Abstract
Spinach and Broccoli are fluorogenic RNA aptamers that bind and activate the fluorescence of DFHBI, a mimic of the green fluorescence protein chromophore. Although Spinach/Broccoli-DFHBI complexes exhibit high fluorescence in vitro, their fluorescence is considerably lower in cells. Here we used an in silico screen to identify BI, a DFHBI derivative that binds Broccoli with higher affinity and leads to markedly higher Broccoli fluorescence in cells compared to other DFHBI-related ligands. We show that BI prevents thermal unfolding of Broccoli at 37°C in mammalian cells, leading to substantially more folded Broccoli and subsequently more fluorescent Broccoli-BI complexes. Additionally, Broccoli-BI complexes are significantly more photostable due to impaired light-induced photoisomerization, and rapid unbinding of photoisomerized cis-BI. These optimized fluorescence properties enable single mRNA transcripts containing 24 Broccoli aptamers in the 3’UTR to be imaged in cells treated with BI. These studies reveal that small molecule ligands can promote RNA folding and function in cells, and show the feasibility of single mRNA imaging with fluorogenic aptamers.
Keywords: Fluorescent probes, Fluorogenic aptamers, Photostability, RNA folding, Single mRNA imaging
Introduction
Fluorogenic aptamers are RNA sequences that bind otherwise nonfluorescent small molecules and induce their fluorescence while bound to RNA.[1] By tagging an RNA with a fluorogenic aptamer, fluorescence can be genetically encoded into the RNA, and the endogenously expressed tagged RNA can be detected by fluorescence microscopy. Since only the RNA-fluorophore complexes are fluorescent, fluorescence signals derive from tagged RNAs, while the unbound fluorophores are nonfluorescent.
Fluorogenic aptamers have been used for diverse applications, including monitoring transcription rates[1a] or for imaging RNA aggregates such as aggregates of the 5S RNA[1b] or trinucleotide repeat RNA,[3] or mRNA aggregates in stress granules.[4] RNA aggregates are relatively easy to image since they contain a large number of tagged RNAs, resulting in a concentrated signal that can be readily detected by fluorescence microscopy.
In contrast, imaging single mRNA molecules using fluorogenic aptamers is challenging. In the case of the Spinach and Broccoli fluorogenic aptamers, photobleaching occurs rapidly, which limits the total fluorescence that can be generated from an aptamer-tagged mRNA. Thus overcoming photobleaching would substantially enhance the ability of Spinach and/or Broccoli to be used as a tag for imaging single mRNA molecules in living cells.
Spinach and Broccoli bind DFHBI, a small molecule mimic of the fluorophore found in GFP.[1b, 6] Upon irradiation, DFHBI can undergo a light-induced cis-trans isomerization to a low fluorescence trans- form.[5] The trans-fluorophore subsequently unbinds from the aptamer. Fluorescence is only restored when a cis-fluorophore from solution binds the aptamer. The fluorescence output could be increased if (1) the ability of the fluorophore to isomerize to the trans isoform could be suppressed, (2) if the low-fluorescence trans-fluorophore would unbind from the aptamer more quickly, and/or (3) if the rate of rebinding of cis-fluorophore could increase.
Here we describe the structure-guided design of BI, a DFHBI derivative that binds Broccoli and produces markedly increased brightness and photostability of Broccoli in cells. BI functions by promoting Broccoli folding in cells, resulting in a substantial increase in the fraction of Broccoli that is folded in cells. In addition to this effect, BI overcomes the key problems that have resulted in photobleaching of Broccoli in previous experiments. As a result of these improved properties, single mRNAs containing an optimized tandem Broccoli tag can be imaged in live mammalian cells.
Results and Discussion
Structure-guided identification of DFHBI derivatives with higher affinity for Spinach
In order to optimize the fluorescence of Spinach and Broccoli for imaging individual mRNAs, we wanted to reduce photobleaching and develop an approach to accelerate the recovery of fluorescence after photobleaching.[5] Photobleaching of Spinach-DFHBI begins with light-induced isomerization of DFHBI from the cis to the trans form.[5] This results in termination of Spinach fluorescence. The trans-DFHBI remains bound as a low-fluorescence Spinach-DFHBI complex. This causes extends the duration of the low-fluorescence state of Spinach. Next, the trans-DFHBI unbinds. Lastly, fluorescence is restored when a cis-DFHBI fluorophore binds Spinach (Figure 1a).[5]
Figure 1. Structure-guided identification of BI, a fluorophore that leads to higher brightness in Broccoli-expressing mammalian cells.
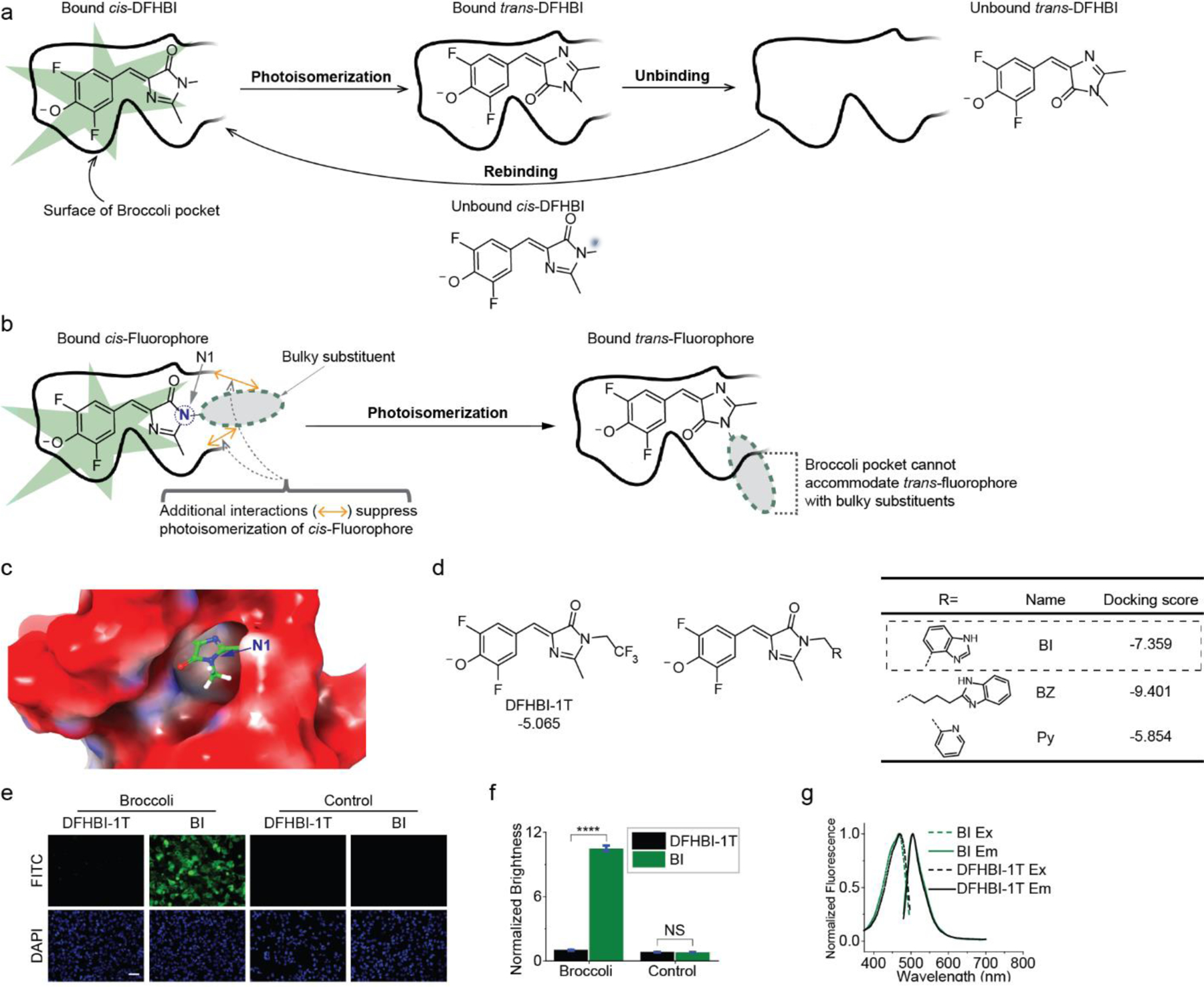
