Abstract
Background: Persistent transmission of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has given rise to a COVID-19 pandemic. Several vaccines, conceived in 2020, that evoke protective spike antibody responses are being deployed in mass public health vaccination programs. Recent data suggests, however, that as sequence variation in the spike genome accumulates, some vaccines may lose efficacy. Methods: Using a macaque model of SARS-CoV-2 infection, we tested the efficacy of a peptide-based vaccine targeting MHC class I epitopes on the SARS-CoV-2 nucleocapsid protein. We administered biodegradable microspheres with synthetic peptides and adjuvants to rhesus macaques. Unvaccinated control and vaccinated macaques were challenged with 1 × 108 TCID50 units of SARS-CoV-2, followed by assessment of clinical symptoms and viral load, chest radiographs, and sampling of peripheral blood and bronchoalveolar lavage (BAL) fluid for downstream analysis. Results: Vaccinated animals were free of pneumonia-like infiltrates characteristic of SARS-CoV-2 infection and presented with lower viral loads relative to controls. Gene expression in cells collected from BAL samples of vaccinated macaques revealed a unique signature associated with enhanced development of adaptive immune responses relative to control macaques. Conclusions: We demonstrate that a room temperature stable peptide vaccine based on known immunogenic HLA class I bound CTL epitopes from the nucleocapsid protein can provide protection against SARS-CoV-2 infection in nonhuman primates.
Keywords: SARS-CoV-2, animal model, macaque, vaccine, MHC class I peptide, T cell
1. Introduction
As of April 2021, over 1.6% of the world population have had confirmed COVID-19 disease and the new case rate is about half a million per day. Less than 2% of the world’s population are vaccinated against SARS-CoV-2 [1]. Confounding factors in the efforts to reach herd immunity to COVID-19 disease include, but are not limited to, the following: (1) the spread of SARS-CoV-2 mutations affecting the efficacy of current iterations of vaccines and therapeutic biologics [2], (2) the speed of SARS-CoV-2 vaccine deployment, development, and manufacture, and (3) in the context of global public health, issues related to vaccine hesitancy, cold supply-chain requirements, the total manufacturing cost per dose, and ease of administration.
We previously described a novel vaccine platform [3,4], which may address many of the above concerns, and now report on its efficacy in a rhesus macaque model of SARS-CoV-2 infection. Macaque models of COVID-19 disease have been previously reported and have served as critical tools for understanding disease pathology and for the development and testing of vaccines and therapeutics [5,6,7,8,9,10,11,12,13,14,15,16,17,18,19,20,21]. While the clinical course of SARS-CoV-2 infection in macaques is milder relative to that observed in humans [6,8,20,21], the macaque model remains the gold standard for preclinical evaluation of COVID-19 vaccines [10,19,22,23,24,25,26,27]. Our overall approach focuses on promoting protective T-cell immunity using synthetic peptides delivered in biodegradable microspheres together with Toll-like receptor (TLR) 4 and 9 adjuvants and differs from current COVID-19 vaccines against spike proteins. The synthetic peptide sequences applied are based on known immunogenic HLA class I bound epitopes that have previously been characterized in either SARS-CoV-1 [28] or SARS-CoV-2 [29,30,31] infections. We provide evidence that application of this vaccine platform in SARS-CoV-2-challenged macaques provides protection from pneumonia-like pathology observed in virally challenged but unvaccinated control non-human primate (NHP) subjects, reduces viral loads as compared to unvaccinated controls, and induces changes in the gene expression patterns in recovered BAL cells consistent with enhanced antigen presentation capacity and markers of T cells.
2. Materials and Methods
2.1. Macaque MHC Class I Typing
The MHC class I genes of a cohort of 15 rhesus macaques (Envigo, Alice, TX, USA) were molecularly typed by the University of Wisconsin–Madison National Primate Research Center. Mamu (Macaca mulatta) MHC class I alleles were typed amplifying the genomic DNA of each subject using a panel of specific primers for exon 2 of all known MHC-A and MHC-B alleles encoded by each subject’s target DNA. Resulting amplicons were sequenced by the Illumina MiSeq method [32]. The primer panel contained specific primer pairs able to amplify all possible MHC-A and MHC-B alleles encoded by each macaque. Sequencing data analysis provided a high-resolution haplotype for the MHC-A and MHC-B alleles carried by each subject. Following analysis, we selected four of the 15 macaques for vaccination based on peptide MHC binding predictions as described below.
2.2. Vaccine Design/Peptide Selection and Manufacture
2.2.1. In Silico Design and Selection of SARS-CoV-2 CTL Epitopes
The overall strategy and rationale for the selection of synthetic peptides used to stimulate potential CTL immune responses in SARS-CoV-2-infected humans has been previously described [3,4]. We selected SARS-CoV-2 nucleoprotein as the target of CTL attack based on the following rationale: (1) survivors of SARS-CoV-1 have shown a memory T-cell response to nucleoprotein at least two years after infection [28], (2) there is >90% amino acid sequence homology between SARS-CoV-1 and SARS-CoV-2 nucleoprotein (the homology for the selected CTL epitopes used in this report is 100%) [33], and (3) in general, there is a lower frequency of mutations resulting in amino acid substitutions (relative to spike protein) that might affect the immunogenicity of the selected CTL epitopes represented by synthetic peptides within the vaccine formulation [34]. The lower mutation frequency may reflect the hypothesis that amino acid substitutions in nucleoprotein may impact viral fitness [35]. We reviewed previous literature and MHC peptide-binding databases [36] and selected five amino sequences representing the SARS-CoV-2 nucleoprotein with predicted strong in vitro affinity for HLA class I molecules [37] and/or documented or predicted immunogenic potential [4,28,29,30,31,33,38,39]. Together this set of peptides yielded potential broad coverage of HLA haplotypes (>90% worldwide) (Supplementary Materials, Table S1A). The predicted binding of this set of peptides was examined within the MHC genotypes of the cohort of 15 rhesus macaques [40] available to us and the best correspondence between selected peptides and the rhesus MHC class I genotype was selected (Supplementary Materials, Table S1B). As the predicted peptide macaque MHC binding coverage for the peptide LLLDRLNQL was incomplete in the available genotypes, we added an additional peptide (ASAFFGMSR) with predicted strong Mamu MHC class I binding to the formulation for a total of six peptides.
2.2.2. Microsphere Preparation and Adjuvant Formulation
The peptide epitopes used in this study were delivered in vivo by intratracheal instillation of a formulation of poly-L-lactide-co-glycolide (PLGA) microspheres containing the corresponding synthetic nine-mer peptides and TLR-9 agonist CpG oligonucleotide adjuvant in a vehicle containing TLR-4 agonist monophosphoryl lipid A (MPLA). The rationale for the choice of the delivery platform and the basic manufacturing scheme used in production has been previously described [3,4,41]. Briefly, room temperature solutions of a synthetic peptide, CpG oligonucleotide, and mannose were mixed with a solution of PLGA in acetone followed by sonication. The formulation was then filtered using a sterile 0.45 micron filter (Pall Corporation, Westborough, MA, USA ), processed through a precision spray-drying device (Buchi Corporation, New Castle, DE, USA), and passed through a drying chamber, with nitrogen gas introduced through a 0.2 micron filter and heated to 65 °C to allow evaporation of the acetone. The dry microsphere stream was analyzed in real time through a laser particle size analyzer (SprayTech, Malvern Instruments, Malvern, PA, USA) before collection (Buchi cyclone dryer) as a dry powder for reconstitution at the time of delivery using a 2% dimethylsufoxide (DMSO) aqueous solution containing MPLA (20 μg/mL). The microspheres and diluent were handled using a BSL-2 non-sterile technique throughout the experiment. Each microsphere contained peptide loaded at approximately 0.1% by weight and CpG 0.01% by weight. Monitoring of the microsphere diameters allowed the production of microspheres with a mean diameter of 10 ± 2 microns. This diameter was selected for formulation to ensure delivery via phagocytosis of no more than 1–4 microspheres per antigen-presenting cell (APC), which have average diameters of 13 microns [3]. cGMP manufacturing protocols were employed using GMP grade synthetic peptides (Peptides International, Louisville, KY, USA), CpG oligodeoxynucleotides (Trilink Biosciences, San Diego, CA, USA), and MPLA (Avanti Polar Lipids, Alabaster, AL, USA). The CpG oligonucleotide and MPLA used in this study were manufactured using the same chemical compositions as equivalent materials used in FDA-approved vaccines. Assessment of thermal stability of the synthetic peptides within the microspheres has been previously reported [3]. Peptide content and structure in microspheres were determined by high-performance liquid chromatography after two months of room temperature storage. We found that over 99% of the peptide was maintained structurally intact.
2.3. Animal Studies
2.3.1. Ethics Statement
The animal research protocols used in this study were performed in strict accordance with the recommendations in the Guide for Care and Use of Laboratory Animals, Eighth Edition (National Academy Press, Washington, DC, USA, 2011). The University of Texas Medical Branch (UTMB) facility where these studies were conducted is accredited by the Association for Assessment and Accreditation of Laboratory Animal Care. The protocols were approved by the UTMB Institutional Animal Care and Use Committee (Protocol Numbers 2,004,051 (natural history/control study) and 2,003,033 (vaccination study)) and complied with the Animal Welfare Act, the U.S. Public Health Service Policy, and other federal statutes and regulations related to animals and experiments involving animals. All hands-on manipulations, including immunizations and biosampling, were performed while animals were sedated via ketamine (5 mg/kg)/dexmedetomidine (0.025 mg/kg) intramuscular injection. All efforts were made to minimize suffering.
2.3.2. Macaques
Adult Indian-origin rhesus macaques (Macaca mulatta, n = 7 (5 male, 2 female), 46–48 months old) or Vietnamese-origin cynomolgus macaques (Macaca fascicularis, n = 1, female, 84 months old), individually identified via unique tattoo, were obtained from Envigo/Covance (Alice, TX, USA). All animals were considered healthy by a veterinarian before being placed on study. Macaques were individually housed in stainless steel nonhuman primate caging equipped with squeeze backs for the duration of the studies. For continuous core body temperature measurements, a DST micro-T implantable temperature logger (Star–Oddi, Gardabaer, Iceland) was surgically implanted into the peritoneal cavity of each animal prior to study initiation; data recording was set to 10 or 15 min intervals for control and vaccinated macaques, respectively. Certified Primate Diet 5048 was provided to the macaques daily. Drinking water (reverse osmosis-purified) was provided ad libitum through an automatic watering system. To promote and enhance the psychological wellbeing of the animals, food enrichment consisting of fresh fruits and vegetables was provided daily. Environmental enrichment including various manipulatives (Kong toys, mirrors, and puzzles) was also provided.
