Significance
Green/red CBCRs are one of the most important CBCR subfamilies owing to their physiological roles in cyanobacteria phylum and optogenetic applications. They are known to utilize a change in the bilin protonation state to cause the marked change in green/red absorption, but the structural bases of the protochromic photocycle are not well understood. Here, we have determined the crystal structure of the chromatic acclimation sensor RcaE of this subfamily in the Pr photoproduct state, identifying a unique conformation of the bilin and its interacting residues. In addition, we provide direct evidence of the protonation state of the bilin via NMR analysis. These findings bring insight to our understanding of the molecular mechanisms underlying the spectral diversity of CBCRs.
Keywords: cyanobacteriochrome, phytochrome, NMR, crystallography
Abstract
Cyanobacteriochromes (CBCRs) are bilin-binding photosensors of the phytochrome superfamily that show remarkable spectral diversity. The green/red CBCR subfamily is important for regulating chromatic acclimation of photosynthetic antenna in cyanobacteria and is applied for optogenetic control of gene expression in synthetic biology. It is suggested that the absorption change of this subfamily is caused by the bilin C15-Z/C15-E photoisomerization and a subsequent change in the bilin protonation state. However, structural information and direct evidence of the bilin protonation state are lacking. Here, we report a high-resolution (1.63Å) crystal structure of the bilin-binding domain of the chromatic acclimation sensor RcaE in the red-absorbing photoproduct state. The bilin is buried within a “bucket” consisting of hydrophobic residues, in which the bilin configuration/conformation is C5-Z,syn/C10-Z,syn/C15-E,syn with the A- through C-rings coplanar and the D-ring tilted. Three pyrrole nitrogens of the A- through C-rings are covered in the α-face with a hydrophobic lid of Leu249 influencing the bilin pKa, whereas they are directly hydrogen bonded in the β-face with the carboxyl group of Glu217. Glu217 is further connected to a cluster of waters forming a hole in the bucket, which are in exchange with solvent waters in molecular dynamics simulation. We propose that the “leaky bucket” structure functions as a proton exit/influx pathway upon photoconversion. NMR analysis demonstrated that the four pyrrole nitrogen atoms are indeed fully protonated in the red-absorbing state, but one of them, most likely the B-ring nitrogen, is deprotonated in the green-absorbing state. These findings deepen our understanding of the diverse spectral tuning mechanisms present in CBCRs.
Phytochromes and cyanobacteriochromes (CBCRs) are members of the phytochrome superfamily of photosensors. They utilize a cysteine-linked linear tetrapyrrole (bilin) chromophore for photo-perception that is bound within a GAF (cGMP phosphodiesterase/adenylyl cyclase/FhlA) domain (1–6). They undergo reversible photoconversion between two distinct light-absorbing states triggered by C15-Z/C15-E photoisomerization of the bilin chromophore (7, 8). Phytochromes are distributed in higher plants, algae, bacteria, and fungi; they typically photoconvert between a red-absorbing C15-Z,anti dark state and a far-red light-absorbing C15-E,anti photoproduct state, although recent studies have revealed substantial spectral diversity in some algal and cyanobacterial phytochromes (9, 10). CBCRs are widely distributed among the cyanobacteria phylum and show marked variation in their absorbing wavelength, spanning the near-ultraviolet to the far-red part of the spectrum (reviewed recently in refs. 4–6). They require only a GAF domain to complete the photocycle, in contrast to phytochromes, which require an extra PHY (phytochrome) domain and, in many cases, a PAS (Per/Arnt/Sim) domain to complete their photocycle (11, 12). The GAF domains of CBCRs have been classified into subfamilies based on the configurations of their dark- and photoproduct-state chromophores, designated by their respective light-absorbing maxima (e.g., blue/green, green/red, etc. subfamilies).
To date, several chemical reactions are known to create the spectral diversity of the CBCR subfamilies, most of which use the chromophore precursor, phycocyanobilin (PCB). The “two-Cys photocycle” mechanism was first proposed in the blue/green subfamily by Rockwell et al. (13), and this mechanism appeared widespread among other violet- and blue-absorbing CBCR subfamilies (14). As confirmed by structural studies, the thiol group of a second Cys residue forms a thioether linkage with the C10 atom, disconnecting bilin conjugation between the B- and C-rings to cause a blue shift in absorption (15, 16). Some CBCR members undergoing two-Cys photocycle isomerize their PCB chromophore to phycoviolobilin (17), which disconnects the π-conjugated system between the A- and B-rings and leads to spectral blue shift in their Cys-free photostates. Subsequently, the “protochromic photocycle” was identified in the green/red subfamily in our previous study (18). In this mechanism, a change in the protonation state of the bilin pyrrole system is responsible for the change in absorption. The deprotonation-induced formation of green-absorbing state has also been observed as an intermediate in the two-Cys photocycle (19). The red/green subfamily harbors a fully protonated chromophore in both photostates, but a twisted conformation of the A- and D-rings is responsible for the blue-shifted photoproduct (8, 20–23); this is known as the “trapped-twist photocycle” (24). Recently, Bandara et al. reported the structure of a far-red–absorbing state of a CBCR that adopts an atypical C15-Z,syn configuration (25). They proposed a model that its far-red absorption maximum arises from a cationic bilin tautomer in which the A-ring is formally di-protonated while the B-ring remains deprotonated.
Cyanobacteria harbor up to several dozen different bilin-binding GAF domains (26), and a single CBCR protein often contains multiple GAF domains (27), suggesting that cyanobacteria utilize a highly complex photosensing system. Although some CBCRs have been shown to regulate the composition of photosynthetic antenna (4, 28–31), cell morphology (32), phototaxis (33), and cell aggregation (34, 35), the physiological role of most CBCRs remains unknown. RcaE and CcaS are members of the green/red CBCR subfamily and optimize the light absorption maxima of the photosynthetic antenna complex phycobilisome via transcriptional regulation of its components in a process called “chromatic acclimation.” RcaE controls the expression of phycobilisome genes via phosphorylation of RcaF and RcaC under red light (4, 18, 27, 28, 36, 37), whereas CcaS controls fewer genes via phosphorylation of the transcriptional factor CcaR under green light (38, 39). The CcaSR system has been applied as one of the most popular photoswitch systems of gene expression in synthetic biology (40–42). RcaE and/or CcaS orthologs are found in ∼15% of all sequenced cyanobacteria genomes, suggesting that they have a physiological impact on survival (43).
Based on pH titration experiments, we previously demonstrated that the bilin-binding GAF domain of RcaE exhibits a protochromic photocycle consisting of a deprotonated C15-Z Pg dark state and a protonated C15-E Pr photoproduct state (18). In this photocycle, green or red absorption is determined by the protonation state of bilin, with C15-Z/C15-E photoisomerization responsible for tuning its pKa (18). Site-directed mutagenesis of RcaE identified the protochromic triad residues, Glu217, Leu249, and Lys261, required for the marked shift in the bilin pKa (18). Resonance Raman spectroscopy of the C15-Z Pg dark state of RcaE (44) was highly consistent with the B-ring–deprotonated bilin system that adopts a C15-Z,anti structure based upon quantum mechanics/molecular mechanics (QM/MM) calculations using the structure of C15-Z,anti Pr dark state of AnPixJ as template (45). However, structural information and direct evidence of the chemical environments of bilin in the green/red CBCR subfamily have not yet been reported, preventing deeper understanding of the molecular basis of the protochromic photocycle.
In this study, we report the crystal structure of the RcaE GAF domain in its Pr photoproduct state, in which its PCB chromophore adopts the C15-E,syn structure never seen previously. We also discovered that PCB is connected to a unique water cluster via hydrogen bond network via Glu217, which could function as a proton exit/influx pathway upon photoconversion. NMR analysis demonstrated that all four pyrrole nitrogen atoms of the bilin are indeed fully protonated in the Pr photoproduct state, whereas one is deprotonated in the Pg state. These findings provide insight into our understanding of the diverse spectral tuning mechanisms in CBCRs.
Results
Crystal Structure of the GAF Domain.
