Significance
Enzymes of the chlorophyll biosynthetic pathway, which bind protoporphyrin and Mg-porphyrins, are susceptible to damage by singlet oxygen production in the presence of light and oxygen. These studies show that heme-derived linear tetrapyrroles (bilins) both stimulate and protect the protoporphyrin-binding CHLH subunit of Mg chelatase, the first committed enzyme of the chlorophyll synthesis, from self-sensitized photodamage and turnover via formation of nonphotosensitizing GENOMES UNCOUPLED 4 (GUN4):bilin:porphyrin adducts, which deliver protoporphyrin to CHLH. GUN4:bilin adducts likely evolved to sustain chlorophyll biosynthesis in an oxic world, accounting for retention of bilin synthesis in nearly all oxygenic photosynthetic species on Earth.
Keywords: heme oxygenase, bilin reductase, reactive oxygen species, photosynthesis, phycocyanobilin
Abstract
Biosyntheses of chlorophyll and heme in oxygenic phototrophs share a common trunk pathway that diverges with insertion of magnesium or iron into the last common intermediate, protoporphyrin IX. Since both tetrapyrroles are pro-oxidants, it is essential that their metabolism is tightly regulated. Here, we establish that heme-derived linear tetrapyrroles (bilins) function to stimulate the enzymatic activity of magnesium chelatase (MgCh) via their interaction with GENOMES UNCOUPLED 4 (GUN4) in the model green alga Chlamydomonas reinhardtii. A key tetrapyrrole-binding component of MgCh found in all oxygenic photosynthetic species, CrGUN4, also stabilizes the bilin-dependent accumulation of protoporphyrin IX-binding CrCHLH1 subunit of MgCh in light-grown C. reinhardtii cells by preventing its photooxidative inactivation. Exogenous application of biliverdin IXα reverses the loss of CrCHLH1 in the bilin-deficient heme oxygenase (hmox1) mutant, but not in the gun4 mutant. We propose that these dual regulatory roles of GUN4:bilin complexes are responsible for the retention of bilin biosynthesis in all photosynthetic eukaryotes, which sustains chlorophyll biosynthesis in an illuminated oxic environment.
Chlorophylls and hemes comprise the core light-harvesting and energy-converting pigments in the photosynthetic apparatus of all oxygenic species and descendants of their nonoxygenic ancestors. Since both classes of tetrapyrroles are pro-oxidants, it is essential that their biosynthetic pathways are tightly regulated in aerobic environments (1, 2). Indeed, the chlorophyll pathway intermediates protoporphyrin IX (PPIX), MgPPIX, and protochlorophyllide (Pchlide) readily generate reactive oxygen species (ROS) via photosensitization of molecular oxygen upon light exposure, resulting in chlorophyll biosynthetic enzyme inactivation, reaction center turnover, antennae photobleaching, and cell death of photosynthetic tissues (3–5). Such photooxidative turnover does not occur in anoxygenic photosynthetic species, which perform photosynthesis in suboxic or anaerobic environments. Because of the light-lability of the photosynthetic apparatus, all oxygenic phototrophs therefore require protective systems to sustain chlorophyll synthesis in the presence of oxygen.
Metabolic regulation of biosynthetic pathways most commonly occurs at the first committed step of the pathway or at branch points that redirect metabolite flow to different products. In nearly every aerobic species examined, heme biosynthesis is regulated by feedback inhibition of the enzyme that yields 5-aminolevulinic acid (ALA), the first committed intermediate of its synthesis (6). Since chlorophyll and heme pathways share ALA as a common precursor, the regulation of ALA levels in photosynthetic species affects both pathways (6, 7). The most obvious targets for selective regulation of the individual pathways are the enzymes that insert iron or magnesium into PPIX, the last common intermediate of the two pathways (Fig. 1A).
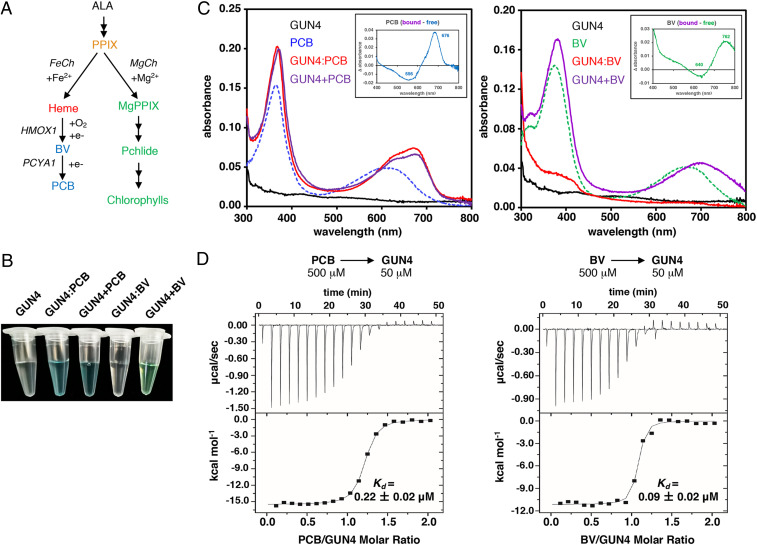
Fig. 1.
CrGUN4 interacts with bilins. (A) Tetrapyrrole pathways in C. reinhardtii. heme, protoheme IX (Heme B); PCYA1, phycocyanobilin:ferredoxin oxidoreductase. (B) Appearance of purified recombinant CrGUN4 apoprotein (GUN4) and those coexpressed in the presence of PCB (GUN4:PCB) or BV (GUN4:BV) in E. coli. Also shown are purified CrGUN4 apoprotein preparations assembled in vitro with equal molar concentrations of PCB (GUN4+PCB) or BV (GUN4+BV). (C) Absorption spectra of free bilins in solution (PCB or BV, 20 μM), purified in vivo GUN4:Bilin adducts (20 μM), and GUN4+Bilin mixtures (20 μM) assembled in vitro. Insets show difference spectra of bound (GUN4+bilin) minus free bilins. (D) ITC analysis of ligands binding by titration of CrGUN4 apoprotein with bilins (Left, PCB; Right, BV). (Upper) Heat response to injections; (Lower) Integrated heats of each injection. The Kd values were derived from nonlinear least-squares analysis of the integration of the heats to a single-site binding model. Values are means ± SD of three technical replicates.
Ferrochelatase (FeCh) catalyzes the insertion of ferrous iron into PPIX to generate heme, the prosthetic group of a wide range of hemoproteins, including key components of the photosynthetic electron transport system. Magnesium chelatase (MgCh) catalyzes the first step committed to the synthesis of chlorophyll (i.e., the ATP-dependent insertion of Mg2+ into PPIX to form MgPPIX) (8). In contrast with the single subunit of FeCh, MgCh consists of three subunits—CHLH, CHLI, and CHLD—all of which display transcriptional and posttranslational regulation in plants and algae (9, 10). Oxygenic phototrophs also evolved a novel tetrapyrrole-binding regulator of MgCh, GENOMES UNCOUPLED 4 (GUN4), which stimulates MgCh activity (11–14). Because of the chlorophyll-deficient phenotype of gun4 mutants in plants, cyanobacteria, and algae especially under high light (11, 15, 16), GUN4 is essential to sustain chlorophyll synthesis in the presence of light and oxygen.
