Abstract
Cholangiocarcinoma (CCA) is a hepatobiliary malignancy associated with steadily increasing incidence and poor prognosis. Ongoing clinical trials are assessing the effectiveness and safety of a few immune checkpoint inhibitors (ICIs) in CCA patients. However, these ICI treatments as monotherapies may be effective for a proportion of patients with CCA. The prevalence and distribution of other immune checkpoints (ICs) in CCA remain unclear. In this pilot study, we screened databases of CCA patients for the expression of 19 ICs and assessed the prognostic significance of these ICs in CCA patients. Notably, expression of immune modulator IDO1 and PD-L1 were linked with poor overall survival, while FASLG and NT5E were related to both worse overall survival and progression-free survival. We also identified immune modulators IDO1, FASLG, CD80, HAVCR2, NT5E, CTLA-4, LGALS9, VTCN1 and TNFRSF14 that synergized with PD-L1 and correlated with worse patient outcomes. In vitro studies revealed that the expression of ICs was closely linked with aggressive CCA subpopulations, such as cancer stem cells and cells undergoing TGF-β and TNF-α-mediated epithelial-to-mesenchymal transition. These findings suggest that the aforementioned IC molecules may serve as potential prognostic biomarkers and drug targets in CCA patients, leading to lasting and durable treatment outcomes.
Keywords: immune checkpoints, EMT, PD-L1, cancer stem cells, cholangiocarcinoma, CD73, Galectin-9, IDO1
1. Introduction
Cholangiocarcinoma (CCA) is the second most common primary liver malignancy. Cholangiocytes, the epithelial cells that line the biliary tree, undergo neoplastic transformation resulting in the formation of CCA [1]. CCA can be categorized as intrahepatic, perihilar, or distal subtypes and accounts for 10–20% of all hepatobiliary malignancies [2]. While potentially curable with surgery if diagnosed at an early stage, the overall clinical outcome of CCA continues to be poor due to its propensity for early local invasion, distant metastasis and high recurrence [3,4]. Furthermore, the majority of patients with CCA are diagnosed at advanced stages, and the administration of chemotherapy has shown limited efficacy [5]. Given the aggressive disease course and lack of targeted treatment options for CCA patients, there is a pressing need for new and efficacious treatment modalities for this malignancy.
Immunotherapy, especially immune checkpoint inhibitors (ICIs), has proven to be an effective therapeutic modality in several chemoresistant malignant diseases including melanoma, renal and non-small cell lung cancers [6]. Immune checkpoints (ICs) maintain self-tolerance and, during an immune response, ICs protect normal tissue from damage [7]. However, tumor cells frequently exploit these ICs, serving as a prominent tumor immune evasion mechanism causing T-cell inactivation and downregulation of T-cell responses [7,8]. Immune checkpoint blockade strategies have been effective in reanimating the T-cell antigen-specific response and associated antitumor effects [8]. Within the tumor microenvironment, ICs may serve as therapeutic targets for the treatment of primary liver malignancies including hepatocellular carcinoma (HCC) and CCA. ICIs hold great promise for HCC and CCA as the deregulation of the immune system contributes to the pathogenesis of these liver malignancies [9,10].
In the past few years, ICIs as monotherapy have elicited a durable and robust antitumor response in only a proportion of cancer patients, with variability both between types of cancers and between patients who share a histological type [11,12]. A trial of monotherapy with anti-PD1 antibody pembrolizumab in phase I/II in CCA patients showed a low response rate (10% to 20%), and little is known of the underlying mechanism of resistance [11,13]. Another phase I trial for CCA patients revealed that the combined treatment with anti-PD-1 antibody nivolumab and cisplatin plus gemcitabine was more effective than nivolumab as monotherapy [14]. Similarly, combining anti-PD-L1 antibody durvalumab with the anti-CTLA4 antibody tremelimumab was more effective than monotherapies in CCA patients [15].
Analysis of immune checkpoint marker testing such as PD-L1 alone for patient selection and predicting response to ICI therapy has proven insufficient in many cancers [16]. Therefore, identifying and characterizing additional predictive biomarkers are of the utmost importance for the selection of a subset of CCA patients who are more likely to respond to ICI therapies. A better understanding of alternative checkpoint pathways may be required to increase the clinical benefits of ICIs in CCA patients. These alternative checkpoints may provide additional targets for rational combinatorial therapies that may enhance the effects of immunotherapy in CCA.
Combining the expression of immune checkpoints with additional biomarkers such as those identifying epithelial-to-mesenchymal transition (EMT) and cancer stem cells (CSCs) are also being considered in the management of some cancers [10,17,18]. EMT is defined by the loss of epithelial properties and the concomitant gain of mesenchymal properties. EMT contributes to the invasion and metastasis of tumor cells and also to immunosuppression [19,20]. A correlation between the EMT phenotype with multiple ICs in many patient tumors has been reported [10,21]. In extrahepatic CCA, a close relationship between EMT and PD-L1 expression has been reported [22]. However, little is known regarding the association of other ICs with an EMT phenotype in CCA. CSCs are also referred to as tumor-initiating or tumor-propagating cells. CSCs represent a small subpopulation of cells within the tumor that contributes to tumor initiation, metastasis and recurrence. CSCs are endowed with self-renewal, pluripotent properties and enhanced resistance to chemotherapy compared to the tumor bulk [23]. The ability of PD-L1 to inhibit cancer stemness in CCA has been demonstrated [24]. Other studies have reported the expression of PD-L1 and PD-L2 in CSCs derived from colon and breast cancers [25]. Little is known regarding the association between expressions of other immune modulators and CCA-related CSC phenotype.
