Abstract
Summary: For rapid visualization of subdural electrodes with respect to cortical and subcortical structures, we describe a novel and fully automated method based on coregistration, normalization, optional cerebellum masking, and volume rendering of 3D MR imaging data taken before and after implantation. The key step employs the skull-stripped preimplantation image as a mask to also remove the skull in the postimplantation image. The extracted brain is presented in 3D with the electrodes directly visible by their susceptibility artifacts. Compared with alternative methods, ours is based on freely available software and does not require manual intervention.
Knowledge of the exact localization of the seizure onset zone is of paramount importance for surgery planning in patients with medically intractable epilepsy. Subdural strip and grid electrodes are used for obtaining cortical recordings of seizure activity and for mapping of eloquent cortical areas in candidates for epilepsy surgery if a noninvasive workup has failed to yield unequivocal data. The information about the exact position of the implanted electrodes on the cortical surface with respect to relevant brain structures is essential for a correct interpretation of the electroencephalographic and mapping data and for the planning of the subsequent surgical procedure (1). The position of subdural electrodes after implantation is estimated so far by a number of methods, including plain skull radiographs, CT, planar 2D MR imaging, 3D reconstruction of the 2D MR imaging data with or without curvilinear reformatting, superimposion of the reconstructed 3D MR images with digital images of the implanted electrodes taken during the implantation, and various combinations of these methods (2–7). We describe a novel, fully automated, and robust method for rapid visualization of subdural strip and grid electrodes both on the convexity and the basal brain surface.
Technique and Patients
MR Imaging Data Acquisition and Processing
3D MR imaging data sets were acquired before and after the implantation of the subdural electrodes (resulting in a “preimplantation image” and a “postimplantation image” for each patient) by using a magnetization-prepared rapid-acquisition gradient-echo (MPRAGE) sequence (9.7 ms/4 ms/1 [TR/TE/excitations]; flip angle, 12°) on a 1.5T scanner (Magnetom Vision; Siemens, Erlangen, Germany). The following parameters were used to obtain isotropic voxels of 1 mm2: matrix of 256 × 256 mm2; 160–180 sections; FOV of 256 mm; and 3D slab thickness of 160–180 mm, depending on head size.
Data sets were transferred in digital imaging and communication (DICOM) format from the MR scanner to a Pentium III 900-mHz PC workstation and converted to ANALYZE format by using the free viewing software MRIcro for Windows (http://www.psychology.nottingham.ac.uk/staff/cr1/mricro.htm) (8). The further image data processing was fully automated by using a batch script for SPM99 (Wellcome Department of Imaging Neuroscience, London, United Kingdom [http://www.fil.ion.ucl.ac.uk/spm]). The script employs standard procedures available within SPM99 and works in three steps (see also Fig 1). The numbers within the figure correspond to the following processing steps:
Fig 1.
