Dubourg G., Million M., Cadoret F., Bilen M., Fenollar F., Levasseur A., Rolain J.M., Fournier P.E., Raoult D. Culturing the human microbiota and culturomics. Nat. Rev. Microbiol. 2018;16:540–550. doi: 10.1038/s41579-018-0041-0. [DOI] [PubMed] [Google Scholar]
- 13.Abusleme L., Dupuy A.K., Dutzan N., Silva N., Burleson J.A., Strausbaugh L.D., Gamonal J., Diaz P.I. The subgingival microbiome in health and periodontitis and its relationship with community biomass and inflammation. ISME J. 2013;7:1016–1025. doi: 10.1038/ismej.2012.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Griffen A.L., Beall C.J., Campbell J.H., Firestone N.D., Kumar P.S., Yang Z.K., Podar M., Leys E.J. Distinct and complex bacterial profiles in human periodontitis and health revealed by 16S pyrosequencing. ISME J. 2012;6:1176–1185. doi: 10.1038/ismej.2011.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lamont R.J., Koo H., Hajishengallis G. The oral microbiota: Dynamic communities and host interactions. Nat. Rev. Microbiol. 2018;16:745–759. doi: 10.1038/s41579-018-0089-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dewhirst F.E., Chen T., Izard J., Paster B.J., Tanner A.C., Yu W.H., Lakshmanan A., Wade W.G. The human oral microbiome. J. Bacteriol. 2010;192:5002–5017. doi: 10.1128/JB.00542-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Endo A., Watanabe T., Ogata N., Nozawa T., Aikawa C., Arakawa S., Maruyama F., Izumi Y., Nakagawa I. Comparative genome analysis and identification of competitive and cooperative interactions in a polymicrobial disease. ISME J. 2015;9:629–642. doi: 10.1038/ismej.2014.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Consortium H.M.P. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nakagawa I., Amano A., Kuboniwa M., Nakamura T., Kawabata S., Hamada S. Functional differences among FimA variants of Porphyromonas gingivalis and their effects on adhesion to and invasion of human epithelial cells. Infect. Immun. 2002;70:277–285. doi: 10.1128/IAI.70.1.277-285.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brogan J.M., Lally E.T., Poulsen K., Kilian M., Demuth D.R. Regulation of Actinobacillus actinomycetemcomitans leukotoxin expression: Analysis of the promoter regions of leukotoxic and minimally leukotoxic strains. Infect. Immun. 1994;62:501–508. doi: 10.1128/IAI.62.2.501-508.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oliveira R.R., Fermiano D., Feres M., Figueiredo L.C., Teles F.R., Soares G.M., Faveri M. Levels of Candidate Periodontal Pathogens in Subgingival Biofilm. J. Dent. Res. 2016;95:711–718. doi: 10.1177/0022034516634619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kumar P.S., Griffen A.L., Moeschberger M.L., Leys E.J. Identification of candidate periodontal pathogens and beneficial species by quantitative 16S clonal analysis. J. Clin. Microbiol. 2005;43:3944–3955. doi: 10.1128/JCM.43.8.3944-3955.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Deng Z.L., Szafranski S.P., Jarek M., Bhuju S., Wagner-Dobler I. Dysbiosis in chronic periodontitis: Key microbial players and interactions with the human host. Sci. Rep. 2017;7:3703. doi: 10.1038/s41598-017-03804-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Perez-Chaparro P.J., McCulloch J.A., Mamizuka E.M., Moraes A., Faveri M., Figueiredo L.C., Duarte P.M., Feres M. Do different probing depths exhibit striking differences in microbial profiles? J. Clin. Periodontol. 2018;45:26–37. doi: 10.1111/jcpe.12811. [DOI] [PubMed] [Google Scholar]
