Abstract
In order to understand the role of intracellular metabolites in cellular processes, it is important to measure the dynamics and fluxes of small molecules in living cells. Although conventional metabolite sensors composed of fluorescent proteins have been made to detect some metabolites, an emerging approach is to use genetically encoded sensors composed of RNA. Because of the ability to rapidly generate metabolite-binding RNA aptamers, RNA-based sensors have the potential to be designed more readily than protein-based sensors. Numerous strategies have been developed to convert the green-fluorescent Spinach or Broccoli fluorogenic RNA aptamers into metabolite-regulated sensors. Nevertheless, red fluorescence is particularly desirable due to low level of red background fluorescence in cells. However, the red fluorescent variant of the Broccoli aptamer, Red Broccoli, does not exhibit red fluorescence in cells when imaged with its cognate fluorophore. It is not known why Red Broccoli is fluorescent in vitro but not in live mammalian cells. Here we develop a new fluorophore, OBI (3,5-difluoro-4-hydroxybenzylidene-imidazolinone-2-oxime-1-benzoimidazole), which binds Red Broccoli with high affinity and makes Red Broccoli resistant to thermal unfolding. We show that OBI enables Red Broccoli to be readily detected in live mammalian cells. Furthermore, we show that Red Broccoli can be fused to a S-adenosyl methionine-binding aptamer to generate a red fluorescent RNA-based sensor that enables imaging of SAM in live mammalian cells. These results reveal a red fluorescent fluorogenic aptamer that functions in mammalian cells and which can be readily developed into red fluorescent RNA-based sensors.
INTRODUCTION
Genetically encoded sensors are important tools that allow researchers to study how metabolites, proteins, or small molecules change over time in individual cells.2-10 These sensors can reveal cell-to-cell variation in intracellular metabolism and can give insights into how various signaling pathways regulate intracellular molecules. A recent advance in genetically encoded sensors was the development of sensors composed of RNA.2, 3 These RNAs comprise a target-binding RNA aptamer fused to a fluorogenic aptamer such as Spinach or Broccoli.1, 11 These aptamers are fused in such a way that binding of the target molecule induces folding, and subsequent fluorescence, of the fluorogenic aptamer. Several approaches have been described to fuse target-binding aptamers to Spinach or Broccoli, two highly related fluorogenic aptamers.2, 4, 13-24 It has recently become possible to use these RNA-based sensors in mammalian cells as a result of new approaches for expressing RNA devices as highly stable circular RNAs.3
A major limitation is that current RNA-based sensors typically exhibit green fluorescence, which is a consequence of being developed using Spinach or Broccoli, which are green-fluorescent fluorogenic aptamers.1, 11 Red fluorescent sensors are highly desirable since cells have very low intrinsic red background fluorescence.25-29 In contrast, green fluorescent probes suffer from the cellular green background fluorescence deriving from endogenous flavins and nicotinamide adenine dinucleotide (NADH), whose levels can change in response to metabolic state.31 Thus, red fluorescent RNA-based sensors would improve the accuracy of intracellular imaging.
So far, red fluorescent RNA-based sensors are not widely used since no red fluorescent fluorogenic aptamers has been shown to be amenable for sensor development in mammalian cells. So far, only Spinach and Broccoli have been shown to be capable of regulation by target-binding aptamers. Spinach and Broccoli bind and activate the green fluorescence of DFHBI (3,5-difluoro-4-hydroxybenzylidene imidazolinone), a mimic of the fluorophore naturally found in green fluorescent proteins. These aptamers contain a U•A•U base triple that forms a major part of the fluorophore-binding pocket.32 This U•A•U base triple is directly connected to a RNA duplex stem whose stability is regulated by target-binding aptamers in most sensors.2, 3, 13 Thus, the Spinach and Broccoli structure can be exploited for RNA-based sensor development.
Here we describe an approach for generating red fluorescent RNA-based sensors. To develop these sensors, we used Red Broccoli, a fluorogenic aptamer that is similar to Broccoli, but binds DFHO (3,5-difluoro-4-hydroxybenzylideneimidazolinone-2-oxime), a mimic of the fluorophore naturally found in DsRed red fluorescent proteins.12 However, Red Broccoli exhibits weak fluorescence that is not detectable in cells. To overcome this, we designed a new fluorophore that binds Red Broccoli with higher affinity, OBI (3,5-difluoro-4-hydroxybenzylidene-imidazolinone-2-oxime-1-benzoimidazole). Treatment with OBI enabled Red-Broccoli-expressing cells to show robust and photostable red fluorescence. We show that Red Broccoli can be fused to an S-adenosyl methionine (SAM)-binding aptamer, resulting in a sensor that exhibits red fluorescence in response to SAM levels in OBI-treated mammalian cells. Overall, this new fluorophore-aptamer pair provides a platform for generating red fluorescent RNA-based sensing tools for mammalian cells.
RESULTS
Comparison of the cellular fluorescence of Broccoli and Red Broccoli.
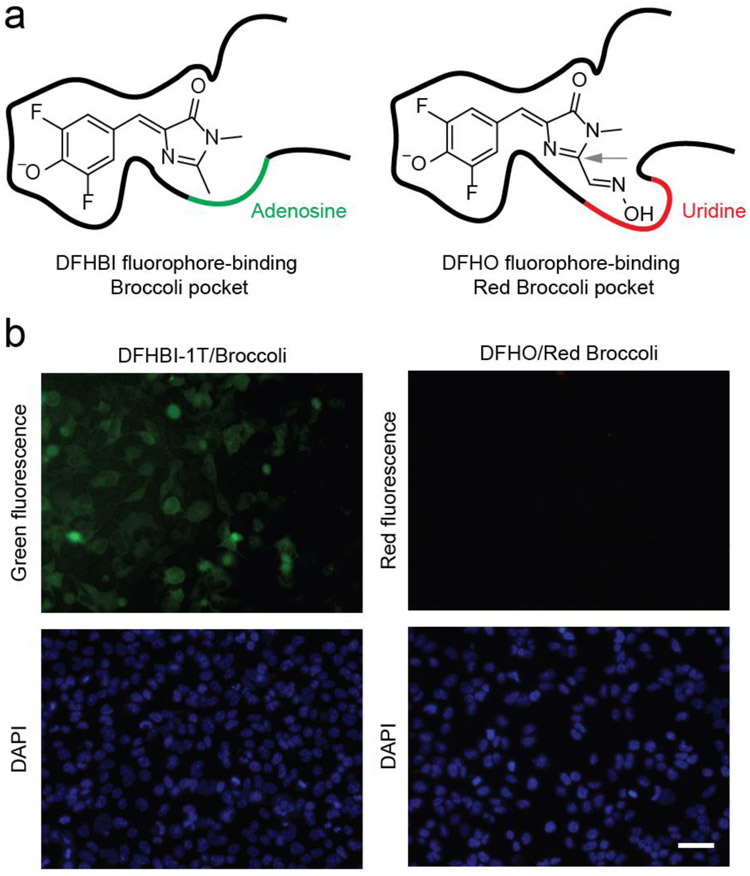
In order to create red fluorescent RNA-based sensors, we chose Red Broccoli as the fluorogenic aptamer. Red Broccoli was previously evolved from Broccoli to bind DFHO, a fluorophore with extended pi-conjugation relative tot he canonical DFHBI ligand of Broccoli.12, 30 Red Broccoli-DFHO complexes therefore exhibit orange fluorescence (Ex = 516 nm; Em = 581 nm) (Table 1). Red Broccoli contains one mutation (A44U) in the fluorophore-binding pocket of Broccoli (Figure 1a and S1), which widens the fluorophore-binding pocket to accommodate the N-oxime moiety of DFHO (Figure 1a).30 Additionally, the U44 appears to interact with the N-oxime, which appears to contribute to the overall red-shifted fluorescence of DFHO (Table 1), compared to the yellow fluorescence of DFHO when bound to Corn (Em = 545 nm).30 Since the fluorophore-binding region of Broccoli and Red Broccoli differ by only one nucleotide (i.e., A44U), the design principles used to generate sensors with Broccoli may also be used for Red Broccoli to create similar RNA-based sensors.
