Significance
Amyloid deposition of α-syn is a hallmark of PD. Heterozygous point mutations of α-syn are associated with the early-onset and rapid progression of fPD. In this study, we found the fibril strain formed by a familial mutant hE46K induced early-onset motor deficit and enhanced α-syn aggregation in vivo. More importantly, we showed through cryogenic electron microscopy and in vivo studies the E46K fibril strain can template wild-type α-syn monomer to form amyloid fibrils, which inherit both the structure and propagation potency of the template. Our work reveals the structural basis underlying the cross-seeding between wild-type and mutant α-syn, underscores the importance of fibril structure in determining α-syn neuropathology, and provides mechanistic understanding of the pathology of E46K-associated fPD.
Keywords: α-synuclein, Parkinson’s diseases, E46K mutant, cryo-electron microscopy, pathology
Abstract
Heterozygous point mutations of α-synuclein (α-syn) have been linked to the early onset and rapid progression of familial Parkinson’s diseases (fPD). However, the interplay between hereditary mutant and wild-type (WT) α-syn and its role in the exacerbated pathology of α-syn in fPD progression are poorly understood. Here, we find that WT mice inoculated with the human E46K mutant α-syn fibril (hE46K) strain develop early-onset motor deficit and morphologically different α-syn aggregation compared with those inoculated with the human WT fibril (hWT) strain. By using cryo-electron microscopy, we reveal at the near-atomic level that the hE46K strain induces both human and mouse WT α-syn monomers to form the fibril structure of the hE46K strain. Moreover, the induced hWT strain inherits most of the pathological traits of the hE46K strain as well. Our work suggests that the structural and pathological features of mutant strains could be propagated by the WT α-syn in such a way that the mutant pathology would be amplified in fPD.
α-Synuclein (α-Syn) is the main component of Lewy bodies, which serve as the common histological hallmark of Parkinson’s disease (PD) and other synucleinopathies (1, 2). α-Syn fibrillation and cell-to-cell transmission in the brain play essential roles in disease progression (3–5). Interestingly, WT α-syn could form fibrils with distinct polymorphs, which exhibit disparate seeding capability in vitro and induce distinct neuropathologies in mouse models (6–10). Therefore, it is proposed that α-syn fibril polymorphism may underlie clinicopathological variability of synucleinopathies (6, 9). In fPD, several single-point mutations of SNCA have been identified, which are linked to early-onset, severe, and highly heterogeneous clinical symptoms (11–13). These mutations have been reported to influence either the physiological or pathological function of α-syn (14). For instance, A30P weakens while E46K strengthens α-syn membrane binding affinity that may affect its function in synaptic vesicle trafficking (14, 15). E46K, A53T, G51D, and H50Q have been found to alter the aggregation kinetics of α-syn in different manners (15–17). Recently, several cryogenic electron microscopy (cryo-EM) studies revealed that α-syn with these mutations forms diverse fibril structures that are distinct from the WT α-syn fibrils (18–26). Whether and how hereditary mutations induced fibril polymorphism contributes to the early-onset and exacerbated pathology in fPD remains to be elucidated. More importantly, most fPD patients are heterozygous for SNCA mutations (12, 13, 27, 28), which leads to another critical question: could mutant fibrils cross-seed WT α-syn to orchestrate neuropathology in fPD patients?
E46K mutation is one of the eight disease-causing mutations on SNCA originally identified from a Spanish family with autosomal-dominant PD (11). E46K-associated fPD features early-onset motor symptoms and rapid progression of dementia with Lewy bodies (11). Studies have shown that E46K mutant has higher neurotoxicity than WT α-syn in neurons and mouse models overexpressing α-syn (29–32). The underlying mechanism is debatable. Some reported that E46K promotes the formation of soluble species of α-syn without affecting the insoluble fraction (29, 30), while others suggested that E46K mutation may destabilize α-syn tetramer and induce aggregation (31, 32). Our previous study showed that E46K mutation disrupts the salt bridge between E46 and K80 in the WT fibril strain and rearranges α-syn into a different polymorph (33). Compared with the WT strain, the E46K fibril strain is prone to be fragmented due to its smaller and less stable fibril core (33). Intriguingly, the E46K strain exhibits higher seeding ability in vitro, suggesting that it might induce neuropathology different from the WT strain in vivo (33).
In this study, we found that human E46K and WT fibril strains (referred to as hE46K and hWT strains) induced α-syn aggregates with distinct morphologies in mice. Mice injected with the hE46K strain developed more α-syn aggregation and early-onset motor deficits compared with the mice injected with the hWT strain. Notably, the hE46K strain was capable of cross-seeding both human and mouse WT (mWT) α-syn to form fibrils (named as hWTcs and mWTcs). The cross-seeded fibrils replicated the structure and seeding capability of the hE46K template both in vitro and in vivo. Our results suggest that the hE46K strain could propagate its structure as well as the seeding properties to the WT monomer so as to amplify the α-syn pathology in fPD.