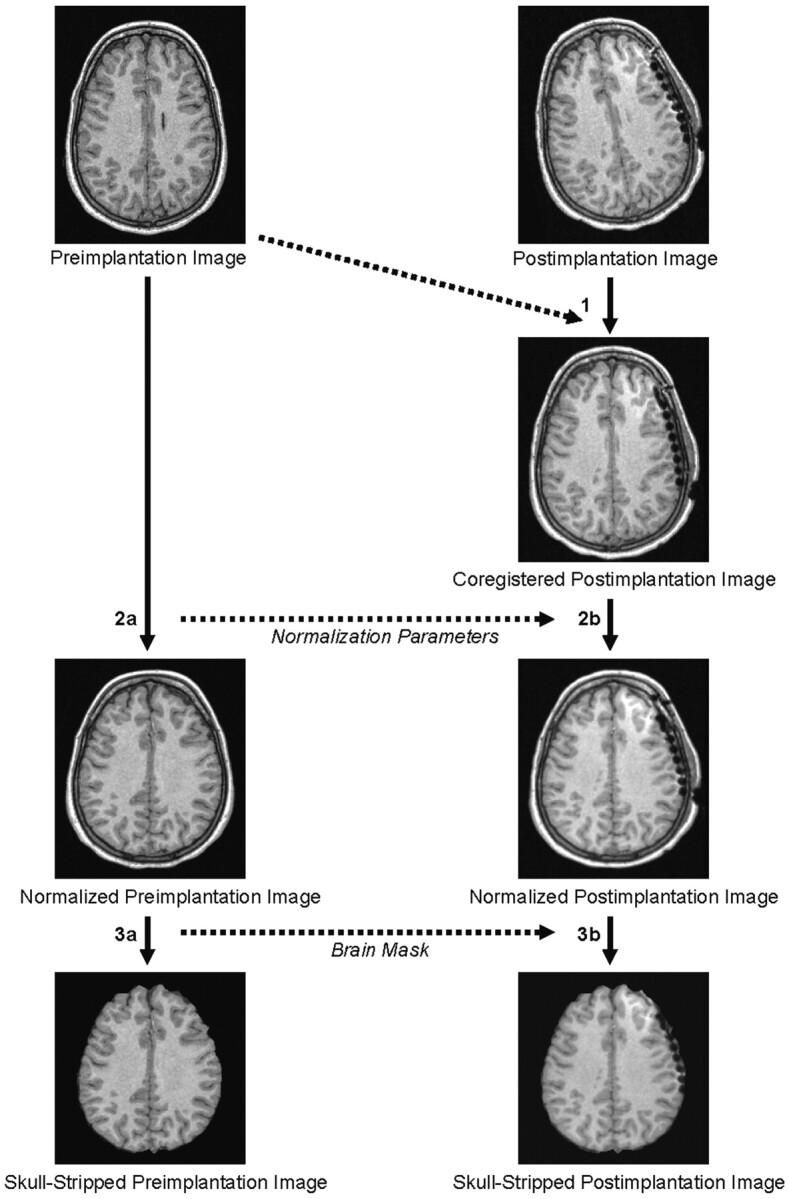
The image data processing is fully automated and consists of the following steps: 1) coregistration of the postimplantation image to the preimplantation image; 2A) normalization of the preimplantation image; 2B) normalization of the postimplantation image based on the transformation parameters derived from the normalization of the preimplantation image; 3A) brain extraction in the preimplantation image; and 3B) brain extraction in the coregistered postimplantation image by using the skull-stripped preimplantation image as a mask.
1) Coregistration—The postimplantation image is fitted to the preimplantation image by using a six-parameter rigid body transformation (9).
2) Normalization—A) The preimplantation image is normalized to the standard brain of the Montreal Neurologic Institute (MNI) included in SPM99 by using the default parameters for normalization (i.e., optimum 12-parameter affine transformation), followed by nonlinear normalization based on 7 × 8 × 7 smooth spatial basis functions; B) simultaneously, the coregistered postimplantation image is also normalized to the MNI brain by using the transformation parameters derived from the normalization of the preimplantation image.
3) Brain extraction—A) Skull stripping in the preimplantation image is done by using the automated brain extraction tool (10) provided as part of the free MRIcro distribution or the FMRIB software library (http://www.fmrib.ox.ac.uk/fsl); B) the brain within the coregistered and normalized postimplantation image is extracted by employing the ImCalc tool of SPM99 and by using the skull-stripped preimplantation image as a mask. To allow for optimal visualization of electrodes on the basal surface of the posterior temporal and/or occipital lobe, this step is optionally combined with the removal of the cerebellum in the skull-stripped postimplantation image by using a standard cerebellum mask based on the “aal” image included in the MRIcro distribution (11).
