Significance
Understanding what determines the distribution of biodiversity remains one of the great challenges in evolutionary biology and ecology. The Northern Hemisphere conifers do not follow a latitudinal diversity gradient and show a unique distribution pattern, that is most species are distributed at middle latitudes. To reveal the mechanisms underlying this pattern, we investigate the spatiotemporal evolution of the largest conifer genus Pinus. We find that the midlatitude region has been an evolutionary museum for Pinus, and particularly, topography has played the most important role in pine diversification. Moreover, our results indicate that pine’s preference of warm and somewhat drier habitats could be helpful for their adaptation to the Anthropocene climate warming, which is important for biodiversity conservation and forest management.
Keywords: Pinus, phylogenomics, spatiotemporal evolution, evolutionary museum, fire adaptation
Abstract
How coniferous forests evolved in the Northern Hemisphere remains largely unknown. Unlike most groups of organisms that generally follow a latitudinal diversity gradient, most conifer species in the Northern Hemisphere are distributed in mountainous areas at middle latitudes. It is of great interest to know whether the midlatitude region has been an evolutionary cradle or museum for conifers and how evolutionary and ecological factors have driven their spatiotemporal evolution. Here, we investigated the macroevolution of Pinus, the largest conifer genus and characteristic of northern temperate coniferous forests, based on nearly complete species sampling. Using 1,662 genes from transcriptome sequences, we reconstructed a robust species phylogeny and reestimated divergence times of global pines. We found that ∼90% of extant pine species originated in the Miocene in sharp contrast to the ancient origin of Pinus, indicating a Neogene rediversification. Surprisingly, species at middle latitudes are much older than those at other latitudes. This finding, coupled with net diversification rate analysis, indicates that the midlatitude region has provided an evolutionary museum for global pines. Analyses of 31 environmental variables, together with a comparison of evolutionary rates of niche and phenotypic traits with a net diversification rate, found that topography played a primary role in pine diversification, and the aridity index was decisive for the niche rate shift. Moreover, fire has forced diversification and adaptive evolution of Pinus. Our study highlights the importance of integrating phylogenomic and ecological approaches to address evolution of biological groups at the global scale.
Understanding what determines species diversity and its distribution pattern is one of the great challenges in evolutionary biology and ecology (1, 2). Gymnosperms and angiosperms are the two major groups of extant seed plants with extreme differences in species diversity and distribution. Compared to angiosperms with ∼300,000 species that dominate most of Earth’s terrestrial ecosystems, extant gymnosperms comprise only 13 families and a little more than 1,000 species (3). Although greatly outnumbered by angiosperms, conifers, the most diverse group of gymnosperms with around 615 extant species worldwide (4), are prominent components in north temperate biomes and constitute over 39% of the world’s forests (5). Unlike woody angiosperms that generally follow a latitudinal diversity gradient (LDG) in the Northern Hemisphere (6, 7), most conifer species are distributed in the mountainous areas at middle latitudes. It remains unknown whether the midlatitude region has been an evolutionary cradle or museum for conifers.
The mechanisms underlying the high species diversity of conifers in the midlatitude region are also poorly studied, while the LDG can be explained by latitudinal variation in ecological limits, diversification rates, and time for species accumulation (2). Sundaram et al. (8) suggested that the formation of conifer hotspots can be attributed to the accumulation of regional diversity over evolutionary time scales in mountainous areas with long-term climate stability, and topographically heterogeneous environments may play a primary role in hotspot formation. Nevertheless, Rueda, Godoy, and Hawkins (7) used a trait-based approach to test the effects of environmental stressors, evolutionary relatedness, and phylogenetic conservatism on conifer distribution in the conterminous United States and found that a strong trade-off between drought and shade tolerance might be the simplest explanation for the distribution. It is of great interest to investigate how evolutionary and ecological factors have worked together to drive the spatiotemporal evolution of conifers in the Northern Hemisphere based on a solid phylogenetic framework and, in particular, whether conifers have retained or shifted their environmental niche space over time.
Pinus provides an ideal example to explore the mechanisms underlying the spatiotemporal evolution of conifers and test whether the midlatitude region is an evolutionary cradle or museum for conifers. This genus, comprising about 113 species, is the largest genus of conifers and, arguably, is the most important genus of trees in the world (4, 9–11). It has a wide distribution in the Northern Hemisphere that coincides with the overall distribution of conifers (4). In addition, Pinus has a long evolutionary history dating back to the late Jurassic or early Cretaceous (12) and therefore participated in the historical dynamics of the Cretaceous and Cenozoic forests. Moreover, many pines have evolved morphological and life history adaptations to fire (13–18), an important ecological factor modulating the terrestrial ecosystem composition and distribution when climate changes (19). To disentangle the mechanisms driving the large-scale distribution of pine species diversity, evolutionary and ecological approaches should be jointly used. However, previous evolutionary studies focused mainly on phylogenetic reconstruction, divergence time estimation, biogeographic history, and fire adaptation of pines at long temporal scales (10, 16, 20–23), while ecological studies often investigated the species distribution dynamics at short temporal scales and ignored historical biogeography (24, 25). The accuracy of any evolutionary or ecological inference is heavily dependent on a fully resolved phylogeny and reliable estimates of species divergence times. Unfortunately, due to the slow evolutionary rates of conifers (3), the phylogenetic relationships of most subsections and species of pines are far from being resolved (10, 20–23, 26–28), which hinders us from studying their spatiotemporal evolution.
In this study, we reconstructed a robust transcriptome-based phylogeny and reestimated divergence times of Pinus based on an extensive sampling of nearly all species of the genus. With this phylogenetic framework, we further investigated the biogeographic history, temporal trend of diversification, and patterns of niche and trait evolution to reveal the factors driving the evolution and spatiotemporal dynamics of Pinus. We aimed to test whether the midlatitude region in the Northern Hemisphere has been an evolutionary cradle or museum for pines and to address whether phylogenetic or ecological conservatism or both have played important roles in shaping the global patterns of pine species richness. We also discussed the evolution of fire syndromes and its importance in the evolution of pine species diversity.
Results
Transcriptome Data Processing and Phylogenetic Analyses.
An average of 68,167 transcripts were de novo assembled for each sample with an average N50 scaffold length (size) of 1,534 base pairs (bp). A summary of the assembly statistics is shown in Dataset S1. For the 1,662 orthogroups (OGs) used in phylogenetic analyses, the coding sequence (CDS) and CDS (first + second) (the first and second codon positions) alignments ranged from 300 to 3,057 bp and from 200 to 2,038 bp in length, respectively. The concatenated CDS and CDS (first + second) alignments consisted of 2,146,620 and 1,431,080 bp, respectively.
The phylogenies inferred from the CDS and CDS (first + second) datasets, using either concatenation or coalescent methods, are highly resolved (ultrafast bootstrap [UFBoot] > 95% and local posterior probability [LPP] > 95% for most nodes) and topologically consistent, except that Pinus nelsonii and a few species within subsections show discordant placements (Fig. 1 and SI Appendix, Figs. S1–S4), which could be attributed to incomplete lineage sorting, hybridization among closely related species, or other unknown factors (22, 27–29).
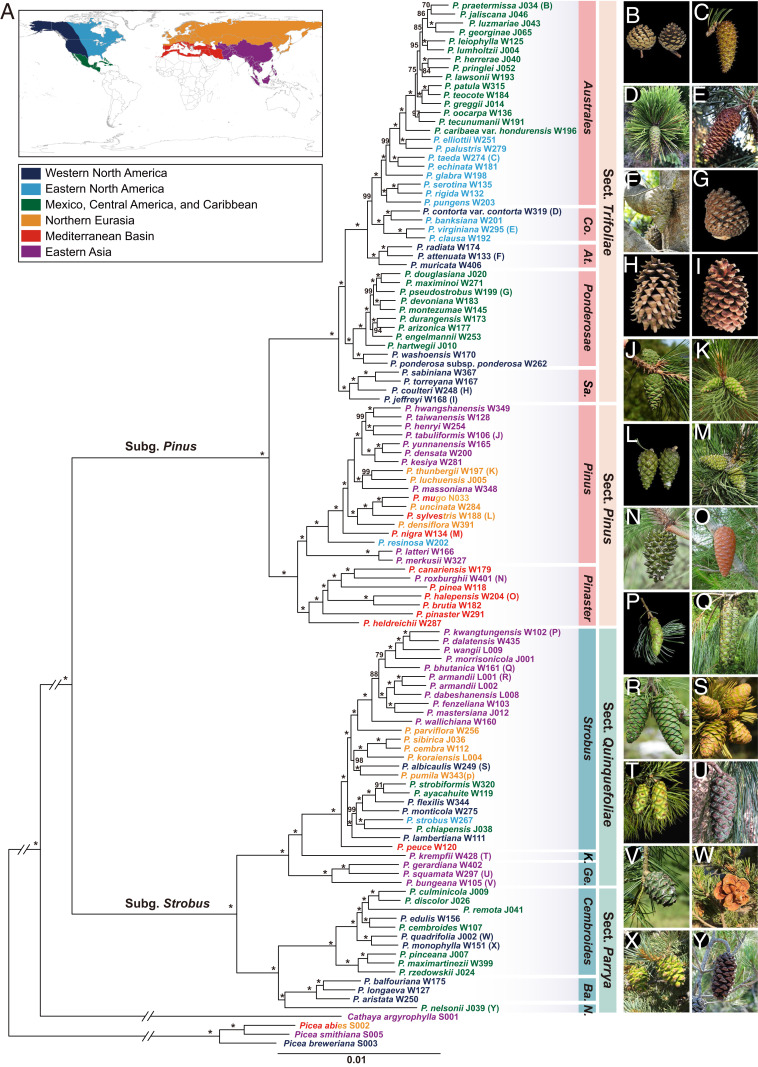
Fig. 1.
