Abstract
RNA-cleaving DNAzymes are widely applied as sensors for detecting metal ions in environmental samples owing to their high sensitivity and selectivity, but their use for sensing biological metal ions in live cells is challenging because constitutive sensors fail to report the spatiotemporal heterogeneity of biological processes. Photocaged DNAzymes can be activated by light for sensing purposes that need spatial and temporal resolution. Studying complex biological processes requires logic photocontrol, but unfortunately all the literature-reported photocaged DNAzymes working in live cells cannot be selectively controlled by light irradiation at different wavelengths. In this work, we developed photocaged DNAzymes responsive to UV and visible light using a general synthetic method based on phosphorothioate chemistry. Taking the Zn2+-dependent DNAzyme sensor as a model, we achieved wavelength-selective activation of photocaged DNAzymes in live human cells by UV and visible light, laying the groundwork for the logic activation of DNAzyme-based sensors in biological systems.
Introduction
Isolated through in vitro selection,1,2 DNAzymes, also called deoxyribozymes or catalytic DNA, are a class of DNA oligonucleotides with protein enzyme-like activities.3−7 A wide variety of reactions can be catalyzed by DNAzymes, including hydrolysis and formation of phosphodiester bonds in nucleic acids,2,8−25 oxidation,26,27 phosphorylation and dephosphorylation,28−33 thymine dimer repair,34 and peptide bond formation.35 Among them, RNA-cleaving DNAzymes often require metal ion cofactors for catalysis and have been widely applied as sensors to detect cognate metal ions with high selectivity.36−56 The detection process is also highly sensitive as signals can be amplified through the metal ion-dependent catalytic process. Signals generated by DNAzymes are analyzed by not only laboratory instruments but also portable devices to enable point-of-use applications.46,57−60 With continuous efforts on the selection of catalytic DNA sequences to cover more and more metal ions across the periodic table, DNAzyme-based sensors have become powerful tools for environmental analysis.50
Compared to in vitro detection, intracellular sensing of metal ions using DNAzymes can reveal the delicate changes of metal ions in cellular processes.61−66 Detection at the cellular or subcellular resolution in a target time window can be achieved by the selective release of sensor molecules. For example, photocaged DNAzymes67−71 containing light-labile or photoswitchable modifications72,73 have enabled the placement of RNA-cleaving activities with a high spatiotemporal resolution.61−66 The photocaging and decaging mechanism ensures that DNAzyme-based sensors are intact prior to their delivery into cells, and the subsequent sensing is only initiated after light irradiation in selected cells or during target cellular processes.74−77 Sequential activation of different DNAzymes can further allow the construction of biological logic gates for more complex and precise sensing applications.36,78 Although photolabile modifications sensitive to light of different wavelengths are readily available,79,80 the wavelength-selective activation of photocaged DNAzyme sensors has never been demonstrated in live cells.
Despite their diverse applications, many photocaged DNAzymes remain complicated to synthesize. Light-responsive modifications are typically incorporated during solid-phase synthesis using noncanonical phosphoramidites.81,82 Although the widely used UV-labile o-nitrophenyl (NP) modification is commercially available as internal or terminal photocleavable linkers in customized oligonucleotide synthesis, NP derivatization on nucleobases81 or ribose61 still needs solid-phase synthesis in laboratories by researchers. Moreover, incorporation of visible-light-labile groups such as 7-diethylaminocoumarin (DEACM) and nitrodibenzofuran requires sophisticated condensation and deprotection procedures.79,83 Therefore, a facile preparation scheme will allow a much broader group of researchers to access and utilize photocaged DNAzyme sensors in their studies.
Phosphorothioate (PS) is a backbone modification in which one of the nonbridging oxygen atoms of the nucleic acid phosphodiester linkage is replaced by a sulfur atom.84 Under mild conditions, PS can be postsynthetically derivatized using arylmethylbromide reagents to introduce many functional groups into oligonucleotides. As demonstrated previously by other researchers and us, chemically modified oligonucleotides85−92 and stimuli-responsive DNAzymes62,93,94 can be prepared using this PS chemistry. DNA oligonucleotides containing PS modifications are commercially available at low cost and can also be easily obtained following the standard solid-phase synthesis protocol simply by replacing the iodine oxidant with sulfurizing reagents.84 The reactions between PS and arylmethylbromide reagents are highly efficient and allow multiple PS sites in one oligonucleotide to be fully derivatized in a single reaction.85−92,94
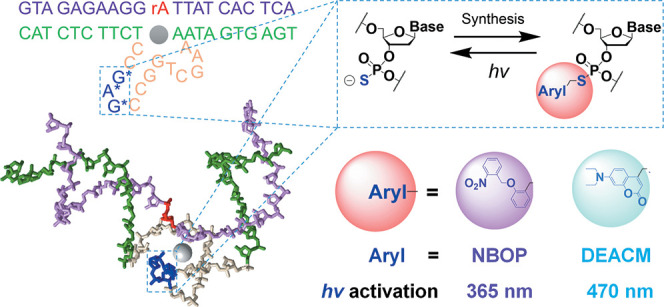
In this work, we synthesized two photocaged DNAzymes bearing 2-(2-nitrobenzyl)oxyphenyl (NBOP) and DEACM modifications through postsynthetic derivatization of PS (Figure 1a,b). We expected their sensor activities to be efficiently caged and wavelength-selectively decaged in live cells as the mechanisms shown in Figure 1c.
Figure 1.
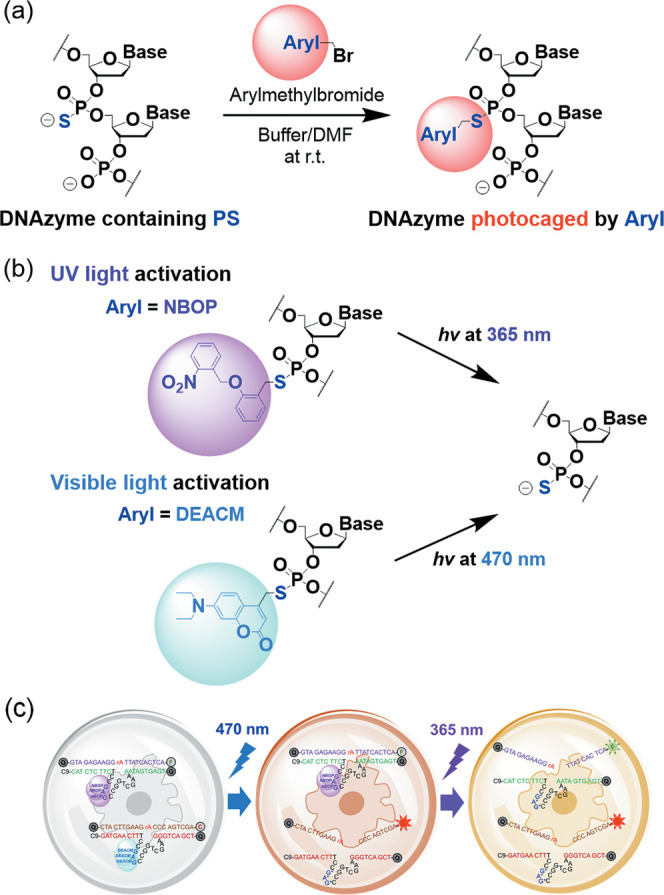
(a) General reaction between PS-containing DNAzyme and arylmethylbromide for synthesizing photocaged DNAzyme. Aryl = aromatic groups; DMF = N,N-dimethylformamide; and r.t. = room temperature (25 °C). (b) NBOP-modified DNAzyme activated by UV light at 365 nm and DEACM-modified DNAzyme activated by visible light at 470 nm. (c) Wavelength-selective activation of two DNAzymes caged by NBOP and DEACM modifications, respectively.