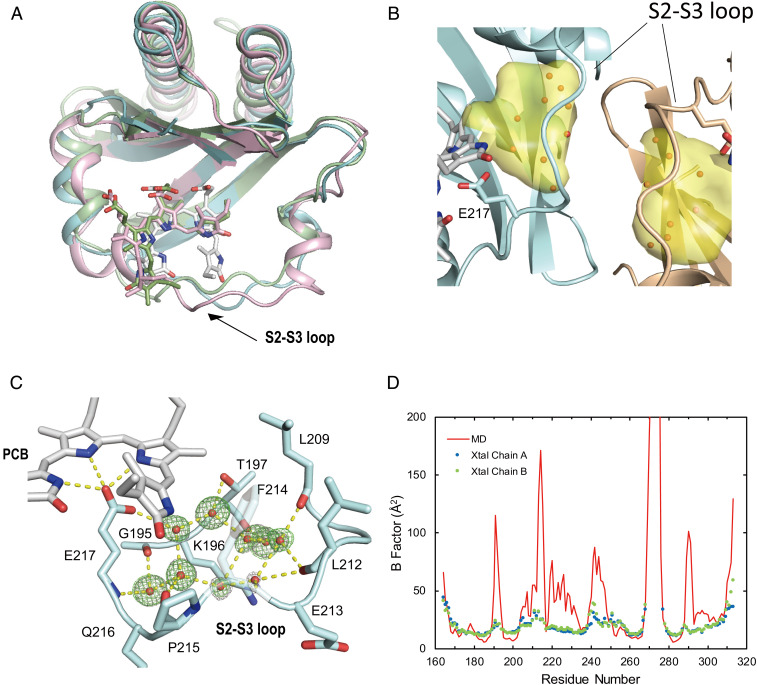
The GAF domain of RcaE was recombinantly expressed in PCB-producing Escherichia coli and purified by Ni-affinity and gel filtration chromatography (46) (Fig. 1A). The protein was crystallized in the Pr photoproduct state and X-ray diffraction data were collected (see Materials and Methods for details). The initial phase was derived by molecular replacement using the coordinates of the TePixJ GAF domain (Protein Data Bank [PDB]: 3VV4) as a search model, and the model was refined to a resolution of 1.63 Å and an R value of 15.8% (free R, 19.9%) (SI Appendix, Table S1). Two protomers whose structures were essentially identical (RMSD for 139 Cα atoms, 0.17 Å) were contained in the asymmetric unit, and molecule A is described below as a representative. Molecule A includes residues 164 through 268 and 277 through 313 and covalently bound PCB. The mainframe of the structure displays a typical GAF fold composed of a five-stranded antiparallel β-sheet and five helices (Fig. 1 B and C). Among the five helices, the short second helix, named H2, is a single turn of a 310 helix (Fig. 1 B and C). The five β-sheet forms aromatic and aliphatic hydrophobic cores on both sides and is backed by the H1 and H5 helices (Fig. 1B). PCB is covalently anchored at a cysteine residue in the H4 helix and buried within the cleft in the GAF domain. The α-face of PCB is covered by residues from the H4 helix (Fig. 1 B and D). The β-face of PCB is covered by a core of the antiparallel β-sheet, which is constituted by strand 4 (S4), strand 5 (S5), and strand 1 (S1) and also a large loop between the S2 through S3 sheets (S2 through S3 loop, residues 203 through 232) that include the H2 and H3 helices (Fig. 1 B and D).
Fig. 1.
Crystal structure of the chromatic acclimation sensor RcaE in the Pr photoproduct state. (A) Domain organization of RcaE. (B) Ribbon drawings of the structure of the GAF domain of RcaE (cyan). The PCB chromophore is depicted in stick representation (gray). The α-face and β-face that account for each side of PCB are indicated by dotted lines. Arrows indicate the direction viewed in E and F. (C) Topology diagram of the secondary structure of the GAF domain of RcaE (cyan). The regions of TePixJ and Slr1393 corresponding to the S2 through S3 loop of RcaE are shown in pink and green, respectively. (D) Fo-Fc map of the PCB chromophore contoured at 2.5σ. The α-face and β-face are indicated by arrows. (E) Close-up view of the PCB-binding site along the arrow labeled E in B. (F) Close-up view shown along the black arrow labeled F in B. The distances between the oxygen atom of the carboxyl group of the Glu217 sidechain and NA, NB, and NC of the PCB are shown in Å. For clarity, H4 has been omitted from E and F.
Detailed Structure of PCB and Its Interacting Residues.
The clear electron density map and Fo − Fc map allowed us to determine the conformations of the PCB chromophore in detail (Fig. 1D). In general, the geometry of the A- to D-rings of the bilin chromophore are each classified by syn/anti conformation and Z/E configuration at the C5, C10, and C15 atoms (64 possible classifications in total). In the Pr structure of RcaE, PCB adopts C5-Z, C10-Z, and C15-E configurations, but remarkably, it adopts all syn conformations. To our knowledge, there is no report of the structure of the bilin-binding protein that harbors a C15-E,syn chromophore. The A-, B-, and C-rings of PCB form a nearly planar structure, but the D-ring is tilted by ∼27° out of the A-B-C plane (Fig. 1D). The bilin conformation with coplaner A- through C-rings and a tilted D-ring has also been reported in the Pr states of phytochromes and other CBCR subfamilies (8, 12, 23, 45). Notably, C15-E,syn bilin in the Pr state of RcaE is more helical compared with the extended C15-Z,anti bilin reported in the other Pr structures. The helical bilin conformation is consistent with the relatively high Soret peak at ∼370 nm in the Pr spectrum of RcaE (47), which is the common characteristic of the green/red CBCR subfamily (18, 38, 39). The hydrophobic lid covering the α-face of PCB is composed of two residues, Leu249 and Phe252 (Fig. 1E). The tilt of the D-ring is supported by hydrophobic interactions with Phe252 on the H4 helix (Fig. 1E). The methyl group of C17 of the D-ring makes Van der Waals contacts with the methyl group of Leu249. The amide group of the D-ring faces the aromatic ring of Phe214 on the S2-S3 loop (Fig. 1 E and F). Electron density was low for the position of the ethyl group of C18 (Fig. 2D), which probably reflects its structural flexibility.
Fig. 2.
A unique porous cavity filled with water molecules in the RcaE GAF domain. (A) Superimposition of the crystal structure of the GAF domain of RcaE (cyan) on those of TePixJ (green) and Slr1393 (pink). The H4 of RcaE and the corresponding region of TePixJ and Slr1393 have been omitted for clarity. (B) The unique porous cavity inside the protein shown as surface representations in yellow. Oxygen atoms of the water molecules in the cavity were shown as red balls. The chain A of the GAF domain is represented as a cyan ribbon. The chain B of a neighboring asymmetric unit is represented as brown. The PCB chromophore are shown as stick models colored in gray. (C) Clustered waters (oxygen) inside the S2 through S3 loop of chain A shown as red balls. Electron density and difference electron density maps are also shown as gray and green meshes contoured at 1.0σ and 3.0σ, respectively. The hydrogen bond network is illustrated by yellow dashed lines. (D) Calculated average B-factors per residue. MD values and values for chain A and chain B obtained from the crystal structure are shown in red, blue, and green, respectively.
The location of PCB against the protein in RcaE is distinct in comparison to other CBCRs. For structural comparison, we chose the GAF domains of TePixJ (PDB:3VV4) and Slr1393 (PDB:5M82), which harbor a bilin chromophore in the C15-E configuration (23, 45). Essentially, the protein part of the GAF domain of RcaE is well fitted to the other GAF domains, but the position of PCB in RcaE is strikingly different. When the structure is viewed with the H4 helix side-up (from the α-face of PCB), PCB is rotated clockwise by one pyrrole ring in RcaE (Fig. 2A). The distances between RcaE and TePixJ with respect to the representative PCB atoms range from 3.1 to 4.1 Å (SI Appendix, Table S2). Similarly, the distances between RcaE and Slr1393 with respect to the representative PCB atoms range from 3.2 to 5.1 Å. Recently, Bandara et al. reported the far-red–absorbing state of the 2551g3 of far-red/orange CBCR subfamily (25), whose GAF domain shows high sequence similarity to that of RcaE. The Pfr dark state of 2551g3 adopts all syn conformation and all Z configuration but also shows a clockwise-rotated location of PCB similar to that seen in RcaE (25).
