Significance
Underground barriers caused by drought or heterogeneous soil components exert mechanical resistance to plants, and plant roots must determine whether to avoid or penetrate these barriers. Yet, very little is known about how roots sense mechanical forces at the molecular level. Using genetics, electrophysiology, and in vivo imaging, we found that Arabidopsis PIEZO1, an ortholog of mammalian Piezo, is required for root penetration and that pzo1 mutants have impaired primary root growth into harder media. These results highlight a conserved function of PIEZOs from plants to mammals.
Keywords: PIEZO, mechanosensation, ion channels, Arabidopsis, root
Abstract
Plant roots adapt to the mechanical constraints of the soil to grow and absorb water and nutrients. As in animal species, mechanosensitive ion channels in plants are proposed to transduce external mechanical forces into biological signals. However, the identity of these plant root ion channels remains unknown. Here, we show that Arabidopsis thaliana PIEZO1 (PZO1) has preserved the function of its animal relatives and acts as an ion channel. We present evidence that plant PIEZO1 is expressed in the columella and lateral root cap cells of the root tip, which are known to experience robust mechanical strain during root growth. Deleting PZO1 from the whole plant significantly reduced the ability of its roots to penetrate denser barriers compared to wild-type plants. pzo1 mutant root tips exhibited diminished calcium transients in response to mechanical stimulation, supporting a role of PZO1 in root mechanotransduction. Finally, a chimeric PZO1 channel that includes the C-terminal half of PZO1 containing the putative pore region was functional and mechanosensitive when expressed in naive mammalian cells. Collectively, our data suggest that Arabidopsis PIEZO1 plays an important role in root mechanotransduction and establish PIEZOs as physiologically relevant mechanosensitive ion channels across animal and plant kingdoms.
Plants extend roots within the soil to access water and nutrients as well as provide stability for the aerial parts of the plant. Underground barriers caused by drought and/or heterogeneous soil components can exert mechanical resistance that alters root extension and penetration (1–3). The root cap at the very tip of the primary root is a dynamic organ that contains different classes of stem cells that divide asymmetrically and is essential for growth through harder media and soils (4). Bending or poking root tips elicits a transient Ca2+ influx with short latency that is blocked by lanthanides, including Gd3+, a nonselective inhibitor of mechanically activated (MA) cation channels (5–7). However, the molecular identity of putative ion channels underlying this response is unknown. Only a few mechanosensitive ion channels have been described in plants (8). MSL8 plays a mechanosensory role in pollen (9), MSL10 is involved in cell swelling (8, 10), OSCA1 has mainly been characterized for its role in osmosensation (11), and OSCA1.3 regulates stomatal closure during immune signaling (12). It has been proposed that MCA1, expressed in the elongation zone but not the root cap, is a stretch-activated calcium permeable ion channel involved in soil penetration; however, evidence for its being a bona fide ion channel capable of detecting mechanical force is lacking (13–15). Recently, it has been shown that mechanosensitive Ca2+ channel activity is dependent on the developmental regulator DEK1; however, whether DEK1 is a pore-forming ion channel has not yet been addressed (16, 17). The genome of Arabidopsis thaliana encodes an ortholog of the mammalian mechanosensitive ion channels PIEZO1 and PIEZO2 (18). Given that PIEZOs play prominent roles in multiple aspects of animal mechanosensation and physiology (19–22), we investigated the role of A. thaliana PIEZO1 (PZO1) in plant mechanosensation. A recent study reported that PZO1 regulated virus translocation within the plant, but its specific role in mechanotransduction was not addressed (23). Here we use genetic tools, electrophysiological methods, and calcium imaging to investigate the role of PZO1 in root mechanosensation.
Results
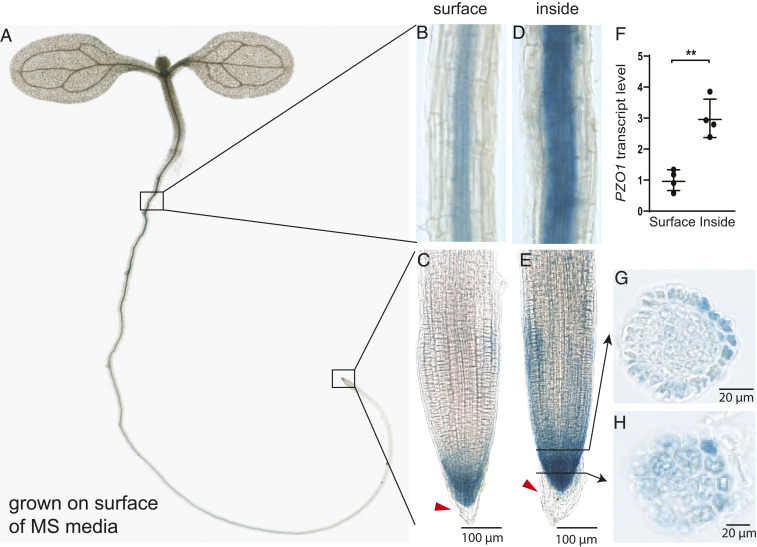
To localize the expression of PZO1 in Arabidopsis, we used PZO1 promoter fused to the reporter gene β-glucuronidase (GUS) and generated two PZO1pro::GUSPlus constructs with different promoter lengths, 823 base pairs (bp) and 2,000 bp. Both constructs showed similar GUS expression, with high levels observed in upper root, both primary and lateral root caps, and pollen grains (Fig. 1 A–C and SI Appendix, Fig. S1). We also detected GUS activity in the root vasculature and in trichromes (plant hairs) (Fig. 1B and SI Appendix, Fig. S1). Cross-sections of root tips revealed expression in lateral root cap (LRC) cells and columella cells (Fig. 1 G and H), which are thought to be important in detecting mechanical forces during root penetration into the soil (4). When plants were grown inside Murashige and Skoog medium (0.5× MS; 0.85% agar [8.5 g/L]) rather than on top of it, higher GUS signal intensity was observed in the upper root and root cap of the seedlings, suggesting expression is enhanced when mechanical stress is applied to roots (Fig. 1 D and E). This increase in GUS activity was confirmed by qRT-PCR; PZO1 expression was threefold higher in plants grown inside MS media (Fig. 1F).
Fig. 1.
Expression pattern of PZO1::GUSPlus reporter line in Arabidopsis root. (A) Expression pattern of the GUS reporter protein under 2,000 bp of the PZO1 promoter in a 7-d-old seedling. (B and C) Expression in the upper root and root tip when the plant is grown on the surface of standard MS media. Red arrowhead indicates that PZO1 is no longer expressed in the oldest root cap cells that are most distal and are known to be sloughed off. (D and E) Expression in the upper root and root tip grown inside the MS media. Black arrows indicate the cross-section of root displayed in G and H. (F) qRT-PCR for PZO1 in upper root of plants grown on the surface (“surface”) or within the MS media (“inside”). **P < 0.01, n = 4 (mean ± SD). (G and H) Cross-sections of the root cap in GUS reporter lines, which indicate expression in columella and LRC cells. The GUS staining images are representative from three different transgenic lines and four to five independent seedlings.