2.4. Immunization, Virus Challenge, Post-Challenge Monitoring and Biosampling
2.4.1. Immunization and ELISPOT Analysis
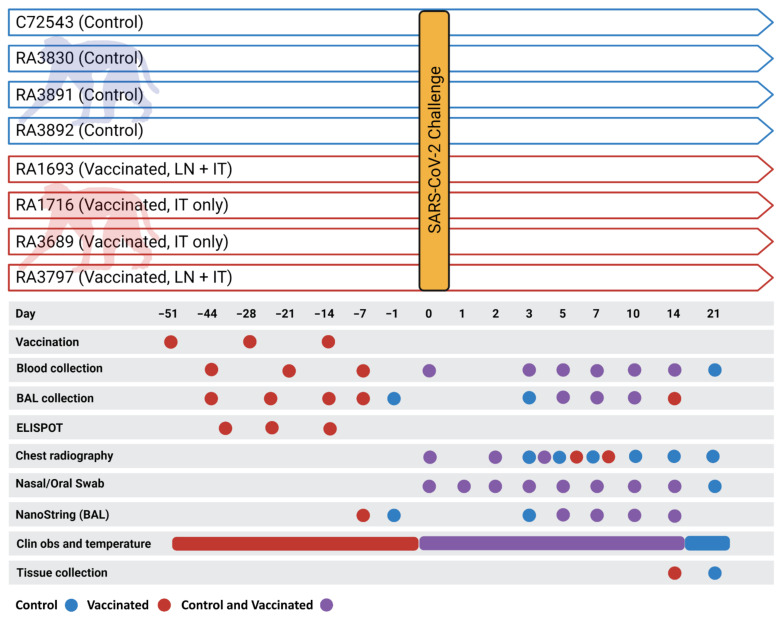
Immunizations were performed on the selected MHC-typed rhesus macaques (n = 4) via ultrasound-guided inguinal lymph node (LN) injection and/or intratracheal instillation (IT). A total of 20 mg of vaccine microsphere preparation in 1 mL was used for each LN injection (two injections/dose/animal) and 100 mg of vaccine microspheres in a 5 mL volume was used for each IT administration. Specifically, on day −51 (51 days prior to virus challenge), two of the macaques (Figure 1) were administered 2 mL of vaccine via LN injection (1 mL per node) and two macaques were administered vaccine via the IT route (5 mls). Subsequent administration of the vaccine, on days −28 and −14, occurred via the IT route only as described previously [42]. On days −44, −21, and −7 (7 days post-vaccination), femoral vein peripheral blood (8 mL) was collected from each animal into a BD Vacutainer® CPT™ Cell Preparation Tube (Becton, Dickinson and Company, Franklin Lakes, New Jersey, USA) with Sodium Heparin (Becton, Dickinson and Company, Franklin Lakes, NJ, USA) and processed to peripheral blood mononuclear cells (PBMCs) per manufacturer instructions. Collected PBMCs were assessed for immunoreactivity via ELISPOT. In brief, ELISPOT assay plates (MabTech Inc., Cincinnati, OH, USA) specific for the detection of primate IFNγ were used according to manufacturer instructions. BAL cell concentrations were adjusted to 1 × 105 cells per mL in a complete growth medium. Diluted BAL cells were dispensed (100 µL/well) into a 96-well plate after which 100 µL of complete growth medium (CGM, negative control), concanavalin A in CGM at 10 µg per well (positive control), and various concentrations of specific (i.e., immunizing) and non-specific peptides (Supplementary Materials, Table S1B) were added. Peptides used for immunization were added to wells at a concentration of 50 µM. All samples were assayed in duplicate. Plates were incubated at 37 °C/5% CO2 for 20–22 h, after which plates were thoroughly washed. Conjugated detection antibody was then added and incubated followed by additional washing. Wells were developed using 3,3′,5,5′-tetramethylbenzidine as a substrate. Counts were performed at Cellular Technology Corporation (Shaker Heights, OH, USA) using an Immunospot Analyzer (CTI, Shaker Heights, Ohio, USA) and all well images were quality-controlled on site. All spot-forming cell counts reported are the result of averaging counts from the duplicate 50 µM immunization-specific peptide wells.
Figure 1.
Legend. Schematic of the experimental protocol. Unvaccinated (control) macaques are represented by blue coloring. Vaccinated macaques are represented by red coloring. Overlapping tasks are represented by purple coloring. Graphic created with BioRender.com.
2.4.2. Virus Challenge
On day 0, macaques were administered 1–5 × 108 TCID50 SARS-CoV-2 (USA_WA1/2020) via combined mucosal atomization (1 mL as delivered using a MAD Nasal™ Intranasal Mucosal Atomization Device (Teleflex, Morrisville, North Carolina, USA) per manufacturer instructions) and intratracheal instillation (4 mL). Intratracheal instillations were performed as described above for delivery of the vaccine. The virus suspension was prepared on the day of challenge from frozen seed stock (kindly provided by Dr. Chien-Te (Kent) Tseng at UTMB) initially generated (one passage) in Vero C1008 (E6) cells (BEI Resources, NR-596, Lot 3956593) from original material provided by the Centers for Disease Control and Prevention in January 2020. Next-generation sequencing confirmed a 100% consensus sequence-level match to the original patient specimen (GenBank accession MN985325.1).
2.4.3. Post-Challenge Monitoring and Chest Radiography
Animals were monitored and scored twice daily for clinical signs of disease, including alterations in activity/appearance (i.e., hunched posture) and food consumption/waste output, and were scored based on general appearance, activity, food consumption, and outward changes in breathing patterns. Prospectively defined criteria that required immediate euthanasia included severe dyspnea and/or agonal breathing and prostate posture/reluctance to move when stimulated. No animals met endpoint criteria during the study. Ventrolateral chest radiography was performed on the days indicated (Figure 1) using a portable GE AMX-4+ computed radiography system (General Electric Company, Boston, Massachussetts, USA) per the manufacturer’s instructions. DICOM data files were independently evaluated by two independent investigators blinded to group assignment with large animal imaging experience via a four-pattern approach (analyses of consolidation, interstitial areas, nodules or masses, and atelectasis).
2.4.4. Biosampling
Blood, nasal cavity samples, and BAL fluid were collected at the indicated times (Figure 1). Femoral vein peripheral blood was collected via Vacutainer® into standard collection tubes containing ethylenediaminetetraacetic acid (EDTA). Hematology was performed on EDTA blood using the Abaxis VETSCAN® HM5 Hematology Analyzer (Abaxis, Inc., Union City, CA, USA). Nasal cavity samples, collected using sterile cotton-tipped medical swabs, were placed into 0.5 mL sterile phosphate-buffered saline (PBS) for viral load analysis. For BAL fluid collection, animals were sedated as previously described and placed in ventral recumbency. The trachea was visualized and cannulated by an appropriately sized rubber feeding tube. Following the placement of the feeding tube, 20 mL of sterile PBS was introduced into the lung and recovered manually through the feeding tube via syringe. This was repeated for a total of 40 mL per animal. The total collected volume from each animal (10–30 mL) was pooled and centrifuged under ambient conditions (10 min at 500× g), after which the supernatant was removed. The resulting cell pellet was resuspended in 2 mL of sterile PBS. From this, 1 mL was used for ELISPOT analysis as described for PBMCs. The remaining volume was used for viral load analysis and gene expression profiling.
2.5. Viral Load Analysis
2.5.1. Infectious Viral Load (TCID50)
Nasal swab and BAL cells suspension samples were serially diluted and incubated with 2 × 104 Vero C1008 (E6) cells (BEI Resources, NR-596, Lot 3956593) in 100 μL of culture medium (MEM/2% FBS) in 96-well flat-bottom plates (n = 5 replicate wells per dilution). Each plate contained negative and positive control wells inoculated with culture medium and diluted virus stock, respectively. Cultures were incubated at 37 °C/5% CO2 for 96 h after which cytopathic effect was measured via microscopic observation. The TCID50/mL value for each sample was calculated as previously described [43].
2.5.2. qRT-PCR
Nasal swab and BAL cell suspension samples (50 μL) were added to TRIzol® LS Reagent (250 μL) and allowed to incubate under ambient conditions for 10 min. Samples were processed to RNA using Zymo Direct-zol™ RNA Mini Prep kits (Zymo Research, Irvine, CA, USA) per manufacturer instructions. RNA samples were analyzed via qRT-PCR targeting the SARS-CoV-2 E gene. The probe (Integrated DNA Technologies, Coralville, IA, USA) was labeled at the 5′-end with fluorophore 9-carboxyfluoroescein (6-FAM) and included an internal quencher (ZEN) and a 3′-end quencher (IowaBlackFQ, IABkFQ). The master mix was prepared by combining forward primer (250 nM, 5′-ACAGGTACGTTAATAGTTAATAGCGT-3′), reverse primer (250 nM, 5′-ATATTGCAGCAGTACGCACACA-3′), and probe (375 nM, 5′-6FAM-ACACTAGCC/ZEN/ATCCTTACTGCGCTTCG-IABkFQ-3′) with 12.5 μL of 2X QuantiFast Probe Mix (QIAGEN, GGermantown, MD, USA), 0.25 μL of 2X QuantiFast RT Mix (QIAGEN), and PCR-grade water (fill to 20 μL). A test sample (5 μL) was added to the master mix, resulting in a final volume of 25 μL per reaction. Real-time analysis was performed using the Bio-Rad CFX96™ Real-Time PCR Detection System. Thermocycling conditions were as follows: step 1, 1 cycle, 50 °C for 10 min; step 2, 1 cycle, 95 °C for 10 min; steps 3–5, 45 cycles, 95 °C for 10 s, 60 °C for 30 s, single read. Negative controls included reaction mixtures without RNA. For quantification purposes, viral RNA extracted from the virus seed stock with a known TCID50/mL titer was used. All qRT-PCR results are expressed as TCID50/mL equivalents.
2.6. Gene Expression Profiling
BAL samples were processed to RNA as described above for qRT-PCR analysis. RNA quantity and quality were assessed using a NanoDrop™ Lite Spectrophotometer (ThermoFisher Scientific, Waltham, MA, USA). Samples, normalized to 20 ng/μL, were analyzed by NanoString Technologies (Seattle, WA, USA) using the nCounter® SPRINT™ Profiler (NanoString Technologies) and gene expression was profiled using the Non-Human Primate Immunology V2 Panel, which contains 754 genes that encompass 17 immune-related signaling pathways with isoform coverage for both Macaca mulatta and Macaca fascicularis. Probe sets that did not cover both Macaca species were eliminated, resulting in a probe set of 730 genes. Raw gene expression data sets received from NanoString Technologies were processed to remove background signals and normalized using the nSolver™ V.3.0 digital analyzer software (NanoString Technologies). Background signal correction was accomplished by subtracting the NanoString negative control genes. Gene expression normalization was performed using the 16 internal reference genes included in the panel.
2.7. Study Termination
At scheduled study termination time points (14 and 21 days post-challenge for vaccinated and control macaques, respectively), animals were humanely euthanized via intravenous administration of a pentobarbital-based euthanasia solution under deep anesthesia followed by bilateral thoracotomy.
2.8. Statistical Analysis
Descriptive statistics were performed using Microsoft Excel. Hypothesis testing was performed by considering the null hypothesis of the absence of an association between the compared variables. The statistical strength of associations of continuous data was tested using Student’s t-testing. Qlucore Omics Explorer 3.5 (Qlucore) and Metascape (Metascape.org) were used to identify the discriminating variables within the NanoString gene expression datasets from BAL sample analysis that were most significantly different between vaccinated and control subjects. The identification of significantly differential variables between the two groups was performed by fitting a linear model for each variable. The set of genes (87 variable genes out of a total of 730 genes) was identified using a p-value of 0.05, at least a threefold change, and a q-value cutoff of 0.1. p-values were adjusted for multiple testing using the Benjamini–Hochberg method [44]. Gene expression data were scaled to a mean = 0 and a variance = 1 before clustering. Hierarchical clustering of gene expression in BAL was performed using a supervised weighted-average-linkage two-comparison approach. The metric used in scaling dendrogram arms was Pearson’s correlation coefficient.