In this pilot study, we sought to identify prognostic immune-modulatory molecules in CCA patients. To this end, we analyzed CCA patient databases from The Cancer Genome Atlas (TCGA) and SurvExpress [26,27]. We correlated the expression of immune-related molecules with patient prognosis. Given that EMT and CSCs have a substantial role in CCA initiation and progression, we assessed the association of immune checkpoint molecule expression in aggressive CCA cell subpopulations such as CSCs and cells undergoing transforming growth factor (TGF)-β1- and tumor necrosis factor (TNF)-α-mediated EMT.
2. Materials and Methods
cBioPortal OncoPrint evaluation of immune checkpoint molecules: cBioPortal OncoPrint (http://cbioportal.org, accessed on 10 May 2021) was used to generate a graphical summary of gene expression changes in immune checkpoint molecules across CCA patient samples. Within cBioPortal, we utilized the Cholangiocarcinoma (TCGA, Firehose Legacy) case set of 51 patients to evaluate gene changes in immune checkpoint genes. CCA patients are represented as columns and immune-modulatory genes are represented as rows. Genomic alterations, including copy number aberrations, changes in gene or protein expressions and mutations, are represented by glyphs and color codes [27].
CCA patient databases: SurvExpress utilizes a gene expression database of different cancers to generate survival analyses of CCA patients (http://bioinformatica.mty.itesm.mx:8080/Biomatec/SurvivaX.jsp, accessed on 10 May 2021). SurvExpress provided a CCA database of 35 patient samples (CHOL-TCGA Cholangiocarcinoma).
Evaluation of immune checkpoint molecules as prognostic biomarkers in CCA patients: SurvExpres was applied to evaluate the relationship between the expressions of 19 immune modulators with the survival of CCA patients based on a Cox regression analysis. The overall survival for CCA patients was estimated by Kaplan–Meier curves. The average intensity of quantile-normalized array data was used for genes with multiple probe sets. Survival and progression-free survival analyses were also performed using a CCA dataset of 36 patients in cBioPortal.
Cell culture and reagents: Prof. Mark Gorrell, Centenary Institute, Australia kindly gifted human CCA cell lines HuCCT-1 and CCLP-1. Human CCA cell line EGI-1 was sourced from Prof. John Mariadason, Olivia Newton-John Cancer Research Institute, Heidelberg, VIC, Australia. MycoAlert tests (ABM, Richmond, BC, Canada) confirmed the mycoplasma-free status of these cell lines. HuCCT-1 was cultured in Roswell Park Memorial Institute (RPMI); 1640 medium (Thermo Fisher Scientific Australia, Scoresby, VIC, Australia) supplemented with 10% fetal bovine serum (FBS) (Gibco, Life Technologies Australia Pty Ltd, Mulgrave, VIC, Australia) and 0.05% Gentamicin (Thermo Fisher Scientific Australia, Scoresby, VIC, Australia). Dulbecco’s modified Eagle’s medium (DMEM) (Thermo Fisher Scientific Australia, Scoresby, VIC, Australia) supplemented with 10% FBS (Gibco, Life Technologies Australia Pty Ltd, Mulgrave, VIC, Australia) and 0.05% Gentamicin (Thermo Fisher Scientific Australia, Scoresby, VIC, Australia) was used to culture CCLP-1. EGI-1 was cultured in DMEM (Thermo Fisher Scientific Australia, Scoresby, VIC, Australia) with 10% FBS (Gibco, Life Technologies Australia Pty Ltd, Mulgrave, VIC, Australia) and 1% penicillin/streptomycin (P/S) (Thermo Fisher Scientific Australia, Scoresby, VIC, Australia). Cells were cultured under a humidified atmosphere with 5% CO2 in the air at 37 °C. The cytokines TGF-β1 and TNF-α were procured from PeproTech, Cranbury, NJ, USA.
3-dimensional sphere enrichment assay: Trypsin-EDTA was used to detach cells grown as monolayers. Cells were suspended in serum-free stem cell medium following the removal of serum with 1 × PBS washes [28]. The serum-free stem cell medium was prepared with DMEM/F12 medium (Thermo Fisher Scientific Australia, Scoresby, VIC, Australia) supplemented with 20 ng/mL recombinant human epidermal growth factor (rhEGF) (PeproTech, Cranbury, NJ, USA), 10 ng/mL recombinant human fibroblast growth factor (rhFGF) (PeproTech, Cranbury, NJ, USA), 2% B27 supplement without vitamin A (Invitrogen, Scoresby, VIC, Australia) and 1% N2 supplement (Invitrogen, Scoresby, VIC, Australia). Then, 5000 cells were plated per well in ultra-low attachment, 6-well plates (Corning, Melbourne, VIC, Australia). Cells were cultured in a humidified atmosphere of 5% CO2 in air at 37 °C for 7 days. The spheres were collected by gentle centrifugation.
RNA extraction and cDNA synthesis: ISOLATE II RNA Mini Kit (Bioline, Eveleigh, NSW, Australia) was used for the purification of RNA [29]. A NanoDrop 2000c spectrophotometer (Thermo Fisher Scientific Australia, Scoresby, VIC, Australia) was used to confirm RNA quantity and purity. Then, 1 µg RNA was reverse transcribed to cDNA with a Bioline SensiFAST cDNA Synthesis Kit (Bioline, Eveleigh, NSW, Australia).