Table 1.
Photophysical and Binding Properties of Red Broccoli-Fluorophore Complexesa
| Fluorophore | Excitation (nm) |
Emission (nm) |
Extinction Coefficient (M−1, cm−1)b |
Quantum Yield |
Brightnessc | Kd (nM) |
|
|---|---|---|---|---|---|---|---|
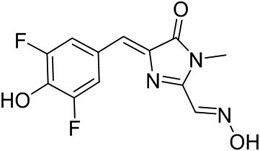 |
DFHO alone | 473 | 561 | 22,000 | 0.00002 | 0.426 | NA |
| Red Broccoli-DFHO | 516 | 581 | 35,000 | 0.34 | 100 | 165 | |
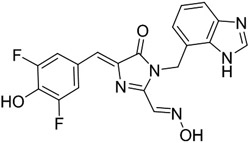 |
OBI alone | 475 | 565 | 3,900 | 0.00004 | 0.151 | NA |
| Red Broccoli-OBI | 541 | 590 | 47,300 | 0.67 | 266 | 23 |
All photophysical and binding properties experiments in Table 1 were measured at 25°C in the buffer containing 40 mM HEPES pH 7.4, 100 mm KCl, 5 mM MgCl2.
The extinction coefficients for Red Broccoli–fluorophores complexes and fluorophores alone were all measured at the maximum excitation wavelength of Red Broccoli–fluorophores complexes in the buffer. The extinction coefficients of the fluorophores alone (i.e., in the absence of Red Broccoli) were also calculated at the excitation wavelength of the Red Broccoli-fluorophore complex since this is the wavelength that is relevant for imaging experiments.
Brightness (extinction coefficient × quantum yield) is relative to Red Broccoli–DFHO.
Figure 1.
Red Broccoli cannot be detected in living cells using DFHO. (a) The fluorophore-binding pocket of Red Broccoli contains a uridine (red curve), while the same position is adenosine (green curve) in Broccoli. In the left panel, Broccoli is shown with bound DFHBI, a mimic of the fluorophore naturally found in green fluorescent proteins.(10) In the right panel, Red Broccoli is schematized with bound DFHO, a mimic of the fluorophore naturally found in DsRed red fluorescent proteins.(31) The C2 position in DFHO (indicated with a gray arrow) contains an additional N-oxime group compared to that in DFHBI that confers red-shifted emission.(31) Additionally, the uridine (indicated with a red curve) in Red Broccoli appears to interact with the N-oxime.(32) (b) Red Broccoli is not readily detected in mammalian cells incubated with DFHO when cells are imaged using fluorescence microscopy. HEK293T cells expressing circular Broccoli RNA were incubated with 10 μM DFHBI-1T (left), resulting in readily detectable green fluorescent cells. Cells expressing circular Red Broccoli RNA in the presence of 10 μM DFHO (right) did not show evidence for readily detectable cellular fluorescence using these specific imaging conditions, microscope, and camera. Images were acquired using a 40× objective with a TRITC red filter cube for Red Broccoli fluorescence, a FITC green filter cube for Broccoli fluorescence, and a DAPI filter cube for Hoechst 33342 staining. Exposure times: 100 ms for the TRITC filter, 100 ms for the FITC filter, and 50 ms for the DAPI filter. Scale bar, 50 μm.
Although Red Broccoli exhibits red fluorescence in vitro12, 30, Red Broccoli does not exhibit sufficient fluorescence to be detectable in DFHO-treated mammalian cells, even when highly expressed as a circular RNA using the Tornado expression system (Figure 1b).3 The inability to detect Red Broccoli is not due to lack of cell permeability or stability of DFHO since the Corn aptamer can be readily visualized in DFHO-treated HEK293 cells.12 Similarly, DFHBI-1T can be used to image the Broccoli aptamer when expressed using the Tornado expression system (Figure 1b and S2). Therefore, if Red Broccoli is to be used for sensor development, its fluorescence properties in cells need to be substantially improved.
The basis for the low brightness of Red Broccoli in cells is not clear, since Red Broccoli-DFHO exhibits an extinction coefficient, quantum yield, and affinity that is similar or superior to Corn-DFHO and Broccoli-DFHBI-1T.12 We therefore considered the possibility that Red Broccoli does not fold well in mammalian cells. Indeed, we recently showed that Broccoli folds poorly in cells.33 We also found that the folding could be markedly improved by switching from its canonical DFHBI ligand to a DFHBI derivative that contains a benzimidazole at the N1 position (BI, referring to the benzimidazole substituent). BI binds to Broccoli with higher affinity than DFHBI, and as a result of the increased folding, results in ~10.5-fold increased fluorescence of Broccoli in cells (Figure S3).33
One mechanism by which BI increases the fraction of folded Broccoli in cells is that Broccoli-BI complexes are highly resistant to thermal denaturation, thus enabling Broccoli to be folded at the 37°C temperature used for live cell imaging.33 An additional benefit of BI is that it exhibits reduced light-induced cis to trans isomerization, which is a photobleaching mechanism for DFHBI.33 Thus, the increased brightness of Broccoli when imaged with BI is due to both improved folding and photostability compared to Broccoli imaged with the parent fluorophore DFHBI.
Rational design OBI, a fluorophore that rescues the fluorescence of Red Broccoli in mammalian cells.