3. Results
3.1. Primary Clinical Outcome
3.1.1. Clinical Signs, Body Temperature Alterations, and Hematology
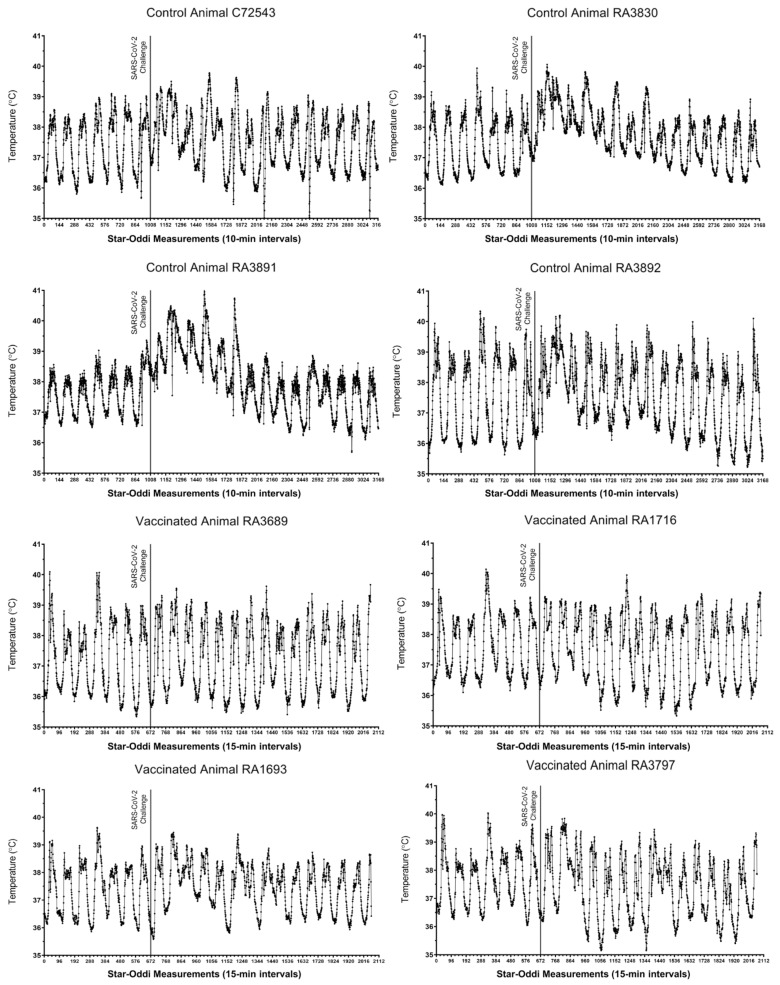
Following SARS-CoV-2 challenge, outward clinical signs measured in control macaques included acute mild lethargy and respiratory distress. All vaccinated animals were normal throughout the post-challenge study period. Core body temperatures, as measured via implanted Star–Oddi DST temperature loggers (Star-Oddi, Reikjavik, Iceland), demonstrated a disruption in the diurnal cycle and mild fever lasting 2–5 days post-challenge in all four control animals (Figure 2, top panels). Conversely, only two of the four vaccinated macaques (RA1693 and RA3797) presented with similar findings, although diurnal cycle disruption was of a shorter duration (1–2 days) and the febrile response was milder (Figure 2, bottom panels). No alterations were measured in the remaining two vaccinated animals. Prior to virus challenge, vaccinated macaques presented with occasional disruptions in the diurnal temperature associated with the vaccination procedure (Supplementary Materials, Figure S1).
Figure 2.
Core body temperature alterations in control and vaccinated macaques following SARS-CoV-2 challenge. For each animal, seven days of pre-challenge baseline temperature measurements are shown. Each tick on the x-axis represents 6 h or 36 individual logger measurements.
Automated hematology analyses were performed on peripheral blood samples (Supplementary Materials, Figure S2). Overall, the number of white blood cells was significantly increased in control subjects on post-challenge days 3 and 5. We observed general lymphopenia in all macaques on day 1 post-challenge. By day 3 post-challenge, lymphocyte counts significantly increased in vaccinated subjects relative to unvaccinated control subjects. Peripheral blood monocyte counts generally peaked on days 1 through 5 in all animals but remained significantly elevated in vaccinated macaques at the end of the study. Neutrophil counts generally rose by day 1 post-challenge in all subjects. There was a transient significant elevation in neutrophils on day 7 post-challenge in vaccinated animals.
3.1.2. Viral Load
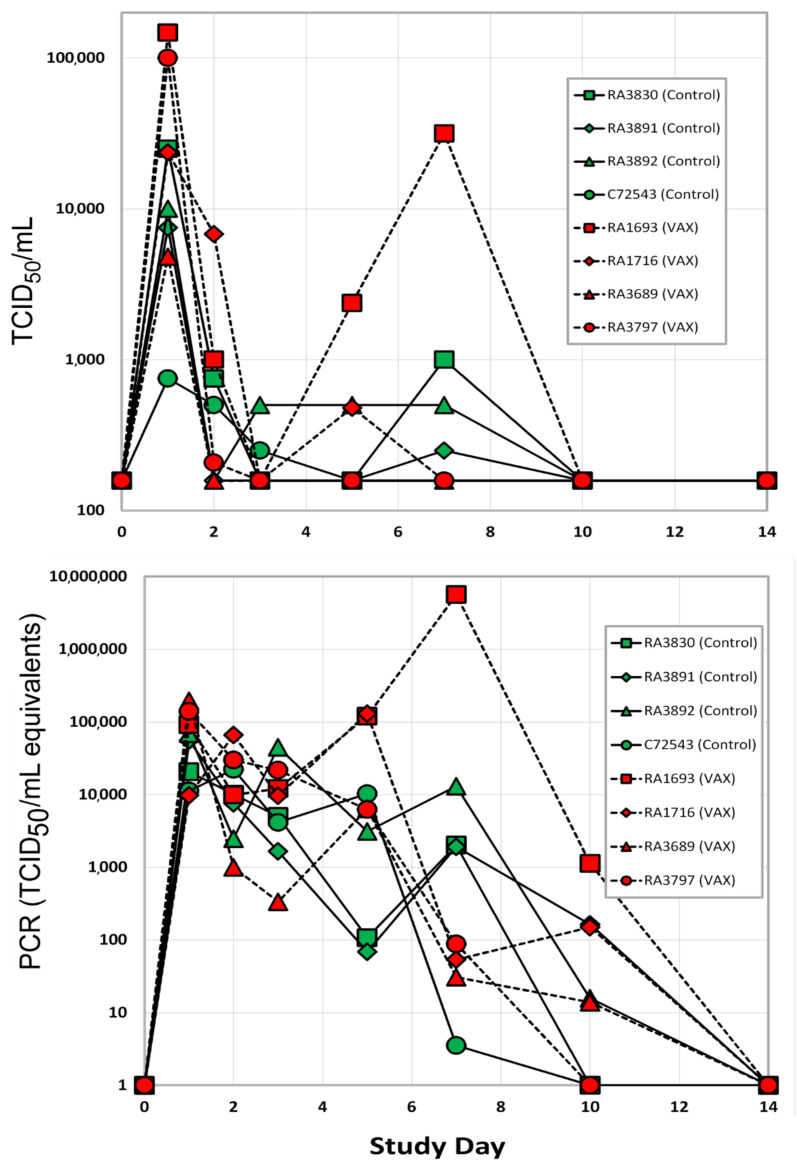
Following SARS-CoV-2 challenge, nasal swab and BAL fluid samples were collected throughout the post-challenge period for analysis of infectious viral load and viral RNA via TCID50 and qRT-PCR assays, respectively. Infectious virus was measured from nasal swabs of control and vaccinated macaques beginning on day 1 post-challenge (Figure 3, top panel). By Day 7, three of the four unvaccinated animals continued to demonstrate infectious viral shedding, albeit at low levels. In contrast, infectious virus could be measured in only one of the vaccinated macaques at the same time point. By 10 days post-challenge, infectious viral loads were undetectable in nasal swab samples from all animals. Viral RNA in nasal swabs generally reflected infectious viral loads. By day 7, three of the four vaccinated animals demonstrated a 100-fold decrease in nasal swab viral RNA relative to the unvaccinated controls (Figure 3, bottom panel). By 14 days post-challenge, SARS-CoV-2 RNA levels were undetectable in nasal swab samples from all subjects. Infectious viral load data were used to calculate an average viral clearance rate post-challenge for each rhesus macaque. The average viral clearance rate from days 2 through 10 was four- to fivefold higher in two of the four vaccinated macaques (RA1693 and RA3797) relative to the unvaccinated controls (Supplementary Materials, Figure S3).
Figure 3.
Viral load in nasal swab samples as measured via TCID50 assay (top) and qRT-PCR (bottom). The LLOD of the plaque assay was 150 units. Red symbols are vaccinated rhesus macaques, control unvaccinated rhesus subjects are shown in green symbols.
3.1.3. Macaque Chest Radiography
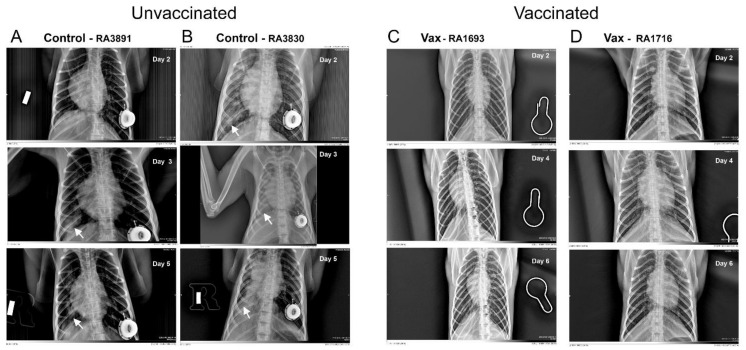
SARS-CoV-2 challenge in unvaccinated controls resulted in mild-to-moderate lung abnormalities, similar to those previously reported for macaques [6,15,16,20,45,46,47,48]. These were predominantly limited to the caudal lung relative to baseline images, peaked 3–5 days post-challenge, and were qualitatively characteristic of subclinical or mild-to-moderate human COVID-19 (e.g., ground-glass opacities with or without reticulation, paving, silhouetting, and/or linear opacities). The mild-to-moderate interstitial pneumonitis seen on the ventrolateral chest radiographs of unvaccinated subjects are consistent with focal infiltrates representing a complex of interstitial macrophages, neutrophils, and plasmacytoid dendritic cells [49]. Abnormalities in control animals resolved by days 10–21. In contrast, vaccinated macaques lacked the appearance of ground-glass opacities in all regions of the lung throughout the study period (Figure 4 and Supplementary Materials, Figures S4–S9). We did observe, however, modest bilateral increases in reticulation in vaccinated macaques on days 3–5, but these abnormalities also resolved by days 10–21. Bronchoalveolar lavage has been reported to affect computerized tomography X-ray results in healthy rhesus macaques [50]. In our study, however, the pattern of reported changes in vaccinated (i.e., healthy) animals on which BAL was performed was more similar to increases in reticulation versus the patchy consolidations observed in the unvaccinated controls.