Quantitative reverse transcription-PCR (qRT-PCR): Applied Biosystems ViiA 7 Real-Time PCR System was used for performing qRT-PCR with Lo-ROX SYBR Green (Bioline, Eveleigh, NSW, Australia) [28]. Briefly, a 3-step cycle of the following conditions, 95 °C for 5 s, 63 °C for 20 s and 75 °C for 20 s was repeated for 40 cycles. Beta-Actin (ActB) was used as the housekeeping gene. Table 1 lists the primers used in this study. The 2ΔΔCt method was used for data analysis. In this 2ΔΔCt method, candidate gene expression was normalized to ActB expression, and copies of target gene per 10,000 copies of ActB was used to present data.
Table 1.
List of qRT-PCR primers.
| Primers | Forward Sequence (5′–3′) | Reverse Sequence (5′–3′) |
|---|---|---|
| ActB | CCAACCGCGAGAAGATGA | CCAGAGGCGTACAGGGATAG |
| CD13 | CAGTGACACGACGATTCTCC | CCTGTTTCCTCGTTGTCCTT |
| CD24 | CACGCAGATTTATTCCAGTGAAAC | GACCACGAAGAGACTGGCTGTT |
| CD44 | CACGTGGAATACACCTGCAA | GACAAGTTTTGGTGGCACG |
| CD90 | AGGACGAGGGCACCTACAC | GCCCTCACACTTGACCAGTT |
| CD133 | GCTTCAGGAGTTTCATGTTGG | GGGGAATGCCTACATCTGG |
| ALDH1A1 | CGGGAAAAGCAATCTGAAGAGGG | GATGCGGCTATACAACACTGGC |
| EpCAM | CCCATCTCCTTTATCTCAGCC | CTGAATTCTCAATGCAGGGTC |
| OCT4 | TTGTGCCAGGGTTTTTGG | ACTTCACCTTCCCTCCAACC |
| SOX2 | ATGGGTTCGGTGGTCAAGT | GGAGGAAGAGGTAACCACAGG |
| NANOG | CTCCAACATCCTGAACCTCAGC | CGTCACACCATTGCTATTCTTCG |
| KLF4 | CATCTCAAGGCACACCTGCGAA | TCGGTCGCATTTTTGGCACTGG |
| ZO1 | GTCCAGAATCTCGGAAAAGTGCC | CTTTCAGCGCACCATACCAACC |
| KRT19 | GGTCAGTGTGGAGGTGGATT | TCAGTAACTCGGACCTGCT |
| E-Cad | AGGCCAAGCAGCAGTACATT | ATTCACATCCAGCACATCCA |
| N-cad | TCCTTGCTTCTGACAATGGA | TTCGCAAGTCTCTGCCTCTT |
| Occludin | TAGTCAGATGGGGGTGAAGG | CATTTATGATGAGCAGCCCC |
| ZEB1 | GGCATACACCTACTCAACTACGG | TGGGCGGTGTAGAATCAGAGTC |
| Slug | TGGTTGCTTCAAGGACACAT | GTTGCAGTGAGGGCAAGAA |
| Fibronectin | CAGTGGGAGACCTCGAGAAG | TCCCTCGGAACATCAGAAAC |
| LGALS9 | ACACCCAGATCGACAACTCCTG | CAAACAGGTGCTGACCATCCAC |
| TNFRSF14 | TTCTCTCAGGGAGCCTCGTCAT | CTCACCTTCTGCCTCCTGTCTT |
| FASLG | CCTTGGTAGGATTGGGCCTG | TCTGGCTGGTAGACTCTCGG |
| TGF-β1 | TACCTGAACCCGTGTTGCTCTC | GTTGCTGAGGTATCGCCAGGAA |
| TNF-α | CCCAGGGACCTCTCTCTAATC | TCTCAGCTCCACGCCATT |
| PD-L1 | GCTGCACTAATTGTCTATTGGGA | AATTCGCTTGTAGTCGGCACC |
| NT5E | TTGGAAATTTGGCCTCTTTG | ACTTCATGAACGCCCTGC |
| VTCN1 | TCTGGGCATCCCAAGTTGAC | TCCGCCTTTTGATCTCCGATT |
Western blot analysis: Western blot analyses were performed as previously described [29]. Briefly, cells were cultured and treated in 6-well plates. RIPA buffer (Thermo Fisher Scientific Australia, Scoresby, VIC, Australia) with Complete (Roche, Australia) and PhosSTOP (Roche, Sydney, NSW, Australia) protease and phosphatase inhibitors were used to lyse cells at 4 °C. Pierce BCA Protein Assay Kit (Thermo Fisher Scientific Australia, Scoresby, VIC, Australia) was utilized to measure total protein concentration. Then, 10 µg of protein was separated by electrophoresis (SDS-PAGE) in a polyacrylamide gel containing sodium dodecyl sulphate (SDS) and transferred to a polyvinylidene difluoride film (PVDF) membrane. Additionally, 5% skim milk in Tris-buffered saline containing 0.1% Tween 20 (TBS-T) was used to block the membranes. Next, the membranes were exposed to primary antibodies at 4 °C overnight. SuperSignal West Femto Maximum Sensitivity Substrate (Thermo Fisher Scientific Australia, Scoresby, VIC, Australia) detected the proteins on the membranes after exposure to HRP-conjugated secondary antibodies. β-Actin was the housekeeping control. Image Quant LAS 500 was used for image capture. Image Studio™ Lite v5.2 software was used for quantification. Table 2 lists the antibodies used in this study.