Results and Discussion
To facilitate wavelength-selective activation of DNAzymes, we chose NBOP and DEACM as the UV- and visible-light-labile modifications, respectively (Figure 1b). We envision that the installation of NBOP or DEACM in the catalytic core would neutralize the negative charges at the PS sites and introduce significant steric hindrance, both of which can perturb the folding and activity of DNAzymes, such as the Zn2+-dependent 8-17 DNAzyme (17Dz),95−97 thereby enabling effective photocaging (Figure 2a).
Figure 2.
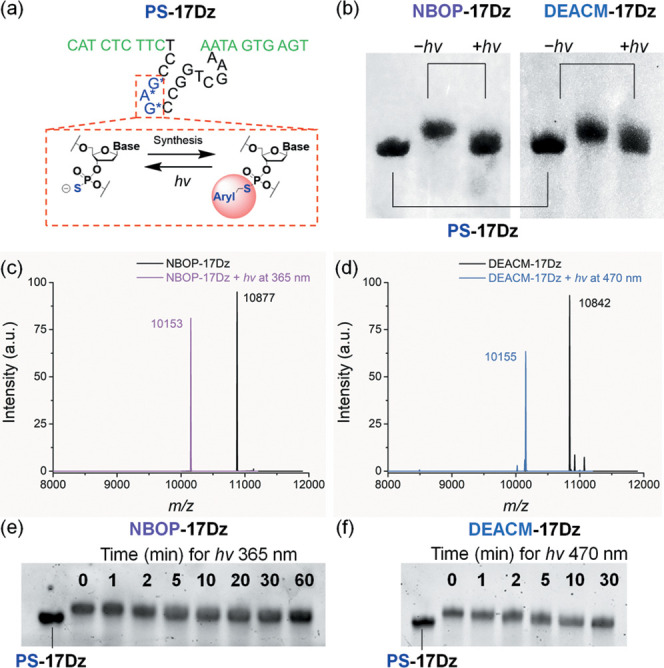
(a) Preparation of photocaged DNAzymes through the postsynthetic derivatization of PS and light-induced decaging. The binding arms of the DNAzyme are colored in green and the catalytic core is colored in black. The blue G*A*G* region contains PS modifications in their phosphorodiester linkages. For NBOP-17Dz, Aryl = NBOP; for DEACM-17Dz, Aryl = DEACM. All three PS sites are subjected to modification. (b) PAGE analyses of PS-17Dz, NBOP-17Dz, and DEACM-17Dz treated with or without light. (c) ESI-MS analyses of NBOP-17Dz before (m/z, found 10 877, NBOP-17Dz calcd 10 876) and after (m/z, found 10 153, PS-17Dz calcd 10 153) light activation. (d) ESI-MS analyses of DEACM-17Dz before (m/z, found 10 842, DEACM-17Dz calcd 10 840) and after (m/z, found 10 155, PS-17Dz calcd 10 153) light activation. (e) Time-dependent decaging of NBOP-17Dz by 365 nm light irradiation. (f) Time-dependent decaging of DEACM-17Dz by 470 nm light irradiation. For all analyses, 10 μM DNAzyme was prepared in sodium phosphate buffer (pH 7.0). Power of light irradiation: 365 nm at 26 mW/cm2 and 470 nm at 13 mW/cm2.
We took 17Dz as the model DNAzyme10,61,65,98 and reacted 17Dz carrying three PS modifications in its catalytic core (PS-17Dz) with 1-(bromomethyl)-2-((2-nitrobenzyl)oxy)benzene (NBOP-Br) and 4-bromomethyl-7-diethylaminocoumarin (DEACM-Br) as the arylmethylbromide reagents, respectively (Figures 1b and 2a). We used three consecutive modifications instead of one or two to fully abrogate the activity of the DNAzymes, according to our previous studies.62,94 When analyzed by polyacrylamide gel electrophoresis (PAGE), the reaction products showed slower electrophoretic mobility than the starting material (Figure 2b), indicating the successful installation of NBOP and DEACM. Electrospray ionization mass spectrometry (ESI-MS) further confirmed the complete derivatization of all the three PS sites (Figure 2c,d).
We then tested whether NBOP and DEACM can be fully removed by light irradiation. NBOP-modified 17Dz (NBOP-17Dz) was treated with light at a wavelength of 365 nm and a power output of 26 mW/cm2. PAGE characterization of the reaction revealed a band of similar mobility to PS-17Dz (Figure 2b), indicating successful light-induced decaging. ESI-MS confirmed that all three NBOP modifications were fully removed (Figure 2c). DEACM-modified 17Dz (DEACM-17Dz) was treated with light at a wavelength of 470 nm and a power output of 13 mW/cm2. Although the maximum absorption of DEACM is at 405 nm, the use of 470 nm is for the optimal balance between photolysis efficiency and selectivity over NBOP.79,83 Complete removal of three DEACM modifications was again confirmed by PAGE and ESI-MS (Figure 2b,d). The photodecaging reactions of both NBOP-17Dz and DEACM-17Dz were time dependent and almost went to completion within 10 min under the assayed conditions (Figure 2e,f).
To quantify the RNA-cleaving activities of the DNAzymes before and after activation, we synthesized a dual-labeled substrate 17S-FQ with a 5′ Iowa Black Quencher and 3′ fluorescein (Figure 3a). In the absence of 17S-FQ cleavage, the quencher is held in close proximity to the fluorophore in the single-stranded DNA, thereby effectively abrogating the fluorescent signal. With an active DNAzyme, cleavage will be introduced at the 3′ end of the ribonucleotide to physically separate the quencher and the fluorophore, significantly enhancing the fluorescence. The RNA-cleaving activities of the DNAzymes were therefore quantified by measuring the fluorescence output, where we considered the maximum fluorescence after the reaction as the 100% cleavage of 17S-FQ. Both 17Dz and PS-17Dz were used as positive controls to show the fast fluorogenic cleavage of 17S-FQ by fully active DNAzymes in the presence of Zn2+ as the cofactor metal ion (Figure 3b,c). In contrast, neither NBOP-17Dz nor DEACM-17Dz showed the RNA-cleaving activity under the same conditions (Figure 3b,c), confirming the successful caging of the sensor function. We then irradiated NBOP-17Dz and DEACM-17Dz with light at wavelengths of 365 and 470 nm, respectively, and measured their RNA-cleaving activities. Fast fluorescence generation was observed for the photocaged DNAzymes, confirming their light-triggered “turn on” (Figure 3b,c). The photocaged DNAzymes were not fully activated, as shown in Figure 3b,c, because of the shorter irradiation time compared with the study in Figure 2e,f, and the shorter irradiation conditions were actually used for cellular studies described in the later paragraphs. We also prepared caged 17Dz modified with NP, a widely used UV-labile group and tested the caging and decaging properties of NP-17Dz. Surprisingly, NP-17Dz was completely inactive, regardless of UV irradiation (Figure S1). The observation may suggest that NP attached to sulfur is more difficult to decage compared to that on oxygen or nitrogen, as reported in literature examples;72,99 therefore, NBOP is essential as an efficient UV-labile group for photocaging PS-containing oligonucleotides.
Figure 3.