As a result of these image processing steps, the skull-stripped postimplantation image can be visualized within MRIcro. Because of their susceptibility artifacts, the locations of electrode contacts are directly visible both in planar MR sections and in the rendered image of the brain’s surface. With the air/surface threshold set to zero, the volume-rendering tool allows the viewer to grasp the electrodes’ spatial relation to the sulcal pattern of the brain and to anatomic landmarks.
Patients
The method was applied to the MR imaging data sets from 22 patients (age range, 4–53 years) with epilepsy in whom subdural strip and/or grid electrodes were implanted from August 2003 to September 2004 at the Epilepsy Center of the University Hospital Freiburg, Freiburg, Germany. Strip and grid electrodes consisted of 4–64 stainless steel contacts with interelectrode distances of 10–15 mm embedded in silastic (Ad-Tech, Racine, WI). Subdural strip electrodes were placed over the lateral convexity, interhemispherically, or below the basal surface of the frontal, temporal, or occipital lobe by means of burr holes. For grid placement, craniotomy with opening of the dura was performed over the cortex area to be recorded.
Results
The acquisition time for one 3D MR imaging data set was 7–8 minutes, depending on head size and slab thickness. The whole image processing of the preimplantation and postimplantation MR data sets took about 10 minutes on a Pentium III 900-mHz workstation and was fully automated and operator-independent. The processed postimplantation images could be viewed immediately by using MRIcro. In each patient evaluated, all electrode contacts of the implanted subdural strip and grid electrodes could be well visualized. A simultaneous examination of the skull-stripped preimplantation and postimplantation 3D MR images in two yoked MRIcro application windows gave an excellent overview of the exact location of subdural strip and grid electrodes and their spatial relationship to anatomical landmarks of the cortex (Figs 2 and 3). In addition, the individual position of each electrode contact, with respect to the presumed epileptogenic lesion, could be exactly depicted by using yoked coregistered planar MR images alongside the 3D MR imaging data. Furthermore, lesions beneath the grid or the brain surface could be highlighted by loading as color-coded overlays and subsequent semitransparent MR imaging volume rendering (Fig 4).
Fig 2.
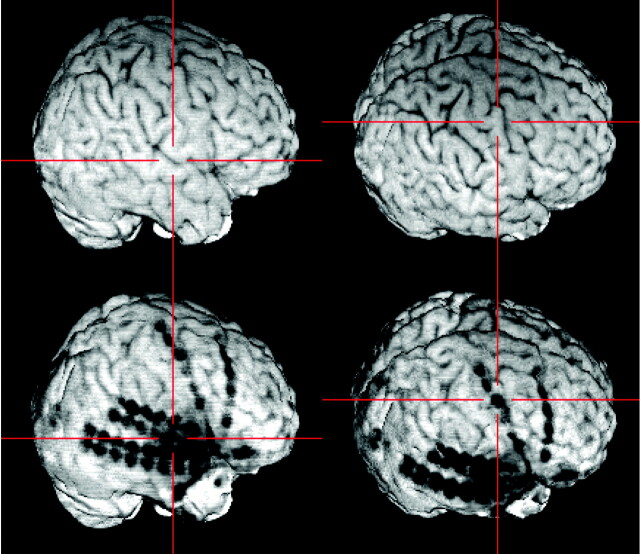
The resulting skull-stripped postimplantation image is visualized within MRIcro. A simultaneous examination of the skull-stripped preimplantation and postimplantation 3D MR images in two yoked MRIcro application windows gives an excellent overview of the exact location of each electrode contact. The viewer is able to grasp the electrodes’ spatial relation to the sulcal pattern of the brain and to anatomic landmarks (e.g., the central sulcus as pointed out here by the crosshairs).
Fig 3.
Subdural strip and double strip electrodes and their relationship to the Sylvian fissure (left) and to the precentral gyrus (right).
Fig 4.
