Significance
Tissue acidification is commonly associated with active inflammation, including within the intestinal mucosa. It remains unclear how such acidification impacts cellular function and adaptation to these changes within the tissue. In the present work, we utilized unbiased gene expression profiling to identify pathways elicited by extracellular acidification. Results from these studies identify a prominent role for cyclic AMP response element-binding protein signaling in the molding of gene expression in intestinal epithelia. This acidification-induced phenotype is translatable from in vitro cell culture systems to in vivo murine models and through to patients with active intestinal inflammation.
Keywords: acidosis, intestinal epithelial cells, mitogen-activated protein kinase, GPR31, CREB phosphorylation
Abstract
Metabolic changes associated with tissue inflammation result in significant extracellular acidosis (EA). Within mucosal tissues, intestinal epithelial cells (IEC) have evolved adaptive strategies to cope with EA through the up-regulation of SLC26A3 to promote pH homeostasis. We hypothesized that EA significantly alters IEC gene expression as an adaptive mechanism to counteract inflammation. Using an unbiased RNA sequencing approach, we defined the impact of EA on IEC gene expression to define molecular mechanisms by which IEC respond to EA. This approach identified a unique gene signature enriched in cyclic AMP response element-binding protein (CREB)-regulated gene targets. Utilizing loss- and gain-of-function approaches in cultured epithelia and murine colonoids, we demonstrate that EA elicits prominent CREB phosphorylation through cyclic AMP-independent mechanisms that requires elements of the mitogen-activated protein kinase signaling pathway. Further analysis revealed that EA signals through the G protein-coupled receptor GPR31 to promote induction of FosB, NR4A1, and DUSP1. These studies were extended to an in vivo murine model in conjunction with colonization of a pH reporter Escherichia coli strain that demonstrated significant mucosal acidification in the TNFΔARE model of murine ileitis. Herein, we observed a strong correlation between the expression of acidosis-associated genes with bacterial reporter sfGFP intensity in the distal ileum. Finally, the expression of this unique EA-associated gene signature was increased during active inflammation in patients with Crohn’s disease but not in the patient control samples. These findings establish a mechanism for EA-induced signals during inflammation-associated acidosis in both murine and human ileitis.
A common but often underappreciated aspect of tissue inflammation is the acidification of the inflammatory microenvironment. While not completely understood, a primary mechanism of tissue acidification includes increased accumulation of lactic acid resulting from enhanced glycolysis. Physiological lactate concentrations are actively maintained in the range of 1.5 to 3 mM in the blood and tissues of healthy individuals (1). During active inflammation, local lactate levels can exceed 10 mM (e.g., in rheumatic synovial fluid) and can increase to as high as 40 mM in some cancerous tissues (1). Other studies have shown that tissue pH can fall to as low as 3 at sites of inflammation (e.g., active inflammatory bowel disease [IBD]) (2, 3). It was observed recently that neutrophils (PMN, polymorphonuclear leukocytes) stimulate the release of lactate, up to 300 μM, from intestinal epithelial cells (IEC) during transepithelial migration (4). With a pKa of 3.8, lactate is present in a solution either as lactic acid at low pH or as the ion salt lactate at higher pH. It is also notable that extracellular lactate serves as more than a byproduct of glycolysis. Indeed, recent studies suggest that lactate can signal to various cell types through G protein-coupled receptors (GPCR) (e.g., GPR81, OR51E1) that transduce signals important in sensing tissue microenvironments (1).
Acute inflammation is associated with the accumulation of large numbers of PMN. Inefficient clearance of PMN at these sites of inflammation can promote bystander tissue damage in diseases such as ulcerative colitis (UC) and Crohn’s disease (CD) (5). It is well established that PMN transepithelial migration is an energetically demanding process which rapidly depletes microenvironmental O2 (6). This depletion of local O2 results in the liberation of large amounts of signaling nucleotides (7), the stabilization of hypoxia-inducible factor (HIF), and an increase in anaerobic glycolysis (8–10). A recent study from our laboratory demonstrated that active PMN transepithelial migration results in the inflammatory acidification of the extracellular space and that adenosine, derived from PMN-associated adenine nucleotides (11), up-regulates the Cl−/HCO3− exchanger SLC26A3, promoting pH homeostasis (4). Despite the significance of tissue acidosis, relatively little is known about the mechanism(s) of acidosis-associated signaling on IEC gene regulation or how the IEC sense extracellular acidosis (EA).
In the current work, we define the mechanisms of signaling and outcomes of EA-induced gene regulation in IEC as well as murine and human tissue. Guided by global RNA sequencing (RNA-seq) profiling to ascertain the transcriptional impact of EA on IEC, we hypothesized that EA imprints a unique fingerprint in IEC dominated by cyclic AMP response element-binding protein (CREB)-associated genes. Validation studies demonstrated acidosis-induced expression of several CREB-associated genes. Such signaling was CREB dependent but independent of elevations in intracellular cyclic AMP (cAMP). Furthermore, GPR31 was identified as the apical surface receptor necessary for acidosis-induced CREB phosphorylation and the expression of acidosis-associated genes. This acidosis-associated, CREB-dependent fingerprint was also observed in the inflamed ileums of TNFΔARE mice and CD patients.
Results
Acidosis Elicits Changes in Intestinal Epithelial Gene Expression.
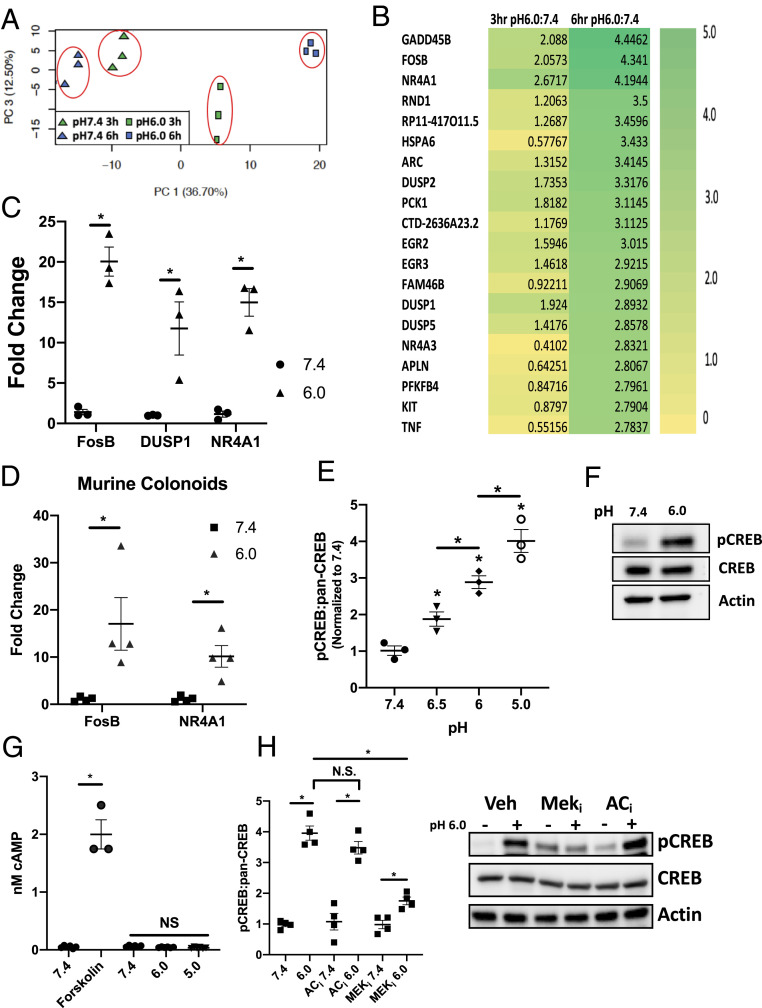
We have previously demonstrated that PMN transepithelial migration results in significant EA. Adaptive pathways elicited by PMN-derived adenosine include the rapid induction of SLC26A3 (4). Here, we examined whether acidosis influenced IEC gene expression independent of other stimuli. To do this, we utilized RNA-seq approaches to profile gene expression in T84 IEC exposed to pH 7.4 or 6.0 Hank’s Balanced Salt Solution with Ca2+ (HBSS+) for periods of 3 or 6 h. As shown in Fig. 1A, principal component analysis revealed prominent separation between treatments and tight clustering within individual treatment groups. The top induced and repressed genes are highlighted in the heatmap displayed in Fig. 1B. To validate the RNA-seq, T84 IEC were independently exposed to acidic HBSS+ (pH 6.0, 6 h), and as shown in Fig. 1C, transcript levels for FosB, DUSP1, and NR4A1 were examined by qPCR. Analysis of the FosB, DUSP1, and NR4A1 transcript revealed a significant increase in expression when exposed to pH 6.0 HBSS+ (20.1 ± 1.8, 11.8 ± 3.28, and 15.0 ± 1.72-fold increase compared to pH 7.4 HBSS+, respectively, each P < 0.001). In addition to FosB, TNF and IL-8 expression were determined after exposure to acidic HBSS+ (pH 6.0, 6 h). As shown in SI Appendix, Fig. S1A, transcript levels for TNF and IL-8 were examined by qPCR, and there was a significant increase in the expression of both genes following expose to pH 6.0 HBSS+ for 6 h (4.3 ± 0.49 and 12.6 ± 1.6-fold increase compared to pH 7.4 HBSS+, respectively, each P < 0.001).
Fig. 1.
Cyclic AMP-independent acidosis-associated CREB phosphorylation. (A) Principal component analysis of acidosis RNA-seq. (B) Top genes induced and repressed by 3 or 6 h exposure to pH 6.0 HBSS+ expressed as log2 fold change (n = 3). (C) Validation of induced FosB, DUSP1, and NR4A1 transcript following exposure to pH 7.4 and 6.0 HBSS+ for 6 h (n = 3). (D) Validation of FosB and NR4A1 transcript expression in C57BL/6 murine colonoids following exposure to pH 7.4 and 6.0 HBSS+ for 6 h (n = 4). (E) Ratio of phospho-CREB to pan-CREB following a 30 min exposure to pH 7.4, 6.5, 6.0, and 5.0 HBSS+ as determined by ELISA (n = 3). (F) Representative Western blot of phospho-CREB and pan-CREB at pH 7.4 and 6.0. (G) Analysis of cAMP in cells exposed to pH 7.4, 6.0, 5.0 HBSS+ or 10 μm forskolin (n = 3). (H) Densitometry analysis and representative Western blot of phospho-CREB and pan-CREB following exposure to pH 7.4 (−) and 6.0 (+) HBSS+ for 30 min in T84 IEC pretreated with 10 μm MEK or 10 μm AC inhibitors (n = 4). Data from each experiment was pooled and expressed as mean ± SEM and the P value determined by t test, *P < 0.01.
To determine the universality of this response and to clarify that this response is not related to the use of a transformed IEC cell line (T84 cells), we replicated these experiments in colonoids derived from C57BL/6 mice. As shown in Fig. 1D and SI Appendix, Fig. S1B, qPCR revealed a significant increase in transcript levels of FosB, NR4A1, and KC [the murine homolog of IL-8 (12, 13)] following exposure to acidic HBSS+ (pH 6.0, 6 h) when compared to pH 7.4 controls (17.0 ± 5.59, 10.2 ± 2.31, and 8.8 ± 2.18-fold increase, respectively, each P < 0.01).
