Abstract
Simple Summary
It is widely thought that tumors are composed of different subpopulations of cancer cells carrying genetic alterations with some of them being common among all cells while others are unique for each subpopulation. This variable genetic profile of tumor cells is a component of what is collectively described as intratumor heterogeneity (ITH). Surviving the immune system and therapies, and establishing metastases are forces of natural selection that act upon ITH and drive tumor evolution and, eventually, the clinical presentation of patients. The aim of this prospective study was to investigate ITH in early-stage operable non-small cell lung cancer. We directly grafted human tumors in immunosuppressed mice and compared the genetic profile of the tumors grown in mice with that of the original human tumors. We identified clinical factors that affected the ability of human tumors to grow as mouse xenografts.
Abstract
Recent advances in sequencing technologies have allowed the in-depth molecular study of tumors, even at the single cell level. Sequencing efforts have uncovered a previously unappreciated heterogeneity among tumor cells, which has been postulated to be the driving force of tumor evolution and to facilitate recurrence, metastasis, and drug resistance. In the current study, focused on early-stage operable non-small cell lung cancer, we used tumor growth in patient-derived xenograft (PDX) models in mice as a fast-forward tumor evolution process to investigate the molecular characteristics of tumor cells that grow in mice, as well as the parameters that affect the grafting efficiency. We found that squamous cell carcinomas grafted significantly more efficiently compared with adenocarcinomas. Advanced stage, patient age and primary tumor size were positively correlated with grafting. Additionally, we isolated and characterized circulating tumor cells (CTC) from patients’ peripheral blood and found that the presence of CTCs expressing epithelial-to-mesenchymal (EMT) markers correlated with the grafting potential. Interestingly, exome sequencing of the PDX tumor identified genetic alterations in DNA repair and genome integrity genes that were under-represented in the human primary counterpart. In conclusion, through the generation of a PDX biobank of NSCLC, we identified the clinical and molecular properties of tumors that affected growth in mice.
Keywords: non-small cell lung cancer, patient-derived xenografts, intratumor heterogeneity, genetic profiling
1. Introduction
Lung cancer is by far the deadliest type of cancer [1] with the majority of cases corresponding to non-small cell lung cancer (NSCLC). The 5-year survival rate of patients with metastatic disease is dismal (6%), while patients with loco-regional disease have a better prognosis depending on the Tumor Node Metastasis (TNM) stage (cancer.gov). While molecular profiling has allowed the identification of several mutations that can be targeted with novel drugs, it has also uncovered extensive intratumor heterogeneity (ITH) [2]. ITH is a common feature of many cancers and has been implicated in disease recurrence and therapeutic response [3].
Mouse models have historically facilitated the study of tumor biology and have been used for the preclinical testing of novel drugs. More recently, the development of patient-derived xenograft (PDX) models has allowed investigators to generate more faithful models of the human disease by engrafting primary tumor specimens directly into immunocompromised mice [4,5]. The reported grafting efficiency varies among studies and tumor types, with melanoma and head and neck carcinoma showing the highest grafting rates while renal cell carcinoma and breast tumors were found at the other end of the scale [6].
In addition to tumor type, other factors, such as the degree of mouse immunodeficiency (i.e., which immunocompromised strain is used) and size of the grafted specimen could also be important factors. Regarding NSCLC, PDXs have been extensively used in preclinical studies targeting mostly EGFR [4,7,8]. Interestingly, published studies have reported a higher grafting efficiency for lung squamous cell carcinomas (LUSC) in comparison to lung adenocarcinomas (LUAD) [9,10]. With the exception of concurrent distant metastases and advanced disease stage [11], most studies fail to identify clinicopathological characteristics, such as age, smoking, or genetic profile, to be associated with the in vivo take rate.
A common observation is the high histological resemblance between the primary tumor and early passage mouse xenografts [4,7,9,12,13]. More recent reports have focused on the comparison of the molecular characteristics of primary tumors and the respective mouse counterparts. While some clonal evolution is identified in the PDX tumors, as indicated by the increased frequency of primary tumor genetic alterations [12,13,14], we are far from understanding how certain subclones within the primary tumor are favored in mice.
The purpose of the current study was to assess how ITH within the primary tumor impacts the grafting efficiency in early-stage NSCLC and how the grafting efficiency correlates with the tumor stage, circulating tumor cell (CTC) count, disease recurrence, progression, and ultimately patient outcome. A consortium of surgeons, oncologists, and pathologists together with scientists in the fields of clinical chemistry, molecular diagnostics, and mouse genetics came together and generated a large panel of fully characterized NSCLC PDXs. Here, we present this panel of PDX models and discuss the molecular and clinical aspects of both the primary and PDX tumors.
2. Materials and Methods
2.1. Patient Recruitment
Eighty-two patients with a median age of 65.5 years (range, 39–86) with histologically documented NSCLC and operable disease were enrolled in the study. All patients gave their written informed consent to participate in the study, which was approved by the Ethics and Scientific Committee of the Metropolitan General hospital (308/28-12-2017). All patients were operated in the “Sotiria” General Hospital (Athens) by the same surgery team (K.P. and K.V.). There were 52 male and 30 female, and 44 (53.7%) of them had an adenocarcinoma (LUAD) histology. The pathological staging revealed that 39 patients had stage I (A and B), 25 had stage II (A and B) and 18 had stage IIIA tumors. Fifty-five patients (67.1%) had no evidence of disease dissemination in resected lymph nodes (N0 disease).
2.2. PDX Generation
All animal experiments were performed according to national and international regulations and were approved by the BRFAA (Biomedical Research Foundation of the Academy of Athens, Athens Greece) ethical committee. NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ (Stock No: 005557; NSG) [15,16] mice were purchased from the Jax repository (Bar Harbor, ME USA) and bred in-house. All xenotransplantations were performed within 6 h from the patient’s surgery due to the close proximity of the hospital and the mouse facility. Tumor specimens up to 5 mm (depending on tissue availability) were transplanted subcutaneously in NSG mice under anesthesia [17]. All fragments were mirror images of tumor segments, which were submitted for molecular analysis by two independent histologists.