Consistent with its proposed role to facilitate delivery of PPIX substrate and release of MgPPIX product, GUN4 binds both tetrapyrroles and also interacts with the PPIX-binding CHLH subunit of MgCh (17–19). Porphyrin binding to GUN4 also promotes its interaction with CHLH and association with chloroplast membranes, the site of chlorophyll biosynthesis (20–22). Whereas the strong up-regulation of GUN4 by light represents a plausible mechanism to redirect PPIX to the chlorophyll pathway (11), GUN4:PPIX complexes are highly photosensitizing, raising questions about its proposed role as a substrate/product chaperone (23). In the present study, we elucidate a GUN4-dependent regulatory mechanism of MgCh activity in the model green alga Chlamydomonas reinhardtii that leverages the ubiquitous presence of linear tetrapyrroles (bilins) in oxygenic phototrophs.
Bilins are linear tetrapyrroles derived from heme by sequential enzymatic reactions of heme oxygenases (HMOX) and ferredoxin-dependent biliverdin reductases (FDBRs) that are found in all oxygenic species (24–26). HMOX are widely distributed oxygen-dependent enzymes that convert heme to biliverdin (BV) with the release of the heme iron and carbon monoxide (24, 27). In cyanobacteria and other eukaryotic algae, FDBR-derived phycobilins function as chromophore precursors of the light-harvesting phycobiliproteins and light-sensing phytochromes, which facilitate phototrophic growth and photoacclimation in fluctuating light environments (28). Although C. reinhardtii lacks both phytochromes and phycobiliproteins, bilins act as a plastid retrograde signal to modulate expression of a small nuclear gene network to detoxify ROS during the diurnal dark-to-light transitions (29). These and follow-up studies have established that bilins are also essential within plastids for proper accumulation of photosystem I (PSI) reaction centers and PSI antennae proteins in daylight (30). In view of the similar chlorophyll-deficient phenotypes of hmox1 and gun4 mutants, such studies strongly implicate bilins to function as regulators of chlorophyll biosynthesis in C. reinhardtii.
The present investigation was undertaken to dissect the molecular basis of bilin regulation of chlorophyll biosynthesis in C. reinhardtii. Our studies establish that CrGUN4 binds bilins with high affinity in vitro, and that bilins stimulate MgCh activity by binding to CrGUN4. Leveraging genetic and chemical complementation analyses, we also show that CrGUN4 and bilins are both required to prevent the light-dependent loss of the MgCh catalytic subunit CrCHLH1 in vivo. Our results support the conclusion that GUN4 evolution is critical to the maintenance of chlorophyll biosynthesis in oxygenic phototrophs by its ability to form complexes with bilins that not only stimulate MgCh activity, but also protect the PPIX-binding CHLH subunit from photooxidative damage and turnover in the presence of light.
Results
CrGUN4 Interacts with Bilins.
Arabidopsis GUN4 has been shown to bind several tetrapyrrole molecules, including heme, PPIX, and MgPPIX (20). For this reason, we first tested whether CrGUN4 could interact with bilins by coexpressing CrGUN4 with cyanobacterial phycocyanobilin (PCB) and BV biosynthesis expression plasmids in Escherichia coli (31). As controls for PCB and BV binding, we respectively coexpressed the Dolichomastix tenuilepis phytochrome (DtenPHY1) and the Deinococcus radiodurans bacteriophytochrome (DrBphP) with PCB and BV plasmids. Cell pellets from PCB cultures of CrGUN4 were blue colored, like those of DtenPHY1 (SI Appendix, Fig. S1A), and affinity-purified CrGUN4 retained the blue color revealing that PCB was tightly bound to the CrGUN4 protein (Fig. 1B). In contrast, unlike cell pellets from BV cultures of DrBphP, CrGUN4 BV cultures were not visibly green colored (SI Appendix, Fig. S1A), nor was affinity-purified CrGUN4 derived therefrom (Fig. 1B). The absorption spectrum of the purified CrGUN4:PCB adduct, as well as that assembled in vitro by mixing apoCrGUN4 with PCB (CrGUN4+PCB), exhibited two maxima at 368∼370 and 670∼674 nm (Fig. 1 C, Left). Addition of BV to affinity-purified CrGUN4 (CrGUN4+BV) yielded a green-colored species with absorption maxima at 380∼382 and 691∼705 nm (Fig. 1 C, Right). By comparison with spectra of PCB and BV in solution, the two absorption maxima of both bilins were significantly red-shifted upon binding to CrGUN4 as documented by the bound- minus free-pigment difference spectra (Fig. 1 C, Insets).
Denaturation studies revealed that both bilins were noncovalently bound to CrGUN4, consistent with the loss of Zn-dependent fluorescence of the CrGUN4 protein after SDS/PAGE (SI Appendix, Fig. S1B) and with the identity of the absorption spectra of the free pigments and the denatured complexes (SI Appendix, Fig. S1 C and D). This contrasts with the retention of zinc-dependent fluorescence of DtenPHY1 following electrophoresis, as evidence of covalent PCB attachment (SI Appendix, Fig. S1B). We also examined the photochemistry of the CrGUN4:PCB adduct. After irradiation with red light (650 ± 20 nm, ∼38 μE for 5 min), only very slight photobleaching occurred and no stable photoproduct was observed (SI Appendix, Fig. S2A). The fluorescence spectrum of the CrGUN4:PCB adduct was nearly indistinguishable from that of PCB in solution (SI Appendix, Fig. S2B), suggesting that the CrGUN4:PCB adduct was not detectably fluorescent. Based on these studies, we conclude that CrGUN4:bilin adducts are noncovalent, nonphotoactive and nonfluorescent.
We next performed spectrophotometric titrations and isothermal titration calorimetry (ITC) measurements to examine the interaction of bilins with CrGUN4 (Fig. 1D). For these studies, we used both PCB and its biosynthetic precursor BV since both bilins are naturally produced in C. reinhardtii plastids (29). For spectrophotometric titrations, we monitored the absorption of premade solutions of PCB or BV to which we added incremental amounts of purified CrGUN4 apoprotein up to 2× molar excess. These measurements showed that both bilins interact with CrGUN4 as revealed by an ∼16∼28% increase in extinction coefficients and small red shifts of both absorption maxima upon binding (SI Appendix, Fig. S2 C and E). Such shifts appeared to saturate at equimolar concentrations for PCB as shown by the difference spectra (SI Appendix, Fig. S2D), thereby implicating formation of a stoichiometric CrGUN4:PCB adduct. However, the difference spectra of CrGUN4:BV suggests that BV may form higher-order complexes with CrGUN4 (SI Appendix, Fig. S2F). By ITC measurements, both bilins gave well-behaved binding isotherms to CrGUN4, revealing dissociation constants (Kd) of 0.22 ± 0.02 μM and 0.09 ± 0.02 μM, for PCB and BV, respectively, that are consistent with 1:1 binding stoichiometry (Fig. 1D).
The Effect of Bilins on PPIX Binding to CrGUN4.