Finally, we examined the need for sustained EA in acidosis-induced gene expression. T84 IEC were exposed to pH 7.4 or 6.0 HBSS+ for 30 min before the media was replaced with 7.4 HBSS+ and the cells incubated for 6 h. As shown in SI Appendix, Fig. S1C, 30 min exposure to low pH did not induce the expression of FosB, NR4A1, or Dusp1, indicating a sustained exposure to EA is required for the induction of acidosis-induced genes.
EA Induces CREB Phosphorylation.
We next sought to determine potential signaling pathways within this RNA-seq data. As a starting point, the RNA-seq data were uploaded to the integrated differential expression and pathway analysis (iDEP) web-based analysis tool (14) and analyzed using the transcriptional regulatory element database (TRED) (15). This analysis identified a significant enrichment of CREB target genes in the data set (P < 0.001). Consistent with this analysis, several of the top induced genes are well-established CREB gene targets (e.g., FosB, NR4A family, PCK1, and the dual specificity protein phosphatase [DUSP] family) (16–18). Based on this observation, we examined CREB phosphorylation following EA. Here, T84 IEC were exposed to increasingly acidic HBSS+ (7.4, 6.5, 6.0, and 5.0) for 30 min and examined for CREB phosphorylation using a phospho-CREB enzyme-linked immunosorbent assay (ELISA). Fig. 1E demonstrates a prominent dose-dependent increase in phospho-CREB with acidification (1.9 ± 0.20, 2.9 ± 0.27, and 4.0 ± 0.32-fold increase compared to pH 7.4 for pH 6.5, 6.0, and 5.0 HBSS+ respectively, ANOVA P < 0.0001). Fig. 1F depicts a representative Western blot, demonstrating significant increases in phospho-CREB at a physiologically relevant pH of 6.0 (3, 19).
EA Does Not Elevate Intracellular cAMP.
It is well established that a common mechanism of CREB phosphorylation is through cAMP signaling, involving both adenylyl cyclase and PKA (20–22). For this reason, intracellular cAMP levels following EA were examined. Interestingly, as shown in Fig. 1G, no increase in intracellular cAMP was evident in IEC exposed to HBSS+ as low as pH 5, whereas positive control (forskolin, 10 μM) rapidly elevated intracellular cAMP when compared to the vehicle control (2.0 ± 0.25 and 0.05 ± 0.021 nM cAMP respectively, P < 0.0001). Phospho-CREB was also examined in T84 IEC pretreated with an adenylyl cyclase inhibitor (2′,5′-Dideoxyadenosine, 10 μM). As shown in Fig. 1H, there is no difference in phospho-CREB when comparing vehicle control T84 IEC and T84 IEC treated with the adenylyl cyclase inhibitor at pH 6.0 (3.8 ± 0.22 and 3.4 ± 0.27, respectively).
Acidosis-Induced CREB Phosphorylation Is Dependent on Mitogen-Activated Protein Kinase Signaling.
cAMP-independent phosphorylation of CREB has been demonstrated to signal through the mitogen-activated protein kinase (MAPK)/ERK pathway (20, 22). Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis of the RNA-seq data revealed a prominent induction of genes associated with the regulation of the MAPK signaling pathway (P < 0.0001). Genes identified in this analysis included members of the DUSP subfamily, DUSP1 and 2. Dusp1 and 2 are negative regulators of the MAPK family, including MEK/ERK, and have been reported to have a role in feedback inhibition of MAPK signaling (23, 24). To examine the role of the MAPK/ERK pathway in acidosis-induced CREB phosphorylation, T84 IEC were pretreated with a MEK1/2 inhibitor (U0126, 10 μM) for 1 h followed by exposure to pH 6.0 HBSS+ for 30 min. As shown in Fig. 1H, cells pretreated with the MEK1/2 inhibitor showed significantly less phospho-CREB following exposure to acidic HBSS+ when compared to vehicle control T84 IEC (1.8 ± 0.13 and 3.8 ± 0.22-fold increase normalized to pH 7.4, respectively, P value <0.001). Observations from the phospho-CREB ELISA were confirmed by Western blot as shown in Fig. 1H. A time course of this response is shown in SI Appendix, Fig. S1D and demonstrated that CREB phosphorylation is observed as early as 10 min following EA, peaks at 30 min, and is diminished at 60 min. The requirement for MEK was evident throughout the time course (SI Appendix, Fig. S1C). The involvement of the MAPK/ERK pathway in acidosis-induced CREB phosphorylation was validated in murine colonoids and revealed that acidosis-induced CREB phosphorylation is likewise MEK1/2 dependent, therefore validating our observations in T84 IEC (SI Appendix, Fig. S1E). Together, these findings identify a prominent role for cAMP-independent CREB phosphorylation elicited by EA.
Acidosis-Induced Gene Expression Is Dependent on MAPK/ERK.
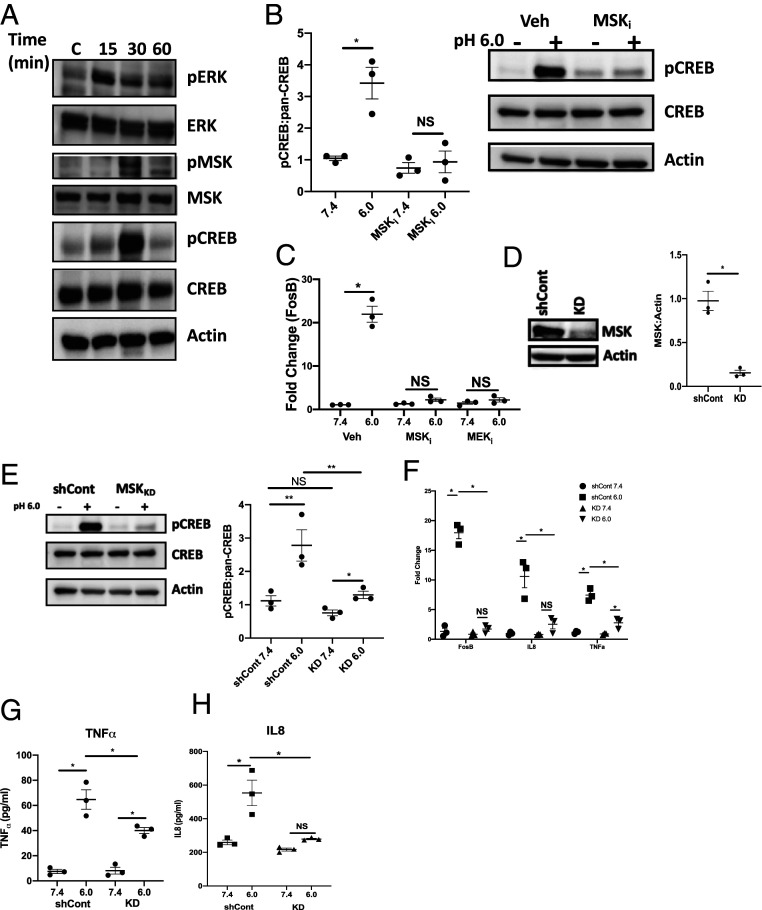
Based on our observation that MEK1/2 inhibitors attenuate CREB phosphorylation, we next aimed to demonstrate activation of the MAPK/ERK pathway under acidic conditions. To examine MAPK/ERK activation, T84 IEC were exposed to pH 6.0 HBSS+ for 0, 15, 30, or 60 min before protein lysates were collected. As shown in Fig. 2A, phospho-ERK was observed 15 min following EA. MSK1, a protein which can be phosphorylated by ERK and can phosphorylate CREB, becomes phosphorylated 30 min following EA (25). As shown in (SI Appendix, Fig. S1F), we observed no significant CREB or ERK phosphorylation beyond 30 min following EA. We extended these studies to determine the significance of MSK1 in acidosis-induced CREB phosphorylation. As shown in Fig. 2B, T84 IEC pretreated for 1 h with a MSK1 inhibitor (SB 747651A dihydrochloride, 1 μM) displayed no increase in phospho-CREB following a 30 min exposure to pH 6.0 HBSS+ when compared to pH 7.4 (0.8 ± 0.30 and 0.9 ± 0.34-fold change, normalized to vehicle control pH 7.4, respectively). We next looked to examine the significance of CREB phosphorylation in the expression of acidosis-associated genes. FosB expression was examined in T84 IEC pretreated with either a MSK1 or MEK1/2 inhibitor for 1 h following exposure to pH 6.0 HBSS+ for 6 h. As shown in Fig. 2C, treatment with either a MSK1 or MEK1/2 inhibitor significantly inhibited expression of FosB under acidic conditions when compared to vehicle controls (1.9 ± 0.37, 1.9 ± 0.56, and 22.2 ± 1.33-fold change, respectively, P < 0.0001). These observations were replicated in murine colonoids. As seen in SI Appendix, Fig. S1G, FosB expression in murine colonoids pretreated with either a MSK1 or MEK1/2 inhibitor was significantly lower than vehicle controls (2.0 ± 0.34, 2.1 ± 0.48, and 13.7 ± 1.94-fold change, respectively, P < 0.0001).
Fig. 2.
MAPK signaling involved in acidosis-associated gene transcription. (A) Western blot depicting phosphorylation of ERK, MSK, and CREB at pH 7.4 or exposed to pH 6.0 for 15, 30, and 60 min. (B) Densitometry analysis and representative Western blot of phospho-CREB and pan-CREB in T84 IEC pretreated with 10 μm MSK1 inhibitor following 30 min exposure to pH 7.4 and 6.0 HBSS+ (n = 3). (C) FosB expression in T84 IEC pretreated with 10 μm MSK or MEK inhibitors following 6 h exposure to pH 7.4 and 6.0 HBSS+ (n = 3). (D) Densitometry analysis and representative Western blot of MSK1 expression in shRNA lentiviral transduced T84 IEC (n = 3). (E) Representative Western blot and densitometry analysis of phospho-CREB and pan-CREB in shCont and MSK1 KD IEC following 30 min exposure to pH 7.4 and 6.0 HBSS+ (n = 3). (F) FosB, IL-8, and TNF-α expression following 6 h exposure to pH 7.4 and 6.0 HBSS+ in shCont and MSK1 KD IEC (n = 3). (G and H) Analysis of secreted TNF (G) and IL-8 (H) following 16 h exposure to pH 7.4 and 6.0 HBSS+ in shCont and MSK1 KD IEC (n = 3). Data from each experiment was pooled and expressed as mean ± SEM and the P value determined by t test or ANOVA, *P < 0.01.
MSK1 Is Required for Acidosis-Associated Gene Transcription.
In an effort to validate the observations made using pharmaceutical inhibitors, we utilized short hairpin RNA (shRNA) to knockdown MSK1 (MSK1 KD) in T84 IEC. As shown in Fig. 2D, Western blot analysis revealed MSK1 was knocked down by 86.4 ± 3.82% in the MSK1 KD IEC compared to vector control (shCont) IEC (P < 0.0001). CREB phosphorylation under acidic conditions was analyzed in MSK1 KD IEC. As shown in Fig. 2E, acidosis-induced CREB phosphorylation (pH 6.0) was significantly attenuated in MSK1 KD IEC compared to shCont IEC (1.3 ± 0.19 and 3.0 ± 0.72-fold change, respectively, normalized to shCont pH 7.4, P < 0.01).
