Significance
Enzyme I (EI) is a central player in bacterial metabolism that controls carbon uptake, virulence, and biofilm formation of bacterial cells. Conserved in gram-positive and gram-negative bacteria but absent in eukaryotes, EI is a promising target for antimicrobial design. EI’s monomer–dimer equilibrium plays a crucial role in regulating enzymatic activity, yet very little is known about the changes in structure, dynamics, and substrate-binding properties undergone upon dimerization. Using a combination of protein engineering and pressure perturbation, we isolated EI’s monomeric state and characterized its structure in solution by NMR spectroscopy. Our study provides an atomic-level description of the structural changes accompanying EI dimerization and opens perspectives for the design of allosteric inhibitors.
Keywords: phosphotransferase system, carbon metabolism, solution NMR, enzyme regulation, high pressure
Abstract
Enzyme I (EI) is a phosphotransferase enzyme responsible for converting phosphoenolpyruvate (PEP) into pyruvate. This reaction initiates a five-step phosphorylation cascade in the bacterial phosphotransferase (PTS) transduction pathway. Under physiological conditions, EI exists in an equilibrium between a functional dimer and an inactive monomer. The monomer–dimer equilibrium is a crucial factor regulating EI activity and the phosphorylation state of the overall PTS. Experimental studies of EI’s monomeric state have yet been hampered by the dimer’s high thermodynamic stability, which prevents its characterization by standard structural techniques. In this study, we modified the dimerization domain of EI (EIC) by mutating three amino acids involved in the formation of intersubunit salt bridges. The engineered variant forms an active dimer in solution that can bind and hydrolyze PEP. Using hydrostatic pressure as an additional perturbation, we were then able to study the complete dissociation of the variant from 1 bar to 2.5 kbar in the absence and the presence of EI natural ligands. Backbone residual dipolar couplings collected under high-pressure conditions allowed us to determine the conformational ensemble of the isolated EIC monomeric state in solution. Our calculations reveal that three catalytic loops near the dimerization interface become unstructured upon monomerization, preventing the monomeric enzyme from binding its natural substrate. This study provides an atomic-level characterization of EI’s monomeric state and highlights the role of the catalytic loops as allosteric connectors controlling both the activity and oligomerization of the enzyme.
The carbohydrate phosphotransferase system (PTS) is a key signal transduction pathway controlling the central carbon metabolism and playing important roles in the regulation of several other cellular functions in bacteria [i.e., chemotaxis, biofilm formation, catabolic gene expression, potassium transport, and inducer exclusion (1–5)]. The PTS comprises a sequential cascade of protein–protein interactions whereby a phosphoryl group originating from phosphoenolpyruvate (PEP) is transferred onto incoming sugars, thereby coupling phosphoryl transfer to active sugar transport across the membrane. There are several sugar-specific branches of the PTS, but all require Enzyme I (EI) to initiate the phosphoryl transfer cascade (3). Binding of PEP to the C-terminal domain of EI (EIC) initiates the PTS by inducing a series of intra- and interdomain conformational rearrangements (6–13) that result in phosphorylation of EI N-terminal domain (EIN) and subsequent transfer of the phosphoryl group to the histidine phosphocarrier protein HPr. Thereafter, the phosphoryl group is transferred from HPr to the sugar-specific enzymes II and ultimately onto the incoming sugar. Interestingly, EI is ubiquitous and one of the best-conserved proteins in both Gram-positive and Gram-negative bacteria and does not share any significant sequence similarity with eukaryotic proteins, making EI a possible target for the development of wide-spectrum antimicrobials (14–19). In addition, owing to its central metabolic function and complex regulatory role, EI is a frequent objective of metabolic engineering efforts (20–24) and a model system for studying the flux of metabolites across different metabolic networks (25, 26).
The functional form of EI is a ∼128-kDa dimer of identical subunits comprising two structurally and functionally distinct domains (27). The EIN domain (residues 1 to 249) contains the site of phosphorylation (His189) and the binding site for HPr. The EIC domain (residues 261 to 575) is responsible for dimerization and contains the binding site for PEP. The EIN and EIC domains are connected by a long helical linker (Fig. 1A) (27). EI undergoes a series of large-scale conformational rearrangements during its catalytic cycle, including the following: 1) a monomer–dimer transition (6, 13), 2) an expanded-to-compact conformational change within EIC (7, 8), and 3) an open-to-close transition describing a reorientation of EIN relative to EIC (8, 9, 11, 12). Binding of PEP to EIC shifts the conformational equilibria toward the catalytically competent dimer/compact/close form and activates the enzyme for catalysis (6, 8). As both oligomeric states are present at physiological concentrations of EI (1 to 10 μM) (28), the monomer–dimer equilibrium of the enzyme has often been suggested to be a major regulatory element for PTS (6, 13). Despite the crucial role of the monomer–dimer equilibrium of EI in regulating bacterial metabolism, very little is known about the changes in structure, dynamics, and substrate-binding properties that EI undergoes upon monomerization. Therefore, understanding the structural changes upon monomerization would 1) reveal how the coupling among intersubunit conformational dynamics mediates allostery and cooperativity in a functionally dimeric protein, 2) provide new insight toward the engineering of new bacterial strains for microbial production of chemicals, and 3) indicate new routes for the discovery of antimicrobials that inhibit EI. Yet, at the high-protein concentration required by standard structural techniques such as X-ray crystallography or NMR spectroscopy, EI is almost exclusively dimeric (dissociation constant, Kd, < 5 μM) (6, 13), which makes the characterization of the monomeric conformation extremely challenging.