Table 2.
List of antibodies.
| Antibodies | Cat. No. | Manufacturer | Antibody Category | Dilution |
|---|---|---|---|---|
| NT5E | ab175396 | Abcam | Primary | 1:6000 |
| LGALS9 | ab227046 | Abcam | Primary | 1:1000 |
| β-Actin | 4967s | Cell Signaling | Primary | 1:4000 |
| Goat anti-mouse HRP | 62-6520 | Invitrogen | Secondary | 1:50,000 |
| Goat anti-rabbit HRP | 65-6120 | Invitrogen | Secondary | 1:50,000 |
Statistical analysis: Kaplan–Meier analysis was used to determine the relationship between immune modulator or EMT or CSC expression and CCA patient survival. A log-rank test was performed, and the p-value for survival analysis was generated [30]. In vitro experiments were repeated at least thrice, and representative results are presented. Using the Kolmogorov–Smirnov Test of Normality, we determined the normal distribution of the in vitro data sets (data not shown). Comparisons of in vitro data were performed with Student’s two-tailed t-test with GraphPad Prism software version 8.00 (GraphPad Software Inc., San Diego, CA, USA). Statistical significance was set at * p < 0.05, ** p < 0.01, *** p < 0.005 and **** p < 0.001.
3. Results
3.1. The Expression of Immune Checkpoint Genes in CCA
To identify ICs involved in CCA immune escape, we examined a panel of 19 immune checkpoint genes previously shown to be associated with patient prognosis in other malignancies. These included both immune-stimulatory and inhibitory genes, namely: FASLG, Galectin-9 (LGALS9), LAG-3, TIM-3 (HAVCR2), VSIR, VTCN1 (B7-H4), IDO-1, TNFRSF9 (CD137), TNFRSF14 (HVEM), TIGIT, CD276 (B7-H3), CD27, PD-L1 (CD274), PD-L2 (PDCD1LG2), NT5E (CD73), CD80, TNFRSF18 (GITR), BTLA and CD28. OncoPrint analysis in cBioPortal was performed to evaluate the expression and any possible genetic changes associated with these ICs in the tumors of CCA patients (n = 51). This patient cohort included 35 patients with intrahepatic cholangiocarcinoma, 9 patients with distal cholangiocarcinoma and 7 patients with perihilar cholangiocarcinoma. Clinical data of the patient cohort is listed in Table 3.
Table 3.
Clinical characteristics of 51 CCA patients.
| CCA Patient ID | Diagnosed Age | Sex | Ablation Embolization Tx Adjuvant | Surgical Margin Resection Status | Adjuvant Postoperative Pharmaceutical Therapy Administered Indicator | American Joint Committee on Cancer Tumor Stage Code | American Joint Committee on Cancer Metastasis Stage Code |
|---|---|---|---|---|---|---|---|
| TCGA-3X-AAV9 | 72 | Male | NO | R0 | YES | T1 | M0 |
| TCGA-5A-A8ZF | NA | NA | NA | NA | NA | NA | NA |
| TCGA-5A-A8ZG | NA | NA | NA | NA | NA | NA | NA |
| TCGA-W7-A93N | NA | NA | NA | NA | NA | NA | NA |
| TCGA-W7-A930 | NA | NA | NA | NA | NA | NA | NA |
| TCGA-W7-A93P | NA | NA | NA | NA | NA | NA | NA |
| TCGA-ZK-AAYZ | NA | NA | NA | NA | NA | NA | NA |
| TCGA-3X-AAVA | 50 | Male | NO | R0 | YES | T1 | M0 |
| TCGA-3X-AAVB | 70 | Female | NO | R0 | NO | T2b | M1 |
| TCGA-3X-AAVC | 72 | Female | NA | R0 | NA | T3 | M0 |
| TCGA-3X-AAVE | 60 | Female | NO | R0 | NO | T1 | M0 |
| TCGA-4G-AAZF | 74 | Male | NO | R0 | NO | T2 | MX |
| TCGA-4G-AAZG | 75 | Female | NO | R0 | NO | T3 | MX |
| TCGA-4G-AAZ0 | 71 | Female | NO | R1 | YES | T2a | M0 |