The key interactions contributing to the location of PCB in the GAF domain of RcaE are hydrogen bonds and/or ion pairs between the carboxylates of the propionic acid of the B-ring and C-ring (hereafter called B-carboxylate and C-carboxylate, respectively). The Tyr227 sidechain forms a hydrogen bond with the B-carboxylate (Fig. 1F), and the Lys261 sidechain makes ion pairs with the B- and C- carboxylates (Fig. 1F). The distance between the Nε atom of the His285 sidechain and C-carboxylate is 2.7 Å, and thus, the His285 sidechain probably also makes an ion pair or hydrogen bond with the C-carboxylate (Fig. 1 E and F). Glu217, the sole acidic interactor with PCB, makes hydrogen bonds with PCB: the distances between one oxygen atom of the carboxyl group of the Glu217 sidechain and NA (A-ring), NB (B-ring), and NC (C-ring) are 2.8 Å, 2.8 Å, and 2.7 Å, respectively (Fig. 1F). Of note, the corresponding distance for ND (D-ring) is 4.4 Å. The other oxygen on the opposite side of Glu217 forms a hydrogen bond with the phenolic oxygen in Tyr187 (Fig. 1F). Furthermore, the aromatic ring of Tyr223 interacts with the methyl group of the B-ring in a perpendicular manner, probably by forming a CH–π interaction (Fig. 1 E and F). The sidechains of Leu209 and Phe214 on the S2 through S3 loop make the part of hydrophobic pocket, which encloses the methyl group of the C-ring and the amide group of the D-ring (Fig. 1F).
Identification of the “Leaky Bucket” Structure.
The hydrophobic residues on the S2 through S3 loop, namely Leu209, Phe214, Phe219, Pro220, and Tyr223, form the wall of the hydrophobic bilin-binding pocket, which surrounds PCB without any direct hydrogen bonds (Fig. 1 E and F). Because the packing of atoms in the hydrophobic region seems loose, we surveyed the cavity inside the GAF domain by using CAVER (48), a program that can detect interior cavities of proteins. Notably, we identified a porous cavity between the C- and D-rings, the S2 through S3 loop, and the S2 strand, with a size of 196.9 Å3 (Fig. 2B, in yellow). This large cavity was filled with an ordered cluster of water molecules with a pentagonal core structure (Fig. 2 B and C) that has been seen in other proteins at their hydrophobic surface by X-ray crystallography (49, 50). However, a similar cavity has not been reported in any other structures of phytochromes or CBCRs including the Pfr dark state of 2551g3 (25). The water molecules are hydrogen bonded with the main chains of Gly195 and Thr197 in the S2 sheet and those of Leu209, Leu212, Phe214, Pro215, and Glu217 in the S2 through S3 loop (Fig. 2C). Although the S2 through S3 loop partly interacts with that of neighboring molecule, the water cluster penetrated into the cavity are not directly involved in the crystal-packing contact (Fig. 2B). We designate the unique structure of the RcaE GAF domain as “leaky bucket,” reflecting both a lid or cover for the bucket and the presence of leaking holes that permit exchange with bulk solvent.
Next, we investigated the dynamic properties of the GAF domain, especially the key interactions between PCB and the protein, the S2 through S3 loop, and the clustered water molecules by molecular dynamics (MD) simulation. A 500 ns MD run at 300 K was performed using the crystal structure as an initial starting geometry and including explicit solvent water molecules. The ion pairs between Lys261 and the B- and C- carboxylates were stable throughout the MD simulation, although the carboxylate rotates to replace each oxygen with the other (SI Appendix, Fig. S3A and Movie S1). Furthermore, the sole acidic interactor Glu217 still maintained interactions with PCB, even though the carboxylate oxygen atoms were in exchange (SI Appendix, Fig. S3B and Movie S1).
The root mean square fluctuation of the backbone C, N, and O atoms over the whole trajectory was calculated and converted to average B-factors, which were used as a measure of structural flexibility. Nearly all of the residues in the GAF domain had relatively low-average B-factors with the exception of residues close to the N and C termini and residues 269 through 276, for which the coordinates are missing in the crystal structure (Fig. 2D). Interestingly, the average B-factor was larger for residues Leu209 to Phe219, corresponding to the middle portion of the S2 through S3 loop, than for other parts of the domain (Fig. 2D). Notably, the experimentally derived B-factors from the X-ray crystallographic analysis showed a similar pattern to that of the MD simulation (Fig. 2D). A trajectory analysis through the 500 ns simulation showed that Glu213, Phe214, and Pro215 underwent a marked conformation change after 340 ns (SI Appendix, Fig. S4A). At 300 ns, the structure of the S2 through S3 loop had essentially retained the initial crystal structure (SI Appendix, Fig. S4B). In the 400 ns snapshot, by contrast, the location of the sidechain of Phe214 had largely shifted to the interior of the H2 helix, a hydrophobic environment made by Met211 (SI Appendix, Fig. S4C). Pro215 had shifted to the vicinity of the D-ring, and the sidechain of Glu213 had swung around. We also investigated the dynamic character of the clustered waters over a very short timescale. Inspection of the trajectory showed that the water molecules forming a hydrogen bond with Glu217 (WAT21 and WAT74, in SI Appendix, Fig. S5) and one of the clustered waters (WAT105, in SI Appendix, Fig. S5) rapidly moved to the outside of the protein within 1 ns (SI Appendix, Fig. S5). These analyses demonstrated the structural flexibility of the S2 through S3 loop, and the rapid exchange of the water molecules between the porous cavity and bulk solvent.
Determination of the Bilin Protonation State by NMR.
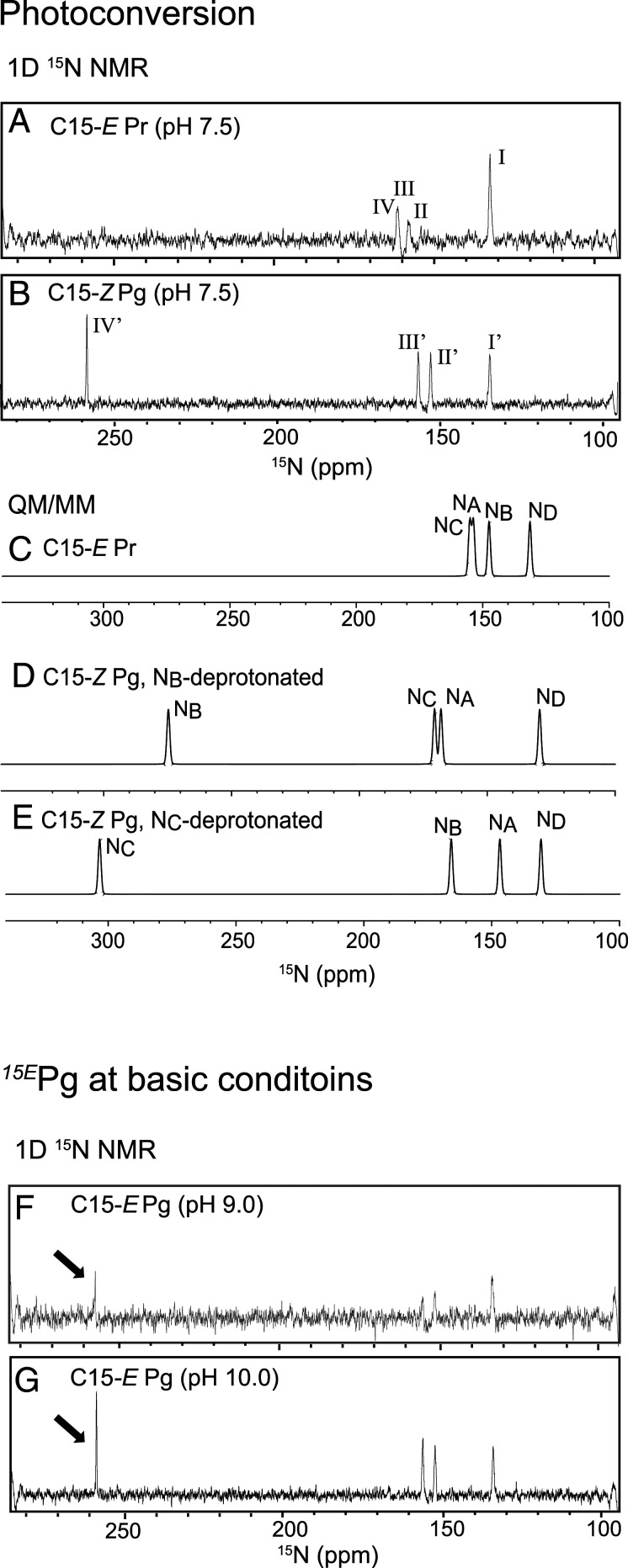
To evaluate the chemical environment of the PCB molecule, especially its protonation state, we performed NMR experiments using isotopically labeled PCB incorporated into the unlabeled GAF domain. First, we tried to detect NH signals of the pyrrole rings by two-dimensional (2D) 1H-15N heteronuclear single quantum correlation (HSQC) experiments. However, we observed only one signal from a pyrrole ring, probably assigned to ND-H, in both the Pr state and the Pg state: namely, (1H,15N) = (7.86, 134.9 ppm) and (1H,15N) = (9.72, 134.4 ppm), respectively, at 30 °C and pH 7.5 (SI Appendix, Fig. S6). This contrasts with previous NMR studies of the red/green CBCR NpR6012g4 (21, 51), in which four 15N-1H signals were observed in both the Pg and the Pr state in 2D 1H-15N HSQC experiments. These results imply that the exchange rates of the NH moieties NA, NB, and NC of PCB are much faster in RcaE than in NpR6012g4.