Figure 4.
Representative chest radiographs of control and vaccinated macaques following SARS-CoV-2 challenge. As shown, control macaques (left columns (A,B)) demonstrated a progression of pulmonary infiltrates during the acute period (days 2–5) of disease post-challenge. In contrast, vaccinated macaques (right columns (C,D)) lacked similar abnormalities. White arrows indicate areas of mild-to-moderate pulmonary infiltrates seen as ground-glass consolidations.
3.2. Secondary Outcomes
Analysis of Gene Expression Patterns in BAL Cells
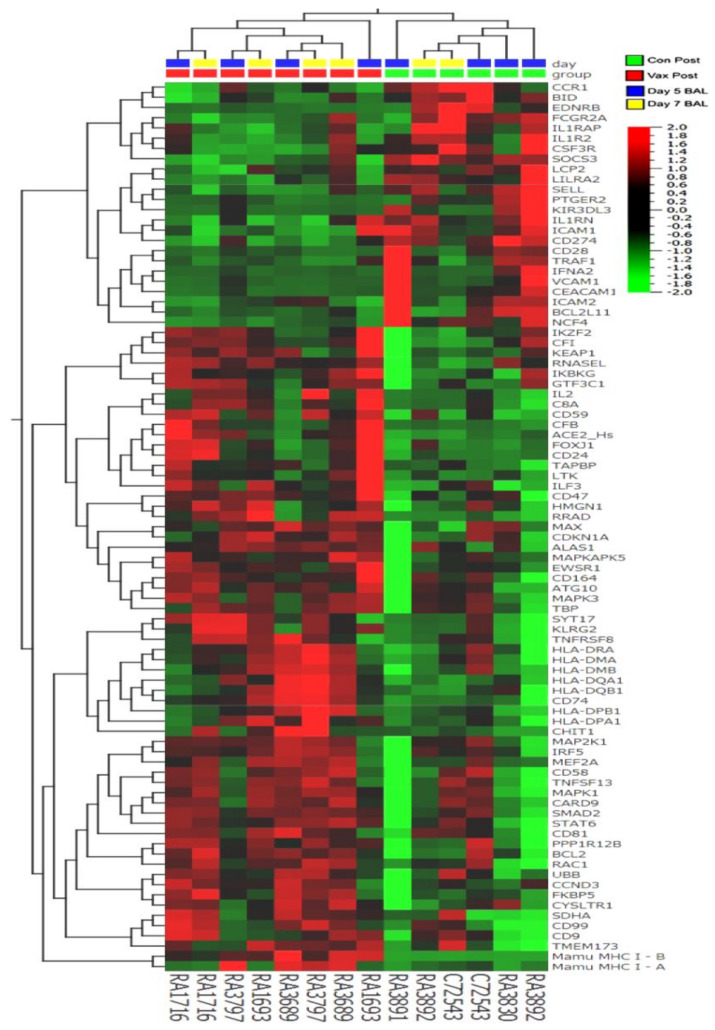
We identified a set of 87 genes in BAL samples collected 5–7 days post-challenge from control animals with statistically significant differential expression (as measured from changes in accumulation of their specific transcripts) versus BAL samples collected from vaccinated animals during the same time points (Figure 5 and Figure 6). We chose to focus on the day 5 and day 7 samples to capture a possible peak of adaptive immune responses to SARS-CoV-2 challenge, as suggested by previous reports [51,52]. Several of the identified differentially regulated genes were of particular interest in the context of adaptive viral T-cell immunity (Table 1 and Table 2). Several differentially regulated immune response genes lying outside the main window of interest (i.e., day 5 alone, day 7 alone, or day 10 alone) were also identified (Table 1 and Table 2 and Supplementary Materials, Figure S10). For example, on day 5 in unvaccinated macaques, we found upregulation of IFIT3 and IL-1RAP. The expression levels of these transcripts have been previously reported to correlate with viral loads in a SARS-CoV-2 rhesus macaque model of COVID-19 disease [5].
Figure 5.
Hierarchical clustering of gene expression in BAL samples collected from control and vaccinated macaques five and seven days post-SARS-CoV-2 challenge. The heatmap shows significantly (p < 0.05) upregulated (red) genes (63 genes > threefold) and downregulated (green) genes (24 genes < 1/3-fold) from a total of 730 genes analyzed using the NanoString Non-Human Primate Immunology V2 Panel and identifies a set of genes possibly associated with protection from SARS-CoV-2 challenge.
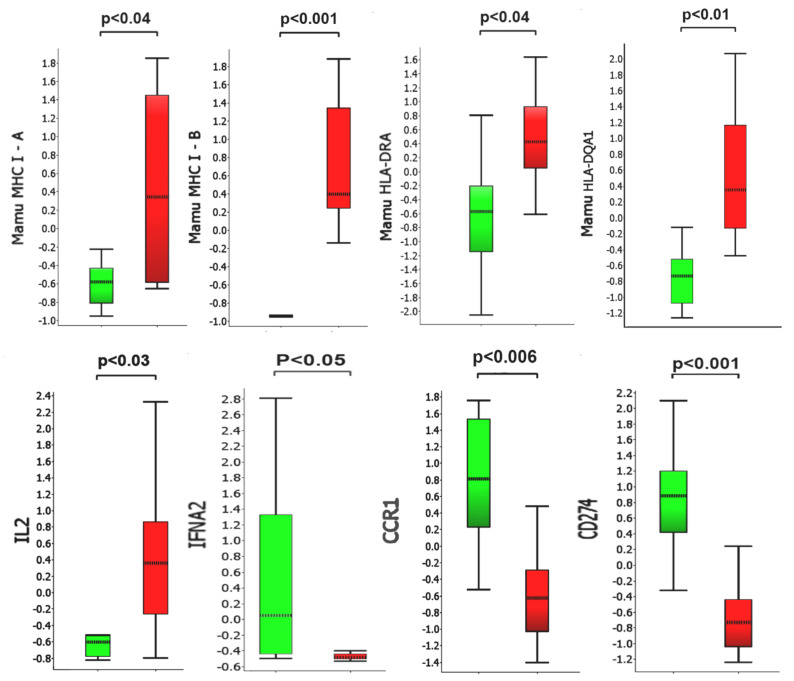
Figure 6.
Comparison of selected transcripts up- or downregulated in collected BAL samples 5–7 days post-challenge. Y-axis values represent fold differences in average scaled counts. Green bars and red bars represent control and vaccinated macaques, respectively. p-values below 0.05 were considered significant.
Table 1.
Differentially regulated genes in BAL cells obtained from SARS-CoV-2-challenged vaccinated versus control macaques.
| Upregulated Transcripts on Days 5 and 7 Post-Challenge 1 | |||||
|---|---|---|---|---|---|
| Gene ID | Function | Distribution | Gene ID | Function | Distribution |
| Mamu MHC1 A | Antigen presentation to CD8+ T cells1 |
Low cell-type specificity 1 | CD8 | Coreceptor for TCR binding to MHC C1 | T cells |
| Mamu MHC1 B | Antigen presentation to CD8+ T cells |
Low cell-type specificity | IL2 | Differentiation/maturation of T cells |
CD4+ and CD8+ T cells |
| TAPBP | MHC class I antigen presentation | Low cell-type specificity | CD81 | Costimulatory signal with CD3 | Low cell-type specificity |
| HLA-DRA 2 | Antigen presentation to CD4+ T cells |
Professional APC | CD9 | Cell adhesion, recognized by CD81 | Low cell-type specificity |
| HLA-DQA1 2 | Antigen presentation to CD4+ T cells |
Professional APC | CD59 | Inhibitor of the complement membrane attack complex | Low cell-type specificity |
| HLA-DQB1 2 | Antigen presentation to CD4+ T cells |
Professional APC | CD24 | Cell adhesion molecule | Eosinophils and B cells |
| CD74 | MHC class II antigen presentation | Professional APC | CD47 | High affinity receptor for thrombospondin-1 | Low cell-type specificity |
| HLA-DMA | MHC class II antigen presentation | Professional APC | CD58 | Ligand of the T-lymphocyte CD2 glycoprotein | Low cell-type specificity |
| HLA-DMB | MHC class II antigen presentation | Professional APC | CD164 | Facilitates adhesion of CD34+ cells | Low cell-type specificity |
| Upregulated Transcripts on Days 5 or 7 Post-Challenge | Upregulated Transcripts on Day 10 Post-Challenge | ||||
| IL17 B | Proinflamatory cytokine | Low cell-type specificity | IL6R | Low affinity receptor for Interleukin 6 | Neutrophil |
| CX3CL1 | Chemotactic for T cells and monocytes | Low cell-type specificity | ABL1 | Tyrosine-protein kinase, role cell growth and survival | Low cell-type specificity |
| CD99 | Facilitates T-cell adhesion | Low cell-type specificity | TYK2 | Tyrosine-protein kinase, initiation of type I IFN signaling | Low cell-type specificity |
Table 2.
Differentially regulated genes in BAL cells obtained from SARS-CoV-2-challenged vaccinated versus control macaques.
| Downregulated Transcripts on Days 5 and 7 Post-Challenge 1 | |||||
|---|---|---|---|---|---|
| Gene ID | Function | Distribution | Gene ID | Function | Distribution |
| IFNA2 | Inhibition of viral replication | Macrophages, eosinophils | CD28 | Provides costimulatory signals required for T-cell activation Receptor for CD80 | T cell |
| CCR1 | C-C chemokine receptor, recruitment of immune effector cells | Macrophages | IL1RAP | Coreceptor with IL1R1 in the IL-1 signaling system |
Neutrophil |
| CD274 | Ligand of PD-1 (PDL1), inhibits expansion of antigen-specific CD8+ T cells and CD4+ helper cells | Monocytes, granulocytes | IL1R2 | Decoy receptor for IL1α and IL1β (IL1B) inhibiting signaling | Macrophage, neutrophils |
| Downregulated Transcripts on Days 5 or 7 Post-Challenge | Downregulated Transcripts on Day 10 Post-Challenge | ||||
| CD80 | Receptor for CD28 and CTLA-4 on T cells | B cells and monocytes, APCs | HLA-DRA2 | Antigen presentation to CD4+ T cells |
Professional APC |
| IFNGR2 | β chain of the gamma interferon receptor | B cells, APCs and neutrophils | HLA-DMA | Antigen presentation to CD4+ T cells |
Professional APC |
| IL8 | C-X-C chemokine for recruitment of neutrophils | Macrophages, epithelial and endothelial cells | LY96 | Confers responsiveness to LPS | Macrophages |
| IL21 | Regulates proliferation of mature B and T cells in response to activating stimuli | Activated CD4+ T cells, NKT cells | CTSC | Cathepsin protease | Macrophages |
| DPP4 | Protease upregulated in SARS-CoV-2 [57] Possible viral entry receptor [58] | T-cell CD2 | TyroBP | Mediates NK cell activation | Macrophages, monocytes |
In BAL samples collected on days 5 and 7, we observed statistically significant upregulation of MHC class I genes, MHC class II and associated accessory genes (CD74 invariant chain, HLA-DM), and T-cell markers (CD8 and IL2) in the vaccinated group relative to the unvaccinated control macaques. We also observed statistically significant downregulation of interferon alpha 2 (IFNA2), the negative regulator of T-cell expansion, PD-L1, the decoy receptor for IL1α and IL1β inhibiting signaling, and FoxJ1, a regulator of Th1 cell activation [53], in the vaccinated group relative to the unvaccinated control macaques.. This pattern suggests enhanced antigen presentation and CD 4/8+ T-cell response capacity in BAL cells from vaccinated macaques relative to the controls. In control animals, we observed upregulation of several genes (CCR1, CSF3R, IFNA2, IL-1RN, IL-1RAP, IL-1R2, and SOXS3) previously reported to be activated during SARS-CoV-2 infection in rhesus macaques [5]. These genes appeared to be downregulated, relative to controls, in vaccinated macaques.
