Significance
Many viral diseases have emerged in recent decades, but prehistoric viral infections remain poorly understood. In some cases, nucleotide sequences of ancient viruses, which infected ancestral animals, have been integrated into their genomes during evolution. Such “molecular fossil records” of viruses help researchers trace past viral infections. Here, we reconstructed the infection history of an RNA virus, the bornavirus, for approximately 100 My in vertebrate evolution, using molecular fossils of ancient bornaviruses. Our analyses using ancient bornaviral sequences from over 100 vertebrate species genomes indicated that bornaviruses infected a broader range of host lineages during their long-term evolution than expected from extant bornaviral host ranges. Our findings highlighted the hidden history of this RNA viral infection over geological timescales.
Keywords: ancient viral infection, endogenous bornavirus-like element, vertebrate evolution, virus–host coevolutionary history, paleovirology
Abstract
Although viruses have threatened our ancestors for millions of years, prehistoric epidemics of viruses are largely unknown. Endogenous bornavirus-like elements (EBLs) are ancient bornavirus sequences derived from the viral messenger RNAs that were reverse transcribed and inserted into animal genomes, most likely by retrotransposons. These elements can be used as molecular fossil records to trace past bornaviral infections. In this study, we systematically identified EBLs in vertebrate genomes and revealed the history of bornavirus infections over nearly 100 My. We confirmed that ancient bornaviral infections have occurred in diverse vertebrate lineages, especially in primate ancestors. Phylogenetic analyses indicated that primate ancestors were infected with various bornaviral lineages during evolution. EBLs in primate genomes formed clades according to their integration ages, suggesting that bornavirus lineages infected with primate ancestors had changed chronologically. However, some bornaviral lineages may have coexisted with primate ancestors and underwent repeated endogenizations for tens of millions of years. Moreover, a bornaviral lineage that coexisted with primate ancestors also endogenized in the genomes of some ancestral bats. The habitats of these bat ancestors have been reported to overlap with the migration route of primate ancestors. These results suggest that long-term virus–host coexistence expanded the geographic distributions of the bornaviral lineage along with primate migration and may have spread their infections to these bat ancestors. Our findings provide insight into the history of bornavirus infections over geological timescales that cannot be deduced from research using extant viruses alone, thus broadening our perspective on virus–host coevolution.
Viral infectious diseases profoundly affect human health, livestock productivity, and ecosystem diversity. Similar to recent viral outbreaks (1), our ancestors were also probably challenged by viral epidemics. Investigations using historical specimens have provided insights into past viral infections, such as the origin of viral infectious diseases (2–6). However, the epidemic history of viruses across hundreds of millions of years is largely unclear.
Endogenous viral elements (EVEs) can be formed by the occasional integration of ancient viral sequences into the host germline genomes (7). EVEs are millions of years old and provide critical information on ancient viruses, such as their host ranges (8, 9), evolutionary timescales (10, 11), or geographical distributions (12). Endogenous bornavirus-like elements (EBLs) are the most abundant viral fossils of RNA viruses found in vertebrate genomes (10, 13, 14). Bornaviruses possess a negative-strand RNA genome and produce several separate messenger RNAs (mRNAs) encoding each of the viral genes (15). Occasional reverse transcription of these mRNAs and insertion of the DNA copy into a germline cell of the animal host by retrotransposons can establish an enduring record of ancient infections. Therefore, EBLs can help us trace the history of ancient bornavirus infections, providing good model systems to study virus–host coevolutionary history over geological timescales.
The family of Bornaviridae consists of three genera: Orthobornavirus, Carbovirus, and Cultervirus (16). Until 2018, only the genus Bornavirus (today Orthobornavirus), which includes viruses that cause immune-mediated neurological diseases in mammals and birds, constituted the family Bornaviridae (17). However, two new genera, Carbovirus and Cultervirus, were established upon discovering novel bornaviral species in carpet pythons and sharpbelly fish samples, respectively (18, 19). Furthermore, these discoveries led to the identification of several novel EBLs classified into these bornaviral genera (18, 20). Since most previous studies have exclusively used orthobornaviruses for EBL detection (10, 13, 14, 21–25), numerous elements similar to carboviruses and culterviruses may remain to be detected and analyzed. Therefore, the current understanding of the history of ancient bornavirus infections is probably incomplete.
In this study, we identified and characterized EBLs derived from three bornaviral genera to reconstruct the long-term history of ancient bornaviral infections comprehensively. Large-scale dating analysis revealed that ancient bornaviral infections have occurred in diverse vertebrate lineages for nearly 100 My. Primate ancestors, in particular, were repeatedly infected with ancient bornaviruses. Phylogenetic analyses clustered EBLs in primate genomes according to their integrated ages, suggesting that bornavirus lineages infecting primate ancestors have changed chronologically. Furthermore, some bornaviral lineages may have coexisted with primate ancestors for tens of millions of years. Interestingly, we found that this long-term virus–host coexistence may have expanded the geographic distributions of the virus and generated new infections in some bat ancestors. Thus, our findings describe virus–host coevolutionary history over geological timescales, which cannot be deduced from research using extant viruses alone.
Results
Systematic Identification of EBLs in Host Genomes.
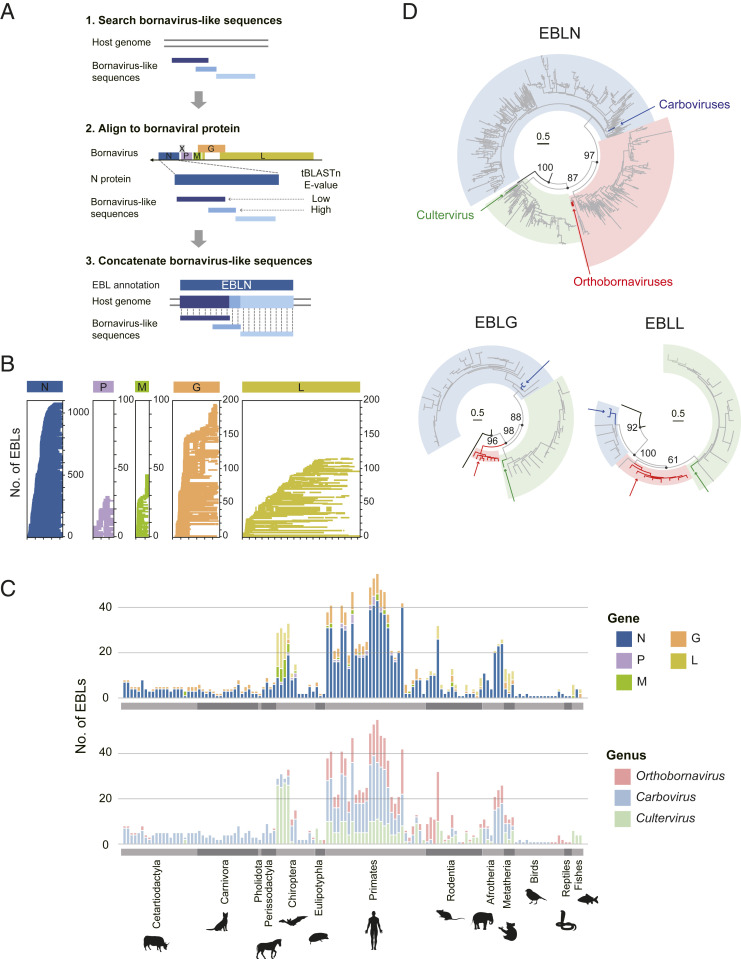
To systematically identify EBLs, we searched for bornavirus-like sequences in the genomic data of 969 eukaryotic species by tBLASTn using bornaviral protein sequences from all genera in the family Bornaviridae as queries (Fig. 1A). Next, we concatenated these sequences based on their location in the host genome and their alignment positions to extant bornaviral proteins because most bornavirus-like sequences were fragmented due to mutation after endogenization (details in Materials and Methods).