Results
The hE46K Strain Leads to Early-Onset Motor Deficit and Distinct α-Syn Aggregates In Vivo.
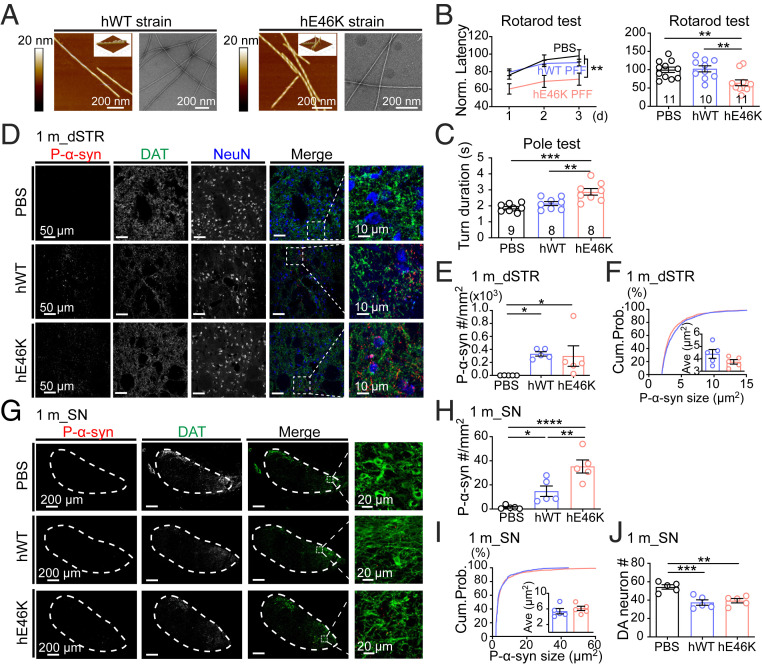
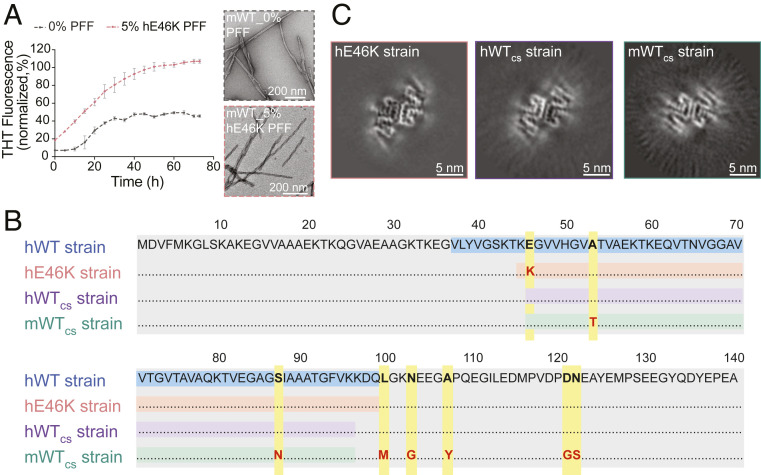
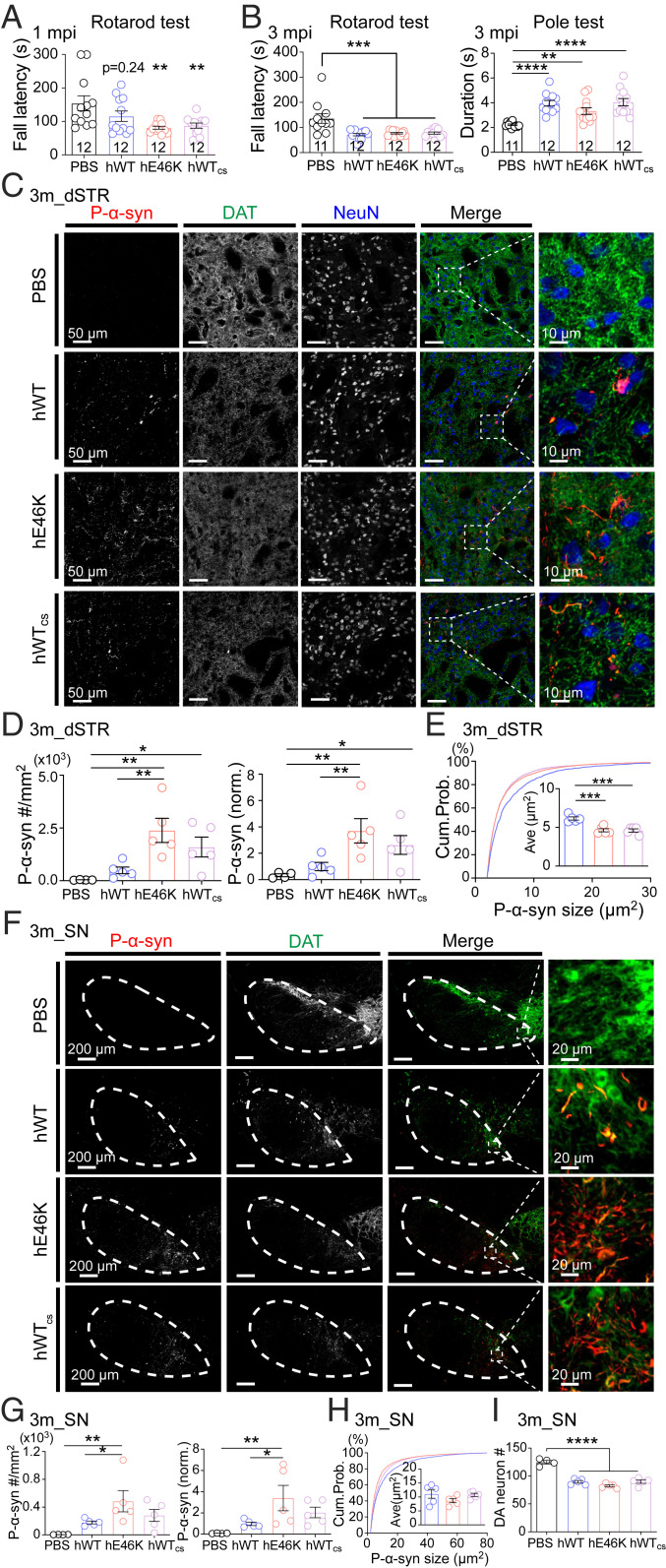
We first prepared the full-length N-acetylated hWT and hE46K α-syn fibrils as described in our previous report (33). Atomic force microscope (AFM) and transmission electron microscopy (TEM) results showed that the hE46K fibril featured a right-handed twist with a fibril width of 10 nm and a periodicity of 64 nm, while the hWT fibril displayed a left-handed twist with a fibril width of 10 nm and a periodicity of 121 nm (Fig. 1A and SI Appendix, Fig. S1 A and B). This confirms the distinct morphologies between the hE46K and hWT fibril strains. To investigate the neuropathology induced by either fibril in vivo, preformed fibril (PFF) of each α-syn strain (SI Appendix, Fig. S1 C–F; also see Methods for detailed preparation of PFFs) was bilaterally inoculated into the dorsal striatum (dSTR) of WT mice (SI Appendix, Fig. S2A). Motor performance was first examined 1 mo postsurgery (SI Appendix, Fig. S2B). Strikingly, mice injected with hE46K PFF performed worse than phosphate-buffered saline (PBS)-injected control mice in both rotarod and pole test, while mice injected with hWT PFF remained normal (Fig. 1 B and C and SI Appendix, Fig. S2C). This suggests that the hE46K strain might induce faster disease progression than the hWT.
Fig. 1.
Mice injected with hE46K PFF develop early-onset motor deficit 1 mo postinoculation. (A) AFM and negative-staining TEM images of the hWT and hE46K fibril strains. The 3D AFM images are in the Top Right corner of the 2D images. (B) Rotarod test for mice injected with PBS (black), hWT PFF (blue), and hE46K PFF (red). The normalized learning curve is on the Left. The final rotarod test after training is on the Right. (C) Time needed for turning up-side-down in pole test. (D) Representative immunofluorescence images of dSTR. Antibodies were used to mark p-α-syn (red), DAT (green), NeuN (blue) (Scale bar, 50 μm). Zoom-in of the merged images are shown on the Right (Scale bar, 10 μm). Quantitative analyses are shown in E, F, and SI Appendix, Fig. S2D. (E) P-α-syn aggregate density. (F) The size cumulative probability curve and the Insert figure shows the average aggregate size of p-α-syn aggregates. (G) Representative immunofluorescence images of SN (Scale bar, 200 μm) (Zoom-in scale bar, 20 μm). Quantitative analyses are shown in H–J and SI Appendix, Fig. S2D. (H) P-α-syn aggregate density. (I) The size cumulative probability curve and the Insert figure shows the average aggregate size of p-α-syn aggregates. (J) DA neuron numbers in SN by counting DAT+ cells. Statistical information is summarized in SI Appendix, Table S1; *P < 0.05; **P < 0.01; ***P < 0.001, ****P < 0.0001.
We further examined α-syn pathology at this time by immunofluorescence staining. In dSTR close to the injected location, both groups of PFF-injected mice developed significant amounts of small α-syn aggregates (Fig. 1 D–F and SI Appendix, Fig. S2D), suggesting that both strains can rapidly and locally induce α-syn aggregation in vivo. Furthermore, the aggregates were observed to spread to the substantia nigra (SN), especially in the medial part of substantia nigra pars compacta (SNpc) (Fig. 1 G–I and SI Appendix, Fig. S2D). Notably, the hE46K PFF induced significantly more α-syn aggregates than the hWT PFF in SN (Fig. 1H). This result suggests that the endogenous α-syn pathology induced by hE46K PFF might propagate more effectively in the synaptically connected brain regions. Additionally, both strains led to moderate but significant dopaminergic (DA) neuron loss in SN (Fig. 1J), indicating a result of α-syn pathology.
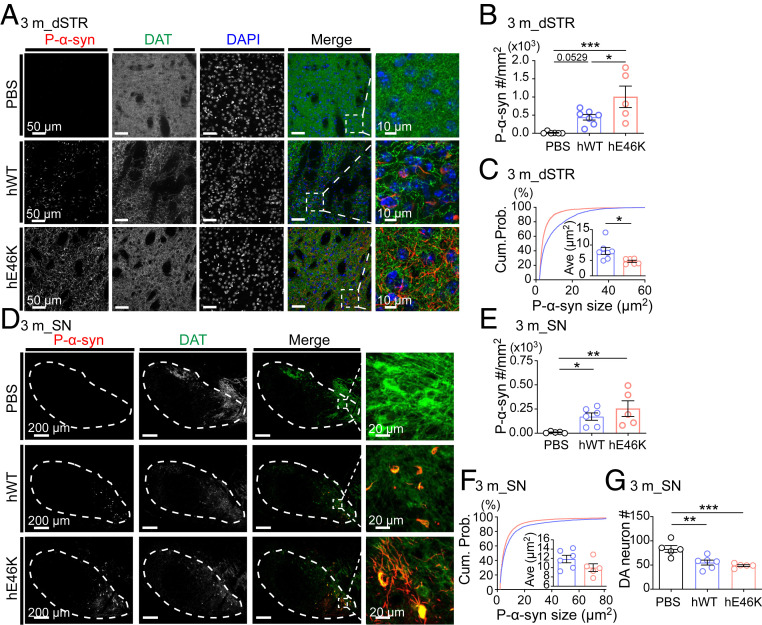
As we examined 3 mo postinjection, mice injected with either strain developed more α-syn aggregates in dSTR than that examined 1 mo postinjection, at the same time the hE46K-injected mice showed higher aggregate density than that of the hWT (Fig. 2 A and B and SI Appendix, Fig. S3A). Interestingly, aggregates induced by hWT PFF were larger in size and mainly localized in somas, while those induced by hE46K were smaller and evenly distributed in the neuropil (Fig. 2 A and C and SI Appendix, Fig. S3B). Spreading of α-syn pathology into the SN, in particular SNpc, was more prominent 3 mo postinjection for both strains (Fig. 2D). The hE46K-injected mice again showed significantly stronger phosphorylated α-syn (p-α-syn) signal (SI Appendix, Fig. S3A) and a trend of higher aggregate density than that of the hWT (Fig. 2 E and F). In addition, we observed that hE46K-injected mice showed a trend of stronger p-α-syn intensity and higher aggregate density than that of the hWT-injected mice in other brain regions (SI Appendix, Fig. S3 C–G).