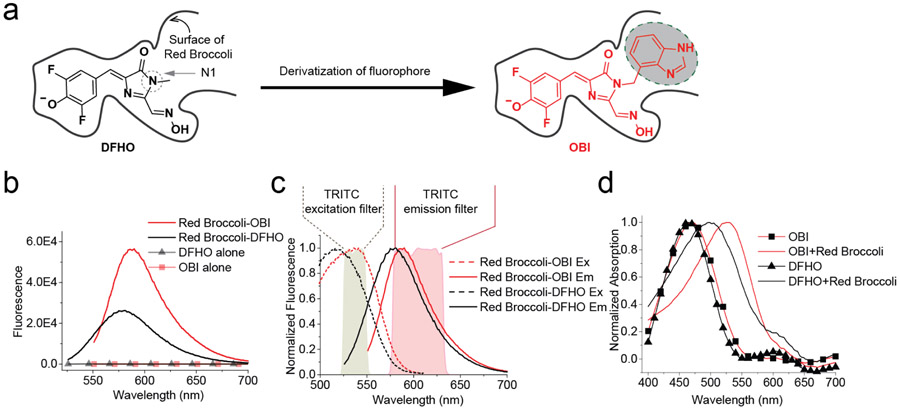
Based on the improved properties of Broccoli-BI complexes, we considered the possibility that lack of cellular Red Broccoli fluorescence could be rescued using a new fluorophore. The benzimidazole moiety in BI is predicted to interact with C33 and A64 of Spinach/Broccoli in a solvent accessible opening of the aptamer (Figure 1a, 2a and Figure S3). These residues are preserved in Red Broccoli (Figure S3). We therefore asked whether a N1-benzimidazole substituent could also be added to DFHO, and make similar interactions with Red Broccoli. If so, this fluorophore would improve the folding of Red Broccoli.
Figure 2.
Rational design of OBI, a novel fluorophore ligand for Red Broccoli. (a) Schematic drawing of DFHO and its derivative OBI. The low cellular fluorescence seen in Figure 1b could reflect the thermal instability of Red Broccoli. To stabilize the Red Broccoli RNA, we designed OBI (red, right), which contains a benzimidazole derivative at the N1 position. The benzimidazole is predicted to make interactions with nucleotides outside the immediate fluorophore-binding pocket, thus potentially providing stabilizing interactions for Red Broccoli. (b) In vitro, DFHO exhibits low brightness it exhibits orange-red fluorescence when complexed with Red Broccoli. Red Broccoli–OBI exhibits 2.66-fold higher overall fluorescence and red-shifted fluorescence compared to Red Broccoli–DFHO (see Table 1). Fluorescence emission spectra of the fluorophore (DFHO or OBI) were obtained in the presence and absence of Red Broccoli. Spectra were measured in a solution containing 1 μM fluorophore and either 0 or 20 μM Red Broccoli. (c) Excitation and emission spectra of DFHO or OBI in a complex with Red Broccoli. Spectra were measured using 20 μM Red Broccoli and 1 μM DFHO or OBI. The spectra of Red Broccoli–OBI more closely aligns with the commonly used TRITC filter compared to the spectra of Red Broccoli–DFHO. (d) The absorbance spectra of DFHO or OBI in the presence or absence of Red Broccoli. OBI shows a large red shift in its absorbance when bound to Red Broccoli. The absorbance spectra were measured in a solution containing 1 μM fluorophore and either 0 and 25 μM Red Broccoli.
We therefore synthesized OBI. OBI comprises DFHO with a benzimidazole substituent at the N1 position of DFHO (Figure 2a). Hydroxybenzylidene dyes, such as OBI, are typically in a cis/trans equilibrium in solution.34, 35 NMR analysis and liquid chromatography (LC) were consistent with >95% cis-OBI with overall high purity (Figure S4).
We first tested whether OBI can be activated by Red Broccoli. As shown in Figure 2b and Table 1, the control fluorophore DFHO showed low fluorescence in the absence of Red Broccoli and 235-fold activation by Red Broccoli. OBI also showed low fluorescence, but a 1762-fold increase in fluorescence upon addition of Red Broccoli (Figure 2b, Table 1). Red Broccoli-OBI exhibits a 2.66-fold increase in overall brightness compared to Red Broccoli-DFHO (Table 1), which is mainly due to the ~2-fold increase in quantum yield and slight increase in extinction coefficient.
Additionally, we noted that OBI exhibits a markedly red-shifted excitation wavelength upon binding Red Broccoli. The Red Broccoli-DFHO complex exhibits an excitation peak at 516 nm, which is red-shifted 41 nm relative to the absorbance peak of DFHO in solution. In contrast, the Red Broccoli-OBI complex exhibits an excitation peak at 541 nm, which is 67 nm red-shifted relative to the OBI absorbance peak (Figure 2c, Table 1). This large red-shift suggests that the benzimidazole causes OBI to interact with Red Broccoli in a different manner than DFHO, potentially resulting in more efficient stacking interactions with the G-quadruplex in Red Broccoli (Figure 2d). Stacking interactions have previously been implicated in the red-shifted excitation spectra of DFHBI once it is bound to Spinach.36
We next examined the ability of Red Broccoli to activate the fluorescence of DFHO and OBI in mammalian cells. For these experiments, we used untransfected HEK293T cells or HEK293T cells expressing circular Red Broccoli using the Tornado expression system.3 Since Red Broccoli bound to DFHO and OBI have different excitation and emission wavelengths (see Table 1), it should be noted that the overall brightness in the microscope is likely to be influenced by the filter cube used for imaging. In these experiments we used a standard TRITC filter cube (Ex: 540 ± 20 nm; Em: 605± 27.5 nm; 565 nm dichroic mirror).