The vaccine formulation containing synthetic peptide cytotoxic T cell epitopes, CpG, and MPLA adjuvants was delivered primarily via intratracheal instillation. While we did not observe any adverse clinical signs of respiratory distress in vaccinated subjects, we examined BAL cell gene expression for lymphokines and cytokines associated with the observed mixed Th1/Th2 patterns that have also been observed in asthma [54] (Supplementary Materials, Figures S11 and S12). In BAL samples collected on days 5 and 7, we did not observe significant differences in expression of IL4, IL5, IL6, IL9, IL10, IL13, CXCL10, CCL5, CCL7, CCL22, CX3CL1, or CXCL1. We did observe a general trend of higher expression for many of the cytokines in BAL cell samples collected from vaccinated macaques pre-challenge compared to unvaccinated pre-challenge controls. This effect was generally transient, diminishing by day 5 post-challenge. This pattern of cytokine and lymphokine expression did not suggest that BAL cells assumed a phenotype associated with a Th2 response.
A major difference between pre-challenge BAL samples was that vaccinated macaques had received prior intratracheal instillation of vaccine formulation containing CpG and MPLA adjuvants. To assess the possible effects and efficacy of our vaccination procedure and formulation, we compared the expression of 60 genes previously reported to be upregulated not by antigenic stimulation, but by adjuvants alone [59,60,61,62,63]. We found 30 out of 60 genes examined to be significantly (p < 0.05) and differentially (>twofold) regulated in BAL cells obtained from vaccinated but unchallenged macaques (day −7) relative to samples from unchallenged control animals (day −1) (Supplementary Materials, Figure S13). This pattern suggests that BAL-associated cells from vaccinated animals were stimulated by the adjuvants in the vaccine formulation. As a further measure of vaccination efficiency, we measured BAL immunoreactivity to the peptides via ELISPOT prior to SARS-CoV-2 challenge (Supplementary Materials, Figure S14). We observed modest immunoreactivity to one of the six CTL peptides (LL9) in three out of the four vaccinated pre-challenge macaques. We did not detect immunoreactivity to the peptide antigens using samples of peripheral blood mononuclear cells (data not shown).
4. Discussion
In our study, SARS-CoV-2 infection in unvaccinated control macaques progressed similar to previous reports using this model. The post-viral challenge period was clinically characterized by (1) two waves of infectious viral particle recovery in the nasal tissues, (2) lymphopenia on day 1 post-challenge in all animals, and (3) progressive development of pneumonia-like infiltrations visible on chest X-rays as “ground-glass-like consolidations”, but few changes associated with human SARS-COV-2 infection, such as loss of appetite, respiratory distress, and vomiting and/or diarrhea. We observed that the kinetics of change in viral loads observed in control rhesus macaques were similar to those previously reported [11]. Additionally, the lung tissue abnormalities revealed by chest radiography were similar in the kinetics of progression and depth to previous reports [6,15,16,20,45,46,47,48]. Together, these observations suggest that SARS-CoV-2 infection in the rhesus model is clinically mild, a conclusion confirming some [6,8,11], but not all [64], previous reports. SARS-CoV-2 infection in microsphere/adjuvant-vaccinated macaques progressed in a pattern different from that of the unvaccinated controls. Specifically, disease was characterized by a trend toward diminished recovery of infectious viral particles from nasal tissues, enhanced recovery of peripheral blood lymphocytes counts, and a significant absence of pneumonia-like infiltrates in the lung. Together, these observations suggest that vaccination conferred some degree of protection against SARS-CoV-2-induced disease.
Supporting this clinical conclusion were our studies of the gene expression profiles in serially harvested BAL cells from SARS-CoV-2-challenged macaques and the immunoreactivity of the BAL cells. In samples collected from vaccinated (but pre-viral challenge) macaque BAL cells, we found distinct changes in gene expression associated with the use of TLR 4 and 9 agonists, including upregulation of CSF1 [61], IRF7 [62], and IL10 [60], suggesting effective delivery of the adjuvant portion of the vaccine formulation. When we examined HLA class I restricted immunoreactivity of the pre-challenge but vaccinated macaque BAL cells towards the vaccinating peptides, we found modest-to-low reactivity towards one of the peptides (LLLDRLNQL) in three of four NHP subjects vaccinated.
Following vaccination and viral challenge, we found evidence of upregulation of both Mamu MHC class I and class II genes in macaque BAL cells relative to unvaccinated viral-challenged macaques. Upregulation of HLA class I and class II molecules in peripheral blood mononuclear cells has been reported following the measles/mumps/rubella (MMR) vaccine in MMR-naïve individuals [65]. Increased signatures of M1-type macrophage APC transcripts in the BAL of SARS-CoV-2-infected rhesus macaques has been previously reported [5]; however, our finding of upregulated expression of MHC class II genes, a hallmark of professional APCs, appears to be unique to the BAL of our vaccinated macaques. Likewise, we found increased upregulation of the IL2 genes in vaccinated relative to unvaccinated macaques. A similar finding has been reported in humans, where higher IL 2 levels distinguish mild/asymptomatic forms of COVID-19 disease from the moderate/severe forms [66]. Following viral challenge in our study (specifically, days 5 to 7), we found downregulation of IFNα2 genes in BAL cells recovered from vaccinated subjects relative to unvaccinated controls. The observation perhaps reflects the decreased viral loads found in vaccinated NHP subjects [5].
While antibodies are a critical component of the protective humoral immune response to pathogens, antibodies that promote disease have been described and categorized as ADE [67] or VAERD, such as those described in MERS-CoV or RSV patients [68,69,70]. Since our vaccine only delivered low-molecular-weight nonameric synthetic peptides, unconjugated to any carrier, we did not expect to generate a significant anti-peptide humoral response. Rather, during the development and testing of the microsphere synthetic peptide COVID 19 vaccine, we were mindful of evoking Th2-biased immune responses, particularly those that occur in the absence of Th1 responses or appropriate T regulatory cell responses [71]. The gene expression patterns of BAL cells obtained from the vaccinated macaques in our study suggested that we did not provoke an unbalanced Th2 response by vaccination.
We note several weaknesses in our approach to demonstrate the efficacy of this experimental vaccine: (1) SARS-CoV-2 infection in rhesus macaques resulted in only mild disease, which appeared to resolve by days 10–14 post-infection. Having noted this in previous reports on the SARS-CoV-2/rhesus model, we tried to induce more severe forms of infection using higher doses of infectious particles than previous reports. Based on clinical signs, we found little effect from the increased dose of the virus on rhesus SARS-CoV-2 disease severity. (2) Due to the limited amount of cells recovered from the BAL, we were unable to perform confirmatory qRT-PCR analysis of the unique gene expression patterns found in vaccinated versus control BAL. Nevertheless, we found that many of our observations had been previously reported from studies in similar models. (3) Not all the macaque chest radiographs were performed on the same day in control versus vaccinated subjects, a reflection of the logistic difficulties in working under BSL ¾ conditions. We are confident, however, that we have captured the chest radiographic abnormalities induced by SAR-CoV-2 infection of macaques (i.e., conspicuous consolidations and infiltrations prevalent in the caudal lobe of the right lung) and adequately shown their absence in vaccinated viral-challenged macaques. (4) We only included one cynomolgus macaque in our control group. This was primarily due to macaque availability at the time the study was conducted. However, this appears to have been both a strength [64] and weakness of the experimental design, as we found similar lung pathology in the cynomolgus macaque, as well as similar patterns of BAL gene expression, as for the control rhesus subjects [72]. (5) Finally, we did not study the effects of adjuvant alone on SARS-CoV-2 infection in the rhesus macaques. Previous experience with this microsphere CTL vaccine platform in a murine Ebola virus model has shown that adjuvant alone is not sufficient to confer protection against lethal virus challenge. Protection was conferred only when the corresponding synthetic CTL peptide epitopes were delivered in the microsphere [4].
We believe this report is the first demonstration of efficacy in a preclinical NHP model of SARS-CoV-2 infection of a synthetic peptide-based vaccine based on known and persistently immunogenic HLA class I bound CTL peptide epitopes of SARS nucleoprotein [28]. The SARS-CoV-2 nucleoprotein genomic sequences have shown significantly reduced mutation rates compared to spike protein. As such, it may represent an additional target for vaccination, perhaps in the context of a booster vaccine for use following SARS-CoV-2 spike protein vaccines based on recombinant protein, mRNA, or adenoviral vectors [73,74,75]. The ready ability to change the sequence of the synthetic peptide HLA class I restricted CTL epitopes used in the system is an attractive feature, given the observed rates of mutation in SARS-CoV-2 as it spreads in the human population in the future. A second potentially attractive feature of this vaccine approach is that it can be delivered by aerosolization to the respiratory mucosa, a route previously demonstrated to efficiently generate lung-dwelling tissue-resident memory T cells [76,77].
5. Conclusions
We demonstrated that rhesus macaques receiving the microsphere vaccine formulation prior to viral challenge are protected from pneumonia-like lung abnormalities that characterize SARS-CoV-2 infection in unvaccinated control macaques. Analysis of gene expression of cells obtained from bronchiolar lavage showed unique signatures consistent with the hypothesis that vaccination with this platform induces a protective T-cell response in viral-challenged macaques.
Acknowledgments
The authors wish to thank Antonella Maffei for her critical review of the manuscript.