Next, to investigate the role of PZO1 in plant physiology and development, we generated two pzo1 CRISPR/Cas9 knockout mutant lines: one in which the entire gene was deleted (referred to as pzo1-5), the other in which the C-terminal half of the gene that encodes the putative channel pore based on its homology to mouse Piezo1 (mPiezo1) was deleted (referred to as pzo1-6). PZO1 has 27% amino acid identity with mPiezo1 with similar overall topology and 38 predicted transmembrane domains (SI Appendix, Fig. S2). We confirmed the lack of PZO1 transcripts in both mutants by PCR and qRT-PCR in samples harvested from the leaves as well as the roots (SI Appendix, Fig. S3). We did not observe any significant difference between wild type (WT) (Col-0) and pzo1 mutants in the growth of roots or aerial parts when grown in MS media or soil.
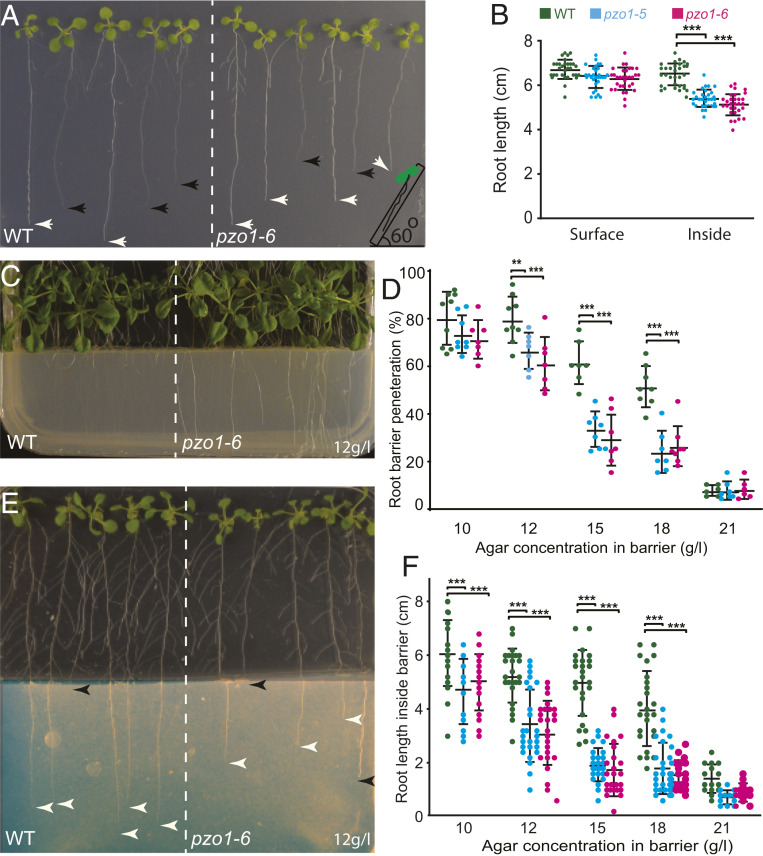
Based on the robust root expression of PZO1, we sought to evaluate its role in root growth. We grew A. thaliana seeds on the surface of the MS media and plates were positioned vertically (at a 90° angle). The length of the seedling roots of WT and the two mutants was not different when grown on top of the MS media (SI Appendix, Fig. S4 A and B). However, when seedling roots grew within the MS media, mutant roots were shorter compared to WT. To confirm the differences observed for root penetration and growth inside the media, we challenged the roots at a 60° plate angle to stimulate growth into the media. Again, we observed that the roots of both mutants were shorter than WT roots (Fig. 2 A and B). To further investigate media penetration, plants were grown at 60° in MS media containing different agar concentrations (in grams per liter: 7, 8.5 [standard], and 10 g/L) mimicking different levels of soil hardness. The root lengths of pzo1 mutants and WT plants were similar in the lowest agar concentration (7 g/L; SI Appendix, Fig. S4C). However, at higher agar concentrations (8.5 and 10 g/L), the average root lengths of both mutants were significantly shorter; for example at 8.5 g/l, the root length of WT was 5.7 ± 0.5 cm (n = 28), while it was 5.2 ± 0.4 cm and 5.2 ± 0.5 cm for pzo1-5 and pzo1-6, respectively (n = 30 to 36). These data show that roots of mutants had shorter lengths than WT in hard medium (SI Appendix, Fig. S4C).
Fig. 2.
Penetration of roots into hard media is compromised in pzo1 mutants. (A) Root length of Arabidopsis roots grown 9 d vertically at 60° to stimulate growth into the MS media. Black arrowheads indicate roots within the MS media and white arrowheads indicate root growth on the surface of MS media. (B) Root length of WT and pzo1 mutants 9 d after germination (n = 30 to 36; mean ± SD). (C) Representative plate of 18-d-old Arabidopsis seedlings challenged by root barriers. The more visible roots have not penetrated the barrier and are on the surface of the MS media. WT and mutants were grown vertically at 90° on normal MS media (0.5× MS + 8.5 g/L agar) before reaching the barrier (0.5× MS + 12 g/L agar). (D) The percentage of roots that penetrate barriers of different concentrations of agar as indicated (n = 8 to 11 plates) and each plate consists of 7 to 12 seedlings. (E) Representative plate of 2-wk-old Arabidopsis seedlings challenged by root barriers indicating the different root growth length in the denser media (imaged brightness was adjusted for clarity). White arrowheads indicate roots within the MS media and black arrowheads indicate root growth on the surface of MS media or roots that did not penetrate the barrier. (F) Root lengths of 2-wk-old seedlings that have penetrated the barriers (indicated) (n = 25 to 28, mean ± SD). The results were analyzed with two-way ANOVA and Tukey’s multiple comparison test. **P < 0.01, ***P < 0.001.
To assess the response of roots to stiff materials that might be encountered during growth, we challenged roots with barriers of varying stiffness consisting of 10, 12, 15, 18, and 21 g/L agar in MS media (Fig. 2D). We plated A. thaliana seeds on the standard agar concentration in MS media (8.5 g/L agar), 2 cm above the barrier. Within 4 to 5 d after germination, seedling roots of all genotypes reached the barrier. At this point, three different scenarios were observed: 1) penetration across the barrier, 2) root coiling and delayed penetration after growing at the interface surface, or 3) no penetration (SI Appendix, Fig. S5 C and D). At a 10 g/L barrier, 80% of WT roots penetrated the harder agar while only 74% and 73% of pzo1-5 and pzo1-6, respectively, were able to penetrate (n = 9). As the agar concentration increased, the barrier penetration phenotype in the mutants became more pronounced. For example, at 15 g/L, the penetration percentage for WT was 58% while only 29% of pzo1-5 and 26% of pzo1-6 roots penetrated (n = 11) (Fig. 2D and Movie S1). Furthermore, mutants showed a delayed penetration with excessive coiling on the barrier surface (SI Appendix, Fig. S5D).
