Significance
Endoplasmic reticulum (ER) stress is a condition triggered by adverse environmental conditions, which in plants include extreme heat, pathogen infection, and drought. These stresses can have a devastating effect on agricultural productivity, and the appropriate responses of plants to stress are therefore critical. Autophagy is a macromolecular degradation pathway that is activated in response to stress, including ER stress, but the mechanism by which it is regulated is still unclear. Here, we describe a modification, persulfidation, of the key autophagy protein ATG18a that affects its binding to membranes and controls its activity. This tight regulation of autophagy therefore allows plants to control the tradeoff between growth and stress tolerance.
Keywords: autophagy, hydrogen sulfide, ER stress, ATG18a, persulfidation
Abstract
Hydrogen sulfide (H2S) is an endogenously generated gaseous signaling molecule, which recently has been implicated in autophagy regulation in both plants and mammals through persulfidation of specific targets. Persulfidation has been suggested as the molecular mechanism through which sulfide regulates autophagy in plant cells. ATG18a is a core autophagy component that is required for bulk autophagy and also for reticulophagy during endoplasmic reticulum (ER) stress. In this research, we revealed the role of sulfide in plant ER stress responses as a negative regulator of autophagy. We demonstrate that sulfide regulates ATG18a phospholipid-binding activity by reversible persulfidation at Cys103, and that this modification activates ATG18a binding capacity to specific phospholipids in a reversible manner. Our findings strongly suggest that persulfidation of ATG18a at C103 regulates autophagy under ER stress, and that the impairment of persulfidation affects both the number and size of autophagosomes.
Macroautophagy (hereafter referred to as autophagy, from the Greek meaning “self-eating”) is a major catabolic process in eukaryotic cells to degrade dysfunctional or unnecessary cellular components, either non-selectively or selectively (1, 2). It has conserved functions in development, cellular homeostasis, and stress responses from yeast to plants and mammals. In plants, autophagy is critically important in many aspects of plant life, including seedling establishment, development, stress resistance, metabolism, and reproduction (1). The autophagy mechanism involves the enclosure of a portion of the cytoplasm into a double membrane vesicle, named an autophagosome. The outer membrane of the autophagosome finally fuses with the vacuole (in yeast and plants) to release the inner autophagic body for hydrolytic degradation of the sequestered cargo. About 40 ATG (autophagy related) genes have been identified in Arabidopsis, which are required for autophagosome formation (3). Autophagy was initially characterized as a bulk degradation pathway induced by nutrient deprivation with a role in nutrient recycling to enable cell survival, but it also contributes to intracellular homeostasis by selectively degrading aggregated proteins, damaged mitochondria, ribosomes, toxic macromolecules, excess peroxisomes, and pathogens to prevent toxicity (4–6). In particular, although the endoplasmic reticulum (ER) is involved in autophagic processes as a source for membranes, it is also the target of a selective type of autophagy, termed reticulophagy or ER-phagy. In plants, this ER-phagy is induced in response to ER stress produced by tunicamycin (TM) or dithiothreitol (DTT) treatments (7) and upon starvation (8). Selective autophagy is mediated by the binding of adaptor proteins, which link a cargo targeted for degradation to the autophagosome machinery (9). These selective autophagy receptors share the feature of interacting with the autophagosome-localized protein ATG8 through an ATG8-interacting motif or a ubiquitin-interacting motif, leading to their recruitment into forming autophagosomes (10–12).
An increasing number of targets for selective autophagy under different stress conditions have emerged in recent years, but the underlying mechanisms of regulation of their degradation are still so far unknown. The activation of bulk and selective autophagy must be tightly controlled by the cellular conditions. In that sense, ATG4 is the only ATG that has been shown to be redox regulated in animal, yeast, algae, and plant systems (13–18). Nevertheless, in the last decade, a growing number of targets involved in autophagy have been shown to be regulated by different posttranslational modifications (PTMs); for example, ATG4b and ATG1 are regulated by S‐nitrosylation and phosphorylation (19, 20). Therefore, the ability of the ATG proteins to interact with a number of autophagic regulators is modulated by different PTMs such as phosphorylation, glycosylation, ubiquitination, and S-nitrosylation (21).
Protein persulfidation is another player in the redox regulation of certain proteins. It is the mechanism for sulfide-mediated signaling and is an oxidative posttranslational modification of cysteine residues caused by hydrogen sulfide (H2S) in which thiolate (–SH) is transformed to a persulfide group (–SSH). Persulfidation of proteins can affect their function, localization inside the cells, stability, and resistance to oxidative stress (22–27). H2S is an endogenously generated gaseous signaling molecule, which has been recently implicated in autophagy regulation both in plants and mammals (28–30).
Analysis of the Arabidopsis des1 mutant, impaired in the cytosolic production of H2S from cysteine, led to the conclusion that H2S acts as an inhibitor of autophagy induced by nutrient deprivation (28). Interestingly, its action is independent of reactive oxygen species (ROS) and nitrogen starvation, and the mechanism of autophagy inhibition by H2S has been proposed to be through persulfidation of specific targets (31). Recently, regulation of the proteolytic activity of ATG4 by persulfidation has been demonstrated in plants (16). Autophagy induced upon nitrogen starvation or osmotic stress was negatively regulated by sulfide, and the mechanism has been explained through persulfidation of C170 of this ATG4 protease, which inhibits proteolytic activity. Collectively, these results suggest that persulfidation may be the molecular mechanism through which sulfide regulates autophagy in plant cells. The susceptibility to persulfidation of the additional ATG-related proteins ATG18a, ATG3, ATG5, and ATG7 was revealed using a high throughput proteomic approach (32), although the role of this modification in these other ATG proteins has not yet been revealed. ATG18a is a core autophagy protein that binds to phosphoinositides (33, 34). It has a seven-bladed β-propeller structure formed by WD40 repeats that bind phosphatidylinositol 3-phosphate (PtdIns(3)P) or phosphatidylinositol (3, 5)-bisphosphate (PtdIns(3,5)P2). These two phosphoinositide-binding sites are located in blades five and six surrounding and sandwiching the conserved L/FRRG motif. ATG18a forms a complex with ATG2, which is involved in autophagosome biogenesis during phagophore expansion (34) and involved in the formation of preautophagosomal structures and lipid recruitment. ATG18a is essential for autophagy under several abiotic stresses, and RNA interference–ATG18a transgenic plants showed an autophagy-defective phenotype during nutrient stress and senescence (35, 36). atg18 mutants show defects in autophagosome formation and display an early senescence phenotype (34). In addition, the ER is a target of autophagy during ER stress in plants, and this ER stress–induced autophagy is dependent on the function of ATG18a (7). Thus, ATG18a is likely to be required for autophagosome formation in Arabidopsis for bulk autophagy and also for reticulophagy during ER stress (7).
In this study, we aimed to clarify the role of sulfide in the regulation of autophagy under ER stress through persulfidation of ATG18a. We found that persulfidation affects ATG18a lipid-binding activity, which in turns regulates the number and size of autophagosomes produced upon ER stress.
Results
Persulfidation of ATG18a Occurs at C103 and Is Sulfide Concentration Dependent.