Fig. 2.
hE46K PFF induces distinct α-syn aggregates compared with hWT PFF in mice 3 mo postinoculation. (A) Representative immunofluorescence images in dSTR. Antibodies were used to label p-α-syn (red), DAT (green) and DAPI (blue) (Scale bar, 50 μm). Zoom-in of the merged images are shown on the Right (Scale bar, 10 μm). Quantitative analyses are shown in B, C, and SI Appendix, Fig. S3A. (B) P-α-syn aggregate density. (C) The size cumulative probability curve (P < 0.0001, Kolmogorov–Smirnov test) and the average aggregate size (Insert). (D) Representative immunofluorescence images in SN (Scale bar, 200 μm) (Zoom-in scale bar, 20 μm). Quantitative analyses are shown in E, F, and SI Appendix, Fig. S3A. (E) P-α-syn aggregate density. (F) The size cumulative probability curve (P < 0.0001, Kolmogorov–Smirnov test) and the average aggregate size (Insert). (G) DA neuron numbers in SN by counting DAT+ cells. Statistical information is summarized in SI Appendix, Table S2; *P < 0.05; **P < 0.01; ***P < 0.001.
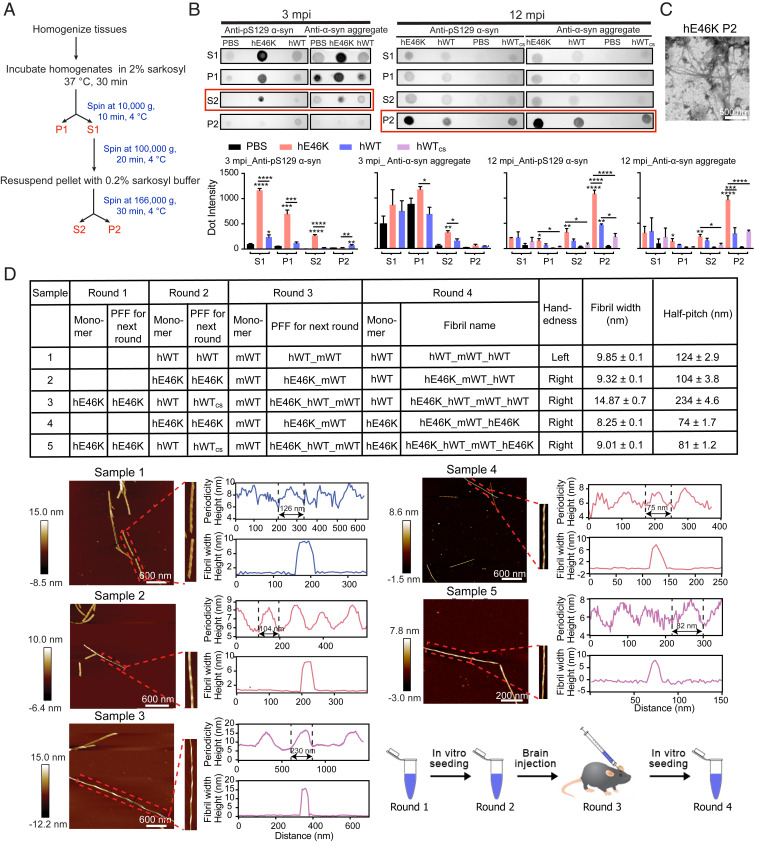
To further assess the morphological differences of the induced α-syn aggregates, we harvested them from the mouse brain (Fig. 3A) and compared them by dot blot. We found that the hE46K PFF induced more α-syn aggregates than the hWT PFF (Fig. 3B), which is consistent with the immunofluorescence imaging. We further used TEM and observed clear fibrils in the ex vivo α-syn aggregates induced by hE46K (Fig. 3C). To obtain a sufficient amount of fibrils for morphological characterization, we used the ex vivo fibrils to seed hWT and hE46K α-syn monomer in vitro, respectively. AFM measurements showed that the fibrils initially induced by the hE46K PFF faithfully inherit the unusual right-handedness of the template after three rounds of cross-seeding (Fig. 3D). Structural alteration was also observed after serial cross-seeding though (Fig. 3D). We will discuss it in the Discussion below. Collectively, these observations suggest that the endogenous mouse α-syn was induced to form different aggregates resembling the injected individual human strains. Compared with the hWT-induced α-syn pathology, that induced by hE46K may spread more efficiently and lead to early onset of motor deficit.
Fig. 3.
Morphological characterization of the induced α-syn deposits harvested from the mouse brain. (A) The experiment flow of the extraction of α-syn deposits from the mouse brain. S, supernatant; P, precipitate. (B) Dot blot images (Top) show the samples obtained from (A) at 3 and 12 mo postinoculation (mpi), respectively. Antibodies: anti-pS129 α-syn (ab51253); anti-α-syn aggregates (ab209538). At 3 mpi, the induced α-syn deposits mainly exist in fraction S2, highlighted with a red box. At 12 mpi, they mainly exist in fraction P2. The statistics of dot blots are shown at the bottom as mean ± SEM, n = 3 individual experiments. One-way ANOVA, *P < 0.05; **P < 0.01; ***P < 0.001, ****P < 0.0001. (C) A TEM image of fraction P2 harvested from the mouse brain injected with hE46K PFF at 12 mpi (Scale bar, 500 nm). (D) AFM measurements of α-syn fibrils seeded with fraction P2 extracted from mouse brain injected with different PFFs at 12 mpi. The table on Top lists the name of monomers and PFFs in each round of fibril formation and the handedness and twist parameters of the final fibrils. Fibril width and half pitch are shown as mean ± SD, n ≥ 5 individual measurements. AFM measurements of the final fibrils are shown Below. Individual fibrils are zoomed in on the Right of each AFM image. Fibril twist periodicity and width are analyzed by using NanoScope Analysis software (version 1.5). The flowchart of serial seeding is shown at the Bottom Right.
The hE46K Strain Cross-Seeds WT α-Syn to Adopt hE46K-Like Structure.
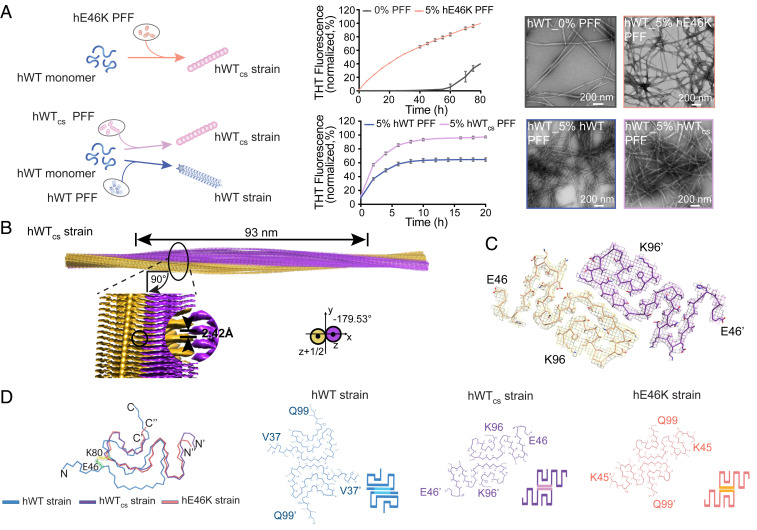
To obtain atomic details of the cross-seeded fibrils, we next directly cross-seeded hWT α-syn monomer with the hE46K PFF in vitro, and the results of the Thioflavin T (ThT) kinetics assay, negative-staining TEM, and gel electrophoresis showed that the hE46K PFF efficiently cross-seeded hWT α-syn to form fibrils (referred to as hWTcs strain) (Fig. 4A and SI Appendix, Fig. S4A). We further determined the near-atomic structure of the hWTcs fibril by using cryo-EM. The three-dimensional (3D) density map of the hWTcs fibril was reconstructed at an overall resolution of 3.8 Å (Fig. 4B, SI Appendix, Fig. S4B, and Table 1). The hWTcs fibril is composed of two protofilaments intertwining along the fibril axis with a right-handed twist (Fig. 4B). The helical rise and twist angle of the hWTcs fibril are 2.42 Å and −179.53°, respectively (Fig. 4B and Table 1). The half pitch of the hWTcs fibril is ∼93 nm (Fig. 4B). Unlike the left-handed twist of all known WT α-syn fibrils, the hWTcs fibril preserves the distinctive right-handed twist of its hE46K template, although the parameters including helical rise and twist angle are slightly different (Fig. 4B, SI Appendix, Fig. S4C, and Table 2).
Fig. 4.