Fig. 1.
Identification of EBLs in vertebrate genomes. (A) Schematic diagram of the procedure to identify EBLs. First, bornavirus-like sequences were detected from host genomes by tBLASTn, using extant bornaviral sequences as queries. Second, the detected bornavirus-like sequences were aligned with corresponding proteins of extant bornaviruses. Third, the bornavirus-like sequences were concatenated based on the host genomic locations and alignment positions with bornaviral proteins. When several bornavirus-like sequences were detected in the same genomic positions in a species genome, the sequence with higher reliability (low E-value score in the tBLASTn search) was used for EBL sequence reconstruction. (B) Alignment coverage plot of EBLs. The scales on the x-axis are marked at intervals of 100 amino acids. The y-axis indicates the number of EBLs identified in this study. (C) Numbers of EBLs in the host genomes. The x-axis indicates the vertebrate species, and the y-axis indicates the number of EBLs identified in the species genome. The bar color shows the bornaviral gene (Upper) or genus from which the EBL originated (Lower). (D) Phylogenetic trees of EBLs and extant bornaviruses. These trees were constructed by the maximum likelihood method using the amino acid sequences of EBLs and extant bornaviral proteins. The branch colors indicate the sequence groups: EBLs (gray), extant nyamivirus used as outgroup (black), extant orthobornaviruses (red), extant carboviruses (blue), and extant cultervirus (green). Colored arrows mark extant bornaviruses. Highlights correspond to the current bornaviral classifications: genus Orthobornavirus (light red), genus Carbovirus (light blue), and genus Cultervirus (light green). Representative supporting values (percent) are shown on branches. The scale bars indicate genetic distances (substitutions per site).
The bornaviral genome encodes six viral proteins: nucleoprotein (N), phosphoprotein (P), matrix protein (M), envelope glycoprotein (G), large RNA-dependent RNA polymerase (L), and accessory protein (X) (15). Previous studies have reported EBLs derived from N, M, G, and L mRNAs, designated EBLN, EBLM, EBLG, and EBLL, respectively (26). Our EBL search identified 1,465 EBLs in 131 vertebrate species, including 1,079 EBLNs, 30 EBLPs, 46 EBLMs, 195 EBLGs, and 115 EBLLs (Fig. 1 B and C). Notably, we identified EBLPs, although these were considered difficult to detect due to methodological limitations, such as low sequence conservation and relatively short gene length of extant bornavirus P genes (26).
We next classified ancient bornaviruses from which EBLs originated at the viral genus level. Since some EBLs were too short to construct reliable phylogenetic trees, we sought to classify all the EBLs into the current bornaviral genera based on sequence similarity scores in the tBLASTn search. However, if there is an unknown genus consisting exclusively of ancient bornaviruses, the method based on sequence similarity with modern viruses may lead to misclassification. To assess this possibility, we first performed phylogenetic analyses using relatively long EBLs and their gene counterparts in modern bornaviruses. Fig. 1D showed that EBLs were clearly divided into three clades corresponding to the current bornaviral classification (16). Therefore, we applied the sequence similarity–based method to classify all the EBLs into current bornaviral genera (details in Materials and Methods). Based on the similarity scores, the EBLs were classified into 364 orthobornaviral, 729 carboviral, and 372 culterviral EBLs (Fig. 1C). Among the 1,465 EBLs, 870 loci were undetectable without using carboviruses and cultervirus as queries, although most previous studies only used orthobornaviral sequences for the EBL searches (10, 13, 14, 21–25). Therefore, the EBL search found numerous previously undetected loci and created a comprehensive dataset for reconstructing the history of bornavirus infections.
Large-Scale Dating Analysis for Bornaviral Integration Ages.
Bornaviral integration ages can be estimated based on the gene orthology (7). Here, we developed a network-based method for determining orthologous relationships among EBLs in our large dataset (SI Appendix, Fig. S1A). Briefly, we first constructed an all-against-all matrix of alignment coverages by pairwise sequence comparison among EBL integration sites. Next, we constructed a sequence similarity network using the matrix and extracted community structures from the network in order to divide the EBLs into groups based on their orthologous relationships. Finally, we manually checked the groupings to avoid inaccurate estimates (details in Materials and Methods and SI Appendix, Figs. S1B and S2).
We divided the 1,465 EBLs into 281 groups by our network-based dating method (Dataset S1). These groupings well reflected the alignment coverage among EBL integration sites (SI Appendix, Fig. S1B). We divided these groups into two categories: 113 groups of “EBLs with orthologs” and 168 groups of “EBLs without orthologs” (Fig. 2A). Each group of “EBLs with orthologs” is composed of loci sharing the same bornaviral integration event, and thus their minimum integration ages can be estimated by the divergent time of their hosts. In contrast, each group of “EBLs without orthologs” consists of a single EBL locus. Such “EBLs without orthologs” may be integrations that occurred after the divergence of hosts from their sister species. Alternatively, the lack of orthologs may simply be a methodological limitation due to the inaccessibility of the genomic data of sister species. For example, only three distantly related species of Eulipotyphla, in which no EBL orthologous relationships were detected, were present in the genomic database used for EBL detection (Fig. 2A). Thus, accumulating genomic data (27, 28) can help estimate integration ages with high accuracy.