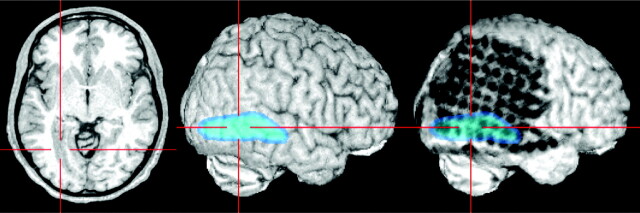
Lesions beneath the grid or the brain surface (here polymicrogyria) can be highlighted by loading as color-coded overlays and subsequent semitransparent volume rendering. The individual position of each electrode contact with respect to the presumed epileptogenic lesion can be exactly depicted by using yoked coregistered planar MR images alongside the 3D MR imaging data.
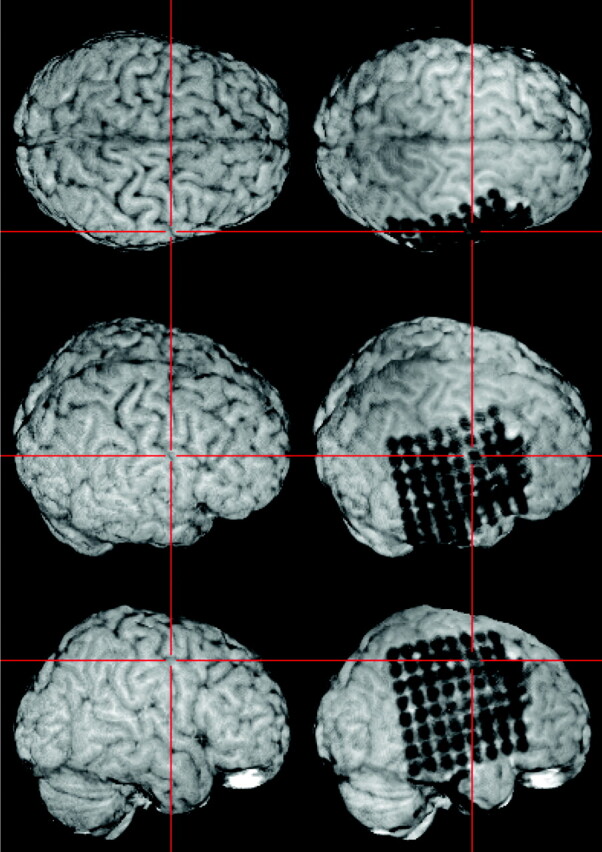
In patients with electrodes on the basal surface of the posterior temporal and/or occipital lobe, the overall view of electrode locations was remarkably improved by masking out the cerebellum in the postimplantation image (Fig 5).
Fig 5.
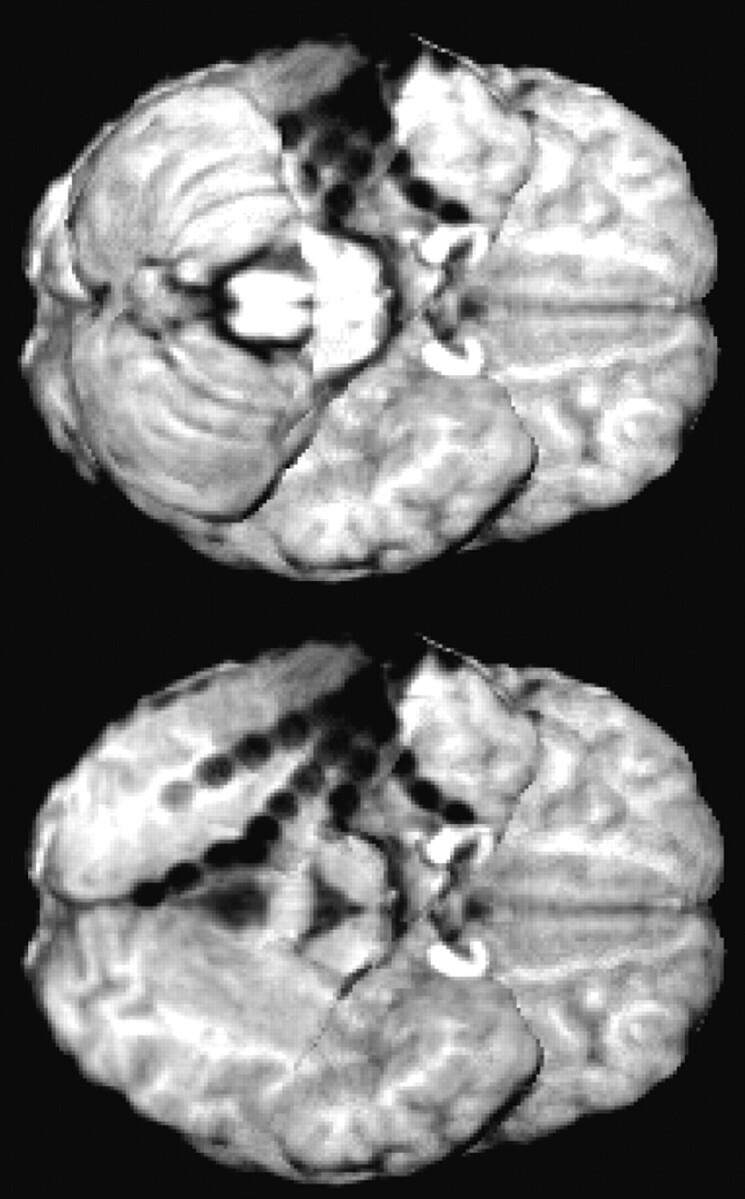
To allow for optimal visualization of electrodes on the posterior basal surface of the brain, the brain extraction (see Fig 1, step 3B) is optionally combined with masking the cerebellum in the skull-stripped postimplantation image.
Discussion
Visualization of implanted electrodes in relation to the brain is essential in the presurgical work-up of candidates for epilepsy surgery.
Principally, a direct visualization of implanted subdural electrodes on the cortical surface should be possible because of their susceptibility artifacts by removal of noncerebral tissue in the 3D MR imaging data set (generally referred to as “skull stripping” or “brain extraction”) and volume rendering of the remaining brain by using a free image viewing and rendering software (e.g., MRIcro for Windows; 8). This approach, however, is impeded by a specific limitation of the brain extraction algorithms, which normally cannot discern between skull and electrode artifacts in T1-weighted MR images. As a result, the electrode artifacts are removed along with non-brain