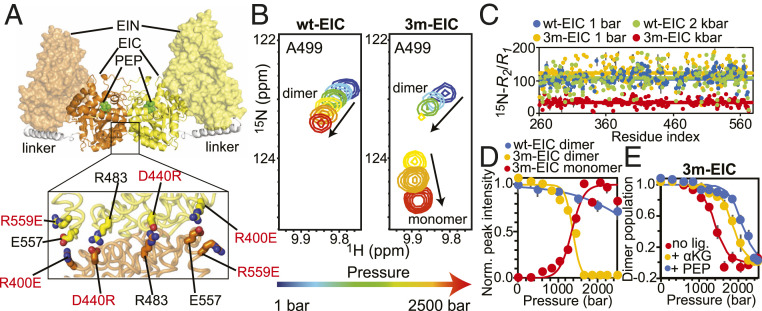
Fig. 1.
A folded EIC monomer at high pressure. (A) Solution structure of the full-length EI dimer. The two subunits are colored orange and yellow, respectively. The EIN and EIC domains are shown as transparent surface and solid cartoon, respectively. The helical linker is colored white. PEP modeled in the active site on EIC is shown as green spheres. In the lower panel, the three single-point mutations introduced at the dimer interface to engineer the 3m-EIC construct are highlighted. (B) Pressure titration for a representative 1H-15N NMR cross-peak that reports on the monomer/dimer equilibrium in wt-EIC (Left) and 3m-EIC (Right). The relationship between cross-peak color and pressure is given by the arrow below the spectra (i.e., blue: 1 bar; red: 2.5 kbar). (C) 15N-R2/R1 data measured for wt-EIC at 1 bar (blue), wt-EIC at 2 kbar (green), 3m-EIC at 1 bar (orange), and 3m-EIC at 2 kbar (red) are plotted versus the residue index. The average R2/R1 value of each construct is shown as a solid line. (D) Normalized signal intensities are averaged over all NMR cross-peaks displaying distinct signals for the monomer and dimer species during the pressure titration on 3m-EIC (i.e., B). The normalized averaged intensities for the wt-EIC dimer (blue circles), 3m-EIC dimer (orange circles), and 3m-EIC monomer (red circles) are plotted versus the external pressure (note that no NMR signal is observable for the wt-EIC monomer in the tested pressure range). The normalized averaged intensities were calculated using 54 peaks form the dimer interface. The error bars are set to 1 SD. The modeling of the data (SI Appendix, Supporting Materials and Methods) is shown as solid curves. Note that the dissociation of the 3m-EIC monomer at high pressure if fully reversible (SI Appendix, Figs. S3 and S4). (E) The fractional population of dimer obtained by pressure titration experiments on 3m-EIC in the absence of ligands (red circles) and in the presence of 20-mM α-KG (orange circles) or PEP (blue circles) is plotted versus the external pressure. The modeled curves (SI Appendix, Supporting Materials and Methods) are shown as solid lines.
Here, we combined protein engineering and hydrostatic pressure perturbation to destabilize the dimeric state of EIC and isolate its monomeric conformation at the high-protein concentrations required for solution NMR experiments. We chose pressure perturbation because hydrostatic pressure shifts the system toward a state that occupies the minimal achievable volume, which is reached by increased water density around charged and polar groups and penetration of water molecules in protein cavities (29–31). Therefore, using kbar pressures, it is feasible to finely tune protein–protein interactions and induce partial or total disassembly of protein oligomers (32, 33). Focusing on EIC, the C-terminal domain of EI responsible for dimerization (Fig. 1A), we first designed three single-point mutations at the dimerization interface of EIC. At atmospheric pressure, the engineered mutant was still able to form an active dimer with a dissociation constant, binding affinity to PEP, and enzymatic activity similar to that of the wild-type enzyme. In high-pressure conditions (≥2 kbar), we observed a complete dissociation of the variant into a stable and well-folded monomer. We then assigned the backbone chemical shift resonances of the monomeric state and obtained an ensemble representation of EIC monomeric conformations in solution by combining residual dipolar couplings with accelerated molecular dynamics (aMD) simulations. Our data show that three catalytic loops which are part of the dimer interface become more disordered in solution upon monomerization. As a result, the monomeric EIC cannot bind to PEP since those loops are too flexible to orient PEP efficiently toward the binding site. This study provides atomic-level characterization of the monomeric EIC conformation, reveals the structural basis for the inactivity of monomeric EI and highlights the ability of high-pressure perturbation to isolate low-population states that are critical for understanding complex biological pathways.
Results
A Stable and Well-Folded Monomer of EIC Can Be Isolated at High Pressure.
In this study, we focused on the EIC domain of EI from a thermophilic organism (Thermoanaerobacter tengcongensis), hereafter called wt-EIC. We chose the thermophilic EIC over its mesophilic homolog because it is more stable in solution and more resilient to structural modifications (34, 35). The full chemical shift assignment of thermophilic EIC is available, and besides differences in active site loop dynamics, its sequence and crystal structure are very close to those of the EIC from Escherichia coli (60% similarity; backbone rmsd = 1 Å) (34, 36, 37). We therefore think that wt-EIC is a good model for studying the dimer-to-monomer transition of EI. Inspection of wt-EIC dimerization interface revealed the presence of salt bridge interactions that stabilize the dimeric state of the enzyme. These include salt bridges between R400 and E557 and between D440 and R483 (Fig. 1A). To destabilize the dimerization interface, we mutated R400 into a glutamic acid residue (R400E) and D440 into an arginine residue (D440R). We further destabilized the interface by introducing a third mutation (R559E), a residue nearby the original salt bridge partner of D400. We first verified if our engineered mutant, henceforth called 3m-EIC, still associates as an active dimer at atmospheric pressure. Analytical ultracentrifugation (AUC) experiments (SI Appendix, Fig. S1 A and B) demonstrate that 3m-EIC associates as a dimer at atmospheric pressure with a Kd comparable to that of the wt-EIC, which is consistent with the observation that solvent-exposed ion pairs (like the salt bridges mutated here) have a modest stability at ambient pressure (38). In addition, enzymatic activity assays (SI Appendix, Fig. S1C) demonstrate that the three engineered mutations don’t affect the ability of the enzyme to bind and hydrolyze PEP. In this respect, it is important to emphasize that PEP hydrolysis catalyzed by EIC proceeds via the same mechanism as the phosphoryl transfer reaction catalyzed by the full-length EI (the only difference being a water molecule acting as the nucleophile instead of the H189 residue from the EIN domain) and that the enzymatic assays in SI Appendix, Fig. S1C are run at experimental conditions in which both wt-EIC and 3m-EIC are fully dimeric.
