Significance
How distinct body regions form along the anterior–posterior axis in vertebrate embryos is a fascinating and incompletely understood developmental process. FAM3B/PANDER is a secreted protein involved in glucose metabolism and type 2 diabetes pathogenesis in mammals, but its receptor has been unknown. Here, we report that FAM3B binds to transmembrane fibroblast growth factor receptors (FGFRs) and activates their downstream signaling pathway. In frog embryos, gain-of-function of FAM3B impairs head development and induces ectopic tail-like structures, whereas loss-of-function of FAM3B promotes head development. FGFR is required downstream of FAM3B for head-to-tail patterning. Our results reveal that FAM3B functions by activating the FGFR pathway in frog embryos and mammalian cells and shed light on its possible role in human diseases.
Keywords: anterior–posterior patterning, embryonic development, Xenopus laevis, FGF signaling, FAM3B cytokine
Abstract
Fibroblast growth factor (FGF)/extracellular signal-regulated kinase (ERK) signaling plays a crucial role in anterior–posterior (A–P) axial patterning of vertebrate embryos by promoting posterior development. In our screens for novel developmental regulators in Xenopus embryos, we identified Fam3b as a secreted factor regulated in ectodermal explants. Family with sequence similarity 3 member B (FAM3B)/PANDER (pancreatic-derived factor) is a cytokine involved in glucose metabolism, type 2 diabetes, and cancer in mammals. However, the molecular mechanism of FAM3B action in these processes remains poorly understood, largely because its receptor is still unidentified. Here we uncover an unexpected role of FAM3B acting as a FGF receptor (FGFR) ligand in Xenopus embryos. fam3b messenger RNA (mRNA) is initially expressed maternally and uniformly in the early Xenopus embryo and then in the epidermis at neurula stages. Overexpression of Xenopus fam3b mRNA inhibited cephalic structures and induced ectopic tail-like structures. Recombinant human FAM3B protein was purified readily from transfected tissue culture cells and, when injected into the blastocoele cavity, also caused outgrowth of tail-like structures at the expense of anterior structures, indicating FGF-like activity. Depletion of fam3b by specific antisense morpholino oligonucleotides in Xenopus resulted in macrocephaly in tailbud tadpoles, rescuable by FAM3B protein. Mechanistically, FAM3B protein bound to FGFR and activated the downstream ERK signaling in an FGFR-dependent manner. In Xenopus embryos, FGFR activity was required epistatically downstream of Fam3b to mediate its promotion of posterior cell fates. Our findings define a FAM3B/FGFR/ERK-signaling pathway that is required for axial patterning in Xenopus embryos and may provide molecular insights into FAM3B-associated human diseases.
In the amphibian Xenopus laevis the anterior–posterior (A–P) axis is determined by concerted actions of maternal and zygotic factors after fertilization (1). Patterning along the A–P axis during the blastula and gastrula stages is dictated by several posteriorizing signals that include fibroblast growth factor (FGF), Wnt, and retinoic acid. These signals promote posterior cell fates while their graded inhibition specifies anterior cell fates (2–4). The broad regions of the A–P axis (head, trunk, and tail) are established by the end of gastrulation and are further elaborated during the somitogenesis stage (2).
FGF signaling is initiated by the binding of ligands to fibroblast growth factor receptor (FGFR) tyrosine kinase receptors. Tyrosine kinase receptor dimerization triggers an intracellular signaling cascade that ultimately leads to MAPK/ERK (mitogen-activated protein kinase/extracellular signal-regulated kinase) activation (5). Activated ERK phosphorylates a myriad of transcription factors to regulate gene expression (5). FGF signaling controls many critical processes during vertebrate early embryogenesis, including gastrulation, mesoderm formation, and A–P axis specification (6). In Xenopus embryos, inhibition of FGF signaling by overexpression of the dominant-negative mutant FGFR1 or treating embryos with the FGFR pharmacological inhibitor SU5402 inhibits trunk and tail development (7–9). Conversely, activation of FGF signaling by injection of FGF4 DNA causes microcephaly and induces ectopic tail-like structures (10, 11).
In the present study, we identify family with sequence similarity 3 member B (FAM3B)—also known as PANDER (pancreatic-derived factor) in the diabetes field—as an FGFR ligand. FAM3B belongs to a family of cytokines consisting of four members, whose cellular functions and mechanisms of action remain incompletely understood (12, 13). FAM3B is implicated in multiple biological processes, among which the best-characterized one is its role as a hormone in glucose and lipid metabolism (13). FAM3B/PANDER protein is secreted together with insulin by pancreatic islet β-cell granules, where its secretion is induced by glucose and is thought to be important in the development of type 2 diabetes (14–19). In knockout mice, Pander mutants have enhanced glucose tolerance, higher body weight, and lower glucose fasting levels due to decreased hepatic glucose production (14, 18). Moreover, FAM3B expression is associated with the progression of multiple types of cancer (20–23). However, the molecular mechanism of FAM3B in these disorders is largely unknown since its receptor has remained unidentified.
In this study we report that FAM3B acts as a ligand of FGF receptor signaling in both Xenopus embryos and mammalian cells. FAM3B binds to FGFR and induces downstream ERK activation. In Xenopus embryos, fam3b is expressed maternally and uniformly in the early embryo and then in the epidermis and regulates A–P axial patterning through the activation of the FGFR/ERK pathway.
Results
Xenopus fam3b Encodes a Secreted Protein.
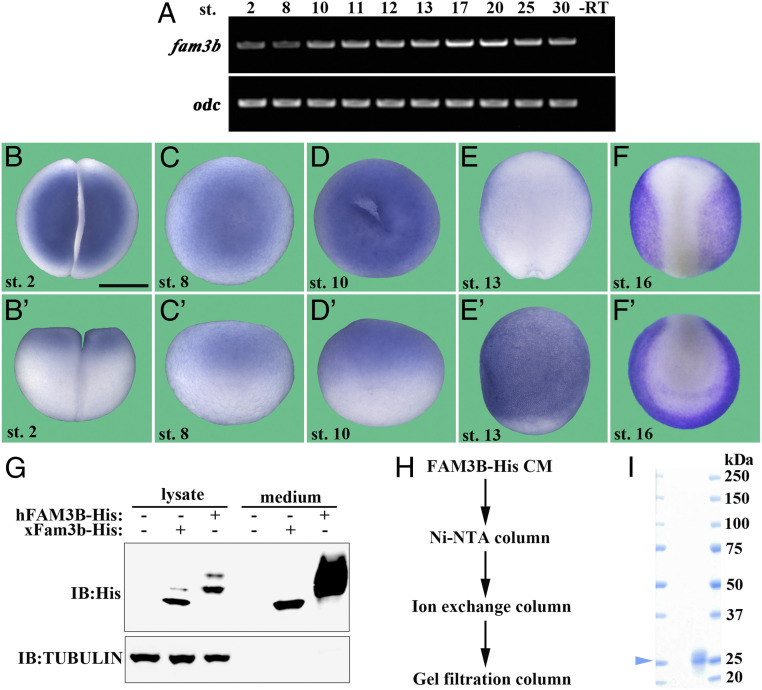
The fam3b gene came to our attention during RNA sequencing (RNA-seq) studies screening for novel regulators of embryonic patterning in Xenopus by various methods (24, 25). X. laevis is allotetraploid (26) and contains two fam3b genes, namely, fam3b.l (longer chromosome) and fam3b.s (short chromosome), which encode two proteins sharing 86% amino acid sequence identity. Our RNA-seq results indicated that fam3b.l was more abundantly expressed than fam3b.s, and therefore we focused our research on fam3b.l (fam3b hereafter). The fam3b messenger RNA (mRNA) levels were high in animal cap ectodermal explants that develop into epidermis and low in dissociated animal caps that develop into neural tissue (SI Appendix, Fig. S1A). Expression was also higher in ventral half-embryos than in dorsal halves, suggesting that it is potentially a ventral or epidermal gene (SI Appendix, Fig. S1B). We then analyzed the spatial and temporal expression pattern of fam3b. RT-PCR revealed that fam3b mRNA was expressed maternally and then showed gradually stronger expression at gastrula to neurula stages, followed by weaker expression at the tailbud stage (Fig. 1A). Whole-mount in situ hybridization (WISH) showed that fam3b mRNA was expressed diffusely throughout the embryo at the two-cell and early stages and in the epidermis at neurula and tailbud stages (Fig. 1 B–F' and SI Appendix, Fig. S1 C–E''). Analysis of protein sequences indicated that FAM3B protein is highly conserved in vertebrates with mouse FAM3B and Xenopus Fam3b proteins sharing 85 and 63% identities to human FAM3B protein, respectively, and mammalian FAM3B homologs harbor a signal peptide with two reported cleavage sites and five conserved cysteine residues (SI Appendix, Fig. S1F) (27). In human FAM3B protein, Cys63-Cys91 and Cys69-Cys229 form two disulfide bridges that are essential for its biological activity (27).