Abbreviations
| ADE | antibody mediated disease enhancement |
| APC | antigen-presenting cell |
| BAL | bronchoalveolar lavage |
| CGM | complete growth medium |
| cGMP | current good manufacturing practice |
| CpG | cytosine triphosphate deoxynucleotide-guanine triphosphate deoxynucleotide oligodeoxynucleotide |
| CTL | cytotoxic T lymphocyte |
| DICOM | digital imaging and communications in medicine |
| DMSO | dimethyl sulfoxide |
| EDTA | ethylenediaminetetraacetic acid |
| ELISPOT | enzyme-linked immune absorbent spot assay |
| HLA | human leucocyte antigen |
| IFNγ | interferon gamma |
| IT | intratracheal |
| LLOD | lower limit of detection |
| LN | lymph node |
| MHC | major histocompatibility complex |
| MPLA | monophosphoryl lipid A |
| NHP | non-human primate |
| PBMC | peripheral blood mononuclear cells |
| PLGA | poly-L-lactide-co-glycolide |
| qRT-PCR | real-time quantitative reverse transcription polymerase chain reaction |
| SARS-CoV-2 | severe acute respiratory syndrome coronavirus 2 |
| TCID50 | fifty-percent tissue culture infective dose |
| Th1 | T helper cell type 1 |
| Th2 | T helper cell type 2 |
| TLR | toll-like receptor |
| VAERD | vaccine-associated enhanced respiratory disease |
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/vaccines9050520/s1, Figure S1. Core body temperature alterations in vaccinated macaques prior to SARS-CoV-2 challenge. For each animal, 35 days of pre-challenge temperature measurements are shown. Each tick on the x-axis represents 12 h or 48 individual logger measurements, Figure S2. Hematological analysis in control and vaccinated macaques challenged with SARS-CoV-2. The counts of white blood cells (WBCs) (upper left panel), the percent of lymphocytes in WBCs (upper right panel), the percent of monocytes in the WBCs (lower left panel), and the percent of neutrophils in the WBCs (lower right panel) were analyzed. An asterisk indicates a statistically significant difference (p < 0.05) between control and vaccinated macaques according to Student’s t-testing, Figure S3. Viral clearance rates in control and vaccinated SARS-CoV-2-challenged macaques. From the total viral loads measurements in nasal swab samples from SARS-CoV-2-challenged macaques, the daily viral clearance rates (i.e., TCID50/mL day n-1 minus TCID50/mL day n) were calculated and averaged over a nine-day period. Red symbols are vaccinated macaque subjects, control unvaccinated macaque subjects are shown with green symbols, Figure S4. Chest radiographs of control rhesus macaque RA3830 following SARS-CoV-2 challenge. As shown, this animal demonstrated a progression of pulmonary infiltrates during the acute period (days 2–5) of disease post-challenge which resolved by study termination (day 21). White arrows indicate areas of mild-to-moderate pulmonary infiltrates seen as ground-glass consolidations, Figure S5. Chest radiographs of control rhesus macaque RA3891 following SARS-CoV-2 challenge. As shown, this animal demonstrated a progression of pulmonary infiltrates during the acute period (days 3–7) of disease post-challenge which resolved by study termination (day 21). White arrows indicate areas of mild-to-moderate pulmonary infiltrates seen as ground-glass consolidations, Figure S6. Chest radiographs of control rhesus macaque RA3892 following SARS-CoV-2 challenge. As shown, this animal demonstrated a progression of pulmonary infiltrates during the acute period (day 3) of disease post-challenge which resolved by study termination (day 21). White arrows indicate areas of mild-to-moderate pulmonary infiltrates seen as ground-glass consolidations, Figure S7. Chest radiographs of control cynomolgus macaque C72543 following SARS-CoV-2 challenge. As shown, this animal demonstrated a progression of pulmonary infiltrates 5–10 days post-challenge which resolved by study termination (day 21). White arrows indicate areas of mild-to-moderate pulmonary infiltrates seen as ground-glass consolidations, Figure S9. Chest radiographs of vaccinated rhesus macaques RA3689 and RA3797 following SARS-CoV-2 challenge. Radiographs are unremarkable, other than showing increased reticulation relative to baseline, appearing on days 2 through 4, clearing on later imaging. In particular, we can note the lack of focal infiltrates or consolidations, Figure S10. Hierarchical clustering of gene expression in BAL samples collected from control and vaccinated macaques on day 5 (left panel), day 7 (middle panel), and day 10 (right panel), five and seven days post-SARS-CoV-2 challenge. Heatmap shows significantly (p < 0.05) upregulated (red) transcripts and downregulated (green) transcripts from a total of 730 genes analyzed using the NanoString Non-Human Primate Immunology V2 Panel, Figure S11. Intratracheal vaccination with the adjuvanted microsphere peptide vaccine did not promote the expression of Th2-type interleukin cytokine transcripts in collected BAL samples relative to levels measured in control macaques. BAL cell gene expression (shown as scaled counts on the y-axis) of cytokines associated with Th2 T-cell responses were plotted by study day (x-axis) for each animal prior to and following the SARS-CoV-2 challenge. No statistically significant differences between control and vaccinated macaques were found, Figure S12. Intratracheal vaccination with the adjuvanted microsphere peptide vaccine did not promote the expression of Th2-type interleukin cytokine transcripts in collected BAL samples relative to levels measured in control macaques. BAL cell gene expression (shown as scaled counts on the y-axis) of cytokines associated with Th2 T-cell responses were plotted by study day (x-axis) for each animal prior to and following the SARS-CoV-2 challenge. No statistically significant differences between control and vaccinated macaques were found, Figure S13. Hierarchical clustering of adjuvant-related transcript expression in BAL samples collected from control and vaccinated macaques prior to virus challenge (indicated as Control Pre and Vax Pre). Gene expression analysis identified differentially regulated genes in BAL samples obtained on day −1 from control animals and day −7 from vaccinated macaques. Heatmap shows significant (p < 0.05) differential expression of a series of genes that had previously been identified (see main text) as being regulated by adjuvants alone. Upregulated (red); downregulated (green). Macaque RA 3830 was not sampled on day −1, Figure S14. Immunoreactivity of BAL-associated cells from vaccinated macaques to immunizing peptides prior to SARS-CoV-2 challenge. Dates shown are assay dates rather than sampling dates. Concanavlin A was used as a positive control. Table S1A: CTL epitope HLA coverage [78], Table S1B: Rhesus Mamu MHC Class I coverage – predicted binding [79].
Author Contributions
All authors made substantial contributions to: (1) the conception and design of the study (R.M.R., S.B., S.C., T.B. (Trevor Brasel), J.E.C., P.L., C.V.H., T.B. (Tikoes Blankenberg) and P.E.H.), the acquisition of data (T.B. (Trevor Brasel), C.M., J.E.C., and C.V.H.), or the analysis and interpretation of data (P.E.H., R.M.R., P.L., T.B. (Trevor Brasel) and J.E.C.); and (2) drafting the article or revising it critically for important intellectual content (P.E.H., P.L., T.B. (Trevor Brasel), S.B., T.H., T.B. (Tikoes Blankenberg), T.M.B., R.C., L.W. and R.M.R.). All authors have read and agreed to the published version of the manuscript.
Funding
This research did not receive any specific external grant from funding agencies.
Institutional Review Board Statement
The protocols were approved by the UTMB Institutional Animal Care and Use Committee (Protocol Numbers 2,004,051 (natural history/control study) and 2,003,033 (vaccination study)) and complied with the Animal Welfare Act, the U.S. Public Health Service Policy, and other federal statutes and regulations related to animals and experiments involving animals.
Informed Consent Statement
Not applicable.
Data Availability Statement
The original DICOM files (>20 MB/file) are available upon request.
Conflicts of Interest
R.M.R., T.B. (Tikoes Blankenberg), S.B., S.C., R.C., T.H., L.W., P.L., and C.V.H. are employees of Flow Pharma, Inc. compensated in cash and stock, and are named inventors on various issued and pending patents assigned to Flow Pharma. Some of these patents pending are directly related to the study presented here. P.L. is a member of Flow Pharma’s Scientific Advisory Board. Thomas Bey (T.B.) is a Flow Pharma stockholder. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflicts of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Ritchie H., Ortiz-Ospina E., Diana B., Mathieu E., Joe H., Bobbie M., Charlie G., Max R. Coronavirus (COVID-19) Vaccinations. [(accessed on 31 January 2021)]; Available online: https://ourworldindata.org/covid-vaccinations.
- 2.Hoffmann M., Arora P., Gross R., Seidel A., Hornich B.F., Hahn A.S., Kruger N., Graichen L., Hofmann-Winkler H., Kempf A., et al. SARS-CoV-2 variants B.1.351 and P.1 escape from neutralizing antibodies. Cell. 2021;184:2384–2393.e12. doi: 10.1016/j.cell.2021.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rubsamen R., Herst C., Lloyd P., Heckerman D. Eliciting cytotoxic T-lymphocyte responses from synthetic vectors containing one or two epitopes in a C57BL/6 mouse model using peptide-containing biodegradable microspheres and adjuvants. Vaccine. 2014;32:4111–4116. doi: 10.1016/j.vaccine.2014.05.071. [DOI] [PubMed] [Google Scholar]
- 4.Herst C., Burkholz S., Sidney J., Sette A., Harris P., Massey S., Brasel T., Cunha-Neto E., Rosa D., Chao W., et al. An effective CTL peptide vaccine for Ebola Zaire Based on Survivors’ CD8+ targeting of a particular nucleocapsid protein epitope with potential implications for COVID-19 vaccine design. Vaccine. 2020;38:4464–4475. doi: 10.1016/j.vaccine.2020.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aid M., Busman-Sahay K., Vidal S.J., Maliga Z., Bondoc S., Starke C., Terry M., Jacobson C.A., Wrijil L., Ducat S., et al. Vascular Disease and Thrombosis in SARS-CoV-2-Infected Rhesus Macaques. Cell. 2020;183:1354–1366.e13. doi: 10.1016/j.cell.2020.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blair R.V., Vaccari M., Doyle-Meyers L.A., Roy C.J., Russell-Lodrigue K., Fahlberg M., Monjure C.J., Beddingfield B., Plante K.S., Plante J.A., et al. Acute Respiratory Distress in Aged, SARS-CoV-2-Infected African Green Monkeys but Not Rhesus Macaques. Am. J. Pathol. 2020;191:274–282. doi: 10.1016/j.ajpath.2020.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bray M., Hatfill S., Hensley L., Huggins J. Haematological, Biochemical and Coagulation Changes in Mice, Guinea-pigs and Monkeys Infected with a Mouse-adapted Variant of Ebola Zaire Virus. J. Comp. Pathol. 2001;125:243–253. doi: 10.1053/jcpa.2001.0503. [DOI] [PubMed] [Google Scholar]