Prior to engraftment, a small piece of the tissue was snap frozen and was later processed for molecular analysis. Mice were kept in pathogen-free conditions throughout their life. All tumors were passaged at least once when they reached a maximum diameter of 1 cm. The growth time from implantation to passage 1 varied from 1 to 4 months. Grafted tumors that failed to grow within 4 months were considered “no-take”. During passage 1, a portion of the PDX tumor was processed for molecular and histological analysis [18] by two pathologists. No distant metastases were ever observed. All early passages were also live-frozen in order to generate a biobank of early passage NSCLC PDXs.
2.3. Histopathological Examination
For the histopathological assessment, we performed hematoxylin (Millipore, Burlington, MA, USA) and eosin (DIAPATH, Martinengo, BG, Italy) (H/E) staining along with immunohistochemical analysis employing anti-TTF1 (SPT24, 1:100 dilution, Leica Biosystems Newcastle Ltd. Balliol Business Park, Benton Ln, Newcastle upon Tyne, United Kingdom) and anti-P40 (BC28, ready to use, Roche, Basel, Switzerland).
2.4. DNA/RNA Extraction
The cellularity of the primary tumor was determined in FFPE tissue using H/E stained sections. Sections (four to five sections at 10 μm each) with a minimum 20% tumor content were selected and used for DNA isolation using the Qiagen FFPE DNA isolation kit (QIAGEN, Hilden, Germany) as per the manufacturer’s instructions and quantified using the Qubit fluorometer using the Qubit BR DNA assay kit (Thermo Fisher Scientific, Waltham, MA, USA).
2.5. Library Preparation and Sequencing
Custom capture probes were designed using SureDesign (Agilent Technologies, Santa Clara, CA, USA) targeting all exons in 58 genes (Table S1). A total of 50–100 ng of DNA from FFPE tissue samples were used for library preparation and sequencing. The library preparation for each sample was done using the SureSelect XT HS Reagent Kit (Agilent Technologies, Santa Clara, CA, USA) according to the manufacturer’s instructions. In brief, pre-enriched adapter-ligated libraries were prepared, and custom capture probes were hybridized to target sequences to allow for sequence enrichment using streptavidin beads. The post-enriched libraries were quantified using the Qubit dsDNA Assay Kit (Thermo Fisher Scientific, Waltham, MA, USA), and the library quality was assessed using the Agilent 2100 bioanalyzer (Agilent Technologies, Santa Clara, CA, USA). The libraries were pooled to equimolar concentrations and sequenced on a NextSeq 500 (Illumina, San Diego, CA, USA).
The NGS panel was developed to cover ~289,255 kbp genomic regions of 58 genes, including the majority of known oncogenes and tumor suppressors in NSCLC and several genes involved in DNA damage response/DNA repair mutations, which could affect the therapeutic decisions. All sequencing was paired-end with read lengths of 150 bp. The average total number of reads across the specimen data sets was 29.8 million. The coverage across the capture regions was assessed using mosdepth [19]. The percentage of base positions at which on-target reads were achieved at depths of 100×, 500×, and 1000× was calculated. For an average specimen, 96.4% of the positions were covered at a depth of 100×, 88.9% at a depth of 500×, and 76.8% at a depth of 1000×.
Sequencing data analysis was performed using bwa 0.7.17-r1188 [20], samtools 1.9 [21], GATK 4.1.2 [22], and ANNOVAR 2018-04-16 [23] in a sequential manner. As cutoff for the analysis, 5% variant allele frequency (VAF) was used and at least 100x depth. Artifacts and common variants were filtered out based on frequency detection in our sample (10%). Differences in the VAF and unique variants in different libraries originating from samples from the patient were identified by direct comparison of the variants’ list.
2.6. Primary and PDX Tumor Comparison
For identification of primary and PDX tumor genetic alterations, the cut-off allele frequency was set at 0.5% for the primary tumor and 5% for the mouse counterpart. Therefore, it was possible to identify genetic alterations that were represented in a large portion of the PDX even if they started from a very low representation in the primary tumor. To exclude passenger mutations in the specific tumor specimen that were coincidentally enriched during the development of the PDX, other genetic alterations in the same primary tumor-PDX pair were used as normalizers. The human tumor cellularity was taken into account, and samples with cellularity < 50 were excluded from the comparison.
2.7. CTC Enrichment for Molecular Analysis
Peripheral blood PB (25 mL) was collected prior to surgery in EDTA tubes after discarding the first 5 mL of blood draw to avoid contamination of skin epithelial cells. CTC enrichment was performed using the size-based microfluidic device Parsortix (ANGLE plc, Guildford, Surrey, UK), which contains a microscope slide sized disposable cassette [24,25]. CTCs were finally harvested in a total volume of 200 μL of PBS, and the total RNA was extracted from the harvested cells using TRIZOL-LS (ThermoFisher Scientific, Waltham, MA, USA). Finally, cDNA synthesis was performed as previously described [26].
2.8. Gene Expression Analysis of CTCs by RT-qPCR
RT-qPCR was performed to evaluate the gene expression of epithelial (CK-8, CK-18, and CK-19), and mesenchymal/EMT (VIM and TWIST1) markers. B2M was used as a reference gene for relative quantification but also for ensuring the presence of amplifiable material in all samples and to avoid false-negative results [27], and RT-qPCR was performed as previously reported [28].
In all patient samples, RT-qPCR data for VIM were normalized in respect to the expression of B2M reference gene by using the 2–ΔΔCq approach. The cut-off value for VIM transcripts was calculated as the mean of signals derived in the healthy donor group plus 2SD. Using this approach, a sample was considered to overexpress VIM based on the fold change of VIM expression in the CTC fraction with respect to the corresponding “CTC” fraction in the group of these healthy individuals.
2.9. Statistical Analysis
Statistical analysis was performed using IBM SPSS Statistics 20 software (IBM Corp., Armonk, New York, USA). The association of PDX groups (PDX+ and PDX–) with categorical variables was evaluated by Fisher’s Exact and Pearson chi-square tests, while the correlation with continuous data was assessed with the non-parametric Mann–Whitney U test for variables with two categories and Kruskal–Wallis tests for variables with more than two categories. Patient death and treatment failure were assessed as clinical end-point events for the survival analysis by Kaplan–Meier curves using the log-rank test.
