Abstract
Objective
To investigate the clinical characteristics and molecular epidemiology of carbapenem-resistant Klebsiella pneumoniae (CRKP) bloodstream infection at a medical center in northeast China, especially after coronavirus disease (COVID-19) pandemic.
Methods
Fifty-one patients were diagnosed with CRKP bloodstream infection between January 2015 and December 2020, among which 42 isolates were available for further study. Species identification and antibiotic susceptibilities were tested with matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) and VITEK 2 systems. Carbapenemase genes, virulence genes and MLST genes were detected by polymerase chain reaction. Moreover, the string test and serum killing assay were performed to evaluate the virulence of the CRKP isolates.
Results
During the six-year period, the detection rate of CRKP in bloodstream infection showed an increasing trend, with the intensive care unit, hematology and respiratory medicine wards mainly affected. Molecular epidemiology analyses showed that KPC-2 was the dominant carbapenemase gene. In addition, the dominant sequence type (ST) of CRKP shifted from ST11 to ST15 strains, which were all sensitive to amikacin in contrast to the ST11 stains. Furthermore, ST15 CRKP strains were positive for the KfuB virulence gene and more resistant to serum killing compared to the ST11 CRKP strains. Nonetheless, the mortality rate of patients infected with ST11 and ST15 CRKP did not show any significant differences.
Conclusion
A shift in the dominant sequence type of CRKP bloodstream infections from ST11 to ST15 was observed during the years 2015–2020. Compared to ST11, the ST15 CRKP strains showed amikacin sensitivity, positivity for KfuB gene, and serum resistance, which may indicate stronger virulence.
Keywords: carbapenem-resistant Klebsiella pneumoniae, sequence type, KPC-2, virulence, drug resistance
Background
Klebsiella pneumoniae (Kpn) is one of the most common Gram-negative pathogens associated with hospital- and community-acquired infections. Kpn can cause clinical infections such as pulmonary infection, urinary tract infection, bloodstream infection (BSI), and surgical site infection.1 In recent years, due to the overuse of antibiotics, the presence of carbapenem resistant Enterobacteriaceae (CRE) has become a serious public health threat, and has been listed as an urgent threat by the United States Centers for Disease Control. According to the results of China Antimicrobial Surveillance Network (CHINET), the resistance rate of Kpn to meropenem increased from 2.9% in 2005 to 26.3% in 2018, which ranked Carbapenem resistant Klebsiella pneumoniae (CRKP) first among the isolated CRE in China.2
CRKP can result in BSI due to CRKP colonization, carbapenem exposure, longer hospitalization time, intensive care unit (ICU) stay, and operation history, which are considered independent risk factors for CRKP-BSI.3,4 CRKP-BSIs are associated with few treatment options, prolonged hospitalization, increased hospital costs, and high mortality rates. A previous meta-analysis showed that the crude mortality rate from CRKP-BSI was 54.3%.5 As for the mechanisms of Kpn resistance to carbapenems, the production of carbapenemases, such as Ambler class A β-lactamases (blaKPC), class B metallo-β-lactamases (blaVIM, blaIMP and blaNDM), and class D β-lactamases (blaOXA-48), are the leading causes. Furthermore, other β-lactamases and efflux pumps may also contribute as resistance mechanisms to carbapenems.6
Previous epidemiology studies have shown that KPC-2 is the widest disseminated carbapenemase in China and the dominant sequence type (ST) is ST11.7 However, China is a country with an extensive geographical distribution and data on the clinical and molecular epidemiology on CRKP-BSI in northeast China, especially during the coronavirus disease 2019 (COVID-19) pandemic, are lacking. Therefore, we conducted this study to evaluate the molecular epidemiology, virulence profiles, and outcomes of CRKP-BSIs at our hospital, which is a medical center in northeast China. This study will provide insight into the clinical management and infection control of CRKP-BSI.
Materials and Methods
Participants and Bacterial Strains
This retrospective study was conducted at the First Affiliated Hospital of China Medical University, a 2249-bed tertiary hospital serving as the medical center of northeast China. Moreover, the Department of Laboratory Medicine at our hospital is also the national clinical research center for laboratory medicine. Based on laboratory-based surveillance, 51 of 491 patients with BSI due to Kpn were identified as CRKP-BSI between January 2015 and December 2020. CRKP was defined as Kpn isolates resistant to imipenem (MICs ≥ 4 μg/mL) or meropenem (MICs ≥ 4 μg/mL). Forty-two strains from 42 patients were available and further experiments were performed.
This present study complied with the Declaration of Helsinki and was approved by the Ethics Committee of the First Affiliated Hospital of China Medical University (Number: 2020075). The ethics committee waived the need for informed consent for the following reasons: (1) this study was retrospective; (2) the patient information in this research was anonymous; (3) the clinical isolates were collected and stored during the routine diagnostic laboratory, and this study had no impact on the patients. The authors stated to confirm the patient data confidentiality.