The hE46K fibril strain cross-seeded hWT α-syn to form fibril with hE46K-like structure. (A) Cross-seeding of hWT monomer with hE46K PFF (Top) and hWTcs PFF (Bottom). The fibril samples at the end of the ThT kinetics assay were imaged by negative-staining TEM (Scale bar: 200 nm). The data are shown as mean ± SD, n = 3 independent samples. (B) Cryo-EM 3D reconstruction density map of the hWTcs fibril strain (PDB ID: 7C1D). Half pitch length, helical rise, and twist angle are indicated. The twist angle is graphically illustrated. The two intertwining protofilaments of the hWTcs strain are colored in yellow and purple, respectively. (C) The top view of the hWTcs fibril strain. One layer of the structure is shown, which consists of two α-syn subunits containing residues 46 to 96. The two subunits are colored in purple and yellow, respectively. (D) Overlay of the subunits of the hWT strain (PDB ID:6A6B), the hWTcs strain (PDB ID:7C1D), and the hE46K strain (PDB ID:6L4S). Residues E46 and K80, which form a salt bridge in the hWT fibril strain, are highlighted in spheres (green, E46; yellow, K80). The structures of the two paring subunits in the three fibril strains are shown with cartoon presentation at the Bottom indicating the different packing of the hWT (blue), hWTcs (purple), and hE46K (orange) structures.
Table 1.
Statistics of cryo-EM data collection and refinement
| Name | hWTcs strain | mWTcs strain |
| PDB ID | 7C1D | n/a |
| EMDB ID | EMD-30269 | EMD-30648 |
| Data Collection | ||
| Magnification | 105,000 | 105,000 |
| Pixel size (Å) | 1.356 | 1.356 |
| Defocus range (μm) | −1.3 to −2.1 | −1.3 to −2.1 |
| Voltage (kV) | 300 | 300 |
| Camera | K2 summit | K2 summit |
| Microscope | Titan Krios | Titan Krios |
| Exposure time (s per frame) | 0.25 | 0.25 |
| Number of frames | 32 | 32 |
| Total dose (e- per Å2) | 35 | 35 |
| Reconstruction | ||
| Micrographs | 317 | 494 |
| Manually picked fibrils | 3,046 | 3,640 |
| Box size (pixel) | 224 | 288 |
| Interbox distance (Å) | 30.4 | 39.1 |
| Segments extracted | 66,850 | 60,161 |
| Segments after Class2D | 63,072 | 60,124 |
| Segments after Class3D | 11,867 | 7,466 |
| Map resolution (Å) | 3.8 | 4.4 |
| FSC threshold | 0.143 | 0.143 |
| Map sharpening B-factor (Å2) | −130.966 | −207.8 |
| Helical rise (Å) | 2.418 | 2.420 |
| Helical twist (°) | −179.53 | −179.55 |
| Refinement | ||
| Initial model used (PDB code) | 6L4S | n/a |
| Map resolution (Å) | 6.7 | n/a |
| FSC threshold | 0.5 | n/a |
| Nonhydrogen atoms | 2,034 | n/a |
| Protein residues | 306 | n/a |
| Ligands | 0 | n/a |
| rmsd Bond lengths | 0.004 | n/a |
| rmsd Bond angles | 0.709 | n/a |
| All-atom clash score | 17.40 | n/a |
| Rotamer outliers (%) | 0.00 | n/a |
| Ramachandran outliers (%) | 0.00 | n/a |
| Ramachandran allowed (%) | 20.41 | n/a |
| Ramachandran favored (%) | 79.59 | n/a |
n/a, not applicable.
Table 2.
Comparison of α-syn fibril parameters
| Name | hE46K fibril | hWTcs fibril | mWTcs fibril |
| PDB ID | 6L4S | 7C1D | — |
| EMDB ID | EMD-0833 | EMD-30269 | EMD-30648 |
| Helical rise (Å) | 2.38 | 2.42 | 2.42 |
| Helical twist (°) | −179.37 | −179.53 | −179.55 |
| Half pitch (nm) | ∼68 | ∼93 | ∼97 |
| Width (nm) | ∼10 | ∼10 | ∼10 |
| Handedness | Right-handed | Right-handed | Right-handed |
The structures of the α-syn subunits are highly similar between the hWTcs and hE46K fibrils with an rmsd. (C-α atoms) of 1.578 Å, which are distinct from that of the hWT fibril (Fig. 4 C and D). Both the hWTcs and hE46K fibrils feature an extended serpentine fold (Fig. 4D). Thus, the structure of the hE46K strain could also be formed by hWT α-syn via cross-seeding. In addition, the data of ThT kinetic assay, TEM, and sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) showed that hWT α-syn monomer formed more fibrils when seeded with the hWTcs strain compared with when seeded with the hWT strain (Fig. 4A and SI Appendix, Fig. S4A), implying that the hWTcs strain may be more potent in inducing α-syn pathologies.
Our in vivo data have indicated that the hE46K strain can overcome the species barrier to cross-seed mWT α-syn (Figs. 1 and 2). Consistently, we observed that the hE46K strain can effectively cross-seed mWT α-syn to form fibrils in vitro (Fig. 5A and SI Appendix, Fig. S4A), despite that, mWT α-syn differs from hE46K α-syn at residues 46, 53, 87, 100, 103, 107, 121, and 122 (Fig. 5B). The structure of the mWTcs fibril was also explored by cryo-EM. The 3D density map of the mWTcs fibril was reconstructed at an overall resolution of 4.4 Å (Fig. 5C, SI Appendix, Fig. S4B, and Table 1). Even though the mWTcs fibril sample did not provide cryo-EM images good enough for model building, the cross-section of the obtained 3D reconstructions clearly showed that the mWTcs fibril displays a highly similar structure of the two pairing subunits as those of the hE46K and hWTcs fibrils (Fig. 5C). The steric zipper–like interfaces between the two pairing subunits are highly conserved in all of the three strains (Fig. 5C). Together, our results showed that the hE46K strain can cross-seed both hWT and mWT α-syn monomer to form the hE46K-like fibril structures. This also supports the dominant role of seeding template rather than primary sequence in determining the fibril polymorphs.
Fig. 5.
Cross-seeding of mWT α-syn monomer with hE46K PFF. (A) ThT kinetic assay (Left) for mWT monomer seeded with 5 mol% hE46K PFF. The data are shown as mean ± SD, n = 3. The samples at the end of the ThT kinetic assay were imaged by negative-staining TEM (Right) (Scale bar, 200 nm). (B) Alignment of the primary sequence of different α-syn strains. The fibril core sequences are highlighted: hWT (blue), hE46K (orange), hWTcs (purple), and mWTcs (green). The point mutations are highlighted in yellow. (C) Cross-sections of the 3D reconstructions of the hE46K, hWTcs, and mWTcs fibril strains (Scale bar, 5 nm).
The hWTcs Strain Inherits the Pathological Properties of the hE46K Strain In Vivo.
Since our structural study revealed that the hWTcs strain shares similar fibril structure as the hE46K strain (Fig. 4D and Table 2), we next asked whether the hWTcs strain could induce comparable pathologies as the hE46K strain in vivo. Again, we injected the PFFs of different strains into the dSTR of WT mice and analyzed the motor function and α-syn pathology at both 1 and 3 mo postinjection. Similar to the hWT and hE46K strains, the hWTcs strain also induced obvious p-α-syn aggregates in both dSTR and SN 1 mo post injection (SI Appendix, Fig. S5 A and B). Both the hE46K and hWTcs-injected mice but not the hWT-injected mice showed impaired rotarod performance at this early time point (Fig. 6A). At 3 mo post injection, all three groups performed similarly worse in both rotarod and pole tests (Fig. 6B). Consistently, all three PFFs induced more than 40% of DA neuron loss in SN (Fig. 6I) and moderate reduction of dopamine active transporter (DAT) signals in dSTR and SN (SI Appendix, Fig. S5 C and D). However, compared with the hWT-injected mice, the hWTcs-injected mice showed a trend of exacerbated α-syn aggregation in both dSTR (Fig. 6D) and SN (Fig. 6G). Furthermore, the hWTcs generally induced small aggregates especially in dSTR, similar to the hE46K but in contrast to the hWT (Fig. 6 E and H). Morphological characterization of the ex vivo deposits confirmed that hWTcs-induced α-syn fibrils share a similar structure as that induced by hE46K but not hWT (Fig. 3). Collectively, these results indicate that pathologically, the hWTcs strain resembles more closely the hE46K than the hWT, supporting the idea that the fibril structure determines its pathology in vivo.
Fig. 6.