3. Results
3.1. Establishment of NSCLC PDXs
Following surgery, sufficient material for grafting in at least one NSG mouse was available from 52 patients. The complete clinical and pathological patient characteristics are provided in Table 1.
Table 1.
Clinical and pathological characteristics of the patients.
| Patient Characteristics | PDX YES (n = 31) | PDX NO (n = 21) | p-Value |
|---|---|---|---|
| Categorical Variables | n (%) | n (%) | |
| Gender | 0.141 a | ||
| Male | 23 (74.2%) | 11 (52.4%) | |
| Female | 8 (25.8%) | 10 (47.6%) | |
| Age | 0.090 a | ||
| <68y b | 10 (34.5%) | 12 (60.0%) | |
| ≥68y | 19 (65.5%) | 8 (40.0%) | |
| Smoking status | 0.497 c | ||
| Current | 19 (61.3%) | 10 (47.6%) | |
| Previous | 6 (19.4%) | 7 (33.3%) | |
| Never | 6 (19.4%) | 4 (19.0%) | |
| Histological type | 0.100 c | ||
| Adenocarcinoma | 10 (32.3%) | 13 (61.9%) | |
| Squamous | 17 (54.8%) | 7 (33.3%) | |
| Large cell or Sarcomatous | 4 (12.9%) | 1 (4.8%) | |
| Clinical stage | 0.040 c | ||
| I | 13 (41.9%) | 10 (47.6%) | |
| II | 6 (19.4%) | 9 (42.9%) | |
| IIIA | 12 (38.7%) | 2 (9.5%) | |
| Tumor stage | 0.038 c | ||
| pT1 | 5 (16.1%) | 9 (42.9%) | |
| pT2 | 15 (48.4%) | 7 (33.3%) | |
| pT3 | 5 (16.1%) | 5 (23.8%) | |
| pT4 | 6 (19.4%) | 0 (0%) | |
| Continuous variables | Median (min–max) | Median (min–max) | |
| Tumor volume (cm3) | 0.070 d | ||
| 33.7 (0.48–572.0) | 14.7 (2.14–245.0) | ||
| Age (years) | 0.030 d | ||
| 71 (56–81) | 64 (40–75) |
a Fisher’s Exact test; b Median age of patients with tumor used in xenotransplantations; c Pearson chi-square; and d Mann–Whitney U test.
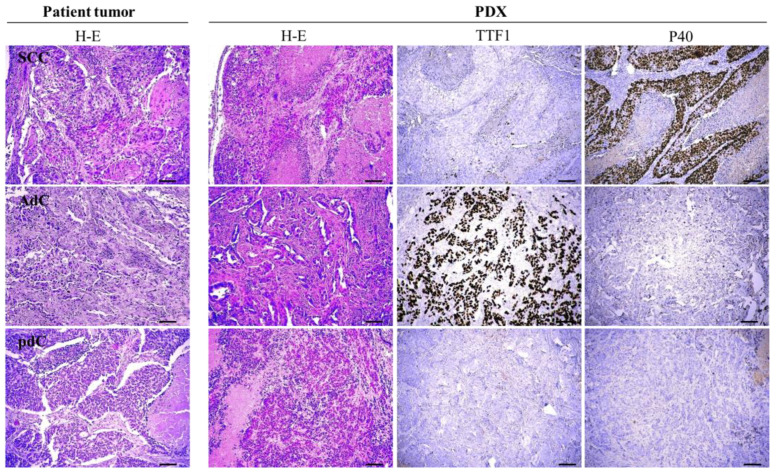
Thirty-one (59.6%) of the 52 specimens yielded successful xenografts, which were passaged at least once before freezing. During the first passage, part of the PDX tumor was fixed in formalin and processed with H/E staining for histopathological characterization in comparison with the corresponding patients’ primary tumors. Microscopical examination revealed that PDXs preserved the histopathological characteristics of the original tumors (Figure 1).
Figure 1.
Histological evaluation of primary tumors and PDX. Images from eosin and hematoxylin staining (H/E) from a representative example of squamous cell carcinoma (SCC), adenocarcinoma (AdC), and a poorly differentiated carcinoma (pdC) showing that the mouse tumor preserved the morphological features of the primary tumor. The squamous cell marker P40 and the adenocarcinoma marker TTF1 were also used on the PDX tumor. Scale bar: 100 μm.
The phenotype of squamous cell carcinoma and adenocarcinoma in the primary tumors was faithfully retained in PDX, verified by the staining pattern of the lung adenocarcinoma (LUAD) marker TTF1 and the lung squamous cell carcinoma (LUSC) marker p40 (Figure 1). Figure 1 also depicts a case of a poorly differentiated primary lesion that fully retained the same differentiation pattern in the mouse.
3.2. Clinical Characteristics and Grafting Efficiency
We performed statistical analysis to identify clinical parameters that could impact the grafting efficiency. All p values are provided in Table 1. Although it did not reach statistical significance, the grafting efficiency was higher among men and current smokers who, as expected, represented the majority of cases. In agreement with reported data [10], adenocarcinomas grafted poorly (p = 0.048) in comparison with the other tumor types (essentially LUSC). According to our experience regarding the generation of PDXs from head and neck squamous cell carcinoma, squamous cell carcinomas, in general, graft with a high efficiency (unpublished data).
The grafting efficiency positively correlated with the tumor/clinical stage, with 6/6 T4 and 12/24 stage IIIA tumors leading to successful grafting in mice, and with advanced age (p = 0.03; Table 1), possibly reflecting late diagnosis and a higher tumor burden. Indeed, the median size of primary tumors with successful grafting in mice was 33.7 cm3 in comparison to the 14.7 cm3 of those that failed to grow in mice (p = 0.07; Table 1). Interestingly, we did not observe any correlation with the disease-free survival or overall survival (Figure S1A,B).
3.3. Correlation between the Presence of CTCs and Grafting
Patient CTCs were detected and characterized based on the expression of epithelial (KRT8, KRT18, and KRT19) and mesenchymal (Vim and Twist1) markers. The gene expression analysis of the above markers demonstrated significant heterogeneity among NSCLC patients. CTCs were detected in 47 out of 52 (90.4%) patients. At least one epithelial marker was detected in 19 out of 52 (36.5%) patients; KRT8 transcripts were detected in 8 out of 47 (17.0%), KRT18 transcripts were detected in 7 out of 47 (14.9%), and KRT19 transcripts were detected in 12 out of 47 (25.5%) samples before surgery (Table 2).
