Abstract
We announce here the first complete chloroplast genome sequence of Tetrastigma planicaule, one important Chinese folk medicinal plant. This complete chloroplast genome is 160,323 bp in length. In total, 131 genes were identified, including 86 protein-coding genes, 37 tRNA genes, and 8 rRNA genes. The results of phylogenetic analysis indicated that Tetrastigma is a separate genus and is closely related to Vitis.
Keywords: Chloroplast genome, Tetrastigma planicaule, Phylogenetic analysis
Tetrastigma planicaule (Hook.) Gagnep. belongs to genus Tetrastigma (Miq.) Planch. (Vitaceae). It is a wild evergreen woody liana, and is mainly distributed in China, India, Laos and Vietnam (Ren et al. 2007). Tetrastigma planicaule is an important Chinese folk medicinal plant which mainly used in the treatment of rheumatoid arthritis, lumbar muscle strain, urticaria and asthma (Chen 2017; Shao et al. 2010). It is also an excellent ornamental plant because of its flat stems and the characteristic of flowering and fruiting on old stems (Zhang et al. 2012). Here, we assembled and characterized the complete chloroplast genome of T. planicaule, which would enrich the gene information and contribute to further study of this plant.
The plant material of T. planicaule was collected from county Daxin (Chongzuo, Guangxi, China; 22.31°N, 106.80°E). Voucher specimens were deposited at the herbarium of Guangxi Forestry Research Institute (Mr Li, zzcx_gfri@163.com) under the number 2020120401, and DNA samples were stored at Guangxi Key Laboratory of Superior Timber Trees Resource Cultivation, Nanning, China. Raw data were generated using the NexteraXT DNA Library Preparation Kit (Illumina, San Diego, CA) with 150 bp paired-end read lengths, then raw sequence reads were edited using the NGS QC Tool Kit v2.3.3. High-quality reads were assembled into contigs using the denovo assembler SPAdes 3.11.0 software (Bankevich et al. 2012). Finally, the complete chloroplast genome was annotated by PGA software (Qu et al. 2019) and submitted to GenBank under the accession number of MW401672.
The total length of complete chloroplast genome of T. planicaule was 160,323 bp, with a total GC content of 37.49%. The complete chloroplast comprised of a large single-copy (LSC) region of 88,181 bp, a small single-copy (SSC) region of 19,096 bp and two inverted repeat (IRS) regions of 26,523 bp. A total of 131 genes were contained in the complete chloroplast genome, including 86 protein-coding genes, 37 tRNA genes and 8 rRNA genes.
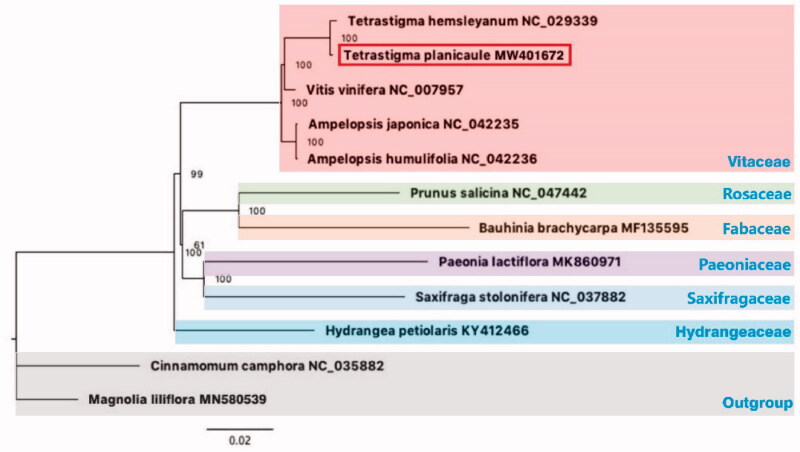
Eleven complete chloroplast genomes of other species were selected to confirm the phylogenetic position of T. planicaule, with Cinnamomum camphora and Magnolia lilifolra used as outgroups. All of these complete chloroplast sequences were aligned by the MAFFT version 7.429 software (Katoh and Standley 2013) and trimmed by TrimAl (Capella-Gutierrez et al. 2009). A maximum-likelihood (ML) tree was inferred by ultrafast bootstrapping with 1000 replicates through IQ-TREE 1.6.12 (Nguyen et al. 2015) based on the TVM + F + R2 nucleotide substitution model, which was selected by ModelFinder (Kalyaanamoorthy et al. 2017) . The result of phylogenetic analysis indicated that Tetrastigma is a separate genus and it is closely related to Vitis (Figure 1). Tetrastigma and Vitis were proved to be sister groups through plastid rbcL DNA sequence (Ingrouille et al. 2002). Our study makes this phylogenetic relationship more clearly, and the results are consistent with previous studies of Yu et al (2019).
Figure 1.
The ML phylogenetic tree based on the complete chloroplast genomes of Tetrastigma planicaule and other 11 species. Numbers near the nodes represent ML bootstrap value.
Funding Statement
This work was supported by the Guangxi Science and Technology Base & Talents’s Special Project under Grant Number [Gui-Science AD17129021].
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in GenBank number MW401672 (https://www.ncbi.nlm.nih.gov/nuccore/MW401672.1/) and SRA number PRJNA715476 (https://www.ncbi.nlm.nih.gov/sra/PRJNA715476).
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capella-Gutierrez S, Silla-Martinez JM, Gabaldon T.. 2009. TrimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 25(15):1972–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen SB. 2017. Studies on the constituents from yao medicine Tetrastigma planicaul. [Master thesis]. Guangxi: Guangxi University of Chinese Medicine. (in Chinese) [Google Scholar]
- Ingrouille MJ, Chase MW, Fay MF, Bowman D, van der Bank M, Bruijn ADE.. 2002. Systematics of vitaceae from the viewpoint of plastid RBCL DNA sequence data. Bot J Linnean Soc. 138(4):421–432. [Google Scholar]
- Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS.. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 14(6):587–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM.. 2013. MAFFT: multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ.. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu XJ, Moore MJ, Li DZ, Yi TS.. 2019. PGA: a software package for rapid, accurate, and flexible batch annotation of plastomes. Plant Methods. 15(1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren H, Wen J.. 2007. Vitaceae. In: Wu ZY, Raven PH, Hong DY, editors. Flora of China, vol. 12. Beijing: Science Press; St. Louis: Missouri Botanical Garden Press; p. 195–208. [Google Scholar]
- Shao JC, He CH, Lei T, et al. 2010. Chemical constituents of Tetrastigma planicaule (Hook.) Gagnep. Chin Pharma J. 45(21):1615–1617. (in Chinese) [Google Scholar]
- Yu X, Tan W, Zhang H, Gao H, Wang W, Tian X.. 2019. Complete chloroplast genomes of ampelopsis humulifolia and ampelopsis japonica: molecular structure, comparative analysis, and phylogenetic analysis. Plants. 8(10):410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XQ, Pan JG, Sun SF.. 2012. Appreciation of vine plants in Shanghai Chenshan Botanical Garden. Garden. (09)36–39. (in Chinese) [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are openly available in GenBank number MW401672 (https://www.ncbi.nlm.nih.gov/nuccore/MW401672.1/) and SRA number PRJNA715476 (https://www.ncbi.nlm.nih.gov/sra/PRJNA715476).