Phylogeny of Pinus. (A) ML tree inferred from the concatenated CDS alignment of 1,662 OGs. The numbers attached to the branches show the UFBoot supports (* for 100%). The 13 subsections of Pinus are listed with some abbreviations, that is, Co. (Contortae), At. (Attenuatae), Sa. (Sabinianae), K. (Krempfianae), Ge. (Gerardianae), Ba. (Balfourianae), and N. (Nelsoniae). Each species is shown in color corresponding to its geographic distribution shown in A. The letters in brackets following species names correspond to the pine images on the right (B–Y).
Our study highly supported two subgenera (Pinus and Strobus), four sections (Trifoliae, Pinus, Quinquefoliae, and Parrya) and 13 subsections (Australes, Contortae, Attenuatae, Ponderosae, Sabinianae, Pinus, Pinaster, Strobus, Krempfianae, Gerardianae, Cembroides, Balfourianae, and Nelsoniae) in the genus Pinus (Fig.1 and SI Appendix, Figs. S1–S4). Among the 11 subsections proposed by a widely recognized study (10), with the exception of Ponderosae and Australes, the other nine subsections were recovered as monophyletic with high support (UFBoot = 100%) (Fig. 1). In particular, the phylogenetic positions of nine subsections (Ponderosae, Australes, Contortae, Strobus, Krempfianae, Gerardianae, Cembroides, Balfourianae, and Nelsoniae), which remained controversial in previous studies (10, 20–22, 27, 28), were well resolved in this study (Fig. 1).
Notably, our results strongly support the monophyly of subsections Sabinianae (30) and Attenuatae (9) in sect. Trifoliae, a strictly North American section. Subsect. Sabinianae includes the Californian big-cone pines, that is, Pinus coulteri, Pinus jeffreyi, Pinus sabiniana, and Pinus torreyana, and subsect. Attenuatae includes three closed-cone (serotinous) species of California and Baja California, that is, Pinus attenuata, Pinus muricata, and Pinus radiata. The two subsections were previously assigned to subsect. Ponderosae and subsect. Australes, respectively (10). In this study, subsect. Sabinianae was sister to the remaining lineages of sect. Trifoliae. This contrasts with previous studies based on chloroplast DNA (10, 20, 21), which placed subsect. Contortae sister to the remaining lineages. Within sect. Quinquefoliae, subsect. Gerardianae diverged first, and the monotypic subsect. Krempfianae with unique, flattened leaves was sister to subsect. Strobus. In addition, the monospecific subsect. Nelsoniae endemic to northeastern Mexico, with several unique features, was resolved as a sister of subsect. Balfourianae in sect. Parrya (Fig. 1).
The transcriptome-based phylogeny was also highly resolved at the species level, which is important for reconstructing the evolutionary and biogeographic histories of the pine genus. For example, we found that in subsect. Gerardianae, Pinus bungeana, a species restricted to the central and northern China, was sister to a clade comprising the other two species of the subsection, that is, the western Himalayan Pinus gerardiana and Pinus squamata, an extremely rare species endemic to northeastern Yunnan, China. Within subsect. Strobus, the European Pinus peuce was sister to a clade composed of two lineages, one comprising the strictly North American white pines and the other including the Eurasian species and Pinus albicaulis from western North America. The subtropical Asian white pines formed a monophyletic group sister to the Japanese Pinus parviflora. Within subsect. Pinus, the two southern Asian species Pinus merkusii and Pinus latteri, which represent the southernmost distribution of the genus close to the equator, branched off first, followed by Pinus resinosa, a species from northeastern North America. Within subsect. Pinaster, a mainly Mediterranean lineage, Pinus canariensis from the Canary Islands was sister to Pinus roxburghii from the Himalayas.
Divergence Time and Ancestral Range Estimation.
The divergence time estimates with or without the fossil of Pinuxylon sp. are very close. In addition, the time estimates based on 16 fossils (SI Appendix, Table S1), following ref. 23, are slightly younger than those based on the four fossils (SI Appendix, Figs. S5 and S6). We found that the older estimates of ref. 23 were caused by the use of the FBD (fossilized birth–death with 21 fossils) method, and their estimates with the ND (node dating with 15 fossils) method were very close to ours. Therefore, we used the estimates based on the four fossils (SI Appendix, Fig. S5). The stem age of Pinus was dated to 155.3 (177 to 135) Ma in the late Jurassic. Its two subgenera were estimated to have diverged at 98.77 (116 to 83) Ma, the four sections at 58.04 to 56.13 Ma in the late Paleocene, and the subsections at 43.99 to 22.84 Ma from the middle Eocene to the early Miocene. About 10% of the extant pine species originated in the late Eocene to Oligocene, and nearly all the others originated in the Miocene (SI Appendix, Fig. S5).
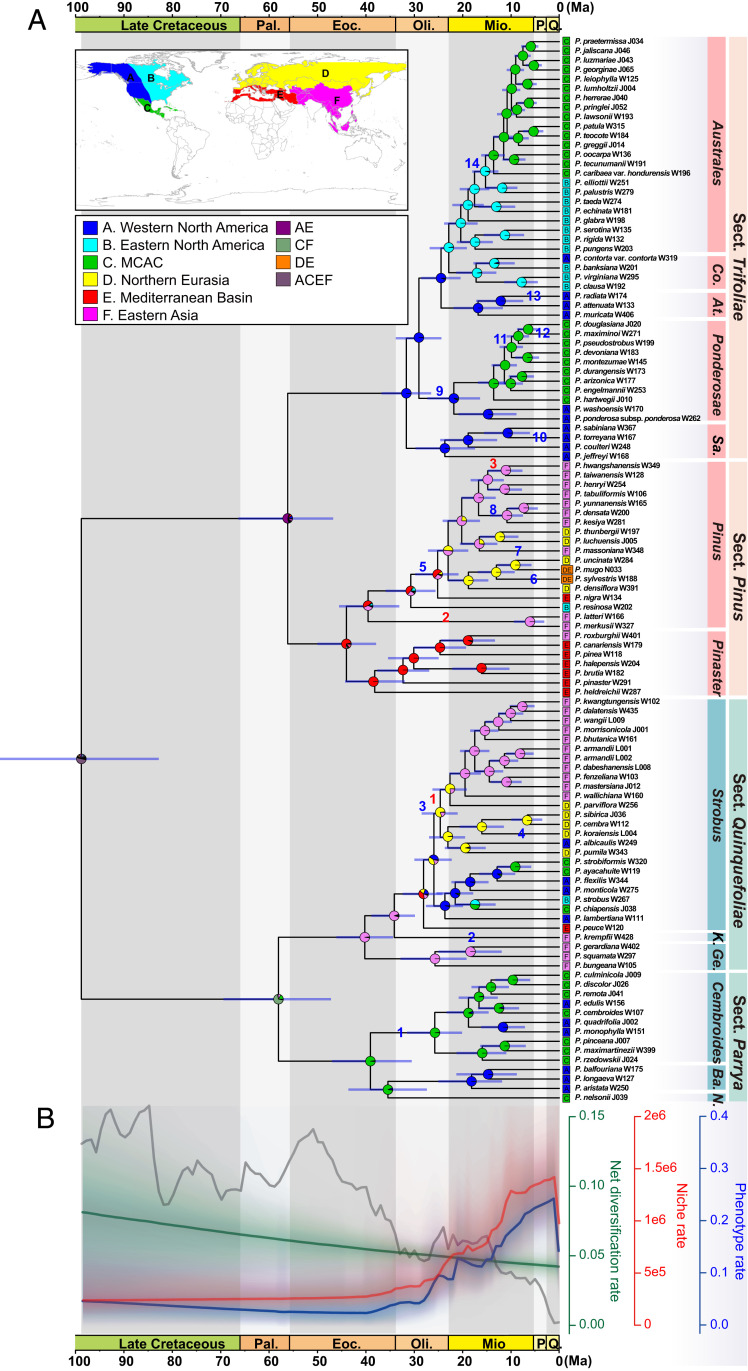
Despite that our data best fit the DIVALIKE+J model (LnL = −114.7, AICc = 235.7) (SI Appendix, Table S2), a similar result was obtained using the DIVALIKE model (SI Appendix, Figs. S7 and S8). According to the ancestral range estimation (Fig. 2A), Pinus widely expanded across the Northern Hemisphere in the late Cretaceous, but it is difficult to determine exactly where the genus originated. Within Pinus, three vicariance events were inferred to have occurred in its early evolutionary history. An initial vicariance event separated subg. Pinus and subg. Strobus in the late Cretaceous. The other two occurred almost simultaneously within the two subgenera during the late Paleogene, leading to the divergence of four sections (Pinus, Trifoliae, Quinquefoliae, and Parrya). Subsequent intercontinental migration and expansion were mainly via the Bering land bridge, except for P. resinosa, which reached eastern North America through the North Atlantic land bridge during the early Oligocene. The biogeographic stochastic mapping (BSM) results indicate that most biogeographical events can be explained by within-area speciation (76%) and dispersals (21%, including 19% events of founder-event speciation). Only a few events involved vicariance (3%) (SI Appendix, Table S3).