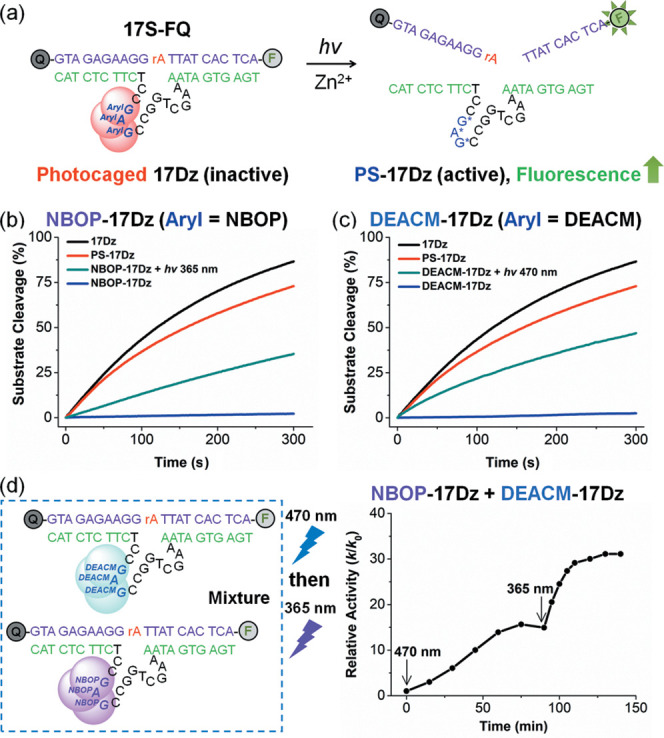
(a) Quantification of photocaged 17Dz’s sensor activity using 17S-FQ in the presence of Zn2+. The DNAzyme substrate 17S-FQ is colored in purple, with the ribonucleotide cleavage site highlighted in red. NBOP-17Dz and DEACM-17Dz can be decaged by light to cleave 17S-FQ. Aryl = NBOP or DEACM. (b and c) Cleavage of 17S-FQ by the indicated DNAzymes. To initiate the reaction, 250 nM 17S-FQ was mixed with 50 nM DNAzyme in 100 mM NaCl and 100 mM MOPS, pH 7.0 at 25 °C. Decaging of NBOP-17Dz was achieved by irradiation with light at 365 nm and 26 mW/cm2 for 5 min prior to incubation with 17S-FQ. DEACM-17Dz was activated by light irradiation at 470 nm and 13 mW/cm2 for 10 min. Active 17Dz and PS-17Dz were included as positive controls. Fluorescence intensities were measured at ex/em = 490/520 nm. The maximum fluorescence after the reaction using 17Dz to cleave 17S-FQ was considered as the intensity representing 100% substrate cleavage. (d) Wavelength-selective activation of NBOP-17Dz and DEACM-17Dz. In the reaction, 250 nM 17S-FQ was incubated with NBOP-17Dz and DEACM-17Dz, both at a concentration of 50 nM, in 100 mM NaCl and 100 mM MOPS, pH 7.0 at 25 °C. Sequential activation of DNAzyme’s Zn2+ sensing activity was achieved using visible and UV light. To obtain clear kinetics, about 25% power output was applied for light irradiation, which corresponds to 6.5 mW/cm2 for UV light and 3.5 mW/cm2 for visible light.
To uncover the mechanism of the photolysis reaction occurred on NBOP-17Dz and DEACM-17Dz, we synthesized their small-molecule mimics, NBOP-PS and DEACM-PS, by reacting diethylthiophosphate with the corresponding arylmethylbromide. The removal of NBOP and DEACM from NBOP-PS and DEACM-PS occurred quickly after light irradiation, yielding diethylthiophosphate as the final product (Figures S2–S7). To further study the photolysis reaction that occurred in oligonucleotides, we synthesized NBOP-modified 15-nt polyT DNA (NBOP-T15) and followed the decaging reaction by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS). We detected MS peaks corresponding to an intermediate o-hydroxybenzyl-T15 and the final product PS-15T after treating NBOP-T15 with UV light (Figure S8), which is consistent with our proposed decaging mechanism through the spontaneous 1,4-elimination (Figure S2).
Using both NBOP-17Dz and DEACM-17Dz, we investigated whether the sequential activation of the two photocaged DNAzymes could be achieved by light irradiation at different wavelengths selectively. As illustrated in Figure 3d, the mixture of NBOP-17Dz and DEACM-17Dz did not cleave 17S-FQ when incubated in the dark. As quantitative cleavage of the substrate occurred within minutes once the DNAzyme activity was unleashed by light using the original settings, we lowered the power output for light irradiation to 25% for more clear kinetics. Upon visible light irradiation at 470 nm, the fluorogenic reaction catalyzed by the activated DNAzyme became faster overtime, reaching a plateau with 15-fold rate enhancement (k/k0) in 60 min. Extended irradiation using light at 470 nm failed to further increase the rate, likely because after 60 min DEACM-17Dz had been fully decaged and the active DNAzyme released from DEACM-17Dz had reached the maximum concentration. We then switched the wavelength of light irradiation to 365 nm and observed that the rate of the fluorogenic reaction started to accelerate again, bringing the rate enhancement (k/k0) to 32-fold within 15 min. No more rate enhancement was observed beyond this point presumably because two photocaged DNAzymes were fully activated by the sequential irradiation of visible and UV light. The 15-fold and 17-fold rate enhancement (k/k0) achieved with visible and UV light irradiation, respectively, suggested minimal cross talk between the triggering mechanisms of DEACM-17Dz and NBOP-17Dz, which is critical to establish their wavelength-selective activation. We did not use 375 nm light irradiation as the first-step activation, because it is known that shorter-wavelength light usually activates longer-wavelength caging groups as well, which has been extensively reported in the literature and hardly avoidable in the experiment.72,79,83,99
With success in wavelength-selective decaging of NBOP-17Dz and DEACM-17Dz demonstrated in vitro, we proceeded to study their light activation inside live cells (Figure 4). The DNAzymes and the substrate were labeled at both 5′ and 3′ ends to minimize their nonspecific degradation in the intracellular environment (Figure 4a). A 4-(4-dimethylaminophenylazo) benzoic acid derivative (Dabyl) as the secondary quencher was installed at the 5′ end of the DNAzymes to further reduce the background fluorescence of the sensors in the absence of cleavage. We first evaluated the intracellular activity of PS-17Dz-DQ. Both 17S-FQ and PS-17Dz-DQ were delivered into HeLa cells through the lipid-mediated transfection. Fluorescence enhancement was detected by confocal microscopy when we supplemented cells with Zn2+ (Figure 4b), suggesting that 17S-FQ was stable in live cells with minimal background fluorescence and the activity of dual-labeled PS-17Dz-DQ was compatible with the cellular environment.
Figure 4.