For the roots that penetrated the various barriers, root length inside the barriers was significantly shorter in the mutants compared to WT (Fig. 2 E and F). For example, at a 12 g/L agar barrier concentration, the root length of WT was 5.2 ± 1.2 cm (n = 28), while it was 3.4 ± 1.3 cm and 3.1 ± 1.2 cm for pzo1-5 and pzo1-6, respectively (n = 25 to 28) (Fig. 2F). The shorter root length observed for mutants became more severe in 15 or 18 g/L agar. Although mutant root coiling at the barrier interface delays root penetration and contributes to the decreased total root length, shorter roots are observed for mutants seeded on media containing 8.5 and 10 g/L agar (SI Appendix, Fig. S4C), indicating that the velocity of root growth is slowed in denser media.
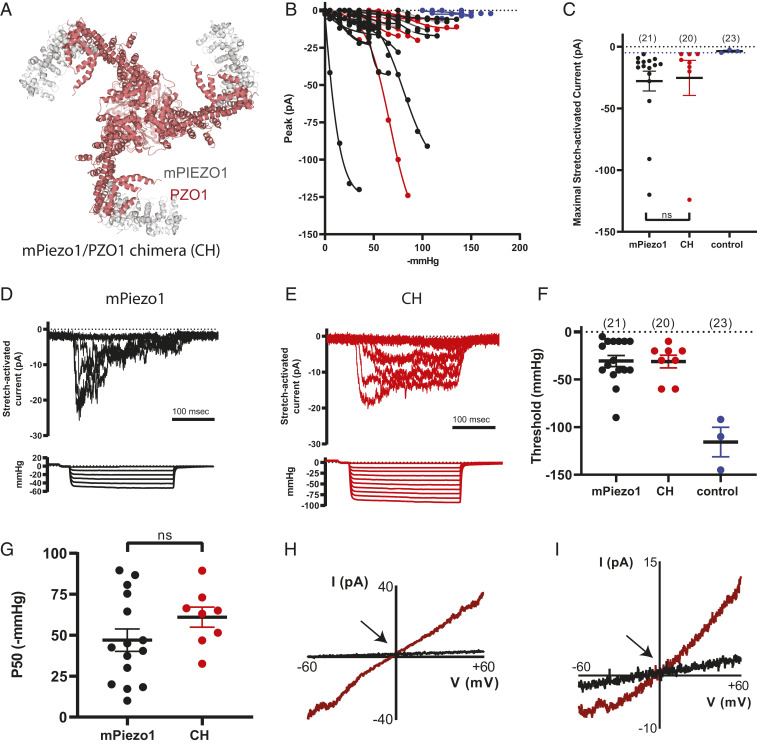
These results implicate a role of PZO1 in mechanosensory processes in plants. To assess whether PZO1 is a mechanosensitive ion channel like its animal homologs, we cloned the full-length coding sequence (7,455 bp) into a mammalian expression vector (Materials and Methods). Transient heterologous expression of PIEZO proteins from various animal species (including mammals and flies) confer robust MA currents (18–20). We heterologously expressed either native or codon-optimized PZO1 in HEK293T Piezo1 knockout (HEK P1KO) cells (24). Neither native nor codon-optimized PZO1 revealed MA currents in two separate assays for mechanotransduction: poking cells with a fire-polished glass pipette (25) and stretching the membrane at the tip of the pipette in cell-attached patch clamp recordings (25). To determine whether the lack of response was due to improper trafficking to the plasma membrane, MYC tags were inserted at five separate predicted extracellular loops of the protein based on homology between transmembrane domains of PZO1 and mPiezo1 (25–27) (SI Appendix, Fig. S2). Immunostaining with an anti-MYC antibody detected PZO1 expression in HEK P1KO cells; however, nonpermeabilized staining revealed that PZO1 did not traffic to the membrane (SI Appendix, Fig. S6). We next generated chimeras (CHs) between mPiezo1 and codon optimized PZO1 in an effort to traffic CHs containing the putative pore domain of PZO1 to the membrane. As the pore region of PIEZOs is located at the C terminus (25–28), we generated seven CHs between mPiezo1 and PZO1 in which the C terminus was derived from PZO1 and the N terminus from mPiezo1 (SI Appendix, Fig. S2). Using an extracellular Myc tag on the N-terminal mouse-derived sequence, we observed that one of the CHs mPiezo1/PZO1 with 49% mouse and 51% PZO1 trafficked to the membrane of HEK P1KO cells (Fig. 3A and SI Appendix, Fig. S6). The structural elements defining the channel pore and those that couple mechanical force to channel activation, including the anchor, clasp, and beam of the CH, are derived from PZO1 (29, 30). Stretch-activated currents (SACs) were observed in 40% of the cell-attached patches recorded from HEK P1KO cells expressing the CH; 76% of patches from mPiezo1-expressing cells revealed SACs. The maximum current elicited (Imax; Fig. 3C), negative pressure thresholds (Fig. 3F), and P50 values (Fig. 3G) were similar for mPiezo1 and CH (SI Appendix, Table S1). Interestingly, inactivation of SAC from CH-expressing cells was apparently abrogated as currents were maintained throughout the entire 250-ms stretch stimulus (Fig. 3E vs. Fig. 3D and SI Appendix, Table S1); the mechanism underlying the decreased ability of the CH to inactivate is unknown. The lower proportion of SAC-expressing patches in the CH is consistent with that observed for mPiezo2 (25, 28). The reversal potential of SACs mediated by mPiezo1 and the CH were similar (Fig. 3 H and I; stretch-induced current is shown in brown), consistent with the CH being a nonselective cation channel. Thus, the CH containing the pore-containing C terminus of PZO1 is activated by a mechanical stimulus, suggesting that the native PZO1 is indeed a nonselective ion channel in plant cells.
Fig. 3.
A chimeric channel that includes putative PZO1 pore sequences is activated by negative pressure applied to cell-attached patches. (A) The region of PZO1 in the CH is highlighted in the mPiezo1 structure; gray is mPiezo1 sequence (577 to 1,185 aa) and red is PZO1 sequence (1,228 to 2,485 aa). The first 577 aa of mPiezo1 are not resolved in the structure. (B) Stimulus-response curves are shown for all mPiezo1 (black) and mPiezo1/PZO1 CH (red) stretch-activated currents (SACs, Vpipette = +80 mV in cell-attached configuration). Small high threshold responses are occasionally observed in HEK P1KO cells transiently transfected with the empty IRES-GFP vector control (blue). The maximal current observed from vector-transfected cells was −5.4 pA (dotted line) and this value is used as a cutoff for identifying mPiezo1- and CH-mediated SAC. (C) Imax is shown for mPiezo1 (black), CH (red), and control cells (blue). (D and E) SACs recorded in a patch from cells expressing either mPiezo1 (D) or CH (E); current amplitudes increase with increasing negative pressure (shown below each family of currents). (F) The negative pressure (mmHg) at which the first response to stretch is observed (threshold) when patches are challenged with −5 mmHg increments is plotted. (G) The pressure producing half-maximal currents (P50, determined using GraphPad Prism) is shown. (H and I) A stretch stimulus eliciting a submaximal response is applied during a voltage ramp protocol in order to record SAC currents in WT mPiezo1- (H) and CH- (I) expressing cells between ±60 mV and determine the apparent reversal potential (Vrev; shown by arrows). ns, no statistical significance.