Fig. 2.
Historical biogeography of Pinus. (A) Ancestral range estimation using BioGeoBEARS with the DIVALIKE+J model. Divergence times were estimated using MCMCTree based on the concatenated CDS dataset (SI Appendix, Fig. S5). Horizontal bars at nodes indicate 95% credible intervals of the divergence time estimates. Pie charts at the nodes indicate relative probabilities of all possible geographic ranges. (B) Net diversification rate (green) and evolutionary rates of niche (red) and phenotypic traits (blue) estimated by BAMM, with color density shading to denote confidence on evolutionary rates through time. The unit of net diversification is species/Ma, and niche and phenotypic rates are unitless. Red numbers along branches represent major ecological niche shifts, and blue numbers along branches represent major phenotype shifts. The gray curve in the background shows the fluctuation of global temperature using the dataset from (94). Co., Contortae; At., Attenuatae; Sa., Sabinianae; K., Krempfianae; Ge., Gerardianae; Ba., Balfourianae; N., Nelsoniae; Pal., Paleocene; Eoc., Eocene; Oli., Oligocene; Mio., Miocene; P., Pliocene; Q., Quaternary; and MCAC represents Mexico, Central America, and Caribbean.
Ancestral Character and Fire Syndrome Reconstructions.
The ancestor of Pinus could have one, two, three, or five needles per fascicle and articulate seed wings. The state three needles per fascicle evolved at least four to five times, and the seed wings were lost (or rudimentary) mainly in subg. Strobus (SI Appendix, Fig. S9 A and B). In addition, the ancestor of Pinus was not fire adapted (avoider) (SI Appendix, Fig. S10). After the split of its two subgenera in the late Cretaceous, the resister (species adapted to low-intensity surface fires by thick bark and lower branch shedding) syndrome initially evolved in subg. Pinus, possibly in the Paleocene, followed by the evader (adults are killed during high-intensity crown fires, but seeds are stored in either soil or the canopy with the trait of serotiny and released after fire disturbance) and endurer (species survived fires through resprouting) strategies that evolved during the Oligocene to the middle Miocene. Although some Eurasian hard pines lost the fire-adapted syndromes, most species of subg. Pinus adapted to different fire intensities. In particular, the evader syndrome mainly occurred in the American sect. Trifoliae and some Mediterranean species of sect. Pinaster. In contrast, in subg. Strobus, most species are not fire adapted, and only eight species, including six from western North America and two from Asia, exhibit the resister fire syndrome (SI Appendix, Fig. S10).
Macroevolutionary Rates.
Net diversification rate estimated by Bayesian Analysis of Macroevolutionary Mixtures (BAMM) (v.2.5.0) (31) showed no shift and decreased slowly from 98.77 (116 to 83) Ma to present in Pinus (Fig. 2B and SI Appendix, Fig. S11 A and D). However, evolutionary rates of both niche and phenotypic traits experienced a strong increase toward the present, beginning around the early Miocene (Fig. 2B and SI Appendix, Fig. S12). In addition, needles per fascicle, fire syndromes, and aridity index are the most important variables by the primary loadings on ordinated phenotype traits and niche data (SI Appendix, Tables S4 and S5). We also found that the evolutionary rate variation had significantly different timing between number of needles per fascicle and fire syndrome. The rate of fire syndrome increased much later, showing a gradual increase from the Eocene (>50 Ma) to the early Miocene and then followed by a quick acceleration to the present. In contrast, the rate of number of needles per fascicle increased from the late Cretaceous (SI Appendix, Fig. S13). Notably, the aridity index was likely decisive for the rate shift of niche evolution. When all the 31 environmental variables were analyzed as a whole, or when the aridity index was analyzed separately, similar results were observed. However, when the aridity index was excluded, the rate of niche evolution was generally constant through time (SI Appendix, Fig. S14).
We also explored the association of diversification rates with global paleotemperature by fitting likelihood models of time and temperature dependence in RPANDA (v.1.9) (32). The linear speciation without extinction with temperature dependence is the best model (AICc = 856.891) (SI Appendix, Tables S6 and S7), and the diversification rate decreased with the decrease of temperature, which was consistent with the results of BAMM (SI Appendix, Figs. S11 and S15).
Niche and Phenotype Conservatism.
The values of Blomberg’s K are 0.358 and 0.354 with P < 0.005, and Pagel’s λ are 0.619 and 0.669 with P < 0.001 for niche and phenotype, respectively (SI Appendix, Table S8). Both measures indicate that significant phylogenetic signals exist in niche and phenotypic traits of Pinus.
Global Patterns of Pine Species Diversity and Mean Divergence Times.
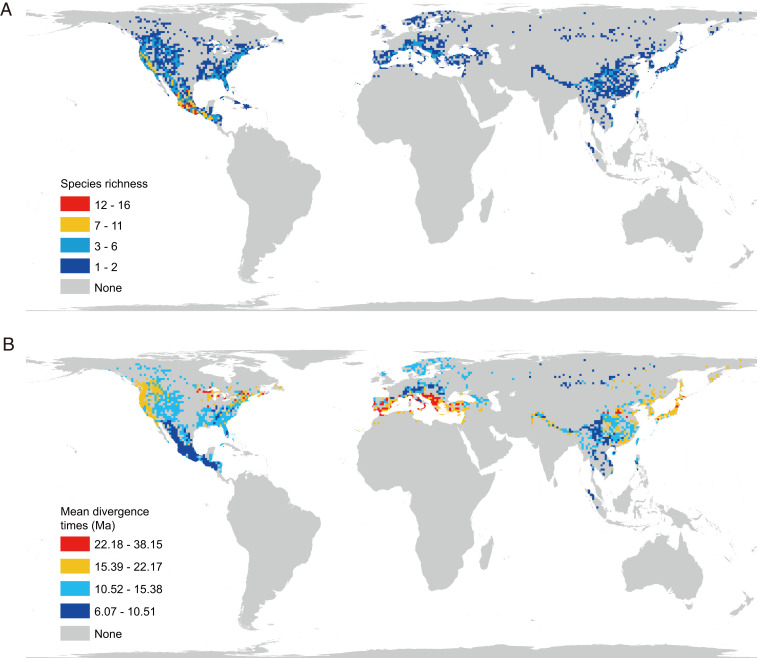
The distribution of grid cell mean species diversity of Pinus showed strong geographical variation with higher diversity in the middle to low latitudes of North America, the Mediterranean, and East Asia in contrast to the lower diversity in the high latitudes of the Northern Hemisphere (Fig. 3A). The highest diversity occurred in Mexico and adjacent areas. In general, species in the middle latitudes, particularly in the Mediterranean and northeastern North America, were much older (38.15 to 15.39 Ma) than those in the lower and higher latitudes such as in Mexico and northern Eurasia (10.51 to 6.07 Ma) (Fig. 3B).
Fig. 3.
The global patterns of species diversity (A) and mean divergence times (B) of pines plotted in grid cells of 100 km × 100 km.
Environmental Variables Driving the Global Distribution of Pine Species Diversity.
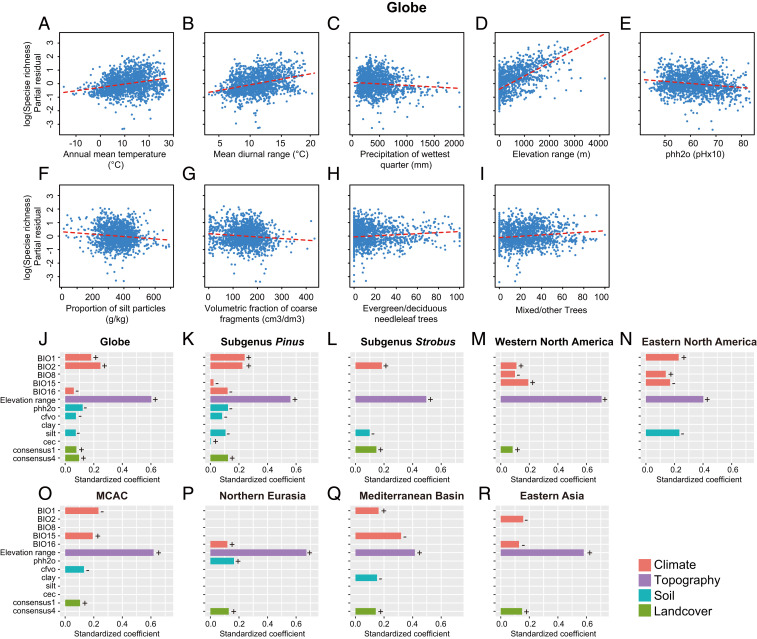
According to the multiple regression analysis, a total of nine environmental variables were significantly correlated with the species richness of global pines (R2 = 0.53) (Fig. 4 A–I and SI Appendix, Fig. S16 A–I and Table S9). The five variables, including annual mean temperature, mean diurnal range, elevation range, evergreen/deciduous needleleaf trees, and mixed/other trees, were positively correlated with species richness, while the other four variables, including precipitation of wettest quarter, soil pH, volumetric fraction of coarse fragments, and proportion of silt particles, were negatively correlated with species richness. Among these variables, topography played the most important role, followed by temperature, soil, and landcover variables, and precipitation was the weakest factor (Fig. 4J and SI Appendix, Table S9). In addition, there was obvious variation in the two subgenera and all regional models (Fig. 4 K–R and SI Appendix, Fig. S17 A–H and Table S9) except that topography was always the strongest predictor of species richness.