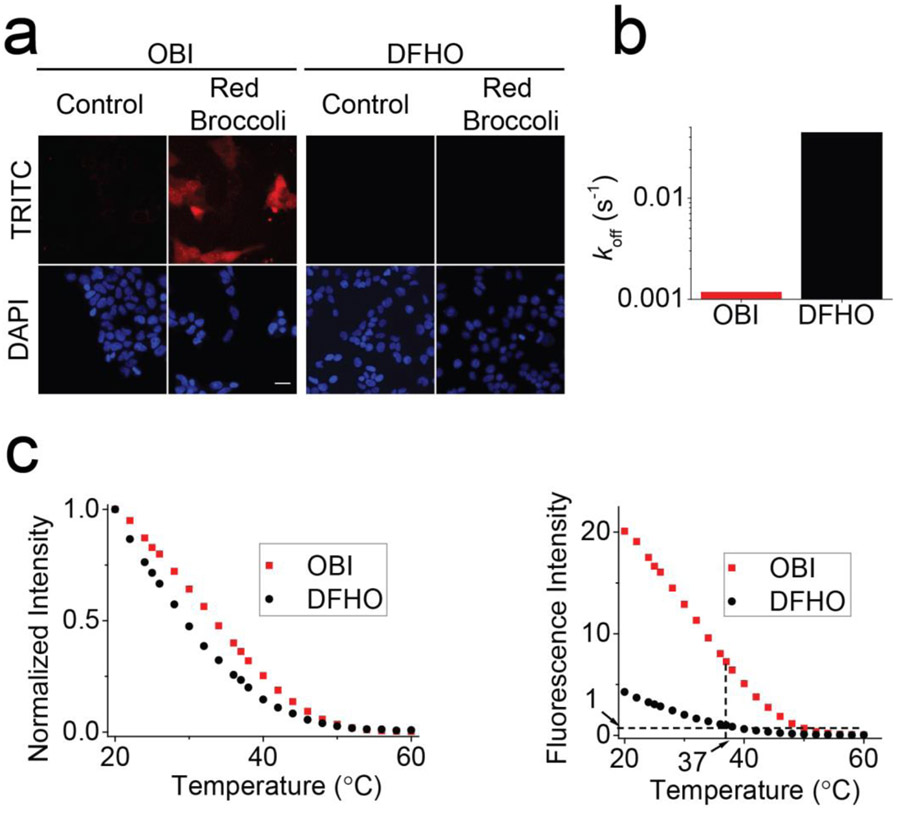
To compare the brightness of DFHO and OBI in Red Broccoli-expressing HEK293T cells, the cells were pretreated for ~1 h with OBI or DFHO (10 μM). When DFHO was added to Red Broccoli-expressing HEK293T cells, minimal fluorescence was detected compared to control HEK293T (Figure 1b). However, the addition of OBI resulted in obvious red fluorescence in Red Broccoli-expressing HEK293T cells (Figure 3a). The background fluorescence levels of OBI and DFHO were similar, as measured in HEK293T cells that do not express Red Broccoli (Figure 3a). Overall, these experiments showed that Red Broccoli is readily detected when OBI is used, but is not detected when DFHO is used.
Figure 3.
OBI stabilizes Red Broccoli folding and enables Red Broccoli to be detected in cells. (a) Incubation of OBI with Red Broccoli-expressing cells results in readily detectable red fluorescence throughput the cytoplasm. HEK293T cells expressing circular Red Broccoli RNA were imaged in the presence of OBI or DFHO (10 μM). Images were acquired with a TRITC filter cube for Red Broccoli fluorescence and a DAPI filter cube for Hoechst 33342 staining. Exposure times: 100 ms for TRITC filter and 10 ms for DAPI filter. Scale bar, 10 μm. (b) OBI exhibits a markedly slower fluorophore-RNA dissociation rate (koff). To determine if OBI exhibits slow unbinding from Red Broccoli, we tested koff. The koff value is 0.0012 s–1 for OBI, which is ~37 times slower than that for DFHO (koff = 0.0443 s–1). (c) Red Broccoli–OBI complexes exhibit higher thermostability than Red Broccoli–DFHO complexes. The fluorescence of a solution containing 20 μM RNA and 1 μM OBI or DFHO was measured in 40 mM HEPES, pH 7.4, 100 mM KCl, and 0.2 mM MgCl2 while gradually increasing the temperature. In the left image, the fluorescence is normalized so that both Red Broccoli–OBI and Red Broccoli–DFHO start at 1.0. Red Broccoli–OBI exhibited an ~5 °C shift in the Tm relative to Red Broccoli–DFHO (left). On the right panel, the absolute fluorescence is presented. Here, it can be seen that the fluorescence of Red Broccoli–OBI is ~7.3-fold higher than that of Broccoli–DFHBI-1T at 37 °C.
We also asked if Red Broccoli and Broccoli could potentially be imaged at the same time using OBI and BI, respectively. However, we noted that OBI is activated by both Red Broccoli and Broccoli in cells and in vitro (Figure S5 and S6, Table S1). Similarly, BI also binds and its fluorescence is activated by both Red Broccoli and Broccoli in cells and in vitro (Figure S5 and S6, Table S1 and S2). Therefore Red Broccoli-OBI and Broccoli-BI are not orthogonal imaging systems.
OBI promotes the folding of Red Broccoli.
We next wanted to understand the basis for the high fluorescence of Red Broccoli imaged with OBI compared to DFHO. Based on our previous findings with Broccoli and BI, the key effects of benzimidazole are: (1) increased binding affinity to Broccoli; (2) suppression of thermal unfolding of Broccoli; and (3) increased photostability.33 As tested below, the beneficial effects of benzimidazole on DFHBI when binding Broccoli also extend to DFHO when binding Red Broccoli.
First, we tested binding affinity by titrating DFHO and OBI with Red Broccoli in vitro. Based on this, we found that OBI binds with a KD of 23 nM (Table 1), compared to 165 nM for DFHO. The increased affinity of OBI is driven primarily by a reduced dissociation rate (Figure 3b). The increased binding affinity of OBI for Red Broccoli compared to DFHO suggests that the benzimidazole substituent in OBI makes contacts with the Red Broccoli.
Second, we tested whether OBI stabilizes and thus increases the amount of folded Red Broccoli in cells. It is important to note that aptamers may fold poorly in cells since they are evolved in vitro, rather than in cells. Thus, even if a fluorophore shows excellent fluorescence properties in vitro, this may not translate to a living cell if the aptamer does not fold well in living cells. Thus, folding can be a major problem that limits brightness for RNA-fluorophore complexes in cells.
To determine if OBI could stabilize the folded form of Red Broccoli, we performed a thermal denaturation analysis of Red Broccoli (20 μM) in the presence of fluorophore (DFHO or OBI, 1 μM) (Figure 3c). These experiments were performed with 0.2 mM MgCl2 since this is the approximate concentration of free Mg2+ in cells.37 Red Broccoli-OBI exhibited a +5°C shift in the Tm relative to Red Broccoli-DFHO (Figure 3c, Left). When OBI and DFHO were compared at 37°C, OBI exhibits ~7.3-fold higher fluorescence than DFHO (Figure 3c, right). Notably, the fluorescence of DFHO with Red Broccoli at 37°C is the similar as OBI or DFHO with Red Broccoli at 60°C. Overall, the low fluorescence of Red Broccoli at 37°C when imaged with DFHO suggests that Red Broccoli is largely unfolded in cells. However, addition of OBI results in a stabilized Red Broccoli-OBI complex that is folded, and thus exhibits readily detectable fluorescence in cells.
Red Broccoli-OBI complexes exhibit increased photostability.
