Significance
Microbes shape human health and disease. Today, there are increasing correlations of gut microbes with specific diseases, but the underlying molecules and mechanism linking the two are not known. This stifles both the understanding of disease causation and the development of suitable treatments. We investigated a dozen patient isolates of Ruminococcus gnavus, a prevalent gut microbe linked to inflammatory bowel disease (IBD). We found that some isolates possess a protective capsule that encourages a symbiotic relationship with the host immune system, while others lack this protective capsule and elicit a robust inflammatory immune response. This work reveals a path by which R. gnavus could be connected to IBD and potential therapeutic interventions.
Keywords: Ruminococcus gnavus, capsular polysaccharide, capsule, inflammation, dendritic cell
Abstract
Active inflammatory bowel disease (IBD) often coincides with increases of Ruminococcus gnavus, a gut microbe found in nearly everyone. It was not known how, or if, this correlation contributed to disease. We investigated clinical isolates of R. gnavus to identify molecular mechanisms that would link R. gnavus to inflammation. Here, we show that only some isolates of R. gnavus produce a capsular polysaccharide that promotes a tolerogenic immune response, whereas isolates lacking functional capsule biosynthetic genes elicit robust proinflammatory responses in vitro. Germ-free mice colonized with an isolate of R. gnavus lacking a capsule show increased measures of gut inflammation compared to those colonized with an encapsulated isolate in vivo. These observations in the context of our earlier identification of an inflammatory cell-wall polysaccharide reveal how some strains of R. gnavus could drive the inflammatory responses that characterize IBD.
Disruptions to the community of microbes that inhabit the human gut are associated with a variety of diseases. Among these, inflammatory bowel diseases (IBD)—mainly Crohn’s disease and ulcerative colitis—are becoming increasingly prevalent (1), and today over 1% of the US population has been diagnosed with IBD (2). Despite its prevalence, the causes of IBD are not known, and the disease is treated symptomatically. Typical treatments involve small molecule anti-inflammatory or immune suppressant drugs or antibodies that target cytokines involved in inflammation (e.g., tumor necrosis factor alpha [TNF-α], IL-12). Even with multiple treatment options, these therapies are not effective for many patients, and many responding patients later develop resistance or diminished responsiveness (3). Over 10% of ulcerative colitis patients and nearly 40% of Crohn’s disease patients eventually require surgical interventions to remove affected tissue (4).
Genetic and environmental factors likely combine to prime the immune system to overrespond to one or several commensal microbes, and this inappropriately up-regulated response results in damage to host tissue. Individuals genetically predisposed to IBD harbor variants in genes for autophagy, anti-inflammatory pathways, and innate immune receptors (5). Environmental factors include the gut microbiota, and surveys of feces and intestinal biopsies have identified potential “pathobionts” (6) such as the adherent-invasive Escherichia coli (7), the fungus Malassezia restricta (8), and the obligate anaerobe Ruminococcus gnavus (9, 10). Intriguingly, R. gnavus transiently blooms in patients with active flares of Crohn’s disease (10) and has been implicated in a variety of other immune disorders such as lupus (11).
R. gnavus is widely distributed at low abundance (roughly 0.1%) in the guts of healthy adults from North America and Europe (12). Whole genome sequencing of R. gnavus isolates coupled with metagenomic analyses revealed two phylogenetically distinct R. gnavus strains that appear differentially enriched in IBD (10). R. gnavus has been widely investigated for potential host interactions. Germ-free mice colonized with R. gnavus have lower levels of IL22-expressing CD4+ T cells and innate lymphoid cells along the gastrointestinal tract and in lymphatic organs compared to germ-free or specific-pathogen–free mice (13). R. gnavus binds directly to a variety of immunoglobin A (IgA) in a “superantigen”-like mode and can be highly IgA-coated regardless of the IgA target (14). R. gnavus effects are not limited to immune responses, as it also decarboxylates the amino acid tryptophan to produce the neurotransmitter tryptamine (15), thereby raising the possibility that this gut microbe may interact with the gut or central nervous system. Importantly, the above studies were limited to the type strain ATCC 29149.
Previously, we showed the cell wall of R. gnavus ATCC 29149 contains a 9 kDa glucose-rhamnose cell-wall polysaccharide (glucorhamnan) that stimulates inflammatory cytokine secretion by innate immune cells in vitro in a TLR4-dependent manner (16). The glucorhamnan biosynthesis genes are present in all sequenced strains of R. gnavus. Since similar rhamnose-rich cell-wall polysaccharides exist in distantly related gram-positive organisms (17) [e.g., Streptococcus (18), Lactococcus (19), and Clostridium (20)], they may be an underappreciated conserved feature of the gram-positive cell wall that is recognized by the innate immune system.
Bacteria produce a variety of extracellular polysaccharides whose nomenclature has not been standardized. Peptidoglycan, the best-known bacterial polysaccharide, forms the bacterial cell wall surrounding the entire cell membrane. Polysaccharides are often named based on their means of cellular attachment: exopolysaccharides are noncovalently associated with the cell and can readily be washed from the cell surface, cell-wall polysaccharides are covalently attached to the peptidoglycan, and lipopolysaccharides (or lipoglycans) can be anchored by a variety of lipids to the cell membrane. Teichoic acids, a major class of polysaccharides found in gram-positive bacteria, are acidic and usually composed of repeating units of glycerol phosphate or ribitol phosphate. The highly negative charge is thought to be important for ion retention. Teichoic acids can be further subdivided into wall or lipoteichoic acids depending on their means of cellular association. Capsular polysaccharides have very high molecular weights (often ≥100,000 Da) and extend far beyond the peptidoglycan and other cell-wall components to produce a thick protective coat around the entire bacterial cell. Capsules, which can often be viewed by microscopy, protect bacteria against desiccation, phages, and immune cells.