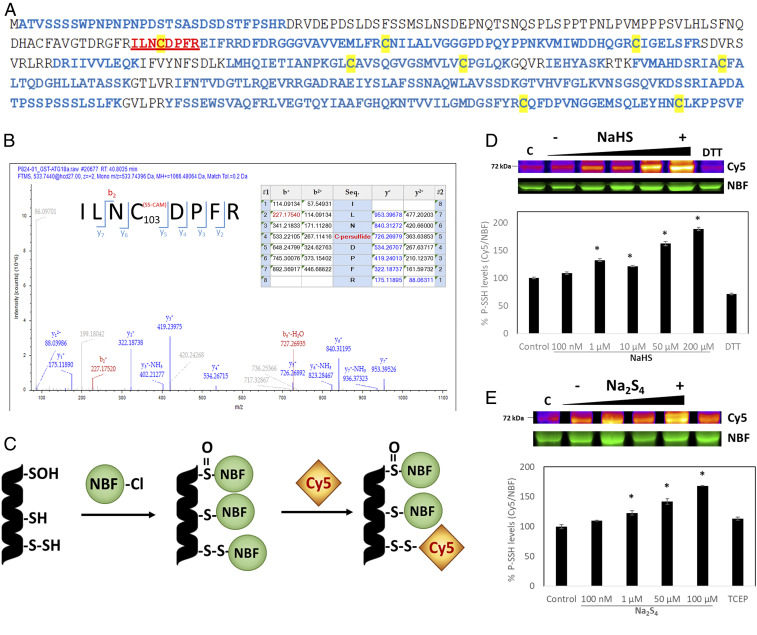
We showed previously in a proteome-scale analysis that Arabidopsis ATG18a is a target for persulfidation (32). To elucidate the target site for persulfidation in ATG18a, we carried out liquid chromatography (LC)–tandem mass spectrometry (MS/MS) analysis on the recombinant purified ATG18a protein. Protein was purified from an Escherichia coli extract and trypsin digested under nonreducing conditions to avoid the reduction of persulfide residues. As disulfide bridges between digested peptides cannot be avoided, two missed cleavages were allowed in the search. The digested peptides were analyzed using LC-MS/MS for a 32-Da mass increase plus carbamidomethylation (SS-CAM) in the fragmentation spectrum. ATG18a was identified with a sequence coverage of 79% (Fig. 1A), and the only peptide showing SS-CAM modification was ILNCDPFR, showing C103 with a persulfide modification (Fig. 1B). The XCorr value from the search with the SEQUEST engine for the ILNC(SS-CAM)DPFR peptide was 2.59. All of the other seven Cys residues were identified in the analysis, but none of them were modified by persulfidation.
Fig. 1.
Mass spectrometry analysis of AtATG18a and in-gel detection of recombinant AtATG18a protein persulfide labeling. (A) The protein was identified with a sequence coverage of 79% highlighted in blue; cysteines are highlighted in yellow, and the peptide containing persulfidated Cys-103 is shown in red and underlined. (B) LC-MS/MS analysis of the tryptic peptide containing Cys-103 of ATG18a. The table inside the spectrum contains the predicted ion types for the modified peptide, and the ions detected in the spectrum are highlighted in red and blue. (C) Scheme of dimedone switch method used to label persulfides with DAz-2/Cy5-alkyne. (D and E) In-gel detection of recombinant AtATG18a protein persulfide labeling, which is represented in fire pseudocoloring to visually enhance the signal. Green fluorescence corresponds to the total protein loaded (NBF-protein adducts). Increasing concentrations of the sulfide donors NaHS (D) and Na2S4 (E) were used and so were two reducing agents, 5 mM TCEP or 25 mM DTT. Control samples were untreated (labeled as C). The ratio of Cy5/NBF signals is used for the quantification represented in the graphs. Data are shown as the mean ± SD of three individual experiments. *P < 0.01 versus control.
To determine whether the modification was sulfide dose dependent, an in-gel detection of persulfidated ATG18a protein was performed. Purified recombinant glutathione S-transferase (GST)-ATG18a was incubated with increasing concentrations (100 nM to 200 µM) of the sulfide donors NaHS and Na2S4 and with two reducing agents tris(2-carboxyethyl)phosphine (TCEP) and DTT. A newly described dimedone switch detection method was then used (37) in which NBF-Cl reacts with all Cys residues and amino groups, giving a characteristic fluorescence (λex = 488 nm) (38), and then persulfide adducts were selectively labeled with Daz-2/Cy5-alkyne (Fig. 1C). Results showed that higher concentrations of either NaHS or Na2S4 produced a more intense Cy5 fluorescent band as a result of a higher level of persulfidation in ATG18a (Fig. 1 D and E). Band quantification demonstrated that protein treated with either sulfide donor at concentrations as low as 1 µM showed a significant increase in protein persulfidation in comparison with untreated protein. Reducing agents efficiently reduced the intensity of the Cy5 fluorescent band, indicating a decrease in ATG18a persulfidation levels when treated with DTT or TCEP. This shows that persulfidation of ATG18a protein is a reversible modification. Free GST protein was used as a negative control to test whether persulfidation of ATG18a was affected by GST persulfidation (SI Appendix, Fig. S1A), and although a band was observed indicating persulfidation, it was not Na2S4 dose dependent. Furthermore, no persulfidated peptides from GST were identified by mass spectrometry. Additionally, no persulfidated band was observed in the mutant purified recombinant ATG18a_C103S (SI Appendix, Fig. S1B).
ER Stress–Induced Autophagy Is Negatively Regulated by Sulfide.
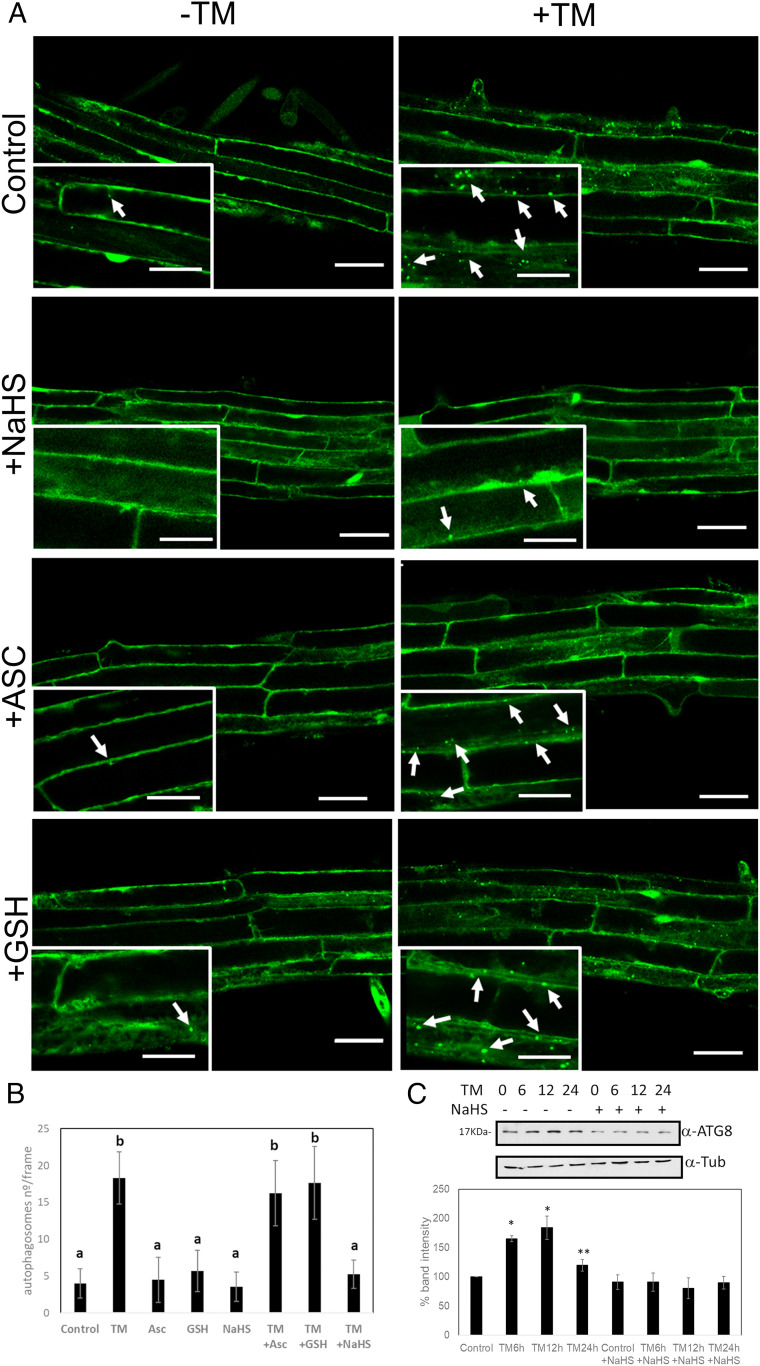
ATG18a has been shown to be critical for ER stress–induced autophagy in Arabidopsis (7). The effect of sulfide on autophagy regulation was therefore tested under ER stress. Arabidopsis transgenic seedlings expressing GFP-ATG8e were treated with TM to induce ER stress and with sulfide to test its role. Numerous autophagosomes were visualized after TM treatment in comparison with the control in which the autophagosomes were very few (Fig. 2A). The number of autophagosomes in the root per frame was quantified manually, and roots treated with TM showed significantly more autophagosomes compared with control roots (Fig. 2B). Treatment with sulfide had no significant effect on the number of autophagosomes in the absence of ER stress. By contrast, seedlings treated with TM and sulfide showed a drastic decrease in the number of autophagosomes compared with TM alone. ER stress can be alleviated by antioxidant treatments (39), and sulfide is able to increase the antioxidant capacity of the cell by inducing the activity of several antioxidant proteins (25, 40, 41). To test if the effect observed was due to the oxidative stress alleviation, treatments with ascorbic acid and glutathione were performed. Compared with control conditions, no effect was observed when treatments were applied under nonstress conditions. In addition, when ER stress was induced with TM, no significant decrease in autophagosomes was detected upon ascorbic acid or glutathione treatments, indicating that the effect of sulfide is not due to its antioxidant properties.