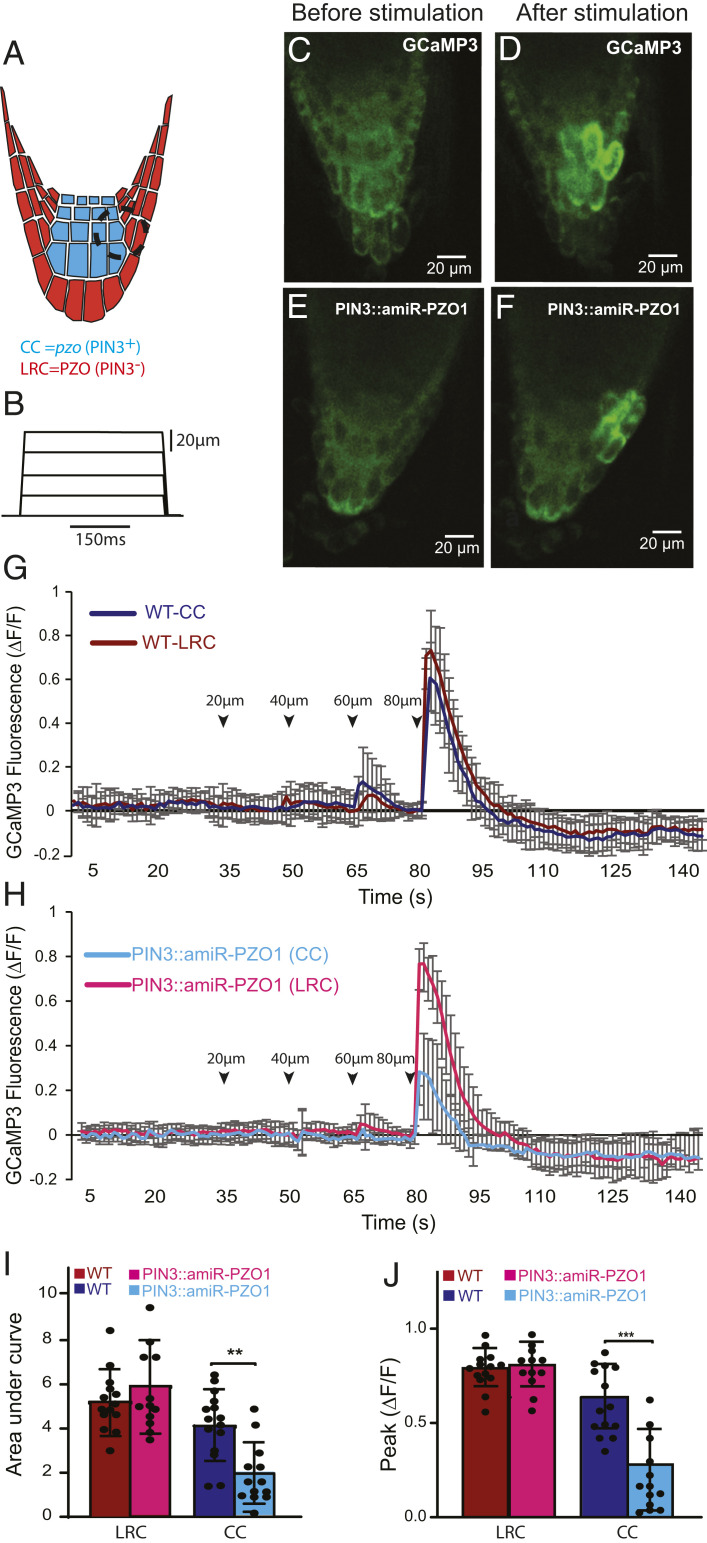
We next investigated whether the observed root growth phenotype could be attributed to compromised PZO1 channel activity in root tips challenged with mechanical forces. To accomplish this, we monitored the calcium response to mechanical stimulation in vivo using a transgenic line expressing a GFP-based Ca2+ indicator (GCaMP3) (31). First, we applied a localized stimulation by a blunt glass pipette to the root cap of WT plants in increments of 20 µm (Fig. 4 A and B). The transient and localized Ca2+ signals appeared in the columella cells and LRC cells starting at an indentation of ∼60 µm, while peak Ca2+ signals were observed at 80 µm of indentation (n = 15) (Fig. 4 C and D and Movie S2). This mechanical indentation did not cause damage to the cells as we evaluated the Ca2+ response after mechanical indentation with nonmechanical stimulation such as cold stress, which has been shown to trigger a Ca2+ response (32). Application of a cold stimulus induced Ca2+ signals in both columella and LRC cells (SI Appendix, Fig. S7). However, at 100-µm indentation and beyond, Ca2+ signals propagated between neighboring cells bidirectionally in a manner similar to a wound-mediated response and were not studied here (31, 33) (Movie S3). To overcome the variability of GCaMP3 signals among different plants, we next generated a model in which PZO1 was knocked down specifically in columella cells by using the artificial PZO1-targeting microRNA driven by the PIN3 promoter (PIN3::amiR-PZO1) (34) (Fig. 4A). This approach provides an internal control within each root tip: mechanical stimulation-dependent Ca2+ responses in columella cells (where PZO1 is knocked down) can be compared with neighboring WT LRC cells. Using this strategy, we observed normal responses in LRC cells but significantly reduced responses in the columella cells at 80 µm of indentation (n = 14, Fig. 4 E–H and Movie S4). Indeed, the area under the curve and the peak of GCaMP3 signals were significantly decreased in the columella but not LRC cells of pzo1 knockdown plants compared to WT plants (Fig. 4 I and J). These data indicate that mechanical stimuli can induce calcium transients in columella cells through PZO1.
Fig. 4.
PZO1 and calcium influx in response to mechanical indentation to the Arabidopsis root tip. (A) PZO1 expression was knocked down in the root cap using an artificial microRNA under the PIN3 promoter (PIN3::amiR-PZO1) such that PZO1 is specifically knocked down in columella cells (CCs) (blue) but PZO1 is still expressed in lateral root cap (LRC) cells (red). Black dashed circle indicates the area of stimulation by the blunt-tip pipette and the ROI for Ca2+ signal intensity measurement. (B) Displacements of the root cap by the stimulating pipette. Mechanical stimulation was initiated 30 to 35 s after GCaMP3 signal recording; a ramp (1 µm/ms) and hold (150 ms) stimulus was applied in 20-µm increments every 15 s. (C and D) Representative image of the GCaMP3 Ca2+ response in a WT root tip before (C) and after (D) a 80-µm mechanical stimulation (images are from Movie S2). (C and F) Representative image of the Ca2+ responses in PZO1 knockdown PIN3::amiR-PZO1 before and after an 80-µm mechanical stimulation (images are from Movie S4). (G) Ca2+ responses in the LRC cells and columella cells in response to mechanical stimulation in 7-d-old seedlings; n = 15 (mean ± SD). Arrowheads indicate the mechanical stimulation (in micrometers). (H) Ca2+ responses after mechanical stimulation in PZO1 knockdown of PIN3::amiR-PZO1 root cap in 7-d-old seedlings. Ca2+ transients are significantly reduced in columella cells compared to LRC cells; n = 14 (mean ± SD). (I) Area under the curve from starting the stimulation by an 80-μm indentation until the GCaMP3 signals returned to baseline (total of 15 s). (J) Maximum peak of fluorescence in WT and PIN3::amiR-PZO1 (pzo1 knockdown) (n = 14 to 15, mean ± SD). **P < 0.01, ***P < 0.001.