Table 2.
CTCs expressing epithelial markers and the grafting efficiency.
| PDX | Total | p Value a | |||
|---|---|---|---|---|---|
| No (−) | Yes (+) | ||||
| KRT18 | Neg | 21 | 19 | 37 | p = 0.009 |
| Pos | 0 | 7 | 7 | ||
| Total | 21 | 26 | 47 | ||
| KRT19 | Neg | 16 | 19 | 34 | p = 1.000 |
| Pos | 5 | 7 | 12 | ||
| Total | 21 | 26 | 47 | ||
| KRT8 | Neg | 17 | 22 | 39 | p = 1.000 |
| Pos | 4 | 4 | 8 | ||
| Total | 21 | 26 | 47 | ||
| Epithelial markers | Neg | 14 | 14 | 28 | p = 0.377 |
| Pos | 7 | 12 | 19 | ||
| Total | 21 | 26 | 47 | ||
a Fisher’s exact test.
Similarly, at least one mesenchymal/EMT marker, was detected in 34 out of 47 (72.3%) patients; VIM transcripts were detected in 27 out of 47 (57.4%) patients, whereas TWIST1 transcripts were detected in 12 (25.5%) patients (Table 3).
Table 3.
CTCs expressing EMT markers and the grafting efficiency.
| PDX | Total | p Value a | |||
|---|---|---|---|---|---|
| No (−) | Yes (+) | ||||
| VIM | Neg | 11 | 9 | 20 | p = 0.231 |
| Pos | 10 | 17 | 27 | ||
| Total | 21 | 26 | 47 | ||
| TWIST1 | Neg | 18 | 17 | 35 | p = 0.179 |
| Pos | 3 | 9 | 12 | ||
| Total | 21 | 26 | 47 | ||
| EMT markers (VIM & TWIST1) | Neg | 8 | 5 | 13 | p = 0.107 |
| Pos | 13 | 21 | 34 | ||
| Total | 21 | 26 | 47 | ||
a Fisher’s exact test.
The grafting efficiency showed a not statistically significant positive trend with CTCs expressing EMT markers TWIST (p = 0.179) and VIM (p = 0.231). The expression of either showed an even stronger trend (p = 0.107) (Table 3). Interestingly, only VIM was detected in healthy donors (HD), the peripheral blood of which was subjected to the same process. Thus, we were able to compare VIM expression in the HD group against surgical samples that did or did not yield PDXs. As Figure S2 indicates, VIM-overexpressing CTCs were found in patients that also grafted successfully.
There was no correlation between the grafting efficiency and the expression of epithelial markers. However, it should be noted that the KRT18 expression, which mainly characterizes LUAD, absolutely correlated with the grafting (7/7 cases, p = 0.009) (Table 3), corresponding to two LUAD, four LUSC, and one large cell carcinoma case. KRT19, on the other hand, which is expressed by both LUAD and LUSC, did not show any correlation with the grafting efficiency [29], despite the fact that it has been shown to be an excellent marker for the detection of lymph node metastases of NSCLC [30].
3.4. Molecular Comparison of PDX and Primary Tumors
Given the well documented ITH, we aimed to investigate whether certain genetic alterations in primary tumor subclones might confer an advantage for efficient grafting in mice. To this direction, we used our custom panel for targeted exome sequencing and generated the genetic alteration profile of the primary tumor and the mouse counterpart. A summary of pathogenic genetic alterations or mutations with unknown clinical significance identified in primary tumors which successfully grafted in mice is provided in Table S2. Assuming that certain subclones are favored during tumor growth in mice, this would lead to an enrichment of certain genetic alterations present only within a subset of tumor cells.
This enrichment would be reflected in the VAF, which would be expected to be higher in the PDX tumor in comparison to the primary one. To identify such cases, the genetic profile of the mouse tumors from passage 1 of the PDXs was compared with the genetic profile of the human counterpart. Interestingly, the most striking examples of mutations that were underrepresented in the primary tumor but were subsequently enriched in the mouse counterpart were found in DNA repair and genome integrity proteins, with the most prominent being TP53 (Table 4).
Table 4.
Molecular differences between primary tumors and PDX.
| CODE | TYPE | GENETIC ALTERATIONS | TUMOR CELLULARITY (%) | VAFPRIM.TUMOR | VAFPDX |
|---|---|---|---|---|---|
| 105 | LUSC |
TP53:c.469G>T:p.V157F KMT2C:c.918T>G:p.Y306* |
90 |
0.057 0.059 |
1 0.062 |
| 110 | LUSC |
TP53:c.413C>T:p.A138V ALK:c.2712T>A:p.H904Q |
85 |
0.432 0.36 |
0.952 0.191 |
| 112 | PLEIO/LUAD | CTNNB1:c.98C>T:p.S33F TP53:c.811G>T:p.E271* STK11:c.487G>T:p.G163C KDR:c.2761delinsAT:p.F921Ifs*13 KDR:c.2757C>G:p.C919W |
80 | 0.49 0.436 0.356 0.316 0.317 |
0.868 0.93 0.895 0.83 0.831 |
| 302 | LUSC | KIT:2508G>T:p.M836I EGFR:c.1150A>T:p.T384S, MEN1:c.184A>G:p.T62A, MET:c.1241A>G:p.D414G |
70 | 0.28 0.392 0.325 0.64 |
0.973 0.993 0.644 0.995 |
| 512 | LUSC |
TP53:c.845G>C:p.R282P CDKN2A:c.316_317T:p.V106Cfs*39 PDGFRA:c.1102G>C:p.E368Q |
75 |
0.336 0.375 0.4 |
0.999 0.998 0.995 |
| 517 | LUSC | RB1:c.852delinsTA:p.I285Nfs*2 | 75 | 0.249 | 0.998 |
| 518 | LUSC | TP53:c.659A>G:p.Y220C | 80 | 0.304 | 0.996 |
| 519 | LCNEC | RB1:c.1953_1954T:p.V654Cfs*3 | 80 | 0.732 | 0.998 |
| 531 | LUSC |
TP53:c.747G>T:p.R249S
CDKN2A:c.262G>T:p.E88* PIK3CA:c.1633G>A:p.E545K |
80 |
0.574
0.416 0.251 |
0.998
0.991 0.337 |
| 546 | LUAD |
KRAS:c.35G>A:p.G12D ATM:c.8851G>A:p.V2951I |
85 |
0.342 0.371 |
0.296
0.997 |
| 551 | LUSC | MTOR c.1333A>G:p.R445G CDKN2A:c.61G>C:p.A21P |
65 | 0.686 0.295 |
0.994 0.966 |
| 568 | LUAD |
TP53:c.880G>T:p.E294*
KRAS:c.34G>T:p.G12C |
65 |
0.09
0.16 |
0.63
0.62 |
| 569 | LUSC |
NF1:c.4662-1G>T TP53:c.994-2A>T; SMAD4:c.1326G>T:p.Q442H |
80 |
0.301
0.274 0.145 |
0.984
0.991 0.953 |
| 571 | LUSC |
CHEK2: c.1114C>T:p.Q372* TP53:c.994-2A>T BRCA2:c.3668A>G:p.H1223R NOTCH1:c.2058_2059T:p.C687Afs*84 |
75 | 0.294 0.287 0.59 0.279 |
0.362 0.952 0.959 0.951 |
Pathogenic events are shown in bold.