| TCGA-4G-AAZR | 74 | Male | NO | R1 | NO | T2a | MX |
| TCGA-4G-AAZT | 62 | Male | NO | R0 | NO | T1 | M0 |
| TCGA-W5-AA2G | 62 | Female | NO | R0 | NO | T1 | M0 |
| TCGA-W5-AA2H | 70 | Female | NO | R0 | YES | T3 | M0 |
| TCGA-W5-AA2I | 66 | Male | NO | R0 | NO | T1 | M0 |
| TCGA-W5-AA2J | 66 | Female | NO | R0 | NO | T4 | M0 |
| TCGA-W5-AA2K | 75 | Female | NO | R0 | NO | T1 | M0 |
| TCGA-W5-AA2M | 49 | Male | NO | R0 | NO | T3 | M1 |
| TCGA-W5-AA2O | 57 | Male | NO | R0 | NO | T1 | M0 |
| TCGA-W5-AA2Q | 68 | Male | NO | R0 | NO | T2b | M0 |
| TCGA-W5-AA2R | 77 | Female | NO | R0 | NO | T1 | M0 |
| TCGA-W5-AA2T | 64 | Female | NO | R0 | YES | T2 | M0 |
| TCGA-W5-AA2U | 78 | Female | NO | R0 | NO | T1 | M0 |
| TCGA-W5-AA2W | 31 | Female | NO | R0 | NO | T2a | M0 |
| TCGA-W5-AA2X | 67 | Male | NO | RX | NO | T2b | M1 |
| TCGA-W5-AA2Z | 29 | Female | NO | R1 | YES | T2 | M0 |
| TCGA-W5-AA30 | 82 | Male | NO | R0 | NO | T1 | M0 |
| TCGA-W5-AA31 | 71 | Male | NO | R0 | NO | T1 | M0 |
| TCGA-W5-AA33 | 60 | Male | NO | R0 | NO | T1 | M0 |
| TCGA-W5-AA34 | 75 | Female | NO | R0 | NO | T1 | M0 |
| TCGA-W5-AA36 | 51 | Female | NO | R0 | YES | T3 | M1 |
| TCGA-W5-AA38 | 55 | Female | NO | R0 | NO | T1 | M0 |
| TCGA-W5-AA39 | 81 | Male | NO | RX | NO | T2 | M0 |
| TCGA-W6-AA0S | 46 | Female | NO | R0 | YES | T1 | Mx |
| TCGA-W6-AA0T | 62 | Female | NO | R0 | YES | T3 | M0 |
| TCGA-WD-A7RX | 71 | Female | NO | RX | NO | T2b | MX |
| TCGA-YR-A95A | 52 | Male | NO | R1 | NO | T2 | M1 |
| TCGA-ZD-A8I3 | 73 | Female | NA | R0 | NA | T2 | MO |
| TCGA-ZH-A8Y1 | 74 | Female | NO | R1 | NO | T3 | MO |
| TCGA-ZH-A8Y2 | 59 | Female | NO | R0 | NO | T1 | MO |
| TCGA-ZH-A8Y3 | 61 | Female | NO | R1 | YES | T2a | MO |
| TCGA-ZH-A8Y4 | 58 | Male | NO | R1 | YES | T1 | MO |
| TCGA-ZH-A8Y5 | 69 | Male | NO | R0 | YES | T3 | M1 |
| TCGA-ZH-A8Y6 | 41 | Female | NA | R0 | NA | T1 | MO |
| TCGA-ZH-A8Y7 | 59 | Male | NO | R1 | NO | T3 | M1 |
| TCGA-ZH-A8Y8 | 73 | Male | NO | R0 | NO | T1 | MO |
| TCGA-ZU-A8S4 | 52 | Male | NO | R0 | YES | T1 | MX |
NA: Clinical data not available for the patient.
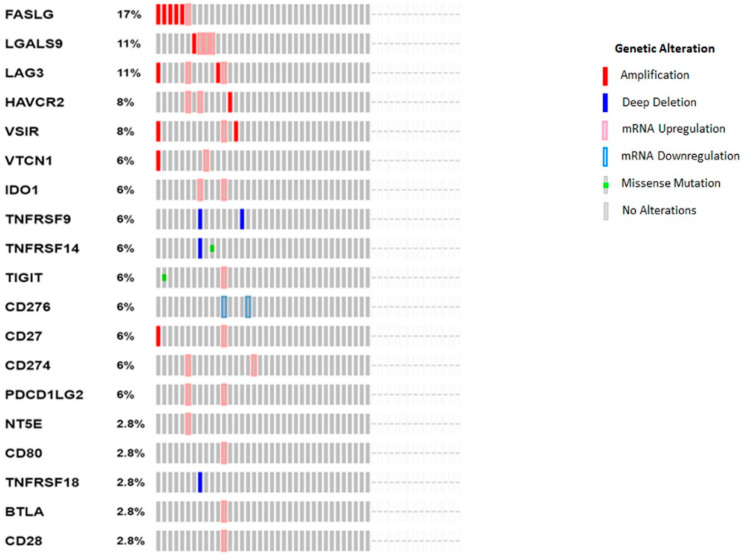
The OncoPrint analyses revealed FASLG (17%) amplification and mRNA upregulation, LGALS9 (11%), LAG3 (11%), HAVCR2 (8%), VSIR (8%), VTCN1 (6%) and CD27 (6%) (Figure 1). mRNA upregulation was identified in IDO1 (6%), CD274 (6%), PDCD1LG2 (6%), NT5E (2.8%), CD80 (2.8%), BTLA (2.8%) and CD28 (2.8%). Briefly, mRNA downregulation was seen for CD276 (6%). Deep deletion was noted in TNFRSF9 (6%) and TNFRSF18 (2.8%). Deep deletion and missense mutation were identified in TNFRSF14 (6%), while missense mutation and mRNA upregulation was noted in TIGIT (6%) (Figure 1). CCA may warrant immunotherapeutic approaches due to increased distribution of immune checkpoint molecules in CCA patients.
Figure 1.
The OncoPrint analyses of CCA patients reveal changes in expression of ICs. Rows and columns represent genes and CCA patients, respectively. Genetic alterations, such as copy number alterations (homozygous deletions and amplifications), mutations and gene expression changes, are represented by glyphs and color codes. The patient order is presented as per alterations.