In view of the strong bilin affinity of CrGUN4, we next investigated the influence of bilins on PPIX binding to CrGUN4. To do so, we first incubated 20 μM CrGUN4 apoprotein with equimolar concentrations of PCB or PPIX followed by addition of the other pigment at the same concentration. A sharp new peak at 408 nm appeared upon mixing PPIX with CrGUN4 (Fig. 2A, compare red dash with solid orange traces), that was coincident with loss of absorption of “free” PPIX in solution at 362 nm. This spectral shift is consistent with the disruption of PPIX dimers, the predominant form of PPIX in aqueous solution (32, 33). Other spectral changes seen in the long wavelength region between 450 and 750 nm indicate formation of a protein-bound PPIX adduct. Upon addition of PCB to this mixture, most of the 408-nm peak disappeared and significant changes also were seen in the region between 450 and 800 nm (Fig. 2A, solid blue trace). Such spectral changes are consistent with formation of the PCB adduct of CrGUN4 and release of free PPIX since the difference spectrum constructed by subtracting the spectrum of a mixture of PPIX and PCB in solution (Fig. 2A, dashed turquoise trace) from that of the ternary mixture spectrum (Fig. 2A, solid blue trace) was nearly identical to that constructed by subtracting the spectrum of PCB in solution from that of the CrGUN4:PCB adduct (compare Insets in Figs. 1 C, Left, and 2A).
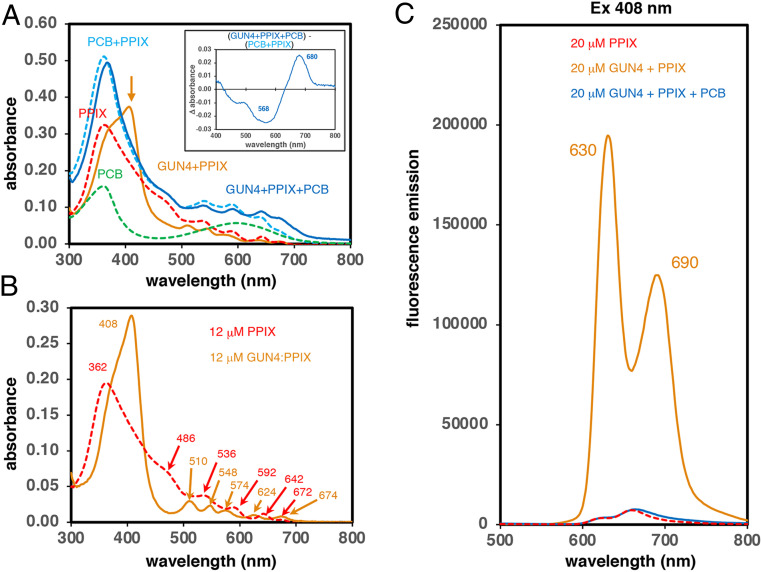
Fig. 2.
PCB addition quenches the strong fluorescence of PPIX adducts of CrGUN4. (A) Absorption spectra of 20 μM free PCB, PPIX, and PCB+PPIX in solution at pH 7.5 are compared with those of binary (CrGUN4+PPIX) and ternary (CrGUN4+PPIX+PCB) mixtures at pH 7.5. Inset shows the difference spectrum constructed by subtracting the spectrum of free PCB+PPIX mixture from that of the ternary (CrGUN4+PPIX+PCB) mixture at pH 7.5. The orange arrow indicates the 408-nm peak in the CrGUN4+PPIX sample. See SI Appendix, Materials and Methods for details. (B) Comparative absorption spectra of 12 μM PPIX and that “calculated” for the CrGUN4:PPIX adduct at pH 7.5 by subtraction of an 8 μM PPIX spectrum from that of 20 μM CrGUN4+PPIX at pH 7.5. Peak wavelengths in nanometers are shown. (C) Comparative fluorescence emission spectra (ex 408 nm) of 20 μM free PPIX, CrGUN4+PPIX and CrGUN4+PPIX+PCB at pH 7.5.
The spectrum of the mixture of equimolar PPIX and CrGUN4 (Fig. 2A, solid orange curve) could be resolved into two components corresponding to 60% bound PPIX (i.e., 12 μM CrGUN4:PPIX adduct) and 40% free PPIX (i.e., 8 μM PPIX). For comparative purposes, the spectra of equimolar solutions of free PPIX and the CrGUN4:PPIX adduct (minus free PPIX) clearly documents the changes in the PPIX environment upon binding to CrGUN4 (Fig. 2B). As shown in Fig. 2C, addition of CrGUN4 to a solution of 20 μM PPIX led to a 40- to 60-fold increase in fluorescence at 630 and 690 nm upon formation of the CrGUN4:PPIX adduct. Moreover, addition of PCB to this mixture reversed this fluorescence increase (Fig. 2C). Together, these data indicate that the fluorescence emission of the CrGUN4:PPIX adduct is quenched upon addition of PCB, reflecting a new electronic environment for PPIX.
To test whether the “released” PPIX remained bound to the newly formed CrGUN4:PCB adduct, we performed side-by-side experiments to generate the CrGUN4 adducts of PPIX and PCB, which were then adsorbed to a Ni2+-NTA matrix. For one-half of each matrix, the bound CrGUN4:PPIX and CrGUN4:PCB adducts were incubated on-column with excess PCB and PPIX, respectively, while the other half were used as controls. After extensive washing, the His-tagged proteins were eluted with 250 mM imidazole. The absorption spectra revealed that both porphyrin and PCB remained bound in the ternary mixtures and that the order of addition did not significantly change the absorption spectra, excepting for light scatter (SI Appendix, Fig. S3A, compare orange and purple traces). Comparison of the spectrum of PPIX dissolved in 250 mM imidazole (SI Appendix, Fig. S3A, dashed red trace) with that of repurified CrGUN4:PPIX (SI Appendix, Fig. S3A, solid red trace), shows that the electronic environment of PPIX is altered upon binding to CrGUN4 (SI Appendix, Fig. S3A). Finally, as shown in SI Appendix, Fig. S3B, the calculated spectrum of the bound PPIX in the CrGUN4:PCB:PPIX ternary complex (SI Appendix, Fig. S3B, solid dark blue trace) could be resolved by subtraction of the spectrum of the CrGUN4:PCB complex from that of the ternary complex (SI Appendix, Fig. S3B, solid orange trace). Taken together, these results show that a tightly bound ternary complex of both tetrapyrroles is produced upon mixing CrGUN4 with PCB and PPIX in either mixing order, so release of free PPIX into a more solvated environment, elicited by PCB binding, is unlikely in light of experiments in SI Appendix, Fig. S3. The similarity of the absorption spectra of free PPIX in solution and bound PPIX in the ternary complex suggests that bound PPIX is present as a dimer similar to that seen in aqueous solutions at these concentrations (32, 33).
CrGUN4 Stimulation of MgCh Activity Is Enhanced by Bilins In Vitro.