We therefore tried to detect 15N signals directly by a one-dimensional (1D) 15N experiment, which does not need a magnetization transfer step from 1H to15N (Fig. 3). Although direct detection of 15N is intrinsically insensitive, we overcame the low sensitivity by using a cryogenic broadband observation probe. Via this strategy, we successfully observed four 15N signals (I, II, III, and IV) within 10 h at 30 °C in the Pr state and four 15N signals (I’, II’, III’, and IV’) in the Pg state (Fig. 3 A and B). The chemical shifts of I, II, III, and IV were 134.9, 160.4, 163.5, and 163.5 ppm, respectively, with III and IV overlapping (SI Appendix, Supporting Text and Fig. S7), and those of I’, II’, III’, and IV’ were 134.4,152.5, 156.3, and 258.1 ppm, respectively. The signals at 130 to 160 ppm corresponded to a protonated Schiff base, while signal IV’ at 258.1 ppm in the Pg state alone showed a large low-field shift, corresponding to a deprotonated Schiff base. This indicates that one nitrogen atom of PCB is deprotonated in the Pg dark state, whereas all nitrogen atoms are protonated in the Pr photoproduct state. We also performed a 1D 15N measurement of the Pr state of RcaE under conditions of pH 9 and pH 10, at which RcaE is converted to the green-absorbing Pg state harboring C15-E PCB (18). Strikingly, the low-field shifted peak at ∼258 ppm was clearly observed even in the C15-E Pg state at both pH 9.0 and pH 10 (Fig. 3 F and G). This result supports the idea that the peak at 258.1 ppm in the Pg state at pH 7.5 is the signal from deprotonated PCB.
Fig. 3.
NMR spectra and quantum mechanical calculation of the 15N chemical shifts of PCB in RcaE. (A and B) Observed spectra after photoconversion at neutral pH. Shown is the 15N 1D NMR spectrum of PCB in the C15-E Pr state (A) and the C15-Z Pg state (B) pt pH 7.5. (C–E) The QM/MM 15N NMR chemical shifts calculated by using the GIAO. (C) Simulated spectrum of the C15-E Pr state of RcaE, in which all nitrogen atoms are protonated. (D) Simulated spectrum of the C15-Z Pg state of RcaE, in which NB is deprotonated and NA, NC, and ND are protonated. (E) Simulated spectrum of the C15-Z Pg state of RcaE, in which NC is deprotonated and NA, NB, and ND are protonated. (F and G) Observed spectra of the C15-E state at alkaline pH. Shown is the 15N 1D NMR spectrum of PCB at pH 9.0 (F) and pH 10 (G).
The assignments of 15N signals are informative for understanding which nitrogen atom is protonated or deprotonated. However, the lack of an NH correlation makes it difficult to establish the assignments experimentally (SI Appendix, Fig. S6). To obtain clues, we performed QM/MM calculations to simulate the 15N NMR spectra (Fig. 3 C–E). For the Pr state, the MD snapshot at 270 ns, which was closest to the averaged structure of the MD trajectory, was used to perform a subsequent geometry optimization. We computed the 15N chemical shifts by using the gauge-independent atomic orbital (GIAO) method (52, 53). The chemical shifts were also calculated for the Pg state. For the latter case, we used two reported Pg models in which C15-Z,anti PCB is deprotonated at NB or NC (39). As shown in Fig. 3 C and D, the 15N 1D spectra of both the Pr (Fig. 3C) and Pg (Fig. 3D) states were well reproduced by the Pr and NB-deprotonated Pg models, respectively. Notably, the chemical shift calculated with the NC-deprotonation (Fig. 3E) deviated from the corresponding experimental value compared to that of NB-deprotonation (Fig. 3D). Further, the calculation in which PCB is deprotonated at NB or NC based on the structure of C15-E,syn crystal structure was also conducted. We constructed initial structures for the deprotonated RcaE from the present C15-E crystal structure. The calculated specta (SI Appendix, Fig. S8 A and B) deviated more from the observed spectrum (Fig. 3G) as compared with the calculated spectra for models in which in C15-Z,anti PCB (Fig. 3 D and E).
On the basis of these results, signals I, II, III, and IV for Pr were assigned to ND, NB, NA, and NC, respectively (or ND, NB, NC, and NA, because III and IV overlapped). On the other hand, signals I’, II’, III’, and IV’ for Pg were assigned to ND, NA, NC, and NB, respectively, suggesting that deprotonation in the Pg state occurs at NB. These QM/MM calculations (Fig. 3 C–E) were performed using an electronic embedding (EE) scheme, in which electrostatic environments of the protein moiety are included in the QM calculations. To explore the effects of these electrostatic environments of the protein including hydrogen bonds, we next performed a QM/MM calculation of the Pr model with a mechanical embedding (ME) scheme, which does not incorporate the electrostatic interactions between the QM and MM regions. A distinct difference between the EE and ME schemes was observed for the calculated spectra (Fig. 3C and SI Appendix, Fig. S8C), indicating that the 15N chemical shifts reflect not only the chromophore structure but also the electrostatic environment of the protein moiety.
Discussion
We have reported a high-resolution (1.63 Å) crystal structure of the bilin-binding GAF domain of the chromatic acclimation sensor RcaE, a representative member of the green/red CBCR subfamily. The PCB molecule adopts a unique C5-Z,syn, C10-Z,syn, and C15-E,syn structure and is buried in the hydrophobic pocket of the GAF domain. We identified a direct hydrogen bond network connecting the NA-NC nitrogen atoms of PCB, the carboxyl group of Glu217, and the pentagon water cluster in a unique cavity within the PCB-binding pocket. MD simulation suggested the rapid exchange of the clustered waters with solvent and the flexibility of the S2 through S3 loop. 1D 15N NMR analysis demonstrated that the four nitrogen atoms of PCB are indeed fully protonated in the Pr photoproduct state, and one of them is deprotonated in the Pg dark state. These findings uncover the structural basis of the protochromic green/red photocycle.
NMR is a powerful approach because it can provide direct information on the chemical environments of PCB, including its protonation state. We used 1D 15N measurements and demonstrated that one of the four NH moieties of PCB is actually deprotonated in the C15-Z Pg dark state created by illumination with red light and also in the C15-E Pg photoproduct state formed under basic pH conditions (Fig. 3). The similarity in the down-filed shift of 15N in the C15-Z Pg and C15-E Pg states suggests that the PCB chromophore undergoes common structural changes by deprotonation, which affects the electronic state of the π-conjugated system of PCB. Comparison of the observed chemical shift of 15N and the calculated chemical shifts using the quantum chemical calculations suggested that deprotonation occurs at the NB nitrogen (Fig. 3 and SI Appendix, Fig. S8), which is consistent with our previous resonance Raman spectroscopy and QM/MM simulation experiment in RcaE (44). Deprotonation in the NB nitrogen is also consistent with solid-state NMR of in vitro–prepared AnPixJ proteins harboring deprotonated PCB fraction for its Pg photoproduct state (54). In 2D 1H-15N HSQC measurement, three of the four NH signals were missing for RcaE (SI Appendix, Fig. S6), whereas all four NH signals were observed for the red/green CBCR NpR6012g4 (21, 51). This difference probably reflects the fast exchange of the proton in the NH moieties of the PCB molecule. Our structure suggested that the three NA-NC pyrrole nitrogens form direct hydrogen bond with Glu217, whereas the ND positioned rather hydrophobic environment (Fig. 1 E and F). Therefore, we speculate that the rapid exchange of the proton occurs in the NA-NC of the PCB. Ultrafast transient absorption spectroscopy suggested the presence of ground-state inhomogeneity in the Pg dark state of RcaE, which was previously interpreted as the existence of different tautomers, namely, NB- and NC-deprotonated PCB (55). Although we did not observe apparent heterogeneity of the Pg state in 1D 15N measurements (Fig. 3), this measurement is less sensitive in detection of the ground-state heterogeneity due to its low S/N signals and changes of the intensity of each peak caused by the protonation state (Fig. 3). We also could not rule out the possibility of that averaging of the signals between NB-deprotonation and NC-deprotonation.