tissue during the brain extraction and are no longer visible on the rendered brain surface after skull stripping. Furthermore, simple removal of noncerebral tissue is not sufficient to visualize electrodes surrounded by brain tissue (i.e., on the basal brain surface between cerebrum and cerebellum). So far, cumbersome, manual, and individually tailored image postprocessing is necessary to depict these electrodes.
Our novel algorithm for rapid and fully automated visualization of subdural electrodes includes the key step of using a coregistered and skull-stripped preimplantation image of the same patient as a mask for the subsequent brain extraction from the postimplantation image. The preimplantation MR imaging data are free of electrode artifacts and can be used for generating normalization parameters (2nd step) and a brain extraction mask (3rd step), which are subsequently applied to the postimplantation images to produce skull-stripped 3D images of superior quality.
The normalization step is necessary because it allows the application of a standardized cerebellum mask. This is required to remove the cerebellum in an automatic fashion (i.e., without cumbersome manual and individually tailored delineation of the cerebellum) to visualize electrodes on the posterior basal brain surface.
A number of other approaches to localize subdural electrodes were reported so far, each of which has its specific advantages and limitations.
Conventional skull radiographs are inexpensive, readily available, and still used to show the position of implanted subdural electrodes. They do not, however, display the underlying brain structures, and the number of different views is restricted. Thus, they provide only gross overview of the position and orientation of subdural electrodes.