It is well established that GUN4 promotes chlorophyll biosynthesis by directly activating MgCh enzymatic activity (11, 17). To explore the impact of bilins on MgCh activity, we next performed in vitro biochemical assays to quantify the production of MgPPIX. A complex enzyme with three subunits, MgCh activity is typically assessed by measuring MgPPIX formation by fluorescence (34, 35) as a function of the concentration of the porphyrin-binding subunit CHLH (9, 36, 37). C. reinhardtii also possesses two CHLH genes, only one of which, CrCHLH1, is highly expressed (30). However, MgCh activity saturates due to the fixed and limiting amount of the other two subunits, CHLI and CHLD. In the absence of CrGUN4, CrCHLH1-dependent MgCh activity is quite low (11). As shown in Fig. 3A, CrCHLH1 displayed sigmoidal kinetics in the presence of excess CrGUN4 with an S0.5 of 206 nM (95% CI 193 to 223 nM) and a Hill coefficient of 3.0 (95% CI 2.6 to 3.4) in both the absence and presence of PCB (P = 0.8). Whereas the presence of CrGUN4 alone afforded modest ∼2-fold stimulation, the effect of PCB on MgCh activity was substantial with Vmax increasing ∼20-fold in the presence of PCB (Fig. 3A) (Vmax = 2.6 nM MgPPIX min−1 per nanomolar CHLD [95% CI 2.5 to 2.8] with PCB versus 0.13 nM MgPPIX min−1 per nanomolar CHLD [95% CI 0.06 to 0.19] without PCB).
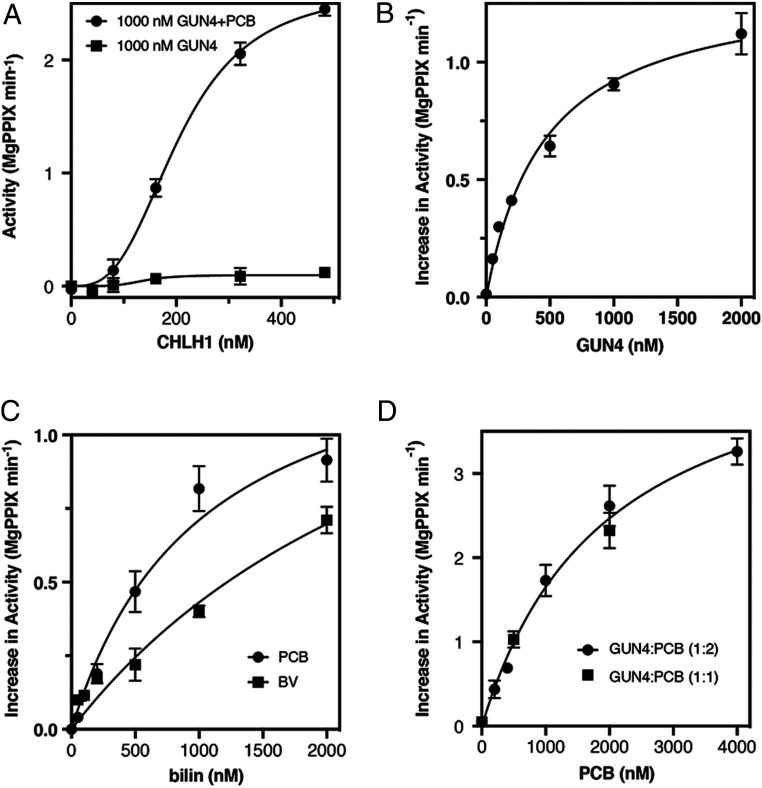
Fig. 3.
CrGUN4 stimulation of MgCh turnover is enhanced by bilins. (A) Comparative kinetics of MgCh activity varying CHLH1 in the presence of GUN4 alone or GUN4 with PCB. [PPIX] = 2 µM. (B) GUN4 stimulation of MgCh at 2,000 nM PCB, 500 nM CHLH1, and 1 µM PPIX, activity at 0 nM GUN4 = 0.013 ± 0.007 MgPPIX min−1. (C) Stimulation of MgCh activity at increasing concentration of bilins (PCB and BV) in the presence of GUN4; 500 nM CHLH1 and 1,000 nM PPIX, activity at 0 nM bilin = 0.037 ± 0.009 MgPPIX min−1. (D) Effect of increasing PCB concentration on MgCh at two different GUN4:PCB ratios with 1,000 nM CHLH1, activity at 0 nM PCB = 0.050 ± 0.018 MgPPIX min−1. Values are means ± SD of three technical replicates in all panels.
In the presence of PCB, the stimulation of MgCh activity by CrGUN4 saturated at a concentration of CrGUN4 similar with that of PCB in the assay (Fig. 3B). These results implicated a stoichiometric CrGUN4:PCB complex to be responsible for MgCh activation, a conclusion consistent with the submicromolar affinity of PCB to CrGUN4 determined by ITC (Fig. 1D). Comparative studies with BV or PCB further revealed that, at saturation, both bilins equally stimulated CrGUN4-dependent MgCh activity (Fig. 3C). While both bilins yielded the same Vmax of 1.46 nM MgPPIX min−1 per nanomolar CHLD (95% CI 1.30 to 1.67), BV afforded a Km of 2,310 nM (95% CI 1,854 to 2,902 nM) that was 2× larger than PCB’s Km of 1,036 nM (95% CI 819 to 1,387 nM). This suggests that the CrGUN4:BV complex is less potent an activator of MgCh than the CrGUN4:PCB complex. However, the stimulation of MgCh activity is more complicated than a simple bilin:GUN4 complex activation implies, since we observed a single saturation curve for MgCh activity plotted versus PCB concentration for two different CrGUN4:PCB stoichiometries (Fig. 3D).
CrCHLH1 Protein Levels Are Reduced in hmox1 and gun4 Mutants.
C. reinhardtii hmox1 and gun4 mutants are both defective in light-dependent growth and chlorophyll accumulation (29, 38). We therefore performed a side-by-side comparison of photoautotrophic (no acetate) and mixotrophic (with acetate) growth of both mutants under various light fluence rates, along with 4A+ WT and both complemented lines as controls (ho1C2, gun4::GUN4-STII #1∼3) (Fig. 4 and SI Appendix, Fig. S4). Significant inhibition of mixotrophic growth of hmox1 was observed under light intensity around 60 μmol photons m−2 s−1, a phenotype that was more exaggerated under photoautotrophic growth conditions, as reported previously (29). In contrast, gun4 displayed a more severe photosynthetic growth deficiency and failed to survive even under ∼30 μmol photons m−2 s−1 light. Genetically complemented lines of both mutants demonstrated similar growth phenotype as 4A+ and ho1C2 control lines (Fig. 4A).
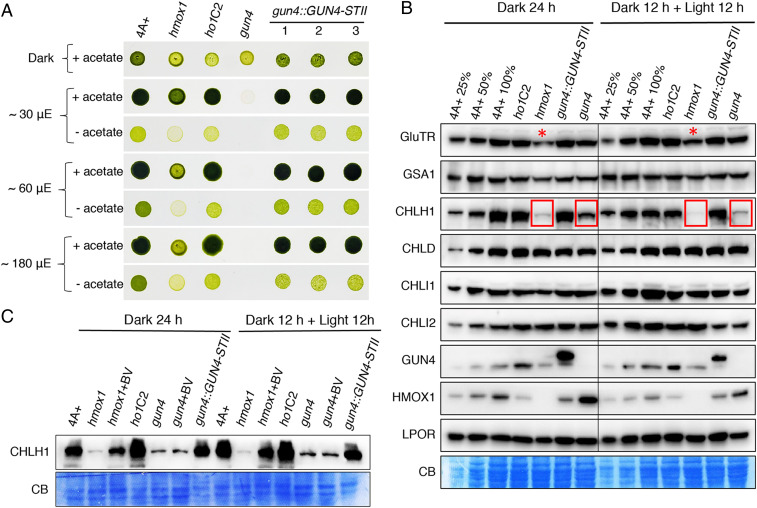
Fig. 4.