Previously, we performed extensive site-directed mutagenesis of residues in the GAF domain of RcaE and investigated the effect of these substitutions on the bilin pKa by monitoring the pH-induced Pg/Pr conversion (18). Taking these results together, we elucidated the detailed roles of each residue in the PCB-binding pocket of RcaE. In the Pr structure, the carboxyl group of Glu217 forms direct hydrogen bonds with the NA-NC pyrrole nitrogen atoms (Fig. 1 E and F). The E217A and E217Q mutants showed the normal Pg formation but were deficient in Pr formation, whereby the upshift in bilin pKa in the Pr state was specifically inhibited (18). By contrast, the E217D mutant of RcaE showed a near normal Pg/Pr conversion and pKa shift (18). Therefore, the role of the carboxyl group of Glu217 is to stabilize the protonation of NH moieties with its negative charge. Although pyrrole nitrogens are hydrogen bonded with carboxylate in other CBCR subfamilies, the uniqueness of RcaE is the presence of the porous cavity in the Pr photoproduct state (Fig. 2C), which is named “leaky bucket” structure. In this structure, the carboxyl group of Glu217 also forms a direct hydrogen bond with the clustered water molecules, which is rapidly exchanging with solvent in the MD simulation (SI Appendix, Fig. S5). In addition, the fast exchanging of the proton in the NA-NC pyrrole nitrogen atoms was implied by the absence of the three of four NH signals in 2D 1H-15N HSQC measurement (SI Appendix, Fig. S6). Therefore, we propose that the leaky bucket structure provides PCB with a route to access solvent waters, which supports the proton transfer and subsequent formation of the Pr photoproduct (Fig. 4, Right). Notably, in this mechanism, proton is supplied by the solvent water, and a proton donor of a specific charged residue is not required.
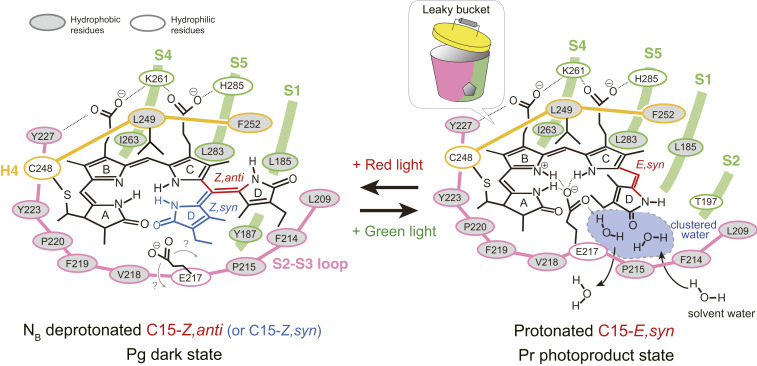
Fig. 4.
A proposed model of the photoconversion of the RcaE GAF domain. In the Pr photoproduct state, PCB adopts C5-Z,syn/C10-Z,syn/C15-E,syn conformation and four pyrrole nitrogen atoms (NA through ND) are protonated. The proton of the PCB is provided by clustered water molecules in the porous cavity (dashed blue area) and stabilized by the negatively charged carboxylate of E217. We designate the unique GAF domain structure as “leaky bucket,” in which the large S2-S3 loop (pink) and the core of the antiparallel β-sheet (green) form a bucket, whereas the hydrophobic L249 and F252 in the H4 (yellow) form a lid. In the Pg dark state, the D-ring adopts C15-Z,anti conformation (red) and NB is deprotonated while the other NA, NC, and ND atoms are protonated. An alternative model with C15-Z,syn (blue) conformation discussed in the main text is also shown. The exact position of E217 in the Pg dark state is unclear.
Upon red light illumination, RcaE undergoes C15-E to C15-Z photoisomerization and subsequent deprotonation of PCB (downshift of the bilin pKa), resulting in formation of the Pg state (18, 38). The E217A and E217Q mutants showed normal pKa value in Pg dark state (18). Therefore, it is less likely that the negatively charged carboxyl group functions as a base and decreases the bilin pKa in the Pg state. The Glu217 loses the regulation of the bilin pKa possibly by altering the position of its carboxyl group in keeping the engagement with certain bilin N-H(s) or carbonyl(s). Alternatively, the carboxyl group of Glu217 may move apart from PCB and toward the protein surface or the porous cavity to interact with water molecules. Notably, our structure shows that Leu249 is positioned toward the α-face of PCB and forms a hydrophobic lid, together with Phe252 (Fig. 1 E and F). The L249H mutant showed a substantial increase in bilin pKa and formed the Pr state even in the C15-Z state, whereas F252H was not deficient in the Pg formation (18). These data suggest that the hydrophobicity of Leu249 is crucial for the deprotonation of PCB in the Pg state. It is known that the pKa values of charged moieties shift in the direction that favors the neutral state in the hydrophobic protein core (56, 57). Therefore, we propose that a “desolvation effect” is the driving force behind the deprotonation of PCB. Lys261 forms ion pairs with both the B- and C-ring carboxylates, together with Tyr227 and His285 (Fig. 1 E and F). Previously, the Y227F and H285Q mutants showed negligible effects on pKa regulation, whereas both K261A and K261M showed substantially decreased pKa in both the Pg and Pr states (18). The presence of the hydrophobic lid of Leu249 and Phe252 in our structure suggests that Lys261 is not likely to function as a proton donor as discussed previously (18) but rather tunes the bilin pKa by adjusting the location of the C15-E,syn PCB molecule in the GAF domain (Fig. 4).
For the PCB structure in the Pg dark state, whose three-dimensional structure is not yet available, we can argue two possible structures: C15-Z,anti and C15-Z,syn (Fig. 4). The extended C15-Z,anti structure for the Pg state of RcaE (Fig. 4, Left) is consistent with the shape of its absorption spectrum having low Soret peak and high green-absorbing peak (18, 38, 39). The C15-Z,anti structure is supported by our recent resonance Raman spectroscopy and QM/MM and MD simulation (44). That study showed that the observed resonance Raman spectra in the Pg state of RcaE were well reproduced by the simulated spectra of C15-Z,anti PCB having the coplanar A- through C-rings in the NB-deprotonated state (44). The C15-Z,anti structure is also supported by the in vitro reconstitution experiment (18). This study showed that apo-RcaE can incorporate a synthetic PCB analog that is sterically locked in the C15-Z,anti structure and forms a photoinert and red-shifted Pg state (18). The structure of C15-Z,anti PCB differs from natural PCB in that 1) it has a bulky D-ring with an additional covalent bridge between C13 of the C-ring and ND of the D-ring, and 2) its C- through D-ring forms a nearly planar structure and is unable to undergo structural changes owing to the presence of dual covalent linkages (58). Incorporation of C15-Z,anti PCB into the apo-protein of our Pr structure would cause a steric clash with Leu209 and Phe214 in the S2 through S3 loop (SI Appendix, Fig. S2). Therefore, the acceptance of the C15-Z,anti PCB suggests the occurrence of a structural rearrangement of the S2 through S3 loop in the Pg dark state. The flexibility of the S2 through S3 loop and the presence of the interior cavity inside the S2 through S3 loop containing a water cluster may enable the structural change of PCB along with C15-E,syn to C15-Z,anti (probably through C15-Z,syn intermediate). In addition, the D-ring may be twisted in the natural Pg state with a less-conjugated bilin π-system, which could explain the red shift of ∼30 nm in the Pg state having coplaner C15-Z,anti locked PCB.
