Significance
Cyanobacteria, which are reliant on photosynthesis for growth, face a fundamental challenge. Replicating the genome takes hours, so the decision to initiate replication must be made at time when conditions may be quite different from the future state of the environment when replication is due to complete. We found that cyanobacteria use their circadian clock to ensure that DNA replication finishes efficiently, both by scheduling initiation early in the day and by directing the accumulation of metabolic resources that support ongoing replication after nightfall. When these circadian protections are removed, replication fails to complete. This study connects genome replication to the ability of a circadian clock to anticipate environmental changes.
Keywords: circadian clock, cyanobacteria, DNA replication, cell cycle, mathematical modeling
Abstract
Disruption of circadian rhythms causes decreased health and fitness, and evidence from multiple organisms links clock disruption to dysregulation of the cell cycle. However, the function of circadian regulation for the essential process of DNA replication remains elusive. Here, we demonstrate that in the cyanobacterium Synechococcus elongatus, a model organism with the simplest known circadian oscillator, the clock generates rhythms in DNA replication to minimize the number of open replication forks near dusk that would have to complete after sunset. Metabolic rhythms generated by the clock ensure that resources are available early at night to support any remaining replication forks. Combining mathematical modeling and experiments, we show that metabolic defects caused by clock–environment misalignment result in premature replisome disassembly and replicative abortion in the dark, leaving cells with incomplete chromosomes that persist through the night. Our study thus demonstrates that a major function of this ancient clock in cyanobacteria is to ensure successful completion of genome replication in a cycling environment.
Circadian clocks, internally generated rhythms in physiology with a ∼24 h period, are found in all domains of life. These clocks allow organisms to coordinate their physiological activities in anticipation of the daily cycle in the external environmental (1–3). Disruption of clocks caused either by mutation or clock–environment mismatch leads to decreased health and reproductive fitness in multiple organisms (4–6). In mammals, risk for age-related diseases such as cancer and cardiometabolic dysfunction is enhanced by circadian disruption (7, 8).
Although much is now understood about the molecular mechanisms that generate rhythms, the origin of these health defects is still incompletely understood. A common target of circadian control shared across many species is the progression of the cell cycle (9–12). In animals, disrupted circadian rhythms are often linked to aberrant cell proliferation and tumorigenesis (13). Successful duplication of the genome is essential for the production of viable progeny. Replicating a bacterial genome can take up to several hours, a timescale over which external illumination from sunlight can change substantially. We therefore speculated that initiation of DNA replication could be a key point of circadian control. The cyanobacterium Synechococcus elongatus, which has the simplest known circadian system, is a powerful model system to address these issues, both because its clock is intimately coupled to cell cycle (9, 14–16) and because clock–environment misalignment has profound effects on reproductive fitness (17). Here, we analyze whether replication is clock-regulated in S. elongatus and the consequences of clock–environment mismatch on DNA replication.
Results
The Circadian Clock Generates Rhythmic DNA Replication in Constant Condition.
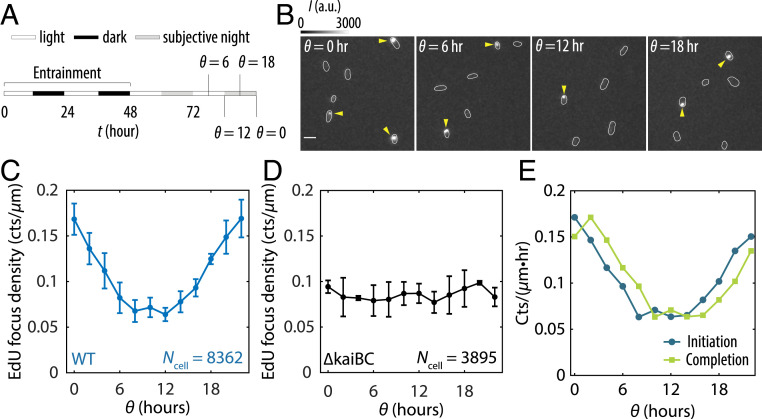
To monitor active DNA replication as a function of internal clock time, we first synchronized cells with two light/dark (LD, 12 h:12 h) pulses to entrain the clock phase and then transferred cells to constant light (LL) (at t = 48 h in Fig. 1A). Since circadian rhythms are self-sustained in constant conditions, we define time modulo 24 h relative to the transfer to LL as , a proxy for the subjective time of the cells (Fig. 1A). Starting from 24 h after transfer to LL, we pulse-labeled cells with 5-ethynyl-2′-deoxyuridine (EdU), a thymidine analog that is incorporated into the genome during DNA replication and is visualized as fluorescent foci via click chemistry (18) (Fig. 1B). We interpret the appearance of a concentrated fluorescent focus as a site of active replication. We typically observe zero or one active replication site per cell (Fig. 1B and SI Appendix, Fig. S1) despite each cell having multiple copies of its chromosome (SI Appendix, Fig. S2), consistent with previous reports that initiation is asynchronous and only a subset of chromosome copies are being actively replicated at any time (19–21).
Fig. 1.
The cyanobacterial circadian clock generates rhythmicity in DNA replication. (A) Schematic diagram showing the entrainment and EdU-labeling procedures. (B) Fluorescence images showing EdU Alexa Fluor 488 foci in wild-type (WT) cells at various : subjective dawn ( = 0 h), subjective midday ( = 6 h), subjective dusk (θ = 12 h), and subjective midnight ( = 18 h). Arrows point to EdU foci. (Scale bar = 3 µm.) (C) EdU densities as a function of in WT cells. The values correspond to the number of active replication events per micrometer of cell length during a 10 min pulse-labeling time window centered at each . (D) EdU densities as a function of in a clock-deletion strain. (E) Initiation and completion rates inferred using Wiener deconvolution and a 2 h replication time window. All error bars are SEMs from three independent experiments.
