Abstract
The DEFECTIVE EMBRYO AND MERISTEMS 1 (DEM1) gene encodes a protein of unknown biochemical function required for meristem formation and seedling development in tomato, but it was unclear whether DEM1’s primary role was in cell division or alternatively, in defining the identity of meristematic cells. Genome sequence analysis indicates that flowering plants possess at least two DEM genes. Arabidopsis has two DEM genes, DEM1 and DEM2, which we show are expressed in developing embryos and meristems in a punctate pattern that is typical of genes involved in cell division. Homozygous dem1 dem2 double mutants were not recovered, and plants carrying a single functional DEM1 allele and no functional copies of DEM2, i.e. DEM1/dem1 dem2/dem2 plants, exhibit normal development through to the time of flowering but during male reproductive development, chromosomes fail to align on the metaphase plate at meiosis II and result in abnormal numbers of daughter cells following meiosis. Additionally, these plants show defects in both pollen and embryo sac development, and produce defective male and female gametes. In contrast, dem1/dem1 DEM2/dem2 plants showed normal levels of fertility, indicating that DEM2 plays a more important role than DEM1 in gamete viability. The increased importance of DEM2 in gamete viability correlated with higher mRNA levels of DEM2 compared to DEM1 in most tissues examined and particularly in the vegetative shoot apex, developing siliques, pollen and sperm. We also demonstrate that gamete viability depends not only on the number of functional DEM alleles inherited following meiosis, but also on the number of functional DEM alleles in the parent plant that undergoes meiosis. Furthermore, DEM1 interacts with RAS-RELATED NUCLEAR PROTEIN 1 (RAN1) in yeast two-hybrid and pull-down binding assays, and we show that fluorescent proteins fused to DEM1 and RAN1 co-localize transiently during male meiosis and pollen development. In eukaryotes, RAN is a highly conserved GTPase that plays key roles in cell cycle progression, spindle assembly during cell division, reformation of the nuclear envelope following cell division, and nucleocytoplasmic transport. Our results demonstrate that DEM proteins play an essential role in cell division in plants, most likely through an interaction with RAN1.
Author summary
Up to half of the genes predicted from genome projects lack a known biological and biochemical function. Many of these genes are likely to play essential roles but it is difficult to reveal their function because minor changes in the genetic sequence can result in lethality and genetic redundancy can obscure analysis. Genome projects predict that flowering plants have at least two DEM genes that encode a protein of unknown cellular and biochemical function. In this paper, we use multiple combinations of dem mutants in Arabidopsis to show that DEM genes are essential for cell division and gamete viability. Interestingly, gamete viability depends not only on the number of functional copies of DEM genes in the gametes, but also on the number of functional copies of DEM genes in the parent plant that produces the gametes. We also show that DEM proteins interact with RAN, a highly conserved protein that controls cell division in all eukaryotic organisms.
Introduction
The life cycle of plants involves the alternation between a diploid sporophytic phase and a haploid gametophytic phase. In flowering plants, the sporophyte is the dominant form representing the adult stage of the life cycle [1]. Cell divisions that generate the sporophytic adult plant, including the flowers and reproductive tissue, are restricted to regions termed meristems [2]. During the reproductive phase, specialized cells within floral organs undergo meiosis to generate haploid spores [3,4]. These haploid spores undergo two or three rounds of mitosis to generate multicellular gametophytes, namely pollen (microgametophytes) and embryo sacs (megagametophytes), but only one cell within the female and male gametophytes contribute to the formation of the zygote during sexual reproduction. While the sporophytes and gametophytes differ vastly in size and patterns of gene expression, they share a requirement for cell division and regulation of the cell cycle. However, mutations that disrupt cell division are often more severe in meiosis and post-meiotic mitosis during gametophyte development in flowering plants [5,6].
Pollen are derived from microspore mother cells that undergo meiosis to generate tetrads of four haploid microspores in a process known as microsporogenesis. Then during microgametogenesis, each microspore undergoes two additional rounds of mitosis to generate mature pollen. The first round of mitosis during microgametogenesis is asymmetric and gives rise to a smaller generative cell enclosed within the larger vegetative cell of pollen. The vegetative cell ceases to undergo further cell division, whereas the generative cell subsequently undergoes a second round of mitosis, which results in the production of two sperm cells contained within the vegetative cell cytoplasm [3,7].
Each embryo sac is derived from a megaspore mother cell, which undergoes meiosis to produce a functional megaspore. However, only one of the four meiotic products gives rise to a functional megaspore, and the other three products of meiosis undergo programmed cell death and degenerate [4]. During megagametogenesis, the functional megaspore undergoes three rounds of mitosis followed by cellularization and differentiation to give rise to a seven-cell embryo sac consisting of an egg cell, two synergid cells, a di-haploid central cell and three antipodal cells. The di-haploid central cell of the megagametophyte is formed by the fusion of two haploid cells during cellularization [8]. Sporophytic tissue surrounds the megagametophyte in a structure termed the ovule. In Arabidopsis, there are multiple ovules contained within each carpel of the flower. After fertilization, each ovule develops into a seed [9].
In dicot plant species, such as tomato and Arabidopsis, there are two DEFECTIVE EMBRYO AND MERISTEMS (DEM) genes, which encode 72 kDa proteins of unknown biochemical function [10]. The DEM1 gene was first identified in tomato, where it is required for organized cell division in the shoot and root meristems [10]. In tomato, DEM1 is highly expressed in a punctate fashion in all meristematic and differentiating regions of the plant, and homozygous dem1 mutants exhibit a loss of the shoot apical meristem, disorganized arrangement of cell patterning in the root meristem and a seedling lethal phenotype [10]. However, it remained to be demonstrated whether DEM1’s primary role was in cell division or alternatively, in defining the identity of meristematic cells in plants.
Here, we show that DEM genes have an essential role in cell division in Arabidopsis. Furthermore, DEM interacts with RAS-RELATED NUCLEAR PROTEIN 1 (RAN1) in a yeast two-hybrid system and in pull-down binding assays, and fluorescent proteins fused to DEM1 and RAN1 co-localize transiently during meiosis and pollen development.
Results
DEM gene family and distantly related homologues
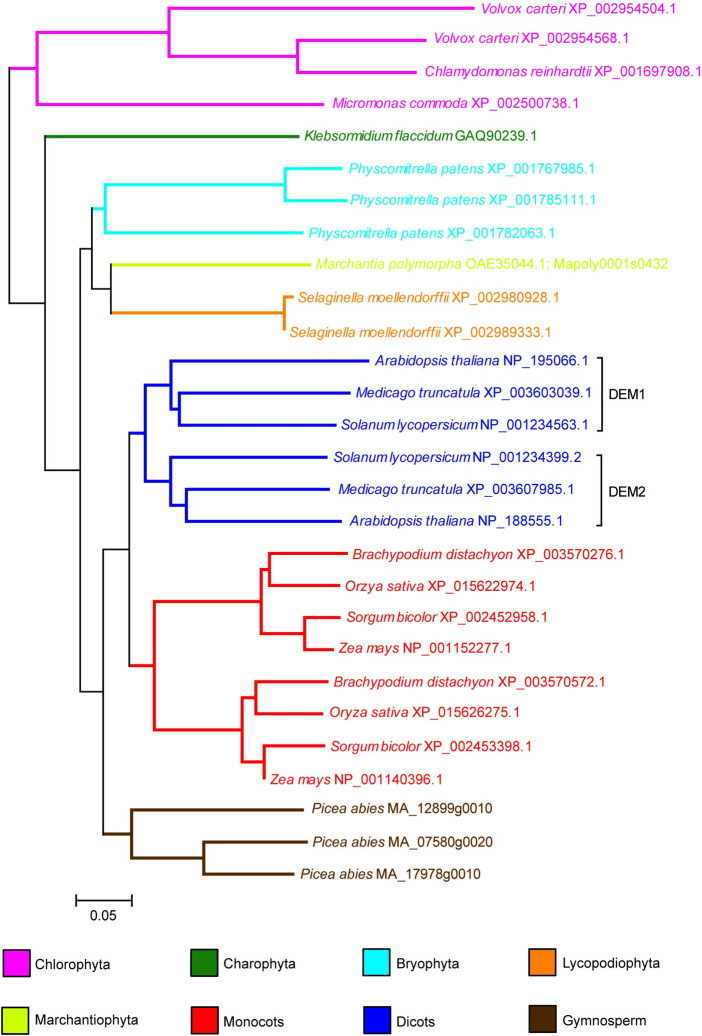
DEM1 and DEM2 are located on chromosome 4 and 2 of Arabidopsis, respectively, and encode proteins that are 63% identical and 79% similar. A phylogenetic tree constructed from an alignment of predicted plant DEM proteins largely mirrors the accepted phylogeny of plants (Fig 1). A gene duplication event in a common ancestor of extant eudicot species formed the DEM1 and DEM2 clades (Fig 1). An independent gene duplication event also occurred in the lineage leading to extant grasses (Fig 1). Similarly, the multiple paralogs of DEM in Physcomitrella (mosses) are more closely related to each other than to DEM proteins in flowering plants (Fig 1), indicating that the common ancestor of land plants possessed a single DEM homologue.
Fig 1. Phylogenetic tree of plant DEM proteins, rooted with the Chlorophyta sequences.
Software and parameters used in the analysis are described in Materials and Methods. The name of the organism is indicated on the tree, followed by the accession number or the Conifer Genome Integrative Explorer gene ID for Picea abies. Scale bar indicates 0.05 substitutions per site.
Database searches revealed the presence of protein sequences with significant similarity to DEM in a wide range of divergent eukaryota, including Protista, Rhodophyta (red algae) and fungi (S1 and S2 Figs). However, DEM-like sequences were not found in the metazoan species analysed except for Rhagoletis zephyria and Papilio xuthus, which belong to the Diptera and Lepidoptera orders, respectively, in the class Insecta (S1 and S2 Figs). Otherwise, most of the DEM protein sequences cluster into their respective taxonomic groups (S1 Fig). These results suggest that the last common ancestor of eukaryotes possessed a DEM gene that has been retained in many lineages but lost in the majority of the metazoans.
Plant DEM proteins are highly conserved throughout their entire length, but particularly in the C-terminal half of the proteins (S2 Fig), and DEM proteins usually contain a predicted N-terminal myristoylation site (S1 Table). Analysis of plant DEM proteins using InterPro [11] showed sequence similarity to the yeast Vacuolar Import and Degradation 27 (VID27) protein [12,13], which contains a predicted WD40/YVTN repeat-like domain. In addition, the area of high conservation between DEM and VID27 protein includes a region that functions as a nuclear rim localization domain in Schizosaccharomyces pombe (accession AB027933) (S3 Fig) [14].
DEM genes are expressed in tissues undergoing cell division
Previous studies using micro-arrays and RNA-seq have shown that both Arabidopsis DEM genes are predominantly expressed in differentiating tissue, but with DEM1 showing highest expression in the early-stage embryos, vegetative shoot apices, developing roots and floral buds, and DEM2 showing highest expression in developing seeds and pollen [15–17].
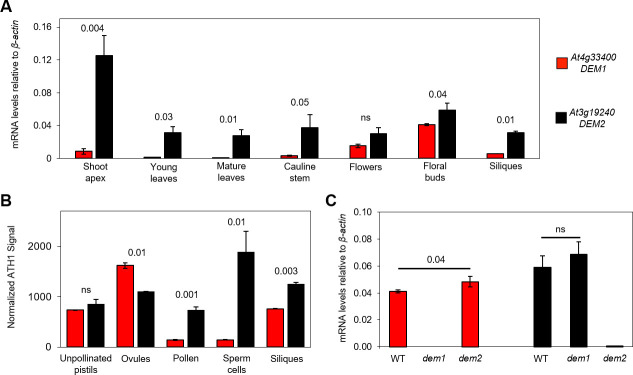
We used quantitative real-time reverse transcriptase PCR (qRT-PCR) to compare DEM1 and DEM2 mRNA levels across a range of tissues in Arabidopsis, and DEM2 mRNA levels were higher than DEM1 mRNA levels in all Arabidopsis tissues examined, although the differences weren’t significant in flowers (Fig 2A). DEM2 mRNA levels were highest in the vegetative shoot apex and floral buds but was detectable in all tissues tested, whereas DEM1 mRNA levels were highest in floral buds but also detectable in flowers, the vegetative shoot apex, siliques and cauline leaves (Fig 2A). Using published microarray data [18–20], we also compared the levels of DEM1 and DEM2 mRNA in reproductive tissues and FACS-purified sperm cells. This analysis showed that DEM2 mRNA levels were much higher than DEM1 mRNA levels in siliques, pollen and sperm cells (Fig 2B). DEM1 and DEM2 mRNA levels in microdissected ovules and unpollinated pistils were more comparable, although significantly higher DEM1 mRNA levels were detected in microdissected ovules (Fig 2B).
Fig 2. DEM1 and DEM2 mRNA levels in various tissues and sperm cells during vegetative and reproductive development.
(A) Quantitative real-time reverse transcriptase PCR (qRT-PCR) was used to examine DEM1 and DEM2 mRNA levels in total RNA extracted from a range of Arabidopsis tissues. Ratios of mRNA levels to that of β-actin mRNA are shown (average ± S.E. for three biological replicates). (B) Microarray-based expression analysis of DEM1 and DEM2 in Arabidopsis reproductive tissues and FACS-purified sperm cells. The figure is based on raw data published by Pina et al. [18], Borges et al. [19] and Boavida et al. [20], and shows mean signal intensities ± S.E. from ATH1 Genechips normalized by the invariant set method, as previously described [19]. Data were derived from two biological replicates for siliques [18], microdissected ovules and unpollinated pistils (UPs) [20], and three biological replicates for pollen and FACS-purified sperm cells [19]. (C) qRT-PCR analysis of DEM1 and DEM2 mRNA levels in total RNA extracted from Arabidopsis wild-type (WT), dem1-2 (dem1) and dem2-2 (dem2) floral buds. Ratios of mRNA levels to that of β-actin mRNA are shown (average ± S.E. for three to six biological replicates). P-values were calculated using one-way ANOVA to indicate significant differences between DEM1 and DEM2 expression in the different tissues and sperm cells of wild-type plants (A, B), or between floral buds of wild-type and dem plants (C).
We also used in situ hybridization to gain a more precise understanding of the expression patterns of DEM1 and DEM2 in differentiating tissues of Arabidopsis. Both genes showed a punctate expression pattern that was most obvious for DEM1 in developing embryos, meristems and differentiating tissues (Fig 3). During early stages of embryogenesis, a strong punctate expression pattern of DEM1 is distributed throughout the embryo, but particularly in cotyledon primordia (Fig 3A–3F). At later stages of embryogenesis, DEM1 expression is limited to regions of the shoot and root meristems and developing vasculature (Fig 3G and 3H). DEM1 mRNA was also observed in vegetative shoot meristems, developing anthers and ovules (Fig 3I, 3K and 3L), but was less detectable in inflorescence meristems (Fig 3J). DEM2 mRNA was also detected in inflorescence meristems, developing anthers and ovules, but was more diffuse throughout these tissues than DEM1 mRNA (Fig 3M and 3N).