By contrast, the C15-Z,syn model for the Pg state of RcaE (Fig. 4, Left) is consistent with the recent crystal structure of the Pfr dark state of 2551g3 of a far-red/orange subfamily, in which the PCB bilin adopts a C5-Z,syn, C10-Z,syn, and C15-Z,syn structure (25). Phylogenetic analysis showed that the far-red–absorbing CBCR subfamily evolved from the green/red subfamily (59). RcaE shares most of residues in the PCB-binding pocket with 2551g3 including Leu249 forming a hydrophobic lid, Tyr227, Arg233, Lys261, and H285 interacting with B- and C-ring carboxylates and Glu217 interacting with the NA-NC pyrrole nitrogens (SI Appendix, Fig. S2). In contrast, residues on the S2 through S3 loop are divergent between RcaE and 2551g3 except for Glu217 (SI Appendix, Fig. S2). In the Pfr state of 2551g3, the A-ring is highly tilted toward the β-face, and there is no porous cavity around the D-ring (25). These structural differences are probably responsible for the distinct spectral sensitivity between the 2551g3 Pfr and RcaE Pg, corresponding to a 200 nm difference in absorption. pH titration showed that the C15-Z,syn PCB is protonated in the Pfr state of 2551g3, in which Glu914 (corresponding to Glu217 in RcaE) forms hydrogen bonds with the four pyrrole nitrogens (NA through ND). Site-directed mutagenesis demonstrated that Glu914 is required for the protonation in the Pfr state (25). From these results, Bandara et al. proposed that the far-red shift of the Pfr dark state of 2551g3 has a di-protonated lactim isomer in the tilted A-ring with the deprotonated NB nitrogen (25). This model has not yet been investigated by vibrational and NMR spectroscopies, but our 1D 15N NMR data agree with the NB-deprotonation model in 2551g3. If the RcaE has a C15-Z,syn structure for the Pg dark state as shown in the 2551g3, PCB undergoes structural changes between C15-Z,syn and C15-E,syn upon photoconversion, which requires less drastic structural changes compared with the C15-Z,anti model. The bilin structure of the C15-Z,syn model is similar to the structure of CpcT, a PCB lyase of phycobiliproteins, that has a deprotonated PCB in all Z,syn conformation (60). The absorption spectrum of CpcT has a characteristic of helical bilin: high Soret peak and low red-absorbing peak around 618 nm (60), which contrasts the absorption spectrum of the Pg state of RcaE. This could suggest the contribution of ring twist on the absorption tuning of the Pg state in the C15-Z,syn model of RcaE.
In summary, we propose a following model for the protochromic green/red photocycle of RcaE (Fig. 4). In the Pr state, PCB adopts a C15-E,syn structure, and the NA-NC pyrrole nitrogen atoms form a direct hydrogen bond with the carboxyl group of Glu217 in β-face. The carboxyl group of Glu217 also forms a direct hydrogen bond with solvent water molecules through the leaky bucket structure, which increases hydrophilicity of the vicinity of PCB and leads to the protonation as well as Pr formation. Red light illumination causes a structural change of PCB from C15-E,syn to C15-Z,anti (or C15-Z,syn), which alters the position of Glu217. The accessibility of PCB with solvent is decreased by the reduced interaction with Glu217 and the hydrophobic interaction with Leu249 in α-face, which leads to the decrease of the bilin pKa. Deprotonation of the NB of PCB causes a change in the π-conjugated system of PCB, resulting in the drastic absorption change from the red to the green region. For reverse reaction, green light illumination causes C15-Z,anti (or C15-Z,syn) to C15-E,syn conversion and protonation of the NB. In this model, the carboxyl group of Glu217 functions as a gate in a proton exit and proton influx pathway to solvent water. We would favor the C15-Z,anti model that is supported by the spectroscopic evidence, but three-dimensional structure determination of RcaE in the deprotonated Pg dark state is required to settle the discussion of the D-ring conformation. This analysis will also refine the structural changes of Glu217 and the contribution of the ring twist in the absorption tuning in our model.
Materials and Methods
Sample Preparation.
We prepared the GAF domain as previously descried (18). In brief, the GAF domain (residues 164 through 313) of RcaE was cloned into plasmid pET-28 (Novagen). The construct was expressed in E. coli strain BL21(DE3) star (Invitrogen) as a fusion protein with a His tag, together with PCB biosynthetic plasmid pKT271 (41). The harvested cells were lysed via sonication. The His tag protein was purified by using a column of Ni-Sepharose 4B (Qiagen) followed by gel filtration using a Superdex 75 column (GE Healthcare) equilibrated with a 10 mM Tris HCl (pH 7.5) buffer containing 50 mM KCl. For crystallization, the protein was further purified by using a HiTrap-SP ion-exchange column (GE Healthcare), equilibrated with a 20 mM Tris HCl (pH 8.0) buffer containing 50 mM KCl and eluted with a gradient of 1M KCl. The purified sample was stored in buffer containing 20 mM Tris HCl (pH 7.5) and 10 mM NaCl.
Crystallization.
Prior to crystallization experiments, the protein solution of RcaE was irradiated with green LED light to adapt it to the Pr photoproduct state (SI Appendix, Fig. S1). Crystallization experiments were carried out under green light, and the crystallization plates were incubated in a dark environment. Crystals of the red-absorbing form were obtained at 293 K via the hanging-drop vapor diffusion method with drops containing of 0.9 μL of protein solution (25 mg/mL in 20 mM Tris HCl, pH 7.5, and 10 mM NaCl) mixed with 0.9 μL of reservoir solution (100 mM Tris HCl, pH 7.2, 200 mM MgCl2, and 27% polyethylene glycol 4000). The blue plate-shaped crystals grew to typical dimensions of 200 × 50 × 20 μm3 within 1 wk.
Data Collection and Structure Refinement.
Data collection was performed at 95 K under green LED light on Nagoya University beamline BL2S1 at the Aichi Synchrotron Radiation Center, Japan (61). The diffraction pattern was indexed, integrated, and scaled by using XDS (62). We confirmed the blue color of the crystal after X-ray irradiation (SI Appendix, Fig. S9). The initial structure was solved by MOLREP (63) as implemented in CCP4 (64) using the TePixJ GAF domain (PDB: 3VV4) as a search model. Refinement and manual correction of the structure were performed by using REFMAC5 (65) and Coot (66), respectively. For analyses of the PCB configurations and the clustered waters, simulated annealing refinement in PHENIX were performed. Parameters for data collection, processing, and structure refinement are listed in SI Appendix, Table S1. Figures were drawn by using PyMOL (PyMOL Molecular Graphics System, Schrödinger, LLC).
NMR.
Uniform labeling of PCB with 15N and 13C was achieved by using M9 minimal medium containing no stable isotope and uniformly labeled 15N and 13C 5-aminolevulinic acid. Prior to plasmid induction, the isotope-enriched 5-aminolevulinic acid was added to M9 medium. The purified sample was dissolved in 50 mM Tris⋅HCl buffer (pH 7.5) containing 50 mM KCl in 95% H2O/5% 2H2O. For pH titration, buffers containing 50 mM CHES (pH 9.0) plus 50 mM KCl, and 50 mM CHES (pH 10.0) plus 50 mM KCl were used. The final concentration of the protein was ∼1 mM. NMR spectra were acquired at 30 °C on a Bruker AVANCEIIIHD 500 instrument equipped with a cryogenic BBFO probe and on a Bruker AVANCEIIIHD 600 instrument equipped with a cryogenic TCI probe. The 15N 1D spectra (NS = 14,336, experimental time ∼10 h) were acquired with 1H decoupling during acquisition. 2D 1H-15N HSQC spectra (NS = 128, experimental time ∼7 h) were acquired by using a standard gradient echo sensitivity improvement pulse sequence with a WATERGATE-Water-flip-back type water suppression technique. Data processing was performed with Topspin ver. 3.5 (Bruker)
MD Simulation.
Both the initial setup and the MD runs were performed with the Amber16 program (67) using an explicit representation of solvent molecules and the ff14SB all-atom force field under periodic boundary conditions. The initial starting geometry was taken from the crystal structure of the GAF domain of RcaE in the Pr state. The whole system was subjected to minimization and heated to 300 K. The system obtained after heating was simulated for 500 ns at 1 atm.
QM/MM Calculation.
The QM/MM calculations were performed by using Gaussian16 (68). The QM region consists of the PCB chromophore, while the remainder of the system was treated as the MM region. The initial geometry was obtained from a snapshot of the MD simulation, and 100 water molecules near the surface of the protein were also included. The QM part of the system was computed at the B3LYP/6-31G* (geometry optimization) or Hartree-Fock/6-311+G(2d,p) (chemical shifts) level of theory. The MM part was described by the Amber force field (69), and the positions of the MM atoms were frozen during geometry optimization. In most cases, an EE scheme that considers the partial charges of the MM region into the QM Hamiltonian was used. The GIAO nuclear magnetic shielding values were calculated, and the chemical shifts for the nitrogen atoms were obtained by subtracting their chemical shielding values from that calculated for NH3.