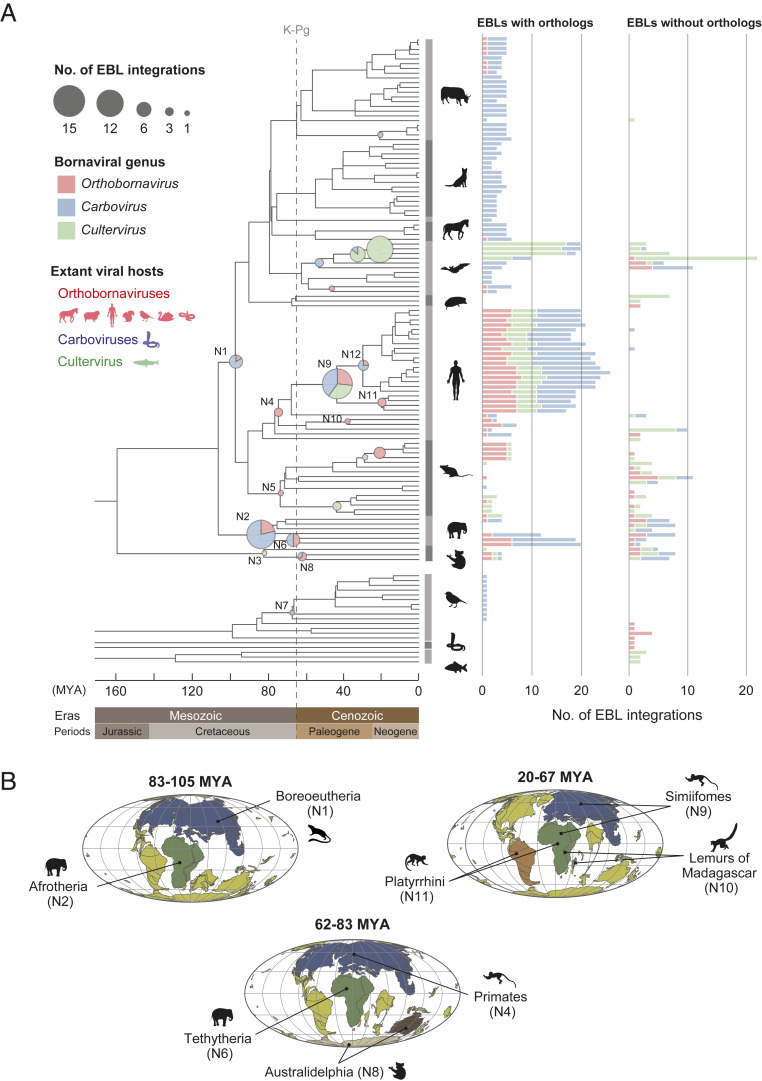
Fig. 2.
History of bornaviral integration events for ∼100 My. (A) Bornaviral integration events during vertebrate evolution. The evolutionary tree of vertebrates was obtained from the TimeTree database. The positions of pie charts on the tree indicate the lower limit ages of bornaviral integration events, and their size shows the number of events in each period. Annotations in the internal nodes on the tree indicate the common ancestors of Boreoeutheria (N1), Afrotheria (N2), Metatheria (N3), Primates (N4), Rodentia (N5), Tethytheria (N6), Passeriformes (N7), Australidelphia (N8), Simiiformes (N9), Lemuroidea (N10), Platyrrhini (N11), and Catarrhini (N12). The right panel shows the numbers of EBLs categorized into “EBLs with orthologs” or “EBLs without orthologs” in each host species. The definitions of these categories are described in the section titled Large-Scale Dating Analysis for Bornaviral Integration Ages. The bar colors show the viral genus as indicated on the left side of the tree. The representative hosts of extant bornaviruses are shown by animal silhouettes on the left side of the tree. (B) Schematic diagram of the geographical distributions of ancient bornaviruses and their hosts. The colored continents, except for yellow, indicate the continents where bornaviral endogenization may have occurred: Laurasia or Eurasia (blue), Africa (green), Antarctica (beige), Australia (dark brown), and South America (brown). The biogeography of hosts during their evolution was cited from previous reports (Dataset S2). Plate tectonic maps were downloaded from Ocean Drilling Stratigraphic Network (ODSN) Plate Tectonic Reconstruction Service (https://www.odsn.de/odsn/services/paleomap/paleomap.html).
Bornaviral Infections Have Occurred since the Mesozoic Era.
Our dating analysis enabled tracing of the history of bornavirus infections back to ∼100 Mya (Fig. 2A). The oldest records of bornaviral infection have been reported in ancestral afrotherians at least 83.3 Mya (10, 23). Here, we found six EBLs that were orthologous among species of Boreoeutheria, suggesting that the oldest bornavirus infections occurred at least 96.5 Mya. Additionally, we identified 18 bornaviral integration events in the Mesozoic era, which occurred in the ancestors of Afrotheria, Tethytheria, Metatheria, Primates, and Rodentia. We also found that a bornavirus integration occurred in ancestral Passeriformes birds during the Mesozoic era, between 66.6 and 82.5 Mya. These results provide strong evidence that bornaviral infections had already occurred in multiple vertebrate lineages in the Mesozoic era.
Ancient Bornaviral Infections in Various Vertebrate Lineages.
We found that ancient bornaviruses infected much broader vertebrate lineages than modern bornaviruses are known to infect (Fig. 2A). Modern orthobornavirus infections have been reported in ungulate animals, shrews, squirrels, humans, a wide range of birds, and garter snakes (17). However, we identified multiple endogenizations of ancient orthobornaviruses in mice, afrotherians, and marsupials, which have not been reported as host species of modern orthobornaviruses (Fig. 2A). In particular, a previous survey of bornaviral reservoirs did not detect orthobornavirus infections in mice (29). Furthermore, the extant carboviruses and cultervirus were detected only in carpet pythons and sharpbelly fish, respectively (18, 19). In contrast, ancient viruses belonging to these genera endogenized in various host lineages, including mammals and birds (Fig. 2A). These results indicate that ancient bornaviruses infected a wider range of vertebrate lineages than the known host ranges of extant bornaviruses, which may reflect consequences that bornaviruses have infected various host lineages during their long-term evolution. We also should note another possibility that our understanding of the extant bornavirus host range may be still insufficient (see Discussion).
Geographical Distributions of Ancient Bornaviral Infections.
We performed integrative analysis of bornaviral endogenizations and mammalian biogeography to infer the geographical distributions of ancient bornavirus infections (Dataset S2). Our results suggested that ancient bornavirus infections occurred in different continents: Laurasia and Africa in the Mesozoic era; Antarctica or Australia around the Cretaceous–Paleogene (K-Pg) boundary; and possibly Eurasia, Africa, or South America in the Cenozoic era (Fig. 2B).
First, we identified EBLs in animals that inhabited Laurasia and Africa in the Mesozoic era (N1, N2, N4, and N6 in Fig. 2B). It has been reported that ancestors of Boreoeutheria and Primates were distributed in Laurasia (30–33), while those of Afrotheria and Tethytheria were present in Africa (30, 31, 34). These results suggested that bornaviral infections spread in Laurasia and Africa during the Cretaceous period. Second, we identified EBLs integrated into the genome of Australidelphia ancestors but not in other marsupials in South America (N8 in Fig. 2B). Since ancestral Australian marsupials are considered to have moved from South America to Australia via Antarctica (35, 36), these bornavirus infections likely occurred in Antarctica or Australia.
Furthermore, we identified bornaviral endogenizations that occurred in ancestral primates in the Cenozoic era. First, we found an EBL integrated into the genome of the ancestor of the Madagascar lemur (N10 in Fig. 2B); however, the EBL was not identified in African galagos, suggesting that the bornavirus infection occurred in Madagascar Island. In contrast, the EBL integration age was estimated at 37.8 to 59.3 Mya (N10 in Fig. 2A), which overlapped with the migration of lemur ancestors from Africa to Madagascar Island around 50 to 60 Mya (37). This overlap presented an alternate possibility that bornaviral endogenization had occurred in African animals before they migrated to Madagascar. Second, we identified several EBLs integrated into the ancestral Platyrrhini genomes (N11 in Fig. 2B). Ancestors of Platyrrhini are presumed to have migrated from Africa to South America during their divergence from Simiiformes (32, 38, 39), thus suggesting evidence of bornavirus infections in these continents (see Discussion).