We explored variations of cell surface polysaccharides among patient isolates of R. gnavus and how these variations impacted immune recognition of this microbe. We found that the type strain ATCC 29149 and phylogenetically related strains of R. gnavus possess a capsular polysaccharide gene cluster, which is absent in a group of more distantly related R. gnavus strains. Strains harboring this gene cluster produce a thick polysaccharide capsule that covers surface cell-wall components—the glucorhamnan, teichoic acid, and peptidoglycan. In contrast, strains that lack this gene cluster do not produce thick capsules. We then show that capsule-producing (“encapsulated”) strains of R. gnavus elicit little to no cytokine production by innate immune cells in vitro, while R. gnavus strains lacking a capsule (“unencapsulated”) induce potent inflammatory cytokine responses. Further, we observe that germ-free mice colonized with an unencapsulated strain of R. gnavus show increased gut inflammation compared to an encapsulated strain, which stimulates a tolerogenic response in the immune system in vivo. Together these observations provide a genetic and mechanistic model that connects some, but not all, R. gnavus strains with inflammation.
Results
Some Strains of R. gnavus Produce a Capsular Polysaccharide.
To understand how phylogenetically distinct strains of R. gnavus differentially interact with the host immune system, we analyzed both their small molecule metabolites and differences in bacterial cell surface factors, such as the glucorhamnan previously described (16). We selected 12 isolates (SI Appendix, Table S1), and during the course of culturing, some strains pelleted readily when centrifuged, while other strains (including ATCC 29149) did not pellet or form easily disturbed pellets (SI Appendix, Fig. S1). This phenotype has previously been attributed to encapsulation by polysaccharides in a variety of bacteria (21, 22). Capsules are often a meshwork of proteins and polysaccharides that form the outermost layer of the bacterial cell, which can prevent individual cells from coming together to form a pellet. This effect could also be seen in the related observation that cells that do not pellet tightly do not settle in culture, even after 1 wk, whereas those strains that pellet readily also settle in culture.
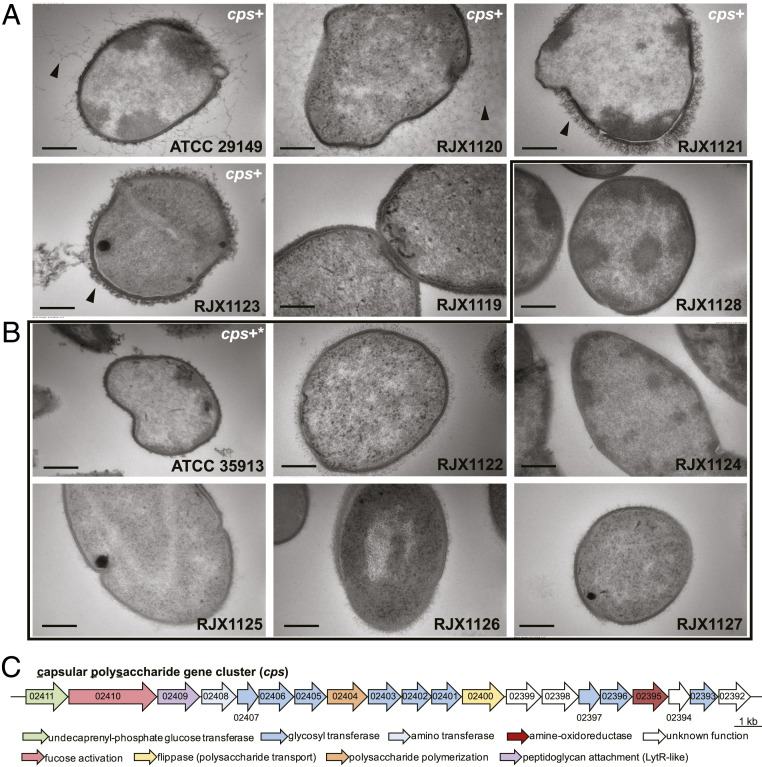
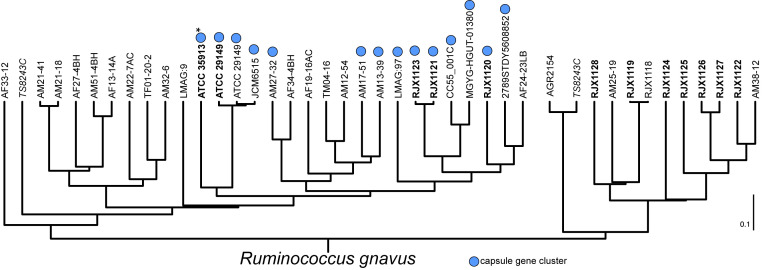
Indeed, by transmission electron microscopy (EM), strains that did not settle or pellet well appeared to be covered with additional material on their cell surface (Fig. 1A and SI Appendix, Fig. S2) compared to strains that settle and pellet readily (Fig. 1B and SI Appendix, Fig. S2). Furthermore, genome inspection of these strains revealed that encapsulated strains possess a conserved gene cluster that is predicted to encode a capsular polysaccharide (cps) (Fig. 1C and SI Appendix, Fig. S3 and Tables S2 and S3). Importantly, other than the glucorhamnan and peptidoglycan gene clusters, this locus is the only polysaccharide biosynthesis gene cluster in common among these strains. The form the capsule takes is highly variable; for instance, ATCC 29149 and RJX1120 have a diffuse capsule that extends far from the cell surface, while RJX1121 and RJX1123 have a compact, thick capsule (Fig. 1A). The cps gene cluster is found in a subset of phylogenetically related strains (Fig. 2) and appears to have been spread through horizontal gene transfer among these strains, as it is nearly identical (>99.9%) across the entire 25-kb locus (compared to a conserved region of the ubiquitous glucorhamnan gene cluster, which is 99.1% identical over 12.5 kb).