Pathologies and early-onset motor deficit induced by hWTcs PFF resemble those induced by hE46K PFF. (A) Rotarod test at 1 mo postinoculation (mpi). (B) Motor performance at 3 mpi examined by rotarod test (Left) and pole test (Right). (C) Representative immunofluorescence images of dSTR at 3 mpi (Scale bar, 50 μm). Zoom-in of the merged images are shown on the Right (Scale bar, 10 μm). Quantitative analyses are shown in D, E, and SI Appendix, Fig. S5C. (D) P-α-syn aggregate density (Left) and normalized p-α-syn intensity (Right) in dSTR at 3 mpi. (E) The size cumulative probability curve (hWT versus hE46K, P < 0.0001; hWT versus hWTcs, P < 0.0001; hE46K versus hWTcs, P = 0.3076; Kolmogorov–Smirnov test), and the average aggregate size (Insert). (F) Representative immunofluorescence images of SN at 3 mpi (Scale bar, 200 μm) (Zoom-in scale bar: 20 μm). Quantitative analyses are shown in G, H, and SI Appendix, Fig. S5D. (G) P-α-syn aggregate density (Left) and normalized p-α-syn intensity (Right) in SN at 3 mpi. (H) The size cumulative probability curve (hWT versus hE46K, P < 0.0001; hWT versus hWTcs, P < 0.0001; hE46K versus hWTcs, P < 0.0001; Kolmogorov–Smirnov test) and the average aggregate size (Insert). (I) DA neuron counts in SN at 3 mpi. Statistical information is summarized in SI Appendix, Table S3; *P < 0.05; **P < 0.01; ***P < 0.001, ****P < 0.0001.
Discussion
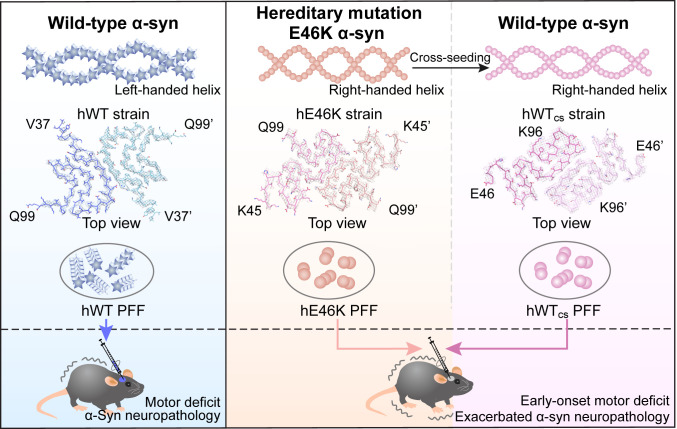
Accumulating evidence showed that different PD-associated familial SNCA mutations may induce α-syn to form fibril strains with polymorphic structures (18–20, 22, 33). Whether and how fibril polymorphism contributes to the exacerbated pathology and early-onset symptoms in fPD remains poorly understood. Our previous study determined the structure of a human α-syn fibril strain with E46K mutation (33). Compared with the hWT strain, this mutant strain is less stable with a higher propensity of fragmentation, and it displays enhanced seeding capability in vitro (33). It has also been reported that E46K mutation may increase the phospholipid binding and filamental assembly of α-syn (34). Consistently, we found here that the hE46K PFF is generally more efficient in inducing α-syn aggregation in vivo than the hWT PFF, and mice injected with the hE46K PFF displayed motor deficit as early as 1 mo postinjection (Fig. 7), which recapitulates the early onset of fPD caused by this missense mutation (11). Given the pivotal role of α-syn in synaptic transmission (35–37), α-syn pathology can impair the function of DA neurons and finally cause neuronal death (38). Suppression of DA neuron activity has been shown to be sufficient to induce PD-like motor deficit (39). Thus, the exacerbated α-syn pathology induced by the hE46K strain might cause DA neuron dysfunction on top of mild cell death, which may be responsible for the early motor symptom. In addition, α-syn pathology has been recently shown to reduce glucocerebrosidase activity, which could expedite the appearance of motor weakness in the absence of DA cell loss (40). The detail mechanism underlying the hE46K-induced early-onset motor deficit will require further investigation.
Fig. 7.
Hypothesis of the exacerbated α-syn neuropathology in heterozygous familial PD. The E46K α-syn strain may pass on its structure and pathology to the WT α-syn to augment the mutant pathology. In vitro fibril structures are presented in this diagram, including the hWT α-syn fibril (PDB ID: 6A6B), the hE46K fibril (PDB ID: 6L4S), and the hWTcs fibril (PDB ID: 7C1D).
Interestingly, with the time-dependent progression of α-syn pathology, we observed generally stronger pathology in all the brain regions examined in the hE46K-injected mice than in the hWT-injected ones. In combination with the enhanced seeding ability of the hE46K strain in vitro (33), these results suggest that the hE46K strain may have stronger seeding and propagating capability in vivo. The underlying mechanism could be potentially explained by the relatively smaller, less stable, and more fragmentation-prone fibril core of the hE46K fibril (24, 33). Previous studies in yeast Sup35 have reported similar findings, where the Sc4 strain is less stable and more fragmentation prone, and also a stronger prion template (41, 42). It is worth noting that recent studies show that the E46K mutation may also form a more stable fibril (19, 22). It will be interesting to explore whether this more stable strain has different seeding and pathological traits in vivo.
Most fPD patients harbored heterozygous SNCA missense mutations (12, 13, 27, 28). In patients carrying E46K mutation, both E46K and WT α-syn coexist (11). The crosstalk between E46K and WT α-syn may play an important role in the disease progression of fPD, which has long been neglected. Here, we show that the hE46K strain exhibits potent cross-seeding ability to template the fibril formation of WT α-syn of both human and mouse origins. The cross-seeded WT strain not only inherits the structural feature of its hE46K template but also induces similar α-syn pathology, which triggers the early-onset motor deficit in mice. These results suggest that the hE46K strain originally formed by E46K mutant α-syn may pass on its structural and pathological features to the WT α-syn and thus to accelerate the disease progression in fPD. Based on this finding in fPD, it is reasonable to hypothesize that pathological somatic mutations of SNCA may possibly spread throughout the brain via a similar mechanism. At the same time, it is worthwhile to note that although the morphology and pathology of the induced fibrils are similar to their templates, they are not the same. The more rounds of seeding were performed, the larger variations of fibril helical parameters were observed (Fig. 3). Strain fidelity during spread is an important topic for the understanding of the progression of neurodegenerative disease and requires further investigation.
Given that the cellular protein concentration of WT and mutant α-syn are presumably similar in the case of heterozygous mutation, an equally important question would be whether the WT α-syn strain can reversely seed the E46K monomer and maintain the structural properties of the WT. We tested this possibility by ThT assay, TEM, and gel electrophoresis (SI Appendix, Fig. S6 A–C). The results showed no effective seeding of the hWT to hE46K monomer. This result is consistent with our previous studies on the structures of hWT and hE46K fibrils showing that hE46K mutation can preclude the formation of the hWT strain by disrupting the salt bridge between E46 and K80 in the WT fibril (SI Appendix, Fig. S6D) (24, 33). In summary, the E46K mutant α-syn strain can pass on its structure and pathology to the WT, which is not applicable reversely. Additionally, it would be interesting to know whether what we found here is common to other fPD-related heterozygous SNCA mutations such as A53T, A53E, and G51D (12, 13, 27, 28). Further study is required to comprehensively understand the fPD α-syn mutations.
Methods
Expression and Purification of Recombinant hWT and hE46K α-Syn.
The preparation of full-length hWT and hE46K α-syn follows the same protocol described previously (24). Briefly, the protein gene encoding was inserted into pET22 vector and coexpressed with yeast N-acetyltransferase complex B in BL21 (DE3) cells to obtain the N-terminally acetylated α-syn protein (43). Protein expression was induced at 37 °C for 4 h with 1 mM isopropyl-1-thio-D-galactopyranoside. Cells were harvested and lysed by sonication, and the supernatant was processed by boiling, streptomycin (20 mg/mL), pH adjustment, and dialysis overnight in turn. Proteins were purified using anion exchange column (GE Healthcare, 17-5156-01) followed by Superdex 75 (GE Healthcare, 28-9893-33) as previously described (24).
Preparation of the hWT and the hE46K α-Syn Fibril Strains.