Complex History of Bornaviral Infections during Primate Evolution: Infections of Distinct Bornaviral Lineages in Each Age.
We found that bornaviruses in the three genera repeatedly endogenized during primate evolution (Fig. 2A). We inferred phylogenetic relationships among ancient bornaviruses using EBLNs, which are the most abundant records among EBLs (Fig. 3 A–C and SI Appendix, Fig. S3), to understand the origin of endogenizations of bornaviral lineages into primate ancestor genomes.
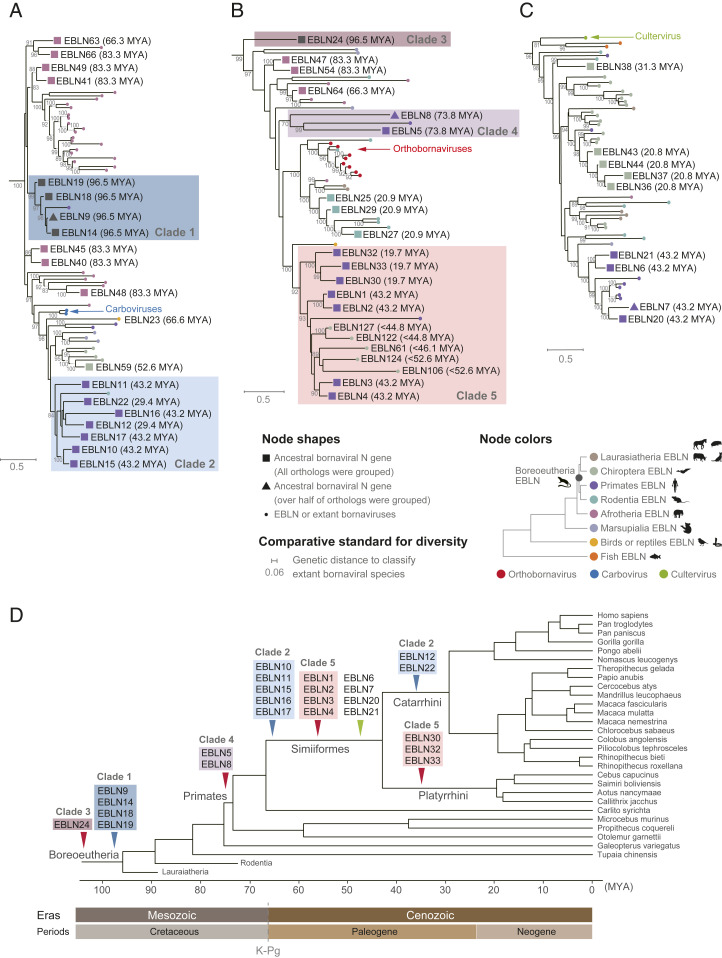
Fig. 3.
Phylogenetic relationships of ancient bornaviruses that infected primate ancestors. (A–C) Phylogenetic analyses of ancient and modern bornaviral N genes. These trees were constructed by the maximum likelihood method using the amino acid sequences of EBLNs and extant bornaviral N proteins of genus Carbovirus (A), Orthobornavirus (B), or Cultervirus (C). Colored arrows mark extant bornaviruses. Square and triangle nodes indicate collapsed clades containing all and over half of the orthologs used in the phylogenetic analyses, respectively. Phylogenetic trees with all expanding nodes are available in SI Appendix, Fig. S3. The node colors indicate the host lineages of ancient bornaviruses or extant bornaviral genera as indicated in the lower right panel. Colored boxes highlight the bornaviral lineages endogenized during primate evolution. Number on the branches are bootstrap values (percent) based on 1,000 replications. The scale bars show genetic distances (substitutions per site). The genetic distance to distinguish extant bornaviral species is shown as the comparative standard for estimating the genetic diversity of ancient bornaviruses. (D) EBLN integration events during primate evolution. Arrowheads indicate the occurrence of ancient bornaviral integrations: orthobornaviral EBLN (red), carboviral EBLN (blue), and culterviral EBLN (green). The colors of highlighted boxes correspond to ancient bornaviral lineages shown in A–C.
We found that several distinct lineages of bornaviruses had sequentially endogenized during primate evolution rather than repeated endogenization of a single bornaviral lineage. For example, the carboviral EBLNs in primate genomes were clearly divided into two viral lineages: the clade 1 viral lineage endogenized in Boreoeutherian ancestors and the clade 2 viral lineage endogenized in Simiiformes and Catarrhini ancestors (Fig. 3 A and D). Furthermore, orthobornaviral EBLNs formed three different clades according to their integration ages (clades 3 to 5 in Fig. 3 B and D). These results suggest that different bornaviral lineages were prevalent in each era during primate evolution.
Next, to infer how diverse bornaviruses endogenized during primate evolution, we calculated the genetic distances between these ancient viral lineages (clades 1 to 5) in our phylogenetic tree (Dataset S3). Using genetic distance for classifying extant species of bornaviruses as a comparative standard, we found that the genetic diversity among these ancient bornaviral lineages was higher than that among extant bornaviral species (Fig. 3 A–C and Dataset S3). Thus, recurrent bornaviral endogenizations during primate evolution may have occurred due to infections of multiple bornaviral lineages comparable to different viral species.
Complex History of Bornaviral Infections during Primate Evolution: Long-Term Virus–Host Coexistence.
In addition to sequential infections of primate ancestors by distinct bornavirus lineages (Fig. 3), we found that some lineages may have established long-term coexistence relationships with the hosts. For example, the clade 2 carboviral lineage repeatedly endogenized in Simiiformes and Catarrhini ancestors between 29.4 and 67.1 Mya (Fig. 3 A and D). Furthermore, endogenizations of the clade 5 orthobornaviral lineage recurred in Simiiformes and Platyrrhini ancestors between 19.7 and 67.1 Mya (Fig. 3 B and D). These results suggest that these bornaviral lineages have coexisted with primate ancestors for tens of millions of years. In summary, we described the complex history of recurrent bornaviral endogenizations during primate evolution, including sequential infections of diverse bornaviral lineages and long-term virus–host coexistence.
Discussion
Snapshots of ancient bornaviral infections have been reported since the discovery of EBLs (10, 13, 14, 21–25); however, the long-term history of bornavirus infections has remained unclear. Here, we systematically identified EBLs in 131 vertebrate species (Fig. 1) and reconstructed the history of bornavirus infections for ∼100 My (Fig. 2). This report comprehensively traces the history of RNA virus infections over geological timescales. Furthermore, phylogenetic analyses suggested chronological changes in infected bornaviral lineages during primate evolution as well as coexistence of some lineages with primate ancestors for tens of millions of years (Fig. 3). Virus–host codivergence alone, which is thought to be as the background of viral evolutionary history (19), is insufficient to explain this mixed pattern. Therefore, our findings suggest that the virus–host coevolutionary relationships have dramatically changed over geological timescales, which complicated the viral evolutionary history.