Next, we investigated the conformational changes experienced by wt-EIC and 3m-EIC under pressure by monitoring the spectral changes in two-dimensional (2D) 1H-15N transverse relaxation-optimized–heteronuclear single quantum coherence spectroscopy (TROSY-HSQC) experiments from 1 bar to 2.5 kbar. When pressurized, protein NMR spectra typically display two types of changes: 1) chemical shift changes and 2) peak intensity changes. The first type of pressure-induced perturbation informs on conformational changes on fast time scale (relative to NMR time scale) typically due to changes in protein surface–water interface and/or small compression of protein native conformations (39). On the other hand, the second type of perturbation (i.e., peak intensity changes) points to major conformational transitions on a slow time scale (e.g., changes in folded/unfolded state populations or monomer/dimer populations) (32, 33, 40). While both wt-EIC and 3m-EIC displayed chemical shift changes under pressure, significant peak intensity changes were only observed for 3m-EIC, suggesting that this variant undergoes major conformational transitions from 1 bar to 2.5 kbar (Fig. 1B). Since the 2D 1H-15N spectrum of 3m-EIC collected at 2 kbar shows no evidence of unfolding (SI Appendix, Fig. S2), the major peak intensity changes observed for this variant were interpreted as pressure-induced dissociation of the native dimer into monomers.
To confirm that our engineered variant is monomeric at 2 kbar, we investigated the picosecond to nanosecond time scale dynamics of wt-EIC and 3m-EIC by measuring the transverse (15N-R2) and longitudinal (15N-R1) relaxation rates at atmospheric and high-pressure conditions (Fig. 1C). The R2/R1 ratio can be used to estimate the protein rotational correlation time (τm) (41). We found that the average R2/R1 ratios measured for wt-EIC were not affected by pressure, with average values of 107 ± 7 and 102 ± 10 at 1 bar and 2 kbar respectively, which translate into a τm ∼25 ns that is close to the value predicted for the EIC dimer based on Stokes’ Law (∼29 ns). At atmospheric pressure, the engineered variant 3m-EIC displays average R2/R1 ratios comparable to that of wt-EIC (R2/R1 = 122 ± 30, for an estimated τm of 26 ns). Remarkably, the relaxation data measured for 3m-EIC at 2 kbar are drastically different from those measured at atmospheric pressure, with an average R2/R1 ratio of 33 ± 4 and a τm ∼13 ns that is consistent with the rotational correlation time predicted for the monomeric EIC (15 ns) (Fig. 1C). Altogether, these relaxation experiments confirm that the dimeric 3m-EIC dissociates under pressure into a stable monomeric conformation that can be isolated at 2 kbar.
Thermodynamics of EIC Dimer-To-Monomer Transition.
Using the collection of 2D 1H-15N TROSY-HSQC spectra collected from 1 bar to 2.5 kbar for wt-EIC and 3m-EIC, we globally fitted the average change of cross-peak intensity as a function of pressure with a simple two-state dimer-to-monomer model. As shown in Fig. 1D for the variant 3m-EIC, both the intensity of the native dimer cross-peaks (in orange) and intensity of new cross-peaks attributed to the monomeric conformation (in red) can be accurately fitted with our model. These results show that the midpoint of the dimer-to-monomer transition occurs for 3m-EIC at around 1.3 kbar and that at 2 kbar, the engineered variant is predominantly (>95%) monomeric. On the other hand, wt-EIC (in blue) is only moderately affected by pressure with at least 70% of the dimeric population subsisting at 2.5 kbar (Fig. 1D). We also compared the stability of 3m-EIC under pressure in the absence and in the presence of either PEP or α-ketoglutarate (α-KG). PEP is the natural substrate of EI in the PTS pathway, while the small-molecule metabolite α-KG acts as an allosteric stimulator or competitive inhibitor depending on the oligomeric state of the enzyme (6). Both PEP and α-KG have been shown to decrease the equilibrium dissociation constant for EIC dimerization by more than 10-fold (4). By fitting the changes of cross-peak intensities from a series of 2D 1H-15N TROSY-HSQC spectra collected from 1 bar to 2.5 kbar for apo 3m-EIC in the absence and the in presence of PEP or α-KG, we extracted the changes in dimerization free energy (ΔG), in partial volume (ΔV), and the extrapolated dimer dissociation constants at atmospheric pressure (Kd) (Fig. 1E and SI Appendix, Table S1).
We found that the Kd of the apo 3m-EIC is very small, about 0.1 nM, which is consistent with the Kd of ∼5 nM previously reported for the E. coli EIC (42) (the slight difference likely reflects the increased stability of the thermophilic EIC compared to its mesophilic homolog). The addition of PEP and α-KG leads to a decrease in Kd by 2 and 3 orders of magnitude, respectively, and a concomitant increase in ΔG by more than ∼1 and ∼2 kcal, respectively (SI Appendix, Table S1). These data confirm that the engineered mutations in 3m-EIC modify the network of interactions at the dimerization interface, rendering it more susceptible to pressure perturbation without drastically affecting its dissociation constant at atmospheric pressure. These results also highlight that the allosteric coupling between the substrate-binding site and the dimerization interface, which is a hallmark of EI activity (6), is still effective in 3m-EIC, as evidenced by the dimer stabilization upon addition of PEP and α-KG.