Fig. 2.
ER stress–induced autophagy is regulated by sulfide in Arabidopsis roots. GFP-ATG8e transgenic plants, 7 d old, were transferred to Murashige and Skoog (MS) liquid medium supplemented with TM to induce ER stress, sulfide (NaHS), ascorbic acid (ASC), and/or glutathione (GSH), or dimethyl sulfoxide (Control). (A) GFP-ATG8e–labeled autophagosomes in root epidermal cells were visualized by confocal microscopy. Arrows indicate GFP-labeled autophagosomes or autophagic bodies. (Insets) Enlargement of confocal microscopy images. (Scale bars, 40 μm for main figure, 20 µm for insets.) (B) Quantification of autophagosomes per frame for each treatment. Values are the mean ± SD (n > 15). Different letters indicate statistically significant differences (ANOVA, Fisher’s least significant difference test, P < 0.05). (C) Immunoblot analysis using anti-ATG8 antibodies in protein extracts from WT Arabidopsis seedlings treated with TM for 6, 12, and 24 h and NaHS. Graph shows the relative band intensity. Bars represent means ± SD (n = 3). *P < 0.01, **P < 0.05.
Another approach was performed in order to corroborate these observations. Wild-type (WT) or GFP-ATG8e–expressing seedlings were treated with TM for 6 h and for a prolonged period of 12 and 24 h and treated with sulfide for 1 h. The effect of sulfide on autophagy was analyzed by quantification of the autophagy marker ATG8 protein by immunoblot using an anti-ATG8 antibody (Fig. 2C) or by analysis of GFP-ATG8e cleavage (SI Appendix, Fig. S2A). Results showed that seedlings incubated with TM induced autophagy with the highest level of induction, ranging from 12 to 24 h of treatment; meanwhile, those treated together with sulfide showed a decrease in autophagy induction, reaching similar levels as observed in the absence of TM (Fig. 2C and SI Appendix, Fig. S2A).
These data suggest that autophagy induced under ER stress is negatively regulated by sulfide to a much greater extent than by antioxidant treatments.
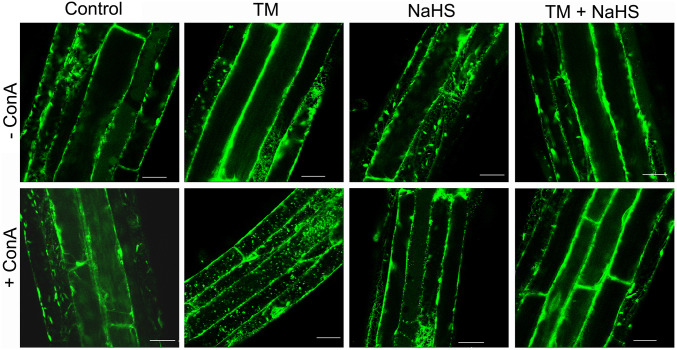
It was shown previously that the ER is degraded by autophagy during ER stress in Arabidopsis (7). Therefore, we studied the effect of sulfide on ER degradation using the fluorescent ER marker protein, GFP-HDEL, under ER stress conditions (42). Confocal microscopy showed the typical ER networks in the cytoplasm in control treatments. Similarly, in the presence of 1 µM concanamycin A (conA) to prevent vacuolar degradation, the vacuole lacked GFP fluorescence in control samples (Fig. 3 and SI Appendix, Fig. S3A). Accumulation of endogenous ATG8 was observed in GFP-HDEL plants treated with TM, while when plants were treated with TM and sulfide, no differences were observed in comparison with control (SI Appendix, Fig. S3 B and C). Similar results were observed in plants treated with sulfide. When plants were treated with TM, a clear increase in GFP-HDEL ER labeling was observed in the vacuole after conA incubation, demonstrating that ER was being degraded in the vacuole. However, plants treated with both TM and sulfide lacked the increase in fluorescence in the vacuoles, showing ER localization similar to the control treatments. To confirm that TM was causing ER stress and that the sulfide does not prevent or alleviate the ER stress, but on the contrary regulates autophagy-specific ER degradation, we assessed bZIP60 splicing in plants treated with TM and sulfide for 6, 12, and 24 h. bZIP60 is an ER stress-responsive transcription factor that is spliced only upon ER stress (43), and the splicing event can be detected by PCR. The results showed that sulfide did not alleviate ER stress, suggesting that the effect observed was specific for autophagy; although ER stress was persistent, autophagy was blocked (SI Appendix, Fig. S2B). Collectively, these results suggest that ER degradation by autophagy is regulated by sulfide.
Fig. 3.
ER degradation by autophagy is regulated by sulfide in Arabidopsis roots. To induce ER stress, 7-d-old GFP-HDEL transgenic plants were transferred to Murashige and Skoog (MS) liquid medium supplemented with TM for 6 h and sulfide (NaHS) for 1 h. Dimethyl sulfoxide was used as solvent (Control). The ER structure in root epidermal cells was visualized by confocal microscopy, representative of 3 to 4 pictures/plant and 4 to 5 plants/treatment. (Scale bars, 40 μm.)
Sulfide Enhances ATG18a Binding to Membranes.
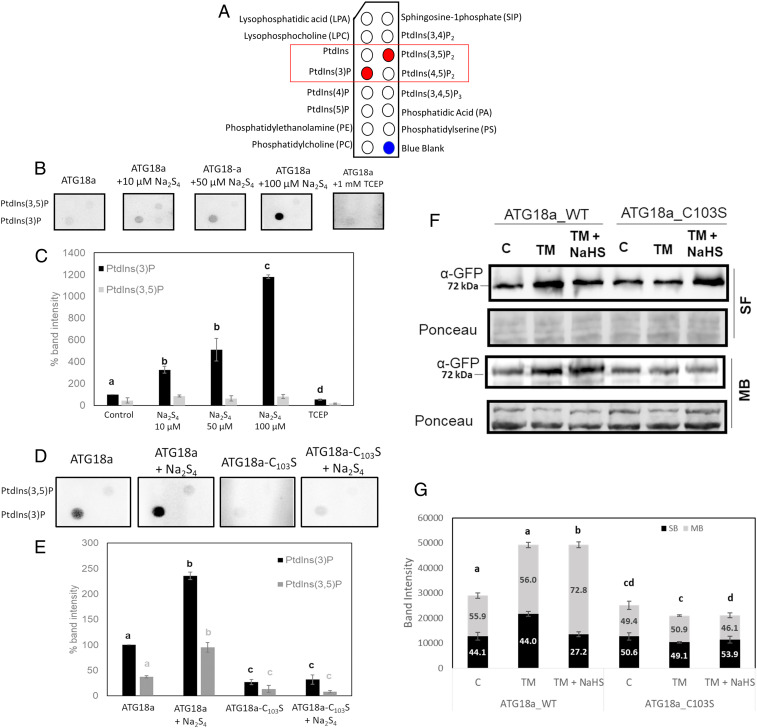
Atg18 protein from Saccharomyces cerevisiae and WIPI3 and WIPI4 from mammals are homologs of ATG18a, and they were reported to bind to PtdIns(3)P and PtdIns(3,5)P2 (33, 44–46). The role of sulfide in the phosphoinositide-binding activity of the ATG18a protein was assayed using phosphoinositide (PIP) strips. PIP strips contain 100 pmol of 15 different lipids (Fig. 4A) spotted onto a cellulose blotting membrane, and protein–lipid–binding specificity can be analyzed easily.