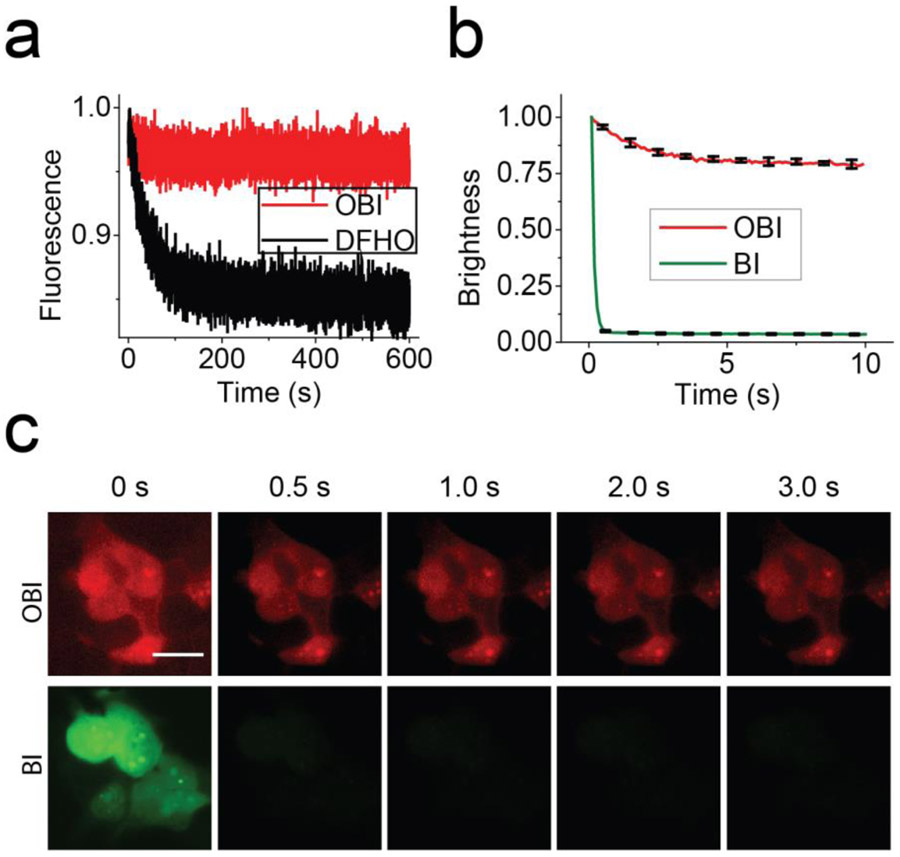
We next tested the photostability of OBI bound to Red Broccoli. To test this, we measured fluorescence in vitro during continuous irradiation in a fluorometer. During these photobleaching experiments, the sample is irradiated through a small window in the cuvette, resulting in minimal irradiation of the majority of the sample. Fluorophores (DFHO or OBI, 1 μM) were preincubated with Red Broccoli (20 μM) in buffer for 30 min, prior to measuring fluorescence. Red Broccoli-DFHO showed a rapid loss of fluorescence (Figure 4a). In contrast, Red Broccoli-OBI showed no obvious loss of fluorescence for the duration of the 600 s time course. Therefore, OBI is markedly more photostable compared to DFHO. Importantly, DFHO is not always rapidly photobleached—its photobleaching can be suppressed by binding to Corn.12 Therefore, unlike Corn, Red Broccoli is unable to suppress the photobleaching of DFHO.12
Figure 4.
Red Broccoli–OBI complexes exhibit increased photostability in cells and in vitro. (a) Red Broccoli–OBI exhibits increased photostability in vitro compared to Red Broccoli–DFHO. To test the in vitro photostability, we examined the fluorescence of Red Broccoli–OBI using continuous illumination in the fluorometer. In these experiments, we used solutions containing 1 μM fluorophore (OBI or DFHO) and 20 μM Red Broccoli. (b) Quantification of in vivo photostability. Red Broccoli–BI loses 50% of its fluorescence signal within ~0.2 s, while the fluorescence of Red Broccoli–OBI maintained >75% of its fluorescence for over 10 s. Error bars indicate s.e.m. for n = 3 cells per condition. (c) OBI exhibits markedly improved photostability in living cells compared to BI. In-cell photostability of Red Broccoli–fluorophores (OBI, and BI 20 μM) was assessed by continuous illumination of cells expressing circular Broccoli at an exposure time of 100 ms for each frame. Photobleaching was performed using light sources of similar intensity: 7.3 mW cm–2 for OBI-incubated cells (TRITC filter cube) and 7.2 mW cm–2 for BI-incubated cells (FITC filter cube). Light intensities were measured using an optical power and energy meter console. Scale bar, 10 μm.
We also tested the photostability of OBI in cells in a microscope. In these experiments, fluorophores were preincubated with circular Red Broccoli-expressing HEK293T cells for 1 h and the fluorescence was quantified using fluorescence microscopy in a selected region of interest in the cytosol of a minimum of ten cells. DFHO was not used in this experiment since DFHO does not exhibit fluorescence in cells expressing Red Broccoli.12, 30
Red Broccoli-expressing cells treated with OBI exhibited high photostability, with a 50% loss in fluorescence seen only after 10 s of continuous illumination (Figure 4b and 4c).
To understand the structural requirements for the photostability of OBI, we used BI, the fluorophore normally used with Broccoli.33 BI is similar to OBI except that it is missing the N-oxime substituent at the C2 position (Figure S2). However, BI also binds Red Broccoli, resulting in green fluorescence (Figure 4b). In the case of BI, ~50% of the cellular fluorescence in circular Red Broccoli-expressing cells was lost in ~0.2 s (Figure 4b and 4c). Thus, the benzimidazole moiety alone is not sufficient to confer photostability. Instead, both the benzimidazole and the N-oxime substituent are needed to make Red Broccoli photostable.
Overall, these experiments demonstrate that OBI binds to Red Broccoli with higher affinity than DFHO, suggesting that the benzimidazole makes additional contacts with Red Broccoli. These additional contacts may account for the ability of OBI to suppress thermal unfolding of Red Broccoli and may suppress the cis-trans isomerization that leads to photobleaching for these fluorophores.34, 35
Live imaging of metabolites in mammalian cells using Red Broccoli-OBI biosensors.