- 8.Chandrashekar A., Liu J., Martinot A.J., Mcmahan K., Mercado N.B., Peter L., Tostanoski L.H., Yu J., Maliga Z., Nekorchuk M., et al. SARS-CoV-2 infection protects against rechallenge in rhesus macaques. Science. 2020;369:812–817. doi: 10.1126/science.abc4776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deng W., Bao L., Liu J., Xiao C., Liu J., Xue J., Lv Q., Qi F., Gao H., Yu P., et al. Primary exposure to SARS-CoV-2 protects against reinfection in rhesus macaques. Science. 2020;369:818–823. doi: 10.1126/science.abc5343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Feng L., Wang Q., Shan C., Yang C., Feng Y., Wu J., Liu X., Zhou Y., Jiang R., Hu P., et al. An adenovirus-vectored COVID-19 vaccine confers protection from SARS-COV-2 challenge in rhesus macaques. Nat. Commun. 2020;11:4207. doi: 10.1038/s41467-020-18077-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McAuliffe J., Vogel L., Subbarao K., Roberts A., Fahle G., Fischer S., Shieh W.-J., Butler E., Zaki S., Claire M.S., et al. Replication of SARS coronavirus administered into the respiratory tract of African Green, rhesus and cynomolgus monkeys. Virology. 2004;330:8–15. doi: 10.1016/j.virol.2004.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McMahan K., Yu J., Mercado N.B., Loos C., Tostanoski L.H., Chandrashekar A., Liu J., Peter L., Atyeo C., Zhu A., et al. Correlates of protection against SARS-CoV-2 in rhesus macaques. Nature. 2020;590:630–634. doi: 10.1038/s41586-020-03041-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mercado N.B., Zahn R., Wegmann F., Loos C., Chandrashekar A., Yu J., Liu J., Peter L., Mcmahan K., Tostanoski L.H., et al. Single-shot Ad26 vaccine protects against SARS-CoV-2 in rhesus macaques. Nature. 2020:1–11. doi: 10.1038/s41586-020-2607-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mukhopadhyay L., Yadav P.D., Gupta N., Mohandas S., Patil D.Y., Shete-Aich A., Panda S., Bhargava B. Comparison of the immunogenicity & protective efficacy of various SARS-CoV-2 vaccine candidates in non-human primates. Indian J. Med. Res. 2020;153:93–114. doi: 10.4103/ijmr.IJMR_4431_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Munster V.J., Feldmann F., Williamson B.N., van Doremalen N., Perez-Perez L., Schulz J., Meade-White K., Okumura A., Callison J., Brumbaugh B., et al. Respiratory disease and virus shedding in rhesus macaques inoculated with SARS-CoV-2. bioRxiv. 2020;585:268–272. doi: 10.1101/2020.03.21.001628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shan C., Yao Y.-F., Yang X.-L., Zhou Y.-W., Gao G., Peng Y., Yang L., Hu X., Xiong J., Jiang R.-D., et al. Infection with novel coronavirus (SARS-CoV-2) causes pneumonia in Rhesus macaques. Cell Res. 2020;30:670–677. doi: 10.1038/s41422-020-0364-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Singh D.K., Singh B., Ganatra S.R., Gazi M., Cole J., Thippeshappa R., Alfson K.J., Clemmons E., Gonzalez O., Escobedo R., et al. Responses to acute infection with SARS-CoV-2 in the lungs of rhesus macaques, baboons and marmosets. Nat. Microbiol. 2021;6:73–86. doi: 10.1038/s41564-020-00841-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Song T.-Z., Zheng H.-Y., Han J.-B., Jin L., Yang X., Liu F.-L., Luo R.-H., Tian R.-R., Cai H.-R., Feng X.-L., et al. Delayed severe cytokine storm and immune cell infiltration in SARS-CoV-2-infected aged Chinese rhesus macaques. Zool. Res. 2020;41:503–516. doi: 10.24272/j.issn.2095-8137.2020.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van Doremalen N., Lambe T., Spencer A., Belij-Rammerstorfer S., Purushotham J.N., Port J.R., Avanzato V., Bushmaker T., Flaxman A., Ulaszewska M., et al. ChAdOx1 nCoV-19 vaccination prevents SARS-CoV-2 pneumonia in rhesus macaques. bioRxiv. 2020;586:578–582. doi: 10.1101/2020.05.13.093195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu P., Qi F., Xu Y., Li F., Liu P., Liu J., Bao L., Deng W., Gao H., Xiang Z., et al. Age-related rhesus macaque models of COVID-19. Anim. Model. Exp. Med. 2020;3:93–97. doi: 10.1002/ame2.12108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zheng H., Li H., Guo L., Liang Y., Li J., Wang X., Hu Y., Wang L., Liao Y., Yang F., et al. Virulence and pathogenesis of SARS-CoV-2 infection in rhesus macaques: A nonhuman primate model of COVID-19 progression. PLoS Pathog. 2020;16:e1008949. doi: 10.1371/journal.ppat.1008949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Corbett K.S., Flynn B., Foulds K.E., Francica J.R., Boyoglu-Barnum S., Werner A.P., Flach B., O’Connell S., Bock K.W., Minai M., et al. Evaluation of the mRNA-1273 Vaccine against SARS-CoV-2 in Nonhuman Primates. N. Engl. J. Med. 2020;383:1544–1555. doi: 10.1056/NEJMoa2024671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gao Q., Bao L., Mao H., Wang L., Xu K., Yang M., Li Y., Zhu L., Wang N., Lv Z., et al. Development of an inactivated vaccine candidate for SARS-CoV-2. Science. 2020;369:77–81. doi: 10.1126/science.abc1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guebre-Xabier M., Patel N., Tian J.-H., Zhou B., Maciejewski S., Lam K., Portnoff A.D., Massare M.J., Frieman M.B., Piedra P.A., et al. NVX-CoV2373 vaccine protects cynomolgus macaque upper and lower airways against SARS-CoV-2 challenge. Vaccine. 2020;38:7892–7896. doi: 10.1016/j.vaccine.2020.10.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang N.-N., Li X.-F., Deng Y.-Q., Zhao H., Huang Y.-J., Yang G., Huang W.-J., Gao P., Zhou C., Zhang R.-R., et al. A Thermostable mRNA Vaccine against COVID-19. Cell. 2020;182:1271–1283.e16. doi: 10.1016/j.cell.2020.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang H., Zhang Y., Huang B., Deng W., Quan Y., Wang W., Xu W., Zhao Y., Li N., Zhang J., et al. Development of an Inactivated Vaccine Candidate, BBIBP-CorV, with Potent Protection against SARS-CoV-2. Cell. 2020;182:713–721.e9. doi: 10.1016/j.cell.2020.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yu J., Tostanoski L.H., Peter L., Mercado N.B., Mcmahan K., Mahrokhian S.H., Nkolola J.P., Liu J., Li Z., Chandrashekar A., et al. DNA vaccine protection against SARS-CoV-2 in rhesus macaques. Science. 2020;369:806–811. doi: 10.1126/science.abc6284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Peng H., Yang L.-T., Wang L.-Y., Li J., Huang J., Lu Z.-Q., Koup R.A., Bailer R.T., Wu C.-Y. Long-lived memory T lymphocyte responses against SARS coronavirus nucleocapsid protein in SARS-recovered patients. Virology. 2006;351:466–475. doi: 10.1016/j.virol.2006.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Peng Y., Mentzer A.J., Liu G., Yao X., Yin Z., Dong D., Dejnirattisai W., Rostron T., Supasa P., Liu C., et al. Broad and strong memory CD4+ and CD8+ T cells induced by SARS-CoV-2 in UK convalescent individuals following COVID-19. Nat. Immunol. 2020;21:1336–1345. doi: 10.1038/s41590-020-0782-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kared H., Redd A.D., Bloch E.M., Bonny T.S., Sumatoh H.R., Kairi F., Carbajo D., Abel B., Newell E.W., Bettinotti M., et al. SARS-CoV-2-specific CD8+ T cell responses in convalescent COVID-19 individuals. J. Clin. Investig. 2021;131:e145476. doi: 10.1172/JCI145476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grifoni A., Weiskopf D., Ramirez S.I., Mateus J., Dan J.M., Moderbacher C.R., Rawlings S.A., Sutherland A., Premkumar L., Jadi R.S., et al. Targets of T Cell Responses to SARS-CoV-2 Coronavirus in Humans with COVID-19 Disease and Unexposed Individuals. Cell. 2020;181:1489–1501.e15. doi: 10.1016/j.cell.2020.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ravi R.K., Walton K., Khosroheidari M. MiSeq: A Next Generation Sequencing Platform for Genomic Analysis. Methods Mol. Biol. 2018;1706:223–232. doi: 10.1007/978-1-4939-7471-9_12. [DOI] [PubMed] [Google Scholar]
- 33.Grifoni A., Sidney J., Zhang Y., Scheuermann R.H., Peters B., Sette A. A Sequence Homology and Bioinformatic Approach Can Predict Candidate Targets for Immune Responses to SARS-CoV-2. Cell Host Microbe. 2020;27:671–680.e2. doi: 10.1016/j.chom.2020.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dimonte S., Babakir-Mina M., Hama-Soor T., Ali S. Genetic Variation and Evolution of the 2019 Novel Coronavirus. Public Health Genom. 2021;10:1–13. doi: 10.1159/000513530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Almazάn F., Galάn C., Enjuanes L. The Nucleoprotein Is Required for Efficient Coronavirus Genome Replication. J. Virol. 2004;78:12683–12688. doi: 10.1128/JVI.78.22.12683-12688.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vita R., Mahajan S., Overton J., Dhanda S.K., Martini S., Cantrell J.R., Wheeler D.K., Sette A., Peters B. The Immune Epitope Database (IEDB): 2018 update. Nucleic Acids Res. 2019;47:D339–D343. doi: 10.1093/nar/gky1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jurtz V.I., Paul S., Andreatta M., Marcatili P., Peters B., Nielsen M. NetMHCpan-4.0: Improved Peptide–MHC Class I Interaction Predictions Integrating Eluted Ligand and Peptide Binding Affinity Data. J. Immunol. 2017;199:3360–3368. doi: 10.4049/jimmunol.1700893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Qin E., Zhu Q., Yu M., Fan B., Chang G., Si B., Yang B., Peng W., Jiang T., Liu B., et al. A complete sequence and comparative analysis of a SARS-associated virus (Isolate BJ01) Chin. Sci. Bull. 2003;48:941–948. doi: 10.1007/BF03184203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Poran A., Harjanto D., Gaynor R.B., Malloy M., Arieta C.M., Rothenberg D.A., Lenkala D., Van Buuren M.M., Addona T.A., Rooney M.S., et al. Sequence-based prediction of SARS-CoV-2 vaccine targets using a mass spectrometry-based bioinformatics predictor identifies immunogenic T cell epitopes. Genome Med. 2020;12:70. doi: 10.1186/s13073-020-00767-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mothé B.R., Southwood S., Sidney J., English A.M., Wriston A., Hoof I., Shabanowitz J., Hunt D.F., Sette A. Peptide-binding motifs associated with MHC molecules common in Chinese rhesus macaques are analogous to those of human HLA supertypes and include HLA-B27-like alleles. Immunogenetics. 2013;65:371–386. doi: 10.1007/s00251-013-0686-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cunha-Neto E., Rosa D.S., Harris P.E., Olson T., Morrow A., Ciotlos S., Herst C.V., Rubsamen R.M. An Approach for a Synthetic CTL Vaccine Design against Zika Flavivirus Using Class I and Class II Epitopes Identified by Computer Modeling. Front. Immunol. 2017;8:640. doi: 10.3389/fimmu.2017.00640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brining D.L., Mattoon J.S., Kercher L., LaCasse R., Safronetz D., Feldmann H., Parnell M.J. Thoracic Radiography as a Refinement Methodology for the Study of H1N1 Influenza in Cynomologus Macaques (Macaca fascicularis) Comp. Med. 2010;60:389–395. [PMC free article] [PubMed] [Google Scholar]