Species Identification and Antimicrobial Resistance Testing
The VITEK 2 automated system and the MALDI TOF MS (bioMérieux, France) were used for isolate identification. Furthermore, 16s rRNA sequencing was performed to confirm the strain identification. The VITEK 2 system was used to test antimicrobial susceptibilities of all isolates and results were interpreted according to the criteria of the Clinical and Laboratory Standards Institute. Resistance to imipenem and meropenem was confirmed using the disc diffusion method or the Etest method. Escherichia coli ATCC 25922 and Pseudomonas aeruginosa ATCC 27853 were used as quality controls for antimicrobial testing.
Detection of Resistance Genes and Virulence Genes
The RAPIDEC CARBA NP assay (bioMérieux) was used for phenotypic detection of carbapenemases. In addition, carbapenemase genes (blaKPC, blaNDM, blaIMP, blaVIM, and blaOXA-48) in all isolates were detected by polymerase chain reaction (PCR). Briefly, DNA was extracted by the boiling method and then subjected to amplification and sequencing.8 Moreover, virulence genes (mrkD, fimH, entB, ybtS, rmpA, aerobactin, IroN, KfuB, wcaG, alls, uge, and magA) and capsular serotype-specific genes for K1, K2, K5, K20, K54, and K57 were also detected as previously described.9,10
Multilocus Sequence Typing (MLST)
Molecular epidemiological characteristics were analyzed by multilocus sequence typing as described in the MLST online database (http://bigsdb.pasteur.fr/klebsiella/klebsiella.html). Seven housekeeping genes (gapA, infB, mdh, pgi, phoE, rpoB, and tonB) were amplified, sequenced, and submitted to the MLST database to define the sequence type of CRKP strains.
String Test and Serum Killing Assay
The string test was conducted, with viscous string >5 mm as positive for the hypermucoviscous phenotype. In addition, the serum killing assay was performed as described previously.11 Serum samples from 10 healthy volunteers were mixed and stored at −80°C for use. A 106 cfu/mL bacteria-containing inoculum of 25 µL was added to 75 µL pooled serum. Then samples were incubated at 37°C, and viable bacteria count was determined by plating on Mueller-Hinton agar at 0, 1, 2, and 3 h, respectively. Isolates were classified as serum sensitive (grade 1 or 2), intermediately sensitive (grade 3 or 4), or serum resistant (grade 5 or 6).
Statistical Analysis
Data were statistically analyzed using SPSS 20.0 and WHONET 5.6 software. Quantitative data are shown as mean ± standard deviation or medians (interquartile range), and were compared using the Student’s t-test or nonparametric test. Qualitative data are presented as rates and were compared using the chi-square or Fisher’s exact test. For all statistical analyses, a p value < 0.05 was considered statistically significant.
Results
General Characteristics of Patients with CRKP-BSI
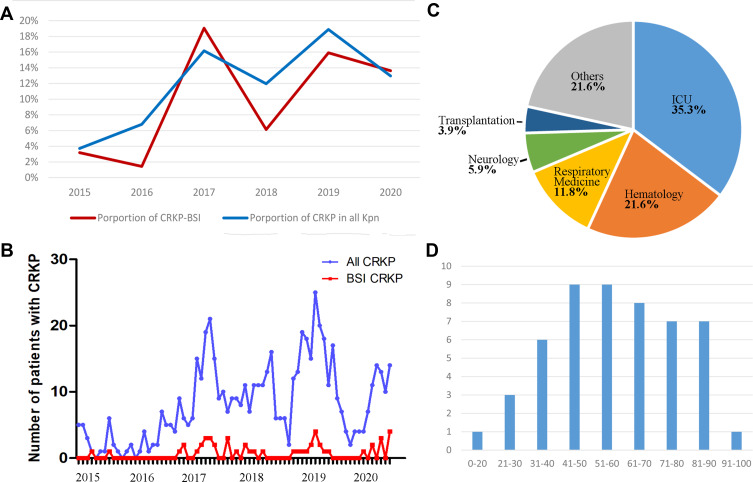
During the six-year period 2015–2020, a total of 491 patients were identified as BSI positive for Kpn, of which 51 patients harbored a CRKP infection (resistance rate = 10.4%). As shown in Figure 1A, the percentage of CRKP-BSI increased gradually, namely, 3.2% in 2015, 1.4% in 2016, 19.1% in 2017, 6.1% in 2018, 15.9% in 2019, and 13.6% in 2020. Furthermore, a high percentage of CRKP-BSI coincided with a high percentage of CRKP in all Kpn isolates, indicating that the CRKP colonization may be associated with BSI of CRKP (Figure 1A and B).