Three Disordered Catalytic Loops Prevent PEP from Binding to EIC in the Monomeric State.
To characterize structural changes associated with the dimer-to-monomer transition of 3m-EIC, we collected backbone amide 1DNH residual dipolar coupling (RDC) data at 1 bar and 2 kbar using Pf1 phage as an alignment medium. Singular value decomposition (SVD) fitting of the 1 bar 1DNH RDCs from well-defined secondary structures to the X-ray coordinates of a single subunit and of the full dimer of wt-EIC (PDB: 2XZ7) (35) yielded R-factors of 27 and 28%, respectively (SI Appendix, Fig. S5, Lower Left). These good correlations between experimental and back-calculated RDC data indicate that the tertiary and quaternary structures of 3m-EIC at 1 bar are similar to those observed in the X-ray structure of wt-EIC. On the other hand, the 1DNH RDCs measured at 2 kbar are in poor agreement with the crystal structure of wt-EIC (R-factor, ∼57%) (SI Appendix, Fig. S5, Lower Right), indicating that monomerization of 3m-EIC upon increasing pressure is coupled to a change in the tertiary fold. As a control, we also measured a set of 1DNH RDCs in the same alignment medium for wt-EIC at 1 bar and 2 kbar. For both datasets, the experimental RDCs from well-defined secondary structures are in good agreement with the ones calculated from the reference X-ray structure (R-factors of 25 and 23%, respectively) (SI Appendix, Fig. S5, Upper), confirming that the tertiary and quaternary folds of wt-EIC are not affected by pressure.
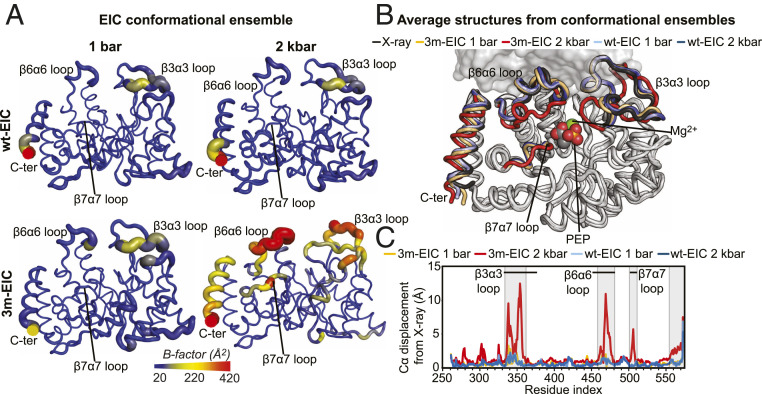
In order to visualize the changes in structure and dynamics that EIC undergoes upon monomerization, we have calculated structural ensembles for wt-EIC and 3m-EIC at 1 bar and 2 kbar by coupling the experimental 1DNH RDC data with aMD simulations. This combined aMD/NMR protocol has been proven successful in generating structural ensembles of dynamical proteins that satisfy solution NMR data (43). Ensembles with 7 and 15 members were necessary to fulfill the complete set (including flexible structural regions) of RDC data measured for wt-EIC at 1 bar (R-factor, ∼23%) and 2 kbar (R-factor, ∼27%), respectively (SI Appendix, Fig. S6, Upper). Similarly, the data measured for 3m-EIC at low and high pressures were satisfied by an 8- (R-factor, ∼27%) and 20-member (R-factor, ∼25%) ensemble, respectively (SI Appendix, Fig. S6, Lower). The modeled structural ensembles are visualized in Fig. 2A. The average structures from each ensemble are compared to the reference X-ray structure of EIC in Fig. 2B. Analysis of the B-factors calculated from the conformational ensembles reveals that dimeric EIC (wt-EIC at 1 bar and 2 kbar and 3m-EIC at 1 bar) is more rigid than monomeric EIC (3m-EIC at 2 kbar) (Fig. 2A). Interestingly, the more pronounced changes in flexibility upon monomerization occur at the C-terminal helix and at the loops comprising the protein-active site (Fig. 2B). These same regions are also the ones displaying the largest conformational changes upon monomerization. Indeed, while the average structures calculated for the wt-EIC at 1 bar, wt-EIC at 2 kbar, and 3m-EIC at 1 bar ensembles are indistinguishable from the X-ray structure of dimeric wt-EIC (Fig. 2C), the average structure obtained for the 3m-EIC ensemble at 2 kbar indicates that formation of monomeric EIC at high pressure is coupled to a conformational transition involving the active site loops α3β3, α6β6, and α7β7 (Fig. 2C). These results are consistent with the changes in C-α chemical shift (ΔC-α) observed in 3m-EIC upon monomerization. Indeed, large ΔC-α values are only observed in the vicinity of the dimer interface and at the active site loops (SI Appendix, Fig. S7), indicating that the structural changes associated with the monomer–dimer equilibrium are highly localized.
Fig. 2.