Fig. 4.
Determinants of pine species diversity. (A–I) Relationships between predictor variables and species richness based on the global multipredictor model across grid cells of 100 km × 100 km. (J–R) The relative importance of four types of variables (climate, topography, soil, and landcover) in explaining species richness based on global, two subgenera, and six regional analyses. The bar plots were implemented in the “ggplot2” package (97) using R (v.3.6.2) (84). The positive and negative correlations are indicated as + and −, respectively. MCAC represents Mexico, Central America, and Caribbean. Abbreviations and explanations of predictor variables refer to SI Appendix, Table S10.
Discussion
Phylogeny of Global Pines and Their Evolution at Middle Latitudes.
A highly resolved species tree with reliable estimates of divergence times is the prerequisite for understanding evolutionary histories and large-scale distribution patterns of worldwide plant genera. Our present study has reconstructed a completely resolved transcriptome-based phylogeny of Pinus comprising almost all extant species using 1,662 OGs. Importantly, the phylogenetic positions of nine out of the 11 widely recognized subsections (Ponderosae, Australes, Contortae, Strobus, Krempfianae, Gerardianae, Cembroides, Balfourianae, and Nelsoniae) of Pinus, which remained controversial in previous studies (10, 20–22, 27, 28), have all been anchored by our phylogenomic analysis (Fig. 1). Compared to previous plant phylogenomic studies at higher taxonomic levels (33, 34), our results indicate that RNA sequencing is also highly efficient for tree of life reconstruction at the species level, even for conifers with large genomes, long generation times, and a slow molecular evolutionary rate (3).
Most extant pine species have an origin in the Neogene in sharp contrast to the ancient origin of the genus. Based on our divergence time estimation, the origin of Pinus was dated to the late Jurassic, but all subsections originated in the middle Eocene to the early Miocene, and ∼90% of extant pine species originated in the Miocene (SI Appendix, Fig. S5). This indicates a rediversification of pines in the Cenozoic, especially in the Neogene. This tempo is also consistent with the Cenozoic global climate changes and tectonic activities. Earth’s climate transitioned from the “greenhouse” warming to the cold “icehouse” beginning at about 34 Ma (35), which promoted a global vegetation shift from old mesic lineages to those better adapted to cooler or drier environments. Many lines of evidence indicate that the post-Eocene climate deterioration shaped the biogeography of all conifer families (36, 37).
Notably, we found that pine species at middle latitudes, particularly in northeastern North America and the Mediterranean Basin, are much older than those at higher or lower latitudes (Fig. 3B and SI Appendix, Fig. S18B). In addition, pine species diversity has greatly increased since the early Cenozoic (SI Appendix, Fig. S15B), but the net diversification rate shows no shift, with a very low extinction rate (Fig. 2B and SI Appendix, Fig. S11C). The above evidence, together with the fact that most pines are distributed at middle latitudes and middle elevations and only a few species occur in boreal and high elevations (SI Appendix, Fig. S18 A and C), leads us to infer that the midlatitude region has acted mainly as an evolutionary museum for global pines. Our inference is consistent with the finding of ref. 8 that mountainous areas with long-term climate stability and heterogeneous environments, rather than biological features, had a major role in fostering lineage accumulation over evolutionary time scales, giving rise to regional diversity hotspots in conifers. However, the rate of net diversification of pines peaked in the Cretaceous and decreased gradually from 98.77 Ma to the present (Fig. 2B). This pattern could also be explained by the ancient cradle model as summarized in ref. 38, although the limited number of extant species in Pinus and the widespread extinction of pine species during the late Cretaceous and early Tertiary may affect the sensitivity of the BAMM analysis, particularly the estimates of speciation and extinction rates. Based on the earliest fossil records, including Pinus yorkshirensis from the early Cretaceous Wealden Formation in Yorkshire, United Kingdom (131 to 129 Ma) (39), and Pinus mundayi from the Lower Cretaceous Chaswood Formation of Nova Scotia, Canada (Valanginian, 140 to 133 Ma) (40), Pinus possibly originated in the middle latitudes of western Europe and eastern North America, which were connected in the early to middle Mesozoic (https://deeptimemaps.com/), but nearly all ancient pines became extinct and most extant pine species originated in the Miocene by rediversification as mentioned earlier (SI Appendix, Fig. S5). The fossil record is consistent with a widespread expansion of pines into many middle-latitude locations from their Eocene refugia during the Oligocene and Miocene (12). Although the middle and high elevations of many mountains at the middle latitudes were covered by ice sheets during the Quaternary ice ages (41), pine species might have survived glaciations by elevational shifts as many other plants (41, 42).
Determinants of Global Patterns of Pine Species Diversity.
Both phylogenetic and ecological conservatisms have played important roles in shaping the global patterns of pine species richness. Significant phylogenetic signals exist in niche and phenotypic traits of Pinus according to the measures of Blomberg’s K and Pagel’s λ (SI Appendix, Table S8). Although the distribution of pine species diversity shows strong geographical variation globally (Fig. 4 A–R), it has an overall high level of species richness at middle latitudes and middle elevations (SI Appendix, Fig. S18 A and C), which is quite different from the widely recognized biogeographic pattern of LDG (2, 43, 44). This distribution pattern might reflect a tendency of Pinus to retain ancestral climatic niche with a preference for warm temperatures. Although our analysis indicates that pines have experienced an acceleration of niche evolution (Fig. 2B), the increase of niche rate was caused mainly by the strong impact of aridity on the diversification of pines from the early Miocene (SI Appendix, Fig. S14). The cold habitats that proliferated in the Neogene likely had less influence on pine diversification, which contrasts with the pattern of Saxifragales, a major angiosperm clade in temperate biomes (45). The polar and elevational expansions of pines were likely constrained by their limited capacity for cold tolerance as indicated by the strong and positive correlation between temperature and species richness (Fig. 4 A, B, and J).
The increasing rates of niche and trait evolution but slowly decreasing net diversification rate imply that ecological divergence has not played a primary role in pine diversification (Fig. 2B). Instead, geography, in particular mountain building, probably had a profound impact, which is also corroborated by the strong positive correlation between topography and species richness at both global and local scales (Fig. 4 A–R). The orogenic events and volcanic activities in the Rocky Mountains, the Himalayas, and Mexican ranges probably have contributed significantly to the diversification and expansion of pines. In addition, for wind-pollinated pines, mountainous topography could be a good explanation for the high proportion of within-area speciation events we detected (SI Appendix, Table S3). It is noteworthy that Mexico and adjacent regions harbor the highest species diversity of pines, but compared to the Mediterranean pines, most of these species have relatively much younger ages (Fig. 3 A and B), suggesting recent diversifications during the mid- to late Miocene. Considering their close phylogenetic affinities to other New World pines, the Mexican species were very likely derived from the southward migrants from both west and east of the middle latitudes following the climatic deterioration of the early Oligocene.
Complex mountain systems usually indicate significant environmental heterogeneity. Our analyses of 31 environmental variables show that pine species richness is significantly positively correlated with five variables (annual mean temperature, mean diurnal range, elevation range, evergreen/deciduous needleleaf trees, and mixed/other trees) (Fig. 4 A, B, D, and H–J and SI Appendix, Fig. S16 and Table S9) but significantly negatively correlated with four variables (precipitation of wettest quarter, soil pH, volumetric fraction of coarse fragments, and proportion of silt particles) (Fig. 4 C, E–G, and J and SI Appendix, Fig. S16 and Table S9), suggesting that beyond a warm environment preference, pines may also prefer a drier habitat and somewhat more acidic soil, especially for subg. Pinus (Fig. 4K and SI Appendix, Fig. S17A and Table S9), despite substantial variation among different regions (Fig. 4 M–R and SI Appendix, Fig. S17 C–H and Table S9). We infer that the hot climate and high precipitation were the main factors limiting the development of pine species diversity in low-latitude regions. This inference is strongly supported by our finding that the aridity index is decisive for the rate shift of niche evolution in Pinus (SI Appendix, Figs. S14 and S19–S22). In Mexico, climate varies greatly, ranging from arid to humid zones, with 56% land area in arid or semiarid lands (northcentral and northwestern Mexico) and 37% in subhumid terrain (temperate forested areas and coastal areas in the Atlantic and Pacific sides) (46). The complex biotic and abiotic factors may well explain why Mexico can sustain the highest pine species richness. From what we learned about the historical dynamics of Pinus, we infer that pines are generally resilient, and their preference of warm and somewhat dry habitats could be helpful for their adaptation to the Anthropocene climate warming, especially at high latitudes and elevations.