CrCHLH1 is deficient in hmox1 and gun4 mutants. (A) Comparative growth of 4A+, hmox1, gun4, cDNA complemented line ho1C2 and genomic DNA complemented lines gun4::GUN4-STII #1∼3 in the dark or under constant white fluorescent light. STII, twin-strep-tag. (B) Immunoblot analyses of representative tetrapyrrole biosynthetic enzymes using corresponding monospecific antibodies. C. reinhardtii cells grown in TAP medium under moderate light (∼30 μmol photons m−2 s−1) were diluted to a density around ∼1 × 106 cells/mL. Half of the cell cultures were acclimated to darkness for 24 h (dark 24 h), whereas the other half were grown in the dark for 12 h and then exposed to light (∼30 μmol photons m−2 s−1) for another 12 h (dark 12 h + light 12 h). Similar amount of protein (∼30 μg) was loaded for each sample. A dilution series (25% and 50%) of the 4A+ WT strain is included for both growth conditions. CB, Coomassie blue staining. Lanes with significant reduction in CrCHLH1 or CrGluTR proteins are indicated with red boxes and asterisks, respectively. (C) Rescue of CHLH1 polypeptide accumulation by BV feeding. Exponential-phase cell cultures grown in darkness were supplemented with or without 20 μM BV.
Previous studies have established that transcripts for tetrapyrrole biosynthetic enzymes in C. reinhardtii were mostly up-regulated in the presence of light and that their levels were either comparable in hmox1 or even elevated in gun4 relative to parental WT strains (14, 30). To further investigate how bilins and CrGUN4 affect accumulation of tetrapyrrole biosynthetic enzymes at the protein level in both light-grown and dark-adapted cultures, we leveraged available and newly developed antibodies to key enzymes of the C. reinhardtii tetrapyrrole pathway. Immunoblot analysis of total cellular protein revealed that levels of CrGSA1, CrCHLD, CrCHLI1, CrCHLI2, and CrLPOR in both hmox1 and gun4 mutants were comparable to those found in 4A+ WT and the genetically complemented control lines (Fig. 4B). In striking contrast, the catalytic subunit of MgCh was dramatically reduced in both hmox1 and gun4 mutants and was restored in complemented lines, indicating that CrHMOX1 and CrGUN4 are both necessary for CrCHLH1 protein accumulation in vivo (Fig. 4B).
CrGUN4 Is Required for BV Rescue of CrCHLH1 and Chlorophyll Accumulation in hmox1 Backgrounds.
We previously showed that bilin-deficient hmox1 mutants could be rescued by feeding exogenous BV (29, 30). We therefore analyzed whether exogenous BV could rescue CrCHLH1 polypeptide accumulation in hmox1 and gun4. In the presence of 20 μM BV, CrCHLH1 levels were significantly restored in hmox1 in both dark-adapted and light-treated cultures (Fig. 4C). In contrast, BV feeding did not restore CrCHLH1 accumulation in the gun4 mutant (Fig. 4C). These results demonstrate that the presence of bilins and CrGUN4 are both required for CrCHLH1 protein stability and accumulation.
To test the role of bilins in CrCHLH1 accumulation and chlorophyll biosynthesis in vivo, we constructed the hmox1gun4 double mutant by crossing hmox1 and gun4 (SI Appendix, Fig. S4A). Compared with hmox1 and gun4 single mutants, the hmox1gun4 double mutant, like gun4, was similarly sensitive to light, and proved viable only under heterotrophic growth conditions in the dark or under dim light (< 30 μmol photons m−2 s−1) (Fig. 5A). The double mutant also exhibited a more pronounced yellow-brown color than gun4. Expression of the WT CrGUN4 allele containing an introduced Strep tag II (GUN4-STII) partially complemented the photosynthetic growth deficiency of the hmox1gun4 double mutant, restoring it to that similar to the hmox1 single mutant (Fig. 5A).
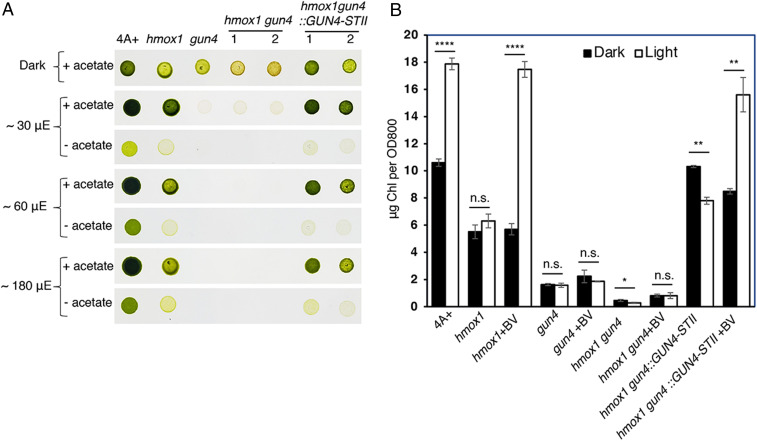
Fig. 5.
BV rescue of CrCHLH1 and chlorophyll accumulation in hmox1 backgrounds requires CrGUN4. (A) Comparative growth of 4A+, hmox1, gun4, hmox1gun4, and two independent genomic DNA complemented lines hmox1gun4::GUN4-STII in the dark or under constant white fluorescent light. STII, Strep-tag II. (B) Chlorophyll accumulation under different growth conditions. Exponential-phase C. reinhardtii cells were either acclimated to darkness for 24 h (dark) or grown in the dark for 12 h and then exposed to light (∼120 μmol photons m−2 s−1) for another 12 h (light). Various mutant strains were supplemented with or without 50 μM BV. Asterisks above pairs of bars indicate significant differences (Student’s t test, *P < 0.05, **P < 0.01, and ****P < 0.0001; n.s., not statistically significant). Values are means ± SD of three biological replicates.
We then measured chlorophyll content in dark-adapted mixotrophic cultures before and after 12-h light exposure that were supplemented with or without 50 μM BV. In contrast with 4A+ WT where chlorophyll levels increased approximately twofold after illumination and in the gun4 complemented line gun4::GUN4-STII (SI Appendix, Fig. S4B), light-dependent chlorophyll accumulation in hmox1, gun4, hmox1gun4, and hmox1gun4::GUN4-STII lines did not occur (Fig. 5B). Consistent with hmox1 rescue by BV ((29)), BV supplementation also rescued light-dependent chlorophyll accumulation in hmox1gun4::GUN4-STII. This recovery was not observed in gun4 or in the hmox1gun4 double mutant, thus indicating that light-dependent chlorophyll accumulation requires both bilins and CrGUN4 (Fig. 5B).