Having validated the impact of MSK1 on CREB phosphorylation in a genetic knockdown model, we turned our attention to acidosis-associated gene transcription. Several acidosis-induced genes identified from the RNA-seq were quantified via qPCR following exposure to pH 6.0 HBSS+ for 6 h in MSK1 KD and shCont IEC. As shown in Fig. 2F, there was no significant increase in either FosB or IL-8 in the MSK1 KD IEC exposed to pH 6.0 HBSS+ for 6 h when compared to pH 7.4 exposed cells (0.8 ± 0.28, 1.7 ± 0.39 and 0.8 ± 0.11, 2.5 ± 0.78-fold change for FosB and IL-8, pH 7.4, 6.0 respectively, normalized to shCont pH 7.4). There was a small but significant increase in TNF-α expression in the MSK1 KD IEC when comparing pH 7.4 and 6.0 exposed cells (0.9 ± 0.09 and 2.8 ± 0.54-fold change, respectively, normalized to shCont 7.4, P < 0.05). However, the fold change in TNF-α was significantly lower in the MSK1 KD IEC when compared to shCont IEC at pH 6.0 (2.8 ± 0.54 and 7.5 ± 0.85-fold change, respectively, normalized to shCont pH 7.4, P < 0.05). The qPCR results for TNF-α and IL-8 were validated by ELISA. shCont and MSK1 KD IEC were treated with pH 7.4 or 6.0 HBSS+ for 24 h. After 24 h, the HBSS+ was collected and analyzed for secreted TNF-α and IL-8. As shown in Fig. 2 G and H, under acidic conditions, MSK1 KD IEC secreted significantly less TNF-α and IL-8 when compared to shCont IEC after 24 h. Following 24 h exposure to acidic HBSS+, the level of secreted TNF-α observed in shCont IEC was 64.5 ± 6.13 pg/mL; this is significantly more than the 40.3 ± 1.83 pg/mL measured in the MSK1 KD IEC (P < 0.01). The concentration of secreted IL-8 measured in shCont IEC under acidic conditions, 553.5 ± 53.9 pg/mL, was significantly more than the 279.3 ± 4.51 pg/mL measured in the MSK1 KD IEC (P < 0.001).
EA Signals through Gαi Coupling.
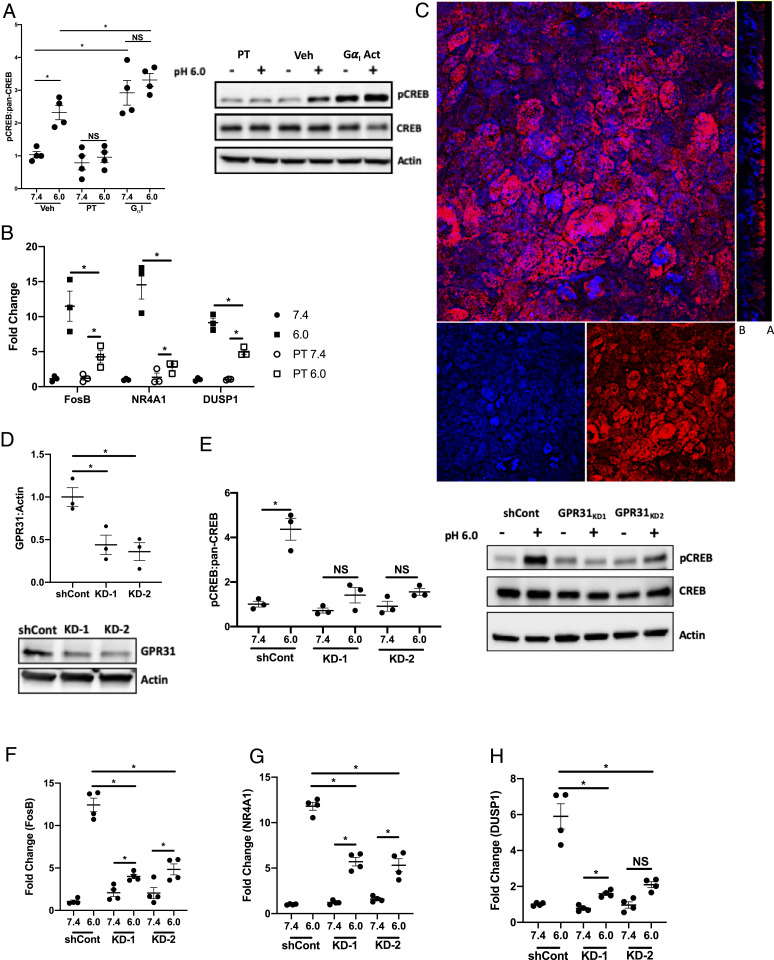
Based on the earlier observation that acidosis-associated CREB phosphorylation was independent of intracellular cAMP elevations and involved the MAPK pathway, we next tested whether the Gαi subunit might transmit signals in IEC following EA. It has been established that activation of the Gαi subunit initiates signaling through the MAPK pathway, specifically MEK and ERK (26). Additionally, signaling through Gαi is independent of cAMP; in fact, activation of Gαi has been shown to inhibit increases in intracellular cAMP (27, 28). To examine the involvement of Gαi in CREB phosphorylation under acidic conditions, T84 IEC were treated with a Gαi inhibitor (pertussis toxin [PT], 1 ng/mL) and a Gαi activator (mastoparan, 1 μM) prior to exposure to acidic HBSS+. As shown in Fig. 3A, Gαi contributes significantly to acidification-associated CREB phosphorylation. T84 IEC treated with PT showed no increases in phospho-CREB following 30 min exposure to pH 6.0 HBSS+ when compared to pH 7.4 HBSS+ (1.0 ± 0.16 and 0.79 ± 0.21-fold change, respectively). Activation of the Gαi subunit with mastoparan resulted in a significant increase in phospho-CREB when compared to vehicle control IEC at pH 7.5 (2.9 ± 0.38 and 1.0 ± 0.095-fold change, respectively, normalized to vehicle control pH 7.4, P < 0.001). There was no significant difference in phospho-CREB when comparing mastoparan-treated IEC exposed to pH 7.4 and 6.0 HBSS+ (2.9 ± 0.38 and 3.3 ± 0.20-fold change, respectively, normalized to vehicle control pH 7.4). A small increase in phospho-CREB was observed when comparing mastoparan-treated IEC and vehicle control IEC exposed to pH 6.0 HBSS+ (3.3 ± 0.20 and 2.3 ± 0.22-fold change, respectively, normalized to vehicle control pH 7.4, P < 0.01). As validation of these results, we examined several other established pH-sensitive surface receptors, TRPV1, 5, and 6 (29–31). T84 cells were pretreated with inhibitors targeting TRPV1, 5, and 6 (10 μm A 425619, 10 μm econazole, and 100 μm 2-aminoethoxydiphenyl borate, respectively) for 1 h before being exposed to pH 7.4 or 6.0 HBSS+ for 30 min. As shown in SI Appendix, Fig. S2A, phospho-CREB was observed after exposure to acidic HBSS+ for 30 min in all cells, with the exception of T84 IEC treated with 1 μm of the MSK1 inhibitor.
Fig. 3.
Acidosis-associated gene regulation through GαI-linked GPR31. (A) Densitometry analysis and representative Western blot of phosphor-CREB and pan-CREB in T84 IEC treated with 1 ng/mL PT or 10 μm mastoparan following 30 min exposure to pH 7.4 and 6.0 HBSS+ (n = 4). (B) FosB, NR4A1, and DUSP1 transcript expression in T84 IEC treated with 1ng/mL PT following 6 h exposure to pH 7.4 and 6.0 HBSS+ (n = 3). (C) Immunofluorescent image of GPR31 expression in T84 IEC; red indicates GPR31 and blue indicates DAPI (100× magnification, compressed z-stack and orthogonal projection, the basal and apical sides are label B and A, respectively). (D) Densitometry analysis and representative Western blot of GPR31 expression in shCont and GRP31 KD T84 IEC (n = 3). (E) Densitometry analysis and representative Western blot of phospho-CREB and pan-CREB in shCont and GPR31 KD T84 IEC following 30 min exposure to pH 7.4 and 6.0 HBSS+. (F and G) FosB (F), NR4A1 (G), and DUSP1 (H) expression in shCont and GPR31 KD T84 IEC following 6 h exposure to pH 7.4 and 6.0 HBSS+ (n = 4). Data from each experiment was pooled and expressed as mean ± SEM and the P value determined by t test, *P < 0.01.
Extending the Gαi studies, we examined the expression of FosB, NR4A1, and DUSP1 under acidic conditions in T84 IEC treated with PT. As shown in Fig. 3B, treatment with PT significantly inhibited acidosis-associated expression of FosB, NR4A1, and DUSP1. When comparing PT-treated cells with vehicle control at pH 6.0, PT treatment significantly inhibited expression of FosB, NR4A1, and DUSP1 (4.3 ± 0.78 and 11.5 ± 1.49, 2.8 ± 0.33 and 14.5 ± 1.43, and 5.0 ± 0.36 and 9.1 ± 0.94-fold change respectively, normalized to vehicle control pH 7.4, P < 0.05).
We also examined the expression of TNF-α and IL-8 under acidic conditions in T84 IEC treated with PT. As shown in SI Appendix, Fig. S2 B and C, treatment with PT significantly inhibited acidosis-associated expression of both TNF-α and IL-8. There was a significant increase in TNF-α in IEC treated with PT when comparing pH 7.4 to 6.0 (1.0 ± 0.11 and 3.3 ± 0.63-fold change, respectively, normalized to vehicle control pH 7.4, P < 0.05). However, when comparing the PT-treated cells with vehicle controls at pH 6.0, PT treatment significantly inhibited expression of the TNF-α transcript (3.3 ± 0.63 and 10.9 ± 1.58-fold change, respectively, normalized to vehicle control pH 7.4, P value < 0.0001). When examining IL-8 expression following exposure to pH 6.0 HBSS+, there was no significant increase in expression in the PT-treated cells when comparing pH 6.0 and 7.4 (2.8 ± 0.33 and 1.2 ± 0.46-fold change, respectively, normalized to vehicle control 7.4). Furthermore, when compared to vehicle controls exposed to pH 6.0, IEC treated with PT expressed significantly less IL-8 (14.5 ± 1.43 and 2.8 ± 0.33-fold change, respectively, normalized to vehicle control 7.4, P < 0.0001).
Loss of MSK Inhibits CREB Phosphorylation following Gαi Activation.
Based on the observation that Gαi is involved in acidosis-associated CREB phosphorylation and gene transcription, we next aimed to determine whether MSK1 was involved. To examine the importance of MSK1 in CREB phosphorylation following Gαi activation, we treated MSK1 KD IEC with the Gαi activator and quantified phospho-CREB. As shown in SI Appendix, Fig. S2D, there was a slight increase in phospho-CREB when comparing MSK1 KD IEC treated with the Gαi activator to untreated MSK1 KD IEC at pH 7.4 (1.4 ± 0.14 and 0.55 ± 0.061, respectively, normalized to shCont pH 7.4, P < 0.01). When comparing the impact of Gαi activation between the MSK1 KD and shCont IEC, we see the loss of MSK1 significantly inhibits Gαi-induced CREB phosphorylation (3.3 ± 0.28 and 1.4 ± 0.14-fold change for shCont and MSK1 KD treated with Gαi activator, respectively, P < 0.0001). Additionally, in the MSK1 KD IEC, pH had no impact on CREB phosphorylation following treatment with the Gαi activator (1.4 ± 0.14 and 1.5 ± 0.23-fold increase for pH 7.4 and 6.0, respectively, normalized to shCont pH 7.4).