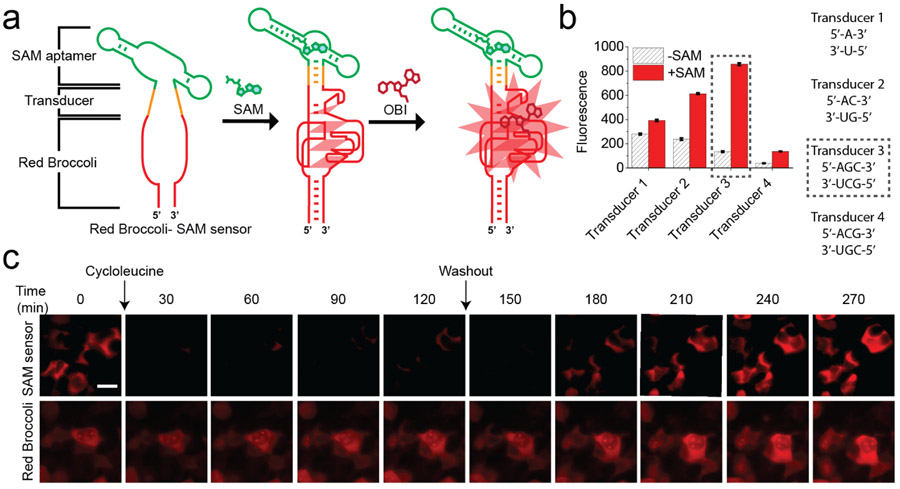
To test whether OBI enables Red Broccoli-based sensor devices in mammalian cells, we expressed a SAM biosensor composed of Red Broccoli and a SAM-binding aptamer derived from a SAM riboswitch.38 For these experiments, we connected Red Broccoli to the SAM-binding aptamer using a transducer domain (Figure 5a and Table S3). The transducer domain is a semi-stable RNA duplex that is unhybridized in the absence of SAM, thus disrupting the Red Broccoli structure.13 However, addition of SAM causes the SAM aptamer to become stabilized, which then brings the strands of the transducer together, resulting in duplex formation and subsequent folding of Red Broccoli.
Figure 5.
OBI enables imaging of endogenous SAM dynamics in mammalian cells using a Red Broccoli-based SAM sensor. (a) Schematic diagram of the Red Broccoli-based SAM sensor used for the experiments in live mammalian cells. The Red Broccoli-based SAM biosensor comprises Red Broccoli (red), a transducer (orange), and a SAM-binding aptamer (green). Sensors are designed so that Red Broccoli folds only when SAM binds to the SAM aptamer. When SAM binds to its aptamer, SAM aptamer folding leads to stabilization of the transducer stem. The stabilized transducer stem then facilitates Red Broccoli folding. (b) Optimization of the SAM biosensor’s transducer stem to increase the signal-to-background for SAM detection. In vitro transcribed Red Broccoli–SAM sensor RNA (1 μM), with each of the indicated transducers, was incubated OBI (10 μM) and vehicle or SAM (0.5 mM) for 1 h at 37 °C. The optimal transducer (Transducer 3) is indicated in a black-dotted box. Error bars indicate s.d. (n = 3 technical replicates). Ex = 541 nm, Em = 590 nm. (c) Detection of intracellular SAM dynamics using the Red Broccoli–OBI sensor. HEK293T cells expressing the Red Broccoli-based SAM biosensor (top row) or Red Broccoli (bottom row) were imaged with OBI (10 μM). Cells were treated with cycloleucine (50 mM) at 0 min, which blocks SAM biosynthesis. The cellular fluorescence rapidly drops after cycloleucine treatment. After 120 min (labeled “Washout”), the media was replaced with cycloleucine-free media. Fluorescence subsequently recovered to baseline levels. In the lower row, cells expressing the control circular Red Broccoli aptamer are shown. Circular Red Broccoli is not responsive to SAM since it lacks the SAM aptamer. No change in fluorescence is seen with any of the treatments. These results demonstrate that the changes in red fluorescence seen in the Red Broccoli-based SAM sensor-expressing cells (top row) reflect specific changes and not toxic effects of cycloleucine treatment. Images are shown at 30 min. Images were acquired with a TRITC filter cube. Exposure time: 500 ms. Scale bar, 20 μm.
We first sought to identify a transducer domain for the Red Broccoli-based SAM biosensor. We tested different transducer sequences with different lengths (Figure 5b and Table S3). These transducers were inserted between Red Broccoli and the SAM aptamer (Figure 5a). The Red Broccoli-based biosensor containing transducer 3 exhibited the largest signal relative to background as measured in vitro using OBI (Figure 5b). Additionally, this sensor showed selective fluorescence activation by SAM compared to S-adenosylhomocysteine, methionine, and adenosine (Figure S7).
We next asked whether the biosensor would detect dynamic changes in SAM levels in mammalian cells. We readily detected Red Broccoli-OBI fluorescence when the biosensor was expressed in HEK293T cells. Within 30 min after application of cycloleucine, a SAM biosynthesis inhibitor, the brightness of cells expressing the Red Broccoli-based SAM biosensor was noticeably reduced, with a complete loss of fluorescence within 2 h (Figure 5c). Control cells expressing the Red Broccoli aptamer without the SAM aptamer showed no change in fluorescence following treatment with cycloleucine (Figure 5c). Therefore, the reduction in fluorescence in cells expressing the SAM biosensor reflects a selective effect of cycloleucine on intracellular SAM levels rather than a non-specific effect on Red Broccoli-OBI fluorescence.
Next, we washed out the cycloleucine to determine whether the drop in intracellular SAM levels was reversible. Following replacement of the media with cycloleucine-free media, fluorescence levels rapidly increased to those observed at the beginning of the experiment (Figure 5c). No obvious change in red fluorescence was observed in cells expressing the control Red Broccoli (Figure 5c). Overall, these data demonstrate that OBI enables a Red Broccoli-based biosensor to function in live cells.
CONCLUSION
Genetically encoded metabolite sensors composed of RNA depend on fluorogenic RNA aptamers to generate metabolite-induced fluorescence in living cells.2 Current designs of these sensors primarily rely on Spinach and the highly related Broccoli aptamers,3, 14, 15 both of which exhibit green fluorescence when they find their cognate ligands, such as DFHBI, a mimic of the fluorophore naturally found in GFP. However, a red fluorogenic aptamer is not available which can be used for sensor development. Here we converted Red Broccoli into a fluorogenic aptamer suitable for sensor development. Red Broccoli was previously only functional in vitro, with no readily detectable fluorescence when expressed in mammalian cells. We find that Red Broccoli can exhibit fluorescence when a new fluorophore is used, OBI. OBI binds with higher affinity than the previously described Red Broccoli ligand DFHO, and markedly suppresses Red Broccoli denaturation induced by thermal melting. Additionally, OBI allows Red Broccoli to be highly photostable in cells.
We showed that Red Broccoli could be fused to a metabolite-binding aptamer, in this case the SAM metabolite-binding aptamer, resulting in a genetically encoded SAM sensor in which SAM levels are reflected by cellular red fluorescence. Overall, these results reveal a new class of genetically encoded RNA-based sensors that produce red fluorescence, thus expanding the abilities for detecting metabolites in cells.