Recombinant hWT and hE46K α-syn (100 μM in 50 mM Tris, pH 7.5, 150 mM KCl, and 0.05% NaN3 buffer) were incubated at 37 °C with constant agitation (900 rpm) in ThermoMixer (Eppendorf) for 7 d, respectively. The fibril yield was calculated as the total amount of α-syn monomer subtracting the amount of residual soluble α-syn after pelleting the fibrils. The pellets were suspended with PBS to 2 μg/μL. The physical state of the fibrils was monitored by TEM. α-Syn PFFs were obtained by sonication with 20% power 15 times (1 s on, 1 s off) on ice by JY92-IIN sonicator. The PFFs were observed again by TEM and measured by AFM to confirm a similar size distribution. Before injected into mice, the same amount of different PFFs was confirmed by SDS-PAGE. For the sample preparation for SDS-PAGE, the PFFs were dissolved in the buffer containing 50 mM Tris, pH 8.0, 150 mM NaCl, 1% Triton X-100, and 2% SDS and boiled for 30 min. The SDS-PAGE gel was stained by Coomassie Brilliant Blue. The band intensities were analyzed by Image Lab 3.0 (Bio-Rad).
hWT and hE46K α-syn fibrils, which were used for the purpose of structural determination by cryo-EM and AFM, were obtained by incubating 100 μM α-syn monomer in the presence of α-syn PFFs (0.5 mol%, relative to α-syn monomer), shaking at 900 rpm at 37 °C for 1 wk. Then, the hWT and hE46K α-syn fibrils were concentrated by centrifugation (14,462 × g, 25 °C, 45 min).
Extraction of Induced α-Syn Deposits from the Mouse Brain.
α-Syn deposits induced by the injected of hWT and hE46K PFFs were basically extracted from the mouse brain following a reported protocol for extraction from the human brain (25). Mouse brains at 3 and 12 mo postinjection were homogenized in 20 vol (v/w) the extraction buffer (10 mM Tris ⋅ HCl, pH 7.5, 0.8 M NaCl, 10% (wt/vol) sucrose, and 1 mM egtazic acid with phosphatase and protease inhibitors), respectively. The homogenate was incubated in the buffer containing 2% sarkosyl at 37 °C for 30 min, followed by centrifugation at 10,000 × g for 10 min. The supernatant was spun at 100,000 × g for 20 min (Supernatant 1, S1). The pellet was suspended with 0.2% sarkosyl and spun at 166,000 × g for 30 min. The sarkosyl-insoluble pellets from the brains 12 mo postinjection were propagated by seeding α-syn monomers. The fibril handedness and twist parameters of the induced fibrils were measured by AFM.
Dot Blot.
As for the supernatant fractions collected from the mouse brain extraction, protein concentration was determined by bicinchoninic acid assay (Thermo Scientific, REF23227). The same number of total proteins for each supernatant sample were spotted onto the nitrocellulose (NC) membrane (Merck, cat: 10600002). As for the sarkosyl-insoluble pellets, 100 μL of 30 mM Tris ⋅ HCl, pH 7.4 was used to resuspend the pellet, and 4 μL of each insoluble fraction was spotted onto the NC membrane. The NC membrane was blocked with 5% bovine serum albumin in TBST buffer (Tris-buffered saline, 0.1% Tween 20) for 1 h at room temperature (RT). The NC membrane was then incubated with primary antibodies: anti-pS129 α-syn (Abcam, ab51253) and anti-α-syn aggregates (Abcam, ab209538), respectively, overnight at 4 °C. After washing three times with the TBST buffer, the NC membrane was incubated with the secondary antibody (goat anti-rabbit IgG H&L, ab6721) conjugated with horseradish peroxidase for 1 h at RT. After washing with the TBST buffer, the film was developed with the enhanced chemiluminescent regent (Shanghai Yisheng Biotechnology, cat: 36208ES60).
Negative-Staining TEM.
The hWT and the hE46K α-syn fibrils were examined by negative-staining electron microscopy. A drop of 5 μL fibril solution was applied to a glow-discharged 200-mesh carbon support film (Beijing Zhongjingkeyi Technology Co., Ltd.). Then, the grid was washed with 5 μL double-distilled water followed by another wash of 5 μL 3% wt/vol uranyl acetate. Then, the grid was stained with 5 μL 3% wt/vol uranyl acetate for 45 s. The excess buffer was removed by filter paper, and the grid was dried by infrared lamp. The sample imaging was applied by Tecnai T12 microscope (FEI Company) operated at 120 KV.
Cryo-EM.
The hWTcs and mWTcs α-syn fibril aqueous solutions were applied to holey carbon grids (Quantifoil R1.2/1.3, 300 mesh) which were glow-discharged, blotted for 6.5 s, and plunge frozen in liquid ethane using FEI Vitrobot Mark IV with 95% humidity at 16 °C. The grids were loaded onto a FEI Titan Krios microscope equipped with a GIF Quantum energy filter (slit width, 20 eV) operated at 300 KV with magnification of 105,000×. The cryo-EM micrographs using super-resolution mode with pixel size 0.678Å were recorded by a Gatan K2 Summit camera, and the defocus values were set from −2.1 to −1.3 μm. All of the micrographs were dose fractionated to 32 frames, and the electron dose rate was set to ∼8 counts per physical pixel per second (∼5.90e− ⋅ s−1 per Å2); the total exposure time is 8 s. Hence, the total dose is 35e− per Å2. Serial EM software was used for all data collection (44).
Image Preprocessing and Helical Reconstruction.
All 32 frames were aligned, summed, and dose weighted by MotionCorr2 and further binned to a pixel size of 1.356 Å (45). CTFFIND4 was used to estimate the defocus values of dose-weighted micrographs (46). Subsequent processes including manual picking, two-dimensional (2D) classification, 3D classification, and 3D autorefine were performed in RELION 3.0 (47).
For the hWTcs dataset, 3,046 filaments were manually picked from 317 micrographs. The extracted segments with a 224-pixel box size were applied to several iterations of 2D classification with a decreasing in-plane angular sampling rate from 12° to 1° and a T = 2 regularization parameter. A helical initial model was reconstructed using selected particles after 2D classification by relion_helix_inimodel2d and applied further as a reference map to perform de novo 3D classification. Symmetry of pseudo-21 was applied according to a staggered arrangement observed in 2D classification. K = 3 was used to gain segments belonging to the same conformation. Three-dimensional auto-refinement was then used to improve the map and postprocessing with a soft mask was used to sharpen the map. The final map was convergence with the rise of 2.418 Å and the twist angle of −179.53°. The overall resolution was reported at 3.80 Å by the gold-standard Fourier shell correlation (FSC) = 0.143 criteria.
For the mWTcs dataset, 3,640 filaments were manually picked from 494 micrographs and then extracted with a box size of 288 pixels. Similarly, with the hWTcs dataset, several iterations of 2D classification were performed to homogenize segments of mWTcs fibrils. A helical initial model was reconstructed using selected particles after 2D classification by relion_helix_inimodel2d and applied further as a reference map to perform de novo 3D classification. Followed by 3D classification with K = 1, several rounds of 3D classification with K = 3 were then performed. After 3D autorefinement and postprocessing with a soft mask generated using relion_mask_create tools, the overall estimated resolution of the final mWTcs reconstruction map is ∼4.4 Å with the rise of 2.42 Å and the twist angle of −179.55°.
Polymorphs of α-Syn Fibrils.
Three thousand and forty-six filaments of the hWTcs strain were selected from 317 micrographs. The filaments contain ∼94% twisted and ∼6% nontwisted filaments. A total of 3,640 filaments of the mWTcs strain were selected from 494 micrographs, which contain ∼99% twisted filaments. Cryo-EM reconstruction was performed for the twisted filaments.
Model Building and Refinement.
A homology model based on the cryo-EM structure (PDB entry code: 6L4S) was built and modified by Coot (48, 49). The model with three adjacent layers (six promoters) was refined using the real-space refinement program in PHENIX (50). The subunit dimers in the middle of three layers were extracted and used as the final model.
Animals.
All procedures were approved by the Institutional Animal Care and Use Committee at the Interdisciplinary Research Center on Biology and Chemistry, Chinese Academy of Science. C57BL/6J mice were either bred in-house or directly purchased from company (Shanghai Ling Chang biotechnology company). Mice were group housed in regular cage on a 12 h:12 h light:dark cycle with ad libitum access to food and water. Male mice, 4 to 6 wk old, were used for all experiments. Only male mice were used to avoid sexual difference in all in vivo experiments.
Stereotaxic Injection of α-Syn PFF.
C57BL/6J male mice, 4 to 6 wk old, were anesthetized with 0.5% isoflurane mixed with 1% O2. α-Syn PFFs (2 µg/µL) were stereotaxically injected into dSTR of both hemispheres at a dose of 0.2 µg/g (body weight). The following coordination was used: +0.2 mm to bregma, ± 2.0 mm from midline, and −2.6 mm from dura. The coordinates of each mouse were scaled by multiplying the scaling factor ζ () to acquire the accurate and consistent injecting location. The injection was done with a 10-μL microsyringe at a speed of 0.5 μL/min for the first 0.2 µL and 0.2 µL/min for the rest. Control mice were bilaterally injected with sterile PBS (1 µL/10 g (body weight)) with the same equipment and speed. After recovery from the anesthetization, mice were transferred back to their home cages with regular housing conditions until the next experiment.