(a) Schematic of the molecular steps that account for photobleaching of Spinach/Broccoli and the resoration of fluorescence. (b) A bulky substituent on the N1 (blue atom) may enable rapid dissociation of trans-fluorophore. Upon isomerization the bulky substituent would cause the fluorophore to no longer fit in Broccoli, causing rapid ejection. (c) Orientation of DFHBI in the Spinach cystal structure.[2] The N1 position faces the solvent-accessible opening. The Spinach surface representation is colored according to electrostatic potential, from red (negative) to blue (positive). (d) Chemical structure and docking score of DFHBI-1T and DFHBI derivatives identified by structure-guided screening. Shown are the top-binding hits. Each of these molecules have higher docking scores that than DFHBI-1T. (e) HEK293T cells expressing circular Broccoli RNA or control mCherry mRNA were imaged in the presence of 10 μM DFHBI-1T or BI. Exposure times: 100 ms for FITC filter, 50 ms for DAPI filter. Scale bar, 100 μm. (f) Quantification of average brightness of Broccoli-expressing cells (left) incubated with 10 μM DFHBI-1T or BI. Background fluorescence (right) measured in cells lacking Broccoli was similar in 10 μM DFHBI-1T or BI-treated cells. Error bars indicate standard error of the mean (s.e.m.) for n=150 cells per condition. ****P < 0.0001. NS, not significant. (g) Excitation and emission spectra of Broccoli bound to either DFHBI-1T (grey) or BI (green). Excitation (Ex, dotted line) and emission (Em, solid line) spectra were measured using in vitro transcribed and purified Broccoli RNA (10 μM) and the indicated fluorophore (0.5 μM) in buffer containing 1 mM MgCl2.
We reasoned that photoisomerization and the persistence of a bound low-fluorescence trans isomer could be resolved with an improved fluorophore that would either (1) be less susceptible to light-induced cis-trans isomerization; or (2) would exhibit rapid unbinding when it isomerizes to the trans-fluorophore.
To suppress cis-trans isomerization, we sought to generate a derivative of DFHBI that would bind with higher affinity. Higher affinity may reflect more contacts with the RNA, which may suppress isomerization (Figure 1b).
To increase the unbinding rate of the trans-fluorophore, we reasoned that a bulky substituent on the imidazolinone ring (Figure 1b) would cause the fluorophore to be rapidly ejected upon isomerization since the trans- fluorophore would not fit in the DFHBI-binding pocket of Spinach. This contrasts with DFHBI, which can remain bound upon isomerization since both the cis- or trans- form can fit in the DFHBI-binding pocket.
To identify DFHBI derivatives that meet these criteria, we virtually screened libraries of DFHBI derivatives for their docking scores in the Spinach fluorophore-binding pocket. This library contained 817 substituents appended to the N1 position of the imidazolinone portion of the DFHBI (Figure 1b). The N1 position faces a solvent-accessible opening in the Spinach aptamer (Figure 1c), so these DFHBI derivatives would be expected to fit into Spinach. In contrast, substituents elsewhere would be unable to fit in the DFHBI-binding pocket of Spinach.[2, 7]
Each substituent contained a minimum of one methylene carbon between the N1 nitrogen and the chemical moieties in the library. We used Glide (grid-based ligand docking with energetics) from Schrödinger.[8] In Glide, a docking score is generated for each molecule based on an energy minimization calculation in which each library member is docked in diverse conformations. The docking score is related to the expected binding affinity.[8a, 8b] We selected three molecules for synthesis and for in vitro binding assays based on their strong docking scores and synthetic accessibility (Figure 1d).
We first asked if the compounds are cell permeable and have low background fluorescence in mammalian cells. We either used HEK293T cells expressing mCherry as a control, or HEK293T cells expressing Broccoli[6] using the Tornado expression system, which results in high level expression of Broccoli as a circular RNA.[9] We used Broccoli since its fluorophore-binding pocket is essentially identical to that of Spinach[10], but it produces improved fluorescence in cells due to a lower dependence on magnesium for folding.[6] Any fluorescence seen in cells lacking Broccoli is by definition background fluorescence, including nonspecific fluorescence activation by cellular constituents.
Cells were pretreated for 2 h with each DFHBI derivative (10 µM) or DFHBI-1T ((Z)-4-(3,5-difluoro-4-hydroxybenzylidene)-2-methyl-1-(2,2,2-trifluoroethyl)-1H-imidazol-5(4H)-one, 10 µM), which exhibits low fluorescence in cells.[11] The different compounds exhibited low background fluorescence in cells lacking Broccoli, similar to DFHBI-1T (Figure 1e, f). When each derivative was applied to circular Broccoli-expressing HEK293T cells, increased fluorescence was seen (Figure 1e).
Surprisingly, one derivative, BI (referring to the benzimidazole substituent), caused a ~10.5-fold increase in cellular fluorescence compared to DFHBI-1T (Figure 1f).
The marked increase in fluorescence was lost when the linker to benzimidazole was lengthened, or if benzimidazole was exchanged with pyridine (Figure S1). Furthermore, derivatives with different linker lengths or attachment points on the benzimidazole (Figure S2a) did not further improve the fluorescence in Broccoli-expressing cells (Figure S2b).
The increased brightness cannot be explained by enhanced photostability since the increased brightness of BI-treated cells can be detected immediately upon imaging. Therefore, we first wanted to understand the mechanism of the unusual degree of fluorescence enhancement seen with BI.