It is surprising that Red Broccoli is not detectable when expressed in mammalian cells using its previous cognate ligand DFHO.12, 30 Red Broccoli contains only one mutation relative to Broccoli, which can be readily detected in mammalian cells using its cognate ligand DFHBI.3, 30 Other mutations are found in Red Broccoli, but these mutations are outside of the fluorophore-binding region and simply involve base pair substitutions in helical regions of the aptamer.12, 30 Nevertheless, one or more of these mutations in Red Broccoli may decrease the overall stability of the aptamer at 37°C, the temperature used for all imaging experiments. Thus, the ability of OBI to bind Red Broccoli in a manner that prevents thermal melting likely explains the ability of OBI to restore the fluorescence of Red Broccoli in mammalian cells. In OBI-treated cells, Red Broccoli can accumulate as a folded aptamer, resulting in readily detectable cellular fluorescence.
OBI likely enhances Red Broccoli thermal stability due to its benzimidazole moiety on the N1 position of DFHO. This substituent is the only difference between OBI and DFHO, and is predicted to make interactions with specific nucleotides on the surface of Red Broccoli.11, 12, 30, 32 These additional interactions likely account for the higher affinity as well as the decreased susceptibility to cis-trans isomerization, the major mechanism that limits the quantum yield and mediates the photobleaching for these ligands.34, 35 Although OBI stabilizes the folded conformation of Red Broccoli, the conformation of the thermally denatured form of Red Broccoli is not known. It may be a subtle conformational change that prevents Red Broccoli from enhancing the quantum yield of its bound fluorophore. OBI is likely to repress this unproductive conformation of Red Broccoli.
It should be noted that the increased photostability of Red Broccoli-OBI complexes compared to Red Broccoli-DFHO complexes might further contribute to the overall increase brightness of OBI-treated Red Broccoli-expressing cells compared to DFHO-treated Red Broccoli-expressing cells.
Red Broccoli is ideal for developing red fluorescent genetically encoded sensors. Red Broccoli is highly similar to Spinach and Broccoli12, 30, and therefore the principles that have been established to develop genetically encoded RNA-based sensors using Spinach and Broccoli2, 14, 15, 22, 24, 39 should be readily applicable to Red Broccoli. Indeed, we showed that a SAM sensor could be developed using Red Broccoli, similar to SAM sensors developed previously using Broccoli.3 We therefore expect that developing red fluorescent sensors should be as straightforward as developing the numerous green fluorescent sensors that have been described previously. These sensors will be useful when co-expressing green fluorescent proteins, or be useful for greater sensitivity due to the substantially reduced endogenous red fluorescence compared to endogenous green fluorescence in cells.27-29, 40
Supplementary Material
ACKNOWLEDGMENT
We thank W. Zhan, H. Zhang, Y. Huang, J. Wu, H. Kim and Q. Hou for useful help and comments. This work was supported by NIH grant R35 NS111631 (S.R.J.) and Starr Cancer Consortium Grant I12–0051.
Footnotes
Supporting Information
Experimental section, synthesis and characterization, DNA sequences, selectivity of RNA sensor
S.R.J. is the co-founder of Lucerna Technologies and has equity in this company. Lucerna has licensed technology related to Spinach and other RNA-fluorophore complexes.
REFERENCES
- 1.Paige JS; Wu KY; Jaffrey SR, RNA Mimics of Green Fluorescent Protein. Science 2011, 333 (6042), 642–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Paige JS; Nguyen-Duc T; Song W; Jaffrey SR, Fluorescence Imaging of Cellular Metabolites with RNA. Science 2012, 335 (6073), 1194–1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Litke JL; Jaffrey SR, Highly efficient expression of circular RNA aptamers in cells using autocatalytic transcripts. Nat. Biotechnol 2019, 37 (6), 667–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Song W; Strack RL; Jaffrey SR, Imaging bacterial protein expression using genetically encoded RNA sensors. Nature Methods 2013, 10 (9), 873–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Patriarchi T; Cho JR; Merten K; Howe MW; Marley A; Xiong W-H; Folk RW; Broussard GJ; Liang R; Jang MJ; Zhong H; Dombeck D; von Zastrow M; Nimmerjahn A; Gradinaru V; Williams JT; Tian L, Ultrafast neuronal imaging of dopamine dynamics with designed genetically encoded sensors. Science 2018, 360 (6396), eaat4422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Qin Y; Dittmer PJ; Park JG; Jansen KB; Palmer AE, Measuring steady-state and dynamic endoplasmic reticulum and Golgi Zn2+ with genetically encoded sensors. Proceedings of the National Academy of Sciences 2011, 108 (18), 7351–7356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tao R; Zhao Y; Chu H; Wang A; Zhu J; Chen X; Zou Y; Shi M; Liu R; Su N, Genetically encoded fluorescent sensors reveal dynamic regulation of NADPH metabolism. Nature methods 2017, 14 (7), 720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cameron WD; Bui CV; Hutchinson A; Loppnau P; Gräslund S; Rocheleau JV, Apollo-NADP+: a spectrally tunable family of genetically encoded sensors for NADP+. Nature methods 2016, 13 (4), 352. [DOI] [PubMed] [Google Scholar]
- 9.Lobas MA; Tao R; Nagai J; Kronschläger MT; Borden PM; Marvin JS; Looger LL; Khakh BS, A genetically encoded single-wavelength sensor for imaging cytosolic and cell surface ATP. Nature communications 2019, 10 (1), 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marvin JS; Shimoda Y; Magloire V; Leite M; Kawashima T; Jensen TP; Kolb I; Knott EL; Novak O; Podgorski K, A genetically encoded fluorescent sensor for in vivo imaging of GABA. Nature methods 2019, 16, 763–770. [DOI] [PubMed] [Google Scholar]