3.2. Immune Biomarkers Prognosticate Poor Survival in CCA Patients
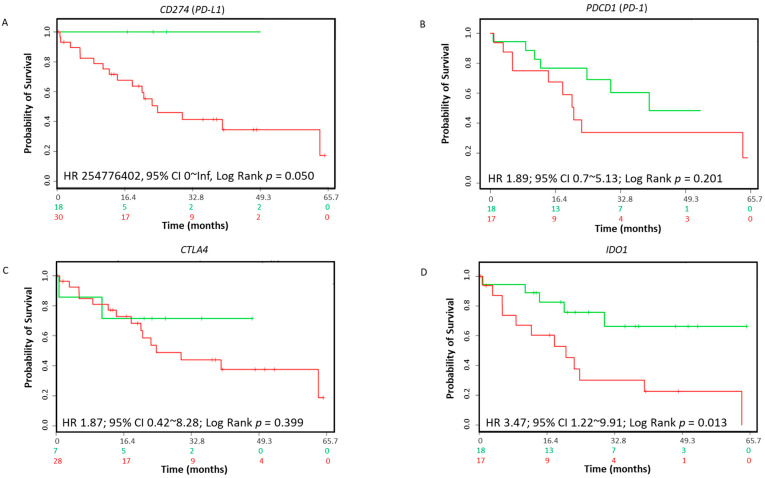
To investigate the prognostic value of ICs in CCA, we evaluated the SurvExpress CCA dataset to evaluate the overall survival in CCA patients. In this cohort of CCA patients, we evaluated whether the expression of putative ICs, PD-L1 (CD274), PD-1 (PDCD1) and CTLA4 were associated with overall survival. No correlation of these ICs with overall survival was observed in CCA patients (Figure 2A–C). Notably, the altered expression of IDO1 (hazard ratio (HR): 3.47; 95% confidence interval (CI): 1.22~9.91; log-rank equal curves p = 0.013) in the 35-CCA-patient cohort was linked with overall worse survival (Figure 2D). Similarly, other immune checkpoint gene expressions showed no correlation with overall CCA patient survival (Table 4). In cBioPortal, the TCGA CCA 36-patient cohort, FASLG (log-rank test p = 1.370 × 10−3) and NT5E (log-rank test p = 2.826 × 10−3) expressions correlated with lower overall survival (Table 5). Similarly, FASLG (log-rank test p = 3.843 × 10−3) and NT5E (log-rank test p = 4.072 × 10−3) expression correlated with lower progression-free survival in the 36 CCA patient cohort (Table 5).
Figure 2.
Association between immune modulators and survival in CCA patients. The Kaplan–Meier survival curves in SurvExpress CCA patients for the gene expression of (A) CD274 (PD-L1), (B) PDCD1 (PD-1), (C) CTLA4 and (D) IDO1. Low-risk groups are indicated in green and high-risk groups are indicated in red. The x-axis presents the study time in months. HR, CI and p-values are shown in the insert.
Table 4.
SurvExpress-based overall survival of 35 CCA patients.
| Immune Modulatory Gene | Risk Groups Hazard Ratio | Confidence Interval | Log-Rank Equal Curves (p-Value) |
|---|---|---|---|
| FASLG | 1.27 | 0.17–9.69 | 0.817 |
| LAG3 | 0.84 | 0.32–2.18 | 0.720 |
| HAVCR2 | 1.87 | 0.71–14.96 | 0.198 |
| VSIR | 1.69 | 0.61–4.68 | 0.309 |
| VTCN1 | 0.87 | 0.33–2.27 | 0.776 |
| TNFRSF9 | 81853258 | 0–∞ | 0.132 |
| TNFRSF14 | 1.39 | 0.53–3.66 | 0.503 |
| TIGIT | 1.37 | 0.5–3.71 | 0.536 |
| CD276 | 0.67 | 0.26–1.75 | 0.414 |
| CD27 | 1.15 | 0.42–3.11 | 0.785 |
| PDCD1LG2 | 1.53 | 0.55–4.21 | 0.409 |
| NT5E | 0.56 | 0.21–1.5 | 0.247 |
| CD80 | 3.41 | 0.45–25.04 | 0.208 |
| TNFRSF18 | 1.92 | 0.73–5.08 | 0.178 |
| BTLA | 1.36 | 0.31–5.97 | 0.686 |
| CD28 | 2.3 | 0.8–6.64 | 0.112 |
Table 5.
cBioPortal-based CCA patient overall survival and progression-free survival.
| Immune Modulatory Gene | Overall Survival Log-Rank Test (p-Value) | Progression-Free Survival Log-Rank Test (p-Value) |
|---|---|---|
| FASLG | 1.370 × 10−3 | 3.843 × 10−3 |
| LGALS9 | 0.286 | 0.759 |
| LAG3 | 0.464 | 0.898 |
| HAVCR2 | 0.095 | 0.225 |
| VSIR | 0.515 | 0.290 |
| VTCN1 | 0.361 | 0.900 |
| IDO1 | 0.450 | 0.242 |
| TNFRSF9 | NA | NA |
| TNFRSF14 | NA | NA |
| TIGIT | 0.515 | 0.290 |
| CD276 | 0.514 | 0.290 |
| CD27 | 0.515 | 0.290 |
| CD274 | 0.054 | 0.109 |
| PDCD1LG2 | 0.464 | 0.898 |
| NT5E | 2.826 × 10−3 | 4.072 × 10−3 |
| CD80 | 0.515 | 0.290 |
| TNFRSF18 | NA | NA |
| BTLA | 0.515 | 0.290 |
| CD28 | 0.515 | 0.290 |
NA: No patient data available with alteration in this gene.
3.3. Association of Clinicopathological Characteristics with Expression of IDO1, FASLG and NT5E in CCA Patients
We next interrogated whether ICs significantly associated with poor prognosis can function as independent risk factors for prognosis prediction in CCA patients. Table 6 shows the relationship between pathological features and expression of IDO1, FASLG and NT5E in CCA patients in the cBioPortal database. Chi-squared tests were used to compare the clinicopathological data. IDO1 expression in CCA patients was associated with neoplasm disease stages II and III (p = 1.874 × 10−3). Patients with altered IDO1 expression was associated with a presence of a risk factors for HCC, including smoking and hepatitis B (p = 0.012).