Loss of GPR31 Inhibits Acidosis-Associated Gene Expression.
We extended these studies to identify the surface receptor associated with the acidosis response observed in our T84 IEC. GPR31, a Gαi-linked GPCR, activates the MEK-ERK1/2 pathway when bound to 12-(S)-hydroxy-5,8,10,14-eicosatetraenoic acid (32). Recently, GPR31 was shown to be activated under acidic conditions in Chinese hamster ovary cells, with the highest level of activation occurring near pH 6.0 (33). Based on these findings, we examined GPR31 expression in T84 IEC and revealed that GPR31 is located primarily on the apical surface of the T84 IEC with little to no expression on the basal surface (Fig. 3C). Having confirmed the presence of GPR31 in the T84 IEC, we transduced the cells with a shRNA targeting GPR31 to create two GPR31 KD lines. As shown in Fig. 3D, GPR31 was knocked down by 56.0 ± 11.38 and 64.0 ± 10.47% in GRP31 KD-1 and 2, respectively, when compared to shCont. Using these GPR31 KD IEC, we examined the impact of GPR31 on acidosis-associated CREB phosphorylation. We observed a significant reduction in phospho-CREB in both KD cell lines when compared to shCont at pH 6.0 (1.4 ± 0.35, 1.6 ± 0.15, 4.5 ± 0.63-fold change, respectively, normalized to shCont 7.4, P < 0.001) (Fig. 3E). Extending these studies, we examined the impact of GPR31 on the expression of FosB, NR4A1, and DUSP1 under acidic conditions. In both GPR31 KD-1 and 2, there was a significant inhibition of acidosis-induced FosB (Fig. 3F), NR4A1 (Fig. 3G), and DUSP1 (Fig. 3H). Following exposure to pH 6.0 HBSS+ for 6 h, we observed a 12.5 ± 1.62, 11.8 ± 1.08, and 5.9 ± 1.35-fold increase in FosB, NR4A1, and DUSP1 in shCont IEC, respectively. Expression of these genes is significantly higher than the 4.0 ± 0.52 and 4.8 ± 1.30, 5.7 ± 0.94 and 5.3 ± 1.42, and 1.6 ± 0.19 and 2.1 ± 0.33-fold increases observed for FosB, NR4A1, and DUSP1 in GPR31 KD-1 and 2, respectively, when normalized to shCont exposed to pH 7.4 HBSS+ (P < 0.01).
We extended these studies and examined the impact of GPR31 on the expression of IL-8 and TNF-α under acidic conditions as well. In both GPR31 KD-1 and 2, there was a significant inhibition of acidosis-induced IL-8 (SI Appendix, Fig. S2E) and TNF-α (SI Appendix, Fig. S2F) RNA expression. Indeed, following exposure to pH 6.0 HBSS+ for 6 h, we observed a 15.4 ± 2.72-fold increase in IL-8 expression in shCont IEC; this is significantly higher than the 4.5 ± 1.39 and 2.9 ± 0.41-fold increase observed in GRP31 KD-1 and KD-2 IEC, respectively, when normalized to shCont exposed to pH 7.4 HBSS+ (P < 0.0001). There was no significant increase in TNF-α in GPR31 KD-1 and 2 IEC when exposed to pH 6.0 HBSS+ for 6 h when compared to pH 7.4 HBSS+ (1.9 ± 0.40 and 1.4 ± 0.36-fold change, respectively, normalized to shCont pH 7.4). When compared to TNF-α expression in shCont IEC exposed to pH 6.0 HBSS+ (7.1 ± 0.93-fold increase, normalized to shCont 7.4), the loss of GPR31 resulted in significantly less TNF-α transcript following a 6 h exposure to pH 6.0 HBSS+ (P < 0.0001). Taken together, these results strongly implicate GPR31 as a surface receptor that relays EA signals to IEC.
Mucosal Acidification Is Associated with Inflammation in TNFΔARE Ileitis.
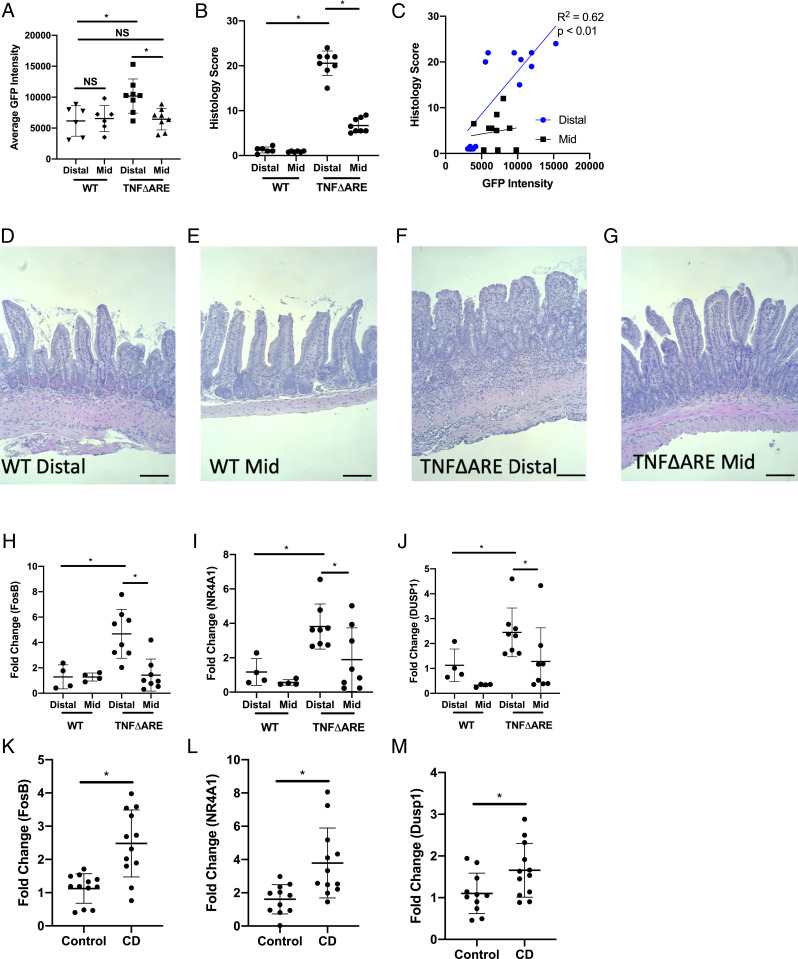
In an effort to better understand the impact of mucosal inflammation on mucosal pH, we constructed an Escherichia coli reporter strain based on the Shewanella oneidensis two-component system SO_4387-SO_4388, which we recently demonstrated is activated by acidic pH (34) (SI Appendix, Fig. S3 A and B). We utilized this E. coli reporter strain to monitor mucosal acidification in the mid- and distal ileum of TNFΔARE and wild-type (WT) mice. In this acid-sensing strain, superfolder green fluorescent protein (sfGFP) expression levels increase from a mean fluorescent intensity (MFI) of 8.2 × 102 at a pH of 7.4 to an MFI of 3.8 × 104 at a pH of 5 (SI Appendix, Fig. S4 A and B). As shown in Fig. 4A, bacteria isolated from the distal ileum of TNFΔARE mice expressed significantly more sfGFP than bacteria isolated from the distal ileum of WT mice, indicative of a more acidic environment (10,183 ± 2,750 versus 6,160 ± 2,461 MFI, respectively, P < 0.01). When comparing bacteria isolated from the distal and mid-ileum of TNFΔARE mice, there is a significant increase in sfGFP fluorescence intensity in the bacteria isolated from the distal ileum (10,183 ± 2,750 and 6,447 ± 1,738 MFI, P < 0.01). There was no significant difference in sfGFP intensity when comparing bacteria isolated from the mid- and distal ileum of WT mice (Fig. 4A).
Fig. 4.
Expression of acidosis-associated genes in inflamed tissue. (A) Flow cytometry analysis of sfGFP intensity in pH reporter E. coli isolated from the mid- and distal ileum of WT and TNFΔARE mice (n = 6 and n = 8 for WT and TNFΔARE mice, respectively). (B) Histological scoring for the mid- and distal ileum collected from WT and TNFΔARE mice (n = 6 and n = 8 for WT and TNFΔARE mice, respectively). (C) Correlation plot between histological score bacterial sfGFP intensity for both the mid- and distal ileum (n = 6 and n = 8 for WT and TNFΔARE mice, respectively). (D–G) Representative histology images for the distal and mid-ileum from WT (D and E) and TNFΔARE (F and G) mice. Black scale bar represents 50 μm. (H–J) Transcript expression of FosB (H), NR4A1 (I), and DUSP1 (J) in the mid- and distal ileum of WT and TNFΔARE mice (n = 4 and n = 8 for WT and TNFΔARE mice, respectively). (K–M) Transcript expression of FosB (K), NR4A1 (L), and DUSP1 (M) in tissue collected from the ileum and colon of healthy controls and CD patients (n = 12). Data from each experiment was pooled and expressed as mean ± SD and the P value determined by t test or ANOVA, *P < 0.01.
Each segment of ileum was scored for inflammation. As shown in Fig. 4B, there was significantly more inflammation in the distal ileum of the TNFΔARE mice when compared to WT mice (20.6 ± 2.7 versus 1.3 ± 0.67, respectively, P < 0.0001). There was some inflammation noted in the mid-ileum of the TNFΔARE mice but far less than the distal ileum (6.7 ± 1.6 and 20.6 ± 2.7, respectively, P < 0.0001). Fig. 4 D–G are representative histology images from the distal and mid-ileum of WT and TNFΔARE mice. The distal ileum showed a significant positive correlation between histologic score and sfGFP intensity (R2 = 0.62, P < 0.01), while the mid-ileum showed no significant correlation (Fig. 4C).
Acidosis-Associated Gene Expression Correlates with Mucosal pH.