Figure 1.
Basic characteristics of patients with Carbapenem resistant Klebsiella pneumoniae bloodstream infection (CRKP-BSI). (A) Trends in the proportion of CRKP-BSI (red) and CRKP in all Kpn isolates (blue) over six years (2015–2020); (B) number of patients with CRKP-BSI (red) and patients with CRKP from any sample type (blue) by month; (C) distribution of CRKP-BSI among different hospital departments; (D) distribution of CRKP-BSI by age group.
Of the 51 patients infected with CRKP, the hospital units involved were the ICU (n = 18, 35.3%), hematology (n = 11, 21.6%), respiratory medicine (n = 6, 11.8%), neurology (n = 3, 5.9%), and transplantation (n = 2, 3.9%) wards, and other wards (n = 11, 21.6%) (Figure 1C). Moreover, 68.6% of the patients were male, with ages ranging from 17 days to 91 years old (Figure 1D, median: 59 years old, interquartile range: 47–74).
Molecular Epidemiology of CRKP
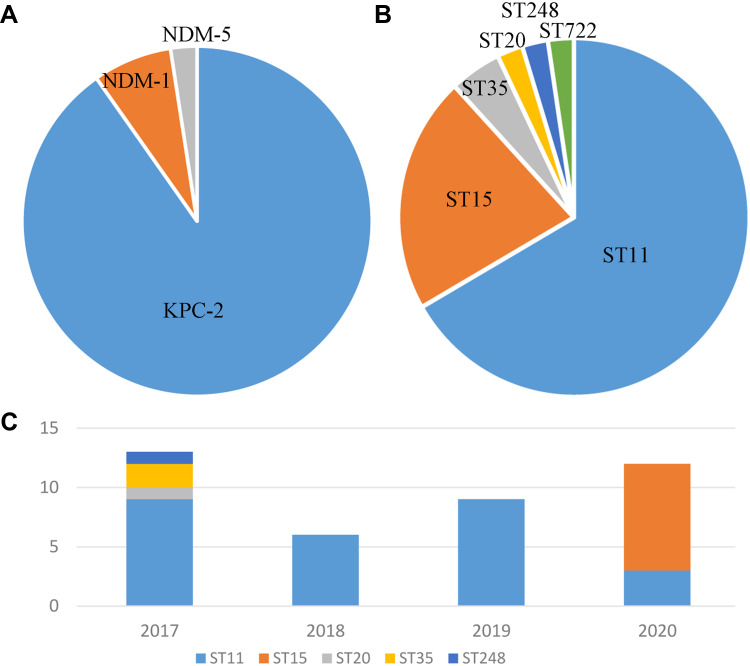
Carbapenemase production was detected in 41 out of 42 CRKP isolates, dominated by KPC-2 (n = 37, 88.1%), followed by NDM-1 (n = 3, 7.1%) and NDM-5 (n = 1, 2.4%). Neither IMP, VIM nor OXA-48 were detected (Figure 2). The 42 CRKP isolates belonged to six STs (Table 1), of which ST11 was predominant (n = 28, 66.7%), followed by ST15 (n = 9, 21.4%), ST35 (n = 2, 4.8%), ST20 (n = 1, 2.4%), ST248 (n = 1, 2.4%), and ST722 (n = 1, 2.4%). It is worth mentioning that ST11 was predominant during the years 2015–2019, whereas ST15 became the predominant sequence type in 2020. Moreover, nine ST15 strains were from four different hospital wards rather than one single ward. As a shift in the sequence type of CRKP was observed in our hospital, the characteristics of ST11 and ST15 CRKP were further analyzed.
Figure 2.
Molecular epidemiology of CRKP-BSI. Distribution of carbapenemase genes (A) and sequence types (B) of the CRKP isolates; (C) distribution of sequence types stratified by year.
Table 1.