Fig 3. DEM1 and DEM2 mRNA expression detected by in situ hybridization in developing embryos and post-embryonic shoot tissues of Arabidopsis ecotype Ws-0.
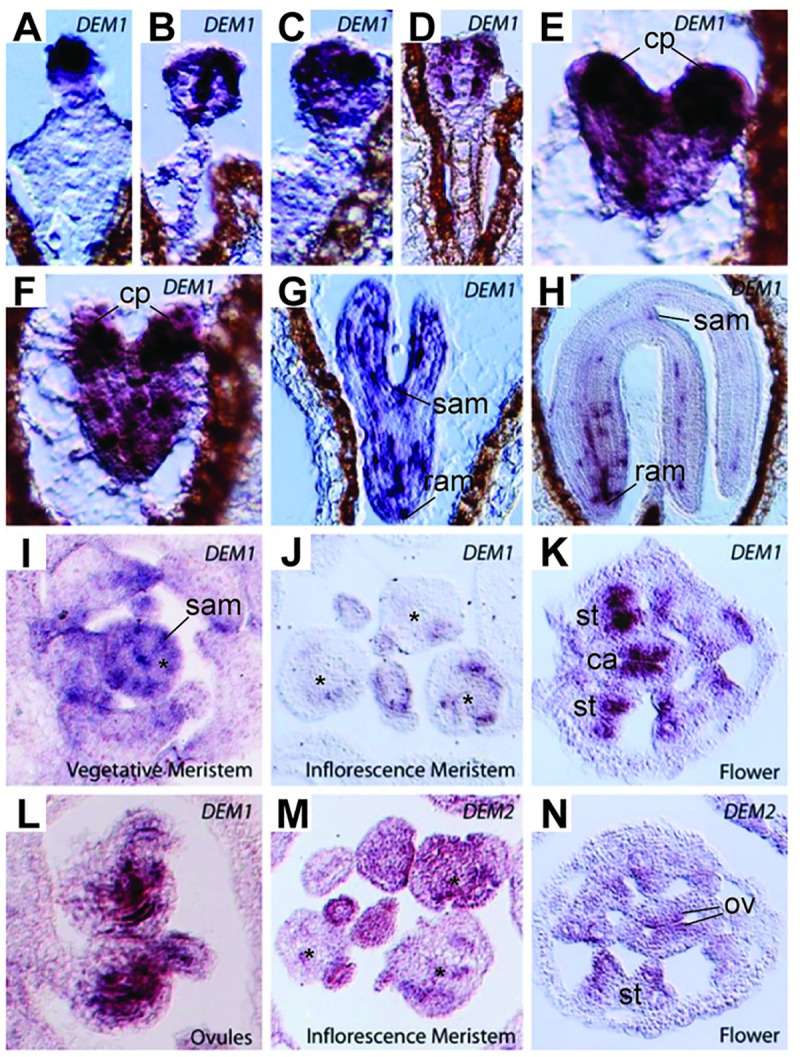
(A-H) Punctate expression of DEM1 in developing embryos. (A-D) Globular stage. (E-F) Heart stage. (G) Torpedo stage. (H) Maturing embryo. (I-J) DEM1 expression in shoot apices is not as clearly punctate as in developing embryos but is clearly expressed in vegetative shoot apical meristem and in some dividing cells. (I) Vegetative meristem. (J) Inflorescence meristem. (K-L) DEM1 expression can clearly be seen in sporophytic tissues of developing anthers and developing ovules. (K) Flower. (L) Ovules. (M-N) DEM2 is expressed in dividing cells of the inflorescence. (M) Inflorescence meristem. (N) Flower. cp, cotyledon primordia; ram, root apical meristem; sam, shoot apical meristem; *, central zone of meristem; st, stamen primordia; ca, carpel primordia; ov, developing ovules.
DEM genes are required for normal male meiosis and pollen viability
To characterize the biological function of the DEM genes in Arabidopsis, we analyzed the phenotype of independent dem1 and dem2 T-DNA insertion mutants in ecotypes Col-0 and Ws-0. Southern blot analysis confirmed the T-DNA insertions in DEM1 and DEM2 (S4A and S4B Fig), and northern blot analysis on floral bud tissue confirmed that dem1 and dem2 mutants in each ecotype lacked the respective mRNA transcript (S4C and S4D Fig). We used qRT-PCR on RNA extracted from floral buds to confirm that dem1 and dem2 mutants in the ecotype Col-0 genetic background lacked the corresponding mRNA transcript (Fig 2C). We also measured DEM1 and DEM2 expression in floral buds of the dem2 and dem1 mutants, respectively, to determine if mutation in one DEM gene could be compensated for by increased expression of the other DEM gene. There was a slight increase in the expression of the other DEM gene in both mutants, however, the increase was only statistically significant for DEM2 expression in the dem1 mutant (Fig 2C).
Plants homozygous for single dem1 or dem2 T-DNA insertion alleles in both Col-0 and Ws-0 ecotypes had no obvious phenotypic defects compared to the wild-type plants (S5A and S5B Fig). Subsequently, for both ecotypes, homozygous dem1 and dem2 plants were crossed to generate F1 plants heterozygous for both genes (DEM1/dem1 DEM2/dem2), and the F2 progeny and subsequent generations were characterized for phenotypic defects that correlated with the number of mutant dem alleles. Strikingly, no homozygous dem1 dem2 double mutants were observed in the F2 or subsequent generations. Furthermore, dem1/dem1 DEM2/dem2 plants were recovered in Col-0 but not in the Ws-0 genetic background. In total, we screened 153 F2 plants in the Ws-0 genetic background and failed to recover either homozygous dem1 dem2 double mutant plants or dem1/dem1 DEM2/dem2 plants.
DEM1/dem1 DEM2/dem2 and DEM1/dem1 dem2/dem2 plants in both genetic backgrounds, and dem1/dem1 DEM2/dem2 plants in the Col-0 background showed no obvious phenotypic defects prior to reproductive development. However, segregation analysis of progeny produced by self-fertilization of these plants showed distortion in favour of the wild-type DEM alleles (S2 and S3 Tables), suggesting that the dem1 dem2 genotype was transmitted through the male and/or female germline at a lower rate than other genotypes.
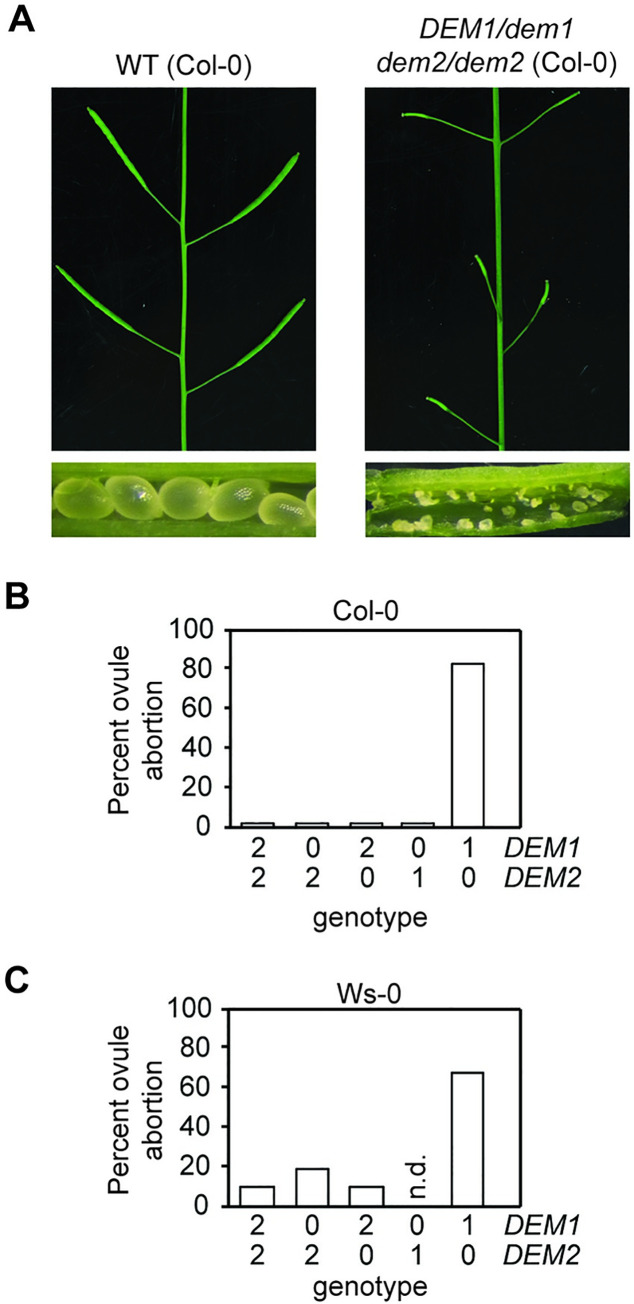
Among genotypes harboring multiple dem alleles, only DEM1/dem1 dem2/dem2 plants exhibited major defects in fertility and had smaller siliques compared to the wild-type plants (Fig 4A). Dissection of mature siliques from the DEM1/dem1 dem2/dem2 plants revealed ovule abortion rates of 60% and 80% in ecotypes Ws-0 and Col-0, respectively (Fig 4B and 4C), suggesting defects in pollen viability and/or ovule development. Indeed, the DEM1/dem1 dem2/dem2 plants showed a high pollen abortion rate of 70–80% (Fig 5A–5C). While self-fertilization of DEM1/dem1 dem2/dem2 plants produced a small amount of viable seed, these plants were completely ineffective in producing progeny when used as males in crosses to wild-type plants (Table 1), which was most likely due to a combination of the high pollen abortion rate, low viability of surviving pollen and ultimately, less viable pollen being attached to the stigma after manual crossing compared to self-pollination.
Fig 4. DEM1 and DEM2 are required for normal seed production in Arabidopsis.
(A-C) Plants harboring only one wild-type DEM1 allele (DEM1/dem1 dem2/dem2) in both the Col-0 and Ws-0 genetic backgrounds exhibited significant ovule abortion. (A) Comparison of wild-type (WT) and DEM1/dem1 dem2/dem2 siliques of Col-0 genetic background. Note the occurrence of degenerated ovules in DEM1/dem1 dem2/dem2 siliques compared to the appearance of normal seeds in WT siliques. (B-C) Percentage of aborted ovules in self-fertilized WT and dem genotypes for Col-0 (B) and Ws-0 (C) genetic backgrounds. At least 200 ovules were assayed, involving at least three separate plants for each genotype. X-axis of (B) and (C) represent the copy number of DEM1 and DEM2 wild-type alleles in each genotype. No dem1/dem1 DEM2/dem2 plants were recovered in the Ws-0 genetic background; n.d., not determined.
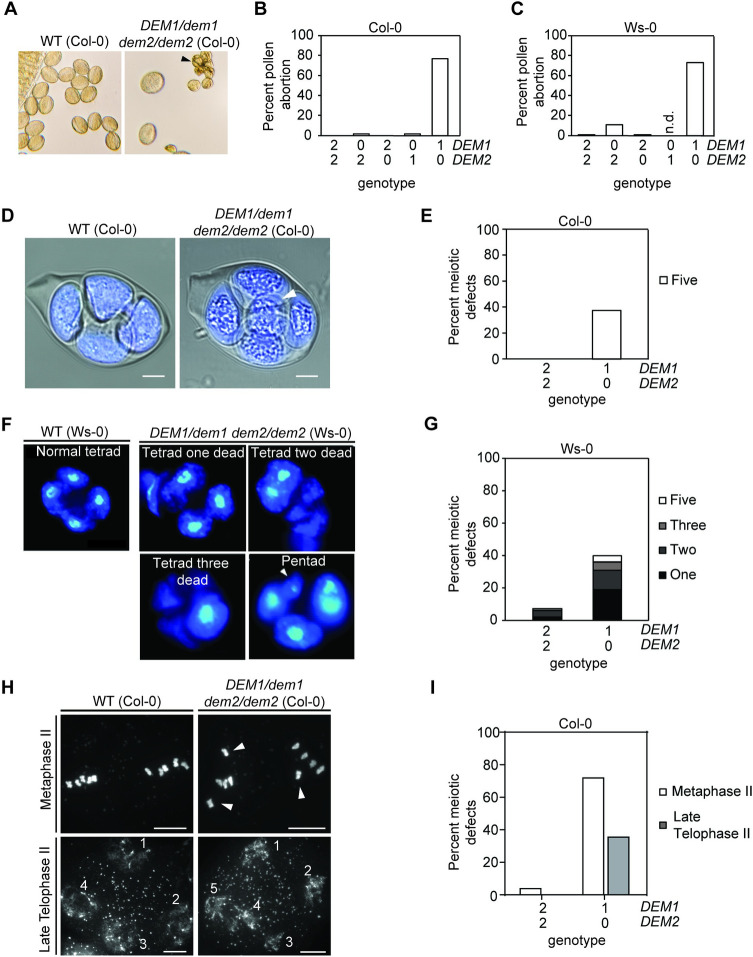
Fig 5. DEM1 and DEM2 are required for normal pollen development and male meiosis in Arabidopsis.
(A) Phenotype of pollen produced by wild-type (WT) and DEM1/dem1 dem2/dem2 plants in the Col-0 genetic background. Dead pollen grains are indicated by a black arrowhead. (B-C) Percentage of aborted mature pollen in WT and dem genotypes for the Col-0 (B) and Ws-0 (C) genetic backgrounds. At least 700 mature pollen in total were assayed, involving at least three separate plants for each genotype. n.d., not determined as dem1/dem1 DEM2/dem2 were not recovered in the Ws-0 genetic background. (D) Confocal image of DAPI-stained tetrads, combined with a DIC imaging to view membrane structures, shows an excess of meiotic products (i.e. a pentad) in a DEM1/dem1 dem2/dem2 plant (ecotype Col-0); an extra product of meiosis of diminished size is highlighted by the white arrowhead. Bar = 5 μm. (E) Percentage abnormal tetrads produced by WT and DEM1/dem1 dem2/dem2 plants in the Col-0 background. At least 80 meiotic events were scored, involving at least three separate plants for each genotype. (F) DAPI-stained defective male tetrads of DEM1/dem1 dem2/dem2 plants in ecotype Ws-0 showing one to three dead microspores, and an extra microspore of diminished size in a pentad (white arrowhead). (G) Percentage of defective tetrads in WT and DEM1/dem1 dem2/dem2 plants of ecotype Ws-0. At least 200 tetrads were assayed for each genotype. (H) DAPI-stained meiotic chromosome spreads revealed misaligned chromosomes (white arrowheads) away from the metaphase plate at metaphase II in DEM1/dem1 dem2/dem2 male meiocytes, and also shown is an example of five daughter cells produced at telophase II in DEM1/dem1 dem2/dem2 male meiocytes (ecotype Col-0). (I) Percentage of male meiocytes showing defects at metaphase II (misaligned chromosomes) and late telophase II (pentads) in wild-type and DEM1/dem1 dem2/dem2 plants in ecotype Col-0. At least 60 meiocytes were assayed, involving at least three separate plants for each genotype. X-axis of (B), (C), (E), (G), and (I) represent the copy number of DEM1 and DEM2 wild-type alleles in each genotype.
Table 1. Transmission of gametes depends on the dosage of functional DEM alleles in both the gametes and the plant that undergoes meiosis to produce the gametes.