The enhanced cellular brightness of Broccoli-BI cannot be fully explained by enhanced photophysical properties
We measured the extinction coefficient and quantum yield of this complex in vitro in a 200 nM solution of Broccoli-fluorophore complex prepared by incubating 200 nM BI or DFHBI-1T with 10 µM in vitro transcribed Broccoli. The fluorescence calculation (i.e., quantum yield × extinction coefficient) show that Broccoli-BI complexes are 1.88-fold brighter than Broccoli-DFHBI-1T complexes at 25°C (Table 1).
Table 1.
Photophysical and Binding Properties of Fluorophore-Broccoli Complexes
| Structure | RNA-Fluorophore | Excitation (nm) |
Emission (nm) |
Extinction Coefficient (M−1, cm−1)[a] |
Quantum Yield |
Brightness[b] |
KD (nM) |
|---|---|---|---|---|---|---|---|
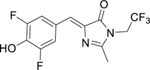 |
DFHBI-1T | 428 | 500 | 35,400 | 0.00041 | 0.121 | N/A |
| Broccoli- DFHBI-1T |
470 | 505 | 28,900 | 0.414 | 100 | 305±39 | |
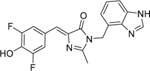 |
BI | 425 | 500 | 42,500 | 0.00048 | 0.171 | N/A |
| Broccoli- BI |
470 | 505 | 33,600 | 0.669 | 188 | 51±5 |
Extinction coefficients were all measured in buffer containing 40 mM HEPES pH 7.4, 100 mM KCl, 1 mM MgCl2. Experiments were performed at 25°C in order to compare with previous studies.[1]
Brightness (extinction coefficient × quantum yield) is relative to Broccoli-DFHBI-1T.
Since this increase in quantum yield is less than the ~10.5-fold increase in brightness of BI compared to DFHBI-1T in cells, the increased quantum yield cannot explain the markedly improved performance of BI in cells.
Additionally, circular Broccoli expression was essentially identical in vehicle-, DFHBI-1T-, and BI-treated cells (Figure S3). Thus, the enhanced brightness in BI-treated cells is not due to increased expression of circular Broccoli.
BI suppresses thermal denaturation of Broccoli
Fluorescence brightness is usually defined as proportional to the product of the extinction coefficient (ɛ) and the quantum yield (ϕ). However, the brightness of an RNA-fluorophore complex involves an additional factor, i.e., the fraction of the RNA that is folded. Thus, for a fluorogenic aptamer, brightness = ɛ × ϕ × % folded. We therefore considered the possibility that BI could increase the fraction of Broccoli that is folded in the cell.
Although Broccoli folding has been measured in vitro[12], measurements of the percent of a folded RNA in cells is not straightforward. Nevertheless, our in vitro studies previously showed that Broccoli and Spinach are not completely folded when synthesized in vitro.[12] Furthermore, like all RNA aptamers, both Broccoli and Spinach are thermally unstable at 37°C, the imaging temperature used in mammalian cells.[3, 6] Thus, folding is a major problem that limits brightness for fluorogenic aptamers in cells.
We therefore performed several tests to determine if BI influences the folding of Broccoli. First, we noted that BI binds with a KD of 51 nM (Table 1 and Figure 2b, c), compared to 305 nM for DFHBI-1T. The increased affinity of BI for Broccoli compared to DFHBI suggests that the benzimidazole moiety in BI makes contacts with the Broccoli RNA, as predicted in the docking study. These additional contacts may create a Broccoli-BI complex that is more thermally stable.
Figure 2. BI stabilizes Broccoli.
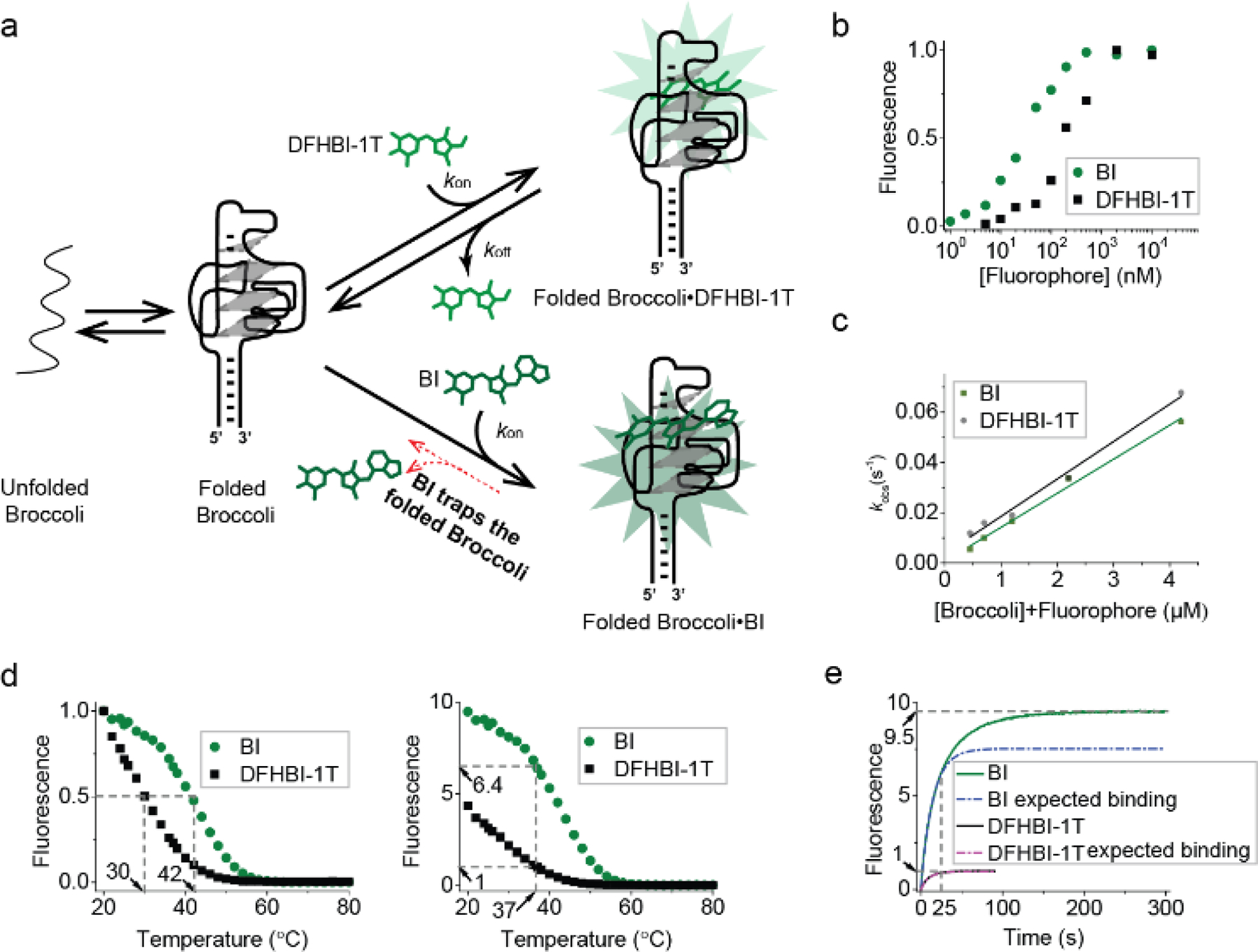
(a) Model for how BI could enhance overall cellular fluorescence by stabilizing Broccoli in folded form. (b) Broccoli (50 nM) was titrated with either DFHBI-1T or BI at 25°C and then fitted using the Hill equation as described previously.[1a] BI (KD= 51 nM), DFHBI-1T (KD= 305 nM). (c) kon and koff were calculated from the linear fit (solid line) of kobs vs. concentration at 25°C. The slope provides kon and the intercept gives koff. kon=14,900 M-1 s-1, koff=0.0036 s-1 for DFHBI-1T and kon=13,600 M-1 s-1, koff=0.0006 s-1. (d) The fluorescence of a solution containing 25 μM RNA and 500 nM BI or DFHBI-1T was measured in 1 mM MgCl2 buffer while gradually increasing the temperature. In the left image, the fluorescence is normalized so that both Broccoli-BI and Broccoli-DFHBI-1T start at 1.0. Broccoli-BI exhibited a ~12°C shift in the Tm relative to Broccoli-DFHBI-1T (left). On the right, the fluorescence is normalized by the fluorescence level of Broccoli-DFHBI-1T at 37°C. (e) Fluorescence intensity upon mixing Broccoli and BI or DFHBI-1T (final concentration: 25 μM Broccoli, 0.5 μM fluorophore) at 37°C (0.2 mM MgCl2 buffer). Upon adding BI to Broccoli, an initial (0–25 sec) rapid increase (k1=0.06771 s-1) in fluorescence was observed (expected binding based on a pseudo-first order binding reaction indicated as a dotted line, the actual observed fluorescence increase is shown in solid color). BI exhibited a continued increase in fluorescence consistent with a second mode of binding (k2=0.02756 s-1) with a plateau at ~200 sec. The rate constants (k1 and k2) were modeled by a single exponential fit in Origin software.