We should note that EBLs can provide only limited snapshots in the enormous diversity of bornaviruses during their long-term evolution. EBLs are the most abundant RNA virus fossils; however, ancient bornavirus sequences would have been rarely fossilized in animal genomes due to a lack of autonomous endogenization ability in bornaviruses. Furthermore, it is also possible that information on ancient bornavirus infections would have been lost during the long-term evolution; for example, EBLs did not reach fixation in the host population or host animals with EBLs have gone extinct. Therefore, we have to keep in mind that EBLs are necessarily incomplete fossil records. These potential limitations in paleovirology would sometimes make it difficult to trace details of ancient virus transmissions. Nonetheless, our systematic analyses using EBLs help us discuss the long-term evolutionary history between bornaviruses and their hosts by obtaining indispensable information of ancient bornaviruses, such as host ranges, timescales, or phylogeny.
Our phylogenetic analyses showed that bornaviruses closely related to the lineage that infected ancestral primates had almost contemporaneously endogenized in the other animals (Fig. 3). For example, carboviruses similar to the clade 1 viral lineage also endogenized in ancestral afrotherians around the late Mesozoic era (EBLN41, 49, 63, and 66 in Fig. 3A). Additionally, carboviruses similar to the clade 2 lineage endogenized in ancestors of Yangochiroptera bats from the late Mesozoic to Cenozoic era (EBLN59 in Fig. 3A). A similar tendency was observed in the orthobornaviral phylogenetic tree (Fig. 3B). Extant orthobornaviruses are no exception; genetically similar viruses infect various host species in mammals, birds, and reptiles (17). These results suggest that bornaviral lineages spread to various hosts in each era and change over time.
By integrating information on host geographical distributions and phylogeny of bornaviruses, we found long-term coexistence of ancient bornaviruses with primate ancestors that may have expanded the viral lineage to other continents. Fig. 3B shows that the clade 5 orthobornaviral lineage has repeatedly endogenized in ancestors of Simiiformes and Platyrrhini. Simiiformes ancestors were reportedly distributed in Eurasia or Africa, while Platyrrhini ancestors likely migrated from Africa to South America during their divergence from Simiiformes (Fig. 2B) (32, 38, 39). These results suggest that the clade 5 viral lineage moved between the continents along with host migrations. Interestingly, viruses in the clade 5 lineage also endogenized in several bat genomes (Fig. 3B), such as Rhinolophus bats (EBLN61), Desmodus bats (EBLN106 and EBLN124), and Miniopterus bats (EBLN122 and EBLN127). Since Rhinolophus and Desmodus bats have reportedly originated in Eurasia and South America, respectively (40), the clade 5 viral lineage may have moved across continents along with primate migrations and further transmitted to these bat ancestors. Thus, long-term virus–host coexistence may have expanded the viral geographic distributions and generated new infections in other hosts. Furthermore, it is also possible that this ancient bornavirus lineage infected other host animals, not only primate and bat ancestors, because the incompleteness of EBLs might have prevented tracing the spread of ancient bornavirus infections. Future investigations, for example, searching for EBLs in genomes extracted from extinct animal specimens (41, 42), would help trace the long-term evolutionary history of bornaviruses in more detail.
We found that the number of bornaviral integration events varied according to the host lineage (Fig. 2A). In nonmammalian vertebrates, we observed low frequencies of bornaviral integrations, consistent with previous studies (24, 25). Furthermore, the numbers of bornaviral endogenizations differ among descendant lineages that diverged from Boreoeutherian ancestors. Remarkably, bornaviral endogenizations rarely occurred in most laurasiatherian lineages but repeatedly occurred in some other host lineages, including Primates, Chiroptera, and Rodentia. Frequency of viral infection, germline integration, and subsequent inheritance may be responsible for these differences. For example, the reverse transcription activities of retrotransposons can affect the integration rate of bornaviral sequences (43). Moreover, the evolutionary constraints and the host population size can also influence the fixation rate of integrated sequences (44). Further studies on the impact of these factors on EBL fixations are necessary to explain such differences.
This study also implies that a large number of EVEs derived from unknown ancient viruses are hidden in the host genomic data. One methodological limitation of paleovirological research is identifying viral fossil records comprehensively. Because the method to identify EVEs involves searching for virus-like sequences in host genomes using modern viral sequences as queries, the detectability of EVEs depends on sequence similarities with extant viruses. Here, we demonstrated that the EBL search using genetically diverse extant bornaviruses provided a large dataset, including previously undetectable loci (Fig. 1). Therefore, further elucidation of extant viral diversity may help clarify ancient viral diversity.
Furthermore, the sequence data of EBLs may be a useful resource for exploring extant viral diversity because metagenomic analyses to detect viral infections also rely on sequence similarities with known viral sequences. Our phylogenetic analyses indicated that EBLNs originated from diverse ancient bornaviruses, and almost all EBLNs formed clades that completely differed from modern ones (Fig. 3). Hence, ancient bornaviruses appear to have been highly divergent and phylogenetically distinct from known modern viruses. Furthermore, these results raise a fascinating question regarding whether viruses genetically similar to EBLs are extinct or have just not been discovered. Future viral metagenomic analyses using EBL sequence data may address this question. Therefore, reusing data between metagenomic analyses for extant viruses and paleovirological investigations may elucidate viral diversity, connecting modern with ancient viral evolution.
In conclusion, we depicted the history of bornavirus infections during vertebrate evolution. Our findings broaden our perspective of virus–host coevolution by providing insight into this RNA viral infection over geological timescales, which cannot be obtained from research using extant viruses alone.
Materials and Methods
Identification of EBLs in Vertebrate Genomic Data.
EBLs were identified by 1) searching for bornavirus-like sequences in the genomes of 969 eukaryotic species, 2) reconstructing EBL sequences, and 3) validating whether these EBL sequences were derived from ancient bornaviruses.
First, bornavirus-like sequences were screened in the Refseq genomic database (version: 20190329), provided by National Center for Biotechnology Information (NCBI) (45) by tBLASTn (version 2.6.0+) (46) with the option “-evalue 0.1,” using sequences in all genus of Bornaviridae as queries (Dataset S4). Second, because most EBLs were detected as fragmented sequences due to mutations occurring after integration, we reconstructed EBL sequences by concatenating sequences if the following conditions were met: 1) detected bornavirus-like sequences were located within 1,000 bp (EBLN, EBLP, EBLM, and EBLG) or 2,000 bp (EBLL) in the host genome, and 2) the order of sequences in the alignment to extant bornaviral proteins were consistent with those in the host genome (Fig. 1A). When more than two bornavirus-like sequences were detected in the same genomic position in a species genome, we preferentially used the sequence with higher reliability (low E-value in the tBLASTn search). The alignment of bornavirus-like sequences and modern bornaviral proteins was conducted using MAFFT (version 7.427) with options “—addfragments” and “—keeplength” (47). Finally, we checked the origin of the EBL candidates based on the bit score obtained from BLASTP (version 2.9.0+) using the Refseq protein database (version: 20200313) and a database consisting of bornaviral protein sequences listed in Dataset S4. If the candidate was more similar to other viral proteins or host proteins other than published EBLs, we removed the sequence from the analysis. After this process, only one EBLL candidate was identified in an insect genome, but we excluded this sequence in subsequent analyses. The concatenation of bornavirus-like sequences yielded over 800 EBL loci equivalent to more than half the length of the intact bornaviral proteins (Fig. 1B).