Fig. 1.
Characterization of the Xenopus fam3b gene and purification of recombinant human FAM3B-His protein. (A) RT-PCR assay showing fam3b expression across different developmental stages. odc was used as a loading control. –RT served as a negative control. (B–F') WISH of fam3b showing its diffuse localization at two-cell (B and B'), blastula (C and C'), and gastrula (D and D') stages (in Xenopus WISH, uniform mRNAs appear stronger in the animal pole due to the accumulation of large yolk platelets in the vegetal pole). At neurula stages, fam3b is localized to the epidermis (E and F'). (Scale bar, 500 μm.) (G) Xenopus fam3b encodes a secreted protein. HEK293T cells were transfected with His-tagged human FAM3B or Xenopus Fam3b constructs. Twenty-four hours after transfection, cells were cultured in serum-free medium for further 20 h. The lysate and medium were subjected to immunoblotting with the indicated antibodies. α-Tubulin served as a loading control and was detected only in the lysate and not in the medium. (H) Flowchart showing purification of His-tagged human FAM3B protein from CM generated from transfected HEK293T cells (Materials and Methods). (I) SDS-PAGE and Coomassie Blue staining showing the purity of recombinant human FAM3B-His protein. Arrowhead indicates the molecular weight of FAM3B protein.
To ascertain that Xenopus Fam3b (xFam3b) and human FAM3B (hFAM3B) were secreted proteins, we analyzed the culture medium of HEK293T cells transfected with carboxyl-terminal His-tagged forms of xFam3b and hFAM3B constructs. Western blots confirmed that both proteins were secreted into the extracellular milieu (Fig. 1G). Fortunately for us, human FAM3B protein was copiously secreted (Fig. 1G, last lane). Using Nickel, ion exchange, and gel filtration columns (Fig. 1H), hFAM3B could be purified to near homogeneity at concentrations suitable for microinjection experiments in Xenopus embryos (Fig. 1I). This purified hFAM3B preparation greatly facilitated our subsequent investigations.
FAM3B Causes Microcephaly and Induces Ectopic Tail-like Structures.
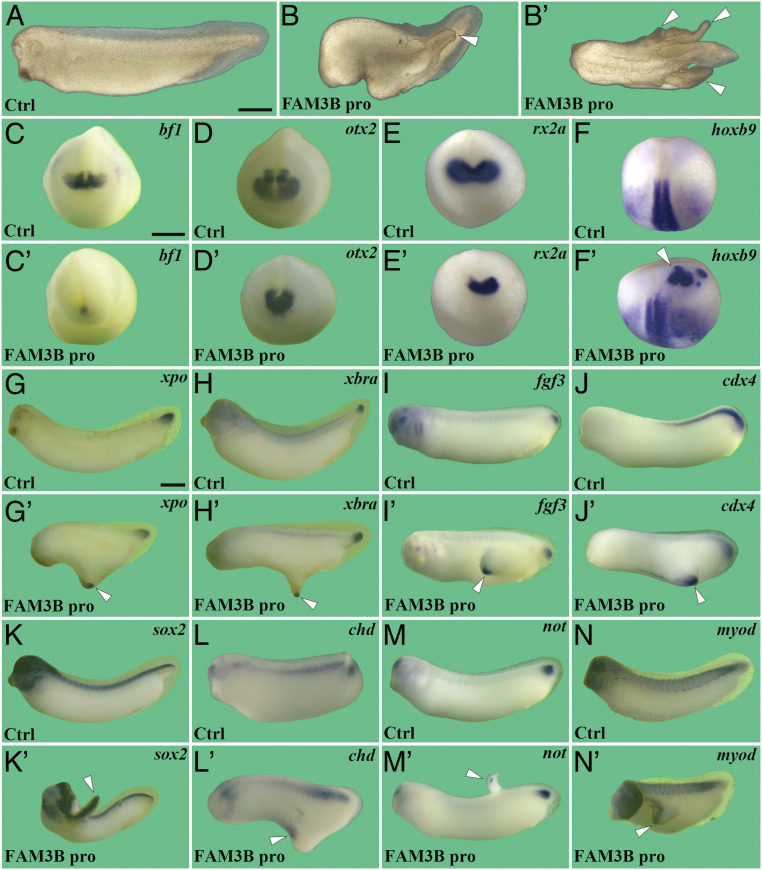
To investigate the physiological relevance of fam3b during early embryonic development, we first performed gain-of-function experiments. Xenopus fam3b mRNA was injected into the animal region of two ventral blastomeres of four-cell-stage Xenopus embryos (SI Appendix, Fig. S2A). Its overexpression led to microcephaly, shortened A–P axes, and induced ectopic tail-like structures, usually a single one, at the expense of head tissues (SI Appendix, Fig. S2 B and C). We next tested the biological activity of purified His-tagged recombinant human FAM3B (rhFAM3B) protein by injecting it into the blastocoele cavity of blastula stage Xenopus embryos (SI Appendix, Fig. S2A'). Injection of rhFAM3B protein (40 nL at 15 µM) resulted in loss of head structures, including cement gland and eyes, and induced multiple tail-like structures at high frequencies (Fig. 2 A and B'). These phenotypic abnormalities were similar to, although stronger than, those caused by Xenopus fam3b mRNA, indicating the functional conservation of this cytokine between Xenopus and human. The formation of multiple tails, as opposed to a single one, may be due to the wider surface area that receives the signal in the blastula cavity. Since embryos injected with rhFAM3B protein exhibited higher phenotype penetrance than those injected with Xenopus fam3b mRNA (97 versus 37% extra tails), we used microinjected rhFAM3B protein to further define the physiological role of FAM3B.
Fig. 2.
FAM3B protein microinjection inhibits head development and induces ectopic tail-like structures. BSA or rhFAM3B protein (40 nL at 15 μM) was injected into the blastocoele cavity of blastula-stage embryos. Embryos were cultured until the neurula or tailbud stages and subjected to phenotypic analysis or WISH as indicated. (A and B') rhFAM3B protein causes microcephaly and short A–P axis and induces ectopic tail-like structures. (A) Lateral view of control. (B) Lateral view. (B') Dorsal view. Arrowheads indicate ectopic tail-like structures. (C–F') rhFAM3B protein decreases expression of the forebrain marker bf1 (C and C'), forebrain–midbrain marker otx2 (D and D'), and eye marker rx2a (E and E') and induces ectopic expression (arrowhead) of the spinal cord marker hoxb9 (F and F'). (G–N') rhFAM3B protein-induced tail-like structures express tailbud markers xpo (G and G'), xbra (H and H'), fgf3 (I and I'), and cdx4 (J and J'), pan neural marker sox2 (K and K'), chordoneural hinge markers chd (L and L') and not (M and M') and somite marker myod (N and N'). Numbers of embryos analyzed were as follows: A, n = 74, 100%; B and B', n = 128, 97% with ectopic tail phenotype; C, n = 37, 100%; C', n = 44, 93%; D, n = 35, 100%; D', n = 42, 97%; E, n = 26, 100%; E', n = 36, 94%; F, n = 32, 100%; F', n = 40, 95%; G, n = 29, 100%; G', n = 41, 95%; H, n = 30, 100%; H', n = 33, 93%; I, n = 27, 100%; I', n = 39, 92%; J, n = 36, 100%; J', n = 40, 92%; K, n = 41, 100%; K', n = 45, 97%; L, n = 34, 100%; L', n = 47, 95%; M, n = 38, 100%; M', n = 42, 88%; N, n = 35, 100%; N', n = 48, 95%. (Scale bars for A and B', C–F', and G–N' all indicate 500 μm.)