Fig. 1.
Some strains of R. gnavus are covered by a capsular polysaccharide. EM of 12 strains of R. gnavus. (A) Strains that do not pellet possess a thick capsule, shown with a black arrow, while (B) strains that pellet readily are not encapsulated, within black outline. Strain numbers are indicated for each sample. (Scale bar, 200 nm.) (C) Encapsulated strains possess a conserved capsular polysaccharide biosynthetic gene cluster (shown in A and B as “cps+”). Genes are numbered following ATCC 29149 locus convention (RUMGNA_02411 through RUMGNA_02392). Strain ATCC 35913 has a defective capsular polysaccharide biosynthesis gene cluster (cps+*) and does not appear encapsulated.
Fig. 2.
Phylogenetic tree based on genomic BLAST of R. gnavus strains shows cps+ strains are phylogenetically distinct. Strains in bold were available for this study. The capsule gene cluster (blue circle) is found in phylogenetically related strains. Note that several genes in the ATCC 35913 cps cluster (marked with *) are disrupted by nonsense mutations likely rendering this cluster nonfunctional. Strain TS8243C (in italics), isolated from a Malawian child (23), has protective roles in malnutrition and has been sequenced twice. These sequences disagree with each other; however, this does not affect our analysis. Dendrogram was generated by NCBI Genome Analysis.
Unencapsulated Strains of R. gnavus Induce Proinflammatory Signals In Vitro.
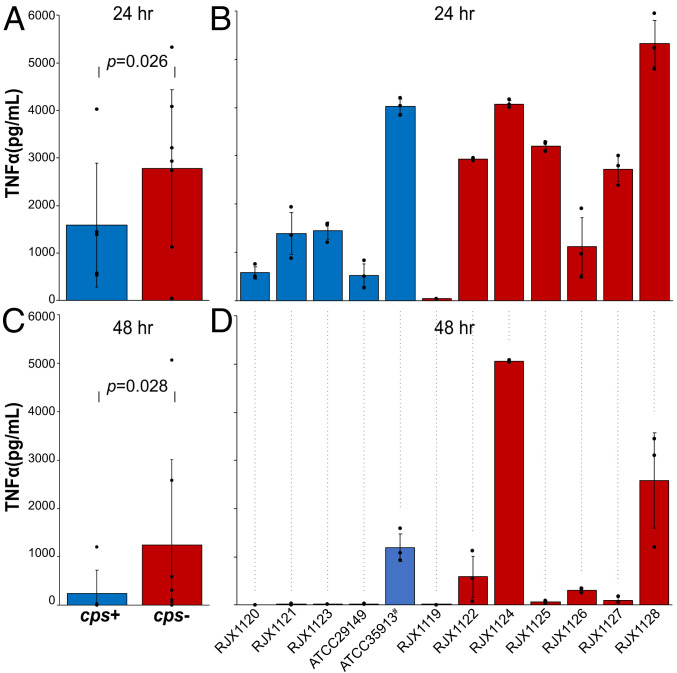
Because the cell surfaces of these strains vary from each other by the presence/absence of a capsule, we hypothesized these strains may be differentially recognized by innate immune cells. To test this hypothesis, murine bone-marrow–derived dendritic cells (mBMDCs) were cultured and exposed overnight to normalized levels of phosphate buffer saline–washed cells of different R. gnavus strains, and cytokine secretion was monitored by ELISA (enzyme-linked immunosorbent assay). We found that encapsulated R. gnavus elicit little to no TNF-α secretion, while strains without a capsule, whose cell walls are exposed, induced robust TNF-α production (Fig. 3). Indeed, TNF-α–inducing activity appears to be inversely proportional to the amount of capsular material on the cell surface. R. gnavus strains ATCC 29149, RJX1120, RJX1121, and RJX1123, which have the most capsular material, elicit virtually no TNF-α response, while strains RJX1128 and RJX1124, which have no capsular material, induce the most potent TNF-α response. Several R. gnavus strains with intermediate capsular material (RJX1122, RJX1125, RJX1126, RJX1127) pellet readily and stimulate moderate TNF-α secretion. Importantly, while these four strains possess thin capsules, they are not made of the capsular polysaccharide encoded by the cps locus, which these isolates lack.
Fig. 3.
Mouse bone-marrow–derived dendritic cells secrete more TNF-α when exposed to unencapsulated strains of R. gnavus. (A) Pooled data and (B) individual cps+ strains (blue) and cps− strains (red) after 24 h of bacterial growth. (C) Pooled data and (D) individual cps+ strains (blue) and cps− strains (red) after 48 h bacterial growth. ATCC 35913 has a nonfunctional cps locus (#).