Two-dimensional MR images principally contain the same information as the 3D MR reconstructions. An implanted electrode with all its contacts can be visualized in a single MR image if it lies flat in the image plane (e.g., an interhemispheral strip electrode in a sagittal MR section). Problems arise in areas where electrodes are bent along the convexity of the underlying cortical surface. In this case, one 2D MR section depicts only a few electrode contacts. Thus, a thorough inspection of many sections in different views is necessary to get a global impression of the implanted electrodes position. The spatial relationships of implanted electrodes to cortical landmarks and to presumed epileptogenic lesion are often difficult to evaluate on single 2D MR images.
The fusion of preimplantation MR imaging data with postimplantation CT data for 3D reconstruction as described by Winkler et al (3) and further developed by Morris et al (7) offers an improved way of localizing implanted subdural electrodes. There are, however, several disadvantages inherent in these methods: the lower spatial resolution of the CT (3-mm sections in the CT scan vs. 1-mm sections in MR imaging), which makes it difficult to fuse the CT and MR imaging data without distortions; the difficulty to delineate implanted electrodes at the base of the skull; and the need for additional radiographic exposure. Finally, the data processing requires manual steps with user interaction, which may be time-consuming (registration of MR and CT data sets by interactive manual transformation [3], electrode segmentation with seed placing by manually identifying each electrode, and segmentation of the brain by using an interactive tracing tool [7]).
Curvilinear reformatting of 3D MR imaging data yields an excellent overview of the position of subdural strip and grid electrodes, particularly with respect to the cortical landmarks or supposed epileptogenic lesions. This procedure, however, also requires manual intervention for interactive delineating the brain surface contour in subsequent MR imaging sections (2).
Intraoperative digital photography in combination with preimplantation 3D MR imaging data can be applied quickly and provides exact localizing information about the position of individual electrode contacts of subdural grids relative to the underlying cortex (5). Unfortunately, this approach is limited to patients in whom craniotomy is performed for electrode placement (i.e., mainly grid electrodes) and to patients with electrodes placed over the lateral convexity of the brain. Electrodes on the basal surface of the brain cannot be depicted. In addition, this method also requires user interaction—namely, plotting manually the location of each electrode contact on the digital photograph of the exposed native cortex and transferring the individual electrode contacts from the photographs to the patient’s brain surface by marking their position on the cerebral convexity with the rendering software (5).
Compared with these methods, our algorithm for visualization of subdural strip and grid electrodes is fast and fully automated, does not require any interactive steps (apart from the decision for optional masking out the cerebellum), and does not implicate any radiographic exposure. Implanted subdural electrodes can be precisely localized on both the convexity and the basal surface of the brain. The location of subdural strip and grid electrodes and their spatial relationship to anatomic landmarks of the cortex, as well as the individual position of each electrode contact with respect to the presumed epileptogenic lesion, can be depicted precisely. A simultaneous examination of the skull-stripped preimplantation and postimplantation 3D MR images in two yoked MRIcro application windows facilitates the planning of the surgical procedure considerably.
The presurgical evaluation of patients in our epilepsy center includes MPRAGE sequences, which require only 7–8 minutes for acquisition and are done routinely before and after implantation of subdural electrodes. Thus, both MR imaging data sets required for this method are readily available for each patient at no additional time and financial expenditure.
Finally, in contrast to other methods proposed, our solution for visualizing subdural electrodes is based on freely available software distributions (i.e., MRIcro and SPM99). The algorithm is simple and comprises only three steps, which can be easily integrated in a batch script for SPM.
If necessary (e.g., for intraoperative navigational purposes) the normalized 3D MR imaging data sets can be easily transferred to their original stereotactic space by reversing the normalization. This can be achieved by employing the deformation toolbox available for SPM99 (12).