Fire-Triggered Speciation and Adaptation of Pines.
Many plants, including pines, have evolved adaptive traits that enable them to persist and reproduce in fire-prone environments (14–16, 47–50). The Cretaceous had a flammable environment, as suggested by abundant fossil charcoal/inertinite records (51, 52) and a markedly higher concentration of atmospheric oxygen (52, 53), and therefore probably created new habitats and facilitated the divergence of the two subgenera of Pinus, which have very different fire-adaptation syndromes (SI Appendix, Fig. S10). The progressive evolution of fire syndromes in pines from fire avoiders to fire embracers, particularly in subg. Pinus, indicates that both fire frequency and intensity greatly increased over time, especially from the early Oligocene to the late Neogene (SI Appendix, Fig. S10), synchronized with the global expansion of grasslands (54). Increasing studies have demonstrated that fire played a pivotal role in the rise of grassland ecosystems, especially the worldwide expansion of C4 grasslands during the Miocene (55–57). This also coincides well with the evolutionary rate of fire syndrome in pines, which began increasing from the early Eocene, followed by an acceleration from the late Neogene toward the present (SI Appendix, Fig. S13). Surprisingly, we find that the rate increase of fire syndrome evolution began much later than the rate increase in needle number per fascicle and dispersal regimes, which occurred around the late Cretaceous (SI Appendix, Fig. S13). This lag could be explained by the evolutionary complexity of fire syndromes, while the needle and dispersal regime traits could react to fire directly. For example, needle traits (e.g., needle number per fascicle, needle length, and density) may affect flammability (58), whereas wind-dispersed seeds increase dispersal and colonization ability (18). Although this inference needs more evidence, clearly fire has acted as a powerful selective force on evolution of both reproductive and vegetative traits of pines and, thus, on pine speciation and adaptation.
Moreover, fire regimes varied greatly in different continents. In both the New World (especially the North American Southwest and Southeast) and the Old World (Mediterranean Basin), more species adapted to intense fire since the early Miocene, likely suggesting increasingly flammable environments there. This view is reinforced by the Miocene evolution of grassland and/or Mediterranean-type ecosystems in these regions (54, 59). In contrast, in East Asia, more fire-avoider species evolved from their fire-adaptive ancestors (SI Appendix, Fig. S10), indicating an environment with less fire activity. This difference could be attributed to the establishment of the East Asian monsoon. Associated with the uplift of the Qinghai–Tibetan Plateau, East Asia progressively transitioned from a zonal climate pattern to a monsoon-dominated pattern arguably beginning at the late Oligocene to the early Miocene, about 25 to 22 Ma (60). This climate transition might have led to three independent rate shifts of niche evolution associated with the diversification of East Asian five-needle pines as well as the divergences of East Asian Pinus taiwanensis/Pinus hwangshanensis and Southeast Asian P. merkusii/P. latteri, respectively (SI Appendix, Figs. S19–S22). This monsoon system has deeply impacted the East Asian biodiversity and vegetation composition, which might have further affected the fire regime. Different fire regimes in the New World and East Asia probably could explain why Pinus reaches its greatest diversity in Mexico and the United States and not in Southwest China, where most of the diversity in the remainder of the pine family is concentrated (61).
Materials and Methods
Sampling, Transcriptome Sequencing, and Data Processing.
A total of 255 accessions representing 112 species of Pinus were included (Dataset S1). Among them, 234 transcriptomes from 107 species were newly generated, and the remaining data were retrieved from the National Center for Biotechnology Information (NCBI). The raw sequencing reads are deposited in the Sequence Read Archive of GenBank (Dataset S1). Cathaya argyrophylla and three spruces, Picea abies, Picea breweriana, and Picea smithiana, were used as outgroups based on the close relationships of the two genera with Pinus (62).
Total RNA was extracted from haploid megametophytes or young leaves with RNAprep Pure Plant Kit (Tiangen). The complementary DNA libraries were prepared using NEBNext Ultra Directional RNA Library Prep Kit for Illumina and used for paired-end sequencing of a 2 × 100 bp run with an Illumina HiSeq 2500 device. Raw reads were checked with FastQC (v.0.11.5) (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/), trimmed using Trimmomatic (v.0.36) (63), and then assembled using Trinity (v.20140717) (64). Redundant transcripts were removed by CD-HIT (v.4.6.5) (65, 66). Putative CDSs were predicted using TransDecoder_r20140704 (https://github.com/TransDecoder/TransDecoder/releases), and the isoform encoding the longest peptide was selected as unigene.
Phylogenetic Analyses.
HaMStR (v.13.2.6) (67) was used to search for single-copy OGs for phylogenetic analyses. After a series of rigorous analyses (SI Appendix, Additional Methods), we finally obtained 1,662 OGs, each of which had a length longer than 100 aligned amino acids and covered at least 110 species. Phylogenomic analyses were performed based on two datasets, that is, CDS and CDS (first + second) of 1,662 OGs, using both concatenation and coalescence methods. For the concatenation analysis, the OGs were concatenated by FASconCAT-G (v.1.04) (68) and used for the maximum likelihood (ML) analysis by IQ-TREE v.2.0-rc1 (69). Branch supports were generated with 1,000 UFBoot replicates (70). For the coalescent analyses, to improve the accuracy of individual gene trees, the sequences with gap characters more than 60% were removed from each OG (71), and then individual gene trees were generated by IQ-TREE v.2.0-rc1 (69) with 1,000 UFBoot replicates (70) under the best substitution model (72). We also contracted branches with very low support (below 10% UFBoot support) from each gene tree to improve the accuracy (73, 74). Based on these individual gene trees, we estimated the species tree in ASTRAL (v.5.7.3) (74) with LPP (75).
Divergence Time Estimation.
The concatenated CDS dataset and corresponding ML tree were used for dating analysis. We used four fossils for the calibration (more details in SI Appendix, Additional Methods). To test the influence of Pinuxylon sp. (76). on divergence time estimates, we also conducted the analysis by excluding this fossil. Considering that Saladin et al. (23) used 12 to 21 fossils (different between the ND and FBD methods) as calibration points to estimate the divergence times of Pinus, we used the 16 fossils following their study with the ND method to calibrate our phylogeny (MCMCTree cannot perform FBD analysis) (SI Appendix, Table S1).
We estimated divergence times using the MCMCTree program in PAML 4.9j (77). The approximate likelihood method was conducted to speed up the likelihood calculation for large datasets (78). The time unit was set to 100 Ma, and a maximum bound for the root was set to 153 Ma based on the oldest fossil of Pinaceae (79). For the minimum and maximum bounds of all calibration points, the default 2.5% tail probability was used. The ML estimates of branch lengths, the gradient vector, and Hessian matrix were calculated using MCMCTree and BASEML (in PAML) programs under the GTR+G substitution models (model = 7). The overall substitution rate (rgene_gamma) and rate-drift parameter (sigma2_gamma) were set as G (1, 33.8) and G (1, 10), respectively. The independent rates model (80) was used to specify the prior of rates, while the posterior times were estimated by using a MCMC algorithm in the MCMCTree program. Two independent MCMC runs were conducted. Each run discarded the first 10 million iterations as burn-in and then sampled every 500 iterations until it gathered 20,000 samples. The stationary state and convergence of each run were checked in Tracer (v.1.7.1) (81) to ensure that all parameters had effective sample sizes (ESS) above 200.
Ancestral Range Estimation.
To trace the biogeographic history of Pinus, an ancestral range estimation was conducted by BioGeoBEARS (v.1.1.2) (82, 83) implemented in R (v.3.6.2) (84). The time-calibrated phylogenetic tree was obtained from PAML 4.9j (77), and outgroups were excluded due to the relict distribution of Cathaya. Six geographical regions were defined based on the study of Badik, Jahner, and Wilson (16), climatic and tectonic histories of continents, and extant distribution patterns of Pinus, including A (western North America), B (eastern North America), C (Mexico, Central America, and Caribbean), D (northern Eurasia), E (Mediterranean Basin, extending to western Asia), and F (eastern Asia, including eastern and southern Asia, from the southernmost of Northeast Asia to Southeast Asia and extending westward along the Himalayas) (Dataset S2). We tested all six models provided by BioGeoBEARS, including a likelihood version of DIVA (“DIVALIKE”), LAGRANGE’s DEC model, and BAYAREA as well as “+J” versions of these models, which consider founder-event speciation (82). The maximum number of areas was set to four. AICc was used for a posteriori test of models. To estimate the number and type of biogeographical events, we also carried out BSM implemented in BioGeoBEARS. Event frequencies were estimated by taking the mean and SD of event counts from 50 BSMs.
Collection of Species Distribution Data and Environmental Variables.