4. Discussion
A personalized approach in cancer therapeutics has long been a noble goal for both clinicians and patients. More sophisticated therapies, often targeting specific mutations are very helpful in this direction. On the other hand, clinical practice has invariably proven that tumor cells develop resistance to targeted therapies [31]. To this point, it is not clear to what extent this therapeutic resistance is intrinsic due to a minority of cells within the tumor that outcompete their less resistant counterparts or whether resistance is an acquired property [31].
Even in the cases of acquired resistance, it is safe to assume that the genetic plasticity of cancer cells and novel acquired genetic alterations help them bypass the toxicity induced by therapeutic agents. On the other hand, tumors with high genetic heterogeneity are much more likely to develop resistance because the combination of genetic events leading to this resistance is easier to achieve. Recent studies have indicated that, even for a single targeted therapy (e.g., EGFR inhibition in NSCLC) a multitude of resistance mechanisms have been described [32,33,34].
Patient-derived xenografts have proven their usefulness in studying various aspects of tumor biology. Not only do they recapitulate the human disease, but they also are almost unlimited sources of biological material. They are particularly useful in preclinical studies for testing new therapeutic regimens [35,36]. In this regard, it is worth mentioning that we have obtained PDXs from tumors carrying EGFR mutations in exons 19 and 20 (Table S2). Patients with EGFR mutations in exons 17–21 have a heterogeneous response to EGFR tyrosine kinase inhibitors as indicated by clinical and experimental data in patients, mouse models, and cell lines [37]. Models, such as the ones described here, could become useful tools in understanding the underlying mechanism for this variable response and in identifying additional biomarkers for clinical use. Moreover, we obtained 4 LUAD PDXs with KRAS mutations (Table S2). In agreement with previous reports [38], KRASG12C is the most common KRAS variant and is currently targetable with clinically-approved covalently-bound inhibitors [39,40]. While clinical trials with various KRASG12C inhibitors are in progress, data from patients and mouse models point to heterogeneous responses and therapeutic resistance as emerging issues [41]. Again, preclinical models, such as those described here, combined with clinical data, will be invaluable tools in understanding the mechanistic aspects of these issues but also to better stratify patients and possibly design improved inhibitors and therapeutic schemes.
Similarly to the metastatic potential and the therapeutic response in humans, the factors affecting the grafting efficiency are largely unknown. In the current study, we asked the question of whether PDX can be useful in studying ITH. Considering that growth in mice imitates metastasis, PDXs provide a platform for fast forwarding tumor evolution, which favors certain tumor cells within a heterogeneous primary tumor. Therefore, a panel of PDXs was developed from patients with early-stage operable NSCLC who had been fully characterized both molecularly and histologically.
The presented data seem to indicate that squamous cell carcinomas grafted more efficiently than adenocarcinomas, a property most likely inherent in squamous cell carcinomas in general. Additionally, the grafting efficiency positively correlated with the tumor and disease stage as well as the primary tumor size. Rather surprisingly, we observed no correlation with disease progression and patient survival; however, this could be attributed mainly to the small numbers of events of the studied cohort due to the patients’ clinical characteristics (mainly with stage I and II presenting a relatively low incidence of disease relapse, as well as the short duration of the follow-up period).
The isolation and characterization of CTCs indicated that CTCs expressing EMT markers were associated with PDX growth, in concordance with the long-known ability of tumor cells that have undergone EMT to escape the primary tumor site and form metastases [42]. Our analysis also revealed that PDX development in mice correlates with the expression of KRT18 in CTCs. Earlier studies suggested a link between CK18 protein expression and unfavorable clinicopathological features and outcomes in squamous cell carcinomas [43]. To our knowledge, this is the first time that the CTC count and specific markers expressed by CTCs have been linked with the grafting efficiency in NSCLC. The biological underpinnings of these findings are of great interest and will fuel future studies.
As expected, primary tumor driver mutations were invariably found in the PDX counterparts. On the other hand, several genetic alterations, which were found at low frequencies in the human specimen, were significantly enriched in mice following successful grafting. The majority of those were in the TP53 locus; however, genetic alterations were also found in the CDKN2A, RB1, and BRCA2, as well as the STK11 locus (also known as LKB1), which is a well-known tumor suppressor in lung cancer [44].
We cannot exclude the possibility that, in certain cases, originally heterozygous mutations reach a variant allele frequency close to 1 due to loss-of-heterozygosity (LOH). LOH has been well documented in NSCLC for the TP53 [45] and the RB1 [46] loci. Moreover, it has been shown that missense single amino acid variations in the DNA binding domain often leads to gain-of-function TP53 proteins, which favor disease progression and metastasis [47,48]. These findings imply that, indeed, there is some kind of cancer cell selection for growth in mice, and that this selection process could favor cells with defects in genome integrity and cell cycle checkpoints.