Clinical Characteristics, Drug Resistance Profiles and Molecular Epidemiology Characteristics of 42 CRKP Isolates
| Isolation No | Gender | Age (Years) | Seperation Date (Year) | Outcomea | AMK | DOX | MIN | SMZ/TMP | CZA | TGC | Carbapenemase Genotype | MLST | Virulence Genes |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | M | 63 | 2015 | Survive | R | R | R | R | S | S | – | ST722 | fimH, uge |
| 2 | M | 48 | 2017 | Survive | R | S | S | S | S | S | KPC-2 | ST11 | fimH, uge, ybts |
| 3 | M | 54 | 2017 | Die | R | S | S | S | S | S | KPC-2 | ST11 | fimH, uge, ybts |
| 4 | M | 54 | 2017 | Die | R | S | S | S | S | S | KPC-2 | ST11 | fimH, uge, ybts |
| 5 | F | 37 | 2017 | Die | R | S | S | S | S | S | KPC-2 | ST11 | fimH, uge, ybts |
| 6 | M | 63 | 2017 | Die | R | I | R | S | S | I | KPC-2 | ST11 | fimH, uge, ybts |
| 7 | M | 44 | 2017 | Survive | S | R | R | R | R | S | NDM1 | ST35 | fimH, uge, ybts, aeroactin, KfuB |
| 8 | F | 81 | 2017 | Die | R | R | R | R | S | S | KPC-2 | ST11 | fimH, uge, ybts |
| 9 | M | 47 | 2017 | Survive | R | S | I | S | S | S | KPC-2 | ST11 | fimH, uge, ybts |
| 10 | M | 29 | 2017 | Survive | S | S | S | R | R | S | NDM1 | ST248 | fimH, uge, ybts, aeroactin, KfuB |
| 11 | M | 59 | 2017 | Survive | S | R | R | R | R | S | NDM1 | ST35 | fimH, uge, ybts, aeroactin |
| 12 | M | 17 days | 2017 | Survive | S | R | R | R | R | R | NDM5 | ST20 | fimH, uge, ybts |
| 13 | M | 27 | 2017 | Die | R | S | S | S | S | S | KPC-2 | ST11 | fimH, uge, ybts |
| 14 | M | 48 | 2017 | Die | R | S | S | S | S | S | KPC-2 | ST11 | fimH, uge, ybts |
| 15 | F | 73 | 2017 | Die | R | R | I | R | S | S | KPC-2 | ST11 | fimH, uge, ybts |
| 16 | F | 38 | 2018 | Survive | R | S | S | S | S | S | KPC-2 | ST11 | fimH, uge, ybts |
| 17 | M | 36 | 2018 | Survive | R | S | I | S | S | S | KPC-2 | ST11 | fimH, uge, ybts, rmpA |
| 18 | F | 33 | 2018 | Survive | R | S | I | S | S | S | KPC-2 | ST11 | fimH, uge, ybts |
| 19 | M | 53 | 2018 | Die | R | S | S | S | S | S | KPC-2 | ST11 | fimH, uge, ybts |
| 20 | F | 76 | 2018 | Die | R | S | I | S | S | S | KPC-2 | ST11 | fimH, uge, ybts |
| 21 | F | 74 | 2018 | Die | R | I | R | S | S | S | KPC-2 | ST11 | fimH, uge, ybts |
| 22 | F | 85 | 2019 | Die | R | R | R | R | S | I | KPC-2 | ST11 | fimH, uge, ybts |
| 23 | M | 59 | 2019 | Survive | R | S | S | S | S | S | KPC-2 | ST11 | fimH, uge, ybts |
| 24 | M | 80 | 2019 | Die | R | R | R | R | S | I | KPC-2 | ST11 | fimH, uge, ybts |
| 25 | M | 90 | 2019 | Die | R | S | S | R | S | S | KPC-2 | ST11 | fimH, uge, ybts |
| 26 | M | 48 | 2019 | Die | R | S | S | R | S | S | KPC-2 | ST11 | fimH, uge, ybts |
| 27 | M | 80 | 2019 | Survive | R | R | R | S | S | S | KPC-2 | ST11 | fimH, uge |
| 28 | M | 50 | 2019 | Die | R | R | R | S | S | S | KPC-2 | ST11 | fimH, uge, ybts |
| 29 | M | 87 | 2019 | Die | R | S | S | R | S | S | KPC-2 | ST11 | fimH, uge, ybts |
| 30 | F | 47 | 2019 | Die | R | R | R | S | S | I | KPC-2 | ST11 | fimH, uge, ybts |
| 31 | M | 63 | 2020 | Die | R | R | R | R | S | S | KPC-2 | ST11 | fimH, uge, ybts |
| 32 | F | 60 | 2020 | Die | R | R | R | R | S | I | KPC-2 | ST11 | fimH, uge, ybts, rmpA |
| 33 | M | 37 | 2020 | Die | S | R | I | S | S | S | KPC-2 | ST15 | fimH, uge, ybts, KfuB |
| 34 | F | 30 | 2020 | Die | R | R | S | S | S | S | KPC-2 | ST11 | fimH, uge, ybts |
| 35 | M | 47 | 2020 | Survive | S | S | S | S | S | S | KPC-2 | ST15 | fimH, uge, ybts, KfuB |
| 36 | M | 62 | 2020 | Die | S | R | S | S | S | S | KPC-2 | ST15 | fimH, uge, ybts, KfuB |
| 37 | F | 31 | 2020 | Die | S | R | R | S | S | S | KPC-2 | ST15 | uge, ybts, KfuB |
| 38 | M | 88 | 2020 | Survive | S | S | S | S | S | S | KPC-2 | ST15 | fimH, uge, ybts, KfuB |
| 39 | M | 78 | 2020 | Survive | S | R | R | S | S | I | KPC-2 | ST15 | fimH, uge, KfuB |
| 40 | M | 62 | 2020 | Survive | S | R | I | S | S | S | KPC-2 | ST15 | fimH, uge, ybts, KfuB |
| 41 | F | 69 | 2020 | Die | S | R | R | S | S | S | KPC-2 | ST15 | fimH, uge, ybts, KfuB |
| 42 | M | 55 | 2020 | Die | S | R | R | S | S | S | KPC-2 | ST15 | fimH, uge, ybts, KfuB |
Note: aSurvive/die within 30 days.