The distribution data of global pine species were collected from literatures (4, 85), online databases (the global biodiversity information facility, https://www.gbif.org/), herbaria (PE, HAST, K, and TAI), and our field work. These datasets were carefully assessed, and some erroneous records (i.e., occurrences in the oceans, ice sheets, and deserts), duplicates, and cultivation records were removed. Finally, a total of 7,725 unique distribution records from 113 species, including 109 species that correspond to those in the phylogenetic analysis (except Pinus discolor, Pinus georginae, and Pinus washoensis) and other four species (Pinus amamiana, Pinus cubensis, Pinus occidentalis, and Pinus tropicalis), were used in this study (Dataset S3).
We collected 31 environmental variables (Dataset S3), including 19 bioclimatic variables and one topographical layer (elevation) (https://www.worldclim.org/data/worldclim21.html) (86), seven soil variables within 30 cm of soil horizon, and two landcover classes (https://www.earthenv.org/landcover) (87) as well as annual potential evapotranspiration and aridity index (https://cgiarcsi.community). The bioclimatic and topographical variables are at a resolution of 2.5 arc minutes, and the others are at a resolution of 30 arc seconds. Mean values of the variables for each species were used in the subsequent evolutionary rate estimation.
Ancestral State Reconstruction of Morphological Traits and Fire Syndromes.
Three important morphological characters related to the adaptation of Pinus, including the number of needles per fascicle (one to seven), seed wing (wingless, adnate, and articulate), and seed dispersal regime (wind, animal, and wind and animal) (Dataset S2), were analyzed in this study. Fire is an important disturbance factor that has generally favored Pinus throughout its natural range in the Northern Hemisphere (88). Rowe (89) divided the response of plants to fire into five strategies and, with the exception of invader, the other four exist in Pinus (16, 88), including avoider (species without any adaptation to fire), resister (species with adaptation to low-intensity surface fires by thick bark and lower branch shedding), endurer (species that survive fires through resprouting), and evader (adults are killed during high-intensity crown fires, but seeds are stored in either the soil or the canopy with the trait of serotiny and released after fire disturbance). Mesquite (v. 3.61) (90) was used to reconstruct the evolutionary histories of the morphological traits and fire syndromes.
Macroevolutionary Rate Estimation.
Diversification rates were estimated with BAMM (v.2.5.0) (31), which uses a reversible-jump MCMC to sample a large number of possible diversification regimes from a given time-calibrated phylogeny. We ran the BAMM analysis using the time-calibrated phylogeny without outgroups obtained from the MCMCTree analysis in PAML 4.9j (77). The MCMC was run for two million generations and sampled every 1,000 generations. Prior values were selected using the “setBAMMpriors” function. Postrun analysis and visualization used the R package BAMMtools (v.2.17) (91) in R (v.3.6.2) (84). The initial 25% of samples of the MCMC run were discarded as burn-in, and the remaining data were assessed for convergence using CODA package (92) in R (v.3.6.2) (84) to ensure that the ESS values are above 200.
To estimate evolutionary rates of niche and phenotypic traits in Pinus, we firstly ordinated all environmental variables and phenotypic data using phylogenetic principal component analysis (PCA) implemented in phytools (v.0.7-70) package (93) with the “phyl.pca” function. Then, we conducted complementary runs using the BAMM trait model on the first axis of the phylogenetic PCA of niche and phenotypic traits data. For niche lability, we ran the analysis for 10 million generations with a 25% burn-in based on the time-calibrated tree obtained from the MCMCTree analysis. For lability in phenotypic traits, we ran the analysis for 200 million generations with a 50% burn-in. Other settings followed those in the diversification analysis.
To explore the effects of global paleotemperature on pine diversification, we also used RPANDA (v.1.9) (32) to fit a series of time- and temperature-dependent likelihood diversification models (SI Appendix, Tables S6 and S7). The global paleotemperature data set was obtained from ref. 94. Speciation and extinction dependences were modeled as all possible combinations of constant, linear, and exponential relationships as well as pure-birth models. We compared the likelihood supports, and the model with the smallest AICc value was chosen as the best diversification model.
Detection of Phylogenetic Signals of Niche and Phenotypic Traits.
Blomberg’s K (95) and Pagel’s λ (96) are two measures of phylogenetic signals. We estimated both for the niche and phenotypic traits of pines using “phylosig” function in the phytools (v.0.7-70) package (93).
Analyses of Global Patterns of Pine Species Diversity and Mean Divergence Times.
To mitigate the effects of area on diversity estimation and potential sampling incompleteness, we projected Behrmann Equal-Area Cylindrical for the entire land of the world and divided the land area into grid cells of 100 km × 100 km. The global distribution dataset above was spatially matched to the grid cells in ArcGIS 10.6, and then the species richness within each grid cell was calculated. To study the global pattern of the divergence times of pines, we extracted the divergence time of each species estimated by molecular dating and calculated the mean divergence time within each grid cell of 100 km × 100 km (Dataset S4).
The Impact of Environmental Variables on the Global Pattern of Pine Species Diversity.
To explore the underlying driving forces of the distribution pattern of pine species diversity, all 31 environmental variables were extracted as mean values within each 100 km × 100 km grid cell (Dataset S4). To reduce collinearity among variables, the initial set of 31 environmental variables was finally reduced to 13 variables with weak pairwise correlation (|r| < 0.7), including five bioclimatic variables (BIO1, annual mean temperature; BIO2, mean diurnal range; BIO8, mean temperature of wettest quarter; BIO15, precipitation seasonality; and BIO16, precipitation of wettest quarter), one topographical variable (elevation range), five soil variables (phh2o, soil pH; cfvo, volumetric fraction of coarse fragments; clay, proportion of clay particles; silt, proportion of silt particles; and cec, cation exchange capacity), and two landcover variables (consensus1, evergreen/deciduous needleleaf trees and consensus4, mixed/other trees) (SI Appendix, Table S10).
Multiple regression models (ordinary least squares) and spatial simultaneous autoregressive models were built for the global pines and for the two subgenera (Pinus and Strobus) and six biogeographic regions of Pinus, with pine species richness as the response variable and 13 environmental variables as predictors (SI Appendix, Table S10). Additional details of the methods are available in SI Appendix, Additional Methods.
Supplementary Material
Acknowledgments
We thank Zheng-Yu Liu (Chongqing Institute of Medicinal Plant Cultivation), Xun Gong (Kunming Institute of Botany, Chinese Academy of Sciences), Chuan Chen (Hangzhou Botanical Garden), Rui-Hui Chen (Taizhou Entry-Exit Inspection and Quarantine Bureau), Jian-Quan Jin, Xiao-Hua Jin, Tian-Gang Gao, and Hui Wu for help in the sample collection. We also thank Royal Botanic Gardens, Kew, Royal Botanic Garden Edinburgh, YunnanYaoshan National Nature Reserve Administration, and the National Herbarium of Mexico for providing some plant materials. We thank Aljos Farjon (Royal Botanic Gardens, Kew) and Denis Filer (University of Oxford) who have contributed the pine distribution data. We thank Hong Du for her assistance in laboratory experiments, Jin-Hua Ran, Yan-Yan Liu (Henan Agricultural University), and Ting-Ting Shen (East China Institute of Technology) for assistance with data analyses as well as Lei Chen and Wu-Bing Xu (Aarhus University) for help with ecological analyses. This study was funded by the National Key Research and Development Program of China (Grant No. 2017YFA0605100), the Strategic Priority Research Program of Chinese Academy of Sciences (CAS) (Grant Nos. XDB31000000, XDA23080000), and Key Research Program of Frontier Sciences, CAS (QYZDJ-SSW-SMC027).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2022302118/-/DCSupplemental.
Data Availability
Transcriptome data have been deposited in the NCBI database under BioProject accession no. PRJNA703422. Alignments, phylogenetic trees, and other files generated during analyses have been deposited at Dryad (https://doi.org/10.5061/dryad.pc866t1nq) (98).