Segregation data from crosses involving dem mutations in the two genetic backgrounds (Col-0 and Ws-0) were analyzed separately and together for significance. The expected segregation of genotypes assuming equal transmission of wild-type DEM and mutant dem alleles is listed in brackets. N, number of progeny scored; P, probability for a chi-square distribution with three degrees of freedom and an expected segregation ratio of 1:1:1:1, or with one degree of freedom and an expected segregation ratio of 1:1. For some crosses, chi-square probabilities were also determined for observed and expected segregation ratios for just dem1 DEM2 versus DEM1 DEM2 gametes, and these probabilities are listed in the third row of the table for these crosses.
| Cross (♀ x ♂) |
N | Genotype of gametes transmitted | P | |||
|---|---|---|---|---|---|---|
|
dem1 dem2 |
DEM1 dem2 |
dem1 DEM2 |
DEM1 DEM2 |
|||
| WT x DEM1/dem1 dem2/dem2a (Col-0) |
0a | n/a | n/a | n/a | n/a | n/a |
| WT x DEM1/dem1 DEM2/dem2 (Col-0) |
58 | 1 (14.5) |
17 (14.5) |
16 (14.5) (20) |
24 (14.5) (20) |
0.0002 0.21 |
| WT x DEM1/dem1 DEM2/dem2 (Ws-0) |
39 | 2 (9.75) |
12 (9.75) |
6 (9.75) (12.5) |
19 (9.75) (12.5) |
0.0007 0.009 |
| WT x DEM1/dem1 DEM2/dem2b (Col-0 and Ws-0) |
97 | 3 (24.25) |
29 (24.25) |
22 (24.25) (32.5) |
43 (24.25) (32.5) |
0.0001 0.009 |
| WT x DEM1/dem1 DEM2/DEM2 (Col-0) |
52 | n/a | n/a | 29 (26) |
23 (26) |
0.41 |
|
DEM1/dem1 dem2/dem2 x WT (Col-0) |
63 | 24 (31.5) |
39 (31.5) |
n/a | n/a |
0.06 |
|
DEM1/dem1 DEM2/dem2 x WT (Col-0) |
48 | 8 (12) |
11 (12) |
9 (12) (14.5) |
20 (12) (14.5) |
0.06 0.04 |
|
DEM1/dem1 DEM2/dem2 x WT (Ws-0) |
24 | 2 (6) |
5 (6) |
6 (6) (8.5) |
11 (6) (8.5) |
0.07 0.23 |
|
DEM1/dem1 DEM2/dem2 x WTb (Col-0 and Ws-0) |
72 | 10 (18) |
16 (18) |
16 (18) (23) |
30 (18) (23) |
0.007 0.04 |
|
DEM1/dem1 DEM2/DEM2 x WT (Col-0) |
33 | n/a | n/a | 16 (16.5) |
17 (16.5) |
0.86 |
a sterile as a male, no progeny was obtained from five separate experiments involving multiple DEM1/dem1 dem2/dem2 plants
bcombined data for Col-0 and Ws-0 backgrounds. n/a, not applicable.
It has been reported that up to 20% of T-DNA insertion mutants of Arabidopsis carry chromosomal translocations, and nearly all translocation lines when crossed to non-transgenic wild type show a pollen abortion rate of ~50% in heterozygous F1 plants [21]. To address the possibility that high pollen abortion rates in DEM1/dem1 dem2/dem2 plants may due to chromosomal translocations in the T-DNA insertion mutants, we assessed pollen abortion rates in double heterozygous DEM1/dem1 DEM2/dem2 F1 plants produced by crossing the single dem1 and dem2 mutants of Col-0. If chromosomal translocations were associated with either of the Col-0 T-DNA mutants, the pollen abortion rate would be expected to be at least 50% in the double heterozygous plants [21]. We assessed 717 pollen grains in total from three different DEM1/dem1 DEM2/dem2 Col-0 plants and found a pollen abortion rate of ~6% compared to the pollen abortion rate of ~70% for DEM1/dem1 dem2/dem2 Col-0 plants (Fig 5B). These data strongly indicate that the high pollen abortion rate observed in DEM1/dem1 dem2/dem2 plants was due to the dem mutations rather than the possibility of chromosomal translocations in the T-DNA mutant lines.
The pollen abortion rate of ~70% in DEM1/dem1 dem2/dem2 plants exceeded the expected number of dem1 dem2 pollen (i.e. 50%; Fig 5A–5C) and indicated that a significant portion of DEM1 dem2 pollen was also aborted. Furthermore, the low fertility of DEM1/dem1 dem2/dem2 plants suggested that the surviving mature pollen also had substantially reduced viability, and that the absence of a wild-type DEM2 allele in the DEM1/dem1 dem2/dem2 sporophyte adversely affected the viability of DEM1 dem2 pollen. Phenotypic analysis of additional dem genotypes revealed that pollen viability was not solely dependent on the pollen’s genotype but also the number of DEM2 alleles in the parent plant that undergoes meiosis. Specifically, segregation distortion against dem1 DEM2 pollen compared to DEM1 DEM2 pollen was observed for DEM1/dem1 DEM2/dem2 plants (P < 0.01) but not for DEM1/dem1 DEM2/DEM2 plants (P = 0.41; Table 1). Thus, in addition to the pollen genotype, pollen viability is dependent on the number of functional DEM2 alleles in the parent plant.
Confocal microscopy on DAPI-stained male tetrads from DEM1/dem1 dem2/dem2 plants of ecotype Col-0 revealed that 38% of tetrads were irregular and had an excess of meiotic products (Fig 5D and 5E). In DEM1/dem1 dem2/dem2 plants of ecotype Ws-0, tetrads with excessive meiotic products were observed, as well as degenerated tetrads with one or more dead microspores (Fig 5F and 5G).
To further investigate the role of DEM in male meiosis, we produced meiotic chromosome spreads of DAPI-stained DEM1/dem1 dem2/dem2 male meiocytes for ecotype Col-0. Interestingly, 72% of the daughter cells produced by meiosis I in DEM1/dem1 dem2/dem2 plants contained at least one chromosome that failed to align on the metaphase plate at metaphase II (Fig 5H and 5I). Furthermore, we confirmed that about a third of male meioses in DEM1/dem1 dem2/dem2 plants of ecotype Col-0 produced five rather than four daughter cells at late telophase II (Fig 5H and 5I). All other phases of meiosis I and II in these DEM1/dem1 dem2/dem2 plants were indistinguishable from wild type (S6 Fig).
As mentioned earlier, dem1/dem1 DEM2/dem2 plants were recovered in Col-0 but not in Ws-0 background, and these Col-0 plants were indistinguishable from the wild-type plants in terms of pollen (Fig 5B) and ovule abortion rates (Fig 4B). Thus, in the Col-0 background at least, a single functional DEM2 allele in dem1/dem1 DEM2/dem2 plants was sufficient for normal vegetative development, fertility and seed production.
In summary, our phenotypic analysis of male reproductive development showed that DEM genes are required for male meiosis, pollen development and male gamete viability, but DEM2 is more important than DEM1. This increased importance of DEM2 correlated with significantly higher levels of DEM2 mRNA compared to DEM1 mRNA in pollen and sperm cells, and in other tissues of the plant (Fig 2).
DEM genes are required for megaspore competitiveness and megagametophyte development
Crosses of DEM1/dem2 DEM2/dem2 females to wild-type males revealed that the dem1 dem2 genotype was transmitted through the female germline to the next generation two to five times less efficiently than DEM1 DEM2, depending on the Col-0 versus Ws-0 genetic background (Table 1). Furthermore, the DEM1 dem2 and dem1 DEM2 gametes were also transmitted through the female germline at a lower frequency than expected for equal segregation of wild-type and mutant alleles (Table 1). Combining the data for Col-0 and Ws-0 together, the results demonstrated that the segregation distortion against dem mutations through the female germline was highly significant (P < 0.01; Table 1). However, despite the reduced co-transmission of dem1 and dem2 alleles, a significant portion of dem1 dem2 genotypes were still transmitted to the next generation (Table 1). Furthermore, at a stage when ovules of wild-type plants were fertilized and contained developing embryos, self-fertilized DEM1/dem1 dem2/dem2 Ws-0 plants had approximately 50% of embryo sacs arrested before the first or second round of post-meiotic mitosis (Fig 6). Thus, high ovule abortion rate observed in DEM1/dem1 dem2/dem2 plants (Fig 4) is most likely due to defects in both pollen (Fig 5A–5C) and embryo sac viability (Fig 6).
Fig 6. DEM1 and DEM2 are required for normal megagametophyte development in Arabidopsis.
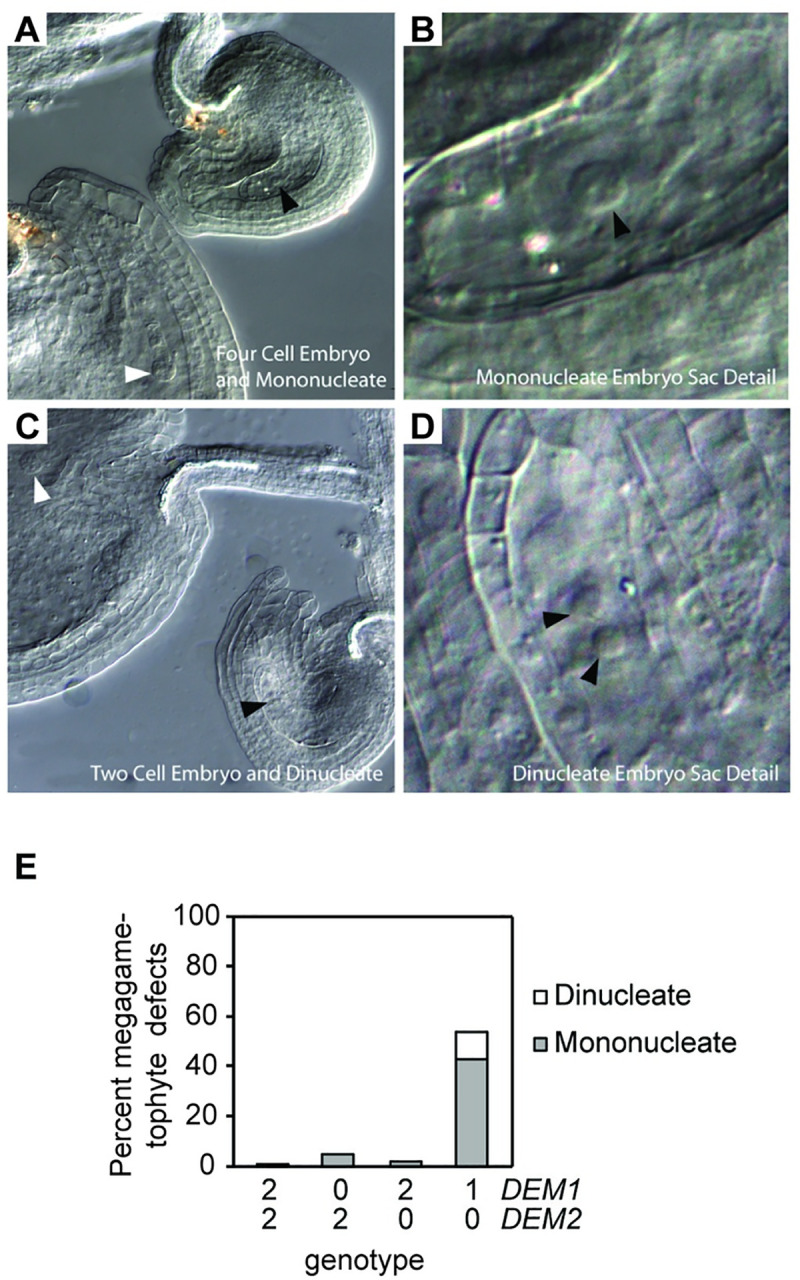
(A-D) Defective female gametophytes produced in DEM1/dem1 dem2/dem2 plants (Ws-0 genetic background). Developing ovules were observed as cleared whole mounts. (A-B) Four cell embryo (white arrowhead) and mononucleate embryo sac (black arrowhead) in two adjacent ovules. (C-D) Two cell embryo (white arrowhead) and di-nucleate embryo sac (black arrowheads) in two adjacent ovules. (E) Percentage of mononucleate and di-nucleate embryo sacs in self-fertilized wild-type and DEM1/dem1 dem2/dem2 plants of ecotype Ws-0. At least 200 ovules were assayed, involving at least three separate plants for each genotype. X-axis represents the copy number of DEM1 and DEM2 wild-type alleles in each genotype.
As was the case for pollen, there was a significant segregation distortion against dem1 DEM2 female gametes compared to DEM1 DEM2 female gametes for DEM1/dem1 DEM2/dem2 plants (P < 0.05) but not for DEM1/dem1 DEM2/DEM2 plants (P = 0.86; Table 1). Therefore, megagametophyte viability is also dependent on the number of functional wild-type DEM2 alleles in both the megagametophyte and in the parent plant that undergoes meiosis to produce the megagametophyte.
Transgenic complementation of gamete viability defects
Despite the PCR genotyping of many hundreds of plants, we never recovered homozygous dem1 dem2 double mutants, most likely due to substantially reduced viability of dem1 dem2 gametes and/or a potentially lethal phenotype of homozygous dem1 dem2 double mutant embryos. As mentioned earlier, dem1/dem1 DEM2/dem2 Col-0 plants showed almost no pollen abortion (Fig 5B), however when self-fertilized, the progeny showed strong segregation distortion in favour of the wild-type DEM2 allele (S3 Table; P < 0.05). To demonstrate transgenic complementation of the gamete viability defects associated with dem mutants, we introduced a wild-type DEM1 transgene into the dem1/dem1 DEM2/dem2 mutant of Col-0. Initially, we transformed the homozygous dem1 mutant with binary vector pUQC10043 carrying the DEM1 transgene linked to the selectable marker gene BAR (S10 Fig), which confers resistance to the herbicide Basta. A T1 plant carrying the DEM1 transgene was selected and crossed to the homozygous dem2 mutant. In the F2 generation from this cross, we identified a dem1/dem1 DEM2/dem2 plant carrying the DEM1 transgene, and then screened the F3 progeny of this plant for Basta resistant dem1 dem2 double mutants. Five of the 23 Basta-resistant F3 progeny that we genotyped with PCR tests were homozygous for both the dem1 and dem2 mutations. The segregation ratio of 5:18 for homozygous dem1 dem2 mutants to other genotypes (dem1/dem1 DEM2/dem2 or dem1/dem1 DEM2/DEM2) fits a 1:3 segregation ratio expected for Mendelian inheritance (P = 0.72). Thus, the DEM1 transgene complemented the gamete viability defects in the dem1/dem1 DEM2/dem2 mutant and allowed the recovery of dem1 dem2 double mutants.
The dem1 dem2 double mutant plants carrying the DEM1 transgene were confirmed by Southern blot analysis (S4B Fig), and these plants showed a very similar phenotype to DEM1/dem1 dem2/dem2 plants (Figs 4, 5B and 5C), i.e., normal vegetative development (S5A Fig) and a high pollen and ovule abortion rate (S5B and S5C Fig). These results suggest that the DEM1 transgene could compensate for the absence of DEM1 but not the absence of DEM2 in the dem1 dem2 double mutant.