We injected rhFAM3B protein into the blastocoele cavity at blastula stage and assayed for various molecular markers at gastrula, neurula and tailbud stages. At gastrula stage, rhFAM3B protein had no effect on expression of the dorsal mesodermal markers chd and gsc (SI Appendix, Fig. S2 D–G). In contrast, rhFAM3B protein, as well as human FGF2 and FGF8B proteins (28, 29), potently expanded the expression of the pan-mesodermal marker xbra in whole embryos (SI Appendix, Fig. S2 J–M) and induced xbra expression in animal cap explants (SI Appendix, Fig. S2N), indicating that FAM3B has mesoderm-inducing activity, but has no effect on the Spemann organizer formation (30, 31). Unlike human FGF2, and to a lesser degree FGF8B, rhFAM3B protein failed to elicit animal cap elongation at concentrations that were sufficient to induce xbra expression in animal caps (SI Appendix, Fig. S2 O–R). There are precedents for proteins that can induce xbra expression without animal cap elongation (32, 33). FAM3B behaves like FGF2 and FGF8B during mesoderm induction, but in general higher concentrations are required.
At the neurula stage, rhFAM3B protein inhibited the expression of anterior neural markers bf1 (forebrain marker) (Fig. 2 C and C'), otx2 (forebrain–midbrain marker) (Fig. 2 D and D'), and rx2a (eye marker) (Fig. 2 E and E') and induced ectopic expression of the posterior marker hoxb9 (Fig. 2 F and F', arrowhead). In contrast, rhFAM3B protein had no effect on the expression of the neural differentiation marker N-tubulin (SI Appendix, Fig. S2 H and I). These results, together with the microcephaly phenotype, indicate that FAM3B promotes posterior patterning.
To examine the nature of the tail-like structures induced by rhFAM3B protein, we analyzed the expression of tissue-specific marker genes in these structures at the tailbud stage. WISH analysis showed that tailbud markers (xpo, xbra, fgf3, and cdx4) (Fig. 2 G–J'), a pan-neural marker (sox2) (Fig. 2 K and K'), chordoneural hinge markers (chd and not) (Fig. 2 L and M'), and a somite marker (myod) (Fig. 2 N and N') were all expressed in the ectopic tail-like structures (indicated by arrowheads), indicating that they were well-patterned ectopic tails.
Taken together, these results suggest that the function of FAM3B is evolutionarily conserved and that its overexpression is sufficient to impair head development and induce ectopic posterior structures with tail characteristics.
Knockdown of fam3b Promotes Anterior Development.
To study the loss-of-function of fam3b, we designed translation-blocking antisense morpholino oligonucleotides (MOs) targeting fam3b.l and fam3b.s (SI Appendix, Fig. S3A). When injected into embryos, fam3b.l MO and fam3b.s MO efficiently blocked the translation of fam3b.l-Flag and fam3b.s-Flag mRNA, respectively (SI Appendix, Fig. S3 B and C).
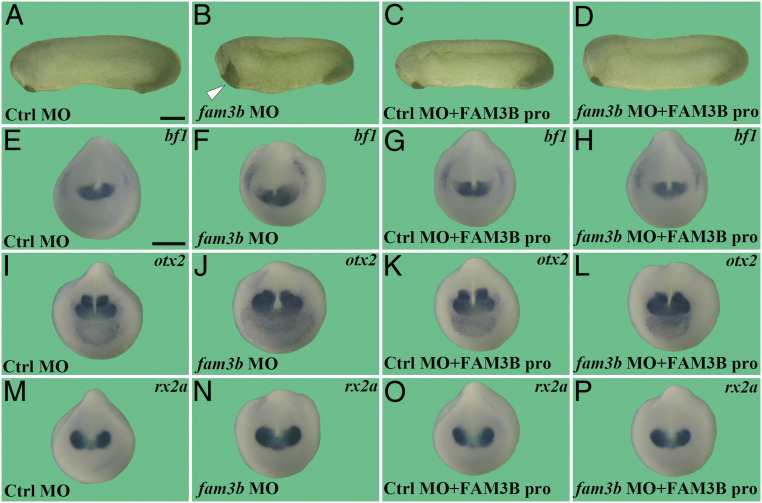
We next examined the knockdown phenotype in vivo by microinjecting fam3b.l and fam3b.s MOs in combination into embryos at the one-cell stage. fam3b knockdown did not significantly change the expression of xbra or hoxb9 (SI Appendix, Fig. S3 D–G). However, compared with control morphants, fam3b morphants exhibited macrocephaly, expanded cement gland, and shortening of the A–P axis (Fig. 3 A and B). Importantly, these defects could be rescued by injection of a low dose (lacking any phenotypic effects on its own) of rhFAM3B protein (40 nL at 3 µM) into the blastocoele cavity (Fig. 3 C and D), indicating that these phenotypes were caused specifically by fam3b depletion. In addition, this indicated that human FAM3B is functionally interchangeable with the endogenous Xenopus Fam3b protein. In line with these phenotypic observations, knockdown of fam3b increased the expression of the anterior neural markers otx2, bf1, and rx2a, which could all be rescued by injection of a low dose of rhFAM3B protein into the blastocoele cavity (Fig. 3 E–P).
Fig. 3.
Knockdown of fam3b expands anterior tissues, and the phenotype is rescued by low amounts of rhFAM3B protein. Embryos were first injected with control MO (120 ng) or fam3b MOs (60 ng fam3b.l MO and 60 ng fam3b.s MO) at the one-cell stage and subsequently BSA or rhFAM3B protein (40 nL at 3 μM) were injected into the blastocoele cavity at blastula stage, as indicated in the panels. Embryos were cultured until tailbud stage for phenotypic analysis or neurula stage for WISH with the indicated neural markers. (A–D) fam3b knockdown leads to enlarged heads and shorter A–P axis, which was rescued by injection of rhFAM3B protein. Arrowhead in B indicates enlarged cement gland. (E–P) fam3b knockdown expands forebrain markers bf1 (E–H), forebrain–midbrain marker otx2 (I–L), and eye marker rx2a (M–P), a phenotype that was rescued by injection of rhFAM3B protein at low doses. Numbers of embryos analyzed were as follows: A, n = 85, 100%; B, n = 96, 92% with enlarged cement glands; C, n = 90, 100%; D, n = 113, 86%; E, n = 53, 100%; F, n = 55, 96%; G, n = 49, 95%; H, n = 60, 88%; I, n = 38, 100%; J, n = 43, 95%; K, n = 50, 96%; L, n = 46, 91%; M, n = 37, 100%; N, n = 47, 93%; O, n = 39, 94%; P, n = 43, 86%. (Scale bars for A–D and E–P indicate 500 μm.)
The anteriorizing effects of fam3b MOs opposed to the phenotypes caused by Xenopus fam3b mRNA and rhFAM3B protein microinjection, indicating a restrictive role of fam3b in anterior development and a supportive role in posterior development.
FGFRs Are Receptors for FAM3B Ligand and Are Required for FAM3B-Induced ERK Activation.
The phenotypes induced by injection of Xenopus fam3b mRNA or rhFAM3B protein strikingly resembled those caused by FGF/FGFR/ERK pathway activation in Xenopus development (7, 11, 34). This was a key realization. Since FAM3B is a secreted protein, we hypothesized that FAM3B might be a ligand of FGF receptor signaling and activate ERK through binding to FGFR.