Fig. 4.
Sulfide enhances ATG18a binding to membranes. (A) Schematic representation of the indicated membrane lipids on a PIP strip. Red dots represent the specific lipids PtdIns(3)P and PtdIns(3,5)P2 for B and D. (B) Binding of recombinant GST-ATG18a proteins to PIP strips. A total of 0.5 µg protein was incubated with an increasing concentration of NaHS and TCEP followed by incubation with PIP strips. Bound protein was detected using anti-GST primary antibody and anti-rabbit–HRP secondary antibody. (C) Spot intensity relative quantification of B. (D) Comparison of binding affinity to PIP strips of 1 µg GST-ATG18a and GST-ATG18a_C103S treated or not with 10 µM Na2S4. (E) Spot intensity relative quantification of D. (F) Immunoblot of soluble (SF) and membrane-bound (MB) fractions of the fusion proteins YFP-ATG18a and YFP-ATG18a_C103S obtained from transgenic lines treated with TM for 6 h and sulfide (NaHS) for 1 h. (G) Relative band intensity quantification of F. In C, E, and G, bars represent means ± SD (n = 3). Different letters indicate statistically significant differences (ANOVA, Tukey test, P < 0.05).
To determine the role of persulfidation in the binding activity of ATG18a, recombinant GST-ATG18a protein was preincubated with increasing concentrations of Na2S4 and TCEP and then subjected to PIP strip binding. Results showed that untreated protein had specificity for PtdIns(3)P, and a weaker binding to PtdIns(3,5)P2 was observed (Fig. 4 B and C). Sulfide enhanced the binding activity to both phosphoinositides, although more specifically to PtdIns(3)P. Binding was sulfide dose dependent, and treatment with 100 µM Na2S4 caused a substantial increase in binding. Treatment with TCEP significantly decreased binding to both phospholipids, demonstrating the reversibility of the sulfide induction of the ATG18a binding activity. The binding of the mutant recombinant protein GST-ATG18a_C103S was also assessed. Both untreated and sulfide-treated protein showed a very significantly reduced binding to these phosphoinositides in comparison with the WT protein (Fig. 4 D and E). Free GST protein was assayed in PIP strips to corroborate that the tag did not affect binding in the recombinant proteins, and results showed that GST had no PtdIns(3)P-binding ability (SI Appendix, Fig. S4). These results suggested that the binding of ATG18a to PtdIns(3)P is significantly and reversibly increased by persulfidation. In addition, a weaker affinity toward PtdIns(3,5)P2 was observed, although the effect of sulfide on this binding is inconclusive.
To confirm this effect in vivo, Δatg18a YFP-ATG18a_WT and Δatg18a YFP-ATG18a_C103S seedlings were treated with TM and sulfide, and a whole protein extract was subjected to cellular fractionation to separate cytosolic proteins and the membrane-bound fraction. The level of enrichment of cellular fractionation was checked using specific markers for membranes (anti-H+ATPase) and cytosol (anti–OASA-1) (SI Appendix, Fig. S5). Treatment with TM increased the level of YFP-ATG18a_WT in comparison with untreated seedlings, and furthermore, the proportion of YFP-ATG18a_WT protein in the membrane-bound fraction was significantly higher in seedlings treated with TM and sulfide than in those treated with only TM (Fig. 4 F and G). This suggests that sulfide treatment increases the proportion of YFP-ATG18a_WT in the membrane fraction. This effect was not observed with the mutant version of the protein, YFP-ATG18a_C103S, in which the proportion of bound protein to membrane was even lower than the control or the TM-treated sample. These in vitro and in vivo experiments suggest that sulfide enhances ATG18a binding to membranes by persulfidation of the C103 residue.
Sulfide Regulates Autophagosome Biogenesis by Persulfidation of ATG18a during ER Stress.
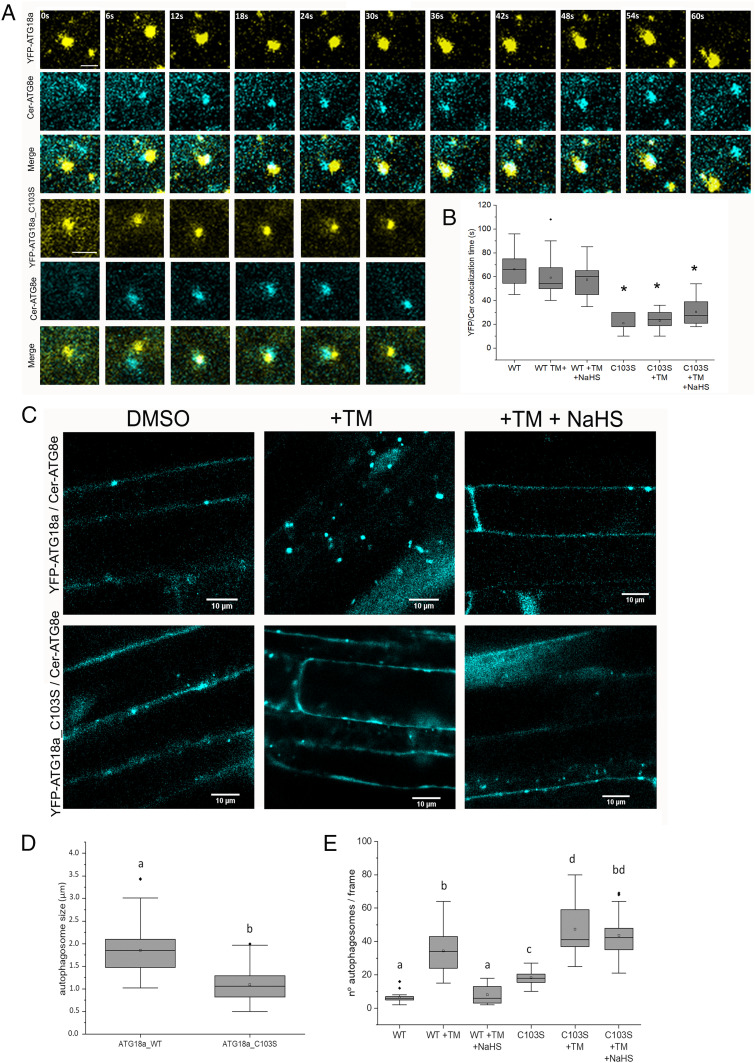
To further examine the subcellular localization of ATG18a and ATG18a_C103S in the plant cell and to test their colocalization with ATG8e and therefore with autophagosomes and their precursors, we generated transgenic plants coexpressing YFP-ATG18a_WT or YFP-ATG18a_C103S and Cerulean-ATG8e in a Δatg18a background. These seedlings were treated with TM and sulfide to study the effect of sulfide during ER stress. By confocal microscopy, ATG18a and ATG8e colocalized, but colocalization time was significantly shorter in those seedlings with the mutant version of ATG18a (Fig. 5A and SI Appendix, Fig. S6). YFP-ATG18a_WT seedlings showed a YFP/CFP colocalization mean time of 66 s, while for YFP-ATG18a_C103S, it was 24 s. Surprisingly, treatments with TM or sulfide did not affect the YFP/Cer colocalization time in comparison with their respective control (Fig. 5B). The shortening in the colocalization time of ATG8e and the mutated version of ATG18a could be indicative of the involvement of C103 of ATG18a in autophagosome biogenesis. To assess whether the mutation in C103 alters the number or size of autophagosomes, we measured these parameters for ATG8-positive puncta within the cells of YFP-ATG18a_WT and YFP-ATG18a_C103S seedlings coexpressing