DEM and RAS-RELATED NUCLEAR PROTEIN 1 (RAN1) interact in vitro and co-localize during male meiosis and pollen development
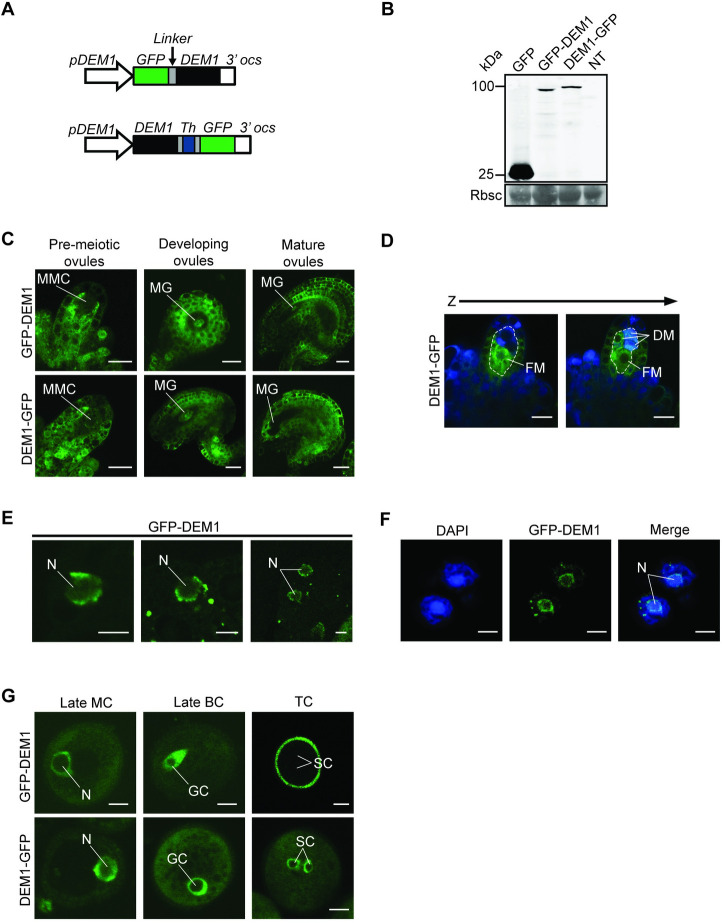
To investigate the intracellular localization of DEM proteins, transgenic lines of Arabidopsis were produced that expressed pDEM1:GFP-DEM1 or pDEM1:DEM1-GFP (Fig 7A). These transgenic lines expressed the full-length fusion proteins of ~100 kDa in size (Fig 7B). Both pDEM1:GFP-DEM1 and pDEM1:DEM1-GFP were expressed in all developing tissues examined, including developing pollen and ovules (Fig 7C–7G) and differentiating root tips (S7 Fig). Furthermore, the fusion proteins localized to cytoplasm and around the nuclear envelope, where they overlapped with DAPI staining of nuclear DNA (Figs 7D, 7F and S7).
Fig 7. GFP-tagged DEM1 is expressed in developing gametophytes and sporophytic tissues of Arabidopsis.
(A) Transgenes used to express DEM1 N-terminal or C-terminal GFP fusion proteins under the control of the DEM1 promoter (pDEM1). The coding sequence for six alanine residues (Linker) were inserted between GFP and DEM1 of pDEM1:GFP-DEM1. Similarly, the pDEM1:DEM1-GFP transgene contained the coding sequence for three alanine residues, followed by a thrombin cleavage site and three more alanine residues (Th). (B) Anti-GFP western blot on floral bud extracts of 35S:GFP (GFP), pDEM1:GFP-DEM1 (GFP-DEM1) and pDEM1:DEM1-GFP (DEM1-GFP) transgenic lines, and a non-transgenic control (NT). Molecular weight is indicated in kDa. The molecular masses of GFP-DEM1 and DEM1-GFP are approximately 100 kDa, and of GFP is 27 kDa. The Coomassie Brilliant Blue-stained (CBB) membrane shows Rubisco (Rbsc) as a loading control. (C) Ubiquitous and punctate GFP-tagged DEM1 expression in ovules prior to meiosis (pre-meiotic ovules), and in developing and mature post-meiotic ovules. In pre-meiotic ovules, very little GFP fluorescence could be seen in the megaspore mother cell (MMC), whereas high expression was observed in the sporophytic cells surrounding the MMC (bar = 10 μm). In developing ovules, high GFP expression is observed in the sporophytic tissue and in the megagametophyte (MG) (bar = 20 μm). In mature ovules, high GFP fluorescence was observed in the sporophytic tissues, but very little expression was seen in the megagametophyte (MG) (bar = 20 μm). (D) Two optical sections (Z-stack) showing high expression of DEM1-GFP in the functional megaspore (FM) of an ovule (bar = 10 μm). DM, dead megaspores. (E) Expression of GFP-DEM1 in male pre-meiotic cells; GFP-DEM1 localized around the nuclear envelope (bar = 5 μm). N, nucleus. (F) Expression of GFP-tagged DEM1 was also detected in meiotic products of male meiosis; two cells of a tetrad are shown in the figure. Note the subcellular localization of GFP-DEM1 around the nuclear envelope relative to the DAPI stained nucleus (N) (bar = 5 μm). (G) Expression of GFP-tagged DEM1 during pollen development (bar = 5 μm). N, nucleus of monocellular pollen; GC, generative cell; SC, sperm cells; MC, monocellular pollen; BC, bicellular pollen; TC, tricellular pollen.
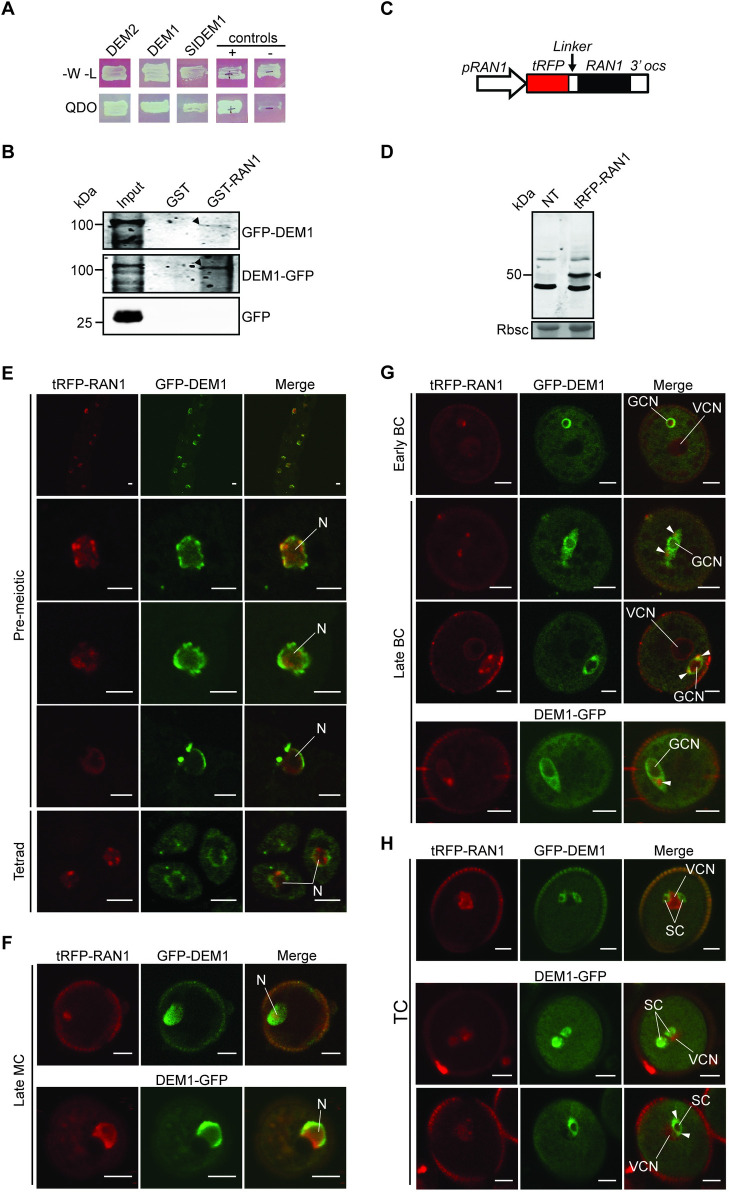
To gain insight into DEM’s molecular function, a yeast two-hybrid system was employed to screen Arabidopsis cDNA libraries for proteins that interact with Arabidopsis DEM2. Six strongly interacting prey vectors were identified (S4 Table), and upon sequencing, were all shown to carry Arabidopsis RAS-RELATED NUCLEAR PROTEIN 1 (RAN1) cDNA sequences (S4 Table). Each of these six RAN1 clones represented one or the other of two unique overlapping clones encoding the C-terminal 92 and 186 amino acids of the predicted 221-amino acid RAN1 protein (S4 Table). The longest RAN1 prey interacted with both Arabidopsis DEMs and full-length tomato DEM1 as bait in the yeast two-hybrid system (Fig 8A).
Fig 8. DEM interacts with RAN1, and the proteins co-localize during male meiosis and pollen development.
(A) RAN1 interacts with Arabidopsis DEM1 and DEM2, and tomato DEM1 (SlDEM1) in the yeast two-hybrid system. Yeast were plated on SD media lacking tryptophan and leucine (-W-L) to select for the bait (DEM) and prey (RAN1) vectors and show equal viability of all strains. Yeast were also plated on quadruple dropout media (QDO) to test for interactions between the bait and the prey. Known interacting (+) and non-interacting (-) controls are also shown. (B) GST-RAN1, but not GST, precipitates 100 kDa GFP-DEM1 and DEM1-GFP from floral bud extracts (black arrowheads). GFP-expressing plants were included to rule out binding of RAN1 to GFP (lower panel). Input represents an anti-GFP western of floral bud extracts from each transgenic line. (C) Transgene used to express RAN1 N-terminal tagRFP (tRFP) fusion protein under the control of the RAN1 promoter (pRAN1). The coding sequence for six glycine residues (Linker) were inserted between tRFP and RAN1. (D) Anti-tRFP western blot of pRAN1:tRFP-RAN1 transgenic and non-transgenic (NT) floral bud extracts. The tRFP-RAN1 is approximately 50 kDa (black arrowhead). The CBB membrane shows Rubisco (Rbsc) as a loading control. (E-H) Expression and co-localization of GFP-tagged DEM1 and tagRFP-RAN1 (tRFP-RAN1) during male meiosis and pollen development in wild-type ecotype Col-0 plants. (E) GFP-DEM1 localized around the nuclear envelope with additional weak signal throughout the nucleus and cytoplasm of pre-meiotic cells and tetrads, whereas tRFP-RAN1 typically localized to the nucleus (N). We examined 18 pre-meiotic cells in total, all 18 cells expressed detectable tagRFP-RAN1 and GFP-DEM1 and showed a similar localization of the fusion proteins. Representative pre-meiotic cells are shown in the figure. (F) Monocellular microspores (MC) show localization of GFP-tagged DEM1 around the nuclear envelope and tRFP-RAN1 to the nucleus, including the nuclear periphery. (G) Bicellular pollen showing co-localization of tRFP-RAN1 and GFP-tagged DEM1 around the nuclear envelope and in extranuclear foci at opposite poles of the generative cell (white arrowheads). (H) In tricellular pollen, GFP-tagged DEM1 was expressed in sperm cells (SC) around the nuclear envelope and throughout the sperm cytoplasm, whereas RFP-tagged RAN1 was primarily expressed in the immediately adjacent vegetative nucleus. In some tricellular pollen, weak RAN1 fluorescence overlapped with strong DEM1 fluorescence in extranuclear foci at opposite poles of sperm cells (lower panel; white arrowheads). We examined 93 pollen in total and 74 (80%) expressed detectable tagRFP-RAN1, 45 (48%) expressed detectable GFP-DEM1 or DEM1-GFP, and 39 (42%) expressed both tagRFP-RAN1 and GFP-DEM1 or DEM1-GFP (E-H). Pollen at the same stage expressing both tagRFP-RAN1 and GFP-DEM1 or DEM1-GFP showed a similar localization of the fusion proteins, and representative pollen are shown in the figure (E-H). Bar = 5 μm. BC, bicellular pollen; TC, tricellular pollen; GCN, generative cell nucleus; VCN, vegetative cell nucleus.
To further investigate a potential interaction between DEM1 and RAN1, GST-RAN1 was expressed and purified from E. coli, and then used as bait in pull-down experiments with floral buds extracts from transgenic lines of Arabidopsis expressing pDEM1:GFP-DEM1 or pDEM1:DEM1-GFP (Fig 8B). The GST-RAN1 pull down assay successfully precipitated the 100 kDa GFP-fusion proteins that corresponded to the molecular weight of full-length GFP-DEM1 and DEM1-GFP (Fig 8B). Furthermore, GST alone did not precipitate GFP-DEM1 or DEM1-GFP, and GST-RAN1 did not precipitate GFP (Fig 8B).
A transgenic line of wild-type Arabidopsis ecotype Col-0 was produced that expressed a pRAN1:tRFP-RAN1 transgene and a full-length ~50 kDa tRFP-RAN1 fusion protein in floral buds (Fig 8C and 8D). The tRFP-RAN1 fusion protein was also detected during microsporogenesis and pollen development, primarily in the nucleus of cells, but also in perinuclear and extranuclear foci in microspores and the generative cell of bicellular pollen, respectively (S8 Fig). To determine if DEM1 and RAN1 had the potential to interact during male meiosis and in developing pollen, the transgene pRAN1:tRFP-RAN1 was co-expressed along with pDEM1:GFP-DEM1 or pDEM1:DEM1-GFP in wild-type ecotype Col-0 (Fig 8E–8H). GFP-tagged DEM1 and tRFP-RAN1 co-localized around the nuclear envelope in pre-meiotic cells, tetrads (Fig 8E) and monocellular microspores (Fig 8F). In early bicellular pollen, GFP-tagged DEM1 and tRFP-RAN1 co-localized around the nuclear envelope of the generative cell, and in late bicellular pollen, co-localized to the aforementioned extranuclear foci at opposite poles of the generative cell (Fig 8G). In tricellular pollen, GFP-tagged DEM1 was expressed in sperm cells around the nuclear envelope and throughout the cytoplasm, whereas RFP-tagged RAN1 was primarily expressed in the immediately adjacent vegetative nucleus (Fig 8H). In some tricellular pollen, weak RAN1 fluorescence also overlapped with strong DEM1 fluorescence in extranuclear foci at opposite poles of sperm cells (Fig 8H, lower panel). No background fluorescence was observed in non-transgenic pre-meiotic cells or gametophytes except for in the pollen coat (S9 Fig).