Dating Analysis For the Integrated Age of EBLs.
To determine orthologous relationships among EBLs, we clustered loci based on the alignment coverages in pairwise sequence comparison between EBL integration sites (SI Appendix, Fig. S1A). First, we extracted the upstream and downstream sequences of EBLs with lengths of 15,000 bp for EBLLs and 10,000 bp for other EBLs. These sequences were trimmed by removing repetitive elements using RepeatMasker (version open-4.0.9) (http://www.repeatmasker.org/) with options “-q,” “-xsmall,” “-a,”“-species,” and RepBase RepeatMasker libraries (version 20181026) (48). Second, we performed the pairwise alignment of these sequences using BLASTN (version 2.9.0+) with the option “evalue 0.1” and constructed an all-against-all matrix for alignment coverage among EBL integration sites. The sequence similarity network was constructed by connecting nodes when their sequence alignment coverage was over 9.0% of the flanking sequence length. Selection of the best criteria to construct a sequence network is described in the next section. The groups were extracted by detecting a community structure using Louvain heuristics. These network analyses were performed using NetworkX (49).
We simultaneously checked the phylogenetic relationships of host species with sequence alignment coverage to correctly estimate EBL ages. The contamination of sequences unrelated to true orthologous relationships leads to overestimation of integration ages, as shown in example 4 in SI Appendix, Fig. S4A. To avoid such issues, when multiple EBL loci were present in the same species genome and the alignment coverage was lower than 50%, we considered these loci as located in different genomic sites and divided them into different groups. Furthermore, the integration ages of older elements tend to be underestimated because the alignment quality among their integration sites may deteriorate following accumulation of sequence changes, such as genomic rearrangement (example 3 in SI Appendix, Fig. S4A). Thus, by examining the phylogenetic relationships of host species and sequence alignment coverage, we combined some groups into EBLG2, EBLL2, EBLL35, or EBLL36 (SI Appendix, Figs. S1B and S2). For example, the EBLG2 group was previously reported to have endogenized into the genome of laurasiatherian ancestors at least 77.0 Mya (18). This group was initially divided into two groups in our analysis, including laurasiatherian and primate loci. However, these groups were connected by low alignment coverage (SI Appendix, Fig. S1B), which led to another hypothesis that these sequences are descendants of the same integration event in the Boreoeutherian ancestor. To test this hypothesis, we confirmed the alignment quality among the EBL integration sites using AliTV (50–52). We found that over 70% sequence similarity covered more than 40% of the alignment by lastz (version 1.04.00) with options “—noytrim,” “—gapped,” and “—strand=both” (51) (SI Appendix, Fig. S2 A–C). Therefore, we combined these groups into the same group. The cases of EBLL2 and EBLL35 were similar to that of EBLG2 (SI Appendix, Fig. S2 B and C). EBLL36 contained tandemly repeated loci at close genomic locations (Dataset S1) because we could not distinguish whether these loci were derived from independent integration events or gene duplications postintegration. Presently, we considered these loci as descendants from the same integration event to avoid overestimating the number of EBL integration events.
After curation, the dates of EBL integration events were assigned according to a vertebrate evolutionary tree from the TimeTree database (53). Each EBL locus was named according to the nomenclature for endogenous retroviruses (54) (Dataset S1). It should be noted that the number of bornaviral integration events was less than that of EBL loci shown in Fig. 1C because redundant sequences in the genomic database used for the EBL search were grouped as the same integration event.
Validation of the Network-Based Dating Method Using Human Transposable Elements.
To validate our dating method, we compared the integration ages of human transposable elements (TEs) estimated based on genomic alignment and those estimated based on network analysis (SI Appendix, Fig. S4). The genomic positions of all human TEs were obtained from the RepeatMasker database (http://www.repeatmasker.org/). First, the orthologs of all human TEs were determined in 18 mammalian genomes by LiftOver (version 357) with the option “-minMatch=0.5,” using genomic alignments provided by the University of California Santa Cruz (UCSC) genome browser (55). Their integration ages were determined by the presence or absence patterns of orthologs. Second, to examine the ortholog detection rate by the network-based dating method for each timescale, we prepared test datasets by random sampling of 100 loci for each timescale based on the dating results using genomic alignment (SI Appendix, Fig. S4 A and B). The details of the network-based dating method are described in the previous section. The estimation of human TE integration ages in the test datasets followed the same strategy, except that the flanked sequences of human TEs were extracted with lengths of 10,000 bp from the soft-masked assembly sequence provided by the UCSC genome browser and repetitive elements detected by the UCSC genome browser procedure were removed from the flanked sequences. Finally, we compared the results between the two methods by checking the following points: predicted ages and detected orthologs (examples are shown in SI Appendix, Fig. S4A).
Furthermore, we evaluated nine different criteria to connect nodes in the sequence similarity network and decided to connect network edges if their sequence alignment coverage was over 9.0% of the flanking sequence length (SI Appendix, Fig. S4 A and C). According to this criterion, the concordant rates between the two methods in predicting ages of Cenozoic TEs were 88.0 to 100.0%, and those of older TEs integrated in the Cretaceous period were 43.0 to 66.0% (SI Appendix, Fig. S4C). The chain files used for LiftOver and the genome assembly sequences are listed in Dataset S5.
Phylogenetic Analysis.
We used amino acid sequences of EBLs with lengths longer than 200 amino acids for EBLL and 100 for EBLN and EBLG. Multiple sequence alignments (MSAs) were constructed by MAFFT with options “—addfragments” and “—keeplength.” MSAs for Fig. 1D were trimmed by excluding sites where over 30% of sequences were gaps, subsequently removing sequences with less than 70% of the total alignment sites. MSA for the EBLN tree (Fig. 3 A–C and SI Appendix, Fig. S3) was trimmed by excluding sites where over 20% of sequences were gaps and subsequently removing sequences with less than 80% of the total alignment sites. Phylogenetic trees were constructed by the maximum likelihood method using IQ-TREE (version 1.6.12) (56). The substitution models were selected based on the Bayesian information criterion score provided by ModelFinder (57): VT+F+G4 for EBLNs, VT+F+G4 for EBLGs, VT+F+R3 for EBLLs (Fig. 1D), and JTT+F+G4 for EBLNs (Fig. 3 A–C and SI Appendix, Fig. S3). The branch supports were measured as the ultrafast bootstrap values given by UFBoot2 (58) with 1,000 replicates. The extant viral sequences used for the phylogenetic analyses are listed in Dataset S6. The ggtree (59) and ETE3 packages (60) were used to visualize the trees.