Although Broccoli folding has been measured in vitro[12], measurements of the percent of a folded RNA in cells is not straightforward. Nevertheless, our in vitro studies previously showed that Broccoli and Spinach are not completely folded when synthesized in vitro.[12] Furthermore, like most RNA aptamers, both Broccoli and Spinach are thermally unstable at 37°C, the imaging temperature for mammalian cells.[3, 6] Thus, misfolding limits brightness for fluorogenic aptamers in cells.
We therefore performed several tests to determine if BI influences the folding of Broccoli. First, we performed a thermal denaturation analysis of Broccoli (25 µM) in the presence of 0.5 µM of DFHBI-1T or BI (Figure 2d). Broccoli fluorescence was measured as the temperature was increased from 20°C and 37°C. While ~77% of Broccoli-DFHBI-1T fluorescence was lost at 37°C, only ~32% Broccoli-BI fluorescence was lost at 37°C. Overall, Broccoli-BI exhibited a ~12°C shift in the Tm relative to Broccoli-DFHBI-1T (Figure 2d, left). Importantly, BI exhibits ~6.4-fold higher fluorescence than DFHBI-1T at 37 °C (Figure 2d, right).
Overall, these data suggest that BI stabilizes Broccoli at 37°C, which therefore results in more Broccoli-BI complexes than Broccoli-DFHBI-1T complexes (Figure 2a). This effect, coupled with the moderate increase in the extinction coefficient and quantum yield relative to DFHBI-1T, may explain the ~10.5-fold increase in brightness of BI in Broccoli-expressing cells.
To further explore if additional mechanisms may mediate the effects of BI, we monitored the time-dependent acquisition of fluorescence after injection of 0.5 µM DFHBI-1T or BI into a cuvette containing a solution of 25 µM Broccoli at 37°C (Figure 2e). Since cells contain ~0.2 mM free Mg2+,[13] these experiments were performed at 37°C in buffer with 0.2 mM MgCl2, in contrast to the other in vitro experiments which used 1 mM MgCl2. As expected, there was a rapid phase of fluorescence increase, reflecting the on-rate of DFHBI-1T or BI. DFHBI-1T showed a plateau in fluorescence by ~25 sec. However, showed a second slow phase of fluorescence increase. The overall rate of fluorescence increase could be modeled with two rate constants, k1 and k2, with k1 representing the initial rate of BI binding, and k2 representing the rate of a further increase in fluorescence (Figure 2e). One possible explanation for this second phase of fluorescence increase is that BI binds a partially folded Broccoli RNA that is normally not able to activate BI fluorescence. However, BI may convert this into a properly folded form. Although a speculative model, this effect may contribute to the overall brightness of BI in Broccoli-expressing cells.
Overall, these data suggest that BI can promote and stabilize the folded form of Broccoli, which may account for the substantial fluorescence enhancement in cells.
Broccoli-BI complexes exhibit increased photostability
To determine if Broccoli-BI complexes also exhibit increased photostability, we measured total fluorescence in HEK293T cells during continuous irradiation in an epifluorescence microscope. As expected, DFHBI-1T (10 µM)-treated cells showed a 50% loss of fluorescence in ~0.6 sec (Figure 3a–b). In contrast, BI-treated cells exhibited increased photostability, with a 50% loss in fluorescence at ~2.9 sec (Figure 3a and 3b).
Figure 3. Broccoli-BI complexes exhibit increased photostability in vivo and in vitro.