Discussion
Plant roots sense physical properties of the soil to either avoid it or penetrate it (3). Here, we report that Arabidopsis PZO1 activity is required for proper root penetration in compacted environments imposing mechanical stresses. PIEZO proteins from numerous animal species are established physiologically relevant MA cation channels (18–20). We present evidence to suggest that PZO1 is functionally conserved as a mechanosensitive ion channel in plant roots. Using calcium imaging we identified at least one cell type in the root cap (columella cells) that requires PZO1 to respond to a mechanical stimulus with increased calcium transients. Mutants in other components of calcium signaling pathways such as cml24 (tch2) show similar growth defects to those reported here for pzo1 (35). The receptor-like kinase FERONIA maintains cell wall integrity through a direct interaction between its extracellular domain and components of the cell wall; it has been proposed to activate a calcium permeable channel whose identity is unknown (36, 37). PZO1 protein might directly alleviate mechanical pressure in columella cells by protecting cell wall integrity and/or by transducing Ca2+ signals to other parts of the root such as the elongation zone. Our findings will enable future research to understand the molecular and cellular pathways involved in mechanotransduction within roots. Our results also suggest that other MA ion channels contribute to barrier penetration since the root growth deficits observed in pzo1 mutants are incomplete. In summary, we provide evidence that PZO1 acts as a mechanosensitive ion channel in root tips: it has appropriate expression in the root, is required for responses to acute mechanical stimulation and for proper root growth, and it forms an ion channel. These results demonstrate that PZO1 mediates a mechanosensory function in Arabidopsis, highlighting a conserved function of PIEZOs from plants to mammals.
Materials and Methods
Plant Material and Growth Conditions.
A. thaliana (Col-0) were soil-grown on a 14-h light/10-h dark cycle (100 μE⋅s−1⋅m−2), 70% humidity at 22 °C. For seedling propagation, seeds were surface sterilized with 70% ethanol for 15 min and washed five times with sterile distilled water. Seeds were grown on plates containing half-strength MS solid medium (0.5× MS) (Sigma-Aldrich, M5524), 0.5 g/L 2-(N-Morpholino)ethanesulfonic acid hydrate, 4-Morpholineethanesulfonic acid (MES hydrate) (Sigma- M2933), pH-adjusted to 5.7, and then supplemented with 8.5 g/L agar (Sigma-Aldrich, A1296). Plates were placed in a growth chamber vertically or as indicated.
Hard Agar Media and Barrier Plates.
First, we poured 160 mL of higher agar concentration in 0.5× MS media into 120-mm × 120-mm × 15-mm plates. After the plates had cured, we cut the solidified media with a sharp sterile blade into four parts. The first and third segments were removed and replaced with standard MS media containing agar at 8.5 g/L concentration with 5-mm thickness.
Generation of PZO1Pro:GUSPlus Plants.
In order to determine tissue-specific PZO1 expression, we used GUSPlus protein under the PZO1 promoter. Since, there is an annotated gene close to PZO1 in the opposite orientation, two different GUSPlus lines containing promoter sizes of 823 bp and 2,000 bp were generated. In order to faster screen the transgenic seeds, we first inserted the FAST (fluorescence-accumulating seed technology) cassette (38) into pCAMBIA-0380 plasmid (CAMBIA), referred to as pCAMBIA-FAST. The FAST cassette was amplified from VSP2Pro:GUSPlus plasmid (39) using forward atgttgggcccggcgcgc-cgagatctTCTAGTAACATAGATGACACC and reverse tggctgcaggtcgacgTCTAG-AGGTACCCGGGATCCAGTGTATGTAGGTATAGTAACATG primers. pCAMBIA-380 was cut by EcoRI and BamHI restriction enzymes (ThermoFisher). Both insert and plasmid were ligated using the Gibson assembly kit (New England Biolabs). The PZO1 promoters were amplified using forward ttgttcatgttactatacctacatacactgtggaaagaaagtaaaggattag and reverse agtagccatgTGGAAACTTTTGTCTTAACG primers and GUSPlus cDNA amplified from VSP2Pro:GUSPlus plasmid using forward aaagtttccaCATGGCTACTACTAAGCATTTG and reverse gtcagatctaccatggtggactcctcttaaCAATTCACACGTGATGGTG primers. pCAMBIA-FAST was cut using BamHI and HindIII. Both inserts and plasmid were ligated by the Gibson assembly kit. Agrobacterium tumefaciens (GV3101)-competent cells were transformed with this plasmid. Seeds that expressed red fluorescence protein (RFP) were selected by fluorescence microscopy. The T3 generation was used for GUS staining experiments.
GUS Staining and Cross-Sectioning.
Transgenic Arabidopsis plants expressing GUS were stained following the protocol described by Jefferson et al. (40). Five- to ten-day-old seedlings or flowers were collected. Samples were stained with 1 mM X-Gluc (Thermo Fisher Scientific, R0851) in a pH 7.0 phosphate buffer containing 10 mM ethylenediaminetetraacetic acid tetrasodium salt dihydrate (EDTA), 0.15 mM potassium ferricyanide, 5 mM potassium ferrocyanide and 10% (vol/vol) Triton X-100 at 37 °C overnight. The tissue was destained with serial dilution of 25%, 50%, 75%, and 95% (vol/vol) ethanol. For root cross-sectioning, the GUS-stained roots were submerged in a box containing Tissue-Tek O.C.T. compound (Sakura FineTek USA) and then frozen in liquid nitrogen. Different layers of the root were sectioned at 15-µm thickness and placed on coverslips.
Generating pzo1 Mutants and Genotyping.