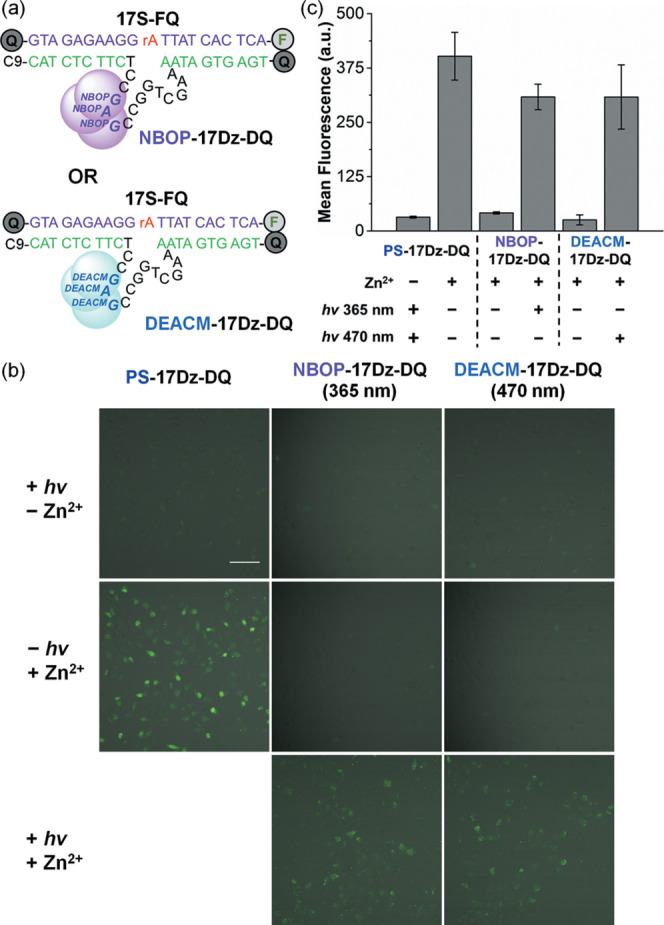
(a) Sequences and modifications of NBOP-17Dz-DQ, DEACM-17Dz-DQ, and 17S-FQ used in the cellular study. C9 is a Spacer 9 modification; F corresponds to fluorescein; Q refers to Dabyl or Iowa Black Quencher. (b and c) Confocal microscopy images (b) and flow cytometry analyses (c) of HeLa cells transfected with 17S-FQ and each of the DNAzymes including PS-17Dz-DQ, NBOP-17Dz-DQ, and DEACM-17Dz-DQ, before and after light irradiation. Fluorescence was only observed upon the cleavage of 17S-FQ by active DNAzyme sensors. Scale bar = 100 μm. For error bars, n = 3. Conditions for light irradiation: 365 nm at 26 mW/cm2 for 5 min and 470 nm at 13 mW/cm2 for 10 min.
Cells transfected with either NBOP-17Dz-DQ + 17S-FQ or DEACM-17Dz-DQ + 17S-FQ showed very weak fluorescence regardless of the Zn2+ supplement (Figure 4b), in agreement with the efficient photocaging effects of NBOP and DEACM modifications. When light irradiation was applied at 365 nm for NBOP-17Dz-DQ and 470 nm for DEACM-17Dz-DQ, strong fluorescent signals were observed, indicating the efficient decaging of the DNAzymes by UV and visible light (Figure 4b). We analyzed the cells transfected with both the substrate and the photocaged DNAzymes using flow cytometry (Figures 4c and S9). The cells carrying 17S-FQ and PS-17Dz-DQ showed mean fluorescence intensities of about 32 in the absence of Zn2+, which were boosted to about 402 once Zn2+ was supplied, reaching more than 12-fold enhancement. The mean fluorescence intensities of about 42 and 26 were detected in the cells transfected with NBOP-17Dz-DQ + 17S-FQ and DEACM-17Dz-DQ + 17S-FQ, respectively, in the presence of Zn2+, corroborating with our microscopy data that photocaged DNAzymes remained inactive prior to light irradiation. Fluorescent signals in these cells were boosted to about 308 and 307 when irradiated, respectively, corresponding to 7.3- and 12-fold activation (Figure 4c).
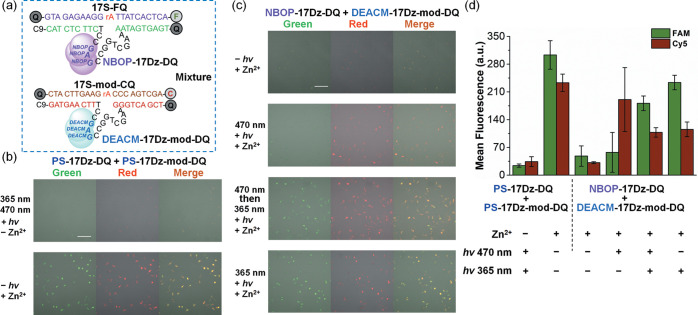
With the intracellular function of photocaged DNAzymes confirmed, we set to investigate whether NBOP-17Dz-DQ and DEACM-17Dz-DQ could be activated in a wavelength-selective manner inside the same cells (Figures 1c and 5a). To distinguish the signals generated by NBOP-17Dz-DQ and DEACM-17Dz-DQ, we modified the substrate-binding arms of the DEACM-caged DNAzyme (DEACM-17Dz-mod-DQ) to recognize a new substrate 17S-mod-CQ. Meanwhile, we switched the fluorophore in the substrate from fluorescein to Cy5 so that we can use different color channels to monitor the RNA-cleaving activities of NBOP-17Dz-DQ and DEACM-17Dz-DQ sensors, respectively. Both PS-17Dz-DQ + 17S-FQ and PS-17Dz-mod-DQ + 17S-mod-CQ displayed intense fluorescence enhancement in HeLa cells upon the addition of Zn2+ (Figure 5b), suggesting that the intracellular functions of the active DNAzyme sensors were not altered by different binding arm sequences or fluorophore modifications. We next cotransfected HeLa cells with NBOP-17Dz-DQ + 17S-FQ and DEACM-17Dz-mod-DQ + 17S-mod-CQ. Minimal fluorescent signals were detected regardless of Zn2+ admission, indicating that the two photocaged DNAzyme sensors were efficiently caged and inactive inside the cells. With visible light irradiation at 470 nm, strong fluorescent signals were detected in the red channel in response to Zn2+ (Figure 5c), suggesting the successful decaging of DEACM-17Dz-mod-DQ. In contrast, no fluorescence enhancement was detected in the green channel (Figure 5c), indicating that the NBOP-17Dz-DQ sensor remained caged under the assayed condition. These results confirmed the wavelength-selective activation of DEACM-17Dz-mod-DQ in the presence of intact NBOP-17Dz-DQ. Subsequent UV irradiation at 365 nm further unleashed green fluorescence by activating NBOP-17Dz-DQ (Figure 5c). The cells transfected with NBOP-17Dz-DQ + 17S-FQ and DEACM-17Dz-mod-DQ + 17S-mod-CQ lighted up in both red and green channels when directly irradiated with UV light, because UV light is of higher energy in nature and can decage both UV-labile NBOP and visible light-labile DEACM, as universally observed in previous photochemical studies.79,80 Flow cytometry analyses corroborated with the confocal microscopy results (Figures 5d and S10). Collectively, we achieved wavelength-selective activation of NBOP-caged and DEACM-caged DNAzyme sensors by sequential application of visible and UV light in live human cells.
Figure 5.
(a) Sequences and modifications of NBOP-17Dz-DQ, DEACM-17Dz-mod-DQ, 17S-FQ, and 17S-mod-CQ used in the cellular study. C is the Cy5 dye. Other labels are the same as those used in Figure 4. The binding arms of DEACM-17Dz-mod-DQ were modified to recognize the substrate 17S-mod-CQ and to eliminate cross talk with NBOP-17Dz-DQ + 17S-FQ. (b) Confocal images of HeLa cells transfected with PS-17Dz-DQ, PS-17Dz-mod-DQ, 17S-FQ, and 17S-mod-CQ under different assay conditions. Both PS-17Dz-DQ and PS-17Dz-mod-DQ were active in cleaving their cognate substrates to release fluorescent signals. (c) Sequential activation of NBOP-17Dz-DQ and DEACM-17Dz-mod-DQ by visible and UV light and global decaging of both DNAzyme sensors by UV light. Green and red fluorescence arose from the cleavage of 17S-FQ and 17S-mod-CQ, respectively, in the presence of active cognate DNAzyme sensors. Scale bars = 100 μm. For error bars, n = 3. Conditions for light irradiation: 365 nm at 56 mW/cm2 for 5 min and 470 nm at 20 mW/cm2 for 10 min. (d) Flow cytometer analyses of samples shown in (b) and (c).