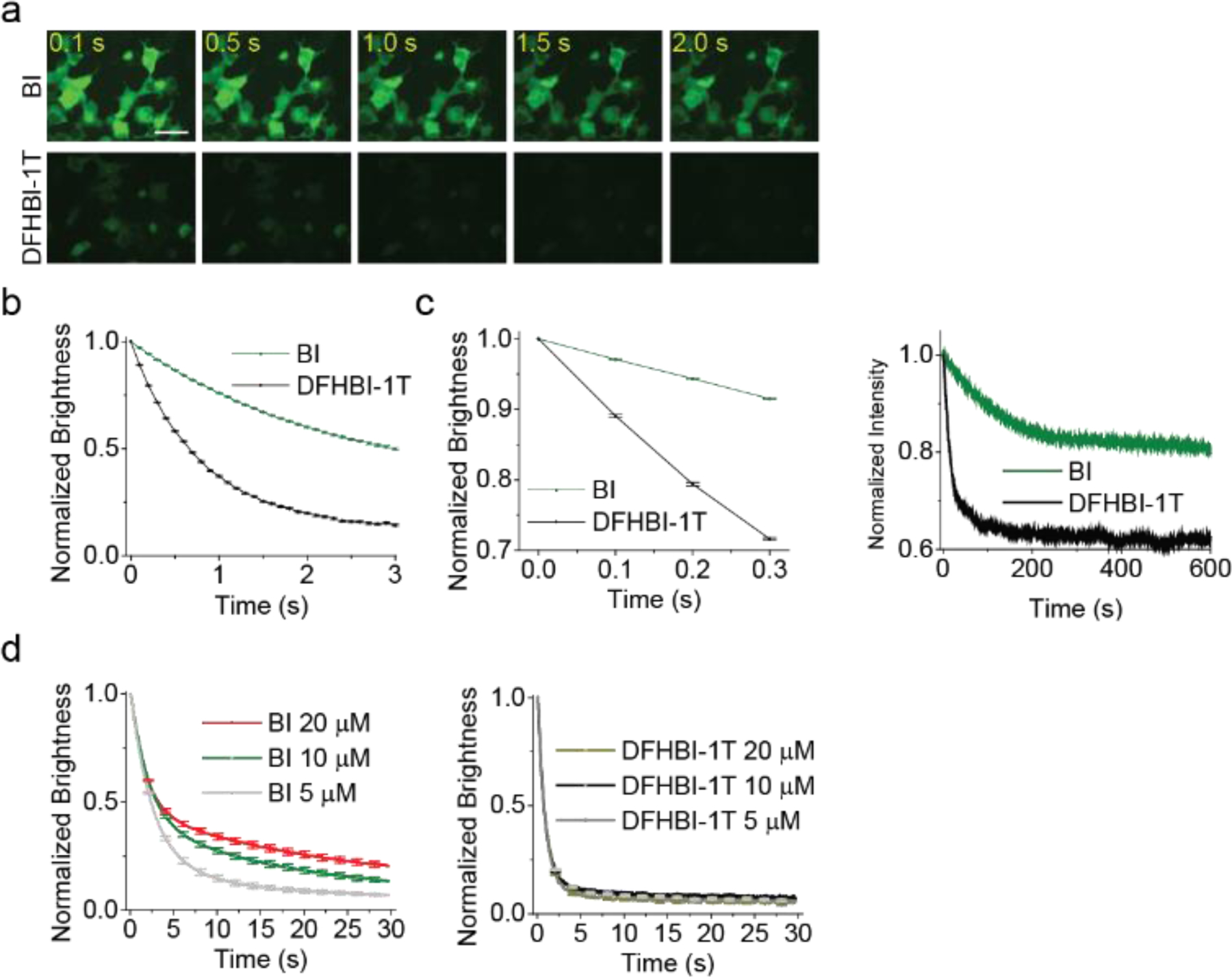
(a) In vivo photostability of Broccoli bound to flurophores (10 μM) was assessed by continuous imaging of Broccoli-expressing cells (100 ms per frame). Scale bar, 50 μm. (b) Quantification of in vivo photostability. Broccoli-DFHBI-1T loses 50% of signal within ~0.6 sec; Broccoli-BI lost 50% of signal within ~2.9 sec. Error bars indicate s.e.m. for n=10 cells per condition. (c) Broccoli-BI exhibits reduced photoisomerization compared to Broccoli–DFHBI-1T. Shown is the initial light-induced rate of fluorescence loss in cells (left) and in vitro (right). In the cellular experiment, flurophore concentration was 10 μM. In the in vitro experiments, we used solutions containing 0.1 μM BI or DFHBI-1T and 1 μM Broccoli. (d) To determine if trans-fluorophore unbinding is rate limiting, we measured photobleaching during continuous illumination of Broccoli-expressing HEK293 cells in a microscope. Fluorescence diminishes over time and reaches a plateau. If unbinding of trans-fluorophore is fast, then the rate-limiting step will be binding of cis-fluorophore from solution. To test this, we added increasing concentrations of fluorophore (5, 10, and 20 μM). Increasing the concentration of BI leads to a corresponding increase in the plateau of fluorescence, suggesting that binding of the fluorophore in solution is rate limiting, not unbinding of trans-BI. In contrast, adding higher concentrations of DFHBI-1T has no effect. This suggests that unbinding of trans-DFHBI is rate limiting. The brightness was computed by measuring the fluorescence signal in ten cells’ area and subtracting background based on ontrol cells not expressing Broccoli. Error bars indicate s.e.m. for n=10 cells per condition.
In both cases, a fluorescence plateau was reached (Figure S4), since fluorescence is restored when cis-fluorophore rebinds the aptamer after ejection of the trans- fluorophore.[5a]
We next asked if the increased photostability reflects reduced photoisomerization of BI. The photoisomerization rate is determined by measuring the initial rate of fluorescence loss upon irradiation[5b] (see Supplementary Note 1) in cells and in vitro. In cells irradiated using an epifluorescence microscope, the photoisomerization rate of Broccoli-BI complexes was 0.283 sec−1, which was ~3.3-fold slower than Broccoli-DFHBI-1T complexes (0.947 sec−1) (Figure 3c, left). In vitro, using a fluorimeter, the photoisomerization rate of Broccoli-BI complexes was 0.008 sec sec−1, which was ~5-fold slower than Broccoli-DFHBI-1T complexes (0.041 sec−1) (Figure 3c, right). Therefore, based on both in-cell and in vitro photostability experiments, BI is less susceptible to photoisomerization than DFHBI-1T when bound to Broccoli.
The other factor that contributes to Broccoli photobleaching is the unbinding rate of trans-DFHBI, the low-fluorescence form of the fluorophore.[5] If trans-fluorophore unbinding rate is slow, then fluorescence cannot be quickly restored by binding cis-fluorophore. In this case, the rate-limiting step for restoring fluorescence is the unbinding of trans-fluorophore. However, if trans-fluorophore unbinding is fast, then the rate-limiting step is the rate of binding of cis-fluorophore.
These two models can be distinguished by measuring if the plateau in fluorescence increases with increasing levels of fluorophore in solution.[5b] The plateau occurs when trans-fluorophore unbinding is balanced by rebinding of cis-fluorophore. By increasing the fluorophore concentration, the binding rate is increased in proportion to the concentration of fluorophore. Therefore, if the plateau in Broccoli fluorescence is increased with increased fluorophore concentration, it demonstrates that the binding rate of the cis-fluorophore is rate limiting. If the plateau is unaffected by the concentration of the fluorophore, then the trans-fluorophore unbinding is the rate-limiting step.
To test this, we measured the fluorescence of Broccoli in HEK293T cells cultured with increasing concentrations of fluorophore. When we tested increasing concentrations of BI, we observed an increase in the fluorescence plateau with increasing concentrations of BI (Figure 3d, left). In contrast, we did not see the change in the plateau with increasing concentrations of DFHBI-1T (Figure 3d, right). This suggests that BI and DFHBI-1T are fundamentally different: when BI is used, rebinding of cis-BI is rate limiting since trans-BI unbinds rapidly. In contrast, when DFHBI-1T is used, unbinding of the trans-DFHBI-1T is rate limiting, and therefore increasing the rebinding rate of cis-DFHBI-1T by increasing DFHBI-1T concentration has no effect.