Unlike the pDEM1:DEM1 transgene (S4 and S5 Figs), pDEM1:DEM1-GFP and pDEM1:GFP-DEM1 transgenes did not allow the recovery of dem1/dem1 dem2/dem2 double mutant plants (S5 Table). However, the DEM1-GFP and GFP-DEM1 transgenes suppressed the extent of pollen and ovule abortion in the DEM1/dem1 dem2/dem2 genotype (S5D Fig), and alleviated the distorted segregation ratio against DEM1/dem1 dem2/dem2 plants compared to DEM1/DEM1 dem2/dem2 plants in progeny of self-fertilized DEM1/dem1 dem2/dem2 plants (S5 Table). Crosses between DEM1/dem1 dem2/dem2 plants carrying the GFP-tagged DEM1 transgenes as males to wild-type female plants produced a limited number of progeny, indicating that the transgenes increased pollen fertility in DEM1/dem1 dem2/dem2 plants (S6 Table). Not all of the progeny produced from these crosses inherited the transgene, suggesting that the GFP-tagged DEM1 transgenes enhanced gamete viability in DEM1/dem1 dem2/dem2 plants by functioning in both pollen and the transgenic parent plant that underwent meiosis to produce the pollen (S6 Table).
Discussion
Prior to our current study, the DEM1 gene of tomato was the only member of the DEM gene family to be genetically characterized in plants [10]. Tomato DEM1 is expressed in all differentiating tissue, and the dem1 mutant of tomato has dysfunctional meristems resulting in a seedling lethal phenotype [10]. However, it was unclear whether DEM1 played a fundamental role in cell division or alternatively, had a more specific role in defining the identity of meristematic cells in tomato. In our current work, we used Arabidopsis as a more tractable genetic model to investigate a possible fundamental role of DEM genes in cell division in plants. In contrast to the dem1 mutant of tomato, both the dem1 and dem2 single mutants of Arabidopsis were indistinguishable from wild type in both vegetative development and reproductive fertility, but a fundamental role for the DEM genes in cell division, gamete viability and fertility was revealed in DEM1/dem1 dem2/dem2 plants carrying mutations in both DEM1 and DEM2. In contrast to DEM1/dem1 dem2/dem2 plants, dem1/dem1 DEM2/dem2 plants showed normal levels of fertility, indicating that DEM2 plays a more important role than DEM1 in gamete viability. The increased importance of DEM2 gamete viability correlated with higher mRNA levels of DEM2 compared to DEM1 in most tissues examined and particularly in the vegetative shoot apex, developing siliques, pollen and sperm (Fig 2).
DEM gene expression in Arabidopsis is consistent with a function in cell division
DEM1 and/or DEM2 transcripts are enriched in tissues undergoing cell division, including the shoot apex, flowers and developing siliques (Fig 2A). The in situ expression pattern of DEM genes in differentiating tissues appeared punctate, particularly for DEM1 (Fig 3), which is typical of cell cycle-regulated genes such as histones, cyclins and cyclin-dependent kinases [22–25]. In support of these findings, DEM1 has been shown previously to be up-regulated in cell cultures during the mitosis phase of the cell cycle [26] and in actively dividing shoot apices [27]. Expression of DEM1 fused to GFP also appeared punctate in differentiating tissues (Figs 7C and S7). Interestingly, GFP-DEM1 and DEM1-GFP were observed in newly formed sperm cells but not in the vegetative cell of pollen, which does not undergo further cell division (Fig 8H). Together, these findings suggest that DEM1 expression is up-regulated immediately prior to, during and just after cell division.
DEM1/dem1 dem2/dem2 mutants have defects in male meiosis, pollen and embryo sac development
DEM1/dem1 dem2/dem2 plants produced defective tetrads with an excess of irregular-sized meiotic products after male meiosis (Fig 5D–5G). Upon performing meiotic chromosome spreads on DEM1/dem1 dem2/dem2 male meiocytes, we found that chromosomes were often misaligned on the metaphase plate at metaphase II (Fig 5H and 5I). Despite the misalignment of chromosomes in ~70% of daughter cells of DEM1/dem1 dem2/dem2 male meiocytes at metaphase II, chromosome segregation during anaphase II and early telophase II appeared normal (S6 Fig). However, we cannot rule out the possibility of anaphase II defects in DEM1/dem1 dem2/dem2 plants because aberrant three-dimensional organisation of chromosomes at anaphase II would be difficult to discern in the chromosome spreads. While the misaligned chromosomes at metaphase II moved towards the poles during meiosis II, they were often not partitioned properly into four distinct daughter nuclei at late telophase II, thereby resulting in the formation of pentads (Fig 5D–5G), and in the case of ecotype Ws-0, also degenerated tetrads (Fig 5F and 5G). In addition to defects in male meiosis and pollen development, we also observed approximately 50% of embryo sacs arrested before the first or second round of post-meiotic mitosis in DEM1/dem1 dem2/dem2 plants (Fig 6).
In contrast to DEM1/dem1 dem2/dem2 plants, dem1/dem1 DEM2/dem2 plants recovered in the Col-0 genetic background had very few aborted pollen (Figs 4B and 5B). These results suggest an increased importance of DEM2 compared to DEM1 particularly in pollen development, which correlated with significantly higher expression of DEM2 compared to DEM1 in pollen and sperm (Fig 2B).
While dem1/dem1 DEM2/DEM2 plants had wild-type phenotypes in both ecotypes, dem1/dem1 DEM2/dem2 plants were not recovered in the Ws-0 background. These results could indicate that a single functional DEM2 allele in dem1/dem1 DEM2/dem2 plants in the Ws-0 background is insufficient for embryo viability, or alternatively, that the dem2 T-DNA insertion allele may interfere with expression of the wild-type DEM2 allele in heterozygous Ws-0 embryos.
Sporophytic and gametophytic expression of DEM contributes to gamete viability
DEM expression during microgametogenesis is essential for pollen viability, as the transmission of the dem1 dem2 genotype through the male germline was almost completely abolished in DEM1/dem1 dem2/dem2 and DEM1/dem1 DEM2/dem2 plants (Table 1). However, DEM1/dem1 DEM2/dem2 plants with one functional copy of DEM2 also transmitted a lower than expected proportion of dem1 DEM2 gametes compared to plants with two functional copies of DEM2 (i.e. DEM1/dem1 DEM2/DEM2; Table 1), suggesting the expression of DEM2 in the diploid parent plant that undergoes meiosis contributes to pollen development. In support of these findings, we showed that DEM1/dem1 dem2/dem2 plants produce a much higher rate of pollen abortion (~70–80%) than expected (50%) if only dem1 dem2 pollen were aborted (Fig 5A–5C). It is therefore likely that in the absence of DEM2, a significant portion of DEM1 dem2 pollen are also aborted in DEM1/dem1 dem2/dem2 plants due to insufficient expression of DEM1 in the diploid microspore mother cell that undergoes meiosis or in other supporting sporophytic tissue around the developing pollen. Consistent with the importance of DEM1 gene expression in the sporophyte in the absence of DEM2, we found that GFP-DEM1 and DEM1-GFP transgenes partially restored pollen fertility in DEM1/dem1 dem2/dem2 plants even when the pollen did not inherit the transgene (S6 Table). One possible explanation for these sporophytic effects is that DEM transcripts or DEM protein produced by the microspore mother cell or other sporophytic cells are passed into the products of meiosis and function during gamete formation.
In the Col-0 genetic background, dem1/dem1 DEM2/dem2 plants were indistinguishable from wild type, whereas DEM1/dem1 dem2/dem2 plants had a wild-type phenotype during vegetative development but showed defects in pollen and embryo sac development (Figs 5, 6 and S5). It is interesting to note that DEM2 mRNA but not the DEM1 mRNA has been found in the phloem [28] and is graft-transmissible [29]. Graft-transmissible mRNAs are often highly expressed [29], and DEM2 mRNAs levels were significantly higher than DEM1 mRNA levels in all tissues and cells examined, except for microdissected ovules and unpollinated pistils (Fig 2). Future research should investigate whether the DEM2 transcript has the capacity to move cell-to-cell during reproductive development.
There was also evidence for both sporophytic and gametophytic expression of DEM genes contributing to embryo sac viability (Table 1 and Fig 6), but the impact of dem mutations on female reproductive development was less severe than on pollen development. DEM1/dem1 DEM2/dem2 plants transmitted the dem1 dem2 genotype through the megagametophyte at a reduced rate, indicating that dem1 dem2 megagametophytes were viable but less likely to become the surviving megaspore following meiosis. The improved viability of dem1 dem2 embryo sacs compared to dem1 dem2 pollen could be due to a larger amount of sporophyte-derived cytoplasm being transferred to the products of female meiosis (i.e. embryo sacs) compared to the products of male meiosis (i.e. pollen).
Potential interaction between DEM and RAN around the nuclear envelope and in extracellular foci
Our results indicate that DEM’s role in cell division during meiosis, and in pollen and embryo sac development, could be mediated through an interaction with RAN1. RAN proteins are members of the RAS superfamily of small GTPase proteins and is highly conserved within eukaryotes [30–32]. In Arabidopsis, three RAN genes (RAN1, RAN2 and RAN3) have been identified with very high sequence identity between themselves and their orthologues in animals and yeasts [31,33,34]. RAN functions as a molecular switch, converting between a guanosine triphosphate (GTP)-bound form (RAN-GTP) and a guanosine diphosphate (GDP)-bound form (RAN-GDP) [35]. The location of these two nucleotide-bound forms of RAN within the cell is crucial to the function of the protein. In animal nuclei, high concentrations of RAN-GTP are generated due to the action of a chromatin-anchored RAN guanine nucleotide exchange factor (RanGEF), which removes GDP from RAN and replaces it with GTP [35,36]. In plants, a functional equivalent of RanGEF is yet to be identified [37,38]. RAN’s intrinsic GTPase activity is very low, but it is strongly activated by RanGTPase activating protein (RanGAP) [39–40]. During interphase in all eukaryotic cells, RanGAP is anchored to the cytoplasmic face of the nuclear envelope and ensures that any RAN-GTP leaving the nucleus is immediately hydrolyzed to RAN-GDP in the cytoplasm [40–43]. The GTPase function of RAN is also influenced by its interaction with RAN binding proteins (RanBPs) at the nuclear envelope [44–48]. Together, the subcellular positioning of these effectors with RAN generates a steep RAN-GTP gradient relative to chromatin and across the nuclear envelope [39,42,48]. This spatial distribution of the RAN-GTP gradient drives many additional processes in eukaryotic cells, including spindle assembly during cell division, reformation of the nuclear envelope following cell division and nucleocytoplasmic transport [32,49–57].
The cell division defects in dem meiocytes, pollen (Fig 5) and embryo sacs (Fig 6), along with disorganized patterns of cell division in meristems of tomato dem1 mutants [10], are consistent with disruption of RAN-associated processes. DEM1 and RAN1 co-localize around the nuclear envelope of pre-meiotic cells and in pollen (Fig 8), and future research should investigate a potential role of DEM proteins in RAN-dependent breakdown of the nuclear envelope, formation of the spindle apparatus during cell division and/or reformation of the nuclear envelope following cell division. Besides co-localizing around the nuclear envelope, DEM1 and RAN1 co-localized transiently in extranuclear foci at opposite poles of the generative and sperm cells of developing pollen, but the nature and function of these foci also requires further investigation. While transient co-localization occurred during male meiosis and pollen development, DEM1 and RAN1 fusion proteins were also observed at unique locations within cells (Fig 8). These differences in subcellular localization of DEM1 and RAN1 suggest that, in addition to their interaction, these proteins are likely to have unique and independent cellular functions.
DEM1 fusion proteins showed an uneven distribution throughout the cytoplasm of differentiating cells in a pattern resembling the endoplasmic reticulum (ER) (Figs 7, 8 and S7) [58]. The nuclear envelope is a double membrane that forms a continuous membrane system with the ER, but in both plants and animals, the nuclear envelope undergoes a cycle of disassembly and reformation during cell division whereas the ER network remains intact [59–60]. Furthermore, Anderson and Hetzer [61] have demonstrated in the metazoan Xenopus that nuclear envelope formation and expansion are driven by chromatin-mediated shaping of the ER network after cell division.
Our transgenic work focusing on DEM1 was initiated prior to our results showing that DEM2 plays a more important role than DEM1 in male gamete viability. However, future work should concentrate on the further characterization of DEM2’s role in meiosis and gamete viability, and the importance of DEM2’s interaction with RAN1 in these processes. Additionally, it will be important to investigate whether the apparent increased importance of DEM2 compared to DEM1 in gamete viability is due to DEM2’s higher expression in sporophytic and gametophytic tissues and cells, or alternatively, due to functional differences between the DEM1 and DEM2 proteins. Given that the DEM genes are expressed in tissues undergoing high rates of cell division, it would also be interesting to assess dem mutants for mitotic phenotypes and changes in mitotic indices in meristems, and for DNA content of somatic cells.
In conclusion, we have shown that DEM genes play a key role in cell division and gamete viability in Arabidopsis, which most likely involves an interaction with RAN1. The presence of DEM homologues in diverse eukaryotic lineages suggests a conserved and critical cell division function of DEM-like genes in many eukaryotes. Alternatively, the region of conservation may relate to an interaction motif or domain of DEM-like proteins, and the biological roles of the proteins in diverse eukaryotic lineages could be distinct. The region of most similarity across eukaryotes corresponds to amino acids 253–458 of Arabidopsis DEM1, and this aligns with amino acids 40–226 of S. pombe DEM-like protein (S3 Fig), which when fused to GFP localizes to the nuclear rim [14]. These conserved amino acids residues may have a function in localizing DEM-like proteins to the nuclear rim, thus allowing DEM-like proteins to conduct biological functions that may be crucial to cell division or other cellular processes in many eukaryotic species.
Materials and methods
Phylogenetic analysis of DEM-like proteins
To identify proteins with similarity to DEM1, BLAST searches [62] using the tomato DEM1 sequence (NP_001234563.1) queried against the NCBI database were undertaken. Protein sequences from sequenced and annotated genomes from various eukaryota lineages were identified for including in multiple sequence alignments. Multiple sequence alignments of protein sequences were undertaken using Multiple Sequence Comparison by Log-Expectation (MUSCLE) from EMBL-EBI with default parameters. Rooted phylogenetic trees were constructed using the Maximum Likelihood method with Bootstrap analysis (1000 replication) and Poisson model in MEGA6 software [63].
Plant handling, growth conditions and transformation
Arabidopsis thaliana genotypes
Two independent T-DNA insertion mutants for both DEM1 (AT4G33400) and DEM2 (AT3G19240) were used in this study. T-DNA insertion mutants dem1-2 (SALK_016893C) and dem2-2 (SALK_070099) in the Columbia (Col-0) ecotype were obtained from the Arabidopsis Biological Resource Center (ABRC, The Ohio State University, Columbus, OH). T-DNA insertion mutants dem1-1 and dem2-1 in the Wassilewskija (Ws-0) ecotype were obtained from the Wisconsin Biotechnology Center [64,65]. Techniques to confirm the presence or absence of T-DNA insertions in DEM1 and DEM2 are described in S1 Text. Details on production of transgenic Arabidopsis Col-0 lines expressing GFP and tagRFP fusion proteins are provided in S1 Text. The oligonucleotides used to genotype Arabidopsis T-DNA insertion mutants are listed in S7 Table.
Arabidopsis growth conditions
Seeds of Arabidopsis were germinated on damp University of California (UC) soil mix or on MS plates. Seeds were then stratified at 4°C for four to five days before growing at 21°C under 16-hour photoperiods of fluorescent lighting (70–80 μmol/m2/s).