Extending these studies, we examined the expression of acidosis-associated genes in the mid- and distal ileums of WT and TNFΔARE mice. As shown in Fig. 4 H–J, there is significant induction of FosB (4.7 ± 1.9-fold increase), NR4A1 (3.8 ± 1.3-fold increase), and DUSP1 (2.4 ± 1.0-fold increase) in the distal ileum of TNFΔARE mice when compared to the distal ileum of WT mice (P < 0.01). When comparing the expression of FosB, NR4A1, and DUSP1 in the mid-ileum of TNFΔARE mice to WT mice, there was no significant increase in any of the genes (1.4 ± 1.3, 1.9 ± 1.8, and 1.3 ± 1.3-fold change, respectively, normalized to WT distal ileum). We next looked to compare gene expression with the sfGFP intensity from the isolated bacteria. As shown in SI Appendix, Fig. S4 A–C, we correlated the expression of FosB (SI Appendix, Fig. S5A), NR4A1 (SI Appendix, Fig. S5B), and DUSP1 (SI Appendix, Fig. S5C) with the average sfGFP intensity of the bacteria from both the distal and mid-ileum. There was a significant positive correlation between the expression of FosB and NR4A1 with the average bacterial sfGFP expression in the distal ileum (R2 = 0.62 and 0.60, respectively, P < 0.01). There was a positive but not significant correlation between the expression of DUSP1 and bacterial sfGFP intensity in the ileum as well (R2 = 0.27, P = 0.08). There was no correlation between the expression of FosB, NR4A1, and DUSP1 in the mid-ileum. These studies were extended to include KC and TNF. KC expression was significantly increased in the distal ileum (13.5 ± 4.8-fold change, P < 0.001) but not the mid-ileum (3.7 ± 2.1-fold change, P < 0.001) of the TNFΔARE mice when compared to WT mice (1.1 ± 0.55 and 2.0 ± 1.6-fold change for the distal and mid-ileum, respectively) (SI Appendix, Fig. S4D). TNF expression was increased in both the distal and mid-ileum of the TNFΔARE; however, the expression of TNF was significantly higher in the distal ileum (16.4 ± 7.6 and 7.2 ± 4.0-fold change, respectively, normalized to the distal and mid-ileum of WT mice, P < 0.01) (SI Appendix, Fig. S4E). Furthermore, we observed a positive correlation between KC expression and bacterial sfGFP intensity in the distal ileum (R2 = 0.55, P < 0.01) but not in the mid-ileum (SI Appendix, Fig. S4F). There was a small but not significant correlation between TNF expression and bacterial sfGFP intensity observed in the distal ileum (R2 = 0.27, P = 0.08) (SI Appendix, Fig. S4G).
We extended these studies to examine the expression of these genes in the colon of mice treated with 3% dextran sodium sulfate (DSS), a chemical irritant which causes colitis in mice. KC was used as a marker of inflammation in these mice, and as shown in SI Appendix, Fig. S6A, mice treated with 3% DSS had a significantly higher expression of KC than water-only controls (134.5 ± 171.1 and 1.3 ± 0.8-fold change, respectively, P < 0.05). In the colon of the DSS-treated mice, we observed significant increases in the transcriptional expression for FosB, Dusp1, and NR4A1 when compared to water-only controls (3.7 ± 2.4, 7.1 ± 7.4, and 15.8 ± 15.6-fold change, respectively, P < 0.05) (SI Appendix, Fig. S6 B–D). Furthermore, when comparing the expression of these three genes to KC, we observed a significant positive correlation between the expression of KC and each of the genes (R2 values 0.31, 0.91, and 0.87 for FosB, Dusp1, an NR4A1, respectively, P < 0.05) (SI Appendix, Fig. S6 E–G).
Increased Expression of Acidosis-Associated Genes in Inflamed CD Patients.
The pH of tissue in patients with IBD can fall to as low as 3 during active inflammation (2, 3). Thus, based on the observations made in the TNFΔARE mice, we extended our studies to compare the expression of acidosis-associated gene expression in tissues collected from healthy controls and active CD patients. Inflamed tissue, as determined during endoscopy, and uninflamed tissue from healthy patients undergoing screening colonoscopy was collected and processed for analysis. Expression of FosB (Fig. 4K), NR4A1 (Fig. 4L), and DUSP1 (Fig. 4M) was determined by qPCR. Expression of all three genes was significantly increased in the inflamed tissue when compared to the uninflamed controls (2.5 ± 1.0, 3.8 ± 2.1, and 1.7 ± 0.64-fold change for FosB, NR4A1, and DUSP1, respectively, normalized to uninflamed controls, P < 0.05). Such findings confirm our findings of acidosis-associated gene expression in cultured epithelia and murine models of mucosal inflammation.
Discussion
Under physiologic conditions, tissues adapt to changes in extracellular pH. An understanding of how tissue senses and responds to inflammatory acidification is a relatively understudied area. Specifically, very little is understood in regards to how IEC sense and transduce acidosis-associated signals. A commonly observed characteristic of many inflammatory sites is tissue acidosis, including active IBD. It has been reported that tissue pH in the colon of individuals with active UC can fall to as low as a pH of 5.0, healthy individuals have colonic pH in the range of 6.8 to 7.4, and extracellular pH can have a significant impact on infiltrating immune cells (2, 35, 36). In a recent study, we showed that PMN transepithelial migration results in significant acidification of the microenvironment and IEC adapt to such changes through the up-regulation of SLC26A3 in an effort to maintain tissue pH homeostasis (4). Here, we examined whether EA independently influenced gene expression in IEC.
Initially, we investigated the impact of extracellular pH on IEC gene transcription. Guided by an unbiased transcriptional profiling of IEC exposed to acidic media, we revealed a significant change in gene expression solely based on lowering the extracellular pH. Such findings that EA influences gene expression are consistent with studies in other cell types (e.g., esophagus and kidney) (37, 38). Extending this analysis, we identified prominent induction of a number of genes that map to CREB-dependent transcriptional regulation. The CREB pathway plays a central role in regulating several inflammatory processes (39–41), and consistent with this premise, we observed a significant induction of TNF-α and IL-8 (and its homolog KC expression in murine colonoids) following exposure to EA. CREB has been shown, for example, to be the dominant transcriptional regulator of TNF-α in IEC (39). While more complicated, CREB has also been shown to cooperatively regulate IL-8 in IEC (42).
To gain insight into the mechanisms involved in acidosis-associated CREB phosphorylation, we presumed a cAMP-dependent induction of CREB. To our surprise, lowering extracellular pH did not elevate intracellular cAMP. This observation in and of itself is quite unexpected given that the predominant pH signaling pathways observed in other tissue types are cAMP dependent (30, 43–45). Based on this observation, we pursued cAMP-independent pathways of CREB phosphorylation. Following EA, we observed phosphorylation of both ERK1/2 and MSK1, both of which are involved in cAMP-independent CREB phosphorylation (46). This observation was further supported with the KEGG analysis which revealed significant up-regulation in genes associated with the MAPK pathway, specifically the DUSP family of proteins, which play a significant role in regulating MAPK signaling (23, 24). Using both pharmacologic and shRNA approaches, we demonstrated the importance of both MAPK signaling and MSK1 in not only CREB phosphorylation but also acidosis-associated gene transcription. ShRNA-mediated knockdown of MSK1 resulted in a loss of acidosis-induced TNF-α and IL-8 expression, implicating a MSK1-CREB axis as a primary transcriptional relay for acidosis-associated responses in IEC.
The observation that acidosis-associated CREB phosphorylation in T84 IEC is independent of both cAMP and calcium channels is of particular interest given that the best characterized pH-sensitive surface receptors operate through either cAMP or involve calcium signaling, that is, CaM kinase and calmodulin (30, 43–45, 47). An alternative mechanism for the activation of the MAPK pathway is through the activation of the Gαi subunit (48, 49). Consistent with earlier work, we observed a significant increase in phospho-CREB after activation of Gαi through the use of mastoparan. When we treated MSK1 KD cells with mastoparan, we observed a significant reduction in phospho-CREB. There was still a slight increase in phospho-CREB when comparing mastoparan-treated MSK1 KD IEC to untreated MSK1 KD IEC; however, we attribute this to the fact that MSK1 is not completely lost in the KD cell line. Furthermore, when Gαi was inhibited by PT, we observed a significant reduction in both acidosis-associated phospho-CREB and the induction of TNF-α and IL-8 under acidic conditions.
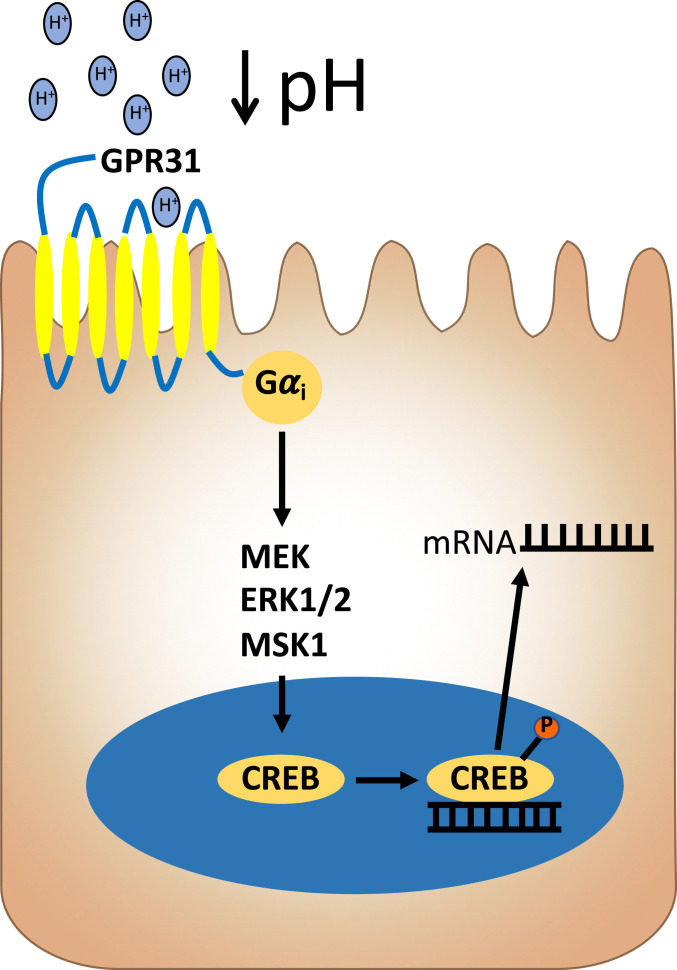
In a search for the surface sensor for low pH in IEC, we focused on GPCRs that signal through Gαi. This search brought us to GPR31, as it has been reported to activate under acidic environments and signals through Gαi (33). Originally identified as the high-affinity receptor for 12(S)-HETE (50), GPR31 signaling has been shown to promote liver damage in ischemia–reperfusion models (32, 51), decrease macrophage efferocytosis (52), and induce dendrite protrusion of CX3CR1+ mononuclear cells in response to lactate and pyruvate in the small intestine (53). GPR31 expression on colonic epithelia has been described in human colon biopsies and appears to be increased in colon cancer (54). Using immunofluorescence and confocal microscopy, we confirmed that GPR31 is expressed in T84 IEC and revealed a primarily apical localization, which would place GPR31 in an ideal location to “sense” luminal pH. A loss of GPR31 significantly inhibited the phosphorylation of CREB at both a pH of 6.5 and 6.0, indicating that GPR31 is involved in sensing a wide range of pH in T84 IEC. As observed in PT-treated cells and the MSK1 KD cell lines, a loss of GPR31 also significantly inhibited the acidosis-induced expression of both IL-8 and TNF-α RNA. Based on these observations, we have put forth the model outlined in Fig. 5. As mentioned above, while GPR31 is increased in colon cancer (54), it is not likely that GPR31 represents a tumor-only observation, as we demonstrated that primary murine colonoids track in parallel to T84 cells with regard to acidosis sensing and CREB-regulated gene expression. In addition, we do not know if 12(S)-HETE may play a role in our model. It is possible, for example, that lowering extracellular pH could activate 12-lipoxygenase to liberate extracellular 12(S)-HETE, providing an autocrine circuit to GPR31 signaling. While no clear evidence exists for such a circuit, 12(S)-HETE is strongly associated with inflammatory environments where acidosis dominates (e.g., ischemia–reperfusion) (55). Based on observations made in this study and the existing literature, we believe that the function of GPR31 is to act as sensor in the early stages of the inflammatory response. Under homeostatic conditions, GPR31 is inactive; however, when the tissue becomes perturbed, GPR31 is activated by the decrease in pH and increased lactate associated with neutrophil transmigration or the presence of invading bacteria. Once activated, GPR31 initiates a signal which promotes proinflammatory signaling by the IEC. IEC have developed a mechanism to maintain pH homeostasis, that is, SLC26A3, during inflammation and would prevent the sustained activation of GPR31. However, during chronic inflammation, these compensatory mechanisms are altered or lost, and the tissue becomes chronically acidified. Under these circumstances, the activation of GPR31 would be sustained, resulting in the acidosis fingerprint observed in this study. Additional work will be necessary to define these principles.