Because the number of chromosome copies in a cell are roughly proportional to its length, we analyzed the linear density of EdU foci as a function of clock time (Fig. 1C and SI Appendix, Fig. S3A). This analysis revealed that the probability of a chromosome undergoing replication is rhythmic even in constant conditions, with peak replication near subjective dawn ( = 0 h) and the trough near subjective dusk ( = 12 h). Deleting the kaiBC genes eliminated rhythmic replication (Fig. 1D and SI Appendix, Fig. S3B), confirming that this effect is caused by the circadian clock.
Monitoring the appearance of newly replicated DNA at specific locations on the chromosome, we estimate that it takes ∼2 h to replicate a chromosome under our growth condition (SI Appendix, section IV and Figs. S4 and S5) (19). Assuming peaks of replication initiation and completion are separated by 2 h, the initiation and completion rates can be inferred from the rhythms in EdU foci (Materials and Methods and SI Appendix, section V), which shows that initiation events are suppressed, and most ongoing replications are completed in anticipation of nightfall (Fig. 1E).
The Circadian Clock Schedules Rhythmic Assembly of Replisomes.
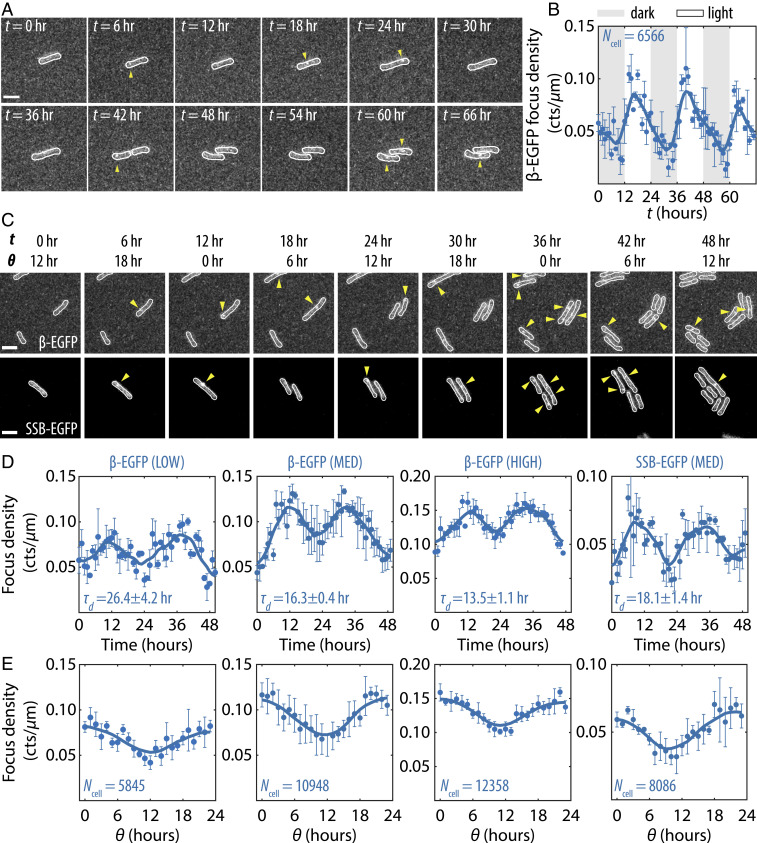
To monitor the dynamics of DNA replication in live cells, we made fluorescent reporters for several replisome components by fusing enhanced green fluorescent protein (EGFP) at their native loci. Among these, the DNA clamp (β-clamp) and the single-stranded DNA-binding protein (SSB), which have the highest stoichiometries at the replisome (22), were bright enough to be detected. When recruited to the replisome, β-EGFP and SSB-EGFP produced detectable fluorescent foci that we tracked in live cells using time-lapse microscopy. We observed that under LD cycles the appearance of β-EGFP foci is highly rhythmic, with a burst of replication activity occurring shortly after lights-on, followed by a gradual decay (Fig. 2 A and B). However, the rhythmicity in replisome assembly is not contingent on an external cue, as we saw clock-dependent, rhythmic appearance and disappearance of fluorescent foci under constant conditions as well (Fig. 2 C and D and SI Appendix, Fig. S6), with peaks and troughs of the EGFP time series closely matching those of EdU incorporation (Fig. 2E). The appearance and disappearance of discrete EGFP foci coincident with nucleotide polymerization implies that the clock regulates replication initiation by controlling the assembly of replisome, as opposed to modulating the progression rate of existing forks.
Fig. 2.
Circadian clock schedules rhythmic assembly of the replisome. (A) Film strips from time-lapse imaging experiments showing β-EGFP foci in wild-type (WT) background as a function of time in LD cycle condition. (All scale bars = 3 µm.) (B) Average β-EGFP focus density (counts/µm) in WT background in LD as a function of time. Solid lines are seven-point running average, and error bars are SEMs from three independent experiments. (C) Film strips showing β-EGFP and SSB-EGFP foci in WT background as a function of time and in LL condition. (D) Average β-EGFP and SSB-EGFP focus densities as a function of time in LL, under different illumination levels (LOW, MED, and HIGH). is the doubling time of each condition (mean ± SD). Error bars are SEMs from two to three independent experiments. (E) Average β-EGFP and SSB-EGFP focus densities as a function of .
S. elongatus can grow with a wide range of doubling times, and hence time needed to replicate the genome, depending on the intensity of illumination (15, 23). To check whether the coordination of replication is primarily clock-controlled, we varied light intensity to achieve different cell cycle times. As the doubling time is shortened, the baseline level of β-EGFP foci increases, but the amplitude of its oscillation and its period remain relatively unchanged (Fig. 2 D and E). Furthermore, because the transcripts for both β-clamp and SSB have not been found to be highly rhythmic (24), it suggests that rhythmic assembly of the replisome is due to regulation of the initiation process itself, not to rhythms in the abundance of replisome components.
DNA Replication Is Suppressed in the Dark in a Clock-Dependent Manner.