Arabidopsis crossing and progeny analysis
For crossing of Arabidopsis, sepals, petals and anthers were removed from suitable flowers of the female parent. Anthers were obtained from newly opened flowers from the male plant and pollen was transferred to the pistil on the carpel of the female parent by direct physical contact. Emasculation controls that were not pollinated were included in crossing experiments as controls. Specific plant genotypes were identified by PCR genotyping.
Arabidopsis seed collection
Seeds were collected after the plants were fully desiccated and turned brown. Siliques were gently crushed and the released seeds were sifted through a tea strainer to remove silique debris. Seeds were stored in 1.5 ml Eppendorf tubes with a small hole in the lid (made with a 0.45 mm needle) and placed in containers with silica gel desiccant to keep the seeds dry and prevent contamination by fungi or bacteria.
Arabidopsis transformation
The Agrobacterium T-DNA binary vectors used in this study are shown in S10 Fig. A single colony of Agrobacterium tumefaciens strain GV3101 containing the appropriate T-DNA binary vector was inoculated into 5 ml LB (Luria-Bertani) medium (1% tryptone, 0.5% yeast extract, 1% NaCl; w/v) with 50 μg/ml gentamicin, 50 μg/ml kanamycin and 50 μg/ml rifampicin and pre-cultured for 48 hours at 28°C. About 1 ml of pre-culture was then inoculated into 200 ml LB containing the same antibiotics. The 200 ml culture was incubated for 24 hours at 28°C and centrifuged at 5000 rpm at 4°C for 15 minutes in a Beckman JA-10 rotor.
The floral dip method [66] was used to transform Arabidopsis. The Agrobacterium pellet was resuspended in 500 ml of 5% (w/v) sucrose containing 187.5 μl of Silwet L-77 (Lehle Seeds, USA). Stems with open floral buds of Arabidopsis were dipped and mildly agitated in the Agrobacterium solution for five seconds. The plants were then covered for 24 hours to maintain high humidity.
Selection of transgenic Arabidopsis plants
Depending on the plant selectable marker, T1 plants were selected in soil for Basta resistance or MS plates for kanamycin resistance. For selecting T1 plants in soil, T1 seeds collected from the transformed plants were evenly spread over a damp UC soil mix and stratified for four to five days at 4°C. After germination, the seedlings were sprayed with Basta herbicide (0.04% w/v) at two-day intervals. To select for kanamycin resistance, sterile T1 seeds were plated on MS plates containing 75 μg/ml kanamycin, stratified for four to five days at 4°C and then incubated vertically in the growth room.
In situ hybridization
In situ hybridization experiments were performed on Arabidopsis wild-type ecotype Ws-0, as described by Jackson [67] and Vielle-Calzada et al. [68]. Antisense DIG-labelled RNA was produced to full-length DEM1 (AT4G33400) and DEM2 (AT3G19240) cDNAs, which were ordered from the Arabidopsis Biological Resource Center (clones G2D10T7 and ATTS5957, respectively).
RNA extraction, gel blot analysis and quantitative real-time reverse transcriptase PCR (qRT-PCR)
For Northern gel blots, total RNA was extracted from Arabidopsis closed floral buds younger than stage 13 [69] as previously described [70–71]. Bundles of fresh or frozen Arabidopsis closed floral buds were collected and crushed with a mortar and pestle under liquid nitrogen into fine powder without any pre-treatment. Total RNA was further purified to remove contaminants using 1-butanol [70]. Northern gel blots were conducted as described previously [71]. For qRT-PCR analysis, Arabidopsis plants (ecotype Columbia) were grown to the four-leaf stage (shoot apex tissue, young and mature leaves) or to the mature plant stage at 2 weeks after flowering (cauline stems, flowers, floral buds and developing siliques) at 24°C day/20°C night and photoperiods of 8 hours (four-leaf stage plants) or 16 hours (mature plants) light (170 μE m-2 s-1). At least three independent samples from each tissue were collected from between three and 100 plants to provide replicate RNA samples for statistical analysis of qRT-PCR assays. Tissue was ground in liquid nitrogen and total RNA was prepared using the SV Total RNA Isolation System (Promega, Madison WI, USA) or TRIzol according to the manufacturer’s instructions (Invitrogen, Life Technologies, Carlsbad, CA, USA). cDNA was produced from 1 μg of total RNA and subsequently used as template (using the equivalent of 20 ng RNA per reaction) for real-time RT-PCR experiments as described previously [72]. The primer pair 5’-ATAACTAGTGGTCCGGTCCATCAC-3’ / 5’-TGGTCATTGCAACGCCTATG-3’ was used for detection of DEM1 transcript levels in various wild-type tissues, except for floral buds (S7 Table). To avoid detection of truncated DEM1 transcripts in the dem1 mutant, the primer pair 5’-CCCCGTCTACTCAGAACTCAGC-3’ / 5’-CGACTTTCAAAATCCACCAT-3’ was used for measuring DEM1 mRNA levels in floral buds of wild-type, dem1 and dem2 plants (S7 Table). The primer pair 5’-CCTTCTCCGGTAACAAATCGC-3’ / 5’-AAAGTCGTTTTCCAGAGAGGCTG-3’ was used for detection DEM2 transcript levels in all genotypes and tissues tested (S7 Table). Expression detected from three β-actin genes of Arabidopsis [72] was used an internal standard to normalise the qRT-PCR data (S7 Table).
DNA extraction and gel blot analysis
High purity genomic DNA was purified and used in Southern gel blot analysis [73] to confirm the presence or absence of T-DNA insertions in DEM1 and DEM2. Radioactive probes were generated as for RNA gel blot analysis.
Gametophyte analysis
Microspores were analyzed by dissecting anthers and staining in 1 μg/ml DAPI (4’, 6-diamino-2-phenylindole), as described elsewhere [74]. Mature pollen was dissected from anthers immediately prior to anthesis and microscopy was used to score the pollen for viability based on appearance (e.g. Fig 5A). Female gametophytes and embryos were analyzed by clearing with Hoyer’s solution [75]. These were then assayed under a ZeissAxioskop 2 compound microscope equipped with a ZeissAxiocam digital camera using florescence, bright field and differential interference contrast (DIC) optics.
Cytology
Meiotic chromosome spreads were performed as described by Ross et al. [76].
Yeast two-hybrid screen
The Matchmaker Library Construction and Screening kit (Clontech Laboratories Inc., California) was used to construct Arabidopsis ecotype Col-0 cDNA libraries and screen them with bait vectors containing the DEM1 and DEM2 cDNAs. Further details are provided in S1 Text.
Expression and purification of GST-RAN1 and protein binding assays
The Arabidopsis RAN1 coding region (AT5G20010) was expressed as a GST-tagged fusion protein in E. coli. Experimental procedures used to express GST-tagged RAN1 and pull-down DEM1-GFP and GFP-DEM1 from transgenic plant extracts are provided in S1 Text.
SDS-PAGE and immunoblotting
Protein samples for immunoblotting were snap-frozen in liquid nitrogen, ground into a fine powder with pestle, briefly thawed with the addition of loading buffer and then treated at ~95°C for 5 minutes. Protein samples were run on 10% SDS-PAGE gels and transferred from the SDS-PAGE to a PVDF membrane (Bio-Rad) following the manufacturer’s protocol. For immunoblotting, anti-His mouse (Amersham Biosciences) or anti-GFP mouse (Roche Applied Science) IgG1 monoclonal primary antibodies were diluted 1000-fold and the secondary Alexa fluor 680 anti-mouse IgG (Invitrogen) were diluted 5000-fold. Blocking reagents were purchased from LI-COR Biosciences and blocking, binding and washing steps were as described in LI-COR protocol document number 988–09288. For visualization of membranes, the Odyssey infra-red scanner was used. Total protein was visualized by staining gels and membranes with Coomassie Brilliant Blue (R250).
Confocal scanning laser microscopy (CSLM) and image analysis
A Zeiss LSM 510 Meta (Carl Zeiss, Germany) confocal laser-scanning microscope (CSLM) was used with a 40 x, 1.3 oil-immersion plan neofluar objective for root tips, ovules, pollen and tetrads. GFP was visualized by excitation with an argon laser at 488 nm and detection with a 505–530 nm band-path filter. RFP was visualized using a HeNe1 laser at 543 nm and detection with a 560 nm long-path filter and DAPI was visualized using a diode laser for 405 nm and detection with a 420–480 nm band-pass filter. The multi-track scan mode was used for co-localization images. Samples for confocal microscopy were dissected and placed in PBS buffer with 10% (v/v) glycerol and kept on ice prior to viewing. For staining nuclei, 2.5 μg/ml DAPI was used and samples kept on ice for 1–5 minutes prior to viewing.
Statistical analysis
All statistical tests were implemented in RStudio using custom R scripts. Multiple corrections of generated p-values were carried out using the Benjamini & Hochberg method.
Supporting information
Software and parameters used in the analysis are described in Materials and Methods. The name of the organism is indicated on the tree, followed by the accession number. Scale bar indicates 0.05 substitutions per site. The plant DEM proteins represented in this tree (Viridiplantae) are the same as those used in Fig 1.
(TIF)
Multiple alignments on full-length DEM and DEM-like protein sequences were undertaken using the Multiple Sequence Comparison by Log-Expectation (MUSCLE) tool from EMBL-EBI and visualized using the Multiple Sequence Alignment Viewer from NCBI. Plant DEM homologues are highly conserved with increased level of identity in the C-terminal half of the proteins. Grey boxes represent amino acids in agreement with the consensus of the multiple sequence alignment.
(TIF)
Pairwise sequence alignment of NP_195066.1 and BAA87237.1 was conducted using the LALIGN tool from EMBL-EBI.
(TIF)
(A) Gene structure of DEM1 and DEM2, along with T-DNA insertions (open triangles), HindIII restriction sites (H3), and location of probes (grey boxes) used for DNA and RNA gel blot analysis. dem1-1 and dem2-1 are T-DNA insertion alleles in Ws-0 ecotype, while dem1-2 and dem2-2 are T-DNA insertion alleles in ecotype Col-0. (B) DNA gel blots on replicate DNA extractions confirmed T-DNA insertions in dem1 and dem2 mutants in Ws-0 and Col-0 genetic backgrounds, and in a dem1 dem2 double mutant (Col-0) complemented with a DEM1 transgene driven by its own promoter pDEM1 (dem1+2 comp). DNA blots for Ws-0 lines were hybridized to the DEM1 3’ probe (upper panel LHS), whereas the blots for the Col-0 lines were hybridized to the DEM1 5’ probe (upper panels RHS). DNA gel blots were all hybridized to the DEM2 5’ probe (lower panel). The T-DNA insertion alleles for the Col-0 dem1 and dem2 mutants are indicated by arrowheads. (C) RNA gel blot analysis of floral buds using the DEM1 and DEM2 5’ probes confirmed T-DNA knock-out of both endogenous dem genes, and over-expression of the pDEM1:DEM1 transgene in the complemented Col-0 dem1 dem2 double mutant (dem1+2 comp). Lower panel shows the 25S rRNA loading control. The two lanes for the complemented Col-0 dem1 dem2 double mutant in (B) and (C) represent two separate plants of a transgenic line that was phenotypically normal during vegetative development and partially fertile. (D) RNA gel blot analysis of floral buds using the DEM1 3’ probe confirmed the absence of full-length transcripts in Ws-0 dem1 plants.
(TIF)
(A) Image showing 6-week old plants of wild type (WT) plants, dem1 and dem2 single mutants, dem1/dem1 DEM2/dem2, DEM1/dem1 dem2/dem2, and dem1 dem2 double mutant complemented with pDEM1:DEM1 transgene (dem1+2 comp) in Col-0 genetic background. (B) Normal seed production was observed in dem1 and dem2 single mutants and in dem1/dem1 DEM2/dem2 plants, but not in dem1/dem1 dem2/dem2 double mutant plants complemented with a pDEM1:DEM1 transgene (dem1+2 comp). (C) Percent ovule, pollen and tetrad defects in DEM1/dem1 dem2/dem2 plants (dem) and dem1/dem1 dem2/dem2 double mutant complemented with a pDEM1:DEM1 transgene (dem1+2 comp). At least 200 ovules, 500 pollen grains and 70 tetrads were assayed for each genotype. (D) Partial complementation of ovule and pollen abortion rates in DEM1/dem1 dem2/dem2 (dem) plants hemizygous for the pDEM1:GFP-DEM1 (GFP-DEM1) or pDEM1:DEM1-GFP (DEM1-GFP) transgene. Non-transgenic DEM1/dem1 dem2/dem2 (dem) and wild-type (WT) plants were included as controls. At least 200 ovules and 700 pollen grains were assessed for each genotype.
(TIF)
(A) Pachytene. (B) Diakinesis. (C) Metaphase I. (D) Anaphase I. (E) Telophase I. (F) Anaphase II. (G) Early telophase II. Bar = 10μm.
(TIF)
(A) Root tips of transgenic seedlings expressing GFP-DEM1 (left) or DEM1-GFP (right); bar = 20 μm. (B) DAPI-stained root tip cells expressing GFP-DEM1 or DEM1-GFP showing cytoplasmic and nuclear envelope localization; bar = 10 μm. (C) Localisation of GFP-DEM1 and DAPI in daughter cells of a root tip epidermal cell that has just divided. (D-E) Root tip expressing GFP:DEM1. (E) Higher magnification of sub-cellular localization of GFP:DEM1 in root tips corresponding to inset in (D), showing expression of GFP:DEM1 relative to the nucleus. Bar represents 50 μm for panels (A-D), and 10 μm for panels (E). Left-hand side, centre and right-hand side panels correspond to DAPI-stained, GFP and merged DAPI/GFP images, respectively (B-E). Images are representative of at least five independent transgenic lines for each transgene.
(TIF)
DAPI staining and localization of tRFP-RAN1 at various stages of microgametophyte development. In tetrads and free monocellular microspores (MC), tRFP-RAN1 predominantly co-localized with DAPI in the nucleus (N), but was concentrated in peripheral nuclear foci, particularly in early MC microgametophytes (white arrowhead). In early to mid-stage bicellular (BC) pollen, tRFP-RAN1 was mainly located in the generative cell nucleus (GCN) and weak expression was detected in the vegetative cell nucleus (VCN). In late BC pollen, tRFP-RAN1 was concentrated in extranuclear foci adjacent to the generative cell nucleus (white arrowhead; GCN), and a weak signal was detected in the vegetative cell nucleus. In tricellular (TC) pollen, tRFP-RAN1 was predominantly located in the vegetative cell nucleus (VCN). Bar = 5 μm.
(TIF)
(A) GFP auto-fluorescence was not detected in pre-meiotic and mature ovules. (B) GFP auto-fluorescence was detected in the pollen coat, but not inside pollen grains. (C) GFP auto-fluorescence was not detected in pre-meiotic cells or tetrads. (D) RFP and GFP auto-fluorescence was not detected in the DAPI stained nuclei of pre-meiotic cells, monocellular and bicellular pollen. Auto-fluorescence of RFP and particularly GFP was detected in the pollen coat of monocellular and bicellular pollen. Bar = 5 μm.