- 11.Filonov GS; Moon JD; Svensen N; Jaffrey SR, Broccoli: Rapid Selection of an RNA Mimic of Green Fluorescent Protein by Fluorescence-Based Selection and Directed Evolution. J. Am. Chem. Soc 2014, 136 (46), 16299–16308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Song W; Filonov GS; Kim H; Hirsch M; Li X; Moon JD; Jaffrey SR, Imaging RNA polymerase III transcription using a photostable RNA-fluorophore complex. Nat. Chem. Biol 2017, 13 (11), 1187–1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.You M; Litke JL; Jaffrey SR, Imaging metabolite dynamics in living cells using a Spinach-based riboswitch. Proc. Natl. Acad. Sci. U. S. A 2015, 112 (21), E2756–E2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kellenberger CA; Wilson SC; Sales-Lee J; Hammond MC, RNA-Based Fluorescent Biosensors for Live Cell Imaging of Second Messengers Cyclic di-GMP and Cyclic AMP-GMP. J. Am. Chem. Soc 2013, 135 (13), 4906–4909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kellenberger CA; Chen C; Whiteley AT; Portnoy DA; Hammond MC, RNA-Based Fluorescent Biosensors for Live Cell Imaging of Second Messenger Cyclic di-AMP. J. Am. Chem. Soc 2015, 137 (20), 6432–6435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu R; Karunanayake Mudiyanselage APKK; Shafiei F; Zhao B; Bagheri Y; Yu Q; McAuliffe K; Ren K; You M, Genetically Encoded Ratiometric RNA-Based Sensors for Quantitative Imaging of Small Molecules in Living Cells. Angew. Chem. Int. Ed 2019, 58 (50), 18271–18275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang XC; Wilson SC; Hammond MC, Next-generation RNA-based fluorescent biosensors enable anaerobic detection of cyclic di-GMP. Nucleic Acids Res. 2016, 44 (17), 139–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kellenberger CA; Hammond MC, In Vitro Analysis of Riboswitch–Spinach Aptamer Fusions as Metabolite-Sensing Fluorescent Biosensors. In Methods Enzymol., Elsevier: 2015; Vol. 550, pp 147–172. [DOI] [PubMed] [Google Scholar]
- 19.Yu QK; Shi J; Mudiyanselage A; Wu R; Zhao B; Zhou M; You MX, Genetically encoded RNA-based sensors for intracellular imaging of silver ions. Chem. Commun 2019, 55 (5), 707–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mudiyanselage A; Wu R; Leon-Duque MA; Ren KW; You MX, "Second-generation" fluorogenic RNA-based sensors. Methods 2019, 161, 24–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sun ZN; Nguyen T; McAuliffe K; You MX, Intracellular Imaging with Genetically Encoded RNA-Based Molecular Sensors. Nanomaterials 2019, 9 (2), 1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bose D; Su YC; Marcus A; Raulet DH; Hammond MC, An RNA-Based Fluorescent Biosensor for High-Throughput Analysis of the cGAS-cGAMP-STING Pathway. Cell Chemical Biology 2016, 23 (12), 1539–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Karns K; Vogan JM; Qin Q; Hickey SF; Wilson SC; Hammond MC; Herr AE, Microfluidic Screening of Electrophoretic Mobility Shifts Elucidates Riboswitch Binding Function. J. Am. Chem. Soc 2013, 135 (8), 3136–3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang XC; Wilson SC; Hammond MC, Next-generation RNA-based fluorescent biosensors enable anaerobic detection of cyclic di-GMP. Nucleic Acids Res. 2016, 44 (17). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ermakova YG; Bilan DS; Matlashov ME; Mishina NM; Markvicheva KN; Subach OM; Subach FV; Bogeski I; Hoth M; Enikolopov G; Belousov VV, Red fluorescent genetically encoded indicator for intracellular hydrogen peroxide. Nature Communications 2014, 5 (1), 5222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang Z; Yan L; Zhang L; Chen Y; Li H; Zhang J; Zhang Y; Li X; Xu B; Fu X, Ultra bright red AIE dots for cytoplasm and nuclear imaging. Polymer Chemistry 2014, 5 (24), 7013–7020. [Google Scholar]
- 27.Fan Y; Makar M; Wang MX; Ai H.-w., Monitoring thioredoxin redox with a genetically encoded red fluorescent biosensor. Nat. Chem. Biol 2017, 13 (9), 1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ermakova YG; Bilan DS; Matlashov ME; Mishina NM; Markvicheva KN; Subach OM; Subach FV; Bogeski I; Hoth M; Enikolopov G, Red fluorescent genetically encoded indicator for intracellular hydrogen peroxide. Nature communications 2014, 5 (1), 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li Y; Tsien RW, pHTomato, a red, genetically encoded indicator that enables multiplex interrogation of synaptic activity. Nat. Neurosci 2012, 15 (7), 1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Filonov GS; Song W; Jaffrey SR, Spectral Tuning by a Single Nucleotide Controls the Fluorescence Properties of a Fluorogenic Aptamer. Biochemistry 2019, 58 (12), 1560–1564. [DOI] [PubMed] [Google Scholar]
- 31.Blacker TS; Mann ZF; Gale JE; Ziegler M; Bain AJ; Szabadkai G; Duchen MR, Separating NADH and NADPH fluorescence in live cells and tissues using FLIM. Nature Communications 2014, 5 (1), 3936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Warner KD; Chen MC; Song W; Strack RL; Thorn A; Jaffrey SR; Ferre-D'Amare AR, Structural basis for activity of highly efficient RNA mimics of green fluorescent protein. Nat. Struct. Mol. Biol 2014, 21 (8), 658–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li X; Kim H; Litke J; Wu J; Jaffrey S, Fluorophore-promoted RNA folding and photostability enable imaging of single Broccoli-tagged mRNAs in live mammalian cells. Angew. Chem. Int. Ed 2020, 59, 4511–4518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Han KY; Leslie BJ; Fei J; Zhang J; Ha T, Understanding the Photophysics of the Spinach–DFHBI RNA Aptamer–Fluorogen Complex To Improve Live-Cell RNA Imaging. J. Am. Chem. Soc 2013, 135 (50), 19033–19038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang P; Querard J; Maurin S; Nath SS; Le Saux T; Gautier A; Jullien L, Photochemical properties of Spinach and its use in selective imaging. Chemical Science 2013, 4 (7), 2865–2873. [Google Scholar]
- 36.Huang H; Suslov NB; Li N-S; Shelke SA; Evans ME; Koldobskaya Y; Rice PA; Piccirilli JA, A G-quadruplex–containing RNA activates fluorescence in a GFP-like fluorophore. Nat. Chem. Biol 2014, 10, 686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dai L-J; Quamme GA, Intracellular Mg2+ and magnesium depletion in isolated renal thick ascending limb cells. The Journal of clinical investigation 1991, 88 (4), 1255–1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cho EJ; Lee J-W; Ellington AD, Applications of Aptamers as Sensors. Annual Review of Analytical Chemistry 2009, 2 (1), 241–264. [DOI] [PubMed] [Google Scholar]
- 39.Kellenberger CA; Hammond MC, In Vitro Analysis of Riboswitch-Spinach Aptamer Fusions as Metabolite-Sensing Fluorescent Biosensors. In Riboswitches as Targets and Tools, BurkeAguero DH, Ed. 2015; Vol. 550, pp 147–172. [DOI] [PubMed] [Google Scholar]
- 40.Acharya A; Bogdanov AM; Grigorenko BL; Bravaya KB; Nemukhin AV; Lukyanov KA; Krylov AI, Photoinduced Chemistry in Fluorescent Proteins: Curse or Blessing? Chem. Rev 2017, 117 (2), 758–795. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.