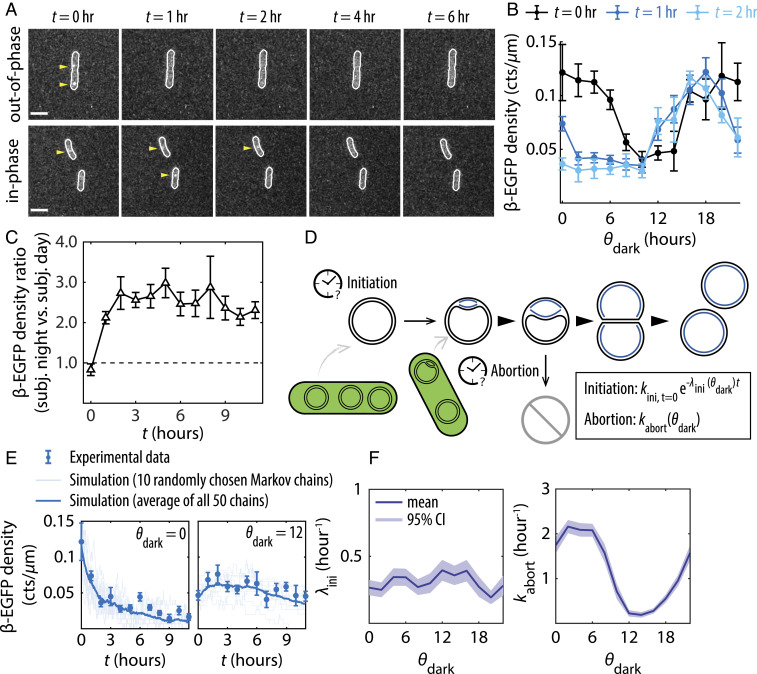
For autotrophic cyanobacteria, night is a prolonged period of energy restriction. Previous work in S. elongatus has shown that the clock also generates rhythms in glycogen storage and the expression of metabolic enzymes in anticipation of dusk, which serve to prepare the cell for the night (4, 25). Completing chromosome replication represents a significant biosynthetic and energetic burden on the cell at night, where millions of nucleotides must be polymerized without further carbon fixation. We thus speculated that the clock’s role in metabolically and transcriptionally preparing cells for nightfall might be needed to allow replication forks to proceed in the dark. We sought to determine the effect of violating the expected temporal program by initiating darkness at different points throughout the clock cycle.
We denote the clock time at the onset of darkness as and refer to conditions with and as “out-of-phase” and “in-phase,” respectively. Because unexpected darkness causes phase shifts in the clock (26, 27), we focus on the impact of a single dark pulse on replication and do not attempt to infer the clock state after the perturbation. We transferred cells from LL to the dark at 12 time points during the clock cycle ( = 0, 2, …, 22 h) (SI Appendix, Fig. S7) and monitored β-EGFP foci every hour in the dark (Fig. 3A and SI Appendix, Figs. S8 and S9). We also monitored EdU incorporation for cells transferred to the dark at subjective dawn ( = 0) and subjective dusk ( = 12) (SI Appendix, Fig. S10). In all cases, we observed a decrease in β-EGFP and EdU foci as the night progressed, suggesting that either replication initiation was inhibited in the dark, ongoing replications were terminated, or a combination of both. However, the rate of decay depends on the internal clock time at the onset of dark (): Cells transferred into dark near subjective dawn ( ∼0 h) experienced the most rapid drop in replication activity in the first 2 hr of darkness (Fig. 3B). In general, cells entering darkness in the subjective night (12 ≤ ≤ 22 h) showed sustained replication activity in the dark compared to cells entering darkness in the subjective day (0 ≤ ≤ 10 h) (Fig. 3 B and C and SI Appendix, Fig. S8B), and this difference is removed by deletion of the kai genes (SI Appendix, Fig. S9). In other words, although cells in the middle of subjective day and the middle of subjective night had similar levels of on-going replication when the lights were turned off, ( = 0 h in Fig. 3C), the cells in a clock state that anticipated nightfall had much higher levels of sustained replication in the dark. This suggests that proper clock state when entering the dark is needed to support replication in the night.
Fig. 3.
The ability to sustain DNA replication in the dark depends on the clock state at the onset of dark. (A) Fluorescence images showing β-EGFP foci at various time points (t) in the dark, for cells subject to a dark pulse either out-of-phase () or in-phase ( = 12) with respect to their entrainments. (Scale bars = 3 µm.) (B) β-EGFP densities as a function of during the first 2 h of dark (t = 0, 1, 2 h). (C) The ratio of β-EGFP densities between cells in the subjective nighttime state (12 h ≤ < 24 h) and those in the subjective daytime state (0 h ≤ < 12 h) when transferred to the dark, plotted as a function of time in the dark. (D) Schematic diagram of the model used to simulate replication events in the dark. (E) Simulated replication events plotted against experimentally collected β-EGFP data in dark. (F) Best-fit initiation decay rate and best-fit abortion rate plotted as a function of . All β-EGFP error bars are SEMs from five independent experiments totaling ∼27,000 cells.
Kinetic Analysis Suggests Higher Probabilities of Replication Failure in the Dark for Cells in the Subjective Morning States at the Onset of Dark.
We consider two possible mechanisms for the clock-dependence of decreased replication in the dark: clock-dependent suppression of new initiation events and clock-dependent failure of existing replication forks. These two scenarios make different quantitative predictions about the time course of detected replisomes in the dark. Intuitively, the steady state level of replisomes in the dark reflects the balance of initiation versus completion and replication failure. In a given time series, a rapid approach to steady state reflects the timescale of either completion or replication failure. Thus, kinetic analysis can make predictions about the relative contribution of these effects from the data. To make this intuition precise, we built a computational model of initiation suppression and replicative abortion (Fig. 3D) and examined whether the rate of either pathway exhibited circadian dependence under the constraint of experimental data (Materials and Methods and SI Appendix, section VI).