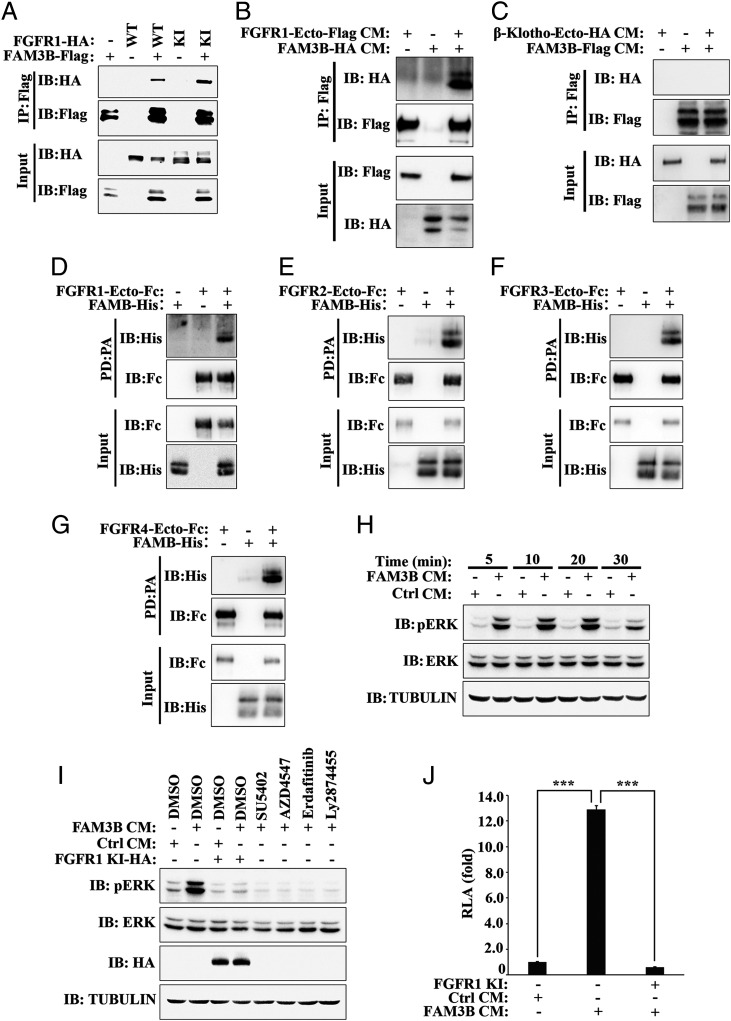
We initially tested this by microinjecting the mRNAs encoding FAM3B-Flag or full-length FGFR1-HA into different cells of Xenopus embryos after completion of the two-cell division (SI Appendix, Fig. S4A). FAM3B protein bound to full length human FGFR1 in Xenopus gastrula extracts (Fig. 4A). Similarly, a kinase-inactive mutant form of human FGFR1 (FGFR1 KI-HA), in which the ectodomain is intact (35), also bound to FAM3B (Fig. 4A).
Fig. 4.
FAM3B binds to FGFR and activates ERK through FGFR. (A) FAM3B binds to full-length FGFR1 in Xenopus embryos. Two-cell-stage embryos were injected with FAM3B-Flag mRNA (400 pg) and wild type (WT) or kinase-inactive mutant (KI) FGFR1-HA mRNA (400 pg) separately into different blastomeres. Gastrula-stage embryos were harvested for immunoprecipitation with Flag beads followed by immunoblotting with Flag and HA antibodies. Total protein expression was confirmed by immunoblotting of the input. (B) FAM3B binds to the ectodomain of FGFR1 in solution. CM for FAM3B-HA and ectodomain of FGFR1 (FGFR1-Ecto-Flag) were combined and allowed to bind, followed by immunoprecipitation with Flag beads and subsequent immunoblotting with Flag and HA antibodies. Protein expression in CM was confirmed by immunoblotting of the input. (C) FAM3B does not bind to β-Klotho ectodomain. CM for the ectodomain of β-Klotho (β-Klotho-Ecto-HA) and FAM3B-Flag were combined and allowed to bind. Total protein expression in the CM was confirmed by immunoblotting of the input. This experiment serves as a negative control for binding to the extracellular domain of FGFRs. (D–G) FAM3B binds to FGFR ectodomain in vitro. Purified rhFAM3B protein (1 μg) was incubated with Fc-tagged human FGFR ectodomain protein (1 μg), including FGFR1-Ecto-Fc (D), FGFR2-Ecto-Fc (E), FGFR3-Ecto-Fc (F), and FGFR4-Ecto-Fc (G) as indicated. Then the protein mixture was subjected to pull-down with Protein A magnetic beads and subsequent immunoblotting with Fc and His antibodies. Protein expression was confirmed by immunoblotting of the input. PD: pull-down; PA: Protein A magnetic beads. (H) FAM3B CM rapidly activates ERK. HEK293T cells were serum starved for 24 h and treated with serum-free control or FAM3B CM for the indicated times. Cells were harvested for immunoblotting with pERK and total ERK antibodies. α-Tubulin served as a loading control. (I) FAM3B CM-induced ERK activation is blocked by FGFR inhibition. HEK293T cells transfected with FGFR1 KI or not were serum starved for 24 h and pretreated with FGFR inhibitors SU5402 (20 μM), AZD4547 (1 μM), Erdafitinib (1 μM), or Ly2874455 (1 μM) for 2 h. Then cells were stimulated with serum-free control CM or FAM3B CM in the presence of these inhibitors for 20 min and harvested for immunoblotting with the indicated antibodies. α-Tubulin served as a loading control. (J) FAM3B CM activated an FGF reporter derived from the mouse Dusp6 promoter (40), which was blocked by FGFR1 inhibition. HEK293T cells were transfected with FGF Luciferase reporter together with Renilla and FGFR1 KI as indicated. Twenty hours after transfection, cells were serum starved for 20 h, followed by treatment with serum-free control or FAM3B CM for 8 h. Experiments were performed in triplicate. Data are mean ± SD. Statistical significance was assessed by unpaired two-tailed Student’s t test. ***P < 0.001. RLA, Relative Luciferase Activity.
Microinjection of rhFAM3B protein, like FGF2 or FGF8B proteins, into the blastocoele cavity activated endogenous ERK phosphorylation at the early gastrula stage (SI Appendix, Fig. S4B). rhFAM3B protein-induced ERK activation was also observed at the midgastrula and neurula stages, and this ERK activation was blocked by SU5402, indicating that it was dependent on FGFR activity (SI Appendix, Fig. S4C). ERK activation also took place in the presence of the protein synthesis inhibitor cycloheximide (CHX) (SI Appendix, Fig. S4C) at a concentration previously shown to inhibit protein synthesis in Xenopus embryos (36, 37), suggesting that the effect of rhFAM3B protein on ERK was transcription independent.
Using conditioned medium (CM) from transfected HEK293T cells, we showed that FAM3B bound to the ectodomain of FGFR1 (Fig. 4B) (5). As a negative control, we used the ectodomain of β-Klotho, a coreceptor required for endocrine FGFs binding to FGFR (5), which failed to bind (Fig. 4C). Since FGF growth factors signal through multiple FGFRs (5), we tested whether FAM3B could directly bind to all four FGFRs in vitro. Indeed, purified rhFAM3B protein could be pulled down by commercially available FGFR-Ecto-Fc protein fusions, but not by Fc protein, indicating that the binding is specific and FAM3B is a bona fide ligand for FGFR (Fig. 4 D–G and SI Appendix, Fig. S4D). In this experiment, rhFAM3B-His and each of the four FGFR-Ecto-Fc proteins were purified, which indicates that the ligand–receptor interaction is direct, without requiring any additional cofactors.
In HEK293T cells, addition of human FAM3B CM, but not control CM, robustly increased ERK phosphorylation within 5 min, and ERK activation was sustained for 30 min (Fig. 4H). Importantly, FAM3B CM-induced ERK activation could be abrogated by treatment with SU5402 or three other FGFR inhibitors: AZD4547, Erdafitinib, and Ly2874455 (Fig. 4I) (38, 39). Likewise, transfection with FGFR1 KI (35) also blocked FAM3B CM-stimulated ERK activation (Fig. 4I). ERK was also activated by transfection of human FAM3B DNA into HEK293T cells, while overexpression of a mutant human FAM3B (C91A/C229A), which fails to form disulfide bridges, had no effect (SI Appendix, Fig. S4E). ERK activation by transfected human FAM3B DNA, as was the case in protein addition experiments, was also blocked by the FGFR inhibitors SU5402, AZD4547, Erdafitinib, or Ly2874455 (SI Appendix, Fig. S4F). Finally, human FAM3B CM activated a FGF Luciferase reporter gene derived from the mouse Dusp6 promoter (40), and this was abrogated by overexpression of dominant-negative FGFR1 KI (Fig. 4J).