Discussion
Bilins are linear tetrapyrroles derived from heme that play a retrograde signaling function in C. reinhardtii when exported out of the chloroplast (28, 29). The targets of bilins in C. reinhardtii, which lacks cytosolic phytochromes, is presently unknown; however, their export promotes accumulation of a small set of transcripts of genes implicated in oxygen metabolism (29). Since bilins themselves are the product of oxygen metabolism, it makes sense that their production would be a proxy for the presence of light and oxygen. In this investigation, we establish that bilins also perform additional roles within chloroplasts to sustain chlorophyll synthesis.
Bilins Perform Dual Roles within Chloroplasts to Maintain Robust Chlorophyll Biosynthesis in the Presence of Light and Oxygen.
Previous studies have established that Arabidopsis GUN4 stimulates MgCh activity by binding PPIX or MgPPIX, the respective substrate and product of MgCh (11). Our studies establish that CrGUN4 can also bind bilins, the presence of which leads to >20-fold enhancement of MgCh turnover beyond that with CrGUN4 alone. While the mechanism of this enhancement requires further study, we conclude that the CrGUN4:PCB:PPIX ternary complex is responsible for this enhancement possibly by promoting the rate of CrCHLH1 loading with its PPIX substrate. We hypothesize that PCB remains bound throughout catalysis, thereby ensuring an adequate supply of chlorophyll to sustain phototrophic growth and to replace damaged photosystems.
Despite its strong transcriptional induction by light, the PPIX-binding CrCHLH1 subunit of MgCh nearly disappears in the absence of either bilins or CrGUN4. Our biochemical and genetic data establish that this phenomenon reflects the lack of CrGUN4:PCB adducts because CrCHLH1 is nearly absent in both hmox1 and gun4 mutants, and the presence of CrGUN4 is required for BV rescue of the CrCHLH1 deficiency in hmox1 backgrounds. Hence, CrGUN4:bilin complexes perform dual roles in C. reinhardtii chloroplasts, as activators of MgCh activity and as factors that stabilize CrCHLH1 accumulation.
The reduced activity of MgCh in hmox1 and gun4 mutants is also expected to lead to overaccumulation of PPIX, a strongly photodynamic molecule that readily generates singlet oxygen (1O2), thereby promoting oxidative damage of the photosynthetic apparatus as has been observed in both mutants (30, 38). Presumably, triplet chlorophyll production in an anaerobic environment harmlessly decays, making GUN4 unnecessary in chlorophyll-containing anaerobic phototrophs. Our studies clearly show that bilin binding to the CrGUN4:PPIX adduct effectively quenches the excited state of the latter, which would inhibit PPIX triplet state formation and subsequent 1O2 generation. Based on this knowledge, we hypothesize that the interaction between the ternary CrGUN4:PCB:PPIX complex and CrCHLH1 facilitates transfer of PPIX substrate to CrCHLH1 while also protecting the resulting CrCHLH1:PPIX complex from photosensitized inactivation and turnover. We conclude that this protective function of bilins via GUN4 binding provides a solid rationale for the retention of bilin biosynthesis in all oxygenic phototrophs from cyanobacteria to all extant eukaryotic phototrophic species (26, 39). In future studies, it will be of great interest to evaluate whether bilins other than PCB can support a similar GUN4-dependent photoprotective role in other photosynthetic species to sustain MgCh activity in the presence of light and oxygen. Understanding the mechanism of damaged CHLH1 turnover in C. reinhardtii also remains an important objective of such studies.
Additional GUN4-Related Proteins Are Found in Cyanobacteria and Other Nonphotosynthetic Bacteria.
The first committed step of chlorophyll biosynthesis is catalyzed by MgCh, which is present in both anoxygenic and oxygenic phototrophic organisms. In contrast with oxygenic phototrophs, anoxygenic photosynthetic bacteria, such as Rhodobacter capsulatus, lack GUN4 orthologs (11, 40). Instead, they possess BchJ, which functions as an MgPPIX chaperone (41). It is notable that many cyanobacteria also possess other GUN4-related proteins that are associated with N-terminal extensions with unknown functions (16, 17). The biological roles of these and other GUN4-related proteins found in actinobacteria, alphaproteobacteria, and archaea (see the PFAM database, http://pfam.xfam.org/family/PF05419) are presently unknown. We speculate that these GUN4-related domains are bilin-binding regions that might also function to protect their translationally fused partners from ROS. In this regard, the presence of a gene that encodes a GUN4-CHLH chimera in the dinoflagellate Symbiodinium microadriaticum (locus OLP92483.1; http://refuge2020.reefgenomics.org) and transcriptome evidence in other dinoflagellates are consistent with this hypothesis (42).
Regulation of Chlorophyll Biosynthesis in Plants Differs from that in Chlorophytes.
In Arabidopsis thaliana, regulation of tetrapyrrole biosynthesis occurs mainly through transcriptional and posttranslational control of glutamyl-tRNA reductase (GluTR), the first committed enzyme of heme and chlorophyll pathways (7, 43). Bilins strongly impact these processes as well via phytochrome-mediated stimulation of photosynthesis-associated nuclear gene expression, which affects both heme and chlorophyll pathways in plants (1). Heme affects the interaction between GluTR and the GluTR-binding protein (GBP) by exposing GluTR for Clp protease-mediated protein degradation (44), and dark-accumulated Pchlide also inhibits GluTR activity in plants (3, 45). Although GBP is widespread in plants, C. reinhardtii contains a GBP-like protein with only 23% homology to Arabidopsis GBP (44, 46).
The evidence that Pchlide does not accumulate in darkness in C. reinhardtii due to the presence of dark-operative protochlorophyllide oxidoreductase (47, 48), and that chlorophyte algae, such as C. reinhardtii lack phytochromes, underscores the differences between the regulation of chlorophyll synthesis in higher plants and chlorophytes. Indeed, the absence of CrGUN4 leads to a significant accumulation of PPIX in C. reinhardtii, whereas the increase in PPIX was much less pronounced in the Arabidopsis gun4 mutant (12, 38, 49). This suggests that plants possess distinct mechanisms to protect against photodynamic inactivation of enzymes of the chlorophyll biosynthetic pathway which remain to be identified.
Bilins Are Potent Antioxidants that Must Be Localized near the Site of 1O2 Generation.
In the C. reinhardtii gun4 mutant, accumulation of PPIX accompanies transcriptional up-regulation of 1O2-responsive gene glutathione peroxidase 5 (GPX5) during the dark-to-light transition, implicating a key role for CrGUN4 in mitigating ROS-induced stress in membranes (14). While PPIX is generated in plastids, its membrane solubility allows it to rapidly distribute into cytoplasmic-facing membranes of the C. reinhardtii cells when overproduced. PPIX generation of 1O2 in membranes is most likely to lead to lipid peroxidation, which is reversed by GPX5. Thus, the bilin-based retrograde signaling pathway is well suited to deal with cytosolic ROS stress that is triggered at dawn (29). CrGUN4-dependent mitigation of ROS production in the chloroplast and this retrograde signaling pathway are connected by bilins. The interdependency of chlorophyll and bilin synthesis therefore appears to be fundamental for the survival of photosynthetic species in an oxygen-rich environment. While this signaling pathway could be mediated by a bilin-dependent photoreceptor like phytochrome that was previously named chlorochrome (30), it is also possible that cytosolic proteins, which bind both bilins and released PPIX like GUN4, would even be more effective at preventing 1O2 production in the first place. Such proteins have not been identified, but remain a potential innovation whose nature is likely to have been explored more than once.