Taken together, the overall increase in photostability of Broccoli-BI complexes reflects (1) its reduced susceptibility to photoisomerization and (2) its rapid unbinding upon photoisomerization, allowing Broccoli fluorescence to be restored quickly by cis-BI in solution. These findings explain the different plateaus seen when using BI and DFHBI-1T: since both DFHBI-1T and BI show similar rebinding rates (Figure 2c), the difference in the plateaus is due to their different photoisomerization rates and different rates of unbinding of the trans-fluorophore.
Notably, the plateau seen in vitro (Figure 3c, right) and in cells (Figure 3d) is different due to the different ways in which the experiments were performed. The in vitro experiments were performed in a cuvette in which only a small part of the sample in the cuvette is exposed the excitation light. In cells, fluorescence was tested in a microscope using a light source that irradiates the entire cell. Since this leads to photoisomerization throughout the entire cell, the loss of fluorescence is more substantial, leading to a lower plateau (Figure 3d).
A concatenated Broccoli tag for imaging single mRNA
We next wanted to determine if these brighter and more photostable Broccoli-BI complexes would enable imaging of individual mRNAs in mammalian cells. We developed an RNA tag comprising multiple consecutive Broccoli aptamers, analogous to mRNA imaging tags comprising 24–48 MS2 hairpins incorporated into mRNA 3’UTRs, which is currently the most common approach for single mRNA imaging.[14] In this approach, mRNAs become fluorescently tagged since the MS2 hairpins bind a MS2 RNA coat protein (MCP) fused to a fluorescent protein.[14b]
Each Broccoli aptamer was incorporated into F30, an RNA scaffold that promotes folding of Broccoli (Figure S5a and S6a).[15] We used “dimeric Broccoli” (dBroccoli), which comprises two Broccoli aptamers placed end-to-end (Figure S5a and S7a). Therefore, F30 with one dBroccoli in each arm contains the equivalent of four Broccoli aptamers (F30–2xdBroccoli) (Figure S5a).
Exact sequence repeats can cause recombination or deletion during plasmid propagation.[16] Additionally, identical hairpin sequences misfold due to “inter-aptamer hybridization”.[12b] We therefore assembled two separate F30–2xdBroccoli sequences with a different F30 variant and different Broccoli aptamers, along with different linkers between each Broccoli and F30 (Figure S6b, 7 and Tables S2–3).
For each tag tested, we used these two F30–2xdBroccoli sequences as a minimal unit, which contains 8 Broccoli aptamers. We then made tags comprising 1, 2, or 3 consecutive copies of this unit, i.e., (F30–2xdBroccoli)2, (F30–2xdBroccoli)4, and (F30–2xdBroccoli)6. The fluorescence intensity of the tag containing 24 Broccoli aptamers was 15.3-fold brighter than a single Broccoli (Figure S5b and S9). A previous study used 64 Spinach repeats but only achieved a 16-fold of fluorescence increase.[17] The use of the F30 folding scaffold and/or sequence variants for F30 and Broccoli may allow the fluorescence of the tag to more linearly correspond to the number of Broccoli aptamers.[12b]
We next expressed a mCherry mRNA with a 3’UTR tagged with (F30–2xdBroccoli)6 or (F30–2xdBroccoli)12, corresponding to 24 or 48 Broccoli aptamers (Figure S10). We first asked whether the Broccoli-tagged transcripts were full-length or degraded into Broccoli-containing intermediates. We performed northern blotting using total cellular RNA 48 h after transfection with a plasmid expressing the mCherry transcript tagged with increasing numbers of Broccoli aptamers. In each case, a single band corresponding to the full-length mRNA was detected (Figure S11).
We next imaged transcripts in living cells incubated with 10 µM DFHBI-1T or BI. No fluorescent puncta were seen in COS7 cells treated with DFHBI-1T (Figure S10b). However, when cells were treated with BI, we observed mobile fluorescent puncta with transcripts containing (F30–2xdBroccoli)6 or (F30–2xdBroccoli)12 (24 or 48 Broccoli equivalents) (Figure S10b and Supplemental Movie 1, 2). These data suggest that BI enables imaging of Broccoli-tagged mRNAs in mammalian cells.
Broccoli-tagged mRNAs exhibit physiologic localizations in live cells
We next asked if mRNA containing the concatenated Broccoli tags exhibit the expected cytoplasmic localization and mobility normally seen with mRNA in cells. For these experiments, we used a mCherry mRNA containing a β-actin 3’UTR sequence (Figure 4a), followed by a 24X Broccoli tag [(F30–2xBroccoli)6]. We used a 3’ UTR sequence from the β-actin transcript since this is commonly used for mRNA imaging by MS2-GFP system and confers targeting to neuronal dendrites.[14b] After the plasmid was transfected in COS7 cells, mobile puncta were readily detected in the cytosol, with a few puncta seen in the nucleus (Figure 4b). Similar results were seen with a 48X Broccoli tag [(F30–2xBroccoli)12] (Figure S12).
Figure 4. Imaging and localization of Broccoli-tagged β-actin mRNA in mammalian cells incubated with BI.
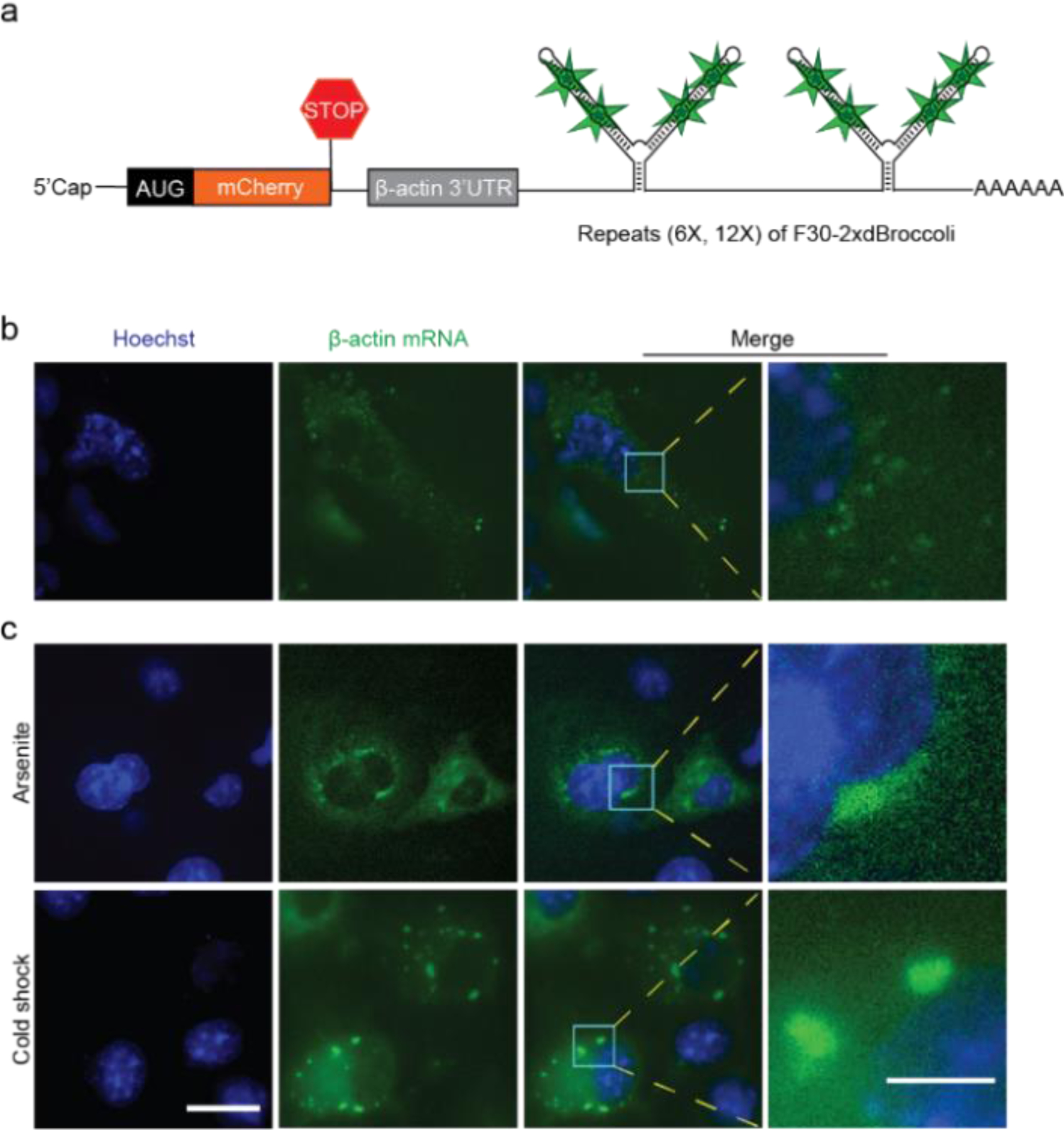