Abbreviations: AMK, amikacin; DOX, doxycycline; MIN, minocycline; SMZ/TMP, sulfamethoxazole/trimethoprim; CZA, ceftazidime/avibactam; TGC, tigecycline.
In vitro Susceptibilities of CRKP Isolates
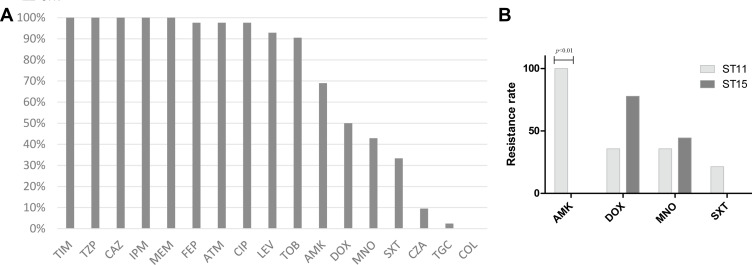
All CRKP isolates were resistant to more than three classes of antibiotics, and showed high resistance (≥90%) to ticarcillin/clavulanic acid, piperacillin/tazobactam, ceftazidime, imipenem, meropenem, cefepime, aztreonam, ciprofloxacin, levofloxacin, and tobramycin (Figure 3A). Fortunately, resistance rates to the other antimicrobial agents were not as high: amikacin (69.0%), doxycycline (50.0%), minocycline (42.9%), sulfamethoxazole/trimethoprim (SXT, 33.3%), ceftazidime/clavulanic acid (CZA, 9.5%), tigecycline (2.4%), and colistin (0.0%).
Figure 3.
In vitro susceptibilities of the CRKP isolates. (A) Antimicrobial resistance patterns of 42 CRKP isolates; (B) comparison of drug resistance rate between ST11 and ST15 CRKP.
Abbreviations: TIM, ticarcillin-clavulanate; TZP, piperacillin-tazobactam; CAZ, ceftazidime; IPM, imipenem; MEM, meropenem; FEP, cefepime; ATM, aztreonam; CIP, ciprofloxacin; LEV, levofloxacin; TOB, tobramycin; AMK, amikacin; DOX, doxycycline; MNO, minocycline; SXT, trimethoprim-sulfamethoxazole; CZA, ceftazidime-avibactam; TGC, tigecycline; COL, colistin.
Further analysis was performed to compare the drug resistance profiles of ST11 and ST15 CRKP strains. As they were all sensitive to CZA, tigecycline, and colistin, we compared drug resistance rates to amikacin, doxycycline, minocycline, and SXT (Figure 3B). ST11 isolates were all resistant to amikacin, while ST15 isolates were all sensitive to amikacin. The resistance rates of ST11 and ST15 CRKP to doxycycline, minocycline and SXT did not show any significant differences.