Monomerization affects the structure and dynamics of the EIC active site. (A) Sausage representation of the aMD/RDC conformational ensembles generated for wt-EIC at 1 bar (Top Left), wt-EIC at 2 kbar (Top Right), 3m-EIC at 1 bar (Bottom Left), and 3m-EIC and 2 kbar (Bottom Right). The cartoons are colored according to the B-factor, as indicated by the color bar. B-factors were calculated using the formula , where Bi and Ui are the B-factor and mean-square displacement of atom i, respectively. (B) The average structures from the conformational ensembles are superimposed on the crystal structure of wt-EIC. The ensembles were calculated for a single subunit of the EIC dimer (SI Appendix, Supporting Materials and Methods). The second subunit from the crystal structure is displayed as a transparent gray surface as a reference. PEP and Mg2+ (not used in the ensemble calculation) are shown as spheres in the EIC active site. The C-terminal helix and the α3β3, α6β6, and α7β7 loops are colored black, light blue, dark blue, orange, and red for the X-ray structure, wt-EIC at 1 bar, wt-EIC at 2 kbar, 3m-EIC at 1 bar, and 3m-EIC at 2 kbar, respectively. (C) The C-α displacement from the X-ray structure calculated for the average structure of wt-EIC at 1 bar (light blue), wt-EIC at 2 kbar (dark blue), 3m-EIC at 1 bar (orange), and 3m-EIC at 2 kbar (red) is plotted versus the residue index. The residues experiencing large C-α displacement upon monomerization are boxed. The location on the α3β3, α6β6, and α7β7 is indicated by black horizontal lines.
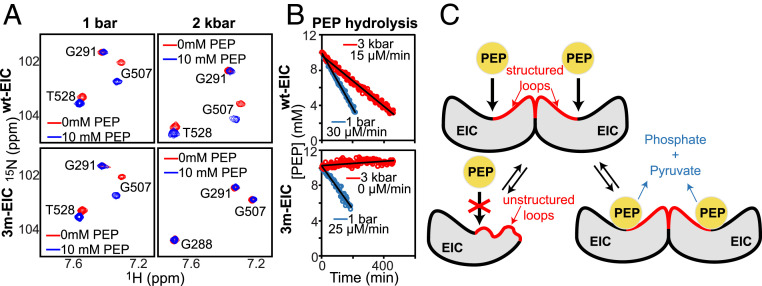
Altogether, these calculations reveal that while the dimer-to-monomer transition of 3m-EIC preserves the overall fold of the monomeric subunit in EIC dimer, it is accompanied by significant changes in local structure and dynamics at the PEP binding site. In particular, the partial unfolding of the catalytic loops α3β3, α6β6, and α7β7 (Fig. 2) suggests that the monomeric EIC cannot bind PEP because those loops are too flexible to orient PEP efficiently toward the binding site. To verify this prediction, we compared the 2D 1H-15N TROSY-HSQC spectra acquired for 3m-EIC at 1 bar and 2 kbar in the absence and in the presence of 10 mM PEP. As expected, we observed a significant chemical shift perturbation at atmospheric pressure for the residues located at the binding sites (Fig. 3A). On the other hand, no chemical shift perturbation was observed for 3m-EIC at 2 kbar (Fig. 3A), indicating that EIC cannot bind to its natural substrate in the monomeric state. Consistent with these data, we found that wt-EIC can still hydrolyze PEP in high-pressure conditions, while 3m-EIC is completely inactive at high pressure (Fig. 3B), confirming that EIC is unable to bind and subsequently degrade PEP in the monomeric state.
Fig. 3.
Monomeric EIC does not bind nor hydrolyze PEP. (A) A selected region of the 1H-15N TROSY spectrum of wt-EIC at 1bar (Top Left), wt-EIC at 2 kbar (Top Right), 3m-EIC at 1 bar (Bottom Left), and 3m-EIC at 2 kbar (Bottom Right) acquired in the absence (red) and in the presence (blue) of 10-mM PEP is shown. Note that monomerization of 3m-EIC at 2 kbar generates large shifts for T528 and G288 cross-peaks. (B) Rate of the PEP hydrolysis reaction catalyzed by 150-μM wt-EIC (Top) and 3m-EIC (Bottom) at 65 °C and an external pressure of 1 bar (blue) and 3 kbar (red). A pressure of 3 kbar was used to ensure that the monomer–dimer equilibrium of 3m-EIC is fully shifted toward the monomeric species. (C) Schematic representation of the effect of monomerization on the structure and biological activity of EIC. The catalytic loops α3β3, α6β6, and α7β7, which form a large portion of the dimer interface, are largely disordered in monomeric EIC and become ordered upon dimerization.
Discussion
Sparsely populated states involved in conformational selection and enzyme catalysis play a crucial role in many biological pathways, but the low population and transient nature of these states make them invisible to most structural biology techniques (44). Solution NMR spectroscopy has emerged as a method of choice for characterizing low-lying states at an atomic level, with multiple experiments dedicated to the indirect detection and characterization of transient intermediate states (39, 45–49). In this work, we focused on the monomeric intermediate state of EI, the enzyme responsible for initiating the phosphoryl transfer cascade in the PTS pathway. The high dimer affinity of EI (Kd < 5 μM) renders the structural characterization of the monomeric state extremely challenging. Yet, obtaining an atomic-level description of this intermediate state is crucial to fully understand the molecular mechanisms underlying EI catalytic activity and regulation. Here, we combined protein engineering and pressure perturbation to shift the dimer–monomer equilibrium of EIC in order to isolate and determine the structural ensemble of the monomeric state in solution. We found conditions at which EIC is predominantly monomeric (>95%), and we were therefore able to directly measure backbone 1DNH RDCs of EIC monomeric state without relying on indirect detection.