(TIF)
T-DNA binary vectors described in this study are all derived from the binary vectors pUQC477 and pUQC214 [71]. 35S represents the Cauliflower Mosaic Virus 35S promoter. The vector pUQC10332 is a derivative of pUQC214 with the 35S:GFP transgene removed, and it has a single NotI site upstream of the BAR selectable marker for inserting additional transgenes. pUQC10847 was constructed by removing the 35S:BAR sequence from pUQC477 [71], leaving a NPTII gene driven by the Agrobacterium NOPALINE SYNTHASE promoter (NOS) as a kanamycin selectable marker linked to a NotI site (for cloning in additional transgenes). OCS 3’, OCTOPINE SYNTHASE 3’ terminator.
(TIF)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
This work was supported by Australian Research Council grants DP120103966 and DP150104048 to B.J.C.; DP0559710 to B.K., T.G.A.L. and B.J.C.; DP160100892 to J.L.B.; FL180100109 to B.K. (https://www.arc.gov.au/grants); and a LabEx Saclay Plant Sciences-SPS grant ANR-10-LABX-0040-SPS to R.M. (https://www6.inrae.fr/saclay-plant-sciences_eng/Innovation-and-partnerships/Saclay-Plant-Sciences-SPS-LabEx-Innovation-booster). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Qiu Y-L, Taylor AB, McManus HA. Evolution of the life cycle in land plants. Journal of Systematics and Evolution. 2012;50(3):171–94. 10.1111/j.1759-6831.2012.00188.x [DOI] [Google Scholar]
- 2.Andres F, Coupland G. The genetic basis of flowering responses to seasonal cues. Nature Reviews Genetics. 2012;13(9):627–39. Epub 2012/08/18. 10.1038/nrg3291 . [DOI] [PubMed] [Google Scholar]
- 3.McCormick S. Control of male gametophyte development. The Plant Cell. 2004;16 Suppl:S142–53. Epub 2004/03/24. 10.1105/tpc.016659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yadegari R, Drews GN. Female gametophyte development. The Plant Cell. 2004;16 Suppl:S133–41. Epub 2004/04/13. 10.1105/tpc.018192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu J, Qu LJ. Meiotic and mitotic cell cycle mutants involved in gametophyte development in Arabidopsis. Molecular Plant. 2008;1(4):564–74. Epub 2008/07/01. 10.1093/mp/ssn033 . [DOI] [PubMed] [Google Scholar]
- 6.Brukhin VB, Jaciubek M, Bolanos Carpio A, Kuzmina V, Grossniklaus U. Female gametophytic mutants of Arabidopsis thaliana identified in a gene trap insertional mutagenesis screen. The International Journal of Developmental Biology. 2011;55(1):73–84. Epub 2011/03/23. 10.1387/ijdb.092989vb . [DOI] [PubMed] [Google Scholar]
- 7.Twell D. Male gametogenesis and germline specification in flowering plants. Sexual Plant Reproduction. 2011;24(2):149–60. Epub 2010/11/26. 10.1007/s00497-010-0157-5 . [DOI] [PubMed] [Google Scholar]
- 8.Yang WC, Shi DQ, Chen YH. Female gametophyte development in flowering plants. Annual Review of Plant Biology. 2010;61:89–108. Epub 2010/03/03. 10.1146/annurev-arplant-042809-112203 . [DOI] [PubMed] [Google Scholar]
- 9.Shi DQ, Yang WC. Ovule development in Arabidopsis: progress and challenge. Current Opinion in Plant Biology. 2011;14(1):74–80. Epub 2010/10/05. 10.1016/j.pbi.2010.09.001 . [DOI] [PubMed] [Google Scholar]
- 10.Keddie JS, Carroll BJ, Thomas CM, Reyes ME, Klimyuk V, Holtan H, et al. Transposon tagging of the Defective embryo and meristems gene of tomato. The Plant Cell. 1998;10(6):877–88. Epub 1998/06/23. 10.1105/tpc.10.6.877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Finn RD, Attwood TK, Babbitt PC, Bateman A, Bork P, Bridge AJ, et al. InterPro in 2017-beyond protein family and domain annotations. Nucleic Acids Research. 2017;45(D1):D190–D9. Epub 2016/12/03. 10.1093/nar/gkw1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wysocki R, Roganti T, Van Dyck E, de Kerchove D’Exaerde A, Foury F. Disruption and basic phenotypic analysis of 18 novel genes from the yeast Saccharomyces cerevisiae. Yeast. 1999;15(2):165–71. Epub 1999/02/25. . [DOI] [PubMed] [Google Scholar]
- 13.Regelmann J, Schule T, Josupeit FS, Horak J, Rose M, Entian KD, et al. Catabolite degradation of fructose-1,6-bisphosphatase in the yeast Saccharomyces cerevisiae: a genome-wide screen identifies eight novel GID genes and indicates the existence of two degradation pathways. Molecular Biology of the Cell. 2003;14(4):1652–63. Epub 2003/04/11. 10.1091/mbc.e02-08-0456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ding DQ, Tomita Y, Yamamoto A, Chikashige Y, Haraguchi T, Hiraoka Y. Large-scale screening of intracellular protein localization in living fission yeast cells by the use of a GFP-fusion genomic DNA library. Genes to Cells. 2000;5(3):169–90. Epub 2000/04/12. 10.1046/j.1365-2443.2000.00317.x . [DOI] [PubMed] [Google Scholar]
- 15.Schmid M, Davison TS, Henz SR, Pape UJ, Demar M, Vingron M, et al. A gene expression map of Arabidopsis thaliana development. Nature Genetics. 2005;37(5):501–6. Epub 2005/04/05. 10.1038/ng1543 . [DOI] [PubMed] [Google Scholar]
- 16.Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ. An "Electronic Fluorescent Pictograph" browser for exploring and analyzing large-scale biological data sets. PloS One. 2007;2(8):e718. Epub 2007/08/09. 10.1371/journal.pone.0000718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Klepikova AV, Kasianov AS, Gerasimov ES, Logacheva MD, Penin AA. A high resolution map of the Arabidopsis thaliana developmental transcriptome based on RNA-seq profiling. The Plant Journal. 2016;88(6):1058–70. Epub 2016/08/24. 10.1111/tpj.13312 . [DOI] [PubMed] [Google Scholar]
- 18.Pina C, Pinto F, Feijo JA, Becker JD. Gene family analysis of the Arabidopsis pollen transcriptome reveals biological implications for cell growth, division control, and gene expression regulation. Plant Physiology. 2005;138(2):744–756. Epub 2005/06/01. 10.1104/pp.104.057935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Borges F, Gomes G, Gardner R, Moreno N, McCormick S, Feijo JA, et al. Comparative transcriptomics of Arabidopsis sperm cells. Plant Physiology. 2008;148(2):1168–81. Epub 2008/08/01. 10.1104/pp.108.125229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boavida LC, Borges F, Becker JD, Feijo JA. Whole genome analysis of gene expression reveals coordinated activation of signaling and metabolic pathways during pollen-pistil interactions in Arabidopsis. Plant Physiology. 2011;155(4):2066–2080. Epub 2011/02/11. 10.1104/pp.110.169813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Clark KA, Krysan PJ. Chromosomal translocations are a common phenomenon in Arabidopsis thaliana T-DNA insertion lines. Plant Journal. 2010;64(6):990–1001. Epub 2011/11/17. 10.1111/j.1365-313X.2010.04386.x ; PMCID: PMC3079379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koning AJ, Tanimoto EY, Kiehne K, Rost T, Comai L. Cell-specific expression of plant histone H2A genes. The Plant Cell. 1991;3(7):657–65. Epub 1991/07/01. 10.1105/tpc.3.7.657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fobert PR, Gaudin V, Lunness P, Coen ES, Doonan JH. Distinct classes of cdc2-related genes are differentially expressed during the cell division cycle in plants. The Plant Cell. 1996;8(9):1465–76. Epub 1996/09/01. 10.1105/tpc.8.9.1465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gaudin V, Lunness PA, Fobert PR, Towers M, Riou-Khamlichi C, Murray JA, et al. The expression of D-cyclin genes defines distinct developmental zones in snapdragon apical meristems and is locally regulated by the Cycloidea gene. Plant Physiology. 2000;122(4):1137–48. Epub 2000/04/12. 10.1104/pp.122.4.1137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang W, Wightman R, Meyerowitz EM. Cell cycle control by nuclear sequestration of CDC20 and CDH1 mRNA in plant stem cells. Molecular Cell. 2017;68(6):1108–19.e3. Epub 2017/12/12. 10.1016/j.molcel.2017.11.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Menges M, Hennig L, Gruissem W, Murray JA. Cell cycle-regulated gene expression in Arabidopsis. The Journal of Biological Chemistry. 2002;277(44):41987–2002. Epub 2002/08/10. 10.1074/jbc.M207570200 . [DOI] [PubMed] [Google Scholar]
- 27.Lopez-Juez E, Dillon E, Magyar Z, Khan S, Hazeldine S, de Jager SM, et al. Distinct light-initiated gene expression and cell cycle programs in the shoot apex and cotyledons of Arabidopsis. The Plant Cell. 2008;20(4):947–68. Epub 2008/04/22. 10.1105/tpc.107.057075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Deeken R, Ache P, Kajahn I, Klinkenberg J, Bringmann G, Hedrich R. Identification of Arabidopsis thaliana phloem RNAs provides a search criterion for phloem-based transcripts hidden in complex datasets of microarray experiments. The Plant Journal. 2008;55(5):746–59. Epub 2008/05/20. 10.1111/j.1365-313X.2008.03555.x . [DOI] [PubMed] [Google Scholar]
- 29.Thieme CJ, Rojas-Triana M, Stecyk E, Schudoma C, Zhang W, Yang L, et al. Endogenous Arabidopsis messenger RNAs transported to distant tissues. Nature Plants. 2015;1(4):15025. Epub 2015/01/01. 10.1038/nplants.2015.25 . [DOI] [PubMed] [Google Scholar]
- 30.Quimby BB, Dasso M. The small GTPase Ran: interpreting the signs. Current Opinion in Cell Biology. 2003;15(3):338–44. Epub 2003/06/06. 10.1016/s0955-0674(03)00046-2 . [DOI] [PubMed] [Google Scholar]
- 31.Vernoud V, Horton AC, Yang Z, Nielsen E. Analysis of the small GTPase gene superfamily of Arabidopsis. Plant Physiology. 2003;131(3):1191–208. Epub 2003/03/20. 10.1104/pp.013052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Clarke PR, Zhang C. Spatial and temporal coordination of mitosis by Ran GTPase. Nature Reviews Molecular Cell Biology. 2008;9(6):464–77. Epub 2008/05/15. 10.1038/nrm2410 . [DOI] [PubMed] [Google Scholar]
- 33.Haizel T, Merkle T, Pay A, Fejes E, Nagy F. Characterization of proteins that interact with the GTP-bound form of the regulatory GTPase Ran in Arabidopsis. The Plant Journal. 1997;11(1):93–103. Epub 1997/01/01. 10.1046/j.1365-313x.1997.11010093.x . [DOI] [PubMed] [Google Scholar]
- 34.Ma L, Hong Z, Zhang Z. Perinuclear and nuclear envelope localizations of Arabidopsis Ran proteins. Plant Cell Reports. 2007;26(8):1373–82. Epub 2007/05/29. 10.1007/s00299-007-0367-y . [DOI] [PubMed] [Google Scholar]
- 35.Bischoff FR, Ponstingl H. Catalysis of guanine nucleotide exchange on Ran by the mitotic regulator RCC1. Nature. 1991;354(6348):80–2. Epub 1991/11/07. 10.1038/354080a0 . [DOI] [PubMed] [Google Scholar]
- 36.Renault L, Kuhlmann J, Henkel A, Wittinghofer A. Structural basis for guanine nucleotide exchange on Ran by the regulator of chromosome condensation (RCC1). Cell. 2001;105(2):245–55. Epub 2001/05/05. 10.1016/s0092-8674(01)00315-4 . [DOI] [PubMed] [Google Scholar]
- 37.Brown BA, Cloix C, Jiang GH, Kaiserli E, Herzyk P, Kliebenstein DJ, et al. A UV-B-specific signaling component orchestrates plant UV protection. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(50):18225–30. Epub 2005/12/07. 10.1073/pnas.0507187102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lonhienne TG, Forwood JK, Marfori M, Robin G, Kobe B, Carroll BJ. Importin-beta is a GDP-to-GTP exchange factor of Ran: implications for the mechanism of nuclear import. The Journal of Biological Chemistry. 2009;284(34):22549–58. Epub 2009/06/25. 10.1074/jbc.M109.019935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bischoff FR, Klebe C, Kretschmer J, Wittinghofer A, Ponstingl H. RanGAP1 induces GTPase activity of nuclear Ras-related Ran. Proceedings of the National Academy of Sciences of the United States of America. 1994;91(7):2587–91. Epub 1994/03/29. 10.1073/pnas.91.7.2587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Becker J, Melchior F, Gerke V, Bischoff FR, Ponstingl H, Wittinghofer A. RNA1 encodes a GTPase-activating protein specific for Gsp1p, the Ran/TC4 homologue of Saccharomyces cerevisiae. The Journal of Biological Chemistry. 1995;270(20):11860–5. Epub 1995/05/19. 10.1074/jbc.270.20.11860 . [DOI] [PubMed] [Google Scholar]
- 41.Seewald MJ, Korner C, Wittinghofer A, Vetter IR. RanGAP mediates GTP hydrolysis without an arginine finger. Nature. 2002;415(6872):662–6. Epub 2002/02/08. 10.1038/415662a . [DOI] [PubMed] [Google Scholar]
- 42.Klebe C, Bischoff FR, Ponstingl H, Wittinghofer A. Interaction of the nuclear GTP-binding protein Ran with its regulatory proteins RCC1 and RanGAP1. Biochemistry. 1995;34(2):639–47. Epub 1995/01/17. 10.1021/bi00002a031 . [DOI] [PubMed] [Google Scholar]
- 43.Xu XM, Meulia T, Meier I. Anchorage of plant RanGAP to the nuclear envelope involves novel nuclear-pore-associated proteins. Current Biology: CB. 2007;17(13):1157–63. Epub 2007/06/30. 10.1016/j.cub.2007.05.076 . [DOI] [PubMed] [Google Scholar]
- 44.Bischoff FR, Krebber H, Smirnova E, Dong W, Ponstingl H. Co-activation of RanGTPase and inhibition of GTP dissociation by Ran-GTP binding protein RanBP1. The EMBO Journal. 1995;14(4):705–15. Epub 1995/02/15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yokoyama N, Hayashi N, Seki T, Pante N, Ohba T, Nishii K, et al. A giant nucleopore protein that binds Ran/TC4. Nature. 1995;376(6536):184–8. Epub 1995/07/13. 10.1038/376184a0 . [DOI] [PubMed] [Google Scholar]
- 46.Cho HK, Park JA, Pai HS. Physiological function of NbRanBP1 in Nicotiana benthamiana. Molecules and Cells. 2008;26(3):270–7. Epub 2008/05/31. . [PubMed] [Google Scholar]
- 47.Yasuda K, Sugiura K, Takeichi T, Ogawa Y, Muro Y, Akiyama M. Nuclear envelope localization of Ran-binding protein 2 and Ran-GTPase-activating protein 1 in psoriatic epidermal keratinocytes. Experimental Dermatology. 2014;23(2):119–24. Epub 2014/01/21. 10.1111/exd.12324 . [DOI] [PubMed] [Google Scholar]
- 48.Kim SH, Roux SJ. An Arabidopsis Ran-binding protein, AtRanBP1c, is a co-activator of Ran GTPase-activating protein and requires the C-terminus for its cytoplasmic localization. Planta. 2003;216(6):1047–52. Epub 2003/04/11. 10.1007/s00425-002-0959-2 . [DOI] [PubMed] [Google Scholar]