In the model, we assume that the rate of new initiation decays as a function of time in the dark with a first-order decay constant , and replication can abort stochastically with a rate constant . We allow both and to potentially depend on the clock state at lights-out (), which enables the model to discover potential clock-dependent regulation. Values of and for different clock states were then estimated by fitting the model to the data using Markov chain Monte Carlo.
Fitting produced reasonable agreement between the model prediction and experimental data (Fig. 3E and SI Appendix, Fig. S11). The best-fit parameter values predict that the probability of failed replication in the dark, , has a strong clock-dependence and can be appreciable. In contrast, the rate of initiation decay in the dark, , need not be rhythmic, suggesting that the influence of the clock is to minimize the rate of replication abortion when the clock state anticipates nightfall. The circadian dependence of cannot be removed from the model without producing large discrepancies between the model prediction and experimental data (SI Appendix, Fig. S12). The best-fit value peaks around subjective morning (0 ≤ ≤ 6 h) (Fig. 3F), indicating that ongoing replication events have the highest chance of aborting if lights are turned off during this time window. Thus, the model predicts that successfully completing DNA replication in the night requires the ability to predict the timing of nightfall.
Unexpected Dark Pulse Causes Replisome Disassembly and Aborted Replication in the Dark.
Inspired by these model predictions, we sought direct experimental evidence for replication failure in the dark. We investigated two opposite conditions, where the cells were given 12 h of darkness either at subjective dusk (= 12 h, “in-phase”) or at subjective dawn (= 0 h, “out-of-phase”).
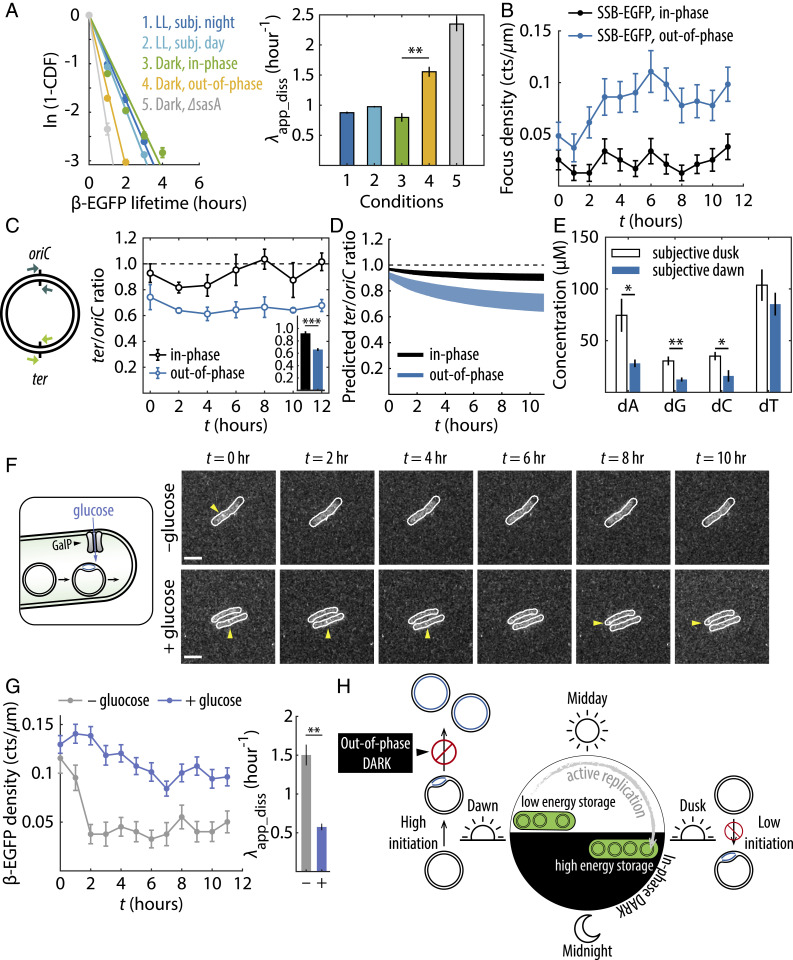
First, we probed how the binding/unbinding kinetics of β-clamp was affected in each case, as replicative abortion should lead to increased dissociation of β-clamp from the replication fork. We quantified and compared the apparent dissociation rate constant () of β-EGFP under different growth conditions by fitting log-survival functions of β-EGFP that measure the time it took for each individual β-EGFP signal to disappear (Fig. 4A). We found that when the dark pulse was in-phase, the value of β-EGFP in the dark was similar to LL conditions (compare the green bin to blue bins in Fig. 4A). On the other hand, when cells were subject to an unexpected, out-of-phase dark pulse, the apparent dissociation rate constant increased significantly (yellow bin in Fig. 4A). Experiments with cells from all 12 groups showed that the apparent dissociation rate constant of β-EGFP is rhythmic, where β-EGFP appeared to dissociate much faster when the clock state is not near subjective dusk (SI Appendix, Fig. S13). This effect was even more pronounced when the clock output histidine kinase SasA was deleted (∆sasA, gray bin in Fig. 4A), a mutation which strongly represses expression of dusk-peaking genes (28, 29). These results together indicate that the β-clamp more rapidly dissociates from the replication fork in case of clock–environment mismatch.
Fig. 4.