TNF-α secretion differences are likely due to the cell-surface differences between the strains, since the effect becomes more pronounced the older the bacterial cultures are. Bacterial cells harvested at 24 h of growth show less of a difference (P value = 0.026) between encapsulated and unencapsulated strains (Fig. 3 A and B), while bacterial cells harvested at 48 h of growth show a profound difference with nearly no stimulation by encapsulated strains (Fig. 3 C and D). Correspondingly, by EM, the capsules appear thicker at this later time point.
Importantly, there are two strains of R. gnavus tested that initially appear to defy the trend that cps+ strains do not induce TNF-α, while cps− strains induce TNF-α secretion. First, the strain ATCC 35913 induces a potent TNF-α secretion by mBMDCs. While this strain possesses the cps gene cluster, closer inspection reveals that five genes in the cluster are interrupted by nonsense mutations (SI Appendix, Fig. S4). Indeed, by EM, this strain does not appear to be encapsulated (Fig. 1B). Another seeming outlier is strain RJX1119, which does not possess the cps gene cluster, nor does it induce a potent TNF-α response. RJX1119 does not settle or pellet well, and EM shows that this strain uniquely forms chains of cells (SI Appendix, Fig. S2), which presumably alter the way it acts in bulk and potentially alter its presentation to innate immune cells. Unlike the other RJX strains in this study, RJX1119 was not isolated from an IBD patient; rather, it was isolated from an infant genetically predisposed to type 1 diabetes, who had been treated with antibiotics (24). In sum, R. gnavus strains that harbor an intact cps locus produce a capsule and do not stimulate TNF-α secretion by mBMDCs, in contrast to strains that lack the cps locus or a capsule.
Structural Features of Capsular Polysaccharide from RJX1120 Indicate the cps Locus Likely Encodes the Capsule.
In order to link the capsule phenotype and the cps gene cluster, capsular polysaccharide was isolated and purified from cps+ strain RJX1120 for structural analysis. Monosaccharide analysis revealed the capsule to be composed of glucose, N-acetyl-quivosamine, and N-acetyl-galactosamine in roughly a 6:2:1 stoichiometry (SI Appendix, Fig. S5 and Table S4). This is in accordance with the cps gene cluster, which contains nine glycosyltransferases—one for each of the monosaccharides in the repeating unit. Furthermore, analytical size-exclusion chromatography showed the purified capsular polysaccharide to elute in the void volume of the column, indicating its molecular weight is at least 100 kDa (SI Appendix, Fig. S6), as would be expected for capsular polysaccharides that extend far beyond the cell wall. Unfortunately, due to its large molecular weight, significant peak broadening, even at temperatures of 75 °C, prohibited further structure determination of the capsular polysaccharide by NMR.
Capsular Polysaccharide Blocks mBMDC Response to Intact R. gnavus.
We next sought to understand mechanistically how the capsular polysaccharide encoded by the cps locus may be preventing proinflammatory cytokine secretion by mBMDCs. To test the role of the major toll-like receptors involved in recognizing bacterial cell-surface molecules, mBMDCs were generated from tlr2−/− or tlr4−/− mice and exposed to PBS-washed, intact R. gnavus isolates grown for 48 h. Regardless of genetic background, the TNF-α secretion levels were unaffected (SI Appendix, Fig. S7). Further, the capsule does not appear to inhibit secretion of TNF-α through a signaling mechanism, as mBMDCs cotreated with either a cps+ strain (SI Appendix, Fig. S8) or pretreated with purified capsular polysaccharide failed to inhibit cytokine secretion by a cps− strain (SI Appendix, Fig. S9). Taken together, this indicates that the capsular polysaccharide may physically block immune-cell access to proinflammatory molecules that make up the deeper layers of the R. gnavus cell wall.
Unencapsulated Strains of R. gnavus Induce Proinflammatory Signals in Monocolonized Mice.
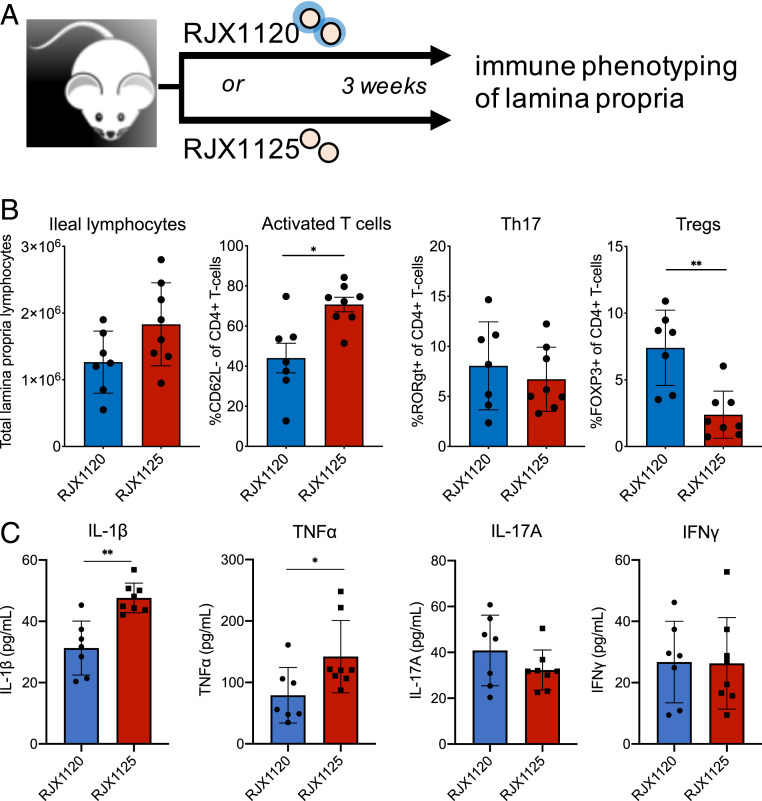
The observation that cps+ R. gnavus are relatively tolerogenic to immune cells in vitro compared to cps− R. gnavus prompted us to ask if the immune system in vivo responds to these strains differently. To understand the in vivo function of these strains, germ-free mice were colonized with either the cps+ strain RJX1120 or the cps− strain RJX1125. Each strain colonized the mouse equally (SI Appendix, Fig. S10), and after 3 wk, mice were killed, and lymphocytes from the lamina propria were isolated and interrogated for differences in cytokine and immune-cell levels (Fig. 4A). We observed a trend toward accumulation of immune cells in the lamina propria of mice colonized with RJX1125, and a higher percentage of the T cells were activated based upon surface expression of CD62L (Fig. 4B). The cps+ R. gnavus-colonized mice also had significantly higher levels of FOXP3+-regulatory T cells, which exert anti-inflammatory effects by tempering the activities of proinflammatory immune cells (Fig. 4B), indicating induction of these tolerance responses may be mediated by the presence of cps genes. Lamina propria lymphocytes also secreted increased levels of the key proinflammatory cytokines IL-1β and TNF-α in RJX1125-colonized mice compared to the RJX1120 colonized mice (Fig. 4C). Taken together, these results indicate that the mouse immune system recognizes and responds to an encapsulated strain and an unencapsulated strain of R. gnavus differently and that the unencapsulated strain evokes a proinflammatory immune response in this gnotobiotic, monocolonized setting.