Behavioral Tests.
All behavioral trainings and tests were carried out between Zeitgeber time 4 to 8 (4 to 8 h post light on). Mice were transferred to the behavioral room at least 1 h prior to the test for sufficient habituation to the testing environment. Final results were presented as absolute or normalized data (normalized to the PBS mice of each independent experiment if multiple batches of experiments were performed and compiled). Two-way ANOVA followed by Tukey post hoc was used for comparing the rotarod learning curve. One-way ANOVA followed by Tukey post hoc for pairwise multiple comparison between groups was used for all other statistical analysis. Only significant or close to significant pairwise comparisons were labeled in the figures. The level of significance was set at P < 0.05. *P < 0.05; **P < 0.01; ***P < 0.001, ****P < 0.0001.
Rotarod Test.
All mice were trained on the rotarod (YLS-4C, Sansbio) for three consecutive days. Each day, mice received two trials with constant speed (5 rpm/min) and the third one with accelerating speed from 0 to 40 rpm/min. Each trial lasts for 2 min with 3 min as interval. After a day of rest, mice were tested twice continuously on the rotarod expediting from 0 to 40 rpm/min in 1.5 min, then remained at 40 rpm/min for another 3.5 min. The maximum rotating speed and the total duration on the rotarod were recorded and used for final analysis. The rotarod was cleaned with 75% alcohol and air dried between each trial.
Open Field Test.
The open field test chamber (40 × 40 cm) was kept in a light/sound-proof box with a camera hanging on the ceiling. Light inside the chamber was kept at 30∼35 lx. After habituation, mice were placed in the center of the open field chamber and allowed to explore for 10 min. Their activities were monitored and recorded by the camera and analyzed by EthoVision XT (Noldus11.5). The chamber was completely cleaned with 75% alcohol and air dried between each trial.
Pole Test.
Metal rod (length = 50 cm, optical diameter = 1 cm) wrapped with medical tape was used for this test. After habituation, mice were placed on top of the rod and their heads were guided toward the ground. All mice were trained to climb down the rod once before testing for five times continuously to measure the climb-down duration. Each test lasted for no longer than 60 s. After then, mice were allowed to rest in their homecages for 30 min before five consecutive tests for the turn duration. To do that, mice were placed on top of the rod with their heads facing upward, the time used to turn their heads downward was recorded. For data analysis, four of the shortest durations from both tests were averaged to estimate the climb-down and turn duration, respectively. Total duration was the sum of climb-down and turn duration.
Immunofluorescence Staining.
Mice were anesthetized with isoflurane vapor and transcranially perfused with 10 mL ice-cold 1× PBS followed by 40 mL 4% paraformaldehyde (PFA). The whole brain was removed and kept in 4% PFA at least overnight before sectioning into 50-μm coronal slices with vibratome (Leica VT 1000S). Brain slices were kept in cryoprotectant solution (30% sucrose, 20% ethylene glycol, and 50% 1× PBS) at −20 °C. Right before staining, slices were rinsed with 1× PBS followed by 2 h incubation at RT with blocking solution: 5% normal goat serum (Sigma) and 0.1% Triton X-100 in 1× PBS. Then, slices were incubated in primary antibody diluted in blocking solution at 4 °C overnight. Slices were then rinsed in rinse buffer: 1× PBS with 0.1% Tween 20, 10 min/time for three times and once more with 1× PBS. Then, slices were incubated in blocking buffer containing second antibody for 2 h at RT. After rinsing in rinse buffer for 10 min × 3 and once more with 1× PBS, slices were incubated in 1× PBS with 1:5,000 4′,6-diamidino-2-phenylindole (DAPI) (Sigma) for 5 min at RT. The rinsing steps were repeated again before the slices were mounted on glass slide and air dried overnight. Mounting medium (Prolong gold antifade reagent, Invitrogen) was used to preserve fluorescence signal. Coronal brain slices by cryosection were also used for some sets of experiments. Fixed whole brain was merged in 30% sucrose–containing 1× PBS for 3 d before snap frozen in optimal cutting temperature compound (OCT, Tissue Freezing Medium, Leica) and stored in a −80 °C freezer. Next, 30-μm coronal slices were sectioned with Leica cryostat (model: CM3050s-1-1-1) and mounted on polylysine-coated glass slides. Slices were stored in a −80 °C freezer before staining. DAT staining was used to label DA neurons and their neurites. DAPI and neuronal nucleus marker (NeuN) staining were used as a reference of brain structure and served as general procedural quality control. The staining protocol was the same as describe above. Antibodies used include: phospho-α-synuclein (S129) monoclonal rabbit antibody (1:250, Abcam, ab51253); DAT monoclonal rat antibody (1:500, Abcam, ab5990); NeuN polyclonal chicken antibody (1:1,000, Millipore, ABN91); goat anti-rabbit and goat anti-rat IgG Alexa Fluor 568 (1:1,000, Abcam, ab175471), Alexa Fluor 647 (1:150, Abcam, ab150159), and goat anti-chicken Alexa Fluor 488 (1:1,000, Thermo Fisher, A-11039).
Confocal Imaging.
Fluorescence images were acquired by a spinning disk microscope (Andor). Lasers of different wavelengths, including 630-, 561-, 488-, and 405-nm, were used respectively to excite Fluor 647, 568, 488, and DAPI respectively and sequentially. Imaging settings were kept constant throughout individual experiment. A 20× (NA = 0.75) air or 40× (NA = 1.25) water immersion objective was used for imaging. The image resolution is 0.6 µm:0.6 µm/pixel (x:y) for 20× and 0.15 µm:0.15 µm/pixel (x:y) for 40×. Stack images were acquired for all brain regions. Step size (µm) and thickness (µm) of each brain region were as followed: SN: 0.5, 1; Striatum: 1, 4 or 5; Others: 1, 4.
Image Analysis.
Images were batched analyzed by Image J and Imaris (Oxford Instruments). First, all images were projected in z direction with max intensity followed by background subtraction. For all brain regions except SN, the whole view was analyzed for signal intensity for all channels. To visualize and analyze SN, montage images were used, and SN was manually selected by referring to the mouse brain atlas. With same background signal subtraction and same contrast transformation for each data set, the mean intensity of p-α-syn and DAT were measured. P-α-syn aggregates were selected automatically by using particle analysis module in ImageJ (auto black and white contrast, particle size between 2 and 80 µm). Aggregate density (#/1 mm2) and size distribution were calculated and reported. DAT+ DA neurons in SNpc were automatically selected and counted by the Spot module of Imaris. Briefly, after manually selecting the soma of one typical DAT+ cell as template, Imaris automatically selects all spherical structures with 12-µm diameter, which avoids most of the nonsoma DAT+ neurites signal. Then, the “Quality” filter was applied. The quality lower threshold was set between 36.4 and 872, judged by subjective observation to include all DAT+ somas and exclude noise. The distribution and average of quality lower threshold used in each group were similar. To report the immunofluorescence results, the mean p-α-syn signal intensity was normalized to hWT, while the mean DAT signal intensity was normalized to PBS control. For different batches of experiments, groups were normalized to their own control before combination. Raw value of p-α-syn aggregate density and DA neuron count were reported.
For all figures, the reported “n” represents the number of animals. Statistical analysis was performed by Prism V8.4 software (GraphPad Software, Inc.). For comparison among multiple groups, the ordinary one-way ANOVA followed by Fisher’s least significant difference post hoc test was used. Kolmogorov–Smirnov test was used to compare the cumulative distribution. All datasets were presented as mean ± SEM. Only significant or close to significant pairwise comparisons were labeled in the figures. The level of significance was set at P < 0.05. *P < 0.05; **P < 0.01; ***P < 0.001, ****P < 0.0001.
Supplementary Material
Acknowledgments
This work was supported by the Major State Basic Research Development Program (Grant No. 2019YFE0120600 to C.L.), the National Natural Science Foundation (NSF) of China (Grant No. 91853113 and 31872716 to C.L., and 31800709 and 32070963 to K.-W.H.), the Science and Technology Commission of Shanghai Municipality (Grant No. 18JC1420500 to C.L.), the "Eastern Scholar" project supported by the Shanghai Municipal Education Commission to D.L., The Shanghai Municipal Science and Technology Major Project (Grant No. 2019SHZDZX02 to C.L. and K.-W.H.), the Shanghai Science and Technology Committee (Grant No. 20XD1425000 to C.L.). We acknowledge the cryo-EM platform of Peking University for providing facility support in cryo-EM.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2012435118/-/DCSupplemental.
Data Availability
All data and procedures are provided in the manuscript and SI Appendix.