References
- 1.Pennisi E., What determines species diversity? Science 309, 90 (2005). [DOI] [PubMed] [Google Scholar]
- 2.Pontarp M., et al., The latitudinal diversity gradient: Novel understanding through mechanistic eco-evolutionary. Trends Ecol. Evol. 34, 211–223 (2019). [DOI] [PubMed] [Google Scholar]
- 3.Wang X. Q., Ran J. H., Evolution and biogeography of gymnosperms. Mol. Phylogenet. Evol. 75, 24–40 (2014). [DOI] [PubMed] [Google Scholar]
- 4.Farjon A., Filer D., An Atlas of the World’s Conifers: An Analysis of their Distribution, Biogeography, Diversity and Conservation Status (Brill, Leiden, 2013). [Google Scholar]
- 5.Armenise L., Simeone M. C., Piredda R., Schirone B., Validation of DNA barcoding as an efficient tool for taxon identification and detection of species diversity in Italian conifers. Eur. J. For. Res. 131, 1337–1353 (2012). [Google Scholar]
- 6.Kerkhoff A. J., Moriarty P. E., Weiser M. D., The latitudinal species richness gradient in New World woody angiosperms is consistent with the tropical conservatism hypothesis. Proc. Natl. Acad. Sci. U.S.A. 111, 8125–8130 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rueda M., Godoy O., Hawkins B. A., Spatial and evolutionary parallelism between shade and drought tolerance explains the distributions of conifers in the conterminous United States. Glob. Ecol. Biogeogr. 26, 31–42 (2017). [Google Scholar]
- 8.Sundaram M., et al., Accumulation over evolutionary time as a major cause of biodiversity hotspots in conifers. Proc. Biol. Sci. 286, 20191887 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Price R., Liston A., Strauss S., “Phylogeny and systematics of Pinus” in Ecology and Biogeography of Pinus, Richardson D. M., Ed. (Cambridge University Press, Cambridge, 1998), pp. 49–68. [Google Scholar]
- 10.Gernandt D. S., Geada López G., Ortiz García S., Liston A., Phylogeny and classification of Pinus. Taxon 54, 29–42 (2005). [Google Scholar]
- 11.Farjon A., A Handbook of the World’s Conifers (Brill, Leiden, 2017). [Google Scholar]
- 12.Millar C. I., “Early evolution of pines” in Ecology and Biogeography of Pinus, Richardson D. M., Ed. (Cambridge University Press, Cambridge, 1998), pp. 69–91. [Google Scholar]
- 13.Richardson D. M., et al., Human impacts in pine forests: Past, present, and future. Annu. Rev. Ecol. Evol. Syst. 38, 275–297 (2007). [Google Scholar]
- 14.He T., Pausas J. G., Belcher C. M., Schwilk D. W., Lamont B. B., Fire-adapted traits of Pinus arose in the fiery Cretaceous. New Phytol. 194, 751–759 (2012). [DOI] [PubMed] [Google Scholar]
- 15.Keeley J. E., Ecology and evolution of pine life histories. Ann. For. Sci. 69, 445–453 (2012). [Google Scholar]
- 16.Badik K. J., Jahner J. P., Wilson J. S., A biogeographic perspective on the evolution of fire syndromes in pine trees (Pinus: Pinaceae). R. Soc. Open Sci. 5, 172412 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lamont B. B., He T., Yan Z., Fire as a pre-emptive evolutionary trigger among seed plants. Perspect. Plant Ecol. Evol. Syst. 36, 13–23 (2019). [Google Scholar]
- 18.Salazar-Tortosa D., Saladin B., Zimmermann N. E., Castro J., Rubio de Casas R., The evolution of seed dispersal is associated with environmental heterogeneity in Pinus. Perspect. Plant Ecol. Evol. Syst. 41, 125464 (2019). [Google Scholar]
- 19.Bond W. J., Woodward F. I., Midgley G. F., The global distribution of ecosystems in a world without fire. New Phytol. 165, 525–537 (2005). [DOI] [PubMed] [Google Scholar]
- 20.Eckert A. J., Hall B. D., Phylogeny, historical biogeography, and patterns of diversification for Pinus (Pinaceae): Phylogenetic tests of fossil-based hypotheses. Mol. Phylogenet. Evol. 40, 166–182 (2006). [DOI] [PubMed] [Google Scholar]
- 21.Parks M., Cronn R., Liston A., Separating the wheat from the chaff: Mitigating the effects of noise in a plastome phylogenomic data set from Pinus L. (Pinaceae). BMC Evol. Biol. 12, 100 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hao Z. Z., Liu Y. Y., Nazaire M., Wei X. X., Wang X. Q., Molecular phylogenetics and evolutionary history of sect. Quinquefoliae (Pinus): Implications for Northern hemisphere biogeography. Mol. Phylogenet. Evol. 87, 65–79 (2015). [DOI] [PubMed] [Google Scholar]
- 23.Saladin B., et al., Fossils matter: Improved estimates of divergence times in Pinus reveal older diversification. BMC Evol. Biol. 17, 95 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Z., Brown J. H., Tang Z., Fang J., Temperature dependence, spatial scale, and tree species diversity in eastern Asia and North America. Proc. Natl. Acad. Sci. U.S.A. 106, 13388–13392 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fragnière Y., et al., Fighting their last stand? A global analysis of the distribution and conservation status of gymnosperms. J. Biogeogr. 42, 809–820 (2015). [Google Scholar]
- 26.Hernández-León S., Gernandt D. S., Pérez de la Rosa J. A., Jardón-Barbolla L., Phylogenetic relationships and species delimitation in pinus section trifoliae inferrred from plastid DNA. PLoS One 8, e70501 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gernandt D. S., et al., Multi-locus phylogenetics, lineage sorting, and reticulation in Pinus subsection Australes. Am. J. Bot. 105, 711–725 (2018). [DOI] [PubMed] [Google Scholar]
- 28.Montes J. R., et al., Phylogenetics of Pinus subsection Cembroides Engelm. (Pinaceae) inferred from low-copy nuclear gene sequences. Syst. Bot. 44, 501–518 (2019). [Google Scholar]
- 29.Wang B., Wang X. R., Mitochondrial DNA capture and divergence in Pinus provide new insights into the evolution of the genus. Mol. Phylogenet. Evol. 80, 20–30 (2014). [DOI] [PubMed] [Google Scholar]
- 30.Little J. E. L., Critchfield W. R., Subdivisions of the Genus Pinus (pines) (United States Department of Agriculture, 1969). [Google Scholar]
- 31.Rabosky D. L., Automatic detection of key innovations, rate shifts, and diversity-dependence on phylogenetic trees. PLoS One 9, e89543 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morlon H., et al., RPANDA: An R package for macroevolutionary analyses on phylogenetic trees. Methods Ecol. Evol. 7, 589–597 (2016). [Google Scholar]
- 33.Ran J. H., Shen T. T., Wang M. M., Wang X. Q., Phylogenomics resolves the deep phylogeny of seed plants and indicates partial convergent or homoplastic evolution between Gnetales and angiosperms. Proc. Biol. Sci. 285, 20181012 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Leebens-Mack J. H.et al.; One Thousand Plant Transcriptomes Initiative , One thousand plant transcriptomes and the phylogenomics of green plants. Nature 574, 679–685 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu Z., et al., Global cooling during the eocene-oligocene climate transition. Science 323, 1187–1190 (2009). [DOI] [PubMed] [Google Scholar]
- 36.Leslie A. B., et al., Hemisphere-scale differences in conifer evolutionary dynamics. Proc. Natl. Acad. Sci. U.S.A. 109, 16217–16221 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pittermann J., Stuart S. A., Dawson T. E., Moreau A., Cenozoic climate change shaped the evolutionary ecophysiology of the Cupressaceae conifers. Proc. Natl. Acad. Sci. U.S.A. 109, 9647–9652 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Couvreur T. L., Forest F., Baker W. J., Origin and global diversification patterns of tropical rain forests: Inferences from a complete genus-level phylogeny of palms. BMC Biol. 9, 44 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ryberg P. E., et al., Reconsidering relationships among stem and crown group Pinaceae: Oldest record of the genus Pinus from the early Cretaceous of Yorkshire, United Kingdom. Int. J. Plant Sci. 173, 917–932 (2012). [Google Scholar]
- 40.Falcon-Lang H. J., Mages V., Collinson M., The oldest Pinus and its preservation by fire. Geology 44, 303–306 (2016). [Google Scholar]
- 41.Hewitt G., The genetic legacy of the Quaternary ice ages. Nature 405, 907–913 (2000). [DOI] [PubMed] [Google Scholar]
- 42.Davis M. B., Shaw R. G., Range shifts and adaptive responses to Quaternary climate change. Science 292, 673–679 (2001). [DOI] [PubMed] [Google Scholar]
- 43.Hillebrand H., On the generality of the latitudinal diversity gradient. Am. Nat. 163, 192–211 (2004). [DOI] [PubMed] [Google Scholar]
- 44.Mannion P. D., Upchurch P., Benson R. B. J., Goswami A., The latitudinal biodiversity gradient through deep time. Trends Ecol. Evol. 29, 42–50 (2014). [DOI] [PubMed] [Google Scholar]
- 45.Folk R. A., et al., Rates of niche and phenotype evolution lag behind diversification in a temperate radiation. Proc. Natl. Acad. Sci. U.S.A. 116, 10874–10882 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Valdez R., Guzman-Aranda J. C., Abarca F. J., Tarango-Arambula L. A., Sanchez F. C., Wildlife conservation and management in Mexico. Wildl. Soc. Bull. 34, 270–282 (2006). [Google Scholar]
- 47.Gómez-González S., Torres-Díaz C., Bustos-Schindler C., Gianoli E., Anthropogenic fire drives the evolution of seed traits. Proc. Natl. Acad. Sci. U.S.A. 108, 18743–18747 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Keeley J. E., Pausas J. G., Rundel P. W., Bond W. J., Bradstock R. A., Fire as an evolutionary pressure shaping plant traits. Trends Plant Sci. 16, 406–411 (2011). [DOI] [PubMed] [Google Scholar]