Cer-ATG8e. Average size was measured for n > 200 puncta over three independent experiments for each condition using ImageJ (47). Results showed that the mutated version of ATG18a decreased the size of autophagosomes compared with the WT version (Fig. 5 C and D and SI Appendix, Fig. S7), suggesting that the impairment of ATG18a for persulfidation could affect the average size of autophagosomes. Furthermore, as expected, YFP-ATG18a_WT seedlings showed an increase in Cer-ATG8e puncta number when treated with TM, which was reverted after sulfide treatment (Fig. 5E). However, untreated YFP-ATG18a_C103S seedlings showed an increase in autophagosome number in comparison with YFP-ATG18a_WT, which was even higher upon treatment with TM. However, sulfide treatment had no significant effect on the number of puncta in YFP-ATG18a_C103S seedlings under ER stress, with no decrease seen. To confirm the effect of YFP-ATG18a_C103S on the extent of autophagy in comparison with YFP-ATG18a_WT, ATG8 levels were measured by immunoblot assay. Results showed that YFP-ATG18a_C103S seedlings had increased levels of ATG8 compared with WT in control conditions (SI Appendix, Fig. S8 A and B). In addition, TM treatment increased ATG8 levels in both genotypes, but sulfide treatment was only able to decrease ATG8 levels in YFP-ATG18a_WT seedlings. Furthermore, autophagy flux was measured under nitrogen starvation stress in both genotypes, and results showed that ATG8 levels were in general higher in YFP-ATG18a_C103S than YFP-ATG18a_WT, while the ratio of free CFP/CFP-ATG8 was smaller in YFP-ATG18a_C103S (SI Appendix, Fig. S8 C and D). However, under this stress condition, the C103S mutation on ATG18a did not affect the negative regulation of autophagy by sulfide. These results suggest that persulfidation of ATG18a at C103 regulates autophagy under ER stress but not under nitrogen starvation, and the impairment of persulfidation affects both the number and size of autophagosomes.
Fig. 5.
Time-lapse confocal fluorescence microscopy of autophagosomes in roots from 1-wk-old seedlings treated with TM and sulfide (NaHS). (A) Confocal images for colocalization of YFP-ATG18a and YFP-ATG18a_C103S with Cer-ATG8e. (Scale bars, 2 μm.) (B) Analysis of YFP/Cerulean colocalization time. The duration of colocalization is represented in seconds. (n > 15). (C) Confocal images for Cer-ATG8e in the double transgenic plants YFP-ATG18a/Cer-ATG8e and YFP-ATG18a_C103S/Cer-ATG8e after treatment with TM and NaHS. (D) Autophagosome size analysis measured with ImageJ, n = 200. (E) Autophagosome number quantification counted manually with ImageJ, n = 15. Values are the mean ± SD. Different letters indicate statistically significant differences (ANOVA, Fisher’s least significant difference test, P < 0.05).
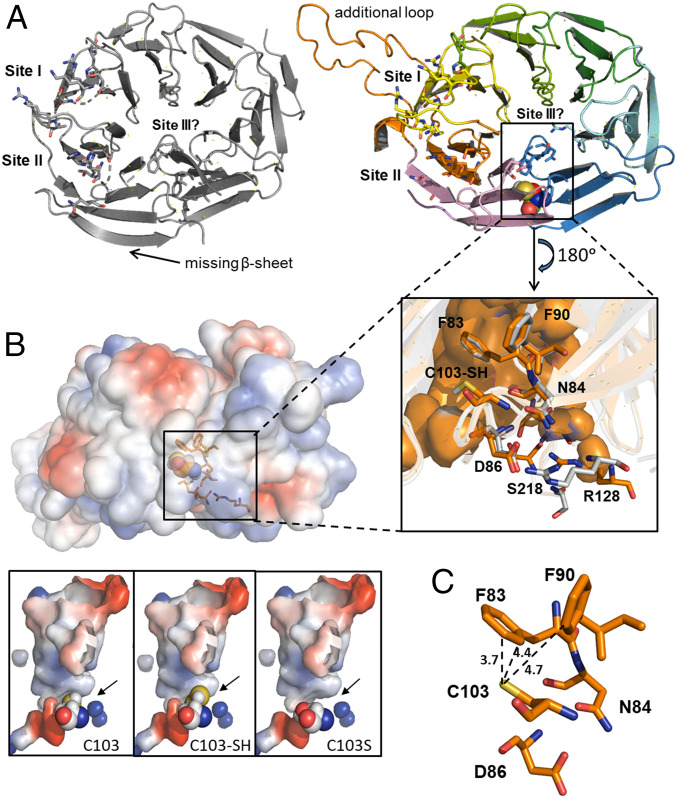
Persulfidation of ATG18a Affects the Cavity Containing C103 and the Surrounding Electrostatic Interaction Network.
To examine the impact of the persulfidation posttranslational modification of C103 in the interaction between AtATG18a and phosphoinositide molecules, we have performed three-dimensional (3D) homology modeling and structural alignment using the Homo sapiens HsWIPI3–ATG2A:WIR peptide complex (48). AtATG18a shares up to 43.95% sequence identity with HsWIPI3 (E-value: 2.3 × 10-23), with conserved residues covering the full sequence. In particular, two highly conserved but not identical phosphoinositide-binding sites (site I and site II) and the potential lipid-binding site III (46) are shown (Fig. 6A). The 3D structural alignment revealed the lack of a β-sheet and the presence of a long, flexible extension in AtATG18a compared with HsWIPI3 (Fig. 6A). This long, flexible loop is negatively charged and likely affects the charge distribution around binding site I compared with HsWIPI3 (SI Appendix, Fig. S9). This finding might negatively influence the binding of PtdIns(3,5)P2 to AtATG18a site I, explaining the weaker binding observed (Fig. 4). It is worth mentioning that site I and site II differ in lipid affinity. Site I tends to recognize preferentially PtdIns(3,5)P2, whereas site II interacts with PtdIns(3)P (46).
Fig. 6.
Predicted structure of AtATG18a. (A) Crystal structure of human WIPI3 (Protein Data Bank identification: 6KRL; Left) and the predicted structural model of Arabidopsis ATG18a (Right). The proposed lipid-binding sites and the cavity site of C103 are highlighted. (Inset) Residues of AtATG18a surrounding the active C103 site (F83, N84, D86, and F90) and participating in binding site III (R128 and S218). The equivalent residues in HsWIPI3 are in gray. (B) Representation of surface electrostatic potential distribution in AtATG18a structural model and zoomed into the putative conformation of the active site, showing with spheres the position of C103, the persulfidation C103-SH, and the mutation C103S. Positively and negatively charged regions are depicted in blue and red, respectively. (C) Zoom into the putative conformation of the active site showing distance (Å) between the catalytic residue C103 and F83 and F90 in AtATG18a.