Fig. 5.
Model of acidosis sensing and signaling in IEC. A proposed mechanism for acidosis sensing and signaling through GPR31 in IEC is shown. Extracellular acidification commonly associated with tissue damage and inflammation elicits signaling in IEC that is mediated by GPR31 and relayed through cAMP-independent CREB activation.
Having established a mucosal fingerprint for acidosis-mediated gene expression, we investigated mucosal acidification and the expression of acidosis-associated genes in an in vivo murine model of ileitis, the TNFΔARE mouse model (56). TNFΔARE mice overexpress systemic TNF-α and develop debilitating arthritis and chronic ileitis by 8 to 10 wk of age. The disease has been documented to be most severe in the distal 4 cm of the ileum. Using histological scoring, we confirmed that the distal ileum of TNFΔARE mice was more inflamed than a segment of ileum 6 to 12 cm from the cecum. Consistent with this observation, we showed that transcription expression of proinflammatory cytokines, TNF and KC, are both significantly increased in the distal ileum of the TNFΔARE mice. We constructed an E. coli reporter strain based on the Shewanella oneidensis two-component system SO_4387-SO_4388, which we recently demonstrated is activated by acidic pH (34), to monitor the relative acidity of the mid- and distal ileum. This strain of E. coli reports pH as a sfGFP intensity signature, where sfGFP increases sigmoidally as pH decreases. We previously utilized a similar approach to sense thiosulfate levels in a DSS model of murine intestinal inflammation (57). With this pH reporting E. coli strain, we demonstrated that there is significant mucosal acidification in the distal ileum, where inflammation is highest, of the TNFΔARE mouse. Having established significant mucosal acidification in the distal ileum, we turned our attention to the expression of acidosis-associated genes in these tissues. There was significant up-regulation of the CREB target genes FosB, NR4A1, and DUSP1 in the distal ileum of the TNFΔARE mouse but not in the mid-ileum when compared to WT controls. Furthermore, we identified a significant positive correlation between the expression of FosB and NR4A1 and the sfGFP intensity of bacteria isolated from the distal ileum. These results were reproduced in a DSS model of murine colitis, where we observed significant increases in the expression of FosB, NR4A1, and DUSP1 in DSS-treated mice when compared to water-only controls.
Having observed significant induction of acidosis-associated genes that correlated with mucosal acidification in the TNFΔARE mouse in conjunction with previous studies that revealed tissue acidosis in patients with active IBD (2, 3), we extended this analysis to tissue derived from active IBD patients. This analysis demonstrated a prominent induction of FosB, NR4A1, and DUSP1 with active IBD. Such a strong correlation in expression patterns between cultured cells, murine tissue, and active IBD lead us to conclude that our profile of acidosis target genes, including FosB, NR4A1, and DUSP1, strongly reflect the profile of inflammatory acidification.
Taken together, these results offer not only molecular insight into the influence of acidosis on IEC gene regulation but also reveal a previously undescribed CREB-dependent pathway for sensing extracellular pH in IEC. Our results highlight an acidosis-associated transcriptional imprinting response that could serve to be important as both a biomarker of active inflammation as well as a template for novel therapeutic drug development.
Materials and Methods
Cell Culture.
T84 (#CCL-248; American Type Culture Collection [ATCC]) human epithelial cell line was obtained from ATCC and was cultured in 95% air with 5% CO2 at 37 °C according to instructions provided by ATCC. Low passage (<20) cells were cultured for 7 to 10 d to obtain confluent cell monolayers determined by light microscopy. HBSS+ (Sigma-Aldrich with 10 mM Hepes (Thermo Fisher Scientific) was adjusted to pH 7.4, 6.5, 6.0, or 5.0 using 10 N HCl. T84 cells were treated with the compounds and concentrations listed in Table 1. Cells were pretreated with inhibitors for 30 min before the start of experiments.
Table 1.
List of pharmacological agents
| Compound | Target | Concentration | Source |
| U0126 | MEK1/2 | 10 μm | Tocris |
| SB 747651A dihydrochloride | MSK1 | 1 μm | Tocris |
| 2′,5′-Dideoxyadenosine | Adenylyl cyclase | 10 μm | Sigma-Aldrich |
| Mastoparan | GαI Activator | 1 μm | Tocris |
| Pertussis Toxin | GαI Inhibitor | 1 ng/mL | Tocris |
| A 425619 | TRPV1 | 10 μm | Tocris |
| Econazole | TRPV5 | 10 μm | Sigma-Aldrich |
| 2-aminoethoxydiphenyl borate | TRPV6 | 100 μm | Tocris |
Mouse colonoids created from mouse IEC were isolated from the colons of C57BL/6 mice as previously described (58). Briefly, colonoids were suspended in Matrigel (Corning) and plated into 6-well plates; each well contains four 25-μL Matrigel bubble overlaid with 1 mL complete media. Complete mouse colonoid media, containing WNT3a, EGF, Noggin, and R-Spondin (WENR), was prepared as previously described (59, 60).
Plasmids encoding MSK1, TRCN0000001494, GPR31, TRCN0000008879, and nontargeting shRNA were purchased from the Functional Genomics Facility in Denver, Colorado. To produce lentiviral vectors, lentiviral plasmids with the target shRNA were transduced into HEK293T cells together with second-generation packaging plasmids (psPAX2 and pMD2.G) following previously published procedures (61). For viral infection of T84 cells, 500 μL viral supernatant was combined with 500 μL cell culture media and was placed on 60 to 70% confluent T84 cells in a 60-mm dish along with 10 ug/mL polybrene and incubated for 24 h. After 24 h, the viral supernatant was replaced with fresh media and incubated for another 24 h. Media containing 6 µg/mL puromycin was added to the cells. After 7 d of selection, cells were collected and assayed for MSK1 and GPR31 knockdown via Western blot, described below.
To measure cAMP, an MSD Cyclic AMP Assay Kit (MESO Scale Diagnostics) was utilized. Briefly, T84 cells grown in 6-well plates were treated with HBSS+ at pH 7.4, 6.0, or 5.0 for 30 min. T84 were washed with HBSS+, trypsonized, and resuspended in HBSS+ at a concentration of 1.0 × 106 cells/mL. A total of 20,000 cells were lysed in the provided plates and cAMP quantified. Technical triplicates were used for each sample. IL-8 and TNF-α were measured using a human ProInflammatory-4 II tissue culture kit following manufacturers protocols (MESO Scale Diagnostics). Briefly, T84 cells were cultured for 16 hr in HBSS+ at pH 7.4 and 6.0. The supernatant was collected and analyzed for IL-8 and TNF-α. Technical triplicates were used for each sample. Each experiment was performed in a biological triplicate using different a cell passage for each replicate.
Construction of the pH Reporter E. coli Strain.
A previously described acidic pH-sensing E. coli strain was modified to facilitate its use in vivo (34). This strain expresses an engineered, acidic pH-activated two-component system sensor consisting of SO_4387 and SO_4388REC -PsdRDBD along with an sfGFP reporter to measure sensor activation (SI Appendix, Fig. S3A). Here, SO_4387 is an acidic pH-activated sensor kinase, and SO_4388REC-PsdRDBD is an engineered version of the transcription-activating response regulator SO_4388 wherein the native DNA-binding domain has been swapped for that of PsdR, enabling this two-component system to activate a well-characterized output promoter with a large dynamic range. In our previously published strain, the expression of SO_4387 and SO_4388REC -PsdRDBD was controlled by small-molecule–inducible promoters that are not suitable for in vivo work. To enable its use in mouse studies, a constitutive sensor was constructed in which constitutive promoters from the Anderson promoter library control SO_4387 and SO_4388REC -PsdRDBD expression. Two rounds of optimization were performed to identify an optimal set of constitutive promoters for SO_4387 and SO_4388REC -PsdRDBD expression in E. coli BW28357 (62) as indicated by a high fold change in sfGFP signal between cells cultured at pH 6 and pH 8 (SI Appendix, Fig. S3 B and C).
To measure pH response, 3 mL overnight Luria–Bertani (LB) cultures were inoculated from glycerol stocks. Overnight cultures (10 µL) were used to inoculate 3 mL precultures in M9 media (1× M9 salts, 2 mM MgSO4, 100 µM CaCl2, 0.4% [vol/vol] glucose, and 0.2% [vol/vol] casamino acids). After a 2-h incubation, precultures were diluted to OD600 = 0.0001 in 3 mL buffered M9 media (50 mM Hepes and 50 mM 2-morpholinoethanesulfonic acid hydrate) adjusted to pH 6 or pH 8. After a 5-h incubation, cultures were placed in an ice bath to arrest growth, and cell fluorescence was measured by flow cytometry on a BD FACScan instrument. FlowCal software and Rainbow Calibration beads (Spherotech, RCP-30-5A) were used to process and normalize flow cytometry data to mean equivalents of fluorescein units (63). The fold-change response to pH was calculated by first subtracting the mean fluorescence of an sfGFP negative control to account for cell autofluorescence and then dividing the mean fluorescence at pH 6 to that at pH 8 for each strain. All culturing steps were performed at 37 °C with 250 rpm shaking. A total of 35 µg/mL chloramphenicol (Alfa Aesar B20841) and 100 µg/mL spectinomycin (Gold Biotechnology S-140-25) were supplemented as needed for plasmid maintenance.
Animal Studies.
The B6.129P-TnfΔARE (TNFΔARE) strain was previously described (56), and WT littermates were bred in house. All animals were handled according to protocols approved by the Institutional Animal Care and Use Committee. TNFΔARE and WT mice that are 10 wk old were given 5 g/l streptomycin in drinking water for 48 h. After 48 h, the mice were gavaged with 100 μL of K-12 E. coli containing the pH reporter construct, described above, in LB at an OD600 of ∼1. 24 h after gavage, the mice were euthanized. Postmortem, the distal 5 cm of the ileum (distal ileum) and a segment of ileum 6 to 12 cm from the cecum (mid-ileum) were collected by blunt dissection. A portion of the mid- and distal ileum was fixed in 10% buffered-formalin (Sigma-Aldrich) prior to paraffin imbedding and staining for histology. A second portion was placed in RNAlater (Sigma-Aldrich) and used for RNA isolation. The remaining ∼4-cm segment was placed in a 15-mL conical with 5 mL ice-cold phosphate-buffered saline (PBS) containing 5 mM ethylenediaminetetraacetic acid and 15 mM Hepes and shaken vigorously for 15 min at 4 C. The conical was than centrifuged for 5 min at 500 × g at 4 °C. Following centrifugation, 4 mL of the supernatant containing the bacteria was transferred to a 15-mL conical containing 1 mL of 5% paraformaldehyde (PFA) for a final concentration of 1% PFA. The bacteria were incubated in 1% PFA for 10 min before the conical was centrifuged for 5 min at 5,000 × g at 4 °C. The supernatant was discarded and the bacteria resuspended in PBS. The bacteria were washed three times before being resuspended in 500 μL PBS and analyzed for sfGFP MFI by flow cytometry.