Fig. 4.
An encapsulated and an unencapsulated R. gnavus strain differentially stimulate the mouse immune system in monoassociation. (A) Scheme for mouse monoassociation experiments. (B) RJX1125 (cps−) induces more lymphocytes in the lamina propria, of which an increased proportion are CD4+-activated T cells, while RJX1120 (cps+) induces more regulatory T cells in this population. (C) Immune cells from RJX1125-colonized mice secrete higher levels of IL-1β and TNF-α in the lamina propria compared to RJX1120-colonized mice. *P ≤ 0.05, **P ≤ 0.005.
Discussion
Finding the causes of and cures for microbiome-implicated diseases requires establishing the molecular mechanisms connecting microbes to disease. To date, sequencing studies of IBD patients have shown compelling correlational data implicating several species in disease. But the mechanisms responsible for these correlations are largely lacking in animal IBD models and are completely absent of patients. Here, we investigated one gut bacterium strongly implicated in IBD, R. gnavus. We found that clinical isolates exhibit differences in their cell-surface structures, and these differences correspond to divergent host immune responses. Some strains possess a protective capsular polysaccharide, which covers deeper, conserved layers of the proinflammatory bacterial cell wall and is tolerogenic, while R. gnavus strains that lack this protective capsule have their cell walls exposed and elicit robust immune responses both in vitro and in in vivo monocolonized mouse experiments.
Beyond IBD, R. gnavus has been associated to other immune-related diseases: childhood asthma (25), infant eczema (26), spinal arthritis (27), and lupus (11). Intriguingly, the group that showed R. gnavus is associated with lupus (11) also found that lupus patients with active disease produce serum IgG antibodies to a cell wall lipoglycan present on one of eight strains of R. gnavus they examined, strain CC55_001C. Furthermore, serum IgG specific to double-stranded DNA (dsDNA), which are thought to be pathologic in nephritic lupus, are cross-reactive to this strain of R. gnavus—potentially directly to its lipoglycan. Since strain CC55_001C possesses the cps gene cluster described here, the capsule may well be the lipoglycan, suggesting a potential role in lupus.
Because many of the above studies used the 16S rRNA gene for relative abundance analysis, they were able to observe an association to R. gnavus as a species. However, these studies do not resolve whether strain-level variability exists as was done with metagenome sequencing for the Prospective Registery in IBD Study at MGH (PRISM) IBD cohort (10). Taking a functional approach to define strain-level differences among R. gnavus isolates, we have found a molecule (the capsular polysaccharide) and its corresponding genetic locus (cps locus). These, in combination with other features of R. gnavus, represent a distinct molecular hypothesis to interrogate for R. gnavus and its association with IBD. Future studies sequencing isolate genomes (or metagenomes) will show if similar strain differences also exist in non-IBD diseases in which R. gnavus has been implicated, or if this is unique to IBD.
Our finding strain differences in encapsulation of R. gnavus is reminiscent of the seminal observations made by Frederick Griffith in the 1920s. Griffith demarcated clinical isolates of Streptococcus pneumoniae as either avirulent “rough” strains that lack a capsule or virulent “smooth” strains that possess a capsule (28). Furthermore, he was able to transform avirulent rough strains into virulent smooth strains by incubating them with a “transforming principle” later used to prove that DNA was the genetic code (29). For S. pneumoniae, virulent smooth strains possess a capsule that protects them from clearance by the host immune system. This is the typical paradigm of virulence: a microbe gains genes that impart pathogenic behavior. However, for R. gnavus, this paradigm may be flipped: some strains appear to have acquired the capsule gene cluster, which imparts commensal behavior and encourages a tolerogenic relationship with its human host.
Previously, other human gut microbes have been shown to possess tolerogenic capsules. Perhaps the best known is polysaccharide A from Bacteroides fragilis (30, 31), which induces regulatory T-cell proliferation and IL-10 production. Deletion or interruption of the capsule of several Bifidobacterium species has also been shown to lead to potent immune activation (32, 33). These studies show that capsules play tolerogenic roles for individual members of the human gut microbiota. Our study shows that clinical isolates of R. gnavus differ in whether they possess a tolerogenic capsule.