Table 6.
Relationship between expression of IDO1, FASLG and NT5E and clinical parameters in CCA patients.
| Clinical Attribute | p = 1.874 × 10−3 | p = 0.937 | p = 0.969 | |||
|---|---|---|---|---|---|---|
| Neoplasm disease stage American Joint Committee on Cancer Code | Altered IDO1 | Unaltered | Altered FASLG | Unaltered | Altered NT5E | Unaltered |
| Stage I | 0% | 55.88% | 20% | 58.06% | 100% | 51.43% |
| Stage II | 50% | 23.53% | 60% | 19.35% | 0% | 25.71% |
| Stage III | 50% | 0% | 0% | 3.23% | 0% | 2.86% |
| Stage IV | 0% | 5.88% | 0% | 6.45% | 0% | 5.71% |
| Stage IVA | 0% | 5.88% | 20% | 3.23% | 0% | 5.71% |
| Stage IVB | 0% | 8.82% | 0% | 9.68% | 0% | 8.57% |
| p = 0.739 | p = 0.018 | p = 3.492 × 10−3 | ||||
| Metastasis stage American Joint Committee on Cancer Code | Altered IDO1 | Unaltered | Altered FASLG | Unaltered | Altered NT5E | Unaltered |
| M0 | 100% | 76.47% | 60% | 80.65% | 0% | 80% |
| M1 | 0% | 14.71% | 0% | 16.13% | 0% | 14.29% |
| MX (No information available) | 0% | 8.82% | 40% | 3.23% | 100% | 5.71% |
| p = 0.665 | p = 0.041 | p = 0.041 | ||||
| Neoplasm disease lymph node stage American Joint Committee on Cancer Code | Altered IDO1 | Unaltered | Altered FASLG | Unaltered | Altered NT5E | Unaltered |
| N0 | 0% | 70.59% | 40% | 77.42% | 0% | 74.29% |
| N1 | 74.29% | 14.71% | 20% | 12.9% | 0% | 14.29% |
| NX (No information available) | 14.29% | 14.71% | 40% | 9.68% | 100% | 11.43% |
| p = 0.301 | p = 0.937 | p = 0.922 | ||||
| Tumor stage American Joint Committee on Cancer Code | Altered IDO1 | Unaltered | Altered FASLG | Unaltered | Altered NT5E | Unaltered |
| T1 | 0% | 55.88% | 20% | 58.06% | 100% | 51.43% |
| T2 | 50% | 14.71% | 0% | 19.35% | 0% | 17.14% |
| T2a | 0% | 5.88% | 40% | 0% | 0% | 5.71% |
| T2b | 0% | 11.76% | 40% | 6.45% | 0% | 11.43% |
| T3 | 50% | 11.76% | 0% | 16.13% | 0% | 14.29% |
| p = 0.049 | p = 1.046 × 10−3 | p = 3.565 × 10−3 | ||||
| Neoadjuvant therapy type administered prior to resection | Altered IDO1 | Unaltered | Altered FASLG | Unaltered | Altered NT5E | Unaltered |
| YES | 0% | 2.94% | 0% | 2.86% | 0% | 2.86% |
| NO | 100% | 97.06% | 100% | 97.14% | 100% | 97.14% |
| p = 0.012 | p = 1.00 | p = 0.998 | ||||
| History hepatocellular risk factor | Altered IDO1 | Unaltered | Altered FASLG | Unaltered | Altered NT5E | Unaltered |
| Hepatitis B and smoking | 50% | 0% | 60% | 6.67% | 0% | 26.47% |
| No history of primary risk factors | 50% | 58.82% | 20% | 63.33% | 100% | 55.88% |
| Other risk factors (cirrhosis/diabetes mellitus/NAFLD/ smoking/ulcerative colitis) | 0% | 41.1% | 20% | 30% | 0% | 17.65% |
| p = 0.569 | p = 0.910 | p = 0.930 | ||||
| Gender | Altered IDO1 | Unaltered | Altered FASLG | Unaltered | AlteredNT5E | Unaltered |
| Male | 50% | 44.12% | 40% | 45.16% | 100% | 44.12% |
| Female | 50% | 55.88% | 60% | 54.84% | 0% | 55.88% |
A positive association between FASLG expression was associated with the neoplasm disease lymph node stage in CCA patients (p = 0.041), where 20% of patients with altered FASLG expression were in the American Joint Committee on Cancer (AJCC) lymph node stage N1. Moreover, 60% of CCA patients with FASLG expression were in the AJCC metastasis stage M0 exhibiting no metastasis (p = 0.018). However, 40% of patients with altered FASLG expression were in the MX stage with no information available on metastasis status. All patients with NT5E expression were in MX and NX of AJCC metastasis and lymph node stages, respectively, where no information was available on metastasis or lymph node status.
IDO1- (p = 0.049), FASLG- (p = 1.046e-3) and NT5E-expressing (p = 3.565e-3) CCA patients were not administered neoadjuvant therapy prior to resection. Expression of these genes was not associated with clinical parameters such as vascular invasion, gender and AJCC tumor stage code T1–T3.