Virulence Analyses
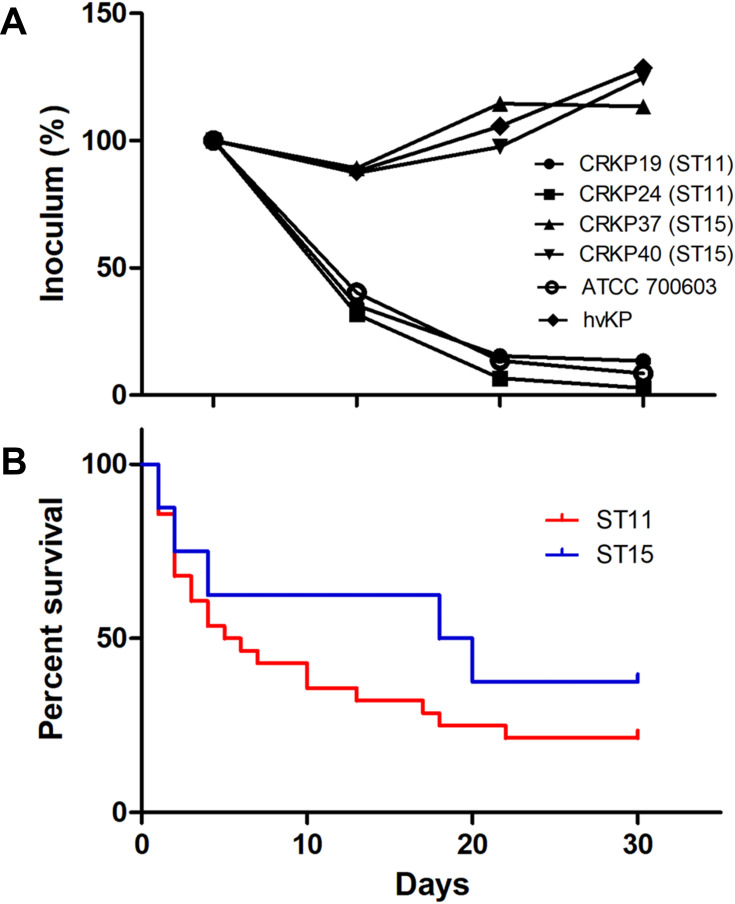
Virulence characteristics of ST11 and ST15 CRKP was further assessed. None of the CRKP isolates were positive for the string test or for common serotypes belonging to K1, K2, K5, K20, K54, or K57. For virulence genes, 100%, 97.6%, 92.9%, 26.2%, 4.8%, and 4.8% of the isolates were positive for uge, fimH, ybtS, KfuB, rmpA and aerobactin genes respectively. All strains were found to be negative for mrkD, entB, IroN, wcaG, alls, and magA genes. Furthermore, all ST15 CRKP isolates were positive for KfuB virulence gene whereas ST11 isolates were all negative. The serum killing assay showed that ST15 CRKP isolates were more resistant to serum killing compared to the ST11 CRKP strains (Figure 4A).
Figure 4.
Virulence characteristics and clinical outcomes of ST11 and ST15 CRKP strains. (A) Serum killing assay of four representative isolates of ST11 and ST15 CRKP, with a hypervirulent Kpn (hvKP) isolate as the positive control; (B) Kaplan–Meier survival estimates of patients with ST11 (red) and ST15 (blue) CRKP-BSI.
Clinical Outcomes
The 7- and 30-day mortality rates of the CRKP-BSI were 38.1% and 61.9% respectively. None of the patients infected with NDM-producing CRKP died; instead, 70.3% of the patients infected with KPC-2-producing CRKP died within 30 days. In addition, the mortality rate of patients infected with ST11 and ST15 CRKP did not show any significant differences (Figure 4B). As for treatment options, over half of the patients received combination treatment, with tigecycline plus carbapenems or colistin the most commonly prescribed.
Discussion
CRKP infection has become an important healthcare-associated threat all over the world, among which BSI of CRKP received particular attention due to its high mortality. Currently data on the clinical characteristics and molecular epidemiology on CRKP-BSI in northeast China are still limited. Therefore, this present study was performed to investigate the molecular epidemiology, virulence characteristics and clinical outcomes of CRKP-BSI in a regional Chinese hospital. Our study found that the dominant KPC-2 producing ST11 CRKP strain shifted to KPC-2 producing ST15 CRKP during the years 2015–2020. Compared with ST11, the ST15 CRKP strains were characterized by high sensitivity to amikacin, positivity for the KfuB virulence gene and resistance to serum killing.
The overall detection rate of CRKP-BSI was 10.4%, with an increasing trend during the years 2015 to 2020. This phenomenon was in agreement with previous CHINET surveillance. Furthermore, the high detection rate of CRKP in BSI coincided with the high percentage of CRKP in all Kpn, which indicated that CRKP-BSI may be associated with CRKP colonization.12 As the ICU, hematology and respiratory medicine wards were the three departments mainly affected, intensive surveillance as well as effective infection control measures are needed to control the transmission of CRKP, especially among high-risk individuals from high-risk departments.13,14
Furthermore, molecular epidemiological studies have demonstrated that KPC-2 was the predominant type of carbapenemase in our hospital, followed by NDM-1 and DNM-5. CRKP isolates with KPC and NDM carbapenemase showed different drug resistance patterns, just as suggested in a previous study.15 Moreover, none of the patients infected with NDM-producing CRKP died within 30 days. Notably, ST11 was the most common sequence type of CRKP during the years 2015 to 2019, whereas ST15 became the predominant type in 2020. The shift in the sequence type of CRKP-BSI in our hospital may be attributed to the COVID-19 epidemic in China at the beginning of 2020; fewer patients were admitted to the medical wards and more exhaustive environmental disinfection could be performed. Subsequently, ST15 CRKP colonization occurred and spread rapidly throughout our hospital in a yet unidentified manner.