- 25.Macovei L., McCafferty J., Chen T., Teles F., Hasturk H., Paster B.J., Campos-Neto A. The hidden ‘mycobacteriome’ of the human healthy oral cavity and upper respiratory tract. J. Oral Microbiol. 2015;7:26094. doi: 10.3402/jom.v7.26094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shah H.N., Collins M.D. Proposal for reclassification of Bacteroides asaccharolyticus, Bacteroides gingivalis, and Bacteroides endodontalis in a new Genus, Porphyromonas. Int. J. Syst. Bacteriol. 1988;38:128–131. doi: 10.1099/00207713-38-1-128. [DOI] [Google Scholar]
- 27.Hespell R.B., Canale-Parola E. Amino acid and glucose fermentation by Treponema denticola. Arch. Mikrobiol. 1971;78:234–251. doi: 10.1007/BF00424897. [DOI] [PubMed] [Google Scholar]
- 28.Tanner A.C., Listgarten M.A., Ebersole J.L., Strzempko M.N. Bacteroides forsythus sp. nov. a slow-growing, Fusiform Bacteroides sp. from the human oral cavity. Int. J. Syst. Bacteriol. 1986;36:213–221. doi: 10.1099/00207713-36-2-213. [DOI] [Google Scholar]
- 29.Kirst M.E., Li E.C., Alfant B., Chi Y.Y., Walker C., Magnusson I., Wang G.P. Dysbiosis and alterations in predicted functions of the subgingival microbiome in chronic periodontitis. Appl. Environ. Microbiol. 2015;81:783–793. doi: 10.1128/AEM.02712-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yamanaka T., Yamane K., Furukawa T., Matsumoto-Mashimo C., Sugimori C., Nambu T., Obata N., Walker C.B., Leung K.P., Fukushima H. Comparison of the virulence of exopolysaccharide-producing Prevotella intermedia to exopolysaccharide non-producing periodontopathic organisms. BMC Infect. Dis. 2011;11:228. doi: 10.1186/1471-2334-11-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Singh A., Wyant T., Anaya-Bergman C., Aduse-Opoku J., Brunner J., Laine M.L., Curtis M.A., Lewis J.P. The capsule of Porphyromonas gingivalis leads to a reduction in the host inflammatory response, evasion of phagocytosis, and increase in virulence. Infect. Immun. 2011;79:4533–4542. doi: 10.1128/IAI.05016-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miller D.P., Hutcherson J.A., Wang Y., Nowakowska Z.M., Potempa J., Yoder-Himes D.R., Scott D.A., Whiteley M., Lamont R.J. Genes Contributing to Porphyromonas gingivalis Fitness in Abscess and Epithelial Cell Colonization Environments. Front. Cell Infect. Microbiol. 2017;7:378. doi: 10.3389/fcimb.2017.00378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Settem R.P., Honma K., Nakajima T., Phansopa C., Roy S., Stafford G.P., Sharma A. A bacterial glycan core linked to surface (S)-layer proteins modulates host immunity through Th17 suppression. Mucosal Immunol. 2013;6:415–426. doi: 10.1038/mi.2012.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Griffiths G.S. Formation, collection and significance of gingival crevice fluid. Periodontol. 2000. 2003;31:32–42. doi: 10.1034/j.1600-0757.2003.03103.x. [DOI] [PubMed] [Google Scholar]
- 35.Guo Y., Nguyen K.A., Potempa J. Dichotomy of gingipains action as virulence factors: From cleaving substrates with the precision of a surgeon’s knife to a meat chopper-like brutal degradation of proteins. Periodontol. 2000. 2010;54:15–44. doi: 10.1111/j.1600-0757.2010.00377.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ishihara K., Miura T., Kuramitsu H.K., Okuda K. Characterization of the Treponema denticola prtP gene encoding a prolyl-phenylalanine-specific protease (dentilisin) Infect. Immun. 1996;64:5178–5186. doi: 10.1128/IAI.64.12.5178-5186.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Baima G., Iaderosa G., Citterio F., Grossi S., Romano F., Berta G.N., Buduneli N., Aimetti M. Salivary metabolomics for the diagnosis of periodontal diseases: A systematic review with methodological quality assessment. Metabolomics. 2021;17:1. doi: 10.1007/s11306-020-01754-3. [DOI] [PubMed] [Google Scholar]