3.4. Coordinate Expression of PD-L1 (CD274), PD-1 and CTLA-4 and Immune Checkpoint Genes in CCA Patients
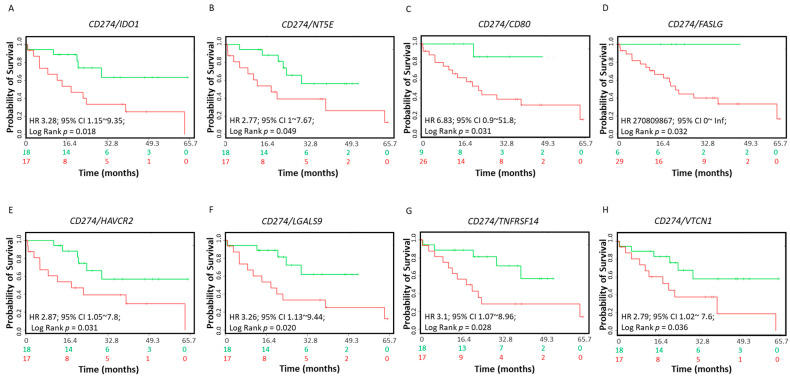
Varying degrees of clinical responses to ICI treatments, including anti-PD-L1, anti-PD-1 or anti-CTLA-4, have been noted in different tumor types. Research has focused on identifying predictive biomarkers to stratify patients to increase the rate of response to ICI therapies. In CCA, the coordinated expression of other ICs with PD-L1, PD-1 and CTLA-4 have not been previously explored. We evaluated whether the coordinate expression of ICs with PD-L1, PD-1 and CTLA-4 showed survival benefits. Coordinate expression of IDO1 (HR: 3.28, CI: 1.15~9.35, log-rank equal curves p = 0.018), CD73 (HR: 2.77, CI: 1~7.57, log-rank equal curves p = 0.040), CD80 (HR: 6.83, CI: 0.9~51.8, log-rank equal curves p = 0.031), FASLG (HR: 270809867, CI: 0~inflog-rank equal curves p = 0.032), HAVCR2 (HR: 2.87, CI: 1.05~7.8, log-rank equal curves p = 0.039), LGALS9 (HR: 3.25, CI: 1.13~9.44, log-rank equal curves p = 0.020), TNFRSF14 (HR: 3.1, CI: 1.07~8.96, log-rank equal curves p = 0.037) and VTCN1 (HR: 2.79, CI: 1.02~7.6, log-rank equal curves p = 0.036) showed significantly worse overall survival when combined with PD-L1 (Figure 3A–H). The other 10 ICIs in combination with PD-L1 did not show a significant association with survival in CCA patients (Supplementary Materials Table S1).
Figure 3.
Relationship of immune modulators in combination with CD274 (PD-L1) and survival in CCA patients. The Kaplan–Meier survival curves in SurvExpress CCA patients for the gene expression of (A) CD274/IDO1, (B) CD274/NT5E, (C) CD274/CD80, (D) CD274/FASLG, (E) CD274/HAVCR2, (F) CD274/LGALS9, (G) CD274/TNFRSF14 and (H) CD274/VTCN1. Low-risk groups are indicated in green and high-risk groups are indicated in red. The x-axis presents the study time in months. HR, CI and p-values are shown in the insert.
Combining the expression of TIGIT (HR: 3.43, CI: 1.15~9.96, log-rank equal curves p = 0.016), TNFRSF14 (HR: 3.06, CI: 1.06~8.84, log-rank equal curves p = 0.029), CTLA4 (HR: 3.02, CI: 1.08~8.45, log-rank equal curves p = 0.027) and FASLG (HR: 2.99, CI: 1.06~8.4, log-rank equal curves p = 0.003) with PD-1 (PDCD1) showed worse overall survival (Supplementary Materials Figure S1A–D). IDO1 (HR: 3.99, CI: 1.39~11.44, log-rank equal curves p = 0.005) and PD-L1 (HR: 270809867, CI: 0~inf, log-rank equal curves p = 0.032) revealed worse overall survival in combination with CTLA-4 (Supplementary Materials Figure S1E,F).
3.5. Anchorage-Independent Three-Dimensional CCA Spheres Express Embryonic Stemness and CSC Markers
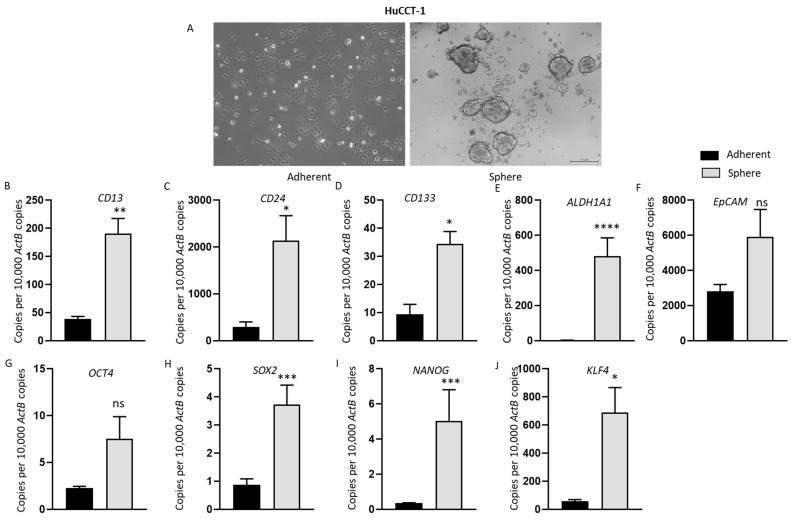