This could be an indication that high-speed tumor evolution fueled by defects in the DNA repair and genome surveillance machineries might be the driving force in mouse grafting. Interestingly, previous studies focusing on tumor evolution during PDX growth did not identified genetic alterations in the aforementioned genes being favored in mice [12,13,14]. On the other hand, the panel of genes assessed in this study was enriched in genes encoding proteins involved in DNA damage response and DNA repair as well as cell cycle regulation.
This is one primary limitation in this study, i.e., assessing the mutation status of a rather short list of genes. Thus, more detailed molecular characterization in the form of whole exome sequencing of the described primary and PDX tumors would likely provide a more complete picture. In fact, whole exome coverage and the assessment of structural variants in the form of fusions and copy number variations (events that our approach cannot detect) would likely identify other gain- and loss-of-function genetic events favoring tumor growth in mice. Moreover, the single cell genomics of tumor cells and/or CTCs would also be valuable in such studies.
5. Conclusions
Herein, we presented the generation and characterization of a PDX panel from early-stage operable NSCLC. We found correlations between the tumor growth in mice and a number of clinicopathological characteristics, including the tumor grade, stage, and size. By analyzing CTCs, we uncovered a positive correlation between CTCs expressing EMT markers and the epithelial marker KRT18 and the ability of the respective primary tumor to grow in mice. More importantly, we identified a positive selection in mice of cancer cells/subclones carrying genetic alterations, mostly in TP53, but also in other genome surveillance and cell cycle control proteins.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/cancers13102446/s1, Figure S1. Survival curves of patients operated with early-stage non-small cell lung cancer. Figure S2. VIM expression in healthy and NSCLC samples. Table S1: Panel of genes assessed with targeted exome sequencing; Table S2: Molecular profile of successfully grafted primary tumors.
Author Contributions
Z.K. generated all the PDX models; A.V., P.M. and E.I.A. performed the sequencing and analyzed sequence data. M.A. and E.L. performed the CTC isolation and characterization; I.S.P., E.P. and I.V. performed the pathology evaluation; K.P. and K.V. performed all lung surgeries; A.M. performed statistical analysis; V.G. and A.K. (Athanasios Kotsakis) designed the study and analyzed data; A.K. (Apostolos Klinakis) designed the study, analyzed data and wrote the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This project has received funding from: the Hellenic Foundation for Research and Innovation (HFRI) and the General Secretariat for Research and Technology (GSRT), under Grant Agreement No. 1964 and the EC Horizon 2020 (grant No. 801347) to A.K. (Apostolos Klinakis).
Institutional Review Board Statement
The study was conducted according to the guidelines of the Declaration of Helsinki and approved by the Ethics and Scientific Committee of the Metropolitan General hospital (308/28-12-2017).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study. Written informed consent has been obtained from the patients to publish this paper.
Data Availability Statement
The data presented in this study are available in Table 4 and Table S2.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Postmus P.E., Kerr K.M., Oudkerk M., Senan S., Waller D.A., Vansteenkiste J., Escriu C., Peters S. Early and locally advanced non-small-cell lung cancer (NSCLC): ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann. Oncol. 2017;28:iv1–iv21. doi: 10.1093/annonc/mdx222. [DOI] [PubMed] [Google Scholar]
- 2.De Sousa V.M.L., Carvalho L. Heterogeneity in Lung Cancer. Pathobiology. 2018;85:96–107. doi: 10.1159/000487440. [DOI] [PubMed] [Google Scholar]
- 3.Marino F.Z., Bianco R., Accardo M., Ronchi A., Cozzolino I., Morgillo F., Rossi G., Franco R. Molecular heterogeneity in lung cancer: From mechanisms of origin to clinical implications. Int. J. Med. Sci. 2019;16:981–989. doi: 10.7150/ijms.34739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fichtner I., Rolff J., Soong R., Hoffmann J., Hammer S., Sommer A., Becker M., Merk J. Establishment of Patient-Derived Non-Small Cell Lung Cancer Xenografts as Models for the Identification of Predictive Biomarkers. Clin. Cancer Res. 2008;14:6456–6468. doi: 10.1158/1078-0432.CCR-08-0138. [DOI] [PubMed] [Google Scholar]
- 5.Zhang X.-C., Zhang J., Li M., Huang X.-S., Yang X.-N., Zhong W.-Z., Xie L., Zhang L., Zhou M., Gavine P., et al. Establishment of patient-derived non-small cell lung cancer xenograft models with genetic aberrations within EGFR, KRAS and FGFR1: Useful tools for preclinical studies of targeted therapies. J. Transl. Med. 2013;11:168. doi: 10.1186/1479-5876-11-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goto T. Patient-Derived Tumor Xenograft Models: Toward the Establishment of Precision Cancer Medicine. J. Pers. Med. 2020;10:64. doi: 10.3390/jpm10030064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kita K., Fukuda K., Takahashi H., Tanimoto A., Nishiyama A., Arai S., Takeuchi S., Yamashita K., Ohtsubo K., Otani S., et al. Patient-derived xenograft models of non-small cell lung cancer for evaluating targeted drug sensitivity and resistance. Cancer Sci. 2019;110:3215–3224. doi: 10.1111/cas.14171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dong X., Guan J., English J.C., Flint J., Yee J., Evans K., Murray N., Macaulay C., Ng R.T., Gout P.W., et al. Patient-Derived First Generation Xenografts of Non–Small Cell Lung Cancers: Promising Tools for Predicting Drug Responses for Personalized Chemotherapy. Clin. Cancer Res. 2010;16:1442–1451. doi: 10.1158/1078-0432.CCR-09-2878. [DOI] [PubMed] [Google Scholar]