(a) Schematic of β-actin mRNA tagged with six repeated F30–2xdBroccoli units [(F30–2xdBroccoli)6] downstream of a portion of the 3’UTR of β-actin (shown in gray). (b) The (F30–2xdBroccoli)6-tagged β-actin mRNA was expressed in COS7 cells and imaged (10 μM BI) (c) supplemented with arsenite (500 μM, 60 minutes) or subjected to cold shock (10°C, 30 min) for induction of stress granules. Images were acquired after 1 h of arsenite treatment or 30 min of cold shock. Exposure times: 500 ms for FITC filter. Scale bars, 20 μm (Left), 5 μm (Right).
Next, we asked if Broccoli-tagged mRNA exhibits degradation rates normally expected for single mRNAs, which typically exhibit half-lives of just a few hours.[18] We therefore added actinomycin D, a transcription inhibitor. We found that nearly 50% of the puncta were lost at ~2.5 h, indicating a half-life that is consistent with single mRNA molecules (Figure S13).
We next imaged mRNA prior to and during stress granule formation induced by arsenite or cold shock. For these experiments, we used a mCherry mRNA containing a β-actin 3’UTR sequence, followed by a 24 Broccoli tag [(F30–2xBroccoli)6]. Prior to the application of arsenite or cold shock, fluorescent green puncta were observed throughout the cell. However, upon application of arsenite (500 µM) or cold shock (10°C), the puncta aggregated into larger foci consistent with stress granules within 60 min (Figure 4c). These data suggest that puncta in untreated cells may behave like single mRNAs that relocalize to granules in response to stress.
As a fourth test, we compared puncta generated using the Broccoli-BI system to puncta generated with the more common MS2-GFP system.[14b] Puncta seen when expressing MS2-tagged mRNA reflect single mRNA.[14a,16,19] To image these puncta, we co-transfected a plasmid expressing an mCherry mRNA containing 24X MS2 hairpins, and a plasmid expressing and GFP-MCP into U2OS cells. As expected, puncta were readily detectable in the cytoplasm (Figure S14).
We first compared the fluorescent puncta size of mRNAs tagged with the MS2 tag or the 24X Broccoli tag [(F30–2xBroccoli)6]. We found that the puncta size of most Broccoli-tagged mRNAs is similar to the puncta size of MS2-tagged mRNAs (Figure S15). Therefore, the size of fluorescent puncta observed using the Broccoli-BI method is consistent with single mRNAs.
We next used Gaussian analysis of puncta intensity, which has previously been used to determine if fluorescence signals represent single mRNAs.[20] We compared the intensity distribution of puncta, and found both MS2-tagged and Broccoli-tagged mRNA follow a single Gaussian distribution in puncta intensity, which again suggests that the puncta are predominantly single mRNA transcripts in cells (Figure S16).
Conclusions
In order to improve the photostability of Broccoli and Spinach in cells, we computationally screened a library of DFHBI derivatives for binding to Spinach, an RNA aptamer that is highly similar to Broccoli. One molecule, BI, exhibited marked increases in Broccoli fluorescence in cells. We find that the mechanism is due to an ability of BI to promote and stabilize folding of Broccoli at the relatively high (37°C) imaging temperatures. Broccoli-BI complexes also exhibit improved photostability, which reflects a reduced ability of BI to undergo light-induced cis-trans isomerization. We also find that BI is rapidly ejected when it is isomerized, allowing efficient replacement with cis-BI and restoration of fluorescence. These combined properties results in a markedly improved Broccoli imaging tag, allowing single mRNA to be imaged when tagged with 24 consecutive Broccoli aptamers in their 3’UTRs.
The unexpected and marked increase of in-cell fluorescence in BI-treated Broccoli-expressing cell is due to the ability of BI to stabilize the folded form of Broccoli. Our data suggests that Broccoli is not well folded in cells at 37°C. BI also appears to promote folding of Broccoli (see Figure 2e), suggesting that BI can bind a partially folded Broccoli intermediate and convert this Broccoli to the folded form. Thus, the major effect of BI is to increase the total amount of folded Broccoli in cells, thus explaining the large increase in intracellular fluorescence.
The ability of BI to stabilize Broccoli structure is likely due to the additional contacts that the benzimidazole makes with Spinach. The improved affinity of BI, compared to DFHBI-1T, supports the idea that BI makes additional contacts with Spinach or Broccoli.
The additional contacts of BI with Broccoli likely also explain the enhanced photostability of Broccoli-BI complexes. We find that the light-induced isomerization rate is slower, thus resulting in more fluorescence output before photobleaching.
BI also overcomes an additional problem of DFHBI: the slow off-rate of the photoisomerized trans-fluorophore. We find that trans-DFHBI-1T unbinding is slow and rate-limiting for the restoration of Broccoli fluorescence. In contrast, unbinding of trans-BI is rapid and not rate limiting. The bulky nature of the benzimidazole moiety may make the trans-BI unable to be accommodated in the Broccoli fluorophore binding pocket, causing its rapid ejection upon photoisomerization. This allows rebinding of cis-BI to be the rate-limiting step, rather than unbinding of trans-BI. Since rebinding is relatively rapid (kon=13,600 M−1 s−1), the Broccoli aptamer does not have long dark states caused by bound cis-BI, in contast to DFHBI-1T.