Conclusions
We prepared photocaged DNAzyme sensors using a facile scheme through postsynthetic derivatization of PS. Using this method, we synthesized NBOP-17Dz and DEACM-17Dz for UV and visible light activations, respectively. The RNA-cleaving activities of two photocaged DNAzymes were activated upon light irradiation both in vitro and in live human cells. We also successfully demonstrated the wavelength-selective activation of NBOP-17Dz and DEACM-17Dz sensors in a sequential manner in the cells. To the best of our knowledge, this is the first example of photocaged DNAzyme sensors that can undergo wavelength-selective activation in live cells. In addition, the PS chemistry reported in this work can be generally applied to prepare chemically modified DNA oligonucleotides of other functionalities. With a facile preparation route, we envision that our work will enable many researchers to easily access and utilize chemically modified DNA oligonucleotides in their research, including, but not limited to, photocaged DNAzymes for logic control of biosensing and gene expression in live cells.
Acknowledgments
We are grateful for the financial support from the National Natural Science Foundation of China (nos. 21675097, 21621003, and 22074076). We thank Yanli Zhang at the Imaging Core Facility, Technology Center for Protein Science, Tsinghua University, for her assistance with confocal microscopy and Pengcheng Jiao at the Center of Biomedical Analysis, Tsinghua University, for his assistance with flow cytometry.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.1c00976.
Experimental details and additional figures (Figures S1–S14) (PDF)
Author Present Address
Lu Xiao – Stauffer I, Department of Chemistry, Stanford University, Stanford, CA 94305, USA.
Author Contributions
# X.X. and L.X. contributed equally.
Author Contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
The authors declare no competing financial interest.
Supplementary Material
References
- Bartel D. P.; Szostak J. W. Isolation of new ribozymes from a large pool of random sequences. Science 1993, 261, 1411–1418. 10.1126/science.7690155. [DOI] [PubMed] [Google Scholar]
- Breaker R. R.; Joyce G. F. A DNA enzyme that cleaves RNA. Chem. Biol. 1994, 1, 223–229. 10.1016/1074-5521(94)90014-0. [DOI] [PubMed] [Google Scholar]
- Breaker R. R. DNA enzymes. Nat. Biotechnol. 1997, 15, 427–431. 10.1038/nbt0597-427. [DOI] [PubMed] [Google Scholar]
- Fiammengo R.; Jaschke A. Nucleic acid enzymes. Curr. Opin. Biotechnol. 2005, 16, 614–621. 10.1016/j.copbio.2005.10.006. [DOI] [PubMed] [Google Scholar]
- Schlosser K.; Li Y. F. Biologically inspired synthetic enzymes made from DNA. Chem. Biol. 2009, 16, 311–322. 10.1016/j.chembiol.2009.01.008. [DOI] [PubMed] [Google Scholar]
- Silverman S. K. Deoxyribozymes: Selection design and serendipity in the development of DNA catalysts. Acc. Chem. Res. 2009, 42, 1521–1531. 10.1021/ar900052y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman S. K. Catalytic DNA: Scope, applications, and biochemistry of deoxyribozymes. Trends Biochem. Sci. 2016, 41, 595–609. 10.1016/j.tibs.2016.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breaker R. R.; Joyce G. F. A DNA enzyme with Mg2+-dependent RNA phosphoesterase activity. Chem. Biol. 1995, 2, 655–660. 10.1016/1074-5521(95)90028-4. [DOI] [PubMed] [Google Scholar]
- Carmi N.; Shultz L. A.; Breaker R. R. In vitro selection of self-cleaving DNAs. Chem. Biol. 1996, 3, 1039–1046. 10.1016/S1074-5521(96)90170-2. [DOI] [PubMed] [Google Scholar]
- Li J.; Lu Y. A highly sensitive and selective catalytic DNA biosensor for lead ions. J. Am. Chem. Soc. 2000, 122, 10466–10467. 10.1021/ja0021316. [DOI] [Google Scholar]
- Santoro S. W.; Joyce G. F.; Sakthivel K.; Gramatikova S.; Barbas C. F. RNA cleavage by a DNA enzyme with extended chemical functionality. J. Am. Chem. Soc. 2000, 122, 2433–2439. 10.1021/ja993688s. [DOI] [PubMed] [Google Scholar]
- Liu J. W.; Brown A. K.; Meng X. L.; Cropek D. M.; Istok J. D.; Watson D. B.; Lu Y. A catalytic beacon sensor for uranium with parts-per-trillion sensitivity and millionfold selectivity. Proc. Natl. Acad. Sci. U. S. A. 2007, 104, 2056–2061. 10.1073/pnas.0607875104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollenstein M.; Hipolito C.; Lam C.; Dietrich D.; Perrin D. M. A highly selective DNAzyme sensor for mercuric ions. Angew. Chem., Int. Ed. 2008, 47, 4346–4350. 10.1002/anie.200800960. [DOI] [PubMed] [Google Scholar]
- Gu H. Z.; Furukawa K.; Weinberg Z.; Berenson D. F.; Breaker R. R. Small, highly active DNAs that hydrolyze DNA. J. Am. Chem. Soc. 2013, 135, 9121–9129. 10.1021/ja403585e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang P.-J.-J.; Lin J.; Cao J.; Vazin M.; Liu J. Ultrasensitive dnazyme beacon for lanthanides and metal speciation. Anal. Chem. 2014, 86, 1816–1821. 10.1021/ac403762s. [DOI] [PubMed] [Google Scholar]
- Torabi S. F.; Wu P.; McGhee C. E.; Chen L.; Hwang K.; Zheng N.; Cheng J.; Lu Y. In vitro selection of a sodium-specific dnazyme and its application in intracellular sensing. Proc. Natl. Acad. Sci. U. S. A. 2015, 112, 5903–5908. 10.1073/pnas.1420361112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saran R.; Liu J. W. A silver DNAzyme. Anal. Chem. 2016, 88, 4014–4020. 10.1021/acs.analchem.6b00327. [DOI] [PubMed] [Google Scholar]
- Shen Z. F.; Wu Z. S.; Chang D. R.; Zhang W. Q.; Tram K.; Lee C.; Kim P.; Salena B. J.; Li Y. F. A catalytic DNA activated by a specific strain of bacterial pathogen. Angew. Chem., Int. Ed. 2016, 55, 2431–2434. 10.1002/anie.201510125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W. H.; Vazin M.; Yu T. M.; Ding J. S.; Liu J. W. In vitro selection of chromium-dependent DNAzymes for sensing chromium(III) and chromium(VI). Chem. - Eur. J. 2016, 22, 9835–9840. 10.1002/chem.201601426. [DOI] [PubMed] [Google Scholar]