Three limitations have to be mentioned: as well as the other visualization techniques, our method can be impeded by epidural or subdural hematoma with mass effect and local brain impression, because the mask for skull stripping is derived from the preimplantation MR imaging data, which are no longer congruent with the postimplantation MR imaging data in this case. Furthermore, the susceptibility artifacts induced by the electrodes can go along with small local field deformations of the MR image. In our experience with 22 patients, however, this never caused a problem for determining the position of an electrode contact in relation to the sulcal pattern of the brain. Finally, volume rendering is less suitable for visualization of interhemispheral electrodes, which is still best achieved with planar sagittal MR images.
Conclusion
We present a fully automated, easy-to-use, and fast method for improved localization of subdural electrodes comprising coregistration, normalization, optional masking of the cerebellum, and volume rendering of 3D MR imaging data sets taken before and after implantation. Compared with alternative visualization methods, it is based on freely available software and does not require manual intervention. The method is simple and can be easily integrated in a batch script for SPM. The resulting 3D MR-images delineate the exact location of subdural strip and grid electrodes and their spatial relationship to anatomic landmarks of the cortex as well as to presumed epileptogenic lesions thus allowing a better planning of the neurosurgical procedure.
References
- 1.Behrens E, Zentner J, van Roost D, et al. Subdural and depth electrodes in the presurgical evaluation of epilepsy. Acta Neurochir (Wien) 1994;128:84–87 [DOI] [PubMed] [Google Scholar]
- 2.Schulze-Bonhage AH, Huppertz HJ, Comeau RM, et al. Visualization of subdural strip and grid electrodes using curvilinear reformatting of 3D MR imaging data sets. AJNR Am J Neuroradiol 2002;23:400–403 [PMC free article] [PubMed] [Google Scholar]
- 3.Winkler PA, Vollmar C, Krishnan KG, et al. Usefulness of 3-D reconstructed images of the human cerebral cortex for localization of subdural electrodes in epilepsy surgery. Epilepsy Res 2000;41:169–178 [DOI] [PubMed] [Google Scholar]
- 4.Noordmans HJ, van Rijen PC, van Veelen CW, et al. Localization of implanted EEG electrodes in a virtual-reality environment. Comput Aided Surg 2001;6:241–258 [DOI] [PubMed] [Google Scholar]
- 5.Wellmer J, von Oertzen J, Schaller C, et al. Digital photography and 3D MRI-based multimodal imaging for individualized planning of resective neocortical epilepsy surgery. Epilepsia 2002;43:1543–1550 [DOI] [PubMed] [Google Scholar]
- 6.Silberbusch MA, Rothman MI, Bergey GK, et al. Subdural grid implantation for intracranial EEG recording: CT and MR appearance. AJNR Am J Neuroradiol 1998;19:1089–1093 [PMC free article] [PubMed] [Google Scholar]
- 7.Morris K, O’Brien TJ, Cook MJ, et al. A computer-generated stereotactic “virtual subdural grid” to guide resective epilepsy surgery. AJNR Am J Neuroradiol 2004;25:77–83 [PMC free article] [PubMed] [Google Scholar]
- 8.Rorden C, Brett M. Stereotaxic display of brain lesions. Behav Neurol 2000;12:191–200 [DOI] [PubMed] [Google Scholar]
- 9.Ashburner J, Friston K. Multimodal image coregistration and partitioning: a unified framework. Neuroimage 1997;6:209–217 [DOI] [PubMed] [Google Scholar]
- 10.Smith SM. Fast robust automated brain extraction. Hum Brain Mapp 2002;17:143–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tzourio-Mazoyer N, Landeau B, Papathanassiou D, et al. Automated anatomical labeling of activations in SPM using a macroscopic anatomical parcellation of the MNI MRI single-subject brain. Neuroimage 2002;15:273–289 [DOI] [PubMed] [Google Scholar]
- 12.Ashburner J, Hutton C, Frackowiak R, et al. Identifying global anatomical differences: deformation-based morphometry. Hum Brain Mapp 1998;6:348–357 [DOI] [PMC free article] [PubMed] [Google Scholar]