DSS Model of Colitis.
C57BL/6 mice (The Jackson Laboratory) were bred in house. All animals were handled according to protocols approved by the Institutional Animal Care and Use Committee. Gender, age, and weight-matched mice were used in DSS studies. DSS (36,000 to 50,000 molecular weight; MP Biomedicals) was added to drinking water (3%) for 5 d before being replaced with water. Mice were monitored daily for weight loss over 7 d. Postmortem colons were harvested by blunt dissection and the distal 2 cm collected. The tissue was placed in TRIzol and homogenized with a tissue grinder before being processed as described below. Four mice were used for each experimental group, and the experiment was repeated twice.
Histological Scoring.
A blinded reviewer scored sections of the mid- and distal ileum from both WT and TNFΔARE mice as previously described (64). Briefly, the reviewer scored the entire sections of ileum for active and chronic inflammation and villus architecture. Markers for active inflammation included presence of neutrophils within the lamina propria and submucosal space and mucosal erosion/ulceration. Markers of chronic inflammation included increased mononuclear inflammatory cells and separation of the crypt base from the muscularis mucosa. The three scores were combined to give the final histological score.
RNA-seq and Analysis.
T84 cells were plated on 6-well plates (Corning) and grown to confluency. T84 cells were washed with HBSS+ before being treated with HBSS+ at pH 7.4 or 6.0 for 3 or 6 h. The cells were collected in TRIzol (Thermo Fisher Scientific) for RNA isolation. Messenger RNA sequencing was done in a biological triplicate using an Illumina HiSeq 4000 platform. Expression data were normalized to fragments per kilobase of transcript per million, and the differentially expressed genes were determined by the DESeq2 package of R software using default values. Significantly differentially expressed genes were defined by log2 fold change > 1 and adjusted P value < 0.05. KEGG and TRED analysis utilized a false discovery rate cutoff of 0.1 and minimum a fold change of 2.
Transcriptional Analysis.
TRIzol reagent (Invitrogen) was used to isolate RNA from T84 cells, murine colonoids, and tissue samples following manufactures instructions. Complementary DNA (cDNA) was reverse transcribed using the iScript cDNA Synthesis Kit (Bio-Rad) using 1 μg RNA. PCR analysis was performed using SYBR Green (Applied Biosystems) using the primers in Table 2. Each experiment was performed in biological triplicate.
Table 2.
List of qPCR Primers
| Gene target | Forward primer | Reverse primer |
| FosB (Human) | 5′-GCTGCAAGATCCCCTACGAAG-3′ | 5′-ACGAAGAAGTGTACGAAGGGTT-3′ |
| NR4A1 (Human) | 5′-ATGCCCTGTATCCAAGCCC-3′ | 5′-GTGTAGCCGTCCATGAAGGT-3′ |
| DUSP1 (Human) | 5′-ACCACCACCGTGTTCAACTTC-3′ | 5′-TGGGAGAGGTCGTAATGGGG-3′ |
| TNF (Human) | 5′-CCTCTCTCTAATCAGCCCTCTG-3′ | 5′-GAGGACCTGGGAGTAGATGAG-3′ |
| IL8 (Human) | 5′-TTTTGCCAAGGAGTGCTAAAGA-3′ | 5′-AACCCTCTGCACCCAGTTTTC-3′ |
| β-actin (Human) | 5′-GCACTCTTCCAGCCTTCCTTCC-3′ | 5′-CAGGTCTTTGCGGATGTCCACG-3′ |
| FosB (Mouse) | 5′-CCTCCGCCGAGTCTCAGTA-3′ | 5′-CCTGGCATGTCATAAGGGTCA-3′ |
| NR4A1 (Mouse) | 5′-CTTCGGCGTCCTTCAAGTTTG-3′ | 5′-GGCTGGAAGTTGGGTGTAGA-3′ |
| DUSP1 (Mouse) | 5′-GTTGTTGGATTGTCGCTCCTT-3′ | 5′-TTGGGCACGATATGCTCCAG-3′ |
| TNF (Mouse) | 5′-CCCTCACACTCAGATCATCTTCT-3′ | 5′-GCTACGACGTGGGCTACAG-3′ |
| KC (Mouse) | 5′-CTGGGATTCACCTCAAGAACATC-3′ | 5′-CAGGGTCAAGGCAAGCCTC-3′ |
| β-actin (Mouse) | 5′-AACCCTAAGGCCAACCGTGAA-3′ | 5′-TCACGCACGATTTCCCTCTCA-3′ |
Protein Analysis and Immunofluorescence.
Whole cell lysates were extracted into radioimmunoprecipitation assay buffer with protease and phosphatase inhibitors (Roche). Lysates were spun at max rpm for 45 min at 4 °C. Supernatant was collected and quantified for normalization using a Pierce BCA Protein Assay Kit (Thermo Fisher Scientific). A 4× Laemmli sample buffer (Bio-Rad) with 2-mercaptoethanol was added to the samples. Samples were boiled for 5 min, and 10 μg lysate was added to 10% Mini-PROTEAN Precast Gels (Bio-Rad). The gels were run at 120 Volts for ∼1 h using Bio-Rad Western blot equipment. The protein was transferred to polyvinylidene difluoride using a Trans-Blot Turbo RTA Kit (Bio-Rad). Western blotting of these lysates was performed using the antibodies in Table 3. Phospho-CREB and total CREB was analyzed using a Human Phospho-CREB (Ser133) and total CREB ELISA kit (RayBiotech). Each experiment was performed in biological triplicate.
Table 3.
List of antibodies
| Protein target | Host species | Source |
| Phospho-CREB (Ser133) | Rabbit | Cell Signaling Technology |
| CREB (48H2) | Rabbit | Cell Signaling Technology |
| Phospho Erk1/2 (Thr202/Tyr204) | Rabbit | Cell Signaling Technology |
| ERK1/2 | Rabbit | Cell Signaling Technology |
| Phospho MSK1 (Thr581) | Rabbit | Cell Signaling Technology |
| MSK1 | Rabbit | Cell Signaling Technology |
| GPR31 (C-terminal) | Rabbit | Abcam |
| Actin | Rabbit | Abcam |
To localize GPR31 expression in IEC, T84 cells were plated on small 0.33-cm2 permeable supports (0.4 μm pore, Corning) until confluent as measured by transepithelial resistance. Inserts were washed with HBSS+ and fixed with iced methanol for 30 min at −20 °C. After fixation, inserts were briefly washed with PBS (Thermo Fisher Scientific). Inserts were blocked in PBS containing 10% goat serum (Jackson ImmunoResearch) at room temperature for 1 h. Inserts were stained with rabbit polyclonal anti-GRP31 followed by Alexa Fluor 568 secondary Ab (Invitrogen) and counter stained with ProLong Gold Antifade with DAPI (Thermo Fisher Scientific). Inserts were imaged using an Olympus FV1000 FCS/RICS confocal microscope equipped with 405, 488, and 543 nm laser. Images were captured every 0.5 μm.
Patient Sample Analysis.
This study was approved by the University of Colorado Anschutz Medical Campus institutional review board, IRB# 14-2012. Patient biopsy samples were collected for the biobank repository at the University of Colorado Crohn’s and Colitis Center. Patient consent was obtained prior to the procedure. During the consent process, patients were informed on how the tissue would be used after collection. Endoscopic disease status and location were noted by the physician who obtained the biopsy samples. A total of 12 healthy controls and 12 inflamed CD samples (collected from both the ileum and colon) were analyzed by qRT-PCR analysis.
Supplementary Material
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2023871118/-/DCSupplemental.
Data Availability
RNA-seq data have been deposited in the Gene Expression Omnibus (GSE154982) (65).
References
- 1.Pucino V., Bombardieri M., Pitzalis C., Mauro C., Lactate at the crossroads of metabolism, inflammation, and autoimmunity. Eur. J. Immunol. 47, 14–21 (2017). [DOI] [PubMed] [Google Scholar]
- 2.Roediger W. E., Lawson M. J., Kwok V., Grant A. K., Pannall P. R., Colonic bicarbonate output as a test of disease activity in ulcerative colitis. J. Clin. Pathol. 37, 704–707 (1984). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fallingborg J., Christensen L. A., Jacobsen B. A., Rasmussen S. N., Very low intraluminal colonic pH in patients with active ulcerative colitis. Dig. Dis. Sci. 38, 1989–1993 (1993). [DOI] [PubMed] [Google Scholar]
- 4.Cartwright I. M., et al., Adaptation to inflammatory acidity through neutrophil-derived adenosine regulation of SLC26A3. Mucosal Immunol. 13, 230–244 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Parkos C. A., Neutrophil-epithelial interactions: A double-edged sword. Am. J. Pathol. 186, 1404–1416 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Colgan S. P., Dzus A. L., Parkos C. A., Epithelial exposure to hypoxia modulates neutrophil transepithelial migration. J. Exp. Med. 184, 1003–1015 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weissmüller T., et al., PMNs facilitate translocation of platelets across human and mouse epithelium and together alter fluid homeostasis via epithelial cell-expressed ecto-NTPDases. J. Clin. Invest. 118, 3682–3692 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chin A. C., Parkos C. A., Pathobiology of neutrophil transepithelial migration: Implications in mediating epithelial injury. Annu. Rev. Pathol. 2, 111–143 (2007). [DOI] [PubMed] [Google Scholar]
- 9.Kominsky D. J., Campbell E. L., Colgan S. P., Metabolic shifts in immunity and inflammation. J. Immunol. 184, 4062–4068 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Campbell E. L., et al., Transmigrating neutrophils shape the mucosal microenvironment through localized oxygen depletion to influence resolution of inflammation. Immunity 40, 66–77 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Curtis V. F., et al., Neutrophils as sources of dinucleotide polyphosphates and metabolism by epithelial ENPP1 to influence barrier function via adenosine signaling. Mol. Biol. Cell 29, 2687–2699 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bozic C. R., et al., The murine interleukin 8 type B receptor homologue and its ligands. Expression and biological characterization. J. Biol. Chem. 269, 29355–29358 (1994). [PubMed] [Google Scholar]
- 13.Bozic C. R., et al., Expression and biologic characterization of the murine chemokine KC. J. Immunol. 154, 6048–6057 (1995). [PubMed] [Google Scholar]
- 14.Ge S. X., Son E. W., Yao R., iDEP: an integrated web application for differential expression and pathway analysis of RNA-Seq data. BMC Bioinf. 19, 534 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jiang C., Xuan Z., Zhao F., Zhang M. Q., Tred: A transcriptional regulatory element database, new entries and other development. Nucleic Acids Res. 35, D137–D140 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Impey S., et al., Defining the CREB regulon: A genome-wide analysis of transcription factor regulatory regions. Cell 119, 1041–1054 (2004). [DOI] [PubMed] [Google Scholar]
- 17.Conkright M. D., et al., Genome-wide analysis of CREB target genes reveals a core promoter requirement for cAMP responsiveness. Mol. Cell 11, 1101–1108 (2003). [DOI] [PubMed] [Google Scholar]