Incorrect clock state causes ongoing replication events to abort in the dark. (A, Left) log-survival functions of β-EGFP foci lifetime under various conditions. Solid lines are linear fits through the origin, the negative slopes of which correspond to the apparent dissociation rate constants (Right). Error bars are SEMs bootstrapped from over 2,000 foci trajectories from two to three independent experiments. **P < 0.01 (two-sample t test). (B) SSB-EGFP density time trace in the dark. Error bars are SEMs from two independent experiments. (C) qPCR quantification of ter/oriC ratios in the dark. Error bars are SEMs from four independent experiments. Inset shows ter/oriC ratios averaged throughout the night. ***P < 0.001. (D) Model-predicted ter/oriC ratios assuming 3 to 6 chromosomes per cell (21). (E) dNTP concentrations at subjective dawn and subjective dusk. Error bars are SEMs from four independent experiments. *P < 0.05, **P < 0.01. (F) Fluorescence images showing β-EGFP foci in GalP-expressing cells subject to an out-of-phase dark pulse, without (−) or with (+) glucose supplement. (G) β-EGFP density and apparent dissociation rate constants for GalP-expressing cells. Error bars are SEMs from two (−glucose) or three (+glucose) experiments. (H) Schematic diagram summarizing main findings of this study.
In the case of replication failure, we would expect dissociation of the β-clamp to coincide with persistently exposed single-stranded DNA that will become coated with SSB (30). To test this prediction, we imaged SSB-EGFP foci over the course of the dark pulse. While β-EGFP foci counts decrease rapidly in the dark for the out-of-phase group (Fig. 3B and SI Appendix, Fig. S8), we observed the opposite trend with SSB-EGFP: SSB foci appear as the night progresses only in the out-of-phase condition (Fig. 4B). This increase in SSB foci despite reduced replication activity is consistent with SSB coating and stabilizing exposed gaps on DNA strands due to failure of the replisome to progress (31).
If an incorrect clock state indeed leads to increased replication failure in the dark, then cells from the out-of-phase group will contain incomplete, partially replicated chromosomes. To directly detect incomplete chromosomes, we performed quantitative PCR (qPCR) analysis to quantify the abundance of genomic regions proximal to the replication terminus site (ter) and the origin of replication (oriC) (Materials and Methods). A ter/oriC ratio less than 1 indicates the presence of incomplete chromosomes. qPCR results demonstrated that the in-phase group had higher ter/oriC ratios than the out-of-phase group at all time points in the dark (Fig. 4C), consistent with model predictions (Fig. 4D). The average measured ter/oriC ratio throughout the night for the in-phase group (0.922 ± 0.085, mean ± SEM) suggests nearly all initiated replication events were successfully completed. On the other hand, the average ter/oriC ratio for the out-of-phase group (0.660 ± 0.053, mean ± SEM) was significantly (two-tailed P value = 7 × 10−4) smaller than 1, suggesting a substantial number of unfinished chromosomes. Together, the microscopy and qPCR data confirm the model prediction that replisomes disassemble prior to completion when nightfall occurs at an incorrect clock time, resulting in partially replicated chromosomes.
Metabolic Limitations Underlie Clock-Related Replication Failure.
What prevents replication from finishing in cells with out-of-phase clocks? Previous work in multiple cyanobacterial species have shown that the level of replication activity in the dark is correlated with respiration and glycogen metabolism (32). It has been shown that insufficient glycogen storage at subjective dawn causes a sharp drop in cellular adenosine triphosphate/adenosine diphosphate (ATP/ADP) ratio in the dark (25), which could impede various ATP–hydrolysis-driven processes during replication, including those performed by the helicase, gyrase, and ligase. We speculate that shortages of other metabolites at subjective dawn may also compromise the ability to sustain replication in the dark. For example, transcription of genes in the oxidative pentose phosphate pathway (OPPP), which provide precursors for nucleotide biosynthesis in the dark, peaks at dusk (33). Because the standing pool of deoxynucleotides (dNTPs) in bacteria are generally much lower than the amount required for a single round of chromosome replication (34, 35), without de novo nucleotide synthesis, cells at subjective dawn likely do not have enough dNTPs to finish a newly initiated round of replication. To evaluate this possibility, we measured the dNTP pool for cell cultures at subjective dawn and subjective dusk (Materials and Methods and SI Appendix, section VIII) (36). We found that cells at subjective dawn indeed maintained a lower level of dNTP pool than those at subjective dusk (Fig. 4E), while having about twice as many ongoing replication events (Figs. 1 and 2). Thus, in addition to reduced glycogen stores, the lower dNTP level at subjective dawn may indicate a metabolic constraint on the ability to sustain replication.
Given these metabolic limitations cells experience in the dark, we asked whether supplying cells with an external energy and carbon source could support more replication events. We used a strain expressing the Escherichia coli sugar transporter GalP (37, 38) and supplemented the media with glucose immediately before an out-of-phase dark pulse, effectively enabling nonphotosynthetic growth at night (Fig. 4F). Comparing untreated and glucose-treated cells, the latter exhibited substantially more β-EGFP and EdU foci throughout the night as well as a significantly decreased apparent dissociation rate for β-EGFP (Fig. 4G and SI Appendix, Fig. S14), suggesting that relieving metabolic limitation can prevent clock-dependent replication failure in the dark.
Discussion
Taking these data together, we conclude that the circadian clock acts in two ways to minimize the possibility of incomplete replication in the night (Fig. 4H). First, the clock preferentially schedules replication initiation for the early part of the day allowing most replication events to complete before sunset. Second, the clock evidently conditions the metabolic state of the cell at subjective dusk to allow it to sustain DNA polymerization in the dark, allowing for replication to complete. However, as a result of this temporal program, cells entering the dark at the wrong internal time are unable to complete DNA replication, resulting in premature disassembly of the replisome.
A longstanding conjecture, associated with Pittendrigh’s “flight from light” hypothesis (39), is that ultraviolet (UV) radiation poses a threat to genome integrity that is mitigated by rhythmic control of physiology. Our results indicate that, in S. elongatus, the circadian clock directs genome replication to occur more often in the early part of the day, avoiding initiation near dusk. However, this logic is not necessarily in conflict with the hypothesis that avoiding UV-induced damage is a primary concern. A recent study has shown that S. elongatus also exhibits a circadian rhythm in UV resistance, where increased glycogen breakdown at subjective dusk causes increased susceptibility to UV damage (40). Thus, one plausible scenario is that the need to remain viable during the night requires substantial flux through glycolysis and the pentose phosphate pathway through glycogen metabolism. However, this dusk-associated metabolic state renders cells vulnerable to UV-induced damage, so that there is a trade-off between UV protection and dark tolerance. This vulnerability can be mitigated by the circadian clock by scheduling replication to occur earlier in the day.