EBL Classification according to Current Bornaviral Genera.
EBLs were classified into current bornaviral genera based on the query bornaviral sequence with the lowest E-value in the tBLASTn search. The results of the phylogenetic analysis–based method and similarity score–based method were highly concordant (EBLN: 99.7%, EBLG: 100%, and EBLL: 100%). Thus, we applied the classification method for all EBL loci (Fig. 1C). We could not create reliable phylogenetic trees using EBLP or EBLM due to the small number of sites available for phylogenetic analysis.
Assessment of Genetic Diversity of Ancient Bornaviral Sequences.
We compared the genetic diversity of ancient and extant bornaviruses to infer how diverse bornaviruses endogenized during primate evolution. First, we used the most recent common ancestor of the EBLN orthologs as the ancestral bornaviral N gene to avoid overestimating the sequence diversity of ancient bornaviruses. Next, to provide a comparison standard for interpreting the ancient bornaviral genetic diversity, we calculated the genetic distance for classifying extant bornaviral species in our phylogenetic tree (0.06 substitutions per site) (Fig. 3 A–C and Dataset S3). The genetic distances between nodes in the phylogenetic tree were calculated using the ETE3 toolkit. It should be noted that this is an alternative method, and International Committee on Taxonomy of Viruses (ICTV) classification for extant bornaviral species is based on sequence similarity among intact viral genomes, differences in their host ranges, and phylogenetic analysis using viral proteins (17, 20).
Supplementary Material
Acknowledgments
We thank Dr. Keiko Takemoto (Institute for Frontier Life and Medical Sciences, Kyoto University, Japan) for technical support. We are grateful to Hidenori Nishihara and Jiaqi Wu (School of Life Science and Technology, Tokyo Institute of Technology, Japan), Jumpei Ito (Institute of Medical Science, The University of Tokyo, Japan), Bea Clarise Garcia, Hsien Hen Lin, Koichi Kitao, and Michiko Iwata (Institute for Frontier Life and Medical Sciences, Kyoto University, Japan) for helpful discussions. We thank Editage (https://www.editage.com/) for editing and reviewing this manuscript for English language. This study was supported by Japan Society for the Promotion of Science (JSPS) KAKENHI JP19J22241 (J.K.), JP20H05682 (K.T.) and JP18K19443 (M.H.); The Ministry of Education, Culture, Sports, Science and Technology (MEXT) KAKENHI JP16H06429 (K.T.), JP16K21723 (K.T.), JP16H0643 (K.T.), JP17H05821 (M.H.), and JP19H04833 (M.H.); and the Hakubi project at Kyoto University (M.H.). Computations were partially performed on the supercomputing systems SHIROKANE (Human Genome Center, the Institute of Medical Science, The University of Tokyo, Japan) and the NIG supercomputer (ROIS National Institute of Genetics, Japan).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
See online for related content such as Commentaries.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2026235118/-/DCSupplemental.
Data Availability
The data, materials, and codes are available at https://github.com/Junna-Kawasaki/EBL_2020. The versions of bioinformatics tools are listed in Dataset S7. All other study data are included in the article and/or supporting information.
References
- 1.Grubaugh N. D., et al., Tracking virus outbreaks in the twenty-first century. Nat. Microbiol. 4, 10–19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Düx A., et al., Measles virus and rinderpest virus divergence dated to the sixth century BCE. Science 368, 1367–1370 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Taubenberger J. K., Reid A. H., Krafft A. E., Bijwaard K. E., Fanning T. G., Initial genetic characterization of the 1918 “Spanish” influenza virus. Science 275, 1793–1796 (1997). [DOI] [PubMed] [Google Scholar]
- 4.Mühlemann B., et al., Ancient human parvovirus B19 in Eurasia reveals its long-term association with humans. Proc. Natl. Acad. Sci. U.S.A. 115, 7557–7562 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Duggan A. T., et al., 17th century variola virus reveals the recent history of smallpox. Curr. Biol. 26, 3407–3412 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mühlemann B., et al., Ancient hepatitis B viruses from the bronze age to the medieval period. Nature 557, 418–423 (2018). [DOI] [PubMed] [Google Scholar]
- 7.Aiewsakun P., Katzourakis A., Endogenous viruses: Connecting recent and ancient viral evolution. Virology 479-480, 26–37 (2015). [DOI] [PubMed] [Google Scholar]
- 8.Kryukov K., Ueda M. T., Imanishi T., Nakagawa S., Systematic survey of non-retroviral virus-like elements in eukaryotic genomes. Virus Res. 262, 30–36 (2019). [DOI] [PubMed] [Google Scholar]
- 9.Hayward A., Cornwallis C. K., Jern P., Pan-vertebrate comparative genomics unmasks retrovirus macroevolution. Proc. Natl. Acad. Sci. U.S.A. 112, 464–469 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Katzourakis A., Gifford R. J., Endogenous viral elements in animal genomes. PLoS Genet. 6, e1001191 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aiewsakun P., Katzourakis A., Marine origin of retroviruses in the early Palaeozoic Era. Nat. Commun. 8, 13954 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gifford R. J., et al., A transitional endogenous lentivirus from the genome of a basal primate and implications for lentivirus evolution. Proc. Natl. Acad. Sci. U.S.A. 105, 20362–20367 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Horie M., et al., Endogenous non-retroviral RNA virus elements in mammalian genomes. Nature 463, 84–87 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Horie M., Kobayashi Y., Suzuki Y., Tomonaga K., Comprehensive analysis of endogenous bornavirus-like elements in eukaryote genomes. Philos. Trans. R. Soc. Lond. B Biol. Sci. 368, 20120499 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Briese T., et al., Genomic organization of Borna disease virus. Proc. Natl. Acad. Sci. U.S.A. 91, 4362–4366 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Amarasinghe G. K., et al., Taxonomy of the order mononegavirales: Update 2019. Arch. Virol. 164, 1967–1980 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuhn J. H., et al., Taxonomic reorganization of the family Bornaviridae. Arch. Virol. 160, 621–632 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hyndman T. H., Shilton C. M., Stenglein M. D., Wellehan J. F. X. Jr, Divergent bornaviruses from Australian carpet pythons with neurological disease date the origin of extant Bornaviridae prior to the end-Cretaceous extinction. PLoS Pathog. 14, e1006881 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shi M., et al., The evolutionary history of vertebrate RNA viruses. Nature 556, 197–202 (2018). [DOI] [PubMed] [Google Scholar]
- 20.Rubbenstroth D., et al., “One (1) new genus including one (1) new species in the family Bornaviridae (order Mononegavirales)” (Tech. Rep. 2018.016M, ResearchGate, 2018).