Finally, 1O2 is a potent oxidant of proteins, lipids, and DNA (50) but only when it is generated nearby due to its short intrinsic lifetime. 1O2-mediated protein oxidation usually induces changes of biological properties (i.e., enzyme inactivation and proteolytic degradation) (51). We therefore propose that the reduced accumulation of CHLH1 in hmox1 and gun4 results from oxidative modification caused by PPIX-generated 1O2 in situ. The decreased accumulation of CHLH1 was also observed in the PPIX-accumulating gun4 mutant of cyanobacteria Synechocystis sp. PCC 6803, whereas CHLH protein abundance was not significantly changed in the Arabidopsis gun4 mutant (12, 16). PPIX-bound GUN4 and CHLH were both reported to significantly generate 1O2 and GUN4 was proposed to directly interact with a sensor to deliver the retrograde 1O2 signal (23). Bilins might be essential to fine-tune the distribution of PPIX between GUN4 and CHLH to reduce the GUN4:PPIX-generated 1O2. It will be interesting to test whether or not the 1O2 derived from GUN4:PPIX is detoxified through the bilin-based retrograde signaling or even by bilin molecule itself (30).
Materials and Methods
Detailed materials and methods are provided in SI Appendix. This includes details about Chlamydomonas strains and their growth conditions, generation of hmox1gun4 double mutant and complemented strains, and methods regarding recombinant enzyme production, MgCh enzyme activity assays, bilin and porphyrin preparations, protein extraction and immunoblot analysis, chlorophyll assays, pigment binding assays, and spectrophotometric measurements.
Supplementary Material
Acknowledgments
We thank Dr. Delin Zhang at the Center for Protein Research, Huazhong Agricultural University, for technical support on isothermal titration calorimetry measurements. This work was supported by grants from the National Natural Science Foundation of China Grants 32070268 and 31570233 (to D.D.); Fundamental Research Funds for the Central Universities Program 2662020SKPY007 (to D.D.); NIH Institute of General Medical Sciences R01 GM068552 and R35 GM139598 (to J.C.L.); Office of Basic Energy Sciences, United States Department of Energy DOE DE-FG02-09ER16117 (to J.C.L.); and a Macquarie University Visiting Researcher Fellowship (to J.C.L.).
Footnotes
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2104443118/-/DCSupplemental.
Data Availability
All study data are included in the article and SI Appendix.
References
- 1.Mochizuki N., et al., The cell biology of tetrapyrroles: A life and death struggle. Trends Plant Sci. 15, 488–498 (2010). [DOI] [PubMed] [Google Scholar]
- 2.Shimizu T., Lengalova A., Martínek V., Martínková M., Heme: Emergent roles of heme in signal transduction, functional regulation and as catalytic centres. Chem. Soc. Rev. 48, 5624–5657 (2019). [DOI] [PubMed] [Google Scholar]
- 3.Meskauskiene R., et al., FLU: A negative regulator of chlorophyll biosynthesis in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 98, 12826–12831 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jung S., et al., Toxic tetrapyrrole accumulation in protoporphyrinogen IX oxidase-overexpressing transgenic rice plants. Plant Mol. Biol. 67, 535–546 (2008). [DOI] [PubMed] [Google Scholar]
- 5.Busch A. W. U., Montgomery B. L., Interdependence of tetrapyrrole metabolism, the generation of oxidative stress and the mitigative oxidative stress response. Redox Biol. 4, 260–271 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tanaka R., Tanaka A., Tetrapyrrole biosynthesis in higher plants. Annu. Rev. Plant Biol. 58, 321–346 (2007). [DOI] [PubMed] [Google Scholar]
- 7.Tanaka R., Kobayashi K., Masuda T., Tetrapyrrole metabolism in Arabidopsis thaliana. Arabidopsis Book 9, e0145 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jensen P. E., Gibson L. C., Henningsen K. W., Hunter C. N., Expression of the chlI, chlD, and chlH genes from the Cyanobacterium synechocystis PCC6803 in Escherichia coli and demonstration that the three cognate proteins are required for magnesium-protoporphyrin chelatase activity. J. Biol. Chem. 271, 16662–16667 (1996). [DOI] [PubMed] [Google Scholar]
- 9.Willows R. D., Biosynthesis of chlorophylls from protoporphyrin IX. Nat. Prod. Rep. 20, 327–341 (2003). [DOI] [PubMed] [Google Scholar]
- 10.Gao Y. S., Wang Y. L., Wang X., Liu L., Hexameric structure of the ATPase motor subunit of magnesium chelatase in chlorophyll biosynthesis. Protein Sci. 29, 1040–1046 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Larkin R. M., Alonso J. M., Ecker J. R., Chory J., GUN4, a regulator of chlorophyll synthesis and intracellular signaling. Science 299, 902–906 (2003). [DOI] [PubMed] [Google Scholar]
- 12.Peter E., Grimm B., GUN4 is required for posttranslational control of plant tetrapyrrole biosynthesis. Mol. Plant 2, 1198–1210 (2009). [DOI] [PubMed] [Google Scholar]
- 13.Zhou S., Sawicki A., Willows R. D., Luo M., C-terminal residues of Oryza sativa GUN4 are required for the activation of the ChlH subunit of magnesium chelatase in chlorophyll synthesis. FEBS Lett. 586, 205–210 (2012). [DOI] [PubMed] [Google Scholar]
- 14.Brzezowski P., et al., The GUN4 protein plays a regulatory role in tetrapyrrole biosynthesis and chloroplast-to-nucleus signalling in Chlamydomonas reinhardtii. Plant J. 79, 285–298 (2014). [DOI] [PubMed] [Google Scholar]
- 15.Sobotka R., et al., Importance of the cyanobacterial Gun4 protein for chlorophyll metabolism and assembly of photosynthetic complexes. J. Biol. Chem. 283, 25794–25802 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wilde A., Mikolajczyk S., Alawady A., Lokstein H., Grimm B., The gun4 gene is essential for cyanobacterial porphyrin metabolism. FEBS Lett. 571, 119–123 (2004). [DOI] [PubMed] [Google Scholar]
- 17.Davison P. A., et al., Structural and biochemical characterization of Gun4 suggests a mechanism for its role in chlorophyll biosynthesis. Biochemistry 44, 7603–7612 (2005). [DOI] [PubMed] [Google Scholar]
- 18.Verdecia M. A., et al., Structure of the Mg-chelatase cofactor GUN4 reveals a novel hand-shaped fold for porphyrin binding. PLoS Biol. 3, e151 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen X., et al., Crystal structures of GUN4 in complex with porphyrins. Mol. Plant 8, 1125–1127 (2015). [DOI] [PubMed] [Google Scholar]