- Wang Y. J.; Ngor A. K.; Nikoomanzar A.; Chaput J. C. Evolution of a general RNA-cleaving FANA enzyme. Nat. Commun. 2018, 9, 5067 10.1038/s41467-018-07611-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L.; Kartik S.; Liu B.; Liu J. From general base to general acid catalysis in a sodium-specific DNAzyme by a guanine-to-adenine mutation. Nucleic Acids Res. 2019, 47, 8154–8162. 10.1093/nar/gkz578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang P. J.; De Rochambeau D.; Sleiman H. F.; Liu J. Target self-enhanced selectivity in metal-specific DNAzymes. Angew. Chem., Int. Ed. 2020, 59, 3573–3577. 10.1002/anie.201915675. [DOI] [PubMed] [Google Scholar]
- Ren W.; Huang P.-J.-J.; De Rochambeau D.; Moon W. J.; Zhang J.; Lyu M.; Wang S.; Sleiman H.; Liu J. Selection of a metal ligand modified DNAzyme for detecting Ni2+. Biosens. Bioelectron. 2020, 165, 112285 10.1016/j.bios.2020.112285. [DOI] [PubMed] [Google Scholar]
- Cuenoud B.; Szostak J. W. A DNA metalloenzyme with DNA-ligase activity. Nature 1995, 375, 611–614. 10.1038/375611a0. [DOI] [PubMed] [Google Scholar]
- Flynn-Charlebois A.; Wang Y. M.; Prior T. K.; Rashid I.; Hoadley K. A.; Coppins R. L.; Wolf A. C.; Silverman S. K. Deoxyribozymes with 2′-5′ RNA ligase activity. J. Am. Chem. Soc. 2003, 125, 2444–2454. 10.1021/ja028774y. [DOI] [PubMed] [Google Scholar]
- Travascio P.; Li Y.; Sen D. DNA-enhanced peroxidase activity of a DNA aptamer-hemin complex. Chem. Biol. 1998, 5, 505–517. 10.1016/S1074-5521(98)90006-0. [DOI] [PubMed] [Google Scholar]
- Cheng X.; Liu X.; Bing T.; Cao Z.; Shangguan D. General peroxidase activity of G-quadruplex–hemin complexes and its application in ligand screening. Biochemistry 2009, 48, 7817–7823. 10.1021/bi9006786. [DOI] [PubMed] [Google Scholar]
- Li Y. F.; Liu Y.; Breaker R. R. Capping DNA with DNA. Biochemistry 2000, 39, 3106–3114. 10.1021/bi992710r. [DOI] [PubMed] [Google Scholar]
- Li Y.; Breaker R. R. Phosphorylating DNA with DNA. Proc. Natl. Acad. Sci. U. S. A. 1999, 96, 2746–2751. 10.1073/pnas.96.6.2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W.; Billen L. P.; Li Y. Sequence diversity, metal specificity, and catalytic proficiency of metal-dependent phosphorylating DNA enzymes. Chem. Biol. 2002, 9, 507–517. 10.1016/S1074-5521(02)00127-8. [DOI] [PubMed] [Google Scholar]
- Chandrasekar J.; Silverman S. K. Catalytic DNA with phosphatase activity. Proc. Natl. Acad. Sci. U. S. A. 2013, 110, 5315–5320. 10.1073/pnas.1221946110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh S. M.; Sachdeva A.; Silverman S. K. DNA catalysts with tyrosine kinase activity. J. Am. Chem. Soc. 2013, 135, 14928–14931. 10.1021/ja407586u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camden A. J.; Walsh S. M.; Suk S. H.; Silverman S. K. DNA oligonucleotide 3′-phosphorylation by a DNA enzyme. Biochemistry 2016, 55, 2671–2676. 10.1021/acs.biochem.6b00151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinnapen D. J. F.; Sen D. A deoxyribozyme that harnesses light to repair thymine dimers in DNA. Proc. Natl. Acad. Sci. U. S. A. 2004, 101, 65–69. 10.1073/pnas.0305943101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman S. K. Pursuing DNA catalysts for protein modification. Acc. Chem. Res. 2015, 48, 1369–1379. 10.1021/acs.accounts.5b00090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willner I.; Shlyahovsky B.; Zayats M.; Willner B. DNAzymes for sensing, nanobiotechnology and logic gate applications. Chem. Soc. Rev. 2008, 37, 1153–1165. 10.1039/b718428j. [DOI] [PubMed] [Google Scholar]
- Liu J. W.; Cao Z. H.; Lu Y. Functional nucleic acid sensors. Chem. Rev. 2009, 109, 1948–1998. 10.1021/cr030183i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B. L.; Dong S. J.; Wang E. K. Homogeneous analysis: Label-free and substrate-free aptasensors. Chem. Asian J. 2010, 5, 1262–1272. 10.1002/asia.200900660. [DOI] [PubMed] [Google Scholar]
- Li D.; Song S. P.; Fan C. H. Target-responsive structural switching for nucleic acid-based sensors. Acc. Chem. Res. 2010, 43, 631–641. 10.1021/ar900245u. [DOI] [PubMed] [Google Scholar]
- Ma D. L.; Chan D. S. H.; Man B. Y. W.; Leung C. H. Oligonucleotide-based luminescent detection of metal ions. Chem. Asian J. 2011, 6, 986–1003. 10.1002/asia.201000870. [DOI] [PubMed] [Google Scholar]
- Du Y.; Li B. L.; Wang E. K. “Fitting” makes “sensing” simple: Label-free detection strategies based on nucleic acid aptamers. Acc. Chem. Res. 2013, 46, 203–213. 10.1021/ar300011g. [DOI] [PubMed] [Google Scholar]
- Liang H.; Zhang X. B.; Lv Y. F.; Gong L.; Wang R. W.; Zhu X. Y.; Yang R. H.; Tan W. H. Functional DNA-containing nanomaterials: Cellular applications in biosensing, imaging, and targeted therapy. Acc. Chem. Res. 2014, 47, 1891–1901. 10.1021/ar500078f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y. T.; Ge B. X.; Sen D.; Yu H. Z. Functional DNA switches: Rational design and electrochemical signaling. Chem. Soc. Rev. 2014, 43, 518–529. 10.1039/C3CS60264H. [DOI] [PubMed] [Google Scholar]
- Gong L.; Zhao Z.; Lv Y.-F.; Huan S.-Y.; Fu T.; Zhang X.-B.; Shen G.-L.; Yu R.-Q. DNAzyme-based biosensors and nanodevices. Chem. Commun. 2015, 51, 979–995. 10.1039/C4CC06855F. [DOI] [PubMed] [Google Scholar]
- Wang F.; Liu X. Q.; Willner I. DNA switches: From principles to applications. Angew. Chem., Int. Ed. 2015, 54, 1098–1129. 10.1002/anie.201404652. [DOI] [PubMed] [Google Scholar]
- Li J.; Mo L. T.; Lu C. H.; Fu T.; Yang H. H.; Tan W. H. Functional nucleic acid-based hydrogels for bioanalytical and biomedical applications. Chem. Soc. Rev. 2016, 45, 1410–1431. 10.1039/C5CS00586H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan S.; Wu Y.; Wang L.; Zhan X.; Zhou P. A mini-review on functional nucleic acids-based heavy metal ion detection. Biosens. Bioelectron. 2016, 86, 353–368. 10.1016/j.bios.2016.06.075. [DOI] [PubMed] [Google Scholar]
- McGhee C. E.; Loh K. Y.; Lu Y. DNAzyme sensors for detection of metal ions in the environment and imaging them in living cells. Curr. Opin. Biotechnol. 2017, 45, 191–201. 10.1016/j.copbio.2017.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saidur M. R.; Aziz A. R. A.; Basirun W. J. Recent advances in DNA-based electrochemical biosensors for heavy metal ion detection: A review. Biosens. Bioelectron. 2017, 90, 125–139. 10.1016/j.bios.2016.11.039. [DOI] [PubMed] [Google Scholar]