The residue C103 is located inside a hydrophobic cavity formed by residues 83FNQD86 and F90, which are highly conserved in the WIPI members of the PROPPIN protein family (48). In fact, these residues of AtATG18a are at the exact positions of those of human WIPI3 (Fig. 6 A, Inset). Studies in the homologs WIPI4 and WIPI3 have demonstrated that the equivalent residues N15/N19 and D17/D21, respectively, to N84 and D86 of AtATG18a are essential for ATG2A binding (45, 48). Interestingly, the residue N84 and the neighboring residues R128 and S218, equivalents to R62 and S151 of human WIPI3, respectively, and exposed at the exterior of the protein, have been reported to participate in an additional potential lipid-binding site III (46). Positive charges around this region (Fig. 6B) would govern the electrostatic interaction with other negatively charged partners such as phosphoinositide molecules. Moreover, the comparison of HsWIPI3 with the AtATG18a model reveals an increase of positive charge distribution on the surface of site III in the Arabidopsis protein (SI Appendix, Fig. S9).
Consistent with this, persulfidation of C103 could potentially affect the molecular recognition in its neighboring protein region and consequently the function of AtATG18a. The cysteine persulfidation by the addition of the –SH group to C103 could cause significant conformational changes due to the following: (1) a steric perturbation in the cavity (Fig. 6C), destabilizing the original hydrophobic interactions and promoting new ones, and (2) an effect of negative charges from the deprotonation of the −SSH group in a highly unipolar environment. The additional sulfur atom would contribute with a covalent radius of 1.02 Å, while that of hydrogen is 0.37 Å. It is worth mentioning that the aromatic thiol π hydrogen-bonding interaction is very sensitive to the orientation of the two lone electron pairs on the sulfur atom relative to the π electron cloud of the phenyl ring (49). In addition, it would be reasonable to expect that persulfidation of C103 would introduce negative charges at the active site since the −SSH group most probably would be completely deprotonated at physiological pH (22, 50). The replacement C103S could also destabilize the hydrophobic cavity site but in a different manner. Although the energetic cost of desolvating serine (−OH) is higher than for a cysteine (−SH) residue, the steric effect due to its replacement would be much smaller than that caused by the additional –SH group (Fig. 6B). Altogether, the C103 modification by persulfidation likely could cause a conformational change and intramolecular rearrangement of the cavity, favoring the interaction with additional phosphoinositide molecules at site III and/or increasing the binding affinity of PtdIns(3)P for its target at site II through an allosteric cooperative effect.
Discussion
Autophagy regulation in plants has become an important, although challenging, topic in recent years due to its involvement in plant health and cellular homeostasis, helping plants to tolerate different stresses such as oxidative stress, pathogen infection, nutrient deprivation, and drought (35, 36, 51, 52). It is now well known that ER stress triggers autophagy both in mammals and plants (7, 53) as a protective mechanism against misfolded proteins accumulating in the ER. Thus, a selective autophagy pathway named ER-phagy is activated to recycle accumulated misfolded proteins in the ER in order to restore ER homeostasis (54). In plants, it has been shown that inositol-requiring enzyme 1 (IRE1) is necessary for ER-phagy, and the delivery of ER to the vacuole for degradation under ER stress depends on the ATG18a protein (7). In mammals, recent studies showed that H2S may influence ER stress, playing an important role in many heart, neurological, and respiratory diseases (55). In plants, H2S was shown to negatively regulate bulk autophagy under nutrient deprivation, independently of redox conditions (28, 31). This mechanism was explained by persulfidation of ATG core proteins (16, 32). In this work, we have demonstrated that sulfide also regulates autophagy induced by ER stress, and this regulation was independent of its indirect antioxidant effect. Furthermore, we showed that sulfide played a signaling role in ER degradation by autophagy.
Although little has been reported about the biochemical function of plant ATG18a protein, in yeast and mammals, a function for the Atg2-Atg18 complex in autophagy has been reported, and the binding of Atg18 to PtdIns(3)P is crucial in autophagy induction (56). Posttranslational modifications of ATG18 are known to control its activity. Atg18 in the yeast Pichia pastoris was shown to be phosphorylated, and phosphorylation enhanced its binding affinity to PtdIns(3)P (57). However, this modification in P. pastoris does not affect autophagy activity, while phosphorylation of Arabidopsis ATG18a inhibits its function in autophagy (58). Therefore, posttranslational regulation of ATG18a must be finely controlled and probably triggered depending on the stress to which the plant is subjected. Furthermore, ATG18a is also required for bulk autophagy, and persulfidation could possibly regulate its activity in multiple stresses in addition to ER stress.
In this study, we explored the functional importance of sulfide in regulating ATG18a binding to PtdIns(3)P. Mass spectrometry analysis of recombinant ATG18a showed that only one Cys residue, C103, was persulfidated under our experimental conditions. Protein persulfidation at more than one Cys residue is rarely found in the literature, and the other Cys residues of ATG18a were not detected as modified peptides, indicating that C103 was the only target for persulfidation. Furthermore, this PTM was shown to be H2S dose dependent at very low concentrations of NaHS and reversible by reducing agents, suggesting this modification may have a biological role in plants. We further showed the potential physiological role of the persulfidation of ATG18a by using WT and C103 mutant recombinant proteins in an in vitro assay; one important finding was that persulfidation of ATG18a increased its affinity for PtdIns(3)P, while impairment of persulfidation abolished almost completely this binding to phospholipids. In vivo, sulfide enhanced the fraction of ATG18a bound to membranes, while the mutant ATG18a_C103S was not affected by sulfide treatment, and in general, the ATG18a_C103S fraction bound to membranes was less than that of the WT ATG18a protein.
We observed that the mutant protein ATG18a_C103S, impaired in persulfidation, was unable to bind properly to PtdIns(3)P, and therefore, the colocalization time with an autophagosome marker was shorter than that of the WT protein. Furthermore, autophagosomes in plants expressing only the ATG18a_C103S mutated version had a smaller average size than when the WT counterpart was expressed. However, it seems that plants respond to this situation by increasing the number of autophagosomes. ATG18a forms a complex with ATG2 that is involved in the expansion of the membrane of the autophagosome for phagophore elongation (59); therefore, it makes sense that disturbing the binding affinity of ATG18a for phosphoinositides could affect autophagosome size. In addition, the colocalization time of ATG18a_C103S with autophagosomes is shorter, and its binding affinity to PtdIns(3)P is weaker than the WT ATG18a, resulting in an outcome of smaller autophagosomes, suggesting that C103 is important in regulating the elongation of phagophore membranes. Previous studies in yeast demonstrated that insufficient levels of Atg7 affect both autophagosome size and number (60), and similarly, mutations disturbing Atg3 function also affect autophagosome size (61). The fact that the misfunction of ATG18a has an effect on both autophagosome size (expansion) and number (nucleation) of autophagosomes may indicate that ATG18a could have additional roles in autophagosome formation.
In yeast, it was demonstrated that Atg18 is important for the localization of the Atg2-Atg18 complex to the preautophagosomal structure (PAS) through its PtdIns(3)P-binding ability (62, 63). In fact, some studies showed that autophagosome formation could efficiently progress in atg2Δatg18Δ cells expressing engineered variants of Atg2 that are capable of localizing to the PAS. Engineered yeast expressing Atg2–HG–FYVE, a fusion of Atg2 with the phosphatidylinositol 3‐phosphate–binding FYVE domain, or Atg2–HG–Atg8, expressing Atg2 fused with the core autophagy protein Atg8, allowed a partial recovery of autophagosome formation independently of Atg18 (64). Nevertheless, the partial recovery suggested that Atg18 possesses other functions beyond targeting Atg2 to the PAS. The general structure of this complex is similar to the mammalian homolog ATG2B-WIPI4, suggesting that the overall shape and function is evolutionarily conserved among species (65, 66). Our results obtained from the 3D structural alignment suggested that persulfidation of C103 could modulate the interaction with PtdIns(3)P molecules by inducing a conformational change of the surrounding site that could affect recognition of potential partners such as PtdIns(3)P through the electrostatic interaction network. Therefore, persulfidation of ATG18a could affect autophagy progression by regulating its PtdIns(3)P-binding affinity.