- 20.Adhikari N. D., Orler R., Chory J., Froehlich J. E., Larkin R. M., Porphyrins promote the association of GENOMES UNCOUPLED 4 and a Mg-chelatase subunit with chloroplast membranes. J. Biol. Chem. 284, 24783–24796 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Adhikari N. D., et al., GUN4-porphyrin complexes bind the ChlH/GUN5 subunit of Mg-Chelatase and promote chlorophyll biosynthesis in Arabidopsis. Plant Cell 23, 1449–1467 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kopečná J., et al., Porphyrin binding to Gun4 protein, facilitated by a flexible loop, controls metabolite flow through the chlorophyll biosynthetic pathway. J. Biol. Chem. 290, 28477–28488 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tarahi Tabrizi S., Sawicki A., Zhou S., Luo M., Willows R. D., GUN4-Protoporphyrin IX is a singlet oxygen generator with consequences for plastid retrograde signaling. J. Biol. Chem. 291, 8978–8984 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Takemoto J. Y., Chang C. W. T., Chen D., Hinton G., Heme-derived bilins. Isr. J. Chem. 59, 378–386 (2019). [Google Scholar]
- 25.Sugishima M., Wada K., Unno M., Fukuyama K., Bilin-metabolizing enzymes: Site-specific reductions catalyzed by two different type of enzymes. Curr. Opin. Struct. Biol. 59, 73–80 (2019). [DOI] [PubMed] [Google Scholar]
- 26.Rockwell N. C., Lagarias J. C., Ferredoxin-dependent bilin reductases in eukaryotic algae: Ubiquity and diversity. J. Plant Physiol. 217, 57–67 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Matsui T., Unno M., Ikeda-Saito M., Heme oxygenase reveals its strategy for catalyzing three successive oxygenation reactions. Acc. Chem. Res. 43, 240–247 (2010). [DOI] [PubMed] [Google Scholar]
- 28.Duanmu D., Rockwell N. C., Lagarias J. C., Algal light sensing and photoacclimation in aquatic environments. Plant Cell Environ. 40, 2558–2570 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Duanmu D., et al., Retrograde bilin signaling enables Chlamydomonas greening and phototrophic survival. Proc. Natl. Acad. Sci. U.S.A. 110, 3621–3626 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wittkopp T. M., et al., Bilin-dependent photoacclimation in Chlamydomonas reinhardtii. Plant Cell 29, 2711–2726 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gambetta G. A., Lagarias J. C., Genetic engineering of phytochrome biosynthesis in bacteria. Proc. Natl. Acad. Sci. U.S.A. 98, 10566–10571 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Karns G. A., Gallagher W. A., Elliott W. B., Dimerization constants of water-soluble porphyrins in aqueous alkali. Bioorg. Chem. 8, 69–81 (1979). [Google Scholar]
- 33.Margalit R., Shaklai N., Cohen S., Fluorimetric studies on the dimerization equilibrium of protoporphyrin IX and its haemato derivative. Biochem. J. 209, 547–552 (1983). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Walker C. J., Weinstein J. D., Further characterization of the magnesium chelatase in isolated developing cucumber chloroplasts. Plant Physiol. 95, 1189–1196 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gibson L. C., Willows R. D., Kannangara C. G., von Wettstein D., Hunter C. N., Magnesium-protoporphyrin chelatase of Rhodobacter sphaeroides: Reconstitution of activity by combining the products of the bchH, -I, and -D genes expressed in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 92, 1941–1944 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jensen P. E., Gibson L. C., Hunter C. N., Determinants of catalytic activity with the use of purified I, D and H subunits of the magnesium protoporphyrin IX chelatase from Synechocystis PCC6803. Biochem. J. 334, 335–344 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Karger G. A., Reid J. D., Hunter C. N., Characterization of the binding of deuteroporphyrin IX to the magnesium chelatase H subunit and spectroscopic properties of the complex. Biochemistry 40, 9291–9299 (2001). [DOI] [PubMed] [Google Scholar]
- 38.Formighieri C., Ceol M., Bonente G., Rochaix J. D., Bassi R., Retrograde signaling and photoprotection in a gun4 mutant of Chlamydomonas reinhardtii. Mol. Plant 5, 1242–1262 (2012). [DOI] [PubMed] [Google Scholar]
- 39.Rockwell N. C., Lagarias J. C., Bhattacharya D., Primary endosymbiosis and the evolution of light and oxygen sensing in photosynthetic eukaryotes. Front. Ecol. Evol., 10.3389/fevo.2014.00066 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zappa S., Li K., Bauer C. E., The tetrapyrrole biosynthetic pathway and its regulation in Rhodobacter capsulatus. Adv. Exp. Med. Biol. 675, 229–250 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sawicki A., Willows R. D., BchJ and BchM interact in a 1 : 1 ratio with the magnesium chelatase BchH subunit of Rhodobacter capsulatus. FEBS J. 277, 4709–4721 (2010). [DOI] [PubMed] [Google Scholar]
- 42.Liu H., et al., Symbiodinium genomes reveal adaptive evolution of functions related to coral-dinoflagellate symbiosis. Commun. Biol. 1, 95 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brzezowski P., Richter A. S., Grimm B., Regulation and function of tetrapyrrole biosynthesis in plants and algae. Biochim. Biophys. Acta 1847, 968–985 (2015). [DOI] [PubMed] [Google Scholar]
- 44.Richter A. S., Banse C., Grimm B., The GluTR-binding protein is the heme-binding factor for feedback control of glutamyl-tRNA reductase. eLife 8, e46300 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kauss D., Bischof S., Steiner S., Apel K., Meskauskiene R., FLU, a negative feedback regulator of tetrapyrrole biosynthesis, is physically linked to the final steps of the Mg(++)-branch of this pathway. FEBS Lett. 586, 211–216 (2012). [DOI] [PubMed] [Google Scholar]
- 46.Czarnecki O., et al., An Arabidopsis GluTR binding protein mediates spatial separation of 5-aminolevulinic acid synthesis in chloroplasts. Plant Cell 23, 4476–4491 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gabruk M., Mysliwa-Kurdziel B., Light-dependent protochlorophyllide oxidoreductase: Phylogeny, regulation, and catalytic properties. Biochemistry 54, 5255–5262 (2015). [DOI] [PubMed] [Google Scholar]
- 48.Hunsperger H. M., Randhawa T., Cattolico R. A., Extensive horizontal gene transfer, duplication, and loss of chlorophyll synthesis genes in the algae. BMC Evol. Biol. 15, 16 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mochizuki N., Tanaka R., Tanaka A., Masuda T., Nagatani A., The steady-state level of Mg-protoporphyrin IX is not a determinant of plastid-to-nucleus signaling in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 105, 15184–15189 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Di Mascio P., et al., Singlet molecular oxygen reactions with nucleic acids, lipids, and proteins. Chem. Rev. 119, 2043–2086 (2019). [DOI] [PubMed] [Google Scholar]
- 51.Davies M. J., Singlet oxygen-mediated damage to proteins and its consequences. Biochem. Biophys. Res. Commun. 305, 761–770 (2003). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and SI Appendix.