Tracking the fates of cells during and after dark, we found that both in-phase and out-of-phase cells were able to resume replication and division when lights were turned back on (SI Appendix, Fig. S15 A and B). However, measured from lights-off to 1 h after lights-on, out-of-phase cells that were actively replicating tend to have a smaller cell size gain compared to in-phase ones (SI Appendix, Fig. S15C). Many out-of-phase and actively replicating cells experienced an overall reduction in cell size during this time period, suggesting that the combination of active replication at lights-off and an incorrect clock state can cause a transient disruption to growth. The specific mechanism remains to be investigated, but one possibility is that these cells need to redirect resources meant for biomass production and maintenance to rescue failed replications in the dark.
This study raises several questions for further investigation. For example, although the unexpected dark pulse evidently causes replisome disassembly and abortion of replication (Figs. 3 and 4), it does not appear to have any large, detrimental effect on the subsequent growth of the vast majority of cells (SI Appendix, Fig. S15). A previous study has shown that polyploidy helps S. elongatus to cope with UV damage (41), and it is possible that having multiple chromosomes is also beneficial in the case of darkness-induced replication failure, especially considering that typically only one out of several chromosomes in a cell is actively replicating at a given time (Fig. 1B and SI Appendix, Fig. S1). The relationship between replication and the long-term fitness cost of clock-mutant cells remains to be explored (4, 17). Repeated interruption of replicating chromosomes could be a contributing factor leading to reduced fitness of clock-mutant cells under LD conditions (17). Finally, the arrest of replication forks and persistence of unfinished chromosomes might potentially increase the probability of mutation in clock–environment mismatch conditions (42, 43).
Circadian rhythms are known to exert a pervasive influence on many aspects of physiology. However, processes that are time-consuming and involve an irreversible commitment may be especially important to temporally regulate because the conditions when such a process is initiated may change dramatically before the process completes. Fundamental biophysical constraints on the mechanism of genome replication make this an irreversible, time-consuming, essential process that may have been an important driver in the evolution of circadian rhythms.
Materials and Methods
Strain Construction.
Molecular cloning was performed following standard protocols for S. elongatus PCC 7942 (44). The thymidine kinase plasmid used for EdU pulse-labeling experiments was a gift from the Yoshikawa laboratory (19). β-EGFP and SSB-EGFP plasmids were made by Gibson assembly (45) of the following DNA fragments: an EcoRI-digested pBlueScript II SK (+) plasmid backbone, a 1 kb sequence upstream of β-clamp or SSB, along with the β-clamp or SSB itself, the egfp sequence, a spectinomycin resistance (SpR) cassette, and a 1 kb downstream sequence. The GalP-expression plasmid was made by assembling the sequence of a constitutive pGlnB promoter and the sequence of galp between the SalI and Xbal restriction sites of plasmid pAM1579 (46). Sequences of insertions were verified by Sanger sequencing and were incorporated into the cyanobacterial genome following transformation. Strains, plasmids, and cloning primers used in this study are listed in SI Appendix, Tables S1–S3.
Cell Culture and Microscopy.
S. elongatus cultures were grown in BG11M liquid medium at 30 °C with shaking, under an ambient light intensity of ∼30 µE m−2⋅s−1 provided by cool white fluorescent light bulbs (Philips Alto II). To entrain cells to different circadian phases, cultures of optical density at 750 nm (OD750) ∼0.1 to 0.4 were diluted back to OD750 = 0.1 and transferred to a 96-well plate (∼250 µL/well). On top of the 96-well plate is a custom-made red light-emitting diode (LED) array where individual LEDs were programmed to generate user-defined temporal patterns of illumination for each well (47).
EdU pulse-labeling experiments were performed with an engineered strain that expresses herpes simplex virus type-1 thymidine kinase (TK) from an isopropyl β-D-1-thiogalactopyranoside (IPTG)-inducible promoter (19). No IPTG was added since basal expression of TK from the leaky promoter was sufficient for signal detection. Fixation and permeabilization procedures are described in SI Appendix, section II.
Time-lapse imaging experiments in constant light condition (LL) with the β-EGFP and the SSB-EGFP strains were performed at three different illumination levels provided by a LED (660 nm): LOW ∼16 µE m−2⋅s−1, MED ∼27 µE m−2 ⋅ s−1, and HIGH ∼40 µE m−2 ⋅ s−1. All other time-lapse imaging experiments were performed either at 27 µE m−2 ⋅ s−1 or in the dark.
Cell segmentation, signal detection, and data analysis were performed with custom-written MATLAB and R scripts.
Inferring Replication Initiation and Completion Profiles by Deconvolution.
The experimentally measured EdU versus curve, denoted as here, can be modeled as a convolution product between the replication initiation rate , and a boxcar function with a duration equal to the time it takes to replicate one chromosome (estimated to be 2 h here); that is, . Thus, the replication initiation rate profile can be solved using a Wiener filter (48) as , where denotes the inverse Fourier transform, and , , and , respectively, correspond to the Fourier transform of , the Fourier transform of , and the noise-to-signal power ratio of , all of which can be computed. The replication completion profile is then a 2 h delayed version of the replication initiation profile .
Stochastic Model of DNA Replication in the Dark.
Conceptual and computational details of the model are described in SI Appendix, section VI. Briefly, cells of different clock states () were instantiated at the onset of dark (). The probability of each cell having a preexisting ongoing replication event was drawn from a probability distribution that depended on the clock state at lights-out. In addition, each cell may (or may not) initiate new rounds of replication in the dark, depending on the initiation rate at as well as the initiation decay rate , the latter of which is a parameter set to be determined by the model. The timing of successive initiation events were simulated using the thinning method (49).