The three mutations engineered in 3m-EIC (R400E, D440R, and R559) were designed to change the balance between the electrostatic and hydrophobic interactions at the dimerization interface to make it more sensitive to pressure perturbation. AUC (SI Appendix, Fig. S1 A and B), pressure titration experiments (Fig. 1D), and enzyme kinetic assays (SI Appendix, Fig. S1C) show that the mutations did not affect the ability of 3m-EIC to form an active dimer at atmospheric pressure. The calculated dimerization Kd is very low (∼0.1 nM), which is comparable to the Kd previously reported for the mesophilic EIC (42). We found that the volume change upon monomerization is rather large (ΔV = −153 ± 2 mL/mol), which corresponds to the van der Waals volume of about 12 water molecules. The magnitude of ΔV likely reflects the preferential hydration of polar and charged side chains exposed upon monomerization as well as the potential penetration of water molecules in small cavities near the dimerization interface (29, 33, 50). Remarkably, the pressure titration experiments revealed that upon binding, PEP stabilizes the dimeric state by +2.3 kcal/mol relative to the apo 3m-EIC, which is significantly greater than the effect of α-KG (Fig. 1E and SI Appendix, Table S1). PEP and α-KG bind to the same site in EIC, and the differences between the binding modes of these natural substrates have been explored by solution NMR experiments and docking simulations (4), but the actual differences in terms of dimerization thermodynamics have yet never been described. Our high-pressure NMR experiments provide here quantitative thermodynamic characterization of the allosteric coupling between substrate binding and dimerization. These results show that PEP stabilizes EIC dimeric state significantly more than α-KG does, likely due to the additional interactions between PEP’s phosphate group with the side chains of R296, R332, and K340 (4, 34).
Application of hydrostatic pressure from 1 bar to 2.5 kbar shifts the equilibrium toward the monomeric state of EIC while preserving the overall tertiary structure of the protein (Fig. 1D and SI Appendix, Fig. S2). We found that 3m-EIC becomes fully monomeric at pressure ≥2 kbar, which allowed us to directly determine the structural features of EIC monomeric state by measuring 1DNH RDCs in high-pressure conditions. Comparison of the experimental 1DNH RDCs with the RDCs predicted for a reference X-ray structure demonstrates that the dimer-to-monomer transition of 3m-EIC is accompanied by significant structural changes (SI Appendix, Fig. S5). With the help of all-atom accelerated molecular dynamics simulations, we were then able to generate a 20-member conformational ensemble of the monomeric 3m-EIC that would satisfy the experimental 1DNH RDCs (SI Appendix, Fig. S6). Examination of the conformational ensemble and average structure revealed that while the overall tertiary structure is preserved, three catalytic loops become significantly disordered in the monomeric state (Fig. 2). Indeed, the loops α3β3, α6β6, and to a lesser extent, α7β7, show large structural deviations from the reference X-ray structure, indicative of partial unfolding upon monomerization (Fig. 2C). Since EIC is inactive and unable to bind PEP in the monomeric state (Fig. 3 A and B), these results provide a mechanistic framework explaining why dimerization is strictly required for EI to be active. The catalytic loops α3β3, α6β6, and α7β7 seem to play a key role in establishing the allosteric coupling between the active site and the dimerization interface. Overall, our experiments suggest that dimerization is required for these catalytic loops to be fully structured, which in turn allows the physiological ligands PEP and α-KG to bind to the EI active site (Fig. 3C).
Materials and Methods
wt-EIC was cloned into a pET21a vector incorporating a His-tagged EIN-solubility tag at the N terminus (51). The R400E, D440R, and R559E mutations were introduced using QuikChange II Site-Directed Mutagenesis Kit (Agilent), and the results were verified by DNA sequencing. Expression and purification of uniformly 15N-, 2H/15N-, and 2H/15N/13C-labeled wt-EIC and 3m-EIC were performed as previously described (37). All NMR spectra were acquired at 40 °C on Bruker 600-, 700-, and 800-MHz spectrometers equipped with Z-shielded gradient triple resonance cryoprobes. Hydrostatic pressure in the sample was controlled using a commercial ceramic high-pressure NMR cell and an automatic pump system (Daedalus Innovations). 15N-R1 and 15N-R1ρ experiments for 15N-labeled wt- and 3m-EIC were carried using heat-compensated pulse schemes with a TROSY readout (52). Backbone amide 1DNH RDCs were measured for on 2H,15N-labeled samples of wt- and 3m-EIC at the 700 MHz by taking the difference in 1JNH scalar couplings in aligned and isotropic media. The alignment media employed was phage pf1 (16 mg/mL; ASLA Biotech) (53), and 1JNH couplings were measured using the amide RDCs by TROSY (ARTSY) pulse scheme (54). SVD analysis of RDCs was carried out using Xplor-NIH (55). aMD simulations were run using AMBER 16 (56). For details, see SI Appendix, Supporting Materials and Methods.
Supplementary Material
Acknowledgments
This work was supported by funds from the National Institute of General Medical Sciences R35GM133488 (to V.V.), from the Roy J. Carver Charitable Trust (to V.V.), and by the Intramural Research Program of the NIH, the National Institute of Diabetes and Digestive and Kidney Disease (to R.G.). J.R. acknowledges the Iowa State University College of Liberal Arts and Sciences and the Roy J. Carver Charitable Trust for their support.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2100298118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or SI Appendix.