Taken together, these results in Xenopus embryos and mammalian cells indicate that FAM3B is a direct ligand for FGFRs and that FAM3B activates ERK via FGFRs.
FAM3B Promotes Posterior Development through FGFR in Xenopus Embryos.
Finally, we investigated in vivo whether FAM3B could regulate A–P axial patterning through FGFRs. It has been shown that zebrafish embryos injected with FGFR1 KI mRNA or Xenopus embryos treated with SU5402 display reduced posterior structures (8, 41). In accordance with these previous results, Xenopus embryos injected with FGFR1 KI mRNA or incubated with SU5402 or Erdafitinib or Ly2874455 showed anteriorization (SI Appendix, Fig. S5 A, B, and E–H), phenocopying fam3b morphants. The formation of ectopic tail-like structures and microcephaly induced by rhFAM3B protein injection into the blastocoele cavity was efficiently blocked by injection of FGFR1 KI mRNA or treatment with SU5402, Erdafitinib, or Ly2874455 (SI Appendix, Fig. S5; compare panels C to D and I to J–L). Consistent with these phenotypes and our biochemical analyses in mammalian cells, overexpression of FGFR1 KI (SI Appendix, Fig. S5M) or treatment with SU5402, Erdafitinib, or Ly2874455 reversed rhFAM3B protein-induced ERK activation in Xenopus embryos (SI Appendix, Fig. S5N).
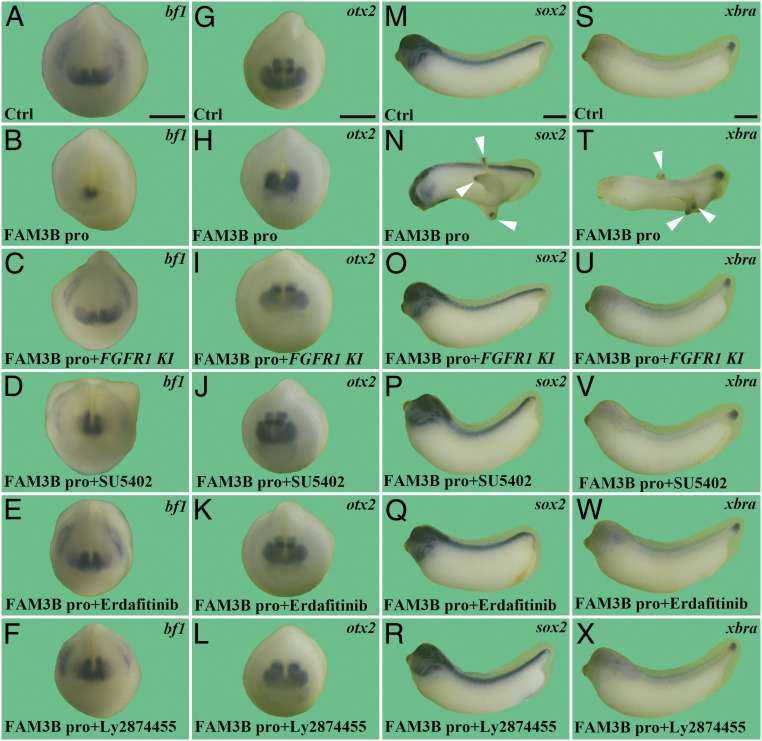
To confirm the epistatic interaction between FAM3B and FGFR in vivo, we examined whether inhibition of FGFR activity could reverse the microcephaly induced by FAM3B protein (Fig. 5 A, B, G, and H) using anterior molecular markers. We found that FGFR1 KI mRNA injection or SU5402, Erdafitinib, or Ly2874455 treatment significantly reversed FAM3B-mediated reduction of bf1 (Fig. 5 A–F) and otx2 (Fig. 5 G–L). In addition, the induction of ectopic tails marked by sox2 (Fig. 5 M–R) and xbra (Fig. 5 S–X) was blocked by dominant-negative FGFR1 KI mRNA or the FGFR inhibitors SU5402, Erdafitinib, or Ly2874455.
Fig. 5.
FAM3B induces (A–P) axial patterning defects through FGFR in Xenopus embryos. Embryos were injected with FGFR1 KI mRNA (100 pg) at the one-cell stage and rhFAM3B protein (40 nL at 15 μM) into the blastocoele cavity at the blastula stage, followed by treatment with SU5402 (20 μM), Erdafitinib (2 μM), or Ly2874455 (20 nM) as indicated. Neurula or tailbud stage embryos were collected for WISH with bf1 (A–F), otx2 (G–L), sox2 (M–R), and xbra (S–X). rhFAM3B protein reduces the head markers bf1 and otx2 and induces sox2 and xbra in ectopic tails, and all phenotypes were rescued by FGFR1 KI mRNA, SU5402, Erdafitinib, or Ly2874455. Arrowheads indicate ectopic tail-like structures. Note that relatively low doses of FGFR1 KI mRNA and FGFR inhibitors were used here. Higher doses of SU5402 (50 to 100 μM) have been shown to cause defective blastopore closure and severe posterior truncation in Xenopus embryos by its own (9, 45). Higher doses of FGFR1 KI mRNA, Erdafitinib, or Ly2874455 had similar blastopore closure effects as those of high dose SU5402. (Scale bars, 500 μm.) Numbers of embryos analyzed were as follows: A, n = 34, 100%; B, n = 65, 98% with microcephaly; C, n = 40, 92%; D, n = 38, 73%; E, n = 41, 85%; F, n = 39, 82%; G, n = 54, 100%; H, n = 49, 100%; I, n = 47, 93%; J, n = 37, 89%; K, n = 56, 91%; L, n = 51, 92%; M, n = 35, 100%; N, n = 63, 95%; O, n = 45, 93%; P, n = 42, 90%; Q, n = 36, 91%; R, n = 40, 95%; S, n = 43, 100%; T, n = 55, 98%; U, n = 44, 90%; V, n = 66, 87%; W, n = 49, 83%; X, n = 53, 92%.
These results indicate that the FAM3B secreted protein acts epistatically upstream of FGFR and promotes posterior development in Xenopus embryos by activating the FGFR pathway.
Discussion
This study suggests three main conclusions. First, fam3b mRNA is initially expressed diffusely in the embryo and then in the epidermis in Xenopus and encodes a secreted protein that acts as an FGFR ligand to promote posterior development. Second, human FAM3B protein is an FGFR ligand that activates the FGFR/ERK pathway through binding to FGFR in human HEK293T cells and in vivo in Xenopus embryos. Third, we provide evidence that FGFRs are the cognate receptors for FAM3B.
These studies were greatly facilitated by the observation that a human FAM3B/PANDER construct His-tagged in the carboxy-terminus was abundantly secreted by HEK293T cells, could be readily purified with Nickel columns, and had high tail-inducing activity after microinjection into the blastocoele cavity of Xenopus embryos.
FAM3B can now be added to the growing list of secreted FGF regulators. Among them, SERPINE2 and Rspo2 are secreted FGF antagonists, whereas FGFBP1, HtrA1, and Pinhead are secreted FGF agonists (11, 42–45). Unlike FAM3B, none of these agonists signals by binding directly to FGFR. The uncanny similarity between the ectopic tail phenotypes described in Xenopus embryos by the laboratory of Edgar Pera for the HtrA1 protease (11) and FAM3B provided the clue that led us to focus our attention on the FGF pathway. As shown in this study, FAM3B directly binds to FGFR and activates the downstream ERK pathway, acting as an FGF ligand. This molecular mechanism is different from those of the other FGF secreted agonists, which do not exert their effects through binding to FGFR. The secreted serine protease HtrA1 was shown to release cell-surface–bound FGFs and stimulate long-range FGF signaling by cleaving proteoglycans (11). FGFBP1 is able to release FGF ligands from extracellular matrix storage and present them to the FGFR and thus potentiates FGF signaling (42). Pinhead activates ERK in an FGFR-dependent manner, but an interaction between Pinhead and FGFR could not be detected, rending the underlying mechanism elusive (44). These different regulators may act in different contexts to tightly control FGF signaling to guarantee required signaling outcomes.