Each preexisting and newly initiated event can be terminated by one of two competing processes: the completion of the chromosome replication (determined by the fork speed) or a replicative abortion event (depending on the abortion rate ), whichever occurs first. Parameter values were searched using Markov chain Monte Carlo, where each chain simultaneously simulates 12 replication time traces corresponding to the 12 β-EGFP time traces measured from experiments. At each iteration, the Poisson-weighted residual sum of squares between the simulated and the experimental time traces were calculated, which determines the probability of the current parameter set being accepted. A total of 50 Markov chains were simulated for 20,000 iterations, and the optimal parameter set, averaged among all 50 chains, are tabulated in SI Appendix, Table S5.
Determination of ter/oriC Ratio by qPCR.
Genomic DNA (gDNA) samples from in-phase and out-of-phase cells were collected at 2 h intervals during the dark pulse. gDNA samples were sheared into ∼500 base pair (bp) fragments using the Covaris S220 sonicator and purified on Qiagen MinElute columns. qPCR reactions were performed on a Bio-Rad CFX384 Real-Time PCR Detection System with Roche LightCycler 480 Sybr Green I Master mix, using the following program: 95 °C for 10 min, and 45 cycles of 95 °C for 10 s, 50 °C for 20 s, 72 °C for 20 s, followed with melting curve analysis. The absolute copy numbers were obtained from calibration curves generated using standard DNA fragments amplified with primers that target an ∼500 bp region encompassing either oriC or ter of the chromosome. qPCR primers are provided in SI Appendix, Table S4.
dNTP Measurements.
A total of 35 mL of cell cultures at subjective dawn and subjective dusk were adjusted to the same of OD750 = 0.7 and harvested. Metabolites were extracted using methanol and chloroform and concentrated overnight using SpeedVac and reconstituted in water (SI Appendix, section VIII). dNTP quantification was performed using primer extension assays following procedures detailed in Purhonen et al. (36).
Supplementary Material
Acknowledgments
We thank Dr. Hirofumi Yoshikawa for the generous gift of the thymidine kinase plasmid. This work was supported by an HHMI Simons Faculty Scholar award and NIH Grant R01 GM107369 to M.J.R.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2022516118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or supporting information.
References
- 1.Dunlap J. C., Molecular bases for circadian clocks. Cell 96, 271–290 (1999). [DOI] [PubMed] [Google Scholar]
- 2.Cohen S. E., Golden S. S., Circadian rhythms in cyanobacteria. Microbiol. Mol. Biol. Rev. 79, 373–385 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Takahashi J. S., Transcriptional architecture of the mammalian circadian clock. Nat. Rev. Genet. 18, 164–179 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Puszynska A. M., O’Shea E. K., Switching of metabolic programs in response to light availability is an essential function of the cyanobacterial circadian output pathway. eLife 6, e23210 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Green R. M., Tingay S., Wang Z. Y., Tobin E. M., Circadian rhythms confer a higher level of fitness to Arabidopsis plants. Plant Physiol. 129, 576–584 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Horn M., et al., The circadian clock improves fitness in the fruit fly, Drosophila melanogaster. Front. Physiol. 10, 1374 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Masri S., Sassone-Corsi P., The emerging link between cancer, metabolism, and circadian rhythms. Nat. Med. 24, 1795–1803 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Scheer F. A., Hilton M. F., Mantzoros C. S., Shea S. A., Adverse metabolic and cardiovascular consequences of circadian misalignment. Proc. Natl. Acad. Sci. U.S.A. 106, 4453–4458 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mori T., Binder B., Johnson C. H., Circadian gating of cell division in cyanobacteria growing with average doubling times of less than 24 hours. Proc. Natl. Acad. Sci. U.S.A. 93, 10183–10188 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Salter M. G., Franklin K. A., Whitelam G. C., Gating of the rapid shade-avoidance response by the circadian clock in plants. Nature 426, 680–683 (2003). [DOI] [PubMed] [Google Scholar]
- 11.Dekens M. P., et al., Light regulates the cell cycle in zebrafish. Curr. Biol. 13, 2051–2057 (2003). [DOI] [PubMed] [Google Scholar]
- 12.Matsuo T., et al., Control mechanism of the circadian clock for timing of cell division in vivo. Science 302, 255–259 (2003). [DOI] [PubMed] [Google Scholar]
- 13.Gaucher J., Montellier E., Sassone-Corsi P., Molecular cogs: Interplay between circadian clock and cell cycle. Trends Cell Biol. 28, 368–379 (2018). [DOI] [PubMed] [Google Scholar]
- 14.Dong G., et al., Elevated ATPase activity of KaiC applies a circadian checkpoint on cell division in Synechococcus elongatus. Cell 140, 529–539 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang Q., Pando B. F., Dong G., Golden S. S., van Oudenaarden A., Circadian gating of the cell cycle revealed in single cyanobacterial cells. Science 327, 1522–1526 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martins B. M. C., Tooke A. K., Thomas P., Locke J. C. W., Cell size control driven by the circadian clock and environment in cyanobacteria. Proc. Natl. Acad. Sci. U.S.A. 115, E11415–E11424 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Woelfle M. A., Ouyang Y., Phanvijhitsiri K., Johnson C. H., The adaptive value of circadian clocks: An experimental assessment in cyanobacteria. Curr. Biol. 14, 1481–1486 (2004). [DOI] [PubMed] [Google Scholar]
- 18.Salic A., Mitchison T. J., A chemical method for fast and sensitive detection of DNA synthesis in vivo. Proc. Natl. Acad. Sci. U.S.A. 105, 2415–2420 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Watanabe S., et al., Light-dependent and asynchronous replication of cyanobacterial multi-copy chromosomes. Mol. Microbiol. 83, 856–865 (2012). [DOI] [PubMed] [Google Scholar]