Since homozygote seeds (insertion in exon) from Salk T-DNA lines were unavailable, we used CRISPR/Cas9 gene editing technology to generate pzo1-5 (full-length deletion) and pzo1-6 (C-terminal deletion) knockout mutants (41–43). Both pzo1 mutants were generated in the pNano65 Ca2+ indicator background (44) and these lines and pNano65 WT were used for all experiments except Ca2+ imaging experiments. Guide RNA 4 and 6 were used for generating a whole deletion in the PZO1 gene, and guide RNA 4 and 5 were used for generating a C-terminal deletionfrom the LGYL motif to the end of the PZO1 gene (1,253 aa to 2,485 aa). The target sequences for the whole deletion in PZO1 were target 4 CCCTCTGCTCTAGCCGCGTACA and target 6 AGGTGCAATATAGTGAACGAGG (protospacer adjacent motif sites are bold). Targets for the partial mutants were target 4 and target 5 CCCAGGTGTAGAATGTCATACT. Primers for genotyping are as follows: PZO1-GT1 TCTGCCACATTCCCACTCAG, PZO1-GT2 GGTTTAGCCATTTCTCGGCG, PZO1-GT3 CGGGAGTGTTGGCTTGGTAT, and PZO1-GT4 CCGCCACGTAAGTTAGCTCT. The pzo1-5 mutants were genotyped with the primer pair PZO1-GT1 and PZO1-GT2, which generates a PCR product of 1,300 bp in mutants. This primer pair could not amplify WT genomic DNA due to the large size of the fragment. To determine zygosity of pzo1-5 mutants, we used the PZO1-GT1 + PZO1-GT4, which amplifies a 949-bp product from WT DNA, but could not amplify a band in the homozygous mutant. For the C-terminal deletion mutant pzo1-6, we used PZO1-GT1 and PZO1-GT3, which would generate a fragment of about 1,000 bp in mutants. PCR of PZO1-GT1 + PZO1-GT4 was performed to determine the zygosity of pzo1-6 as well.
Generating pzo1 Knockdown Mutant.
We used atMIR390 microRNA (45) to knockdown PZO1 in columella cells. We synthesized atMIR390 microRNA that included the PZO1 targeting sequence of TGCAGTTGCTCGGTCTTCCGA- and was amplified using the forward TTTTTGTCCCTTCAAGTATAGGGGGGA-AAAAAAGGTAG and reverse tcttaaagcttggctgcaggGAGACTAAAGATGAGA-TCTAATC primers. PIN3 promoter was amplified using the forward cggcgcgccgaattcccgggAATTTTATTGCATATAGTGTGTTTATTAAATG and reverse TTTTTTCCCCCCTATACTTGAAGGGACAAAAATGGAAAAC primers. pCAMBIA-1380 plasmid was cut by BamHI and SalI restriction enzymes, and the cut plasmids and both fragments were ligated using the Gibson assembly kit. A. tumefaciens, GV3101-competent cells were then transformed by this plasmid. GCaMP3 plants were transformed using A. tumefaciens bacteria. MS media containing 10 mg/L hygromycin B was used for screening the transgenic seeds. Two different independent T3 transgenic lines were used for Ca2+ imaging. We were not able to measure the efficiency of PZO1 knockdown in the columella cells due to difficulty of isolation of the columella cells from neighboring cells to perform qRT-PCR; thus the efficiency of PZO1 knockdown in the columella cells is unknown.
qRT-PCR.
The upper roots of the Arabidopsis plant grown either inside or on top of the surface of the MS media were harvested and total RNA was extracted using the RNeasy plant mini extraction kit (Qiagen). A total of 1 µg of the total RNA was copied into cDNA with the SuperScript III First-Strand Synthesis according to the manufacturer’s instructions (ThermoFisher). qRT-PCR was performed on 50 ng of cDNA in a final volume of 10 µL according to the SsoAdvanced Universal SYBR Green supermix (Bio-Rad). Ubiquitin-conjugating enzyme (UBC21) At5g25760 was used as a reference gene. Four biological replicates, which were a mixture of three to five plants, were used for each experiment. Primers used were: UBC21 (At5g25760) forward CAGTCTGTGTGTAGAGCTATCATAGCAT and reverse AGAAGATTCCCTGAG-TCGCAGTT and PZO1 (AT2G48060) forward ACGCTCTGATATCCAAATGGT- and reverse ACTTCATCCGTCTGATCCTC.
Cloning PZO1.
The full-length of 7,455 bp of PZO1 was amplified in two parts from Arabidopsis total RNA extracted from roots and leaves. The PZO1 gene was cloned into pCDNA3-1 IRES2-eGFP Zeo+ (PZO1GFP), a mammalian expression vector. The target gene and eGFP are transcribed in the same mRNA, but the proteins are expressed independently through IRES2. Therefore, expression of eGFP does not affect the functionality of the protein. We also generated a human codon-optimized PZO1 (Genewiz) for expression in mammalian cell culture (HEK P1KO cells).
Generating Myc-Tagged Plasmids.
The sites of Myc-tag insertion were chosen based on homology alignments between mPiezo1 and PZO1 and the tag was shown to work for these experiments in the extracellular loop of mPiezo1. The position of the Myc tag is highlighted in SI Appendix, Fig. S2. The sequence of Myc tag (GAACAAAAACTTATTTCTGAAGAAGATCTG) was inserted in the primers. The Myc-tag insertion was cloned into pCDNA3-1 IRES2-eGFP Zeo+ (PZO1 GFP). Myc tags 1 to 3 were inserted in codon-optimized PZO1 and Myc 4 and 5 in the native codon of PZO1. All plasmids were generated by the Gibson assembly kit (NEB) according to the manufacturer’s instructions. The sequences of the primers are in SI Appendix, Table S2.
Generating Chimeras.
The chimeras (CH) between mPiezo1 and PZO1 were generated by swapping specific regions highlighted in SI Appendix, Fig. S2. The N terminus of all CHs was derived from mPiezo1, and the C terminus was derived from a mammalian codon-optimized PZO1. The junction between CHs was chosen based on conserved amino acid regions between mPiezo1 and PZO1. We amplified the fragments using Q5 polymerase (NEB). pCDNA3-1 IRES2-eGFP Zeo+ plasmid was cut by BamHI and EcoRI, followed by ligation of plasmid and fragments using the Gibson assembly kit (NEB). The ligated plasmids were transformed into XL-gold competent cells and colonies were screened. All positive CH clones were verified by full-length DNA sequencing.
Electrophysiological Characterization of Arabidopsis PIEZO (PZO1), mPiezo1, and the CH.
HEK P1KO cells were transiently transfected with PZO1, mPiezo1, mPiezo1/PZO1 CH, or empty vector using Lipofectamine 2000 per manufacturer’s instructions and allowed to settle on 12-mm diameter Poly-D-Lysine-coated coverslips as previously described (24). Electrophysiological recordings were made 2 to 3 d after transfection using cell-attached patch clamp methods to determine their responsiveness to membrane stretch using a High Speed Pressure Clamp HSPC-1 (ALA Instruments) (25, 46). Pipettes had resistances of 1.4 to 2.6 MΩ when filled with 130 mM NaCl, 5 KCl, 1 CaCl2, 1 MgCl2, 10 TEA-Cl, and 10 HEPES (pH 7.3 with NaOH). Cells were exposed to a high KCl bath solution during recording to depolarize the membrane potential (140 KCl, 1 MgCl2, 10 glucose, 10 HEPES [pH 7.3 with KOH]). Negative pressure steps (250 ms in duration) were applied after 6 s at +5 mmHg followed by 20 ms at 0 mmHg; steps of increasing negative pressure in −5 mmHg increments were applied every 15 s. For SAC recordings, the Multiclamp700A feedback resistor used was either 5 or 50 GΩ depending on the size of the currents. To determine the apparent reversal potential (Vrev), a stretch stimulus eliciting a submaximal response was applied during a voltage ramp protocol in order to record SAC currents between ±60 mV. The apparent Vrev determined from this voltage ramp protocol was validated in the same patch with a typical voltage step protocol. Whole cell recordings were made as described for PZO1-transfected cells 2 to 3 d after transfection (19).