Loss-of-function morpholino experiments indicated that FAM3B/PANDER is required to activate FGF receptor signaling during A–P axial patterning of Xenopus embryos. fam3b knockdown shortened the A–P axis and expanded cephalic tissues marked by bf1 and otx2. These effects were specific, as they could be rescued by microinjecting a low dose of rhFAM3B protein into the blastocoele cavity. However, fam3b knockdown did not appear to cause posterior defects; this may be due to redundancy from other factors such as FGF3, FGF4, FGF8, HtrA1, and Pinhead (6, 11, 44) that could compensate for the loss of FAM3B during posterior development. Xenopus fam3b is expressed uniformly during cleavage stages and then in epidermis, distinct from FGF ligands that are predominantly expressed in mesodermal and neural tissues (46). We identified fam3b as a gene highly expressed in epidermal animal cap explants but expressed at very low levels in dissociated animal cap cells that develop into anterior neural tissue (25). We hypothesize that FAM3B may also play a role in skin development, although this has not been investigated here.
We note that the regulatory role of FAM3B in A–P axial patterning of Xenopus embryos does not seem to be conserved in the mouse. Fam3b/Pander knockout mice are viable and do not display early embryonic A–P development defects and instead exhibit enhanced glucose tolerance and hepatic insulin sensitivity, suggesting an important role for this gene in the development of type 2 diabetes (19). There are precedents for such drastic differences between phenotypes of mutant mice and developing Xenopus embryos. For example, Rspo2 regulates mesoderm formation via Wnt and FGF signaling in Xenopus, while this role is not conserved in mouse (45, 47, 48). Another case is that of angiopoietin-like 4 (Angptl4), a secreted protein that promotes notochord formation by antagonizing Wnt signaling in Xenopus, yet regulates triglyceride metabolism in mutant mice (49–51).
The human genome encodes 22 FGF family proteins that can be arranged into canonical FGFs, intracellular FGFs, and endocrine FGFs (5). In contrast to canonical FGFs that control cell proliferation, differentiation, and survival and intracellular FGFs that serve as cofactors for voltage-gated sodium channels and other molecules, the endocrine FGFs play major roles in adult homeostasis, regulating phosphate, bile acid, and carbohydrate and lipid metabolism (5, 52). In this sense, FAM3B might be considered as a member of the endocrine FGF-like signals due to its role in glucose and lipid metabolism in mammals (13). However, the molecular function of FAM3B is different from those of endocrine FGFs, for we did not detect an interaction between FAM3B and β-Klotho, which acts as a coreceptor for endocrine FGFs binding to FGFR (5). It is currently unknown whether a coreceptor modulates the binding of FAM3B to FGFR, which will require further investigation. However, since we have shown that purified recombinant FGFR-Ecto-Fc and purified FAM3B bind to each other in vitro, a coreceptor should not be a requirement. Further, FAM3B, as well as other members of FAM3 family has been proposed to represent a class of signaling molecules with a unique structure that is distinct from those of other cytokines and growth factors (27, 53).
While here we have demonstrated physiological relevance by showing that the FAM3B/FGFR/ERK pathway is crucial for A–P axial patterning during Xenopus embryonic development, our findings probably have broader pathophysiological implications. First, FAM3B has been suggested to be a causal gene for type 2 diabetes. In humans and mice, increased circulating FAM3B/PANDER levels are associated with hyperglycemia, pancreatic β-cell dysfunction, and insulin resistance (54–58). Second, FAM3B has been reported to act as an oncogene that, when expressed, promotes the progression of several types of cancer, including colon cancer, prostate cancer, esophageal cancer, and gastric cancer (20–23). However, the mechanism of action of FAM3B in these disorders remained elusive and controversial largely due to the unknown identity of the receptor for FAM3B.
Here we have identified FGFRs 1 to 4 as cognate receptors for FAM3B and unveiled that FAM3B activates the FGFR/ERK pathway. Given the prominent role of FGF receptor signaling in type 2 diabetes and cancer (5, 59), our findings may shed light on the pathogenic mechanism of FAM3B in these disorders. Moreover, the FGFR inhibitors AZD4547, Erdafitinib, and Ly2874455, which are in clinical trials for FGF-addicted cancer (38, 39), efficiently blocked FAM3B-induced ERK activation, raising the possibility that these inhibitors may also serve as potential drugs for FAM3B-associated human diseases. Our findings provide a mechanistic insight into the molecular function of the previously orphan ligand FAM3B and may open therapeutic approaches for type 2 diabetes and cancer associated with this versatile cytokine.
Materials and Methods
Embryo Manipulations.
X. laevis frogs were purchased from the Nasco Company. Embryos were obtained through in vitro fertilization and cultured in 0.1× Marc’s Modified Ringers (25) and staged according to Nieuwkoop and Faber (60).
Cloning.
Full-length X. laevis fam3b.l and fam3b.s were cloned from complementary DNA (cDNA) of stage 13 X. laevis embryos. Human FAM3B cDNA clone was obtained from GeneScript (catalog no. OHu15165). Epitope-tagged constructs for X. laevis fam3b.l, fam3b.s, and human FAM3B were generated by inserting the full length of the respective genes into pCS2 vectors containing carboxyl-terminal HA or Flag or 6× His tags suitable for both mRNA in vitro synthesis and mammalian expression.
mRNA Synthesis and Morpholinos.
For in vitro mRNA synthesis, pCS2-fam3b.l-HA, pCS2-fam3b.l-3×Flag, pCS2-fam3b.s-3×Flag, pCS2-FAM3B-3×Flag, pCS2-FGFR1-HA, and pCS2-FGFR1-KI-HA were linearized with Not1 and transcribed with SP6 RNA polymerase using the mMESSAGE mMACHINE SP6 Transcription Kit (Thermo Scientific, catalog no. AM1340). Translation-blocking fam3b MOs were designed and synthesized by Gene Tools. The fam3b.l MO was 5′ TCGACAGGAACCTGAAGTTGACCAT 3′. The fam3b.s MO was 5′ TAGGCAGGAACCGTAAGTTGACCAT 3′. The amounts of mRNAs or MOs injected per embryo are indicated in the figure legends.
Protein Purification.
Serum-free human FAM3B CM derived from HEK293T cells transfected with pCS2-FAM3B-His construct was first subjected to purification with HisPur Ni-NTA Spin Purification Kit (Thermo Scientific, catalog no. 88228). Briefly, the column was equilibrated with Equilibration Buffer (1× phosphate-buffered saline [PBS], 10 mM imidazole). FAM3B CM was added to the column and allowed to go through by flow gravity. The column was washed three times with Washing Buffer (1× PBS, 25 mM imidazole). The protein was eluted with Elution Buffer (1× PBS, 250 mM imidazole). Imidazole was removed with Zeba Spin Desalting Columns (Thermo Scientific, catalog no. 89889). FAM3B-His protein was further purified with ion exchange chromatography using Mono S cation exchange chromatography column 5/50 GL (Cytiva, catalog no. 17516801) and gel filtration chromatography using Superdex 75 Increase 10/300 GL (Cytiva, catalog no. 29148721) following the manufacturer’s instructions. FAM3B-His protein was concentrated using Amicon Ultra centrifugal filter-10K (Millipore, catalog no. UFC501024). Protein concentration was measured with the BCA Protein Assay Kit (Thermo Scientific, catalog no. PI23225), and the purity of the protein was analyzed by sodium dodecyl sulfate/polyacrylamide gel electrophoresis (SDS/PAGE) and Coomassie blue staining. Finally, purified recombinant human FAM3B-His protein in PBS (pH 7.4) aliquots were snap-frozen in liquid nitrogen and stored at −80 °C.
Protein Injection.