- 43.Ramakrishnan M.A. Determination of 50% endpoint titer using a simple formula. World J. Virol. 2016;5:85–86. doi: 10.5501/wjv.v5.i2.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hochberg Y., Benjamini Y. More powerful procedures for multiple significance testing. Stat. Med. 1990;9:811–818. doi: 10.1002/sim.4780090710. [DOI] [PubMed] [Google Scholar]
- 45.Finch C.L., Crozier I., Lee J.H., Byrum R., Cooper T.K., Liang J., Sharer K., Solomon J., Sayre P.J., Kocher G., et al. Characteristic and quantifiable COVID-19-like abnormalities in CT- and PET/CT-imaged lungs of SARS-CoV-2-infected crab-eating macaques (Macaca fascicularis) bioRxiv. 2020;14 doi: 10.1101/2020.05.14.096727. [DOI] [Google Scholar]
- 46.Lu S., Zhao Y., Yu W., Yang Y., Gao J., Wang J., Kuang D., Yang M., Yang J., Ma C., et al. Comparison of nonhuman primates identified the suitable model for COVID-19. Signal Transduct. Target. Ther. 2020;5:1–9. doi: 10.1038/s41392-020-00269-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xie L., Zhou Q., Liu S., Wu Q., Ji Y., Zhang L., Xu F., Gong W., Melgiri N.D., Xie P. Normal Thoracic Radiographic Appearance of the Cynomolgus Monkey (Macaca fascicularis) PLoS ONE. 2014;9:e84599. doi: 10.1371/journal.pone.0084599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Silverman S., Morgan J.P. Thoracic radiography of the normal rhesus macaque (Macaca mulatta) Am. J. Veter. Res. 1980;41:1704–1719. [PubMed] [Google Scholar]
- 49.Singh D.K., Ganatra S.R., Singh B., Cole J., Alfson K.J., Clemmons E., Gazi M., Gonzalez O., Escobedo R., Lee T.-H., et al. SARS-CoV-2 infection leads to acute infection with dynamic cellular and inflammatory flux in the lung that varies across nonhuman primate species. bioRxiv. 2020:136481. doi: 10.1101/2020.06.05.136481. [DOI] [Google Scholar]
- 50.Maaskant A., Meijer L., Bakker J., van Geest L., Zijlmans D.G., Langermans J.A., Stammes M.A. Bronchoalveolar lavage affects thorax computed tomography of healthy and SARS-CoV-2 infected rhesus macaques (Macaca mulatta) bioRxiv. 2021 doi: 10.1101/2021.02.04.429761. Preprint. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rabenstein H., Behrendt A.C., Ellwart J.W., Naumann R., Horsch M., Beckers J., Obst R. Differential Kinetics of Antigen Dependency of CD4+ and CD8+ T Cells. J. Immunol. 2014;192:3507–3517. doi: 10.4049/jimmunol.1302725. [DOI] [PubMed] [Google Scholar]
- 52.Trobaugh D.W., Yang L., Ennis F.A., Green S. Altered effector functions of virus-specific and virus cross-reactive CD8 + T cells in mice immunized with related flaviviruses. Eur. J. Immunol. 2010;40:1315–1327. doi: 10.1002/eji.200839108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lin L., Spoor M.S., Gerth A.J., Brody S.L., Peng S.L. Modulation of Th1 Activation and Inflammation by the NF-B Repressor Foxj1. Science. 2004;303:1017–1020. doi: 10.1126/science.1093889. [DOI] [PubMed] [Google Scholar]
- 54.Fitzpatrick A.M., Higgins M., Holguin F., Brown L.A.S., Teague W.G., National Institutes of Health/National Heart. Blood Institute’s Severe Asthma Research The molecular phenotype of severe asthma in children. J. Allergy Clin. Immunol. 2010;125:851–857.e18. doi: 10.1016/j.jaci.2010.01.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Uhlén M., Fagerberg L., Hallström B.M., Lindskog C., Oksvold P., Mardinoglu A., Sivertsson Å., Kampf C., Sjöstedt E., Asplund A., et al. Proteomics. Tissue-based map of the human proteome. Science. 2015;347:1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 56.Uhlen M., Karlsson M.J., Zhong W., Tebani A., Pou C., Mikes J., Lakshmikanth T., Forsström B., Edfors F., Odeberg J., et al. A genome-wide transcriptomic analysis of protein-coding genes in human blood cells. Science. 2019;366:eaax9198. doi: 10.1126/science.aax9198. [DOI] [PubMed] [Google Scholar]
- 57.Amati F., Vancheri C., Latini A., Colona V.L., Grelli S., D’Apice M.R., Balestrieri E., Passarelli C., Minutolo A., Loddo S., et al. Expression profiles of the SARS-CoV-2 host invasion genes in nasopharyngeal and oropharyngeal swabs of COVID-19 patients. Heliyon. 2020;6:e05143. doi: 10.1016/j.heliyon.2020.e05143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Raj V.S., Mou H., Smits S.L., Dekkers D.H.W., Müller M.A., Dijkman R., Muth D., Demmers J.A.A., Zaki A., Fouchier R.A.M., et al. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature. 2013;495:251–254. doi: 10.1038/nature12005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Salyer A.C.D., David S.A. Transcriptomal signatures of vaccine adjuvants and accessory immunostimulation of sentinel cells by toll-like receptor 2/6 agonists. Hum. Vaccines Immunother. 2018;14:1686–1696. doi: 10.1080/21645515.2018.1480284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Olafsdottir T., Lindqvist M., Harandi A.M. Molecular signatures of vaccine adjuvants. Vaccine. 2015;33:5302–5307. doi: 10.1016/j.vaccine.2015.04.099. [DOI] [PubMed] [Google Scholar]
- 61.Mosca F., Tritto E., Muzzi A., Monaci E., Bagnoli F., Iavarone C., O’Hagan D., Rappuoli R., De Gregorio E. Molecular and cellular signatures of human vaccine adjuvants. Proc. Natl. Acad. Sci. USA. 2008;105:10501–10506. doi: 10.1073/pnas.0804699105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lampe A.T., Puniya B.L., Pannier A.K., Helikar T., Brown D.M. Combined TLR4 and TLR9 agonists induce distinct phenotypic changes in innate immunity in vitro and in vivo. Cell. Immunol. 2020;355:104149. doi: 10.1016/j.cellimm.2020.104149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.An H., Xu H., Yu Y., Zhang M., Qi R., Yan X., Liu S., Wang W., Guo Z., Qin Z., et al. Up-regulation of TLR9 gene expression by LPS in mouse macrophages via activation of NF-κB, ERK and p38 MAPK signal pathways. Immunol. Lett. 2002;81:165–169. doi: 10.1016/S0165-2478(02)00010-X. [DOI] [PubMed] [Google Scholar]
- 64.Fahlberg M.D., Blair R.V., Threeton B.M., Golden N., Datta P.K., Roy C.J., Bohm R.P., Maness N.J., Fischer T., Rappaport J., et al. Cellular events of acute, resolving or progressive COVID-19 in SARS-CoV-2 infected non-human primates. Nat. Commun. 2020;11:6078. doi: 10.1038/s41467-020-19967-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dhiman N., Bonilla R.G., Jacobson R.M., O’Kane D., Poland G.A. Differential HLA Gene Expression in Measles Vaccine Seropositive and Seronegative Subjects: A Pilot Study. Scand. J. Infect. Dis. 2003;35:332–336. doi: 10.1080/00365540310007105. [DOI] [PubMed] [Google Scholar]
- 66.Tjan L.H., Furukawa K., Nagano T., Kiriu T., Nishimura M., Arii J., Hino Y., Iwata S., Nishimura Y., Mori Y. Early differences in cytokine production distinguish severity of COVID-19. J. Infect. Dis. 2021;223:1145–1149. doi: 10.1093/infdis/jiab005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Katzelnick L.C., Gresh L., Halloran M.E., Mercado J.C., Kuan G., Gordon A., Balmaseda A., Harris E. Antibody-dependent enhancement of severe dengue disease in humans. Science. 2017;358:929–932. doi: 10.1126/science.aan6836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chin J., Magoffin R.L., Shearer L.A., Schieble J.H., Lennette E.H. Field evaluation of a respiratory syncytial virus vaccine and a trivalent parainfluenza virus vaccine in a pediatric population. Am. J. Epidemiol. 1969;89:449–463. doi: 10.1093/oxfordjournals.aje.a120957. [DOI] [PubMed] [Google Scholar]
- 69.Kim H.W., Canchola J.G., Brandt C.D., Pyles G., Chanock R.M., Jensen K., Parrott R.H. Respiratory syncytial virus disease in infants despite prior administration of antigenic inactivated vaccine. Am. J. Epidemiol. 1969;89:422–434. doi: 10.1093/oxfordjournals.aje.a120955. [DOI] [PubMed] [Google Scholar]
- 70.Rauh L.W., Schmidt R. Measles Immunization with Killed Virus Vaccine. Serum Antibody Titers and Experience with Exposure to Measles Epidemic. Am. J. Dis. Child. 1965;109:232–237. doi: 10.1001/archpedi.1965.02090020234007. [DOI] [PubMed] [Google Scholar]
- 71.Georas S.N., Guo J., De Fanis U., Casolaro V. T-helper cell type-2 regulation in allergic disease. Eur. Respir. J. 2005;26:1119–1137. doi: 10.1183/09031936.05.00006005. [DOI] [PubMed] [Google Scholar]
- 72.Salguero F.J., White A.D., Slack G.S., Fotheringham S.A., Bewley K.R., Gooch K.E., Longet S., Humphries H.E., Watson R.J., Hunter L., et al. Comparison of rhesus and cynomolgus macaques as an infection model for COVID-19. Nat. Commun. 2021;12:1260. doi: 10.1038/s41467-021-21389-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Logunov D.Y., Dolzhikova I.V., Zubkova O.V., Tukhvatullin A.I., Shcheblyakov D.V., Dzharullaeva A.S., Grousova D.M., Erokhova A.S., Kovyrshina A.V., Botikov A.G., et al. Safety and immunogenicity of an rAd26 and rAd5 vector-based heterologous prime-boost COVID-19 vaccine in two formulations: Two open, non-randomised phase 1/2 studies from Russia. Lancet. 2020;396:887–897. doi: 10.1016/S0140-6736(20)31866-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhu F.-C., Guan X.-H., Li Y.-H., Huang J.-Y., Jiang T., Hou L.-H., Li J.-X., Yang B.-F., Wang L., Wang W.-J., et al. Immunogenicity and safety of a recombinant adenovirus type-5-vectored COVID-19 vaccine in healthy adults aged 18 years or older: A randomised, double-blind, placebo-controlled, phase 2 trial. Lancet. 2020;396:479–488. doi: 10.1016/S0140-6736(20)31605-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Folegatti P.M., Ewer K.J., Aley P.K., Angus B., Becker S., Belij-Rammerstorfer S., Bellamy D., Bibi S., Bittaye M., Clutterbuck E.A., et al. Safety and immunogenicity of the ChAdOx1 nCoV-19 vaccine against SARS-CoV-2: A preliminary report of a phase 1/2, single-blind, randomised controlled trial. Lancet. 2020;396:467–478. doi: 10.1016/S0140-6736(20)31604-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Rakhra K., Abraham W., Wang C., Moynihan K.D., Li N., Donahue N., Baldeon A.D., Irvine D.J. Exploiting albumin as a mucosal vaccine chaperone for robust generation of lung-resident memory T cells. Sci. Immunol. 2021;6:eabd8003. doi: 10.1126/sciimmunol.abd8003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.van Doremalen N., Purushotham J., Schulz J., Holbrook M., Bushmaker T., Carmody A., Port J., Yinda K.C., Okumura A., Saturday G., et al. Intranasal ChAdOx1 nCoV-19/AZD1222 vaccination reduces shedding of SARS-CoV-2 D614G in rhesus macaques. bioRxiv. 2021:426058. doi: 10.1101/2021.01.09.426058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bui H.-H., Sidney J., Dinh K., Southwood S., Newman M.J., Sette A. Predicting population coverage of T-cell epitope-based diagnostics and vaccines. BMC Bioinform. 2006;7:153. doi: 10.1186/1471-2105-7-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Fukami-Kobayashi K., Shiina T., Anzai T., Sano K., Yamazaki M., Inoko H., Tateno Y. Genomic evolution of MHC class I region in primates. Proc. Natl. Acad. Sci. USA. 2005;102:9230–9234. doi: 10.1073/pnas.0500770102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The original DICOM files (>20 MB/file) are available upon request.