- 18.Volakakis N., et al., NR4A orphan nuclear receptors as mediators of CREB-dependent neuroprotection. Proc. Natl. Acad. Sci. U.S.A. 107, 12317–12322 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roediger W. E., et al., Luminal ions and short chain fatty acids as markers of functional activity of the mucosa in ulcerative colitis. J. Clin. Pathol. 35, 323–326 (1982). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang H., Xu J., Lazarovici P., Quirion R., Zheng W., cAMP response element-binding protein (CREB): A possible signaling molecule link in the pathophysiology of schizophrenia. Front. Mol. Neurosci. 11, 255 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yan K., Gao L. N., Cui Y. L., Zhang Y., Zhou X., The cyclic AMP signaling pathway: Exploring targets for successful drug discovery (Review). Mol. Med. Rep. 13, 3715–3723 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ichiki T., Role of cAMP response element binding protein in cardiovascular remodeling: Good, bad, or both? Arterioscler. Thromb. Vasc. Biol. 26, 449–455 (2006). [DOI] [PubMed] [Google Scholar]
- 23.Liu C., et al., Dual-specificity phosphatase DUSP1 protects overactivation of hypoxia-inducible factor 1 through inactivating ERK MAPK. Exp. Cell Res. 309, 410–418 (2005). [DOI] [PubMed] [Google Scholar]
- 24.Shah S., King E. M., Chandrasekhar A., Newton R., Roles for the mitogen-activated protein kinase (MAPK) phosphatase, DUSP1, in feedback control of inflammatory gene expression and repression by dexamethasone. J. Biol. Chem. 289, 13667–13679 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Arthur J. S., Cohen P., MSK1 is required for CREB phosphorylation in response to mitogens in mouse embryonic stem cells. FEBS Lett. 482, 44–48 (2000). [DOI] [PubMed] [Google Scholar]
- 26.Winitz S., et al., Involvement of Ras and Raf in the Gi-coupled acetylcholine muscarinic m2 receptor activation of mitogen-activated protein (MAP) kinase kinase and MAP kinase. J. Biol. Chem. 268, 19196–19199 (1993). [PubMed] [Google Scholar]
- 27.Manna P. P., Frazier W. A., CD47 mediates killing of breast tumor cells via Gi-dependent inhibition of protein kinase A. Cancer Res. 64, 1026–1036 (2004). [DOI] [PubMed] [Google Scholar]
- 28.Chen J., Iyengar R., Inhibition of cloned adenylyl cyclases by mutant-activated Gi-alpha and specific suppression of type 2 adenylyl cyclase inhibition by phorbol ester treatment. J. Biol. Chem. 268, 12253–12256 (1993). [PubMed] [Google Scholar]
- 29.Yoshida A., et al., TRPV1 is crucial for proinflammatory STAT3 signaling and thermoregulation-associated pathways in the brain during inflammation. Sci. Rep. 6, 26088 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yeh B. I., Sun T. J., Lee J. Z., Chen H. H., Huang C. L., Mechanism and molecular determinant for regulation of rabbit transient receptor potential type 5 (TRPV5) channel by extracellular pH. J. Biol. Chem. 278, 51044–51052 (2003). [DOI] [PubMed] [Google Scholar]
- 31.Lambers T. T., et al., Extracellular pH dynamically controls cell surface delivery of functional TRPV5 channels. Mol. Cell Biol. 27, 1486–1494 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang X. J., et al., An ALOX12-12-HETE-GPR31 signaling axis is a key mediator of hepatic ischemia-reperfusion injury. Nat. Med. 24, 73–83 (2018). [DOI] [PubMed] [Google Scholar]
- 33.Mashiko M., Kurosawa A., Tani Y., Tsuji T., Takeda S., GPR31 and GPR151 are activated under acidic conditions. J. Biochem. 166, 317–322 (2019). [DOI] [PubMed] [Google Scholar]
- 34.Schmidl S. R., et al., Rewiring bacterial two-component systems by modular DNA-binding domain swapping. Nat. Chem. Biol. 15, 690–698 (2019). [DOI] [PubMed] [Google Scholar]
- 35.Lardner A., The effects of extracellular pH on immune function. J. Leukoc. Biol. 69, 522–530 (2001). [PubMed] [Google Scholar]
- 36.Rajamäki K., et al., Extracellular acidosis is a novel danger signal alerting innate immunity via the NLRP3 inflammasome. J. Biol. Chem. 288, 13410–13419 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rafiee P., et al., Effect of curcumin on acidic pH-induced expression of IL-6 and IL-8 in human esophageal epithelial cells (HET-1A): Role of PKC, MAPKs, and NF-kappaB. Am. J. Physiol. Gastrointest. Liver Physiol. 296, G388–G398 (2009). [DOI] [PubMed] [Google Scholar]
- 38.Nowik M., et al., Genome-wide gene expression profiling reveals renal genes regulated during metabolic acidosis. Physiol. Genomics 32, 322–334 (2008). [DOI] [PubMed] [Google Scholar]
- 39.Taylor C. T., Fueki N., Agah A., Hershberg R. M., Colgan S. P., Critical role of cAMP response element binding protein expression in hypoxia-elicited induction of epithelial tumor necrosis factor-alpha. J. Biol. Chem. 274, 19447–19454 (1999). [DOI] [PubMed] [Google Scholar]
- 40.Kawaguchi M., et al., IL-17F-induced IL-11 release in bronchial epithelial cells via MSK1-CREB pathway. Am. J. Physiol. Lung Cell Mol. Physiol. 296, L804–L810 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Westbom C. M., et al., CREB-induced inflammation is important for malignant mesothelioma growth. Am. J. Pathol. 184, 2816–2827 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Choi E. Y., et al., Transcriptional regulation of IL-8 by iron chelator in human epithelial cells is independent from NF-kappaB but involves ERK1/2- and p38 kinase-dependent activation of AP-1. J. Cell. Biochem. 102, 1442–1457 (2007). [DOI] [PubMed] [Google Scholar]
- 43.Mogi C., et al., Sphingosylphosphorylcholine antagonizes proton-sensing ovarian cancer G-protein-coupled receptor 1 (OGR1)-mediated inositol phosphate production and cAMP accumulation. J. Pharmacol. Sci. 99, 160–167 (2005). [DOI] [PubMed] [Google Scholar]
- 44.Mohapatra D. P., Nau C., Regulation of Ca2+-dependent desensitization in the vanilloid receptor TRPV1 by calcineurin and cAMP-dependent protein kinase. J. Biol. Chem. 280, 13424–13432 (2005). [DOI] [PubMed] [Google Scholar]
- 45.Chen A., et al., Activation of GPR4 by acidosis increases endothelial cell adhesion through the cAMP/Epac pathway. PLoS One 6, e27586 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Deak M., Clifton A. D., Lucocq L. M., Alessi D. R., Mitogen- and stress-activated protein kinase-1 (MSK1) is directly activated by MAPK and SAPK2/p38, and may mediate activation of CREB. EMBO J. 17, 4426–4441 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tomura H., et al., Prostaglandin I(2) production and cAMP accumulation in response to acidic extracellular pH through OGR1 in human aortic smooth muscle cells. J. Biol. Chem. 280, 34458–34464 (2005). [DOI] [PubMed] [Google Scholar]
- 48.Okamoto H., et al., EDG1 is a functional sphingosine-1-phosphate receptor that is linked via a Gi/o to multiple signaling pathways, including phospholipase C activation, Ca2+ mobilization, Ras-mitogen-activated protein kinase activation, and adenylate cyclase inhibition. J. Biol. Chem. 273, 27104–27110 (1998). [DOI] [PubMed] [Google Scholar]
- 49.Gripentrog J. M., Miettinen H. M., Formyl peptide receptor-mediated ERK1/2 activation occurs through G(i) and is not dependent on beta-arrestin1/2. Cell. Signal. 20, 424–431 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Guo Y., et al., Identification of the orphan G protein-coupled receptor GPR31 as a receptor for 12-(S)-hydroxyeicosatetraenoic acid. J. Biol. Chem. 286, 33832–33840 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang F., et al., Ischemia reperfusion injury promotes recurrence of hepatocellular carcinoma in fatty liver via ALOX12-12HETE-GPR31 signaling axis. J. Exp. Clin. Cancer Res. 38, 489 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Manega C. M., et al., 12(S)-Hydroxyeicosatetraenoic acid downregulates monocyte-derived macrophage efferocytosis: New insights in atherosclerosis. Pharmacol. Res. 144, 336–342 (2019). [DOI] [PubMed] [Google Scholar]
- 53.Morita N., et al., GPR31-dependent dendrite protrusion of intestinal CX3CR1+ cells by bacterial metabolites. Nature 566, 110–114 (2019). [DOI] [PubMed] [Google Scholar]
- 54.Rong Y. M., et al., Overexpression of G protein-coupled receptor 31 as a poor prognosticator in human colorectal cancer. World J. Gastroenterol. 24, 4679–4690 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Napolitano M., The role of the 12(S)-HETE/GPR31/12-HETER axis in cancer and ischemia-reperfusion injury. Biochem. Soc. Trans. 47, 743–754 (2019). [DOI] [PubMed] [Google Scholar]
- 56.Kontoyiannis D., Pasparakis M., Pizarro T. T., Cominelli F., Kollias G., Impaired on/off regulation of TNF biosynthesis in mice lacking TNF AU-rich elements: Implications for joint and gut-associated immunopathologies. Immunity 10, 387–398 (1999). [DOI] [PubMed] [Google Scholar]
- 57.Daeffler K. N., et al., Engineering bacterial thiosulfate and tetrathionate sensors for detecting gut inflammation. Mol. Syst. Biol. 13, 923 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mahe M. M., et al., Establishment of gastrointestinal epithelial organoids. Curr. Protoc. Mouse Biol. 3, 217–240 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Finkbeiner S. R., et al., Transcriptome-wide analysis reveals hallmarks of human intestine development and maturation In Vitro and In Vivo. Stem Cell Reports, 10.1016/j.stemcr.2015.04.010 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jones J. C., et al., Cellular plasticity of Defa4Cre-expressing paneth cells in response to notch activation and intestinal injury. Cell. Mol. Gastroenterol. Hepatol. 7, 533–554 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang L., et al., Long noncoding RNA DANCR is a positive regulator of proliferation and chondrogenic differentiation in human synovium-derived stem cells. DNA Cell Biol. 36, 136–142 (2017). [DOI] [PubMed] [Google Scholar]
- 62.Zhou L., Lei X. H., Bochner B. R., Wanner B. L., Phenotype microarray analysis of Escherichia coli K-12 mutants with deletions of all two-component systems. J. Bacteriol. 185, 4956–4972 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Castillo-Hair S. M., et al., FlowCal: A user-friendly, open source software tool for automatically converting flow cytometry data from arbitrary to calibrated units. ACS Synth. Biol. 5, 774–780 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Burns R. C., et al., Antibody blockade of ICAM-1 and VCAM-1 ameliorates inflammation in the SAMP-1/Yit adoptive transfer model of Crohn’s disease in mice. Gastroenterology 121, 1428–1436 (2001). [DOI] [PubMed] [Google Scholar]
- 65.Cartwright I. M. and Colgan S. P., Next generation sequencing of acidosis response in intestinal epithelial cells. Gene Expression Omnibus. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE154982. Deposited 23 July 2020.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq data have been deposited in the Gene Expression Omnibus (GSE154982) (65).