BI has other benefits, including small increases in extinction coefficient and quantum yield relative to DFHBI-1T, which likely further enhance brightness in cells.
Although we focused on Broccoli, the folding issues described here likely affect all RNA aptamers, including fluorogenic aptamers. Our data suggests that a potential solution is to design cognate fluorophores that can bind and promote aptamer folding in cells.
We also developed a 24X Broccoli array for tagging mRNA for imaging. This tag was optimized to reduce repetitive Broccoli sequences, F30 sequences, and linker sequences since repetitive sequences can cause misfolding and recombination and deletions during plasmid propagation. This tag can be used for labeling various mRNAs and imaging their fluorescence in BI-treated cells.
Like the commonly used 24X MS2 hairpin tag, the 24X Broccoli tag that can potentially alter the fate of an mRNA in the cell. Therefore, experiments using RNA imaging tags are best done by comparing two mRNAs that have the same tag, but differ by a putative functional element, or differ because of an experimental treatment that might affect mRNA behavior. Any result should then be validated with the endogenous transcript.
Although the 24X Broccoli tag is slightly longer than the 24X MS2 tag, Broccoli has the benefit of not recruiting fluorescent proteins to the mRNA. Additionally, since the fluorescent proteins usually have nuclear-localization elements, it is possible that the subsequent recruitment of ~24 nuclear localization elements to an mRNA might affect its trafficking behavior.[21] This problem would not be seen when using the Broccoli tag since no protein is recruited (Table S1). Imaging with Broccoli should not affect the detection of nuclear mRNAs, which are usually difficult to detect with the MS2 system due to nuclear-localized fluorescent proteins (Figure S14).
To the best of our knowledge, this is first time that single mRNA molecules have been imaged in mammalian cells using a fluorogenic aptamer and a small fluorophore. We expect that BI will facilitate the use of Broccoli as an imaging tag for mRNA and will be generally useful to increase fluorescence signals for diverse in-cell applications that use Broccoli.
Supplementary Material
Acknowledgments
We thank W. Zhan, S. Suter and J. Moon for useful comments and suggestions, R. Wang (Memorial Sloan-Kettering Cancer Center) for assistance with HRMS. This work was supported by NIH grant R35 NS111631 (S.R.J.).
Footnotes
Supporting information for this article is given via a link at the end of the document.
References
- [1].a) Song W, Filonov GS, Kim H, Hirsch M, Li X, Moon JD, Jaffrey SR, Nat. Chem. Biol 2017, 13, 1187–1194; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Paige JS, Wu KY, Jaffrey SR, Science 2011, 333, 642–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Warner KD, Chen MC, Song W, Strack RL, Thorn A, Jaffrey SR, Ferre-D’Amare AR, Nat. Struct. Mol. Biol 2014, 21, 658–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Strack RL, Disney MD, Jaffrey SR, Nature Methods 2013, 10, 1219–1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Braselmann E, Wierzba AJ, Polaski JT, Chromiński M, Holmes ZE, Hung S-T, Batan D, Wheeler JR, Parker R, Jimenez R, Gryko D, Batey RT, Palmer AE, Nat. Chem. Biol 2018, 14, 964–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].a) Wang P, Querard J, Maurin S, Nath SS, Le Saux T, Gautier A, Jullien L, Chemical Science 2013, 4, 2865–2873; [Google Scholar]; b) Han KY, Leslie BJ, Fei J, Zhang J, Ha T, J. Am. Chem. Soc 2013, 135, 19033–19038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Filonov GS, Moon JD, Svensen N, Jaffrey SR, J. Am. Chem. Soc 2014, 136, 16299–16308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Huang H, Suslov NB, Li N-S, Shelke SA, Evans ME, Koldobskaya Y, Rice PA, Piccirilli JA, Nat. Chem. Biol 2014, 10, 686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].a) Friesner RA, Banks JL, Murphy RB, Halgren TA, Klicic JJ, Mainz DT, Repasky MP, Knoll EH, Shelley M, Perry JK, Shaw DE, Francis P, Shenkin PS, J. Med. Chem 2004, 47, 1739–1749; [DOI] [PubMed] [Google Scholar]; b) Halgren TA, Murphy RB, Friesner RA, Beard HS, Frye LL, Pollard WT, Banks JL, J. Med. Chem 2004, 47, 1750–1759; [DOI] [PubMed] [Google Scholar]; c) Friesner RA, Murphy RB, Repasky MP, Frye LL, Greenwood JR, Halgren TA, Sanschagrin PC, Mainz DT, J. Med. Chem 2006, 49, 6177–6196. [DOI] [PubMed] [Google Scholar]
- [9].Litke JL, Jaffrey SR, Nat. Biotechnol 2019, 37, 667–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Filonov GS, Song W, Jaffrey SR, Biochemistry 2019, 58, 1560–1564. [DOI] [PubMed] [Google Scholar]
- [11].Song W, Strack RL, Svensen N, Jaffrey SR, J. Am. Chem. Soc 2014, 136, 1198–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].a) Strack RL, Song W, Jaffrey SR, Nat. Protoc 2014, 9, 146–155; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Filonov GS, Kam CW, Song W, Jaffrey SR, Chem. Biol 2015, 22, 649–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Dai L-J, Quamme GA, The Journal of clinical investigation 1991, 88, 1255–1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].a) Wu B, Eliscovich C, Yoon YJ, Singer RH, Science 2016, 352, 1430–1435; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Bertrand E, Chartrand P, Schaefer M, Shenoy SM, Singer RH, Long RM, Mol. Cell 1998, 2, 437–445. [DOI] [PubMed] [Google Scholar]
- [15].Filonov GS, Jaffrey SR, Curr. Protoc. Chem. Biol 2016, 8, 1–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Tutucci E, Vera M, Biswas J, Garcia J, Parker R, Singer RH, Nature Methods 2017, 15, 81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Zhang J, Fei J, Leslie BJ, Han KY, Kuhlman TE, Ha T, Sci. Rep 2015, 5, 17295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Yang E, van Nimwegen E, Zavolan M, Rajewsky N, Schroeder M, Magnasco M, Darnell JE, Genome Res 2003, 13, 1863–1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].a) Fusco D, Accornero N, Lavoie B, Shenoy SM, Blanchard J-M, Singer RH, Bertrand E, Curr. Biol 2003, 13, 161–167; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Grünwald D, Singer RH, Nature 2010, 467, 604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].a) Trcek T, Chao JA, Larson DR, Park HY, Zenklusen D, Shenoy SM, Singer RH, Nat. Protoc 2012, 7, 408; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Yamagishi M, Ishihama Y, Shirasaki Y, Kurama H, Funatsu T, Exp. Cell Res 2009, 315, 1142–1147; [DOI] [PubMed] [Google Scholar]; c) Park HY, Buxbaum AR, Singer RH, Methods Enzymol 2010, 472, 387–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Tyagi S, Nature Methods 2009, 6, 331. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