Additional studies are required to understand what role encapsulated and unencapsulated R. gnavus strains play in the guts of IBD patients and, potentially, in other systemic inflammatory diseases. Our work provides a testable model for a molecular understanding of how R. gnavus contributes to inflammatory diseases, with clear genetic and mechanistic components. More broadly, it highlights the ability of reduced-complexity molecular studies to clarify how members of the gut microbiota may shape human health and disease.
Methods
Bacterial Strains and Growth.
ATCC 29149 and ATCC 35913 came from the feces of healthy adults and were purchased from ATCC.
RJX1119 was isolated from an infant treated with antibiotics (24), RJX1120 through RJX1127 were isolated from biopsies from IBD patients, and strain RJX1128 was isolated from feces of an IBD patient (10). Strains were grown anaerobically at 37 °C in defined medium (34).
EM.
Strains R. gnavus were grown in defined medium for 24 and 48 h. Cells were fixed overnight in 2.5% glutaraldehyde/1.25% paraformaldehyde/0.03% picric acid + 10 mM lysine and 0.1% ruthenium red in 0.1 M sodium cacodylate buffer (pH 7.4). Samples were then washed three times in 0.1 M cacodylate buffer, postfixed with 1% osmium tetroxide (OsO4) for 2 h, washed in water twice and in maleate buffer (MB) once, and incubated in 1% uranyl acetate in MB for 1 h followed by one wash in MB, two washes in water, and subsequent dehydration in grades of alcohol (10 min each; 50%, 70%, 90%, 2 × 10 min 100%). The samples were then put in propylene oxide for 1 h and infiltrated overnight in a 1:1 mixture of propylene oxide and Spurr’s Low Viscosity Embedding media. The following day, the samples were embedded in Spurr’s Low Viscosity Embedding media and polymerized at 60 °C for 48 h. Ultrathin sections (about 80 nm) were cut on a Reichert Ultracut-S microtome, picked up onto copper grids stained with lead citrate, and examined in a JEOL 1200EX Transmission electron microscope, and images were recorded with an AMT 2k CCD camera.
Dendrogram.
A dendrogram was generated by genomic BLAST and provided by The National Center for Biotechnology Information (NCBI) Genome resource (https://www.ncbi.nlm.nih.gov/genome/979).
mBMDC Assay.
Bacterial cells were grown in defined medium as above for 24 and 48 h. Cultures were centrifuged and bacterial pellets were resuspended in 1× PBS and corrected to an optical density measured at 600 nm (OD600) of 1.0. Bone marrow was isolated from C57BL/6 mice and grown in the presence of granulocyte-macrophage colony-stimulating factor (GM-CSF) (PreProTech) for 7 d. Once matured, the BMDCs were scraped and plated in 96-well plates and allowed to adhere. After at least 3 h, BMDCs were treated with whole bacterial cells overnight. Additionally, BMDCs were treated with pure capsule preparation overnight and then, the following morning, were treated with lipopolysaccharide (LPS) for 1.5 h. TNF-α production was detected using ELISAs (ThermoFisher) as described previously (16).
Mouse Experiments.
As described previously (35), 6- to 8-wk-old female C57BL/6N germ-free mice (originally purchased from Taconic) were maintained at a gnotobiotic facility at the Broad Institute of MIT and Harvard and used for this study. Mice were housed in cages with five mice per cage and given access to sterilized food and water ad libitum. Mice were allocated to experimental groups randomly. All experimental procedures were conducted under protocols approved by the Institutional Animal Care and use Committee at the Broad Institute.
Bacterial Mixture Preparation and Inoculation.
Anaerobic cultures of R. gnavus from frozen stock were plated on brain heart infusion (BHI) agar for 2 d, and, subsequently, pure cultures were selected from the plate and resuspended into sterile, reduced PBS at a concentration of 109 cells/mL, similar to previously described experiments (35). Bacterial mixtures in PBS were immediately transported to the animal facility, and each mouse was gavaged with 100 μL of the mixture equating to 108 cells per mouse total. The concentration of the mixture in cell per milliliter was carried out using an ultraviolet spectrometer, and gavage doses were confirmed after by back-titering the inocula on BHI agar.
Immune Phenotyping.
As described previously (35), ∼5 cm of the ileum from each mouse was excised, attached fat and Peyer’s patches were removed, and tissues were cut longitudinally to further remove luminal contents by washing with ice-cold PBS with 2% fetal bovine serum (FBS). Epithelial cells were isolated using a PBS buffer containing 1 mM EDTA, 1 mM dithiothreitol (DTT), and 10% FBS, shaking at 37 °C for 15 min. The intact intestinal tissue was resuspended in additional PBS buffer containing 1 mM EDTA, 1 mM DTT, and 10% FBS, shaking at 37 °C for 30 min in order to remove the intraepithelial lymphocytes. Lamina propria lymphocytes were purified by digesting the remaining tissue for 30 min at 37 °C with 1 mg/mL DNase and 0.5 mg/mL collagenase. After digestion, lymphocytes were further purified using a 40% Percoll gradient, resuspended in RPMI 1640 with 10% FBS, and enumerated using a hemocytometer.
Flow Cytometry.