It is noteworthy that previous studies described how sulfide regulates bulk autophagy triggered by nutrient starvation or ABA (16, 31), and in this study, we demonstrate that ER-induced autophagy is also negatively regulated by sulfide. We provide a level of regulation of autophagy in plant systems by sulfide through the persulfidation of ATG18a. Together, our data suggest a model in which the dynamics of ATG18a association with forming autophagosomes is critical for autophagy during ER stress. The reversible persulfidation of ATG18a increases its binding to membranes via association with PtdIns(3)P but potentially delays its release, inhibiting autophagosome maturation. Preventing persulfidation by the mutation of C103 decreases its membrane affinity and localization time to phagophores. As ATG18a is thought to be involved in lipid transfer during autophagosome expansion, this decreased phagophore localization leads to the production of smaller autophagosomes. Autophagosome numbers increase, potentially to compensate for this. In this way, persulfidation of ATG18a may modulate the extent of autophagosome production during stress to ensure an appropriate physiological response.
Materials and Methods
A detailed description of all materials and methodology is included in SI Appendix, Materials and Methods. This includes the expression and purification of recombinant GST-tagged protein, mass spectrometry, persulfide detection in cell lysates, plant genotypes, protein modeling, protein–lipid–binding assay, membrane fractionation, autophagy detection, microscopy, bZIP60 splicing assay, and expression and purification of free GST protein.
Supplementary Material
Acknowledgments
This work was supported in part by the European Union’s Horizon 2020 research and innovation programme under Marie Skłodowska-Curie Grant Agreement No. 834120. We also acknowledge the Project US-1255781 European Regional Development Fund for funding support (to A.A.), Government of Aragón FEDER Grant E35_17R (to I.Y.), Agencia Estatal Investigación European Regional Development Fund Grant PID2019-109785GB-I00 (to C.G.), and NIH Grant 1R01GM120316-01A1 (to D.C.B.). We thank Dr. F.J. Quintero and Dr. J.M. Pardo for providing the anti-H+ATPase antibody.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2023604118/-/DCSupplemental.
Data Availability
All study data are included in the main text and SI Appendix.
References
- 1.Bassham D. C., Function and regulation of macroautophagy in plants. Biochim. Biophys. Acta 1793, 1397–1403 (2009). [DOI] [PubMed] [Google Scholar]
- 2.Bassham D. C., et al., Autophagy in development and stress responses of plants. Autophagy 2, 2–11 (2006). [DOI] [PubMed] [Google Scholar]
- 3.Zientara-Rytter K., Sirko A., To deliver or to degrade–An interplay of the ubiquitin-proteasome system, autophagy and vesicular transport in plants. FEBS J. 283, 3534–3555 (2016). [DOI] [PubMed] [Google Scholar]
- 4.Pankiv S., et al., p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J. Biol. Chem. 282, 24131–24145 (2007). [DOI] [PubMed] [Google Scholar]
- 5.Reggiori F., Komatsu M., Finley K., Simonsen A., Autophagy: More than a nonselective pathway. Int. J. Cell Biol. 2012, 219625 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kraft C., Reggiori F., Peter M., Selective types of autophagy in yeast. Biochim. Biophys. Acta 1793, 1404–1412 (2009). [DOI] [PubMed] [Google Scholar]
- 7.Liu Y., et al., Degradation of the endoplasmic reticulum by autophagy during endoplasmic reticulum stress in Arabidopsis. Plant Cell 24, 4635–4651 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu J., et al., ATI1 (ATG8-interacting protein 1) and ATI2 define a plant starvation-induced reticulophagy pathway and serve as MSBP1/MAPR5 cargo receptors. Autophagy, 10.1080/15548627.2021.1872886 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Johansen T., Lamark T., Selective autophagy mediated by autophagic adapter proteins. Autophagy 7, 279–296 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lei Y., Klionsky D. J., UIM-UDS: A new interface between ATG8 and its interactors. Cell Res. 29, 507–508 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marshall R. S., Hua Z., Mali S., McLoughlin F., Vierstra R. D., ATG8-Binding UIM proteins define a new class of autophagy adaptors and receptors. Cell 177, 766–781.e24 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johansen T., Lamark T., Selective autophagy: ATG8 family proteins, LIR motifs and cargo receptors. J. Mol. Biol. 432, 80–103 (2020). [DOI] [PubMed] [Google Scholar]
- 13.Woo J., Park E., Dinesh-Kumar S. P., Differential processing of Arabidopsis ubiquitin-like Atg8 autophagy proteins by Atg4 cysteine proteases. Proc. Natl. Acad. Sci. U.S.A. 111, 863–868 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pérez-Pérez M. E., Zaffagnini M., Marchand C. H., Crespo J. L., Lemaire S. D., The yeast autophagy protease Atg4 is regulated by thioredoxin. Autophagy 10, 1953–1964 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pérez-Pérez M. E., Lemaire S. D., Crespo J. L., Control of autophagy in Chlamydomonas is mediated through redox-dependent inactivation of the ATG4 protease. Plant Physiol. 172, 2219–2234 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Laureano-Marín A. M., et al., Abscisic acid-triggered persulfidation of the Cys protease ATG4 mediates regulation of autophagy by sulfide. Plant Cell 32, 3902–3920 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pajares M., Cuadrado A., Engedal N., Jirsova Z., Cahova M., The role of free radicals in autophagy regulation: Implications for ageing. Oxid. Med. Cell. Longev. 2018, 2450748 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Scherz-Shouval R., et al., Reactive oxygen species are essential for autophagy and specifically regulate the activity of Atg4. EMBO J. 26, 1749–1760 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pengo N., Agrotis A., Prak K., Jones J., Ketteler R., A reversible phospho-switch mediated by ULK1 regulates the activity of autophagy protease ATG4B. Nat. Commun. 8, 294 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shivaraj S. M., et al., Nitric oxide and hydrogen sulfide crosstalk during heavy metal stress in plants. Physiol. Plant. 168, 437–455 (2020). [DOI] [PubMed] [Google Scholar]
- 21.Xie Y., et al., Posttranslational modification of autophagy-related proteins in macroautophagy. Autophagy 11, 28–45 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Filipovic M. R., Zivanovic J., Alvarez B., Banerjee R., Chemical biology of H2S signaling through persulfidation. Chem. Rev. 118, 1253–1337 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kimura H., Physiological roles of hydrogen sulfide and polysulfides. Handb. Exp. Pharmacol. 230, 61–81 (2015). [DOI] [PubMed] [Google Scholar]
- 24.Aroca A., Gotor C., Bassham D. C., Romero L. C., Hydrogen sulfide: From a toxic molecule to a key molecule of cell life. Antioxidants 9, 621 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aroca Á., Serna A., Gotor C., Romero L. C., S-sulfhydration: A cysteine posttranslational modification in plant systems. Plant Physiol. 168, 334–342 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aroca A., Schneider M., Scheibe R., Gotor C., Romero L. C., Hydrogen sulfide regulates the cytosolic/nuclear partitioning of glyceraldehyde-3-phosphate dehydrogenase by enhancing its nuclear localization. Plant Cell Physiol. 58, 983–992 (2017). [DOI] [PubMed] [Google Scholar]
- 27.Filipovic M. R., Persulfidation (S-sulfhydration) and H2S. Handb. Exp. Pharmacol. 230, 29–59 (2015). [DOI] [PubMed] [Google Scholar]