Supplementary Material
Acknowledgments
We thank Dr. J. Clark Lagarias, Dr. Masahiko Ikeuchi, and Dr. Nathan C. Rockwell for helpful discussions. We thank Dr. Masatsune Kainosho and Dr Teppei Kanaba (Bruker) for helpful discussions on the 15N 1D NMR experiment. This work was supported by JSPS KAKENHI (Grants 19H05645 and 19H05773 to Y.I., 19K06707 to Y.H., and 19H03169 to M.M.). This work is also supported by research grants to Y.H. from the Foundation for the promotion of Ion Engineering and Engineering and Japan Gasoline Co. Scholarship Foundation. The crystallography experiment at BL2S1 was performed under the approval of the Aichi Synchrotron Radiation Center (Proposal No. 2019N5012), and the NMR experiments were performed in part using NMR spectrometers with ultrahigh magnetic fields under the Collaborative Research Program of the Institute for Protein Research, Osaka University, NMRCR-18-05, -19-05, and -20-05, and performed in part at RIKEN of NMR Platform supported by the Ministry of Education, Culture, Sports, Science and Technology, Japan. Some of the computations were performed at the Research Center for Computational Science, Okazaki, Japan.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2024583118/-/DCSupplemental.
Data Availability
The X-ray diffraction dataset and associated model have been deposited in the Research Collaboratory for Structural Bioinformatics PDB (https://www.rcsb.org) under PDB ID code 7CKV. All other study data are included in the article and/or supporting information.
References
- 1.Rockwell N. C., Su Y. S., Lagarias J. C., Phytochrome structure and signaling mechanisms. Annu. Rev. Plant Biol. 57, 837–858 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ikeuchi M., Ishizuka T., Cyanobacteriochromes: A new superfamily of tetrapyrrole-binding photoreceptors in cyanobacteria. Photochem. Photobiol. Sci. 7, 1159–1167 (2008). [DOI] [PubMed] [Google Scholar]
- 3.Auldridge M. E., Forest K. T., Bacterial phytochromes: More than meets the light. Crit. Rev. Biochem. Mol. Biol. 46, 67–88 (2011). [DOI] [PubMed] [Google Scholar]
- 4.Wiltbank L. B., Kehoe D. M., Diverse light responses of cyanobacteria mediated by phytochrome superfamily photoreceptors. Nat. Rev. Microbiol. 17, 37–50 (2019). [DOI] [PubMed] [Google Scholar]
- 5.Fushimi K., Narikawa R., Cyanobacteriochromes: Photoreceptors covering the entire UV-to-visible spectrum. Curr. Opin. Struct. Biol. 57, 39–46 (2019). [DOI] [PubMed] [Google Scholar]
- 6.Villafani Y., Yang H. W., Park Y. I., Color sensing and signal transmission diversity of cyanobacterial phytochromes and cyanobacteriochromes. Mol. Cells 43, 509–516 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Song C., et al., The D-ring, not the A-ring, rotates in Synechococcus OS-B’ phytochrome. J. Biol. Chem. 289, 2552–2562 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lim S., et al., Correlating structural and photochemical heterogeneity in cyanobacteriochrome NpR6012g4. Proc. Natl. Acad. Sci. U.S.A. 115, 4387–4392 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rockwell N. C., et al., Eukaryotic algal phytochromes span the visible spectrum. Proc. Natl. Acad. Sci. U.S.A. 111, 3871–3876 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Song J. Y., et al., Spectral and photochemical diversity of tandem cysteine cyanobacterial phytochromes. J. Biol. Chem. 295, 6754–6766 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu S. H., Lagarias J. C., Defining the bilin lyase domain: Lessons from the extended phytochrome superfamily. Biochemistry 39, 13487–13495 (2000). [DOI] [PubMed] [Google Scholar]
- 12.Essen L. O., Mailliet J., Hughes J., The structure of a complete phytochrome sensory module in the Pr ground state. Proc. Natl. Acad. Sci. U.S.A. 105, 14709–14714 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rockwell N. C., et al., A second conserved GAF domain cysteine is required for the blue/green photoreversibility of cyanobacteriochrome Tlr0924 from Thermosynechococcus elongatus. Biochemistry 47, 7304–7316 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rockwell N. C., Martin S. S., Feoktistova K., Lagarias J. C., Diverse two-cysteine photocycles in phytochromes and cyanobacteriochromes. Proc. Natl. Acad. Sci. U.S.A. 108, 11854–11859 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Burgie E. S., Walker J. M., Phillips G. N. Jr, Vierstra R. D., A photo-labile thioether linkage to phycoviolobilin provides the foundation for the blue/green photocycles in DXCF-cyanobacteriochromes. Structure 21, 88–97 (2013). [DOI] [PubMed] [Google Scholar]
- 16.Cornilescu C. C., et al., Dynamic structural changes underpin photoconversion of a blue/green cyanobacteriochrome between its dark and photoactivated states. J. Biol. Chem. 289, 3055–3065 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ishizuka T., et al., The cyanobacteriochrome, TePixJ, isomerizes its own chromophore by converting phycocyanobilin to phycoviolobilin. Biochemistry 50, 953–961 (2011). [DOI] [PubMed] [Google Scholar]
- 18.Hirose Y., et al., Green/red cyanobacteriochromes regulate complementary chromatic acclimation via a protochromic photocycle. Proc. Natl. Acad. Sci. U.S.A. 110, 4974–4979 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sato T., et al., Protochromic absorption changes in the two-cysteine photocycle of a blue/orange cyanobacteriochrome. J. Biol. Chem. 294, 18909–18922 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Narikawa R., Fukushima Y., Ishizuka T., Itoh S., Ikeuchi M., A novel photoactive GAF domain of cyanobacteriochrome AnPixJ that shows reversible green/red photoconversion. J. Mol. Biol. 380, 844–855 (2008). [DOI] [PubMed] [Google Scholar]
- 21.Rockwell N. C., Martin S. S., Lim S., Lagarias J. C., Ames J. B., Characterization of red/green cyanobacteriochrome NpR6012g4 by solution nuclear magnetic resonance spectroscopy: A hydrophobic pocket for the C15-E,anti chromophore in the photoproduct. Biochemistry 54, 3772–3783 (2015). [DOI] [PubMed] [Google Scholar]
- 22.Wiebeler C., Rao A. G., Gärtner W., Schapiro I., The effective conjugation length is responsible for the red/green spectral tuning in the cyanobacteriochrome Slr1393g3. Angew. Chem. Int. Ed. Engl. 58, 1934–1938 (2019). [DOI] [PubMed] [Google Scholar]
- 23.Xu X., et al., Structural elements regulating the photochromicity in a cyanobacteriochrome. Proc. Natl. Acad. Sci. U.S.A. 117, 2432–2440 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rockwell N. C., Martin S. S., Gulevich A. G., Lagarias J. C., Conserved phenylalanine residues are required for blue-shifting of cyanobacteriochrome photoproducts. Biochemistry 53, 3118–3130 (2014). [DOI] [PubMed] [Google Scholar]
- 25.Bandara S., et al., Crystal structure of a far-red-sensing cyanobacteriochrome reveals an atypical bilin conformation and spectral tuning mechanism. Proc. Natl. Acad. Sci. U.S.A. 118, e2025094118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cho S. M., et al., Genomic survey and biochemical analysis of recombinant candidate cyanobacteriochromes reveals enrichment for near UV/Violet sensors in the halotolerant and alkaliphilic cyanobacterium Microcoleus IPPAS B353. J. Biol. Chem. 290, 28502–28514 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shin H., Ren Z., Zeng X., Bandara S., Yang X., Structural basis of molecular logic OR in a dual-sensor histidine kinase. Proc. Natl. Acad. Sci. U.S.A. 116, 19973–19982 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kehoe D. M., Gutu A., Responding to color: The regulation of complementary chromatic adaptation. Annu. Rev. Plant Biol. 57, 127–150 (2006). [DOI] [PubMed] [Google Scholar]
- 29.Gutu A., Kehoe D. M., Emerging perspectives on the mechanisms, regulation, and distribution of light color acclimation in cyanobacteria. Mol. Plant 5, 1–13 (2012). [DOI] [PubMed] [Google Scholar]
- 30.Hirose Y., et al., Characterization of the genuine type 2 chromatic acclimation in the two Geminocystis cyanobacteria. DNA Res. 24, 387–396 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sanfilippo J. E., Garczarek L., Partensky F., Kehoe D. M., Chromatic acclimation in cyanobacteria: A diverse and widespread process for optimizing photosynthesis. Annu. Rev. Microbiol. 73, 407–433 (2019). [DOI] [PubMed] [Google Scholar]