- Zhou W. H.; Saran R.; Liu J. W. Metal sensing by DNA. Chem. Rev. 2017, 117, 8272–8325. 10.1021/acs.chemrev.7b00063. [DOI] [PubMed] [Google Scholar]
- Peng H. Y.; Newbigging A. M.; Wang Z. X.; Tao J.; Deng W. C.; Le X. C.; Zhang H. Q. DNAzyme-mediated assays for amplified detection of nucleic acids and proteins. Anal. Chem. 2018, 90, 190–207. 10.1021/acs.analchem.7b04926. [DOI] [PubMed] [Google Scholar]
- Zhang J. J. RNA-cleaving DNAzymes: Old catalysts with new tricks for intracellular and in vivo applications. Catalysts 2018, 8, 550. 10.3390/catal8110550. [DOI] [Google Scholar]
- Lake R. J.; Yang Z.; Zhang J.; Lu Y. DNAzymes as activity-based sensors for metal ions: Recent applications, demonstrated advantages, current challenges, and future directions. Acc. Chem. Res. 2019, 52, 3275–3286. 10.1021/acs.accounts.9b00419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L.; Liu J. Catalytic nucleic acids: Biochemistry, chemical biology, biosensors, and nanotechnology. iScience 2020, 23, 100815 10.1016/j.isci.2019.100815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConnell E. M.; Cozma I.; Morrison D.; Li Y. Biosensors made of synthetic functional nucleic acids toward better human health. Anal. Chem. 2020, 92, 327–344. 10.1021/acs.analchem.9b04868. [DOI] [PubMed] [Google Scholar]
- Zhao D.; Yin Q.; Chang Y.; Liu M. Nucleic acid circuits for cell imaging: From the test tube to the cell. Trends Anal. Chem. 2020, 122, 115706 10.1016/j.trac.2019.115706. [DOI] [Google Scholar]
- Mazumdar D.; Liu J. W.; Lu G.; Zhou J. Z.; Lu Y. Easy-to-use dipstick tests for detection of lead in paints using non-cross-linked gold nanoparticle-DNAzyme conjugates. Chem. Commun. 2010, 46, 1416–1418. 10.1039/b917772h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Y.; Lu Y. Using personal glucose meters and functional DNA sensors to quantify a variety of analytical targets. Nat. Chem. 2011, 3, 697–703. 10.1038/nchem.1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H. Z.; Li Y. C.; Ou L. M. L. Reading disc-based bioassays with standard computer drives. Acc. Chem. Res. 2013, 46, 258–268. 10.1021/ar300104b. [DOI] [PubMed] [Google Scholar]
- Huang Y. S.; Fang L. T.; Zhu Z.; Ma Y. L.; Zhou L. J.; Chen X.; Xu D. M.; Yang C. Y. Design and synthesis of target-responsive hydrogel for portable visual quantitative detection of uranium with a microfluidic distance-based readout device. Biosens. Bioelectron. 2016, 85, 496–502. 10.1016/j.bios.2016.05.008. [DOI] [PubMed] [Google Scholar]
- Hwang K.; Wu P. W.; Kim T.; Lei L.; Tian S. L.; Wang Y. X.; Lu Y. Photocaged DNAzymes as a general method for sensing metal ions in living cells. Angew. Chem., Int. Ed. 2014, 53, 13798–13802. 10.1002/anie.201408333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X. Y.; Feng M. L.; Xiao L.; Tong A. J.; Xiang Y. Postsynthetic modification of DNA phosphodiester backbone for photocaged DNAzyme. ACS Chem. Biol. 2016, 11, 444–451. 10.1021/acschembio.5b00867. [DOI] [PubMed] [Google Scholar]
- Wang W. J.; Satyavolu N. S. R.; Wu Z. K.; Zhang J. R.; Zhu J. J.; Lu Y. Near-infrared photothermally activated DNAzyme-gold nanoshells for imaging metal ions in living cells. Angew. Chem., Int. Ed. 2017, 56, 6798–6802. 10.1002/anie.201701325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z. K.; Fan H. H.; Satyavolu N. S. R.; Wang W. J.; Lake R.; Jiang J. H.; Lu Y. Imaging endogenous metal ions in living cells using a DNAzyme-catalytic hairpin assembly probe. Angew. Chem., Int. Ed. 2017, 56, 8721–8725. 10.1002/anie.201703540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z.; Loh K. Y.; Chu Y.-T.; Feng R.; Satyavolu N. S. R.; Xiong M.; Huynh S. M. N.; Hwang K.; Li L.; Xing H.; Zhang X.; Chelma Y. R.; Gruebele M.; Lu Y. Optical control of metal ion probes in cells and zebrafish using highly selective DNAzymes conjugated to upconversion nanoparticles. J. Am. Chem. Soc. 2018, 140, 17656–17665. 10.1021/jacs.8b09867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao R.; Xu L.; Hao C.; Xu C.; Kuang H. Circular polarized light activated chiral satellite nanoprobes for the imaging and analysis of multiple metal ions in living cells. Angew. Chem., Int. Ed. 2019, 58, 3913–3917. 10.1002/anie.201814282. [DOI] [PubMed] [Google Scholar]
- Liu Y.; Sen D. Light-regulated catalysis by an RNA-cleaving deoxyribozyme. J. Mol. Biol. 2004, 341, 887–892. 10.1016/j.jmb.2004.06.060. [DOI] [PubMed] [Google Scholar]
- Ting R.; Lermer L.; Perrin D. M. Triggering DNAzymes with light: A photoactive c8 thioether-linked adenosine. J. Am. Chem. Soc. 2004, 126, 12720–12721. 10.1021/ja046964y. [DOI] [PubMed] [Google Scholar]
- Keiper S.; Vyle J. S. Reversible photocontrol of deoxyribozyme-catalyzed RNA cleavage under multiple-turnover conditions. Angew. Chem., Int. Ed. 2006, 45, 3306–3309. 10.1002/anie.200600164. [DOI] [PubMed] [Google Scholar]
- Lusic H.; Young D. D.; Lively M. O.; Deiters A. Photochemical DNA activation. Org. Lett. 2007, 9, 1903–1906. 10.1021/ol070455u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards J. L.; Seward G. K.; Wang Y. H.; Dmochowski I. J. Turning the 10-23 DNAzyme on and off with light. ChemBioChem 2010, 11, 320–324. 10.1002/cbic.200900702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klan P.; Solomek T.; Bochet C. G.; Blanc A.; Givens R.; Rubina M.; Popik V.; Kostikov A.; Wirz J. Photoremovable protecting groups in chemistry and biology: Reaction mechanisms and efficacy. Chem. Rev. 2013, 113, 119–191. 10.1021/cr300177k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Protti S.; Ravelli D.; Fagnoni M. Wavelength dependence and wavelength selectivity in photochemical reactions. Photochem. Photobiol. Sci. 2019, 18, 2094–2101. 10.1039/C8PP00512E. [DOI] [PubMed] [Google Scholar]