- 21.Belyi V. A., Levine A. J., Skalka A. M., Unexpected inheritance: Multiple integrations of ancient bornavirus and ebolavirus/marburgvirus sequences in vertebrate genomes. PLoS Pathog. 6, e1001030 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mukai Y., Horie M., Tomonaga K., Systematic estimation of insertion dates of endogenous bornavirus-like elements in vesper bats. J. Vet. Med. Sci. 80, 1356–1363 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kobayashi Y., et al., Exaptation of bornavirus-like nucleoprotein elements in afrotherians. PLoS Pathog. 12, e1005785 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cui J., et al., Low frequency of paleoviral infiltration across the avian phylogeny. Genome Biol. 15, 539 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gilbert C., et al., Endogenous hepadnaviruses, bornaviruses and circoviruses in snakes. Proc. Biol. Sci. 281, 20141122 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Horie M., Tomonaga K., Paleovirology of bornaviruses: What can be learned from molecular fossils of bornaviruses. Virus Res. 262, 2–9 (2019). [DOI] [PubMed] [Google Scholar]
- 27.Feng S., et al., Dense sampling of bird diversity increases power of comparative genomics. Nature 587, 252–257 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zoonomia Consortium , A comparative genomics multitool for scientific discovery and conservation. Nature 587, 240–245 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hilbe M., et al., Shrews as reservoir hosts of borna disease virus. Emerg. Infect. Dis. 12, 675–677 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Springer M. S., Meredith R. W., Janecka J. E., Murphy W. J., The historical biogeography of Mammalia. Philos. Trans. R. Soc. Lond. B Biol. Sci. 366, 2478–2502 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nishihara H., Maruyama S., Okada N., Retroposon analysis and recent geological data suggest near-simultaneous divergence of the three superorders of mammals. Proc. Natl. Acad. Sci. U.S.A. 106, 5235–5240 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Springer M. S., et al., Macroevolutionary dynamics and historical biogeography of primate diversification inferred from a species supermatrix. PLoS One 7, e49521 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bloch J. I., Silcox M. T., Boyer D. M., Sargis E. J., New Paleocene skeletons and the relationship of plesiadapiforms to crown-clade primates. Proc. Natl. Acad. Sci. U.S.A. 104, 1159–1164 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gheerbrant E., Schmitt A., Kocsis L., Early African fossils elucidate the origin of embrithopod mammals. Curr. Biol. 28, 2167–2173.e2 (2018). [DOI] [PubMed] [Google Scholar]
- 35.Nilsson M. A., et al., Tracking marsupial evolution using archaic genomic retroposon insertions. PLoS Biol. 8, e1000436 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Eldridge M. D. B., Beck R. M. D., Croft D. A., Travouillon K. J., Fox B. J., An emerging consensus in the evolution, phylogeny, and systematics of marsupials and their fossil relatives (Metatheria). J. Mammal. 100, 802–837 (2019). [Google Scholar]
- 37.Poux C., et al., Asynchronous colonization of madagascar by the four endemic clades of primates, tenrecs, carnivores, and rodents as inferred from nuclear genes. Syst Biol. 54, 719–730 (2005). [DOI] [PubMed] [Google Scholar]
- 38.Jaeger J. J., Marivaux L., Paleontology. Shaking the earliest branches of anthropoid primate evolution. Science 310, 244–245 (2005). [DOI] [PubMed] [Google Scholar]
- 39.Bond M., et al., Eocene primates of South America and the African origins of new world monkeys. Nature 520, 538–541 (2015). [DOI] [PubMed] [Google Scholar]
- 40.Teeling E. C., et al., A molecular phylogeny for bats illuminates biogeography and the fossil record. Science 307, 580–584 (2005). [DOI] [PubMed] [Google Scholar]
- 41.van der Valk T., et al., Million-year-old DNA sheds light on the genomic history of mammoths. Nature 591, 265–269 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Orlando L., et al., Recalibrating Equus evolution using the genome sequence of an early Middle Pleistocene horse. Nature 499, 74–78 (2013). [DOI] [PubMed] [Google Scholar]
- 43.Horie M., Tomonaga K., Non-retroviral fossils in vertebrate genomes. Viruses 3, 1836–1848 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Johnson W. E., Origins and evolutionary consequences of ancient endogenous retroviruses. Nat. Rev. Microbiol. 17, 355–370 (2019). [DOI] [PubMed] [Google Scholar]
- 45.O’Leary N. A., et al., Reference sequence (RefSeq) database at NCBI: Current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 44, D733–D745 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Camacho C., et al., BLAST+: Architecture and applications. BMC Bioinformatics 10, 421 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Katoh K., Standley D. M., MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 30, 772–780 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bao W., Kojima K. K., Kohany O., Repbase Update, a database of repetitive elements in eukaryotic genomes. Mob. DNA 6, 11 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hagberg A. A., Schult D. A., Swart P. J., “Exploring network structure, dynamics, and function using NetworkX” in Proceedings of the 7th Python in Science Conference (SciPy2008), Varoquaux G., Vaught T., Millman J., Eds. (Pasadena, CA, 2008), pp. 11–15. [Google Scholar]
- 50.Ankenbrand M. J., Hohlfeld S., Hackl T., Förster F., Ali T. V., AliTV—interactive visualization of whole genome comparisons. PeerJ Comput. Sci. 3, e116 (2017). [Google Scholar]
- 51.Harris R. S., Improved Pairwise Alignment of Genomic Dna (The Pennsylvania State University, 2007). [Google Scholar]
- 52.Stajich J. E., et al., The Bioperl toolkit: Perl modules for the life sciences. Genome Res. 12, 1611–1618 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kumar S., Stecher G., Suleski M., Hedges S. B., TimeTree: A resource for timelines, timetrees, and divergence times. Mol. Biol. Evol. 34, 1812–1819 (2017). [DOI] [PubMed] [Google Scholar]
- 54.Gifford R. J., et al., Nomenclature for endogenous retrovirus (ERV) loci. Retrovirology 15, 59 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Haeussler M., et al., The UCSC genome browser database: 2019 update. Nucleic Acids Res. 47, D853–D858 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nguyen L.-T., Schmidt H. A., von Haeseler A., Minh B. Q., IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kalyaanamoorthy S., Minh B. Q., Wong T. K. F., von Haeseler A., Jermiin L. S., ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 14, 587–589 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hoang D. T., Chernomor O., von Haeseler A., Minh B. Q., Vinh L. S., UFBoot2: Improving the ultrafast bootstrap approximation. Mol. Biol. Evol. 35, 518–522 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yu G., Smith D. K., Zhu H., Guan Y., Lam T. T. Y., ggtree: An r package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods Ecol. Evol. 8, 28–36 (2017). [Google Scholar]
- 60.Huerta-Cepas J., Serra F., Bork P., ETE 3: Reconstruction, analysis, and visualization of phylogenomic data. Mol. Biol. Evol. 33, 1635–1638 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data, materials, and codes are available at https://github.com/Junna-Kawasaki/EBL_2020. The versions of bioinformatics tools are listed in Dataset S7. All other study data are included in the article and/or supporting information.