- 32.Bordowitz J. R., Montgomery B. L., Photoregulation of cellular morphology during complementary chromatic adaptation requires sensor-kinase-class protein RcaE in Fremyella diplosiphon. J. Bacteriol. 190, 4069–4074 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wilde A., Mullineaux C. W., Light-controlled motility in prokaryotes and the problem of directional light perception. FEMS Microbiol. Rev. 41, 900–922 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Enomoto G., Ni-Ni-Win, Narikawa R., Ikeuchi M., Three cyanobacteriochromes work together to form a light color-sensitive input system for c-di-GMP signaling of cell aggregation. Proc. Natl. Acad. Sci. U.S.A. 112, 8082–8087 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Enomoto G., Ikeuchi M., Blue-/Green-Light-Responsive cyanobacteriochromes are cell shade sensors in red-light replete niches. iScience 23, 100936 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kehoe D. M., Grossman A. R., Similarity of a chromatic adaptation sensor to phytochrome and ethylene receptors. Science 273, 1409–1412 (1996). [DOI] [PubMed] [Google Scholar]
- 37.Kehoe D. M., Grossman A. R., New classes of mutants in complementary chromatic adaptation provide evidence for a novel four-step phosphorelay system. J. Bacteriol. 179, 3914–3921 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hirose Y., Shimada T., Narikawa R., Katayama M., Ikeuchi M., Cyanobacteriochrome CcaS is the green light receptor that induces the expression of phycobilisome linker protein. Proc. Natl. Acad. Sci. U.S.A. 105, 9528–9533 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hirose Y., Narikawa R., Katayama M., Ikeuchi M., Cyanobacteriochrome CcaS regulates phycoerythrin accumulation in Nostoc punctiforme, a group II chromatic adapter. Proc. Natl. Acad. Sci. U.S.A. 107, 8854–8859 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tabor J. J., Levskaya A., Voigt C. A., Multichromatic control of gene expression in Escherichia coli. J. Mol. Biol. 405, 315–324 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schmidl S. R., Sheth R. U., Wu A., Tabor J. J., Refactoring and optimization of light-switchable Escherichia coli two-component systems. ACS Synth. Biol. 3, 820–831 (2014). [DOI] [PubMed] [Google Scholar]
- 42.Castillo-Hair S. M., Baerman E. A., Fujita M., Igoshin O. A., Tabor J. J., Optogenetic control of Bacillus subtilis gene expression. Nat. Commun. 10, 3099 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hirose Y., et al., Diverse chromatic acclimation processes regulating phycoerythrocyanin and rod-shaped phycobilisome in cyanobacteria. Mol. Plant 12, 715–725 (2019). [DOI] [PubMed] [Google Scholar]
- 44.Osoegawa S., et al., Identification of the deprotonated pyrrole nitrogen of the bilin-based photoreceptor by Raman spectroscopy with an advanced computational analysis. J. Phys. Chem. B 123, 3242–3247 (2019). [DOI] [PubMed] [Google Scholar]
- 45.Narikawa R., et al., Structures of cyanobacteriochromes from phototaxis regulators AnPixJ and TePixJ reveal general and specific photoconversion mechanism. Proc. Natl. Acad. Sci. U.S.A. 110, 918–923 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mukougawa K., Kanamoto H., Kobayashi T., Yokota A., Kohchi T., Metabolic engineering to produce phytochromes with phytochromobilin, phycocyanobilin, or phycoerythrobilin chromophore in Escherichia coli. FEBS Lett. 580, 1333–1338 (2006). [DOI] [PubMed] [Google Scholar]
- 47.Falk H., The Chemistry of Linear Oligopyrroles and Bile Pigments (Springer-Verlag, New York, 1989), p. 416. [Google Scholar]
- 48.Jurcik A., et al., CAVER analyst 2.0: Analysis and visualization of channels and tunnels in protein structures and molecular dynamics trajectories. Bioinformatics 34, 3586–3588 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Teeter M. M., Water structure of a hydrophobic protein at atomic resolution: Pentagon rings of water molecules in crystals of crambin. Proc. Natl. Acad. Sci. U.S.A. 81, 6014–6018 (1984). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nakasako M., Water-protein interactions from high-resolution protein crystallography. Philos. Trans. R. Soc. Lond. B Biol. Sci. 359, 1191–1204, discussion 1204–1206 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rockwell N. C., Martin S. S., Lim S., Lagarias J. C., Ames J. B., Characterization of red/green cyanobacteriochrome NpR6012g4 by solution nuclear magnetic resonance spectroscopy: A protonated bilin ring system in both photostates. Biochemistry 54, 2581–2600 (2015). [DOI] [PubMed] [Google Scholar]
- 52.Wolinski K., Hinton J. F., Pulay P., Efficient implementation of the gauge-independent atomic orbital method for NMR chemical shift calculations. J. Am. Chem. Soc. 112, 8251–8260 (1990). [Google Scholar]
- 53.Cheeseman J. R., Trucks G. W., Keith T. A., Frisch M. J., A comparison of models for calculating nuclear magnetic resonance shielding tensors. J. Chem. Phys. 104, 5497–5509 (1996). [Google Scholar]
- 54.Song C., et al., A red/green cyanobacteriochrome sustains its color despite a change in the bilin chromophore’s protonation state. Biochemistry 54, 5839–5848 (2015). [DOI] [PubMed] [Google Scholar]
- 55.Gottlieb S. M., et al., Primary photodynamics of the green/red-absorbing photoswitching regulator of the chromatic adaptation E domain from Fremyella diplosiphon. Biochemistry 52, 8198–8208 (2013). [DOI] [PubMed] [Google Scholar]
- 56.Robinson A. C., Castañeda C. A., Schlessman J. L., García-Moreno E. B., Structural and thermodynamic consequences of burial of an artificial ion pair in the hydrophobic interior of a protein. Proc. Natl. Acad. Sci. U.S.A. 111, 11685–11690 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kougentakis C. M., Skerritt L., Majumdar A., Schlessman J. L., The properties of buried ion pairs are governed by the propensity of proteins to reorganize. bioRxiv [Preprint] (2020). 10.1101/2020.02.03.932012 (Accessed 4 February 2020). [DOI]
- 58.Kaori N., et al., Total syntheses of sterically locked phycocyanobilin derivatives bearing a 15Z-anti or a 15E-anti CD-ring component. Bull. Chem. Soc. Jpn. 83, 1309–1322 (2010). [Google Scholar]
- 59.Rockwell N. C., Martin S. S., Lagarias J. C., Identification of cyanobacteriochromes detecting far-red light. Biochemistry 55, 3907–3919 (2016). [DOI] [PubMed] [Google Scholar]
- 60.Zhou W., et al., Structure and mechanism of the phycobiliprotein lyase CpcT. J. Biol. Chem. 289, 26677–26689 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Watanabe N., et al., Protein crystallography beamline BL2S1 at the Aichi synchrotron. J. Synchrotron Radiat. 24, 338–343 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kabsch W., Xds. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vagin A., Teplyakov A., Molecular replacement with MOLREP. Acta Crystallogr. D Biol. Crystallogr. 66, 22–25 (2010). [DOI] [PubMed] [Google Scholar]
- 64.Winn M. D., et al., Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67, 235–242 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Murshudov G. N., et al., REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D Biol. Crystallogr. 67, 355–367 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Emsley P., Lohkamp B., Scott W. G., Cowtan K., Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Case D.et al., Amber 16 (University of California, San Francisco, CA, 2016). [Google Scholar]
- 68.Frisch M. J.et al., Gaussian 16 Rev. C.01 (Gaussian, Inc., Wallingford, CT, 2016). [Google Scholar]
- 69.Cornell W. D., et al., A second generation force field for the simulation of proteins, nucleic acids, and organic molecules. J. Am. Chem. Soc. 117, 5179–5197 (1995). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The X-ray diffraction dataset and associated model have been deposited in the Research Collaboratory for Structural Bioinformatics PDB (https://www.rcsb.org) under PDB ID code 7CKV. All other study data are included in the article and/or supporting information.