References
- 1.Spillantini M. G., et al., Alpha-synuclein in Lewy bodies. Nature 388, 839–840 (1997). [DOI] [PubMed] [Google Scholar]
- 2.Baba M., et al., Aggregation of alpha-synuclein in Lewy bodies of sporadic Parkinson’s disease and dementia with Lewy bodies. Am. J. Pathol. 152, 879–884 (1998). [PMC free article] [PubMed] [Google Scholar]
- 3.Braak H., Sandmann-Keil D., Gai W., Braak E., Extensive axonal Lewy neurites in Parkinson’s disease: A novel pathological feature revealed by alpha-synuclein immunocytochemistry. Neurosci. Lett. 265, 67–69 (1999). [DOI] [PubMed] [Google Scholar]
- 4.Kim S., et al., Transneuronal propagation of pathologic α-synuclein from the gut to the brain models Parkinson’s disease. Neuron 103, 627–641.e7 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Volpicelli-Daley L. A., et al., Exogenous α-synuclein fibrils induce Lewy body pathology leading to synaptic dysfunction and neuron death. Neuron 72, 57–71 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shahnawaz M., et al., Discriminating α-synuclein strains in Parkinson’s disease and multiple system atrophy. Nature 578, 273–277 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bousset L., et al., Structural and functional characterization of two alpha-synuclein strains. Nat. Commun. 4, 2575 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Peelaerts W., et al., α-Synuclein strains cause distinct synucleinopathies after local and systemic administration. Nature 522, 340–344 (2015). [DOI] [PubMed] [Google Scholar]
- 9.Peng C., et al., Cellular milieu imparts distinct pathological α-synuclein strains in α-synucleinopathies. Nature 557, 558–563 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guo J. L., et al., Distinct α-synuclein strains differentially promote tau inclusions in neurons. Cell 154, 103–117 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zarranz J. J., et al., The new mutation, E46K, of alpha-synuclein causes Parkinson and Lewy body dementia. Ann. Neurol. 55, 164–173 (2004). [DOI] [PubMed] [Google Scholar]
- 12.Lesage S.et al.; French Parkinson’s Disease Genetics Study Group , G51D α-synuclein mutation causes a novel parkinsonian-pyramidal syndrome. Ann. Neurol. 73, 459–471 (2013). [DOI] [PubMed] [Google Scholar]
- 13.Polymeropoulos M. H., et al., Mutation in the alpha-synuclein gene identified in families with Parkinson’s disease. Science 276, 2045–2047 (1997). [DOI] [PubMed] [Google Scholar]
- 14.Rovere M., et al., E46K-like α-synuclein mutants increase lipid interactions and disrupt membrane selectivity. J. Biol. Chem. 294, 9799–9812 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Flagmeier P., et al., Mutations associated with familial Parkinson’s disease alter the initiation and amplification steps of α-synuclein aggregation. Proc. Natl. Acad. Sci. U.S.A. 113, 10328–10333 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Greenbaum E. A., et al., The E46K mutation in α-synuclein increases amyloid fibril formation. J. Biol. Chem. 280, 7800–7807 (2005). [DOI] [PubMed] [Google Scholar]
- 17.Lázaro D. F., et al., Systematic comparison of the effects of alpha-synuclein mutations on its oligomerization and aggregation. PLoS Genet. 10, e1004741 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boyer D. R., et al., Structures of fibrils formed by α-synuclein hereditary disease mutant H50Q reveal new polymorphs. Nat. Struct. Mol. Biol. 26, 1044–1052 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Boyer D. R., et al., The α-synuclein hereditary mutation E46K unlocks a more stable, pathogenic fibril structure. Proc. Natl. Acad. Sci. U.S.A. 117, 3592–3602 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sun Y., et al., Cryo-EM structure of full-length α-synuclein amyloid fibril with Parkinson’s disease familial A53T mutation. Cell Res. 30, 360–362 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guerrero-Ferreira R., et al., Cryo-EM structure of alpha-synuclein fibrils. eLife 7, e36402 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guerrero-Ferreira R., et al., Two new polymorphic structures of human full-length alpha-synuclein fibrils solved by cryo-electron microscopy. eLife 8, e48907 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li B., et al., Cryo-EM of full-length α-synuclein reveals fibril polymorphs with a common structural kernel. Nat. Commun. 9, 3609 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li Y., et al., Amyloid fibril structure of α-synuclein determined by cryo-electron microscopy. Cell Res. 28, 897–903 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schweighauser M., et al., Structures of α-synuclein filaments from multiple system atrophy. Nature 585, 464–469 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ni X., McGlinchey R. P., Jiang J., Lee J. C., Structural insights into α-synuclein fibril polymorphism: Effects of Parkinson’s disease-related C-terminal truncations. J. Mol. Biol. 431, 3913–3919 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nussbaum R. L., Polymeropoulos M. H., Genetics of Parkinson’s disease. Hum. Mol. Genet. 6, 1687–1691 (1997). [DOI] [PubMed] [Google Scholar]
- 28.Pasanen P., et al., Novel alpha-synuclein mutation A53E associated with atypical multiple system atrophy and Parkinson's disease-type pathology. Neurobiol. Aging 35, 2180.e1-5 (2014). [DOI] [PubMed] [Google Scholar]
- 29.Íñigo-Marco I., et al., E46K α-synuclein pathological mutation causes cell-autonomous toxicity without altering protein turnover or aggregation. Proc. Natl. Acad. Sci. U.S.A. 114, E8274–E8283 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Prots I., et al., α-Synuclein oligomers induce early axonal dysfunction in human iPSC-based models of synucleinopathies. Proc. Natl. Acad. Sci. U.S.A. 115, 7813–7818 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dettmer U., et al., Parkinson-causing α-synuclein missense mutations shift native tetramers to monomers as a mechanism for disease initiation. Nat. Commun. 6, 7314 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nuber S., et al., Abrogating native α-synuclein tetramers in mice causes a L-DOPA-responsive motor syndrome closely resembling Parkinson’s disease. Neuron 100, 75–90.e5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhao K., et al., Parkinson’s disease associated mutation E46K of α-synuclein triggers the formation of a distinct fibril structure. Nat. Commun. 11, 2643 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Choi W., et al., Mutation E46K increases phospholipid binding and assembly into filaments of human alpha-synuclein. FEBS Lett. 576, 363–368 (2004). [DOI] [PubMed] [Google Scholar]
- 35.Fortin D. L., et al., Neural activity controls the synaptic accumulation of alpha-synuclein. J. Neurosci. 25, 10913–10921 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Burré J., et al., Alpha-synuclein promotes SNARE-complex assembly in vivo and in vitro. Science 329, 1663–1667 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Abeliovich A., et al., Mice lacking alpha-synuclein display functional deficits in the nigrostriatal dopamine system. Neuron 25, 239–252 (2000). [DOI] [PubMed] [Google Scholar]
- 38.Froula J. M., et al., α-Synuclein fibril-induced paradoxical structural and functional defects in hippocampal neurons. Acta Neuropathol. Commun. 6, 35 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Heo J. Y., et al., Aberrant tonic inhibition of dopaminergic neuronal activity causes motor symptoms in animal models of Parkinson’s disease. Curr. Biol. 30, 276–291.e9 (2020). [DOI] [PubMed] [Google Scholar]
- 40.Henderson M. X., et al., Glucocerebrosidase activity modulates neuronal susceptibility to pathological α-synuclein insult. Neuron 105, 822–836.e7 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tanaka M., Chien P., Naber N., Cooke R., Weissman J. S., Conformational variations in an infectious protein determine prion strain differences. Nature 428, 323–328 (2004). [DOI] [PubMed] [Google Scholar]
- 42.Tanaka M., Collins S. R., Toyama B. H., Weissman J. S., The physical basis of how prion conformations determine strain phenotypes. Nature 442, 585–589 (2006). [DOI] [PubMed] [Google Scholar]
- 43.Johnson M., Coulton A. T., Geeves M. A., Mulvihill D. P., Targeted amino-terminal acetylation of recombinant proteins in E. coli. PLoS One 5, e15801 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mastronarde D. N., Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 152, 36–51 (2005). [DOI] [PubMed] [Google Scholar]
- 45.Zheng S. Q., et al., MotionCor2: Anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rohou A., Grigorieff N., CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 192, 216–221 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.He S., Scheres S. H. W., Helical reconstruction in RELION. J. Struct. Biol. 198, 163–176 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tuttle M. D., et al., Solid-state NMR structure of a pathogenic fibril of full-length human α-synuclein. Nat. Struct. Mol. Biol. 23, 409–415 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Emsley P., Cowtan K., Coot: Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004). [DOI] [PubMed] [Google Scholar]
- 50.Adams P. D., et al., PHENIX: A comprehensive python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data and procedures are provided in the manuscript and SI Appendix.