For immune cell profiling, ∼1,000,000 cells per mouse were stained with fluorochrome-conjugated antibodies against CD45, CD3, CD4, CD8, FOXP3, RORγt, and CD62L (eBioscience), and their populations were analyzed by a CytoFLEX (Beckman-Coulter) using software packages from CellQuest and FlowJo version 12 as described previously (35).
Cytokine Profiling.
As described previously (35), for ex vivo quantification of cytokine secretion, isolated lamina propria lymphocytes were washed with complete tissue culture media (RPMI, 10% FBS, 1% glutamine, and 1% 1:1 penicillin/streptomycin) and cultured in 1 mL of the same media in 24-well plates for 24 h at 37 °C, 5% carbon dioxide. The resulting supernatants were used to determine the amount of each cytokine using murine-specific ELISA kits (BD Biosciences) and a cytometrix bead array (CBA) flex set (BD Biosciences) relative to a standard curve according to the manufacturer’s recommendations.
Capsular Polysaccharide Purification.
The slime layer and cell pellet from 3 L of RJX1120 growth in defined medium was resuspended in lysis buffer (50 mM Tris [pH 7.5], 50 mM MgSO4, 20% sucrose). Cells were lysed with 100 units of mutanolysin (M0991) at 37 °C overnight. To complete lysis, sodium dodecylsulfate was added to 0.5% and stirred at room temperature for 1 h and then at 55 °C for 1 h. Cell debris was removed by centrifugation, and the supernatant, containing liberated polysaccharides, was treated with 1 mg proteinase K and incubated at 37 °C for 3 h to remove protein contaminants. The supernatant was concentrated with a 3-kDa molecular-weight cutoff filter and dialyzed against water extensively. Dialysate was applied to 5 mL HiTrap Q HP pre-equilibrated with 10 mM Tris, pH 8. Flow-through and fractions were collected with increasing concentrations of NaCl. Capsular material was found in the flow-through via H2SO4-phenol staining. Molecular weight of capsular polysaccharide was determined on Agilent HPLC1200 equipped with refractive index detector with a Superdex 200 Increase column and 50 mM NH4OAc, pH 6, as mobile phase. Dextran blue and polyethylene glycols were used as molecular-weight standards.
Capsular Polysaccharide Composition Analysis.
Glycosyl composition analysis was performed by combined gas chromatography/mass spectrometry (GC/MS) of the per-O-trimethylsilyl (TMS) derivatives of the monosaccharide methyl glycosides produced from the sample by acidic methanolysis as described previously (36). An aliquot equal to 0.3 mg sample weight was pipetted from each sample solution. To this, 20 µg inositol was added as internal standard. The solution was then lyophilized to remove water for the methanolysis. Briefly, the samples were heated in 1 M methanolic HCl in a sealed screw-top glass test tube for 18 h at 80 °C. After cooling and removal of the solvent under a stream of nitrogen, the sample was treated with a 2:1:1 ratio mixture of methanol, pyridine, and acetic anhydride at room temperature for 30 min. The solvents were evaporated, and the sample was derivatized with 200 µL Tri-Sil (Pierce) at 80 °C for 30 min. Following extraction with hexane, GC/MS analysis of TMS methyl glycosides was performed on an Agilent 7890A GC interfaced to a 5975C MSD (mass selective detector) using a Supelco Equity-1–fused silica capillary column (30 m × 0.25 mm ID).
Supplementary Material
Acknowledgments
We thank the Electron Microscopy Facility at Harvard Medical School, Aditi Kanaan, and Yue Shu for assistance with bacterial culturing. This work was funded by NIH Grants R01-AT009708 (J.C. and R.J.X.) and F32-GM126650 (M.T.H.). Polysaccharide structural analysis was supported by the Chemical Sciences, Geosciences, and Biosciences Divisions of the Office of Basic Energy Sciences, US Department of Energy Grant (DE-SC0015662) to the Complex Carbohydrate Research Center.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2007595118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or supporting information.
References
- 1.Molodecky N. A., et al., Increasing incidence and prevalence of the inflammatory bowel diseases with time, based on systematic review. Gastroenterology 142, 46–54.e42 (2012). [DOI] [PubMed] [Google Scholar]
- 2.Dahlhamer J. M., Zammitti E. P., Ward B. W., Wheaton A. G., Croft J. B., Prevalence of inflammatory bowel disease among adults aged ≥18 years–United States, 2015. MMWR Morb. Mortal. Wkly. Rep. 65, 1166–1169 (2016). [DOI] [PubMed] [Google Scholar]
- 3.Ben-Horin S., Chowers Y., Review article: Loss of response to anti-TNF treatments in Crohn’s disease. Aliment. Pharmacol. Ther. 33, 987–995 (2011). [DOI] [PubMed] [Google Scholar]
- 4.Frolkis A. D., et al., Risk of surgery for inflammatory bowel diseases has decreased over time: A systematic review and meta-analysis of population-based studies. Gastroenterology 145, 996–1006 (2013). [DOI] [PubMed] [Google Scholar]
- 5.Liu T.-C., Stappenbeck T. S., Genetics and pathogenesis of inflammatory bowel disease. Annu. Rev. Pathol. 11, 127–148 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Khan I., et al., Alteration of gut microbiota in inflammatory bowel disease (IBD): Cause or consequence? IBD treatment targeting the gut microbiome. Pathogens 8, 1–28 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Darfeuille-Michaud A., et al., High prevalence of adherent-invasive Escherichia coli associated with ileal mucosa in Crohn’s disease. Gastroenterology 127, 412–421 (2004). [DOI] [PubMed] [Google Scholar]
- 8.Limon J. J., et al., Malassezia is associated with Crohn’s disease and exacerbates colitis in mouse models. Cell Host Microbe 25, 377–388.e6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Joossens M., et al., Dysbiosis of the faecal microbiota in patients with Crohn’s disease and their unaffected relatives. Gut 60, 631–637 (2011). [DOI] [PubMed] [Google Scholar]