- 38.Dabdoub S.M., Ganesan S.M., Kumar P.S. Comparative metagenomics reveals taxonomically idiosyncratic yet functionally congruent communities in periodontitis. Sci. Rep. 2016;6:38993. doi: 10.1038/srep38993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kato T., Yamazaki K., Nakajima M., Date Y., Kikuchi J., Hase K., Ohno H., Yamazaki K. Oral Administration of Porphyromonas gingivalis Alters the Gut Microbiome and Serum Metabolome. mSphere. 2018;3:5. doi: 10.1128/mSphere.00460-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sakanaka A., Kuboniwa M., Hashino E., Bamba T., Fukusaki E., Amano A. Distinct signatures of dental plaque metabolic byproducts dictated by periodontal inflammatory status. Sci. Rep. 2017;7:42818. doi: 10.1038/srep42818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Grenier D. Nutritional interactions between two suspected periodontopathogens, Treponema denticola and Porphyromonas gingivalis. Infect. Immun. 1992;60:5298–5301. doi: 10.1128/IAI.60.12.5298-5301.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sakanaka A., Takeuchi H., Kuboniwa M., Amano A. Dual lifestyle of Porphyromonas gingivalis in biofilm and gingival cells. Microb. Pathog. 2016;94:42–47. doi: 10.1016/j.micpath.2015.10.003. [DOI] [PubMed] [Google Scholar]
- 43.Saito Y., Fujii R., Nakagawa K.I., Kuramitsu H.K., Okuda K., Ishihara K. Stimulation of Fusobacterium nucleatum biofilm formation by Porphyromonas gingivalis. Oral Microbiol. Immunol. 2008;23:1–6. doi: 10.1111/j.1399-302X.2007.00380.x. [DOI] [PubMed] [Google Scholar]
- 44.Marquis R.E. Oxygen metabolism, oxidative stress and acid-base physiology of dental plaque biofilms. J. Ind. Microbiol. 1995;15:198–207. doi: 10.1007/BF01569826. [DOI] [PubMed] [Google Scholar]
- 45.Barnard J.P., Stinson M.W. Influence of environmental conditions on hydrogen peroxide formation by Streptococcus gordonii. Infect. Immun. 1999;67:6558–6564. doi: 10.1128/IAI.67.12.6558-6564.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sczepanik F.S.C., Grossi M.L., Casati M., Goldberg M., Glogauer M., Fine N., Tenenbaum H.C. Periodontitis is an inflammatory disease of oxidative stress: We should treat it that way. Periodontol. 2000. 2020;84:45–68. doi: 10.1111/prd.12342. [DOI] [PubMed] [Google Scholar]
- 47.Robles A.G., Reid K., Roy F., Fletcher H.M. Porphyromonas gingivalis mutY is involved in the repair of oxidative stress-induced DNA mispairing. Mol. Oral Microbiol. 2011;26:175–186. doi: 10.1111/j.2041-1014.2011.00605.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ballini A., Dipalma G., Isacco C.G., Boccellino M., Di Domenico M., Santacroce L., Nguyen K.C.D., Scacco S., Calvani M., Boddi A., et al. Oral Microbiota and Immune System Crosstalk: A Translational Research. Biology. 2020;9:131. doi: 10.3390/biology9060131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Inchingolo F., Martelli F.S., Gargiulo Isacco C., Borsani E., Cantore S., Corcioli F., Boddi A., Nguyen K.C.D., De Vito D., Aityan S.K., et al. Chronic Periodontitis and Immunity, Towards the Implementation of a Personalized Medicine: A Translational Research on Gene Single Nucleotide Polymorphisms (SNPs) Linked to Chronic Oral Dysbiosis in 96 Caucasian Patients. Biomedicines. 2020;8:115. doi: 10.3390/biomedicines8050115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gaudilliere D.K., Culos A., Djebali K., Tsai A.S., Ganio E.A., Choi W.M., Han X., Maghaireh A., Choisy B., Baca Q., et al. Systemic Immunologic Consequences of Chronic Periodontitis. J. Dent. Res. 2019;98:985–993. doi: 10.1177/0022034519857714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tsukasaki M., Komatsu N., Nagashima K., Nitta T., Pluemsakunthai W., Shukunami C., Iwakura Y., Nakashima T., Okamoto K., Takayanagi H. Host defense against oral microbiota by bone-damaging T cells. Nat. Commun. 