- 49.Wang X, Xu Y, Han Y, Bao S, Du J, Yuan M, et al. Overexpression of RAN1 in rice and Arabidopsis alters primordial meristem, mitotic progress, and sensitivity to auxin. Plant Physiology. 2006;140(1):91–101. Epub 2005/12/20. 10.1104/pp.105.071670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zuccolo M, Alves A, Galy V, Bolhy S, Formstecher E, Racine V, et al. The human Nup107-160 nuclear pore subcomplex contributes to proper kinetochore functions. The EMBO Journal. 2007;26(7):1853–64. Epub 2007/03/17. 10.1038/sj.emboj.7601642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xu XM, Zhao Q, Rodrigo-Peiris T, Brkljacic J, He CS, Muller S, et al. RanGAP1 is a continuous marker of the Arabidopsis cell division plane. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(47):18637–42. Epub 2008/11/18. 10.1073/pnas.0806157105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zang A, Xu X, Neill S, Cai W. Overexpression of OsRAN2 in rice and Arabidopsis renders transgenic plants hypersensitive to salinity and osmotic stress. Journal of Experimental Botany. 2010;61(3):777–89. Epub 2009/12/19. 10.1093/jxb/erp341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rodrigo-Peiris T, Xu XM, Zhao Q, Wang HJ, Meier I. RanGAP is required for post-meiotic mitosis in female gametophyte development in Arabidopsis thaliana. Journal of Experimental Botany. 2011;62(8):2705–14. Epub 2011/02/02. 10.1093/jxb/erq448 . [DOI] [PubMed] [Google Scholar]
- 54.Chen N, Xu Y, Wang X, Du C, Du J, Yuan M, et al. OsRAN2, essential for mitosis, enhances cold tolerance in rice by promoting export of intranuclear tubulin and maintaining cell division under cold stress. Plant, Cell & Environment. 2011;34(1):52–64. Epub 2010/09/10. 10.1111/j.1365-3040.2010.02225.x . [DOI] [PubMed] [Google Scholar]
- 55.Zhang MS, Arnaoutov A, Dasso M. RanBP1 governs spindle assembly by defining mitotic Ran-GTP production. Developmental Cell. 2014;31(4):393–404. Epub 2014/12/03. 10.1016/j.devcel.2014.10.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xu P, Cai W. RAN1 is involved in plant cold resistance and development in rice (Oryza sativa). Journal of Experimental Botany. 2014;65(12):3277–87. Epub 2014/05/03. 10.1093/jxb/eru178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xu P, Zang A, Chen H, Cai W. The small G protein AtRAN1 regulates vegetative growth and stress tolerance in Arabidopsis thaliana. PloS One. 2016;11(6):e0154787. Epub 2016/06/04. 10.1371/journal.pone.0154787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liu Y, Burgos JS, Deng Y, Srivastava R, Howell SH, Bassham DC. Degradation of the endoplasmic reticulum by autophagy during endoplasmic reticulum stress in Arabidopsis. The Plant Cell. 2012;24(11):4635–51. Epub 2012/11/24. 10.1105/tpc.112.101535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Porter KR, Machado RD. Studies on the endoplasmic reticulum. IV. Its form and distribution during mitosis in cells of onion root tip. The Journal of Biophysical and Biochemical Cytology. 1960;7:167–80. Epub 1960/02/01. 10.1083/jcb.7.1.167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Burke B, Ellenberg J. Remodelling the walls of the nucleus. Nature Reviews Molecular cell Biology. 2002;3(7):487–97. Epub 2002/07/03. 10.1038/nrm860 . [DOI] [PubMed] [Google Scholar]
- 61.Anderson DJ, Hetzer MW. Nuclear envelope formation by chromatin-mediated reorganization of the endoplasmic reticulum. Nature Cell Biology. 2007;9(10):1160–6. Epub 2007/09/11. 10.1038/ncb1636 . [DOI] [PubMed] [Google Scholar]
- 62.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Research. 1997;25(17):3389–402. Epub 1997/09/01. 10.1093/nar/25.17.3389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Molecular Biology and Evolution. 2013;30(12):2725–9. Epub 2013/10/18. 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Krysan PJ, Young JC, Tax F, Sussman MR. Identification of transferred DNA insertions within Arabidopsis genes involved in signal transduction and ion transport. Proceedings of the National Academy of Sciences of the United States of America. 1996;93(15):8145–50. Epub 1996/07/23. 10.1073/pnas.93.15.8145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Krysan PJ, Young JC, Sussman MR. T-DNA as an insertional mutagen in Arabidopsis. The Plant Cell. 1999;11(12):2283–90. Epub 1999/12/10. 10.1105/tpc.11.12.2283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. The Plant Journal. 1998;16(6):735–43. Epub 1999/03/09. 10.1046/j.1365-313x.1998.00343.x . [DOI] [PubMed] [Google Scholar]
- 67.Jackson D. In-situ hybridization in plants. Molecular Plant Pathology: A Practical Approach. 1991:163–74. [Google Scholar]
- 68.Vielle-Calzada JP, Thomas J, Spillane C, Coluccio A, Hoeppner MA, Grossniklaus U. Maintenance of genomic imprinting at the Arabidopsis medea locus requires zygotic DDM1 activity. Genes & Development. 1999;13(22):2971–82. Epub 1999/12/02. 10.1101/gad.13.22.2971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Smyth DR, Bowman JL, Meyerowitz EM. Early flower development in Arabidopsis. The Plant Cell. 1990;2(8):755–67. Epub 1990/08/01. 10.1105/tpc.2.8.755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Krebs S, Fischaleck M, Blum H. A simple and loss-free method to remove TRIzol contaminations from minute RNA samples. Analytical Biochemistry. 2009;387(1):136–8. Epub 2009/01/17. 10.1016/j.ab.2008.12.020 . [DOI] [PubMed] [Google Scholar]
- 71.Brosnan CA, Mitter N, Christie M, Smith NA, Waterhouse PM, Carroll BJ. Nuclear gene silencing directs reception of long-distance mRNA silencing in Arabidopsis. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(37):14741–6. Epub 2007/09/06. 10.1073/pnas.0706701104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Schenk P, Kazan K, Manners J, Anderson J, Simpson R, Wilson I, et al. Systemic gene expression in Arabidopsis during an incompatible interaction with Alternaria brassicicola. Plant Physiology. 2003;132(2):999–1010. Epub 2003/06/01. 10.1104/pp.103.021683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Carroll BJ, Klimyuk VI, Thomas CM, Bishop GJ, Harrison K, Scofield SR, et al. Germinal transpositions of the maize element Dissociation from T-DNA loci in tomato. Genetics. 1995;139(1):407–20. Epub 1995/01/01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Johnson-Brousseau SA, McCormick S. A compendium of methods useful for characterizing Arabidopsis pollen mutants and gametophytically-expressed genes. The Plant Journal. 2004;39(5):761–75. Epub 2004/08/19. 10.1111/j.1365-313X.2004.02147.x . [DOI] [PubMed] [Google Scholar]
- 75.Liu CM, Meinke DW. The titan mutants of Arabidopsis are disrupted in mitosis and cell cycle control during seed development. The Plant Journal. 1998;16(1):21–31. Epub 1998/11/10. 10.1046/j.1365-313x.1998.00268.x . [DOI] [PubMed] [Google Scholar]
- 76.Ross KJ, Fransz P, Jones GH. A light microscopic atlas of meiosis in Arabidopsis thaliana. Chromosome Research. 1996;4(7):507–16. Epub 1996/11/01. 10.1007/BF02261778 . [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Software and parameters used in the analysis are described in Materials and Methods. The name of the organism is indicated on the tree, followed by the accession number. Scale bar indicates 0.05 substitutions per site. The plant DEM proteins represented in this tree (Viridiplantae) are the same as those used in Fig 1.
(TIF)
Multiple alignments on full-length DEM and DEM-like protein sequences were undertaken using the Multiple Sequence Comparison by Log-Expectation (MUSCLE) tool from EMBL-EBI and visualized using the Multiple Sequence Alignment Viewer from NCBI. Plant DEM homologues are highly conserved with increased level of identity in the C-terminal half of the proteins. Grey boxes represent amino acids in agreement with the consensus of the multiple sequence alignment.
(TIF)
Pairwise sequence alignment of NP_195066.1 and BAA87237.1 was conducted using the LALIGN tool from EMBL-EBI.
(TIF)
(A) Gene structure of DEM1 and DEM2, along with T-DNA insertions (open triangles), HindIII restriction sites (H3), and location of probes (grey boxes) used for DNA and RNA gel blot analysis. dem1-1 and dem2-1 are T-DNA insertion alleles in Ws-0 ecotype, while dem1-2 and dem2-2 are T-DNA insertion alleles in ecotype Col-0. (B) DNA gel blots on replicate DNA extractions confirmed T-DNA insertions in dem1 and dem2 mutants in Ws-0 and Col-0 genetic backgrounds, and in a dem1 dem2 double mutant (Col-0) complemented with a DEM1 transgene driven by its own promoter pDEM1 (dem1+2 comp). DNA blots for Ws-0 lines were hybridized to the DEM1 3’ probe (upper panel LHS), whereas the blots for the Col-0 lines were hybridized to the DEM1 5’ probe (upper panels RHS). DNA gel blots were all hybridized to the DEM2 5’ probe (lower panel). The T-DNA insertion alleles for the Col-0 dem1 and dem2 mutants are indicated by arrowheads. (C) RNA gel blot analysis of floral buds using the DEM1 and DEM2 5’ probes confirmed T-DNA knock-out of both endogenous dem genes, and over-expression of the pDEM1:DEM1 transgene in the complemented Col-0 dem1 dem2 double mutant (dem1+2 comp). Lower panel shows the 25S rRNA loading control. The two lanes for the complemented Col-0 dem1 dem2 double mutant in (B) and (C) represent two separate plants of a transgenic line that was phenotypically normal during vegetative development and partially fertile. (D) RNA gel blot analysis of floral buds using the DEM1 3’ probe confirmed the absence of full-length transcripts in Ws-0 dem1 plants.
(TIF)
(A) Image showing 6-week old plants of wild type (WT) plants, dem1 and dem2 single mutants, dem1/dem1 DEM2/dem2, DEM1/dem1 dem2/dem2, and dem1 dem2 double mutant complemented with pDEM1:DEM1 transgene (dem1+2 comp) in Col-0 genetic background. (B) Normal seed production was observed in dem1 and dem2 single mutants and in dem1/dem1 DEM2/dem2 plants, but not in dem1/dem1 dem2/dem2 double mutant plants complemented with a pDEM1:DEM1 transgene (dem1+2 comp). (C) Percent ovule, pollen and tetrad defects in DEM1/dem1 dem2/dem2 plants (dem) and dem1/dem1 dem2/dem2 double mutant complemented with a pDEM1:DEM1 transgene (dem1+2 comp). At least 200 ovules, 500 pollen grains and 70 tetrads were assayed for each genotype. (D) Partial complementation of ovule and pollen abortion rates in DEM1/dem1 dem2/dem2 (dem) plants hemizygous for the pDEM1:GFP-DEM1 (GFP-DEM1) or pDEM1:DEM1-GFP (DEM1-GFP) transgene. Non-transgenic DEM1/dem1 dem2/dem2 (dem) and wild-type (WT) plants were included as controls. At least 200 ovules and 700 pollen grains were assessed for each genotype.
(TIF)
(A) Pachytene. (B) Diakinesis. (C) Metaphase I. (D) Anaphase I. (E) Telophase I. (F) Anaphase II. (G) Early telophase II. Bar = 10μm.
(TIF)
(A) Root tips of transgenic seedlings expressing GFP-DEM1 (left) or DEM1-GFP (right); bar = 20 μm. (B) DAPI-stained root tip cells expressing GFP-DEM1 or DEM1-GFP showing cytoplasmic and nuclear envelope localization; bar = 10 μm. (C) Localisation of GFP-DEM1 and DAPI in daughter cells of a root tip epidermal cell that has just divided. (D-E) Root tip expressing GFP:DEM1. (E) Higher magnification of sub-cellular localization of GFP:DEM1 in root tips corresponding to inset in (D), showing expression of GFP:DEM1 relative to the nucleus. Bar represents 50 μm for panels (A-D), and 10 μm for panels (E). Left-hand side, centre and right-hand side panels correspond to DAPI-stained, GFP and merged DAPI/GFP images, respectively (B-E). Images are representative of at least five independent transgenic lines for each transgene.
(TIF)
DAPI staining and localization of tRFP-RAN1 at various stages of microgametophyte development. In tetrads and free monocellular microspores (MC), tRFP-RAN1 predominantly co-localized with DAPI in the nucleus (N), but was concentrated in peripheral nuclear foci, particularly in early MC microgametophytes (white arrowhead). In early to mid-stage bicellular (BC) pollen, tRFP-RAN1 was mainly located in the generative cell nucleus (GCN) and weak expression was detected in the vegetative cell nucleus (VCN). In late BC pollen, tRFP-RAN1 was concentrated in extranuclear foci adjacent to the generative cell nucleus (white arrowhead; GCN), and a weak signal was detected in the vegetative cell nucleus. In tricellular (TC) pollen, tRFP-RAN1 was predominantly located in the vegetative cell nucleus (VCN). Bar = 5 μm.
(TIF)
(A) GFP auto-fluorescence was not detected in pre-meiotic and mature ovules. (B) GFP auto-fluorescence was detected in the pollen coat, but not inside pollen grains. (C) GFP auto-fluorescence was not detected in pre-meiotic cells or tetrads. (D) RFP and GFP auto-fluorescence was not detected in the DAPI stained nuclei of pre-meiotic cells, monocellular and bicellular pollen. Auto-fluorescence of RFP and particularly GFP was detected in the pollen coat of monocellular and bicellular pollen. Bar = 5 μm.
(TIF)
T-DNA binary vectors described in this study are all derived from the binary vectors pUQC477 and pUQC214 [71]. 35S represents the Cauliflower Mosaic Virus 35S promoter. The vector pUQC10332 is a derivative of pUQC214 with the 35S:GFP transgene removed, and it has a single NotI site upstream of the BAR selectable marker for inserting additional transgenes. pUQC10847 was constructed by removing the 35S:BAR sequence from pUQC477 [71], leaving a NPTII gene driven by the Agrobacterium NOPALINE SYNTHASE promoter (NOS) as a kanamycin selectable marker linked to a NotI site (for cloning in additional transgenes). OCS 3’, OCTOPINE SYNTHASE 3’ terminator.
(TIF)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Data Availability Statement
All relevant data are within the manuscript and its Supporting Information files.