References
- 1.Deutscher J., et al., The bacterial phosphoenolpyruvate:carbohydrate phosphotransferase system: Regulation by protein phosphorylation and phosphorylation-dependent protein-protein interactions. Microbiol. Mol. Biol. Rev. 78, 231–256 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Postma P. W., Lengeler J. W., Jacobson G. R., Phosphoenolpyruvate:carbohydrate phosphotransferase systems of bacteria. Microbiol. Rev. 57, 543–594 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clore G. M., Venditti V., Structure, dynamics and biophysics of the cytoplasmic protein-protein complexes of the bacterial phosphoenolpyruvate: Sugar phosphotransferase system. Trends Biochem. Sci. 38, 515–530 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Venditti V., Ghirlando R., Clore G. M., Structural basis for enzyme I inhibition by α-ketoglutarate. ACS Chem. Biol. 8, 1232–1240 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Doucette C. D., Schwab D. J., Wingreen N. S., Rabinowitz J. D., α-Ketoglutarate coordinates carbon and nitrogen utilization via enzyme I inhibition. Nat. Chem. Biol. 7, 894–901 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nguyen T. T., Ghirlando R., Venditti V., The oligomerization state of bacterial enzyme I (EI) determines EI’s allosteric stimulation or competitive inhibition by α-ketoglutarate. J. Biol. Chem. 293, 2631–2639 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Venditti V., Clore G. M., Conformational selection and substrate binding regulate the monomer/dimer equilibrium of the C-terminal domain of Escherichia coli enzyme I. J. Biol. Chem. 287, 26989–26998 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Venditti V., Tugarinov V., Schwieters C. D., Grishaev A., Clore G. M., Large interdomain rearrangement triggered by suppression of micro- to millisecond dynamics in bacterial Enzyme I. Nat. Commun. 6, 5960 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Venditti V., Schwieters C. D., Grishaev A., Clore G. M., Dynamic equilibrium between closed and partially closed states of the bacterial Enzyme I unveiled by solution NMR and X-ray scattering. Proc. Natl. Acad. Sci. U.S.A. 112, 11565–11570 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Suh J.-Y., Cai M., Clore G. M., Impact of phosphorylation on structure and thermodynamics of the interaction between the N-terminal domain of enzyme I and the histidine phosphocarrier protein of the bacterial phosphotransferase system. J. Biol. Chem. 283, 18980–18989 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Teplyakov A., et al., Structure of phosphorylated enzyme I, the phosphoenolpyruvate:sugar phosphotransferase system sugar translocation signal protein. Proc. Natl. Acad. Sci. U.S.A. 103, 16218–16223 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schwieters C. D., et al., Solution structure of the 128 kDa enzyme I dimer from Escherichia coli and its 146 kDa complex with HPr using residual dipolar couplings and small- and wide-angle X-ray scattering. J. Am. Chem. Soc. 132, 13026–13045 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Patel H. V., Vyas K. A., Savtchenko R., Roseman S., The monomer/dimer transition of enzyme I of the Escherichia coli phosphotransferase system. J. Biol. Chem. 281, 17570–17578 (2006). [DOI] [PubMed] [Google Scholar]
- 14.Kok M., Bron G., Erni B., Mukhija S., Effect of enzyme I of the bacterial phosphoenolpyruvate: Sugar phosphotransferase system (PTS) on virulence in a murine model. Microbiology (Reading) 149, 2645–2652 (2003). [DOI] [PubMed] [Google Scholar]
- 15.Edelstein P. H., Edelstein M. A., Higa F., Falkow S., Discovery of virulence genes of Legionella pneumophila by using signature tagged mutagenesis in a guinea pig pneumonia model. Proc. Natl. Acad. Sci. U.S.A. 96, 8190–8195 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hava D. L., Camilli A., Large-scale identification of serotype 4 Streptococcus pneumoniae virulence factors. Mol. Microbiol. 45, 1389–1406 (2002). [PMC free article] [PubMed] [Google Scholar]
- 17.Nguyen T. T., Venditti V., An allosteric pocket for inhibition of bacterial Enzyme I identified by NMR-based fragment screening. J Struct Biol X 4, 100034 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jones A. L., Knoll K. M., Rubens C. E., Identification of Streptococcus agalactiae virulence genes in the neonatal rat sepsis model using signature-tagged mutagenesis. Mol. Microbiol. 37, 1444–1455 (2000). [DOI] [PubMed] [Google Scholar]
- 19.Lau G. W., et al., A functional genomic analysis of type 3 Streptococcus pneumoniae virulence. Mol. Microbiol. 40, 555–571 (2001). [DOI] [PubMed] [Google Scholar]
- 20.Chubukov V., et al., Engineering glucose metabolism of Escherichia coli under nitrogen starvation. npj Syst. Biol. Appl. 3, 16035 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yan Q., Fong S. S., Increasing carbon source uptake rates to improve chemical productivity in metabolic engineering. Curr. Opin. Biotechnol. 53, 254–263 (2018). [DOI] [PubMed] [Google Scholar]
- 22.Gosset G., Improvement of Escherichia coli production strains by modification of the phosphoenolpyruvate:sugar phosphotransferase system. Microb. Cell Fact. 4, 14 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Flores N., Xiao J., Berry A., Bolivar F., Valle F., Pathway engineering for the production of aromatic compounds in Escherichia coli. Nat. Biotechnol. 14, 620–623 (1996). [DOI] [PubMed] [Google Scholar]
- 24.Zampieri M., Hörl M., Hotz F., Müller N. F., Sauer U., Regulatory mechanisms underlying coordination of amino acid and glucose catabolism in Escherichia coli. Nat. Commun. 10, 3354 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Long C. P., Au J., Sandoval N. R., Gebreselassie N. A., Antoniewicz M. R., Enzyme I facilitates reverse flux from pyruvate to phosphoenolpyruvate in Escherichia coli. Nat. Commun. 8, 14316 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shimizu K., Matsuoka Y., Regulation of glycolytic flux and overflow metabolism depending on the source of energy generation for energy demand. Biotechnol. Adv. 37, 284–305 (2019). [DOI] [PubMed] [Google Scholar]