- 10.Hall A. B., et al., A novel Ruminococcus gnavus clade enriched in inflammatory bowel disease patients. Genome Med. 9, 103(2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Azzouz D., et al., Lupus nephritis is linked to disease-activity associated expansions and immunity to a gut commensal. Ann. Rheum. Dis. 78, 947–956 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Qin J.et al.; MetaHIT Consortium , A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464, 59–65 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Geva-Zatorsky N., et al., Mining the human gut microbiota for immunomodulatory organisms. Cell 168, 928–943.e11 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bunker J. J., et al., B cell superantigens in the human intestinal microbiota. Sci. Transl. Med. 11, eaau9356 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Williams B. B., et al., Discovery and characterization of gut microbiota decarboxylases that can produce the neurotransmitter tryptamine. Cell Host Microbe 16, 495–503 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Henke M. T., et al., Ruminococcus gnavus, a member of the human gut microbiome associated with Crohn’s disease, produces an inflammatory polysaccharide. Proc. Natl. Acad. Sci. U.S.A. 116, 12672–12677 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mistou M. Y., Sutcliffe I. C., van Sorge N. M., Bacterial glycobiology: Rhamnose-containing cell wall polysaccharides in gram-positive bacteria. FEMS Microbiol. Rev. 40, 464–479 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Linzer R., Reddy M. S., Levine M. J., Structural studies of the rhamnose-glucose polysaccharide antigen from Streptococcus sobrinus B13 and 6715-T2. Infect. Immun. 50, 583–585 (1985). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sadovskaya I., et al., Another brick in the wall: A rhamnan polysaccharide trapped inside peptidoglycan of Lactococcus lactis. mBio 8, e01303-17(2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Watanabe Y., Misaki H., Structure of a new rhamnose polysaccharide, clostrhamnan, isolated from antitumor hot water extract of clostridium saccharoperbutylacetonicum cells. Agric. Biol. Chem. 51, 931–932 (1987). [Google Scholar]
- 21.Sellin M., Håkansson S., Norgren M., Phase-shift of capsule expression in group B streptococci type III. Dev. Biol. Stand. 85, 245–249 (1995). [PubMed] [Google Scholar]
- 22.Chatzidaki-Livanis M., Coyne M. J., Roche-Hakansson H., Comstock L. E., Expression of a uniquely regulated extracellular polysaccharide confers a large-capsule phenotype to Bacteroides fragilis. J. Bacteriol. 190, 1020–1026 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Blanton L V.et al., Gut bacteria that prevent growth impairments transmitted by microbiota from malnourished children. Science 351, aad3311 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yassour M., et al., Natural history of the infant gut microbiome and impact of antibiotic treatment on bacterial strain diversity and stability. Sci. Transl. Med. 8, 343ra81 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chua H. H., et al., Intestinal dysbiosis featuring abundance of Ruminococcus gnavus associates with allergic diseases in infants. Gastroenterology 154, 154–167 (2018). [DOI] [PubMed] [Google Scholar]
- 26.Zheng H., et al., Altered gut microbiota composition associated with eczema in infants. PloS One 11, e0166026 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Breban M., et al., Faecal microbiota study reveals specific dysbiosis in spondyloarthritis. Ann. Rheum. Dis. 76, 1614–1622 (2017). [DOI] [PubMed] [Google Scholar]
- 28.Griffith F., The significance of pneumococcal types. J. Hyg. (Lond). 27, 113–159 (1928). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Avery O. T., Macleod C. M., McCarty M., Studies on the chemical nature of the substance inducing transformation of pneumococcal types: Induction of transformation by a desoxyribonucleic acid fraction isolated from pneumococcus type III. J. Exp. Med. 79, 137–158 (1944). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mazmanian S. K., Liu C. H., Tzianabos A. O., Kasper D. L., An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell 122, 107–118 (2005). [DOI] [PubMed] [Google Scholar]
- 31.Mazmanian S. K., Round J. L., Kasper D. L., A microbial symbiosis factor prevents intestinal inflammatory disease. Nature 453, 620–625 (2008). [DOI] [PubMed] [Google Scholar]
- 32.Fanning S., et al., Bifidobacterial surface-exopolysaccharide facilitates commensal-host interaction through immune modulation and pathogen protection. Proc. Natl. Acad. Sci. U.S.A. 109, 2108–2113 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schiavi E., et al., The surface associated exopolysaccharide of Bifidobacterium longum 35624 plays an essential role in dampening host proinflammatory responses and repressing local TH17 responses. Appl. Environ. Microbiol. 82, 7185–7196 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tramontano M., et al., Nutritional preferences of human gut bacteria reveal their metabolic idiosyncrasies. Nat. Microbiol. 3, 514–522 (2018). [DOI] [PubMed] [Google Scholar]
- 35.Brown E. M., et al., Bacteroides-derived sphingolipids are critical for maintaining intestinal homeostasis and symbiosis. Cell Host Microbe 25, 668–680.e7 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Santander J., et al., Mechanisms of intrinsic resistance to antimicrobial peptides of Edwardsiella ictaluri and its influence on fish gut inflammation and virulence. Microbiology (Reading) 159, 1471–1486(2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or supporting information.