It is well known that ST11 is the most common type of CRKP strains prevailing in China. ST11 CRKP stains isolated at our hospital are resistant to amikacin, which differs from the characteristics of the ST15 CRKP strains and is in line with previous reports, which revealed relatively high amikacin resistance rates of ST11 CRKP isolates.16,17 Resistance to amikacin may be associated with genes armA, rmtB, rmtC, aac(3ʹ)-Ia, aac(6ʹ)-Ib, aac(3ʹ)-IIa, aac(3ʹ)-IId, ant (2ʹ)-Ia, and ant(3ʹ)-Ia.7 Consistently, another study also showed lower amikacin resistance rates for ST15 CRKP isolates.18 Based on these findings, different treatment options should be selected for ST11 and ST15 CRKP strains.
In addition, the virulence characteristics of ST11 and ST15 CRKP have been evaluated. None of the CRKP isolates proved to be hypervirulent strains. Regardless, ST15 CRKP isolates were all positive for the KfuB gene whereas ST11 isolates were negative. Furthermore, the serum killing assay also indicated a higher serum resistance of ST15 CRKP over ST11 CRKP strains. Similarly, previous studies have suggested that ST11 CRKP strains were less virulent than other sequence type strains.19–21 Altogether, these results suggested that ST15 CRKP may be characterized by greater virulence, which should come to the attention of clinicians who are to take reasonable treatment measures as soon as possible to improve the prognosis of patients.
In the present study, cases of CRKP-BSI were associated with high mortality, with 7-day and 30-day crude mortality of 38.1% and 61.9% respectively. These findings align with the high crude mortality rates of CRKP-BSI reported in previous studies.5,22 More attention is needed for the early diagnosis of CRKP-BSI and the precise treatment of this life-threatening illness.23 However, the mortality rate of patients infected with ST11 and ST15 CRKP did not show any significant differences, which may be due to the small sample size in our study. Besides, a combination of higher sensitivity to some drugs and stronger virulence of ST15 CRKP may also be the reasons. Hence, further studies with a large sample size are needed.
This study has some limitations. First, since this was a single-center study, whether the differences in drug resistance and virulence patterns of ST11 and ST15 CRKP could represent characteristics of the isolates from the other areas deserves further confirmation. Second, based on the clinical data, the crude mortality of patients infected with ST11 and ST15 CRKP did not show any significant differences and further studies, with larger sample sizes, are warranted for confirmation. Despite these limitations, this is the first study comparing the characteristics of ST11 and ST15 CRKP strains, and provides valuable information for the clinical management of CRKP-BSI.
In conclusion, CRKP isolates of BSI from the medical center in northeast China shifted from a prevalence of ST11 strains to ST15 strains during the years 2015 to 2020. The detected ST15 CRKP strains displayed amikacin sensitivity, positivity of the KfuB virulence gene, and resistance to serum killing, features that differ from the ST11 strains. These findings highlighted the evolution of CRKP in a single hospital and provided clues for the clinical treatment and infection control of CRKP.
Funding Statement
This work was supported by the CAMS Innovation Fund for Medical Sciences (2019-I2M-5-027).
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Wyres KL, Lam MMC, Holt KE. Population genomics of Klebsiella pneumoniae. Nat Rev Microbiol. 2020;18(6):344–359. doi: 10.1038/s41579-019-0315-1 [DOI] [PubMed] [Google Scholar]
- 2.Hu F, Guo Y, Yang Y, et al. Resistance reported from China antimicrobial surveillance network (CHINET) in 2018. Eur J Clin Microbiol Infect Dis. 2019;38(12):2275–2281. doi: 10.1007/s10096-019-03673-1 [DOI] [PubMed] [Google Scholar]
- 3.Li Y, Li J, Hu T, et al. Five-year change of prevalence and risk factors for infection and mortality of Carbapenem-resistant Klebsiella pneumoniae bloodstream infection in a tertiary hospital in North China. Antimicrob Resist Infect Control. 2020;9(1):79. doi: 10.1186/s13756-020-00728-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xiao T, Zhu Y, Zhang S, et al. A retrospective analysis of risk factors and outcomes of Carbapenem-resistant Klebsiella pneumoniae bacteremia in nontransplant patients. J Infect Dis. 2020;221(Suppl 2):S174–S183. doi: 10.1093/infdis/jiz559 [DOI] [PubMed] [Google Scholar]