- 27.Chauvin F., Brand L., Roseman S., Enzyme I: The first protein and potential regulator of the bacterial phosphoenolpyruvate: Glycose phosphotransferase system. Res. Microbiol. 147, 471–479 (1996). [DOI] [PubMed] [Google Scholar]
- 28.Mattoo R. L., Waygood E. B., Determination of the levels of HPr and enzyme I of the phosphoenolpyruvate-sugar phosphotransferase system in Escherichia coli and Salmonella typhimurium. Can. J. Biochem. Cell Biol. 61, 29–37 (1983). [DOI] [PubMed] [Google Scholar]
- 29.Roche J., et al., Cavities determine the pressure unfolding of proteins. Proc. Natl. Acad. Sci. U.S.A. 109, 6945–6950 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grigera J. R., McCarthy A. N., The behavior of the hydrophobic effect under pressure and protein denaturation. Biophys. J. 98, 1626–1631 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chalikian T. V., Macgregor R. B. Jr, Origins of pressure-induced protein transitions. J. Mol. Biol. 394, 834–842 (2009). [DOI] [PubMed] [Google Scholar]
- 32.Alderson T. R., et al., Local unfolding of the HSP27 monomer regulates chaperone activity. Nat. Commun. 10, 1068 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Louis J. M., Roche J., Evolution under drug pressure remodels the folding free-energy landscape of mature HIV-1 protease. J. Mol. Biol. 428, 2780–2792 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dotas R. R., et al., Hybrid thermophilic/mesophilic enzymes reveal a role for conformational disorder in regulation of bacterial enzyme I. J. Mol. Biol. 432, 4481–4498 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Navdaeva V., et al., Phosphoenolpyruvate: Sugar phosphotransferase system from the hyperthermophilic thermoanaerobacter tengcongensis. Biochemistry 50, 1184–1193 (2011). [DOI] [PubMed] [Google Scholar]
- 36.Oberholzer A. E., et al., Crystal structure of the phosphoenolpyruvate-binding enzyme I-domain from the thermoanaerobacter tengcongensis PEP: Sugar phosphotransferase system (PTS). J. Mol. Biol. 346, 521–532 (2005). [DOI] [PubMed] [Google Scholar]
- 37.Dotas R. R., Venditti V., Resonance assignment of the 128 kDa enzyme I dimer from Thermoanaerobacter tengcongensis. Biomol. NMR Assign. 13, 287–293 (2019). [DOI] [PubMed] [Google Scholar]
- 38.Urbauer J. L., Ehrhardt M. R., Bieber R. J., Flynn P. F., Wand A. J., High-resolution triple-resonance NMR spectroscopy of a novel calmodulin peptide complex at kilobar pressures. J. Am. Chem. Soc. 118, 11329–11330 (1996). [Google Scholar]
- 39.Kitahara R., Hata K., Li H., Williamson M. P., Akasaka K., Pressure-induced chemical shifts as probes for conformational fluctuations in proteins. Prog. Nucl. Magn. Reson. Spectrosc. 71, 35–58 (2013). [DOI] [PubMed] [Google Scholar]
- 40.Roche J., et al., Effect of internal cavities on folding rates and routes revealed by real-time pressure-jump NMR spectroscopy. J. Am. Chem. Soc. 135, 14610–14618 (2013). [DOI] [PubMed] [Google Scholar]
- 41.Kay L. E., Torchia D. A., Bax A., Backbone dynamics of proteins as studied by 15N inverse detected heteronuclear NMR spectroscopy: Application to staphylococcal nuclease. Biochemistry 28, 8972–8979 (1989). [DOI] [PubMed] [Google Scholar]
- 42.Patel H. V., et al., Properties of the C-terminal domain of enzyme I of the Escherichia coli phosphotransferase system. J. Biol. Chem. 281, 17579–17587 (2006). [DOI] [PubMed] [Google Scholar]
- 43.Purslow J. A., et al., Active site breathing of human Alkbh5 revealed by solution NMR and accelerated molecular dynamics. Biophys. J. 115, 1895–1905 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Roche J., Royer C. A., Lessons from pressure denaturation of proteins. J. R. Soc. Interface 15, 20180244 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Roche J., Royer C. A., Roumestand C., “Exploring protein conformational landscapes using high-pressure NMR” in Methods in Enzymology, Wand A. J., Ed. (Academic Press, 2019), vol. 614, pp. 293–320. [DOI] [PubMed] [Google Scholar]
- 46.Nguyen L. M., Roche J., High-pressure NMR techniques for the study of protein dynamics, folding and aggregation. J. Magn. Reson. 277, 179–185 (2017). [DOI] [PubMed] [Google Scholar]
- 47.Caro J. A., Wand A. J., Practical aspects of high-pressure NMR spectroscopy and its applications in protein biophysics and structural biology. Methods 148, 67–80 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Roche J., High-pressure NMR of biological systems in solution. eMagRes 8, 121–126 (2019). [Google Scholar]
- 49.Venditti V., Egner T. K., Clore G. M., Hybrid approaches to structural characterization of conformational ensembles of complex macromolecular systems combining NMR residual dipolar couplings and solution X-ray scattering. Chem. Rev. 116, 6305–6322 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rouget J.-B., et al., Size and sequence and the volume change of protein folding. J. Am. Chem. Soc. 133, 6020–6027 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Khatiwada B., Purslow J. A., Underbakke E. S., Venditti V., N-terminal fusion of the N-terminal domain of bacterial enzyme I facilitates recombinant expression and purification of the human RNA demethylases FTO and Alkbh5. Protein Expr. Purif. 167, 105540 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lakomek N.-A., Ying J., Bax A., Measurement of 15N relaxation rates in perdeuterated proteins by TROSY-based methods. J. Biomol. NMR 53, 209–221 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Clore G. M., Starich M. R., Gronenborn A. M., Measurement of residual dipolar couplings of macromolecules aligned in the nematic phase of a colloidal suspension of rod-shaped viruses. J. Am. Chem. Soc. 120, 10571–10572 (1998). [Google Scholar]
- 54.Fitzkee N. C., Bax A., Facile measurement of 1H-15N residual dipolar couplings in larger perdeuterated proteins. J. Biomol. NMR 48, 65–70 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schwieters C. D., Kuszewski J. J., Tjandra N., Clore G. M., The Xplor-NIH NMR molecular structure determination package. J. Magn. Reson. 160, 65–73 (2003). [DOI] [PubMed] [Google Scholar]
- 56.Case D. A., et al., The Amber biomolecular simulation programs. J. Comput. Chem. 26, 1668–1688 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or SI Appendix.