Xenopus embryos at blastula stage were injected into the blastocoele cavity with 40 nL protein solution of 0.1% bovine serum albumin (BSA) alone as controls or together with purified recombinant His-tagged human FAM3B (rhFAM3B) protein or human FGF2 protein (Sino Biological, catalog no. 10014-HNAE) or human FGF8B protein (Sino Biological, catalog no. 16277-HNAE) as described (61).
In Vitro Pull-Down Assay.
Purified recombinant human FAM3B-His protein was incubated with commercially available purified recombinant human IgG1 Fc or FGFR-Ecto-Fc protein (the ectodomain of human FGFRs fused to the Fc region of human IgG1 at the C terminus) from Sino Biological (IgG1 Fc, catalog no. 10702-HNAH; FGFR1-Ecto-Fc, catalog no. 16482-H02H; FGFR2-Ecto-Fc, catalog no. 10824-H03H; FGFR3-Ecto-Fc, catalog no. 16044-H02H; FGFR4-Ecto-Fc, catalog no. 10538-H02H) in binding buffer containing 1× PBS and 0.1% Nonidet P-40 with top-to-bottom rotation at 4 °C for 4 h. Except for a small fraction of input, the mixture was further incubated with Protein A superparamagnetic beads (Invitrogen, catalog no. 10-001-D) with rotation at 4 °C for 1 h. Then the beads were washed with binding buffer four times (10 min per wash without changing tubes) and finally denatured in 2× loading buffer at 100 °C for 5 min followed by SDS-PAGE and Western blot.
Chemical Inhibitors.
FGFR inhibitors SU5402 (catalog no. S7667), AZD4547 (catalog no. S2801), Erdafitinib (catalog no. S8401), and Ly2874455 (catalog no. S7057) were purchased from Selleckchem. CHX was purchased from Cell Signaling Technology (catalog no. 2112S). These chemical inhibitors were used to treat cells and Xenopus embryos at concentrations indicated in the figure legends.
Statistical Analyses.
Statistical significance was assessed by a paired one-tailed Student’s t test in fam3b RPKM level comparison and an unpaired two-tailed Student’s t test in FGF reporter assay. Statistically significant results in all figures are indicated as *P < 0.05 and ***P < 0.001.
Additional methods including additional cloning, whole-mount in situ hybridization, RT-PCR, protein sequence alignment, cell culture and transfection, conditioned medium preparation and secretion assay, Western blots, animal cap, immunoprecipitation, and luciferase reporter assays are available in SI Appendix, SI Materials and Methods.
Supplementary Material
Acknowledgments
This work was supported by grants from the “Young Talent Support Plan” of Xi’an Jiaotong University (YX6J005 to Y.D.); the National Natural Science Foundation of China (32070803 to Y.D. and 31970756 to Q.T.); the Ministry of Science and Technology of China (2019YFA0801403 to Q.T.); and the Beijing Advanced Innovation Center for the Structural Biology (to Q.T.). Work at the University of California Los Angeles (UCLA) was funded by grants from the UCLA Jonsson Cancer Center (P20CA01042); the Cancer Research Coordination Committee (C21CR2039); the Norman Sprague Fund for Molecular Oncology, and the Howard Hughes Medical Institute (to E.M.D.R.).
Footnotes
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2100342118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or SI Appendix.
References
- 1.Heasman J., Patterning the early Xenopus embryo. Development 133, 1205–1217 (2006). [DOI] [PubMed] [Google Scholar]
- 2.Kimelman D., Martin B. L., Anterior-posterior patterning in early development: Three strategies. Wiley Interdiscip. Rev. Dev. Biol. 1, 253–266 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tuazon F. B., Mullins M. C., Temporally coordinated signals progressively pattern the anteroposterior and dorsoventral body axes. Semin. Cell Dev. Biol. 42, 118–133 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carron C., Shi D. L., Specification of anteroposterior axis by combinatorial signaling during Xenopus development. Wiley Interdiscip. Rev. Dev. Biol. 5, 150–168 (2016). [DOI] [PubMed] [Google Scholar]
- 5.Ornitz D. M., Itoh N., The fibroblast growth factor signaling pathway. Wiley Interdiscip. Rev. Dev. Biol. 4, 215–266 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dorey K., Amaya E., FGF signalling: Diverse roles during early vertebrate embryogenesis. Development 137, 3731–3742 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Amaya E., Musci T. J., Kirschner M. W., Expression of a dominant negative mutant of the FGF receptor disrupts mesoderm formation in Xenopus embryos. Cell 66, 257–270 (1991). [DOI] [PubMed] [Google Scholar]
- 8.Delaune E., Lemaire P., Kodjabachian L., Neural induction in Xenopus requires early FGF signalling in addition to BMP inhibition. Development 132, 299–310 (2005). [DOI] [PubMed] [Google Scholar]
- 9.Fletcher R. B., Harland R. M., The role of FGF signaling in the establishment and maintenance of mesodermal gene expression in Xenopus. Dev. Dyn. 237, 1243–1254 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pownall M. E., Tucker A. S., Slack J. M., Isaacs H. V., eFGF, Xcad3 and Hox genes form a molecular pathway that establishes the anteroposterior axis in Xenopus. Development 122, 3881–3892 (1996). [DOI] [PubMed] [Google Scholar]
- 11.Hou S., Maccarana M., Min T. H., Strate I., Pera E. M., The secreted serine protease xHtrA1 stimulates long-range FGF signaling in the early Xenopus embryo. Dev. Cell 13, 226–241 (2007). [DOI] [PubMed] [Google Scholar]
- 12.Zhang X., et al., FAM3 gene family: A promising therapeutical target for NAFLD and type 2 diabetes. Metabolism 81, 71–82 (2018). [DOI] [PubMed] [Google Scholar]
- 13.Wilson C. G., Robert-Cooperman C. E., Burkhardt B. R., PANcreatic-DERived factor: Novel hormone PANDERing to glucose regulation. FEBS Lett. 585, 2137–2143 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhu Y., et al., Cloning, expression, and initial characterization of a novel cytokine-like gene family. Genomics 80, 144–150 (2002). [DOI] [PubMed] [Google Scholar]
- 15.Cao X., et al., Pancreatic-derived factor (FAM3B), a novel islet cytokine, induces apoptosis of insulin-secreting beta-cells. Diabetes 52, 2296–2303 (2003). [DOI] [PubMed] [Google Scholar]
- 16.Cao X., et al., Effects of overexpression of pancreatic derived factor (FAM3B) in isolated mouse islets and insulin-secreting betaTC3 cells. Am. J. Physiol. Endocrinol. Metab. 289, E543–E550 (2005). [DOI] [PubMed] [Google Scholar]
- 17.Yang J., et al., Structure-function studies of PANDER, an islet specific cytokine inducing cell death of insulin-secreting beta cells. Biochemistry 44, 11342–11352 (2005). [DOI] [PubMed] [Google Scholar]
- 18.Moak S. L., et al., Enhanced glucose tolerance in pancreatic-derived factor (PANDER) knockout C57BL/6 mice. Dis. Model. Mech. 7, 1307–1315 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Robert-Cooperman C. E., et al., PANDER transgenic mice display fasting hyperglycemia and hepatic insulin resistance. J. Endocrinol. 220, 219–231 (2014). [DOI] [PubMed] [Google Scholar]
- 20.Li Z., et al., A non-secretory form of FAM3B promotes invasion and metastasis of human colon cancer cells by upregulating Slug expression. Cancer Lett. 328, 278–284 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maciel-Silva P., et al., FAM3B/PANDER inhibits cell death and increases prostate tumor growth by modulating the expression of Bcl-2 and Bcl-XL cell survival genes. BMC Cancer 18, 90 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.He S. L., et al., FAM3B promotes progression of oesophageal carcinoma via regulating the AKT-MDM2-p53 signalling axis and the epithelial-mesenchymal transition. J. Cell. Mol. Med. 23, 1375–1385 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Song C., Duan C., Upregulation of FAM3B promotes cisplatin resistance in gastric cancer by inducing epithelial-mesenchymal transition. Med. Sci. Monit. 26, e921002 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ding Y., et al., Spemann organizer transcriptome induction by early beta-catenin, Wnt, Nodal, and Siamois signals in Xenopus laevis. Proc. Natl. Acad. Sci. U.S.A. 114, E3081–E3090 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ding Y., et al., Bighead is a Wnt antagonist secreted by the Xenopus Spemann organizer that promotes Lrp6 endocytosis. Proc. Natl. Acad. Sci. U.S.A. 115, E9135–E9144 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Session A. M., et al., Genome evolution in the allotetraploid frog Xenopus laevis. Nature 538, 336–343 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johansson P., et al., FAM3B PANDER and FAM3C ILEI represent a distinct class of signaling molecules with a non-cytokine-like fold. Structure 21, 306–313 (2013). [DOI] [PubMed] [Google Scholar]