- Hwang K.; Mou Q.; Lake R. J.; Xiong M.; Holland B.; Lu Y. Metal-dependent DNAzymes for the quantitative detection of metal ions in living cells: Recent progress, current challenges, and latest results on fret ratiometric sensors. Inorg. Chem. 2019, 58, 13696–13708. 10.1021/acs.inorgchem.9b01280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M.; Zhao J.; Chu H.; Mi Y.; Zhou Z.; Di Z.; Zhao M.; Li L. Light-activated nanoprobes for biosensing and imaging. Adv. Mater. 2019, 31, 1804745 10.1002/adma.201804745. [DOI] [PubMed] [Google Scholar]
- Samanta D.; Ebrahimi S. B.; Mirkin C. A. Nucleic-acid structures as intracellular probes for live cells. Adv. Mater. 2020, 32, e1901743 10.1002/adma.201901743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y.; Yang Z.; Lu Y. Photocaged functional nucleic acids for spatiotemporal imaging in biology. Curr. Opin. Chem. Biol. 2020, 57, 95–104. 10.1016/j.cbpa.2020.05.003. [DOI] [PubMed] [Google Scholar]
- Tregubov A. A.; Nikitin P. I.; Nikitin M. P. Advanced smart nanomaterials with integrated logic-gating and biocomputing: Dawn of theranostic nanorobots. Chem. Rev. 2018, 118, 10294–10348. 10.1021/acs.chemrev.8b00198. [DOI] [PubMed] [Google Scholar]
- Schafer F.; Joshi K. B.; Fichte M. A.; Mack T.; Wachtveitl J.; Heckel A. Wavelength-selective uncaging of dA and dC residues. Org. Lett. 2011, 13, 1450–1453. 10.1021/ol200141v. [DOI] [PubMed] [Google Scholar]
- Rodrigues-Correia A.; Weyel X. M. M.; Heckel A. Four levels of wavelength-selective uncaging for oligonucleotides. Org. Lett. 2013, 15, 5500–5503. 10.1021/ol402657j. [DOI] [PubMed] [Google Scholar]
- Liu Q. Y.; Deiters A. Optochemical control of deoxyoligonucleotide function via a nucleobase-caging approach. Acc. Chem. Res. 2014, 47, 45–55. 10.1021/ar400036a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seyfried P.; Heinz M.; Pinter G.; Kloetzner D.-P.; Becker Y.; Bolte M.; Jonker H. R. A.; Stelzl L. S.; Hummer G.; Schwalbe H.; Heckel A. Optimal destabilization of DNA double strands by single-nucleobase caging. Chem.-Eur. J. 2018, 24, 17568–17576. 10.1002/chem.201804040. [DOI] [PubMed] [Google Scholar]
- Menge C.; Heckel A. Coumarin-caged dG for improved wavelength-selective uncaging of DNA. Org. Lett. 2011, 13, 4620–4623. 10.1021/ol201842x. [DOI] [PubMed] [Google Scholar]
- Eckstein F. Phosphorothioates, essential components of therapeutic oligonucleotides. Nucleic Acid Ther. 2014, 24, 374–387. 10.1089/nat.2014.0506. [DOI] [PubMed] [Google Scholar]
- Fidanza J. A.; Ozaki H.; Mclaughlin L. W. Site-specific labeling of DNA sequences containing phosphorothioate diesters. J. Am. Chem. Soc. 1992, 114, 5509–5517. 10.1021/ja00040a004. [DOI] [Google Scholar]
- Lee J. H.; Wernette D. P.; Yigit M. V.; Liu J.; Wang Z.; Lu Y. Site-specific control of distances between gold nanoparticles using phosphorothioate anchors on DNA and a short bifunctional molecular fastener. Angew. Chem., Int. Ed. 2007, 46, 9006–9010. 10.1002/anie.200702569. [DOI] [PubMed] [Google Scholar]
- Tan L. H.; Xing H.; Lu Y. DNA as a powerful tool for morphology control, spatial positioning, and dynamic assembly of nanoparticles. Acc. Chem. Res. 2014, 47, 1881–1890. 10.1021/ar500081k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W.; Wang F.; Ding J.; Liu J. Tandem phosphorothioate modifications for DNA adsorption strength and polarity control on gold nanoparticles. ACS Appl. Mater. Interfaces 2014, 6, 14795–14800. 10.1021/am504791b. [DOI] [PubMed] [Google Scholar]
- Huang P. J. J.; Liu J. W. An ultrasensitive light-up Cu2+ biosensor using a new DNAzyme cleaving a phosphorothioate-modified substrate. Anal. Chem. 2016, 88, 3341–3347. 10.1021/acs.analchem.5b04904. [DOI] [PubMed] [Google Scholar]
- Guo Y.; Zhang J.; Ding F.; Pan G.; Li J.; Feng J.; Zhu X.; Zhang C. Stressing the role of DNA as a drug carrier: Synthesis of DNA-drug conjugates through grafting chemotherapeutics onto phosphorothioate oligonucleotides. Adv. Mater. 2019, 31, 1807533 10.1002/adma.201807533. [DOI] [PubMed] [Google Scholar]
- Zhang J.; Guo Y.; Ding F.; Pan G.; Zhu X.; Zhang C. A camptothecin-grafted DNA tetrahedron as a precise nanomedicine to inhibit tumor growth. Angew. Chem., Int. Ed. 2019, 58, 13794–13798. 10.1002/anie.201907380. [DOI] [PubMed] [Google Scholar]
- Guo Y.; Zhang J.; Pan G.; Choi C. H. J.; Wang P.; Li Y.; Zhu X.; Zhang C. Grafting multi-maleimides on antisense oligonucleotide to enhance its cellular uptake and gene silencing capability. Chem. Commun. 2020, 56, 7439–7442. 10.1039/D0CC02548H. [DOI] [PubMed] [Google Scholar]
- Feng M. L.; Ruan Z. Y.; Shang J. C.; Xiao L.; Tong A. J.; Xiang Y. Photocaged g-quadruplex DNAzyme and aptamer by post-synthetic modification on phosphodiester backbone. Bioconjug. Chem. 2017, 28, 549–555. 10.1021/acs.bioconjchem.6b00646. [DOI] [PubMed] [Google Scholar]
- Xiao L.; Gu C.; Xiang Y. Orthogonal activation of RNA-cleaving DNAzymes in live cells by reactive oxygen species. Angew. Chem., Int. Ed. 2019, 58, 14167–14172. 10.1002/anie.201908105. [DOI] [PubMed] [Google Scholar]
- Brown A. K.; Li J.; Pavot C. M. B.; Lu Y. A lead-dependent DNAzyme with a two-step mechanism. Biochemistry 2003, 42, 7152–7161. 10.1021/bi027332w. [DOI] [PubMed] [Google Scholar]
- Kim H. K.; Rasnik I.; Liu J. W.; Ha T. J.; Lu Y. Dissecting metal ion-dependent folding and catalysis of a single DNAzyme. Nat. Chem. Biol. 2007, 3, 763–768. 10.1038/nchembio.2007.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H. H.; Yu X.; Chen Y. Q.; Zhang J.; Wu B. X.; Zheng L. N.; Haruehanroengra P.; Wang R.; Li S. H.; Lin J. Z.; Li J. X.; Sheng J.; Huang Z.; Ma J. B.; Gan J. H. Crystal structure of an RNA-cleaving DNAzyme. Nat. Commun. 2017, 8, 2006 10.1038/s41467-017-02203-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J.; Zheng W. C.; Kwon A. H.; Lu Y. In vitro selection and characterization of a highly efficient Zn(II)-dependent RNA-cleaving deoxyribozyme. Nucleic Acids Res. 2000, 28, 481–488. 10.1093/nar/28.2.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelliccioli A. P.; Wirz J. Photoremovable protecting groups: Reaction mechanisms and applications. Photochem. Photobiol. Sci. 2002, 1, 441–458. 10.1039/b200777k. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.