In Vivo Ca2+ Imaging.
A. thaliana plants expressing genetically encoded Ca2+ (GCaMP3), obtained from Edward E. Farmer, University of Lausanne, Lausanne, Switzerland were used for in vivo imaging. Images were collected with a Nikon Instruments A1R+ confocal mounted onto an inverted Ti-E microscope. An S Plan Fluor ELWD 20× objective, numerical aperture (NA) 0.45, was used to acquire images at 1 frame/s 1,024 × 512 scan area (frame rate 0.946 ms/frame), 0.62 microns/pixel, pinhole 1.4 AU, laser power 0.2 microwatts out of the objective. A Coherent 488-nm solid-state laser was used for excitation, with a Chroma 525/50 emission filter. Nikon Elements software was used for time lapse intensity measurements. GCaMP3 imaging was recorded 30 to 35 s before applying mechanical stimulation. A day before Ca2+ imaging, 5- to 7-d-old seedlings were transferred onto a 60-mm × 24-mm coverglass covered by 1 to 2 mm of MS media in 0.6% low melting agarose (IBI Scientific). MS media provide nutrients for plant growth and also function as a cushion for seedlings during mechanical indentation to prevent tissue damage. The GCaMP3 plants were excited using a mercury lamp, 488-nm laser, and emission filter of 525/50 nm with an Andoer 897 EM-CCD camera. The GFP signals of several regions of interest (ROI) such as columella cells and lateral root cap cells were analyzed using the NIS-Elements imaging software. Representative images are shown in Fig. 4, after adjusting brightness and contrast for clarity in publication. We used (ΔF/F), the equation ΔF/F = (F − F0)/F0 for analyses of the fluorescence changes. F0 is baseline fluorescence that was calculated from 10 s before stimulation and F is fluorescence of the recording. We estimated the area under the curve using the equation (y1 + y2)/(2*(t2 − t1)), where y is the value of (ΔF/F) and t is each time point of the GCaMP3 recording.
Mechanical Stimulation to Root.
For plant in vivo Ca2+ measurements, mechanical stimulation was achieved using a fire-polished glass pipette (tip diameter 15 to 20 μm) positioned at an angle of 80° to the recorded cells. Downward movement of the probe was driven by a Clampex-controlled piezo-electric crystal microstage (E625 LVPZT Controller/Amplifier; PhysikInstrumente). The probe had a velocity of 1 μm⋅ms−1 during the ramp segment of the command for forward motion and the stimulus was held for 150 ms before releasing the stimulus. To assess the mechanical responses of a cell, the probe was first placed as close to the cell as possible (this distance could vary from plant to plant). We optimized the mechanical stimulation and found that four series of mechanical steps in 20-μm increments every 15 s led to a transient and local Ca2+ response. Longer or more mechanical steps led to Ca2+ fluxes traveling bidirectionally, both sides of the region being poked, resembling damage/wounding in the Ca2+ signaling. To control the mechanical stimulation after mechanical indentation, we applied 20 μL of cold water (4 °C) to the root, ∼1 cm from the root tip.
Statistics.
All results in the main figures and supplementary data with error bars are represented as mean ± SD according to standard methods using Microsoft Excel or GraphPad Prism. The P values were generated with Student’s one-tail unpaired t tests or with two-way ANOVA with Tukey’s multiple comparison test. For qRT-PCR experiments, four biological replicates and three technical replicates were used. The biological replicates are indicated as “n” in the figure legends.
Supplementary Material
Acknowledgments
We thank Dr. Simon Gilroy, Dr. Edward E. Farmer, Dr. Elliot Meyerowitz, and Dr. Ting Li for seeds. We also thank Dr. Kathryn R. Spencer for help with in vivo Ca2+ imaging and Dr. Elizabeth S. Haswell, Dr. Steve Kay, Dr. Julian Schroeder, Haley DeGuzman, Tess Whitwam, Shang Ma, Ivan Radin, Kara Marshall, Yunxiao Zhang, Swetha Murthy, Anant Gharpure, and Viktor Lukacs for discussion and critical reading of the manuscript. Funding was provided by an early and advanced mobility fellowship from the Swiss National Science Foundation (P300PA_164695 and P2LAP3_151727 to S.A.R.M.). Y.Z. was supported by NIH GM114660, and C.G. was supported by a scholarship from the China Scholarship Council. This work was supported by the Howard Hughes Medical Institute and partly supported by National Heart, Lung, and Blood Institute Grant R01HL143297.
Footnotes
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2102188118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or supporting information.
References
- 1.Bengough A. G., McKenzie B. M., Hallett P. D., Valentine T. A., Root elongation, water stress, and mechanical impedance: A review of limiting stresses and beneficial root tip traits. J. Exp. Bot. 62, 59–68 (2011). [DOI] [PubMed] [Google Scholar]
- 2.Clark L. J., Whalley W. R., Barraclough P. B., How do roots penetrate strong soil? Plant Soil 255, 93–104 (2003). [Google Scholar]
- 3.Iijima M., Kato J., Taniguchi A., Combined soil physical stress of soil drying, anaerobiosis and mechanical impedance to seedling root growth of four crop species. Plant Prod. Sci. 10, 451–459 (2007). [Google Scholar]
- 4.Kumpf R. P., Nowack M. K., The root cap: A short story of life and death. J. Exp. Bot. 66, 5651–5662 (2015). [DOI] [PubMed] [Google Scholar]
- 5.Monshausen G. B., Bibikova T. N., Weisenseel M. H., Gilroy S., Ca2+ regulates reactive oxygen species production and pH during mechanosensing in Arabidopsis roots. Plant Cell 21, 2341–2356 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Legué V., et al., Cytoplasmic free Ca2+ in Arabidopsis roots changes in response to touch but not gravity. Plant Physiol. 114, 789–800 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Toyota M., Furuichi T., Tatsumi H., Sokabe M., Cytoplasmic calcium increases in response to changes in the gravity vector in hypocotyls and petioles of Arabidopsis seedlings. Plant Physiol. 146, 505–514 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Basu D., Haswell E. S., Plant mechanosensitive ion channels: An ocean of possibilities. Curr. Opin. Plant Biol. 40, 43–48 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hamilton E. S., et al., Mechanosensitive channel MSL8 regulates osmotic forces during pollen hydration and germination. Science 350, 438–441 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Basu D., Haswell E. S., The mechanosensitive ion channel MSL10 potentiates responses to cell swelling in Arabidopsis seedlings. Curr. Biol. 30, 2716–2728.e6 (2020). [DOI] [PubMed] [Google Scholar]
- 11.Yuan F., et al., OSCA1 mediates osmotic-stress-evoked Ca2+ increases vital for osmosensing in Arabidopsis. Nature 514, 367–371 (2014). [DOI] [PubMed] [Google Scholar]