- 9.Russo M.V., Efaversani A., Egatti S., Ericca D., Gobbo A.E., Eferrero S., Epalleschi A., Evaira V., Ebosari S. A New Mouse Avatar Model of Non-Small Cell Lung Cancer. Front. Oncol. 2015;5:52. doi: 10.3389/fonc.2015.00052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ilie M., Nunes M., Blot L., Hofman V., Long-Mira E., Butori C., Selva E., Merino-Trigo A., Vénissac N., Mouroux J., et al. Setting up a wide panel of patient-derived tumor xenografts of non-small cell lung cancer by improving the preanalytical steps. Cancer Med. 2014;4:201–211. doi: 10.1002/cam4.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee H.W., Lee J.-I., Lee S.J., Cho H.J., Song H.J., Jeong D.E., Seo Y.J., Shin S., Joung J.-G., Kwon Y.-J., et al. Patient-Derived Xenografts from Non–Small Cell Lung Cancer Brain Metastases Are Valuable Translational Platforms for the Development of Personalized Targeted Therapy. Clin. Cancer Res. 2015;21:1172–1182. doi: 10.1158/1078-0432.CCR-14-1589. [DOI] [PubMed] [Google Scholar]
- 12.Morgan K.M., Riedlinger G.M., Rosenfeld J., Ganesan S., Pine S.R. Patient-Derived Xenograft Models of Non-Small Cell Lung Cancer and Their Potential Utility in Personalized Medicine. Front. Oncol. 2017;7:2. doi: 10.3389/fonc.2017.00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jung H.-Y., Kim T.H., Lee J.-E., Kim H.K., Cho J.H., Choi Y.S., Shin S., Lee S.-H., Rhee H., Lee H.K., et al. PDX models of human lung squamous cell carcinoma: Consideration of factors in preclinical and co-clinical applications. J. Transl. Med. 2020;18:1–13. doi: 10.1186/s12967-020-02473-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hao C., Wang L., Peng S., Cao M., Li H., Hu J., Huang X., Liu W., Zhang H., Wu S., et al. Gene mutations in primary tumors and corresponding patient-derived xenografts derived from non-small cell lung cancer. Cancer Lett. 2015;357:179–185. doi: 10.1016/j.canlet.2014.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Coughlan A.M., Harmon C., Whelan S., O’Brien E.C., O’Reilly V.P., Crotty P., Kelly P., Ryan M., Hickey F.B., O’Farrelly C., et al. Myeloid Engraftment in Humanized Mice: Impact of Granulocyte-Colony Stimulating Factor Treatment and Transgenic Mouse Strain. Stem Cells Dev. 2016;25:530–541. doi: 10.1089/scd.2015.0289. [DOI] [PubMed] [Google Scholar]
- 16.Shultz L.D., Lyons B.L., Burzenski L.M., Gott B., Chen X., Chaleff S., Kotb M., Gillies S.D., King M., Mangada J., et al. Human Lymphoid and Myeloid Cell Development in NOD/LtSz-scid IL2R γ Null Mice Engrafted with Mobilized Human Hemopoietic Stem Cells. J. Immunol. 2005;174:6477–6489. doi: 10.4049/jimmunol.174.10.6477. [DOI] [PubMed] [Google Scholar]
- 17.Rampias T., Karagiannis D., Avgeris M., Polyzos A., Kokkalis A., Kanaki Z., Kousidou E., Tzetis M., Kanavakis E., Stravodimos K., et al. The lysine-specific methyltransferase KMT 2C/ MLL 3 regulates DNA repair components in cancer. EMBO Rep. 2019;20:e46821. doi: 10.15252/embr.201846821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Paraskevopoulou V., Bonis V., Dionellis V.S., Paschalidis N., Melissa P., Chavdoula E., Vasilaki E., Pateras I.S., Klinakis A. Notch controls urothelial integrity in the mouse bladder. JCI Insight. 2020;5:e133232. doi: 10.1172/jci.insight.133232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pedersen B.S., Quinlan A.R. Mosdepth: Quick coverage calculation for genomes and exomes. Bioinformatics. 2018;34:867–868. doi: 10.1093/bioinformatics/btx699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li H. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics. 2011;27:2987–2993. doi: 10.1093/bioinformatics/btr509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Van der Auwera G.A., O’Connor B.D. Genomics in the Cloud: Using Docker, GATK, and WDL in Terra. 1st ed. O’Reilly Media, Inc.; Sebastopol, CA, USA: 2020. [Google Scholar]
- 23.Wang K., Li M., Hakonarson H. ANNOVAR: Functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Porras T.B., Kaur P., Ring A., Schechter N., Lang J.E. Challenges in using liquid biopsies for gene expression profiling. Oncotarget. 2018;9:7036–7053. doi: 10.18632/oncotarget.24140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chudziak J., Burt D.J., Mohan S., Rothwell D.G., Mesquita B., Antonello J., Dalby S., Ayub M., Priest L., Carter L., et al. Clinical evaluation of a novel microfluidic device for epitope-independent enrichment of circulating tumour cells in patients with small cell lung cancer. Analyst. 2015;141:669–678. doi: 10.1039/C5AN02156A. [DOI] [PubMed] [Google Scholar]
- 26.Zavridou M., Mastoraki S., Strati A., Koutsodontis G., Klinakis A., Psyrri A., Lianidou E. Direct comparison of size-dependent versus EpCAM-dependent CTC enrichment at the gene expression and DNA methylation level in head and neck squamous cell carcinoma. Sci. Rep. 2020;10:1–9. doi: 10.1038/s41598-020-63055-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Markou A., Tzanikou E., Strati A., Zavridou M., Mastoraki S., Bournakis E., Lianidou E. PIM-1 Is Overexpressed at a High Frequency in Circulating Tumor Cells from Metastatic Castration-Resistant Prostate Cancer Patients. Cancers. 2020;12:1188. doi: 10.3390/cancers12051188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ntzifa A., Strati A., Kallergi G., Kotsakis A., Georgoulias V., Lianidou E. Gene expression in circulating tumor cells reveals a dynamic role of EMT and PD-L1 during osimertinib treatment in NSCLC patients. Sci. Rep. 2021;11:1–12. doi: 10.1038/s41598-021-82068-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nhung N.V., Mirejovský P., Mirejovský T., Melínová L. Cytokeratins and lung carcinomas. Ceskoslovenska Patol. 1999;35:80–84. [PubMed] [Google Scholar]