- 49.Pausas J. G., Evolutionary fire ecology: Lessons learned from pines. Trends Plant Sci. 20, 318–324 (2015). [DOI] [PubMed] [Google Scholar]
- 50.Archibald S., et al., Biological and geophysical feedbacks with fire in the Earth system. Environ. Res. Lett. 13, 033003 (2018). [Google Scholar]
- 51.Scott A. C., The Pre-Quaternary history of fire. Palaeogeogr. Palaeoclimatol. Palaeoecol. 164, 281–329 (2000). [Google Scholar]
- 52.Glasspool I. J., Scott A. C., Phanerozoic concentrations of atmospheric oxygen reconstructed from sedimentary charcoal. Nat. Geosci. 3, 627–630 (2010). [Google Scholar]
- 53.Belcher C. M., et al., Increased fire activity at the Triassic/Jurassic boundary in Greenland due to climate-driven floral change. Nat. Geosci. 3, 426–429 (2010). [Google Scholar]
- 54.Strömberg C. A. E., Evolution of grasses and grassland ecosystems. Annu. Rev. Earth Planet. Sci. 39, 517–544 (2011). [Google Scholar]
- 55.Edwards E. J.et al.; C4 Grasses Consortium , The origins of C4 grasslands: Integrating evolutionary and ecosystem science. Science 328, 587–591 (2010). [DOI] [PubMed] [Google Scholar]
- 56.Hoetzel S., Dupont L., Schefuß E., Rommerskirchen F., Wefer G., The role of fire in Miocene to Pliocene C4 grassland and ecosystem evolution. Nat. Geosci. 6, 1027–1030 (2013). [Google Scholar]
- 57.Karp A. T., Behrensmeyer A. K., Freeman K. H., Grassland fire ecology has roots in the late Miocene. Proc. Natl. Acad. Sci. U.S.A. 115, 12130–12135 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schwilk D. W., Ackerly D. D., Flammability and serotiny as strategies: Correlated evolution in pines. Oikos 94, 326–336 (2001). [Google Scholar]
- 59.Rundel P. W., et al., Mediterranean biomes: Evolution of their vegetation, floras, and climate. Annu. Rev. Ecol. Evol. Syst. 47, 383–407 (2016). [Google Scholar]
- 60.Licht A., et al., Asian monsoons in a late Eocene greenhouse world. Nature 513, 501–506 (2014). [DOI] [PubMed] [Google Scholar]
- 61.Farjon A., World Checklist and Bibliography of Conifers (Kew Publishing, ed. 2, 2001). [Google Scholar]
- 62.Ran J. H., Shen T. T., Wu H., Gong X., Wang X. Q., Phylogeny and evolutionary history of Pinaceae updated by transcriptomic analysis. Mol. Phylogenet. Evol. 129, 106–116 (2018). [DOI] [PubMed] [Google Scholar]
- 63.Bolger A. M., Lohse M., Usadel B., Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Grabherr M. G., et al., Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 29, 644–652 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Li W., Godzik A., Cd-hit: A fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22, 1658–1659 (2006). [DOI] [PubMed] [Google Scholar]
- 66.Fu L., Niu B., Zhu Z., Wu S., Li W., CD-HIT: Accelerated for clustering the next-generation sequencing data. Bioinformatics 28, 3150–3152 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ebersberger I., Strauss S., von Haeseler A., HaMStR: Profile hidden markov model based search for orthologs in ESTs. BMC Evol. Biol. 9, 157 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kück P., Longo G. C., FASconCAT-G: Extensive functions for multiple sequence alignment preparations concerning phylogenetic studies. Front. Zool. 11, 81 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nguyen L. T., Schmidt H. A., von Haeseler A., Minh B. Q., IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hoang D. T., Chernomor O., von Haeseler A., Minh B. Q., Vinh L. S., UFBoot2: Improving the ultrafast bootstrap approximation. Mol. Biol. Evol. 35, 518–522 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sayyari E., Whitfield J. B., Mirarab S., Fragmentary gene sequences negatively impact gene tree and species tree reconstruction. Mol. Biol. Evol. 34, 3279–3291 (2017). [DOI] [PubMed] [Google Scholar]
- 72.Kalyaanamoorthy S., Minh B. Q., Wong T. K. F., von Haeseler A., Jermiin L. S., ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 14, 587–589 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhang C., Sayyari E., Mirarab S., “ASTRAL-III: Increased scalability and impacts of contracting low support branches” in Comparative Genomics, Meidanis J., Nakhleh L., Eds. (Springer International Publishing, Cham, Switzerland, 2017), pp. 53–75. [Google Scholar]
- 74.Zhang C., Rabiee M., Sayyari E., Mirarab S., ASTRAL-III: Polynomial time species tree reconstruction from partially resolved gene trees. BMC Bioinformatics 19 (suppl. 6), 153 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sayyari E., Mirarab S., Fast coalescent-based computation of local branch support from quartet frequencies. Mol. Biol. Evol. 33, 1654–1668 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Meijer J. J. F., Fossil woods from the late Cretaceous Aachen formation. Rev. Palaeobot. Palynol. 112, 297–336 (2000). [DOI] [PubMed] [Google Scholar]
- 77.Yang Z., PAML 4: Phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24, 1586–1591 (2007). [DOI] [PubMed] [Google Scholar]
- 78.dos Reis M., Yang Z., Approximate likelihood calculation on a phylogeny for Bayesian estimation of divergence times. Mol. Biol. Evol. 28, 2161–2172 (2011). [DOI] [PubMed] [Google Scholar]
- 79.Rothwell G. W., Mapes G., Stockey R. A., Hilton J., The seed cone Eathiestrobus gen. Nov.: Fossil evidence for a Jurassic origin of Pinaceae. Am. J. Bot. 99, 708–720 (2012). [DOI] [PubMed] [Google Scholar]
- 80.Rannala B., Yang Z., Inferring speciation times under an episodic molecular clock. Syst. Biol. 56, 453–466 (2007). [DOI] [PubMed] [Google Scholar]
- 81.Rambaut A., Drummond A. J., Xie D., Baele G., Suchard M. A., Posterior summarization in bayesian phylogenetics using Tracer 1.7. Syst. Biol. 67, 901–904 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Matzke N. J., Probabilistic historical biogeography: New models for founder-event speciation, imperfect detection, and fossils allow improved accuracy and model-testing. Front. Biogeogr. 5, 242–248 (2013). [Google Scholar]
- 83.Matzke N. J., Model selection in historical biogeography reveals that founder-event speciation is a crucial process in Island Clades. Syst. Biol. 63, 951–970 (2014). [DOI] [PubMed] [Google Scholar]
- 84.R Core Team , R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing, Vienna, Austria, 2019). [Google Scholar]
- 85.Liu Y. Y., Jin W. T., Wei X. X., Wang X. Q., Cryptic speciation in the Chinese white pine (Pinus armandii): Implications for the high species diversity of conifers in the Hengduan Mountains, a global biodiversity hotspot. Mol. Phylogenet. Evol. 138, 114–125 (2019). [DOI] [PubMed] [Google Scholar]
- 86.Fick S. E., Hijmans R. J., WorldClim 2: New 1-km spatial resolution climate surfaces for global land areas. Int. J. Climatol. 37, 4302–4315 (2017). [Google Scholar]
- 87.Hengl T., et al., SoilGrids250m: Global gridded soil information based on machine learning. PLoS One 12, e0169748 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Agee J. K., “Fire and pine ecosystems” in Ecology and Biogeography of Pinus, Richardson D. M., Ed. (Cambridge University Press, Cambridge, 1998), pp. 193–218. [Google Scholar]
- 89.Rowe J. S., “Concepts of fire effects on plant individual and species” in The Role of Fire in Northern Cicumpolar Ecosystems, Wein R. W., MacLean D. A., Eds. (John Wiley and Sons, New York, 1983), pp. 135–154. [Google Scholar]
- 90.Maddison W., Maddison D., Mesquite: A Modular System for Evolutionary Analysis. Version 3.6.1 (2019). Available at http://www.mesquiteproject.org. Accessed 15 September 2020.
- 91.Rabosky D. L., et al., BAMMtools: An R package for the analysis of evolutionary dynamics on phylogenetic trees. Methods Ecol. Evol. 5, 701–707 (2014). [Google Scholar]
- 92.Plummer M., Best N., Cowles K., Vines K., CODA: Convergence diagnosis and output analysis for MCMC. R News 6, 7–11 (2006). [Google Scholar]
- 93.Revell L. J., phytools: An R package for phylogenetic comparative biology (and other things). Methods Ecol. Evol. 3, 217–223 (2012). [Google Scholar]
- 94.Sun M., et al., Recent accelerated diversification in rosids occurred outside the tropics. Nat. Commun. 11, 3333 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Blomberg S. P., Garland T. Jr, Ives A. R., Testing for phylogenetic signal in comparative data: Behavioral traits are more labile. Evolution 57, 717–745 (2003). [DOI] [PubMed] [Google Scholar]
- 96.Pagel M., Inferring the historical patterns of biological evolution. Nature 401, 877–884 (1999). [DOI] [PubMed] [Google Scholar]
- 97.Wickham H., Ggplot2: Elegant Graphics for Data Analysis (Springer-Verlag, New York, 2016). [Google Scholar]
- 98.Jin W. T., et al., Phylogenomic and ecological analyses reveal the spatiotemporal evolution of global pines. Dryad. 10.5061/dryad.pc866t1nq. Deposited 16 March 2021. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Transcriptome data have been deposited in the NCBI database under BioProject accession no. PRJNA703422. Alignments, phylogenetic trees, and other files generated during analyses have been deposited at Dryad (https://doi.org/10.5061/dryad.pc866t1nq) (98).