- 20.Jain I. H., Vijayan V., O’Shea E. K., Spatial ordering of chromosomes enhances the fidelity of chromosome partitioning in cyanobacteria. Proc. Natl. Acad. Sci. U.S.A. 109, 13638–13643 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen A. H., Afonso B., Silver P. A., Savage D. F., Spatial and temporal organization of chromosome duplication and segregation in the cyanobacterium Synechococcus elongatus PCC 7942. PLoS One 7, e47837 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reyes-Lamothe R., Sherratt D. J., Leake M. C., Stoichiometry and architecture of active DNA replication machinery in Escherichia coli. Science 328, 498–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kondo T., et al., Circadian rhythms in rapidly dividing cyanobacteria. Science 275, 224–227 (1997). [DOI] [PubMed] [Google Scholar]
- 24.Ito H., et al., Cyanobacterial daily life with Kai-based circadian and diurnal genome-wide transcriptional control in Synechococcus elongatus. Proc. Natl. Acad. Sci. U.S.A. 106, 14168–14173 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pattanayak G. K., Phong C., Rust M. J., Rhythms in energy storage control the ability of the cyanobacterial circadian clock to reset. Curr. Biol. 24, 1934–1938 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kiyohara Y. B., Katayama M., Kondo T., A novel mutation in kaiC affects resetting of the cyanobacterial circadian clock. J. Bacteriol. 187, 2559–2564 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johnson C. H., Elliott J. A., Foster R., Entrainment of circadian programs. Chronobiol. Int. 20, 741–774 (2003). [DOI] [PubMed] [Google Scholar]
- 28.Iwasaki H., et al., A kaiC-interacting sensory histidine kinase, SasA, necessary to sustain robust circadian oscillation in cyanobacteria. Cell 101, 223–233 (2000). [DOI] [PubMed] [Google Scholar]
- 29.Gutu A., O’Shea E. K., Two antagonistic clock-regulated histidine kinases time the activation of circadian gene expression. Mol. Cell 50, 288–294 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shereda R. D., Kozlov A. G., Lohman T. M., Cox M. M., Keck J. L., SSB as an organizer/mobilizer of genome maintenance complexes. Crit. Rev. Biochem. Mol. Biol. 43, 289–318 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Buss J. A., Kimura Y., Bianco P. R., RecG interacts directly with SSB: Implications for stalled replication fork regression. Nucleic Acids Res. 36, 7029–7042 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ohbayashi R., et al., Variety of DNA replication activity among cyanobacteria correlates with distinct respiration activity in the dark. Plant Cell Physiol. 58, 279–286 (2017). [DOI] [PubMed] [Google Scholar]
- 33.Vijayan V., Zuzow R., O’Shea E. K., Oscillations in supercoiling drive circadian gene expression in cyanobacteria. Proc. Natl. Acad. Sci. U.S.A. 106, 22564–22568 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bochner B. R., Ames B. N., Complete analysis of cellular nucleotides by two-dimensional thin layer chromatography. J. Biol. Chem. 257, 9759–9769 (1982). [PubMed] [Google Scholar]
- 35.Buckstein M. H., He J., Rubin H., Characterization of nucleotide pools as a function of physiological state in Escherichia coli. J. Bacteriol. 190, 718–726 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Purhonen J., Banerjee R., McDonald A. E., Fellman V., Kallijärvi J., A sensitive assay for dNTPs based on long synthetic oligonucleotides, EvaGreen dye and inhibitor-resistant high-fidelity DNA polymerase. Nucleic Acids Res. 48, e87 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McEwen J. T., Machado I. M., Connor M. R., Atsumi S., Engineering Synechococcus elongatus PCC 7942 for continuous growth under diurnal conditions. Appl. Environ. Microbiol. 79, 1668–1675 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pattanayak G. K., Lambert G., Bernat K., Rust M. J., Controlling the cyanobacterial clock by synthetically rewiring metabolism. Cell Rep. 13, 2362–2367 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pittendrigh C. S., Temporal organization: Reflections of a darwinian clock-watcher. Annu. Rev. Physiol. 55, 16–54 (1993). [DOI] [PubMed] [Google Scholar]
- 40.Kawasaki K., Iwasaki H., Involvement of glycogen metabolism in circadian control of UV resistance in cyanobacteria. PLoS Genet. 16, e1009230 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ohbayashi R., et al., Coordination of polyploid chromosome replication with cell size and growth in a cyanobacterium. mBio 10, e00510-19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Larsen N. B., et al., Stalled replication forks generate a distinct mutational signature in yeast. Proc. Natl. Acad. Sci. U.S.A. 114, 9665–9670 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sedletska Y., Radicella J. P., Sage E., Replication fork collapse is a major cause of the high mutation frequency at three-base lesion clusters. Nucleic Acids Res. 41, 9339–9348 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Golden S. S., Brusslan J., Haselkorn R., Expression of a family of psbA genes encoding a photosystem II polypeptide in the cyanobacterium Anacystis nidulans R2. EMBO J. 5, 2789–2798 (1986). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gibson D. G., et al., Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 6, 343–345 (2009). [DOI] [PubMed] [Google Scholar]
- 46.Andersson C. R., et al., Application of bioluminescence to the study of circadian rhythms in cyanobacteria. Methods Enzymol. 305, 527–542 (2000). [DOI] [PubMed] [Google Scholar]
- 47.Leypunskiy E., et al., The cyanobacterial circadian clock follows midday in vivo and in vitro. eLife 6, e23539 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wiener N., Extrapolation, Interpolation, and Smoothing of Stationary Time Series, with Engineering Applications (Technology Press of the Massachusetts Institute of Technology, Cambridge, 1949). [Google Scholar]
- 49.Lewis P. A. W., Shedler G. S., Simulation of nonhomogeneous Poisson processes by thinning. Nav. Res. Logist. 26, 403–413 (1979). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or supporting information.