- 5.Xu L, Sun X, Ma X. Systematic review and meta-analysis of mortality of patients infected with Carbapenem-resistant Klebsiella pneumoniae. Ann Clin Microbiol Antimicrob. 2017;16(1):18. doi: 10.1186/s12941-017-0191-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Karakonstantis S, Kritsotakis EI, Gikas A. Treatment options for K. pneumoniae, P. aeruginosa and A. baumannii co-resistant to carbapenems, aminoglycosides, polymyxins and tigecycline: an approach based on the mechanisms of resistance to carbapenems. Infection. 2020;48(6):835–851. doi: 10.1007/s15010-020-01520-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liao W, Liu Y, Zhang W. Virulence evolution, molecular mechanisms of resistance and prevalence of ST11 Carbapenem-resistant Klebsiella pneumoniae in China: a review over the last 10 years. J Glob Antimicrob Resist. 2020;23:174–180. doi: 10.1016/j.jgar.2020.09.004 [DOI] [PubMed] [Google Scholar]
- 8.Poirel L, Walsh TR, Cuvillier V, et al. Multiplex PCR for detection of acquired carbapenemase genes. Diagn Microbiol Infect Dis. 2011;70(1):119–123. doi: 10.1016/j.diagmicrobio.2010.12.002 [DOI] [PubMed] [Google Scholar]
- 9.Su S, Zhang J, Zhao Y, et al. Outbreak of KPC-2 Carbapenem-resistant Klebsiella pneumoniae ST76 and Carbapenem-resistant K2 hypervirulent Klebsiella pneumoniae ST375 strains in Northeast China: molecular and virulent characteristics. BMC Infect Dis. 2020;20(1):472. doi: 10.1186/s12879-020-05143-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhan L, Wang S, Guo Y, et al. Outbreak by hypermucoviscous Klebsiella pneumoniae ST11 isolates with carbapenem resistance in a tertiary hospital in China. Front Cell Infect Microbiol. 2017;7:182. doi: 10.3389/fcimb.2017.00182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li D, Liao W, Huang HH, et al. Emergence of hypervirulent ceftazidime/avibactam-resistant Klebsiella pneumoniae isolates in a Chinese Tertiary Hospital. Infect Drug Resist. 2020;13:2673–2680. doi: 10.2147/IDR.S257477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rao K, Seekatz A, Bassis C, et al. Enterobacterales infection after intestinal dominance in hospitalized patients. mSphere. 2020;5(4). doi: 10.1128/mSphere.00450-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bassetti M, Peghin M. How to manage KPC infections. Ther Adv Infect Dis. 2020;7:2049936120912049. doi: 10.1177/2049936120912049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pryor R, Viola-Luqa C, Hess O, et al. Barrier precautions in the era of multidrug pathogens. Curr Treat Options Infect Dis. 2020:1–11. doi: 10.1007/s40506-020-00230-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bush K, Bradford PA. Epidemiology of beta-lactamase-producing pathogens. Clin Microbiol Rev. 2020;33(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu X, Zhang W, Zhao Z, et al. Molecular characterization of Carbapenem-resistant Klebsiella pneumoniae isolates with focus on antimicrobial resistance. BMC Genomics. 2019;20(1):822. doi: 10.1186/s12864-019-6225-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou H, Zhang K, Chen W, et al. Epidemiological characteristics of Carbapenem-resistant Enterobacteriaceae collected from 17 hospitals in Nanjing district of China. Antimicrob Resist Infect Control. 2020;9(1):15. doi: 10.1186/s13756-019-0674-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Han Y, Huang L, Liu C, et al. Characterization of Carbapenem-resistant Klebsiella pneumoniae ST15 clone coproducing KPC-2, CTX-M-15 and SHV-28 spread in an intensive care unit of a Tertiary Hospital. Infect Drug Resist. 2021;14:767–773. doi: 10.2147/IDR.S298515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu Y, Liu PP, Wang LH, et al. Capsular polysaccharide types and virulence-related traits of epidemic KPC-producing Klebsiella pneumoniae isolates in a Chinese University Hospital. Microb Drug Resist. 2017;23(7):901–907. doi: 10.1089/mdr.2016.0222 [DOI] [PubMed] [Google Scholar]
- 20.Liu C, Du P, Zhao J, et al. Phenotypic and genomic characterization of virulence heterogeneity in multidrug-resistant ST11 Klebsiella pneumoniae during inter-host transmission and evolution. Infect Drug Resist. 2020;13:1713–1721. doi: 10.2147/IDR.S243836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhao Y, Zhang S, Fang R, et al. Dynamic epidemiology and virulence characteristics of Carbapenem-resistant Klebsiella pneumoniae in Wenzhou, China from 2003 to 2016. Infect Drug Resist. 2020;13:931–940. doi: 10.2147/IDR.S243032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hussein K, Raz-Pasteur A, Finkelstein R, et al. Impact of carbapenem resistance on the outcome of patients’ hospital-acquired bacteraemia caused by Klebsiella pneumoniae. J Hosp Infect. 2013;83(4):307–313. doi: 10.1016/j.jhin.2012.10.012 [DOI] [PubMed] [Google Scholar]
- 23.Ho VP, Kaafarani H, Rattan R, et al. Sepsis 2019: what surgeons need to know. Surg Infect (Larchmt). 2020;21(3):195–204. doi: 10.1089/sur.2019.126 [DOI] [PMC free article] [PubMed] [Google Scholar]