- 28.Álvarez C., et al., Cysteine-generated sulfide in the cytosol negatively regulates autophagy and modulates the transcriptional profile in Arabidopsis. Plant Cell 24, 4621–4634 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang D., et al., Detection of protein S-sulfhydration by a tag-switch technique. Angew. Chem. Int. Ed. Engl. 53, 575–581 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kundu S., Pushpakumar S., Khundmiri S. J., Sen U., Hydrogen sulfide mitigates hyperglycemic remodeling via liver kinase B1-adenosine monophosphate-activated protein kinase signaling. Biochim. Biophys. Acta 1843, 2816–2826 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Laureano-Marín A. M., Moreno I., Romero L. C., Gotor C., Negative regulation of autophagy by sulfide is independent of reactive oxygen species. Plant Physiol. 171, 1378–1391 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aroca A., Benito J. M., Gotor C., Romero L. C., Persulfidation proteome reveals the regulation of protein function by hydrogen sulfide in diverse biological processes in Arabidopsis. J. Exp. Bot. 68, 4915–4927 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dove S. K., et al., Svp1p defines a family of phosphatidylinositol 3,5-bisphosphate effectors. EMBO J. 23, 1922–1933 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wun C.-L., Quan Y., Zhuang X., Recent advances in membrane shaping for plant autophagosome biogenesis. Front. Plant Sci. 11, 565 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu Y., Xiong Y., Bassham D. C., Autophagy is required for tolerance of drought and salt stress in plants. Autophagy 5, 954–963 (2009). [DOI] [PubMed] [Google Scholar]
- 36.Xiong Y., Contento A. L., Bassham D. C., AtATG18a is required for the formation of autophagosomes during nutrient stress and senescence in Arabidopsis thaliana. Plant J. 42, 535–546 (2005). [DOI] [PubMed] [Google Scholar]
- 37.Zivanovic J., et al., Selective persulfide detection reveals evolutionarily conserved antiaging effects of S-sulfhydration. Cell Metab. 30, 1152–1170.e13 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bernal-Perez L. F., Prokai L., Ryu Y., Selective N-terminal fluorescent labeling of proteins using 4-chloro-7-nitrobenzofurazan: A method to distinguish protein N-terminal acetylation. Anal. Biochem. 428, 13–15 (2012). [DOI] [PubMed] [Google Scholar]
- 39.Malhotra J. D., et al., Antioxidants reduce endoplasmic reticulum stress and improve protein secretion. Proc. Natl. Acad. Sci. U.S.A. 105, 18525–18530 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Aroca A., Gotor C., Romero L. C., Hydrogen sulfide signaling in plants: Emerging roles of protein persulfidation. Front. Plant Sci. 9, 1369 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xie Z.-Z., Liu Y., Bian J.-S., Hydrogen sulfide and cellular redox homeostasis. Oxid. Med. Cell. Longev. 2016, 6043038 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Batoko H., Zheng H. Q., Hawes C., Moore I., A rab1 GTPase is required for transport between the endoplasmic reticulum and golgi apparatus and for normal golgi movement in plants. Plant Cell 12, 2201–2218 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Deng Y., et al., Heat induces the splicing by IRE1 of a mRNA encoding a transcription factor involved in the unfolded protein response in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 108, 7247–7252 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bakula D., et al., WIPI3 and WIPI4 β-propellers are scaffolds for LKB1-AMPK-TSC signalling circuits in the control of autophagy. Nat. Commun. 8, 15637 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Baskaran S., Ragusa M. J., Boura E., Hurley J. H., Two-site recognition of phosphatidylinositol 3-phosphate by PROPPINs in autophagy. Mol. Cell 47, 339–348 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liang R., Ren J., Zhang Y., Feng W., Structural conservation of the two phosphoinositide-binding sites in WIPI proteins. J. Mol. Biol. 431, 1494–1505 (2019). [DOI] [PubMed] [Google Scholar]
- 47.Humphry N. J., Wheatley S. P., Survivin inhibits excessive autophagy in cancer cells but does so independently of its interaction with LC3. Biol. Open 7, bio037374 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ren J., et al., Multi-site-mediated entwining of the linear WIR-motif around WIPI β-propellers for autophagy. Nat. Commun. 11, 2702 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Duan G., Smith V. H., Weaver D. F., Characterization of aromatic-thiol π-type hydrogen bonding and phenylalanine-cysteine side chain interactions through ab initio calculations and protein database analyses. Mol. Phys. 99, 1689–1699 (2001). [Google Scholar]
- 50.Cuevasanta E., et al., Reaction of hydrogen sulfide with disulfide and sulfenic acid to form the strongly nucleophilic persulfide. J. Biol. Chem. 290, 26866–26880 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hanaoka H., et al., Leaf senescence and starvation-induced chlorosis are accelerated by the disruption of an Arabidopsis autophagy gene. Plant Physiol. 129, 1181–1193 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Signorelli S., Tarkowski Ł. P., Van den Ende W., Bassham D. C., Linking autophagy to abiotic and biotic stress responses. Trends Plant Sci. 24, 413–430 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lee W. S., Yoo W. H., Chae H. J., ER stress and autophagy. Curr. Mol. Med. 15, 735–745 (2015). [DOI] [PubMed] [Google Scholar]
- 54.Grumati P., Dikic I., Stolz A., ER-phagy at a glance. J. Cell Sci. 131, jcs217364 (2018). [DOI] [PubMed] [Google Scholar]
- 55.Wang H., Shi X., Qiu M., Lv S., Liu H., Hydrogen sulfide plays an important protective role through influencing endoplasmic reticulum stress in diseases. Int. J. Biol. Sci. 16, 264–271 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Krick R., Tolstrup J., Appelles A., Henke S., Thumm M., The relevance of the phosphatidylinositolphosphat-binding motif FRRGT of Atg18 and Atg21 for the Cvt pathway and autophagy. FEBS Lett. 580, 4632–4638 (2006). [DOI] [PubMed] [Google Scholar]
- 57.Tamura N., et al., Atg18 phosphoregulation controls organellar dynamics by modulating its phosphoinositide-binding activity. J. Cell Biol. 202, 685–698 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang B.et al., Phosphorylation of ATG18a by BAK1 suppresses autophagy and attenuates plant resistance against necrotrophic pathogens. Autophagy, 10.1080/15548627.2020.1810426 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kang S., Shin K. D., Kim J. H., Chung T., Autophagy-related (ATG) 11, ATG9 and the phosphatidylinositol 3-kinase control ATG2-mediated formation of autophagosomes in Arabidopsis. Plant Cell Rep. 37, 653–664 (2018). [DOI] [PubMed] [Google Scholar]
- 60.Cawthon H., Chakraborty R., Roberts J. R., Backues S. K., Control of autophagosome size and number by Atg7. Biochem. Biophys. Res. Commun. 503, 651–656 (2018). [DOI] [PubMed] [Google Scholar]
- 61.Sakoh-Nakatogawa M., Kirisako H., Nakatogawa H., Ohsumi Y., Localization of Atg3 to autophagy-related membranes and its enhancement by the Atg8-family interacting motif to promote expansion of the membranes. FEBS Lett. 589, 744–749 (2015). [DOI] [PubMed] [Google Scholar]
- 62.Kotani T., Kirisako H., Koizumi M., Ohsumi Y., Nakatogawa H., The Atg2-Atg18 complex tethers pre-autophagosomal membranes to the endoplasmic reticulum for autophagosome formation. Proc. Natl. Acad. Sci. U.S.A. 115, 10363–10368 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Obara K., Sekito T., Niimi K., Ohsumi Y., The Atg18-Atg2 complex is recruited to autophagic membranes via phosphatidylinositol 3-phosphate and exerts an essential function. J. Biol. Chem. 283, 23972–23980 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kobayashi T., Suzuki K., Ohsumi Y., Autophagosome formation can be achieved in the absence of Atg18 by expressing engineered PAS-targeted Atg2. FEBS Lett. 586, 2473–2478 (2012). [DOI] [PubMed] [Google Scholar]
- 65.Zheng J. X., et al., Architecture of the ATG2B-WDR45 complex and an aromatic Y/HF motif crucial for complex formation. Autophagy 13, 1870–1883 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chowdhury S., et al., Insights into autophagosome biogenesis from structural and biochemical analyses of the ATG2A-WIPI4 complex. Proc. Natl. Acad. Sci. U.S.A. 115, E9792–E9801 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the main text and SI Appendix.