- 12.Thor K., et al., The calcium-permeable channel OSCA1.3 regulates plant stomatal immunity. Nature 585, 569–573 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kamano S., et al., Transmembrane topologies of Ca2+-permeable mechanosensitive channels MCA1 and MCA2 in Arabidopsis thaliana. J. Biol. Chem. 290, 30901–30909 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shigematsu H., et al., Structural characterization of the mechanosensitive channel candidate MCA2 from Arabidopsis thaliana. PLoS One 9, e87724 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nakagawa Y., et al., Arabidopsis plasma membrane protein crucial for Ca2+ influx and touch sensing in roots. Proc. Natl. Acad. Sci. U.S.A. 104, 3639–3644 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tran D., et al., A mechanosensitive Ca2+ channel activity is dependent on the developmental regulator DEK1. Nat. Commun. 8, 1009 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Amanda D., et al., DEFECTIVE KERNEL1 (DEK1) regulates cell walls in the leaf epidermis. Plant Physiol. 172, 2204–2218 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Coste B., et al., Piezo1 and Piezo2 are essential components of distinct mechanically activated cation channels. Science 330, 55–60 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Murthy S. E., Dubin A. E., Patapoutian A., Piezos thrive under pressure: Mechanically activated ion channels in health and disease. Nat. Rev. Mol. Cell Biol. 18, 771–783 (2017). [DOI] [PubMed] [Google Scholar]
- 20.Kim S. E., Coste B., Chadha A., Cook B., Patapoutian A., The role of Drosophila Piezo in mechanical nociception. Nature 483, 209–212 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ranade S. S., et al., Piezo2 is the major transducer of mechanical forces for touch sensation in mice. Nature 516, 121–125 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Woo S.-H., et al., Piezo2 is the principal mechanotransduction channel for proprioception. Nat. Neurosci. 18, 1756–1762 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang Z., et al., Genetic analysis of a Piezo-like protein suppressing systemic movement of plant viruses in Arabidopsis thaliana. Sci. Rep. 9, 3187 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dubin A. E., et al., Endogenous Piezo1 can confound mechanically activated channel identification and characterization. Neuron 94, 266–270.e3 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Coste B., et al., Piezo1 ion channel pore properties are dictated by C-terminal region. Nat. Commun. 6, 7223 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guo Y. R., MacKinnon R., Structure-based membrane dome mechanism for Piezo mechanosensitivity. eLife 6, e33660 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saotome K., et al., Structure of the mechanically activated ion channel Piezo1. Nature 554, 481–486 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang L., et al., Structure and mechanogating of the mammalian tactile channel PIEZO2. Nature 573, 225–229 (2019). [DOI] [PubMed] [Google Scholar]
- 29.Jiang Y., Yang X., Jiang J., Xiao B., Structural designs and mechanogating mechanisms of the mechanosensitive piezo channels. Trends Biochem. Sci., 10.1016/j.tibs.2021.01.008 (2021). [DOI] [PubMed] [Google Scholar]
- 30.Lewis A. H., Grandl J., Inactivation kinetics and mechanical gating of Piezo1 ion channels depend on subdomains within the cap. Cell Rep. 30, 870–880.e2 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nguyen C. T., Kurenda A., Stolz S., Chételat A., Farmer E. E., Identification of cell populations necessary for leaf-to-leaf electrical signaling in a wounded plant. Proc. Natl. Acad. Sci. U.S.A. 115, 10178–10183 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wilkins K. A., Matthus E., Swarbreck S. M., Davies J. M., Calcium-mediated abiotic stress signaling in roots. Front. Plant Sci. 7, 1296 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Toyota M., et al., Glutamate triggers long-distance, calcium-based plant defense signaling. Science 361, 1112–1115 (2018). [DOI] [PubMed] [Google Scholar]
- 34.Friml J., Wiśniewska J., Benková E., Mendgen K., Palme K., Lateral relocation of auxin efflux regulator PIN3 mediates tropism in Arabidopsis. Nature 415, 806–809 (2002). [DOI] [PubMed] [Google Scholar]
- 35.Wang Y., Wang B., Gilroy S., Wassim Chehab E., Braam J., CML24 is involved in root mechanoresponses and cortical microtubule orientation in Arabidopsis. J. Plant Growth Regul. 30, 467–479 (2011). [Google Scholar]
- 36.Shih H.-W., Miller N. D., Dai C., Spalding E. P., Monshausen G. B., The receptor-like kinase FERONIA is required for mechanical signal transduction in Arabidopsis seedlings. Curr. Biol. 24, 1887–1892 (2014). [DOI] [PubMed] [Google Scholar]
- 37.Ngo Q. A., Vogler H., Lituiev D. S., Nestorova A., Grossniklaus U., A calcium dialog mediated by the FERONIA signal transduction pathway controls plant sperm delivery. Dev. Cell 29, 491–500 (2014). [DOI] [PubMed] [Google Scholar]
- 38.Shimada T. L., Shimada T., Hara-Nishimura I., A rapid and non-destructive screenable marker, FAST, for identifying transformed seeds of Arabidopsis thaliana. Plant J. 61, 519–528 (2010). [DOI] [PubMed] [Google Scholar]
- 39.Mousavi S. A. R., Chauvin A., Pascaud F., Kellenberger S., Farmer E. E., GLUTAMATE RECEPTOR-LIKE genes mediate leaf-to-leaf wound signalling. Nature 500, 422–426 (2013). [DOI] [PubMed] [Google Scholar]
- 40.Jefferson R. A., Kavanagh T. A., Bevan M. W., GUS fusions: Beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6, 3901–3907 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gao X., Chen J., Dai X., Zhang D., Zhao Y., An effective strategy for reliably isolating heritable and Cas9-free Arabidopsis mutants generated by CRISPR/Cas9-Mediated genome editing. Plant Physiol. 171, 1794–1800 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gao Y., et al., Auxin binding protein 1 (ABP1) is not required for either auxin signaling or Arabidopsis development. Proc. Natl. Acad. Sci. U.S.A. 112, 2275–2280 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gao Y., Zhao Y., Self-processing of ribozyme-flanked RNAs into guide RNAs in vitro and in vivo for CRISPR-mediated genome editing. J. Integr. Plant Biol. 56, 343–349 (2014). [DOI] [PubMed] [Google Scholar]
- 44.Choi W.-G., Toyota M., Kim S.-H., Hilleary R., Gilroy S., Salt stress-induced Ca2+ waves are associated with rapid, long-distance root-to-shoot signaling in plants. Proc. Natl. Acad. Sci. U.S.A. 111, 6497–6502 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Carbonell A., et al., New generation of artificial MicroRNA and synthetic trans-acting small interfering RNA vectors for efficient gene silencing in Arabidopsis. Plant Physiol. 165, 15–29 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Murthy S. E., et al., OSCA/TMEM63 are an evolutionarily conserved family of mechanically activated ion channels. eLife 7, e41844 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or supporting information.