2018;9:701. doi: 10.1038/s41467-018-03147-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shi B., Lux R., Klokkevold P., Chang M., Barnard E., Haake S., Li H. The subgingival microbiome associated with periodontitis in type 2 diabetes mellitus. ISME J. 2020;14:519–530. doi: 10.1038/s41396-019-0544-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Martellacci L., Quaranta G., Patini R., Isola G., Gallenzi P., Masucci L. A Literature Review of Metagenomics and Culturomics of the Peri-implant Microbiome: Current Evidence and Future Perspectives. Materials. 2019;12:3010. doi: 10.3390/ma12183010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.The Japanese Society of Periodontology . JSP Clinical Practice Guideline for the Periodontal Treatment. Ishiyaku Publishers; Tokyo, Japan: 2015. [(accessed on 11 May 2021)]. pp. 26–28. Available online: http://www.perio.jp/publication/upload_file/guideline_perio_plan2015_en.pdf. [Google Scholar]
- 55.Tonetti M.S., Greenwell H., Kornman K.S. Staging and grading of periodontitis: Framework and proposal of a new classification and case definition. J. Clin. Periodontol. 2018;45:S149–S161. doi: 10.1111/jcpe.12945. [DOI] [PubMed] [Google Scholar]
- 56.Schmieder R., Edwards R. Quality control and preprocessing of metagenomic datasets. Bioinformatics. 2011;27:863–864. doi: 10.1093/bioinformatics/btr026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kechin A., Boyarskikh U., Kel A., Filipenko M. cutPrimers: A New Tool for Accurate Cutting of Primers from Reads of Targeted Next Generation Sequencing. J. Comput. Biol. 2017;24:1138–1143. doi: 10.1089/cmb.2017.0096. [DOI] [PubMed] [Google Scholar]
- 58.Langmead B., Wilks C., Antonescu V., Charles R. Scaling read aligners to hundreds of threads on general-purpose processors. Bioinformatics. 2019;35:421–432. doi: 10.1093/bioinformatics/bty648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Duran-Pinedo A.E., Chen T., Teles R., Starr J.R., Wang X., Krishnan K., Frias-Lopez J. Community-wide transcriptome of the oral microbiome in subjects with and without periodontitis. ISME J. 2014;8:1659–1672. doi: 10.1038/ismej.2014.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 61.Huson D.H., Auch A.F., Qi J., Schuster S.C. MEGAN analysis of metagenomic data. Genome Res. 2007;17:377–386. doi: 10.1101/gr.5969107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kanehisa M., Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Suzuki S., Kakuta M., Ishida T., Akiyama Y. GPU-Acceleration of Sequence Homology Searches with Database Subsequence Clustering. PLoS ONE. 2016;11:e0157338. doi: 10.1371/journal.pone.0157338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Abubucker S., Segata N., Goll J., Schubert A.M., Izard J., Cantarel B.L., Rodriguez-Mueller B., Zucker J., Thiagarajan M., Henrissat B., et al. Metabolic reconstruction for metagenomic data and its application to the human microbiome. PLoS Comput. Biol. 2012;8:e1002358. doi: 10.1371/journal.pcbi.1002358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Faul F., Erdfelder E., Lang A.G., Buchner A. G*Power 3: A flexible statistical power analysis program for the social, behavioral, and biomedical sciences. Behav. Res. Methods. 2007;39:175–191. doi: 10.3758/BF03193146. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The read data analyzed in this study were deposited in the DNA Data Bank of Japan under the accession numbers DRR285642-DRR285683.