- 30.Bugalho A., Martins C., Dias S.S., Nunes G., Silva Z., Correia M., Gomes M.J.M., Videira P.A. Cytokeratin 19, Carcinoembryonic Antigen, and Epithelial Cell Adhesion Molecule Detect Lung Cancer Lymph Node Metastasis in Endobronchial Ultrasound-Guided Transbronchial Aspiration Samples. Clin. Lung Cancer. 2013;14:704–712. doi: 10.1016/j.cllc.2013.06.004. [DOI] [PubMed] [Google Scholar]
- 31.Rosell R., Karachaliou N., Morales-Espinosa D., Costa C., Molina M.A., Sansano I., Gasco A., Viteri S., Massuti B., Wei J., et al. Adaptive resistance to targeted therapies in cancer. Transl. Lung Cancer Res. 2013;2:152–159. doi: 10.3978/j.issn.2218-6751.2012.12.08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nagano T., Tachihara M., Nishimura Y. Mechanism of Resistance to Epidermal Growth Factor Receptor-Tyrosine Kinase Inhibitors and a Potential Treatment Strategy. Cells. 2018;7:212. doi: 10.3390/cells7110212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Karachaliou N., Codony-Servat J., Bracht J.W.P., Ito M., Filipska M., Pedraz C., Chaib I., Bertran-Alamillo J., Cardona A.F., Molina M.A., et al. Characterising acquired resistance to erlotinib in non-small cell lung cancer patients. Expert Rev. Respir. Med. 2019;13:1019–1028. doi: 10.1080/17476348.2019.1656068. [DOI] [PubMed] [Google Scholar]
- 34.Westover D., Zugazagoitia J., Cho B., Lovly C., Paz-Ares L. Mechanisms of acquired resistance to first- and second-generation EGFR tyrosine kinase inhibitors. Ann. Oncol. 2018;29:i10–i19. doi: 10.1093/annonc/mdx703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Constant S., Huang S., Wisniewski L., Mas L.W.A.C. Advanced Human In vitro Models for the Discovery and Development of Lung Cancer Therapies. In: Vallisuta O., Olimat S., editors. Drug Discovery and Development—From Molecules to Medicine. IntechOpen; Rijeka, Croatia: 2015. [Google Scholar]
- 36.Pompili L., Porru M., Caruso C., Biroccio A., Leonetti C. Patient-derived xenografts: A relevant preclinical model for drug development. J. Exp. Clin. Cancer Res. 2016;35:1–8. doi: 10.1186/s13046-016-0462-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gristina V., Malapelle U., Galvano A., Pisapia P., Pepe F., Rolfo C., Tortorici S., Bazan V., Troncone G., Russo A. The significance of epidermal growth factor receptor uncommon mutations in non-small cell lung cancer: A systematic review and critical appraisal. Cancer Treat. Rev. 2020;85:101994. doi: 10.1016/j.ctrv.2020.101994. [DOI] [PubMed] [Google Scholar]
- 38.Nacchio M., Sgariglia R., Gristina V., Pisapia P., Pepe F., De Luca C., Migliatico I., Clery E., Greco L., Vigliar E., et al. KRAS mutations testing in non-small cell lung cancer: The role of Liquid biopsy in the basal setting. J. Thorac. Dis. 2020;12:3836–3843. doi: 10.21037/jtd.2020.01.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Canon J., Rex K., Saiki A.Y., Mohr C., Cooke K., Bagal D., Gaida K., Holt T., Knutson C.G., Koppada N., et al. The clinical KRAS(G12C) inhibitor AMG 510 drives anti-tumour immunity. Nature. 2019;575:217–223. doi: 10.1038/s41586-019-1694-1. [DOI] [PubMed] [Google Scholar]
- 40.Hallin J., Engstrom L.D., Hargis L., Calinisan A., Aranda R., Briere D.M., Sudhakar N., Bowcut V., Baer B.R., Ballard J.A., et al. The KRASG12C Inhibitor MRTX849 Provides Insight toward Therapeutic Susceptibility of KRAS-Mutant Cancers in Mouse Models and Patients. Cancer Discov. 2020;10:54–71. doi: 10.1158/2159-8290.CD-19-1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dunnett-Kane V., Nicola P., Blackhall F., Lindsay C. Mechanisms of Resistance to KRASG12C Inhibitors. Cancers. 2021;13:151. doi: 10.3390/cancers13010151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schliekelman M.J., Taguchi A., Zhu J., Dai X., Rodriguez J., Celiktas M., Zhang Q., Chin A., Wong C.-H., Wang H., et al. Molecular Portraits of Epithelial, Mesenchymal, and Hybrid States in Lung Adenocarcinoma and Their Relevance to Survival. Cancer Res. 2015;75:1789–1800. doi: 10.1158/0008-5472.CAN-14-2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Menz A., Weitbrecht T., Gorbokon N., Büscheck F., Luebke A.M., Kluth M., Hube-Magg C., Hinsch A., Höflmayer D., Weidemann S., et al. Diagnostic and prognostic impact of cytokeratin 18 expression in human tumors: A tissue microarray study on 11,952 tumors. Mol. Med. 2021;27:1–16. doi: 10.1186/s10020-021-00274-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sanchez-Cespedes M. The role of LKB1 in lung cancer. Fam. Cancer. 2011;10:447–453. doi: 10.1007/s10689-011-9443-0. [DOI] [PubMed] [Google Scholar]
- 45.Mogi A., Kuwano H. TP53 Mutations in Nonsmall Cell Lung Cancer. J. Biomed. Biotechnol. 2011;2011:1–9. doi: 10.1155/2011/583929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tamura K., Zhang X., Murakami Y., Hirohashi S., Xu H.-J., Hu S.-X., Benedict W.F., Sekiya T. Deletion of three distinct regions on chromosome 13q in human non-small-cell lung cancer. Int. J. Cancer. 1997;74:45–49. doi: 10.1002/(SICI)1097-0215(19970220)74:1<45::AID-IJC8>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 47.Oren M., Rotter V. Mutant p53 Gain-of-Function in Cancer. Cold Spring Harb. Perspect. Biol. 2009;2:a001107. doi: 10.1101/cshperspect.a001107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Klinakis A., Rampias T. TP53 mutational landscape of metastatic head and neck cancer reveals patterns of mutation selection. EBioMedicine. 2020;58:102905. doi: 10.1016/j.ebiom.2020.102905. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data presented in this study are available in Table 4 and Table S2.