- 28.Nutt S. L., Dingwell K. S., Holt C. E., Amaya E., Xenopus Sprouty2 inhibits FGF-mediated gastrulation movements but does not affect mesoderm induction and patterning. Genes Dev. 15, 1152–1166 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sivak J. M., Petersen L. F., Amaya E., FGF signal interpretation is directed by Sprouty and Spred proteins during mesoderm formation. Dev. Cell 8, 689–701 (2005). [DOI] [PubMed] [Google Scholar]
- 30.De Robertis E. M., Larraín J., Oelgeschläger M., Wessely O., The establishment of Spemann’s organizer and patterning of the vertebrate embryo. Nat. Rev. Genet. 1, 171–181 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.De Robertis E. M., Kuroda H., Dorsal-ventral patterning and neural induction in Xenopus embryos. Annu. Rev. Cell Dev. Biol. 20, 285–308 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang S., et al., QSulf1, a heparan sulfate 6-O-endosulfatase, inhibits fibroblast growth factor signaling in mesoderm induction and angiogenesis. Proc. Natl. Acad. Sci. U.S.A. 101, 4833–4838 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fletcher R. B., Baker J. C., Harland R. M., FGF8 spliceforms mediate early mesoderm and posterior neural tissue formation in Xenopus. Development 133, 1703–1714 (2006). [DOI] [PubMed] [Google Scholar]
- 34.Böttcher R. T., Pollet N., Delius H., Niehrs C., The transmembrane protein XFLRT3 forms a complex with FGF receptors and promotes FGF signalling. Nat. Cell Biol. 6, 38–44 (2004). [DOI] [PubMed] [Google Scholar]
- 35.Mohammadi M., et al., Identification of six novel autophosphorylation sites on fibroblast growth factor receptor 1 and elucidation of their importance in receptor activation and signal transduction. Mol. Cell. Biol. 16, 977–989 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cascio S., Gurdon J. B., The initiation of new gene transcription during Xenopus gastrulation requires immediately preceding protein synthesis. Development 100, 297–305 (1987). [DOI] [PubMed] [Google Scholar]
- 37.Kurth T., Meissner S., Schäckel S., Steinbeisser H., Establishment of mesodermal gene expression patterns in early Xenopus embryos: The role of repression. Dev. Dyn. 233, 418–429 (2005). [DOI] [PubMed] [Google Scholar]
- 38.Dai S., Zhou Z., Chen Z., Xu G., Chen Y., Fibroblast growth factor receptors (FGFRs): Structures and small molecule inhibitors. Cells 8, 614 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Facchinetti F., et al., Facts and new hopes on selective FGFR inhibitors in solid tumors. Clin. Cancer Res. 26, 764–774 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ekerot M., et al., Negative-feedback regulation of FGF signalling by DUSP6/MKP-3 is driven by ERK1/2 and mediated by Ets factor binding to a conserved site within the DUSP6/MKP-3 gene promoter. Biochem. J. 412, 287–298 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Persaud A., et al., Nedd4-1 binds and ubiquitylates activated FGFR1 to control its endocytosis and function. EMBO J. 30, 3259–3273 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Czubayko F., et al., A secreted FGF-binding protein can serve as the angiogenic switch in human cancer. Nat. Med. 3, 1137–1140 (1997). [DOI] [PubMed] [Google Scholar]
- 43.Acosta H., et al., The serpin PN1 is a feedback regulator of FGF signaling in germ layer and primary axis formation. Development 142, 1146–1158 (2015). [DOI] [PubMed] [Google Scholar]
- 44.Ossipova O., Itoh K., Radu A., Ezan J., Sokol S. Y., Pinhead signaling regulates mesoderm heterogeneity via the FGF receptor-dependent pathway. Development 147, dev188094 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Reis A. H., Sokol S. Y., Rspo2 antagonizes FGF signaling during vertebrate mesoderm formation and patterning. Development 147, dev189324 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lea R., Papalopulu N., Amaya E., Dorey K., Temporal and spatial expression of FGF ligands and receptors during Xenopus development. Dev. Dyn. 238, 1467–1479 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kazanskaya O., et al., R-Spondin2 is a secreted activator of Wnt/beta-catenin signaling and is required for Xenopus myogenesis. Dev. Cell 7, 525–534 (2004). [DOI] [PubMed] [Google Scholar]
- 48.Nam J. S., et al., Mouse R-spondin2 is required for apical ectodermal ridge maintenance in the hindlimb. Dev. Biol. 311, 124–135 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Köster A., et al., Transgenic angiopoietin-like (angptl)4 overexpression and targeted disruption of angptl4 and angptl3: Regulation of triglyceride metabolism. Endocrinology 146, 4943–4950 (2005). [DOI] [PubMed] [Google Scholar]
- 50.Desai U., et al., Lipid-lowering effects of anti-angiopoietin-like 4 antibody recapitulate the lipid phenotype found in angiopoietin-like 4 knockout mice. Proc. Natl. Acad. Sci. U.S.A. 104, 11766–11771 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kirsch N., et al., Angiopoietin-like 4 is a Wnt signaling antagonist that promotes LRP6 turnover. Dev. Cell 43, 71–82.e6 (2017). [DOI] [PubMed] [Google Scholar]
- 52.Li X., The FGF metabolic axis. Front. Med. 13, 511–530 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Goetz R., Mohammadi M., Exploring mechanisms of FGF signalling through the lens of structural biology. Nat. Rev. Mol. Cell Biol. 14, 166–180 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cao X., et al., Elevated circulating level of a cytokine, pancreatic-derived factor, is associated with metabolic syndrome components in a Chinese population. J. Diabetes Investig. 7, 581–586 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shehata M. M., Kamal M. M., El-Hefnawy M. H., El-Mesallamy H. O., Association of serum pancreatic derived factor (PANDER) with beta-cell dysfunction in type 2 diabetes mellitus. J. Diabetes Complications 31, 748–752 (2017). [DOI] [PubMed] [Google Scholar]
- 56.MarElia C. B., Kuehl M. N., Shemwell T. A., Alman A. C., Burkhardt B. R., Circulating PANDER concentration is associated with increased HbA1c and fasting blood glucose in type 2 diabetic subjects. J. Clin. Transl. Endocrinol. 11, 26–30 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang H., et al., Effects of circulating member B of the family with sequence similarity 3 on the risk of developing metabolic syndrome and its components: A 5-year prospective study. J. Diabetes Investig. 9, 782–788 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Koroglu N., et al., Increased pancreatic-derived factor (PANDER) levels in gestational diabetes mellitus. Gynecol. Endocrinol. 35, 866–868 (2019). [DOI] [PubMed] [Google Scholar]
- 59.Katoh M., Therapeutics targeting FGF signaling network in human diseases. Trends Pharmacol. Sci. 37, 1081–1096 (2016). [DOI] [PubMed] [Google Scholar]
- 60.Nieuwkoop P. D., Faber J., Normal Table of Xenopus laevis (Daudin): A Systematic and Chronological Survey of the Development from the Fertilized Egg till the End of Metamorphosis (Garland Publishing, New York, London, 1967). [Google Scholar]
- 61.Pera E. M., Ikeda A., Eivers E., De Robertis E. M., Integration of IGF, FGF, and anti-BMP signals via Smad1 phosphorylation in neural induction. Genes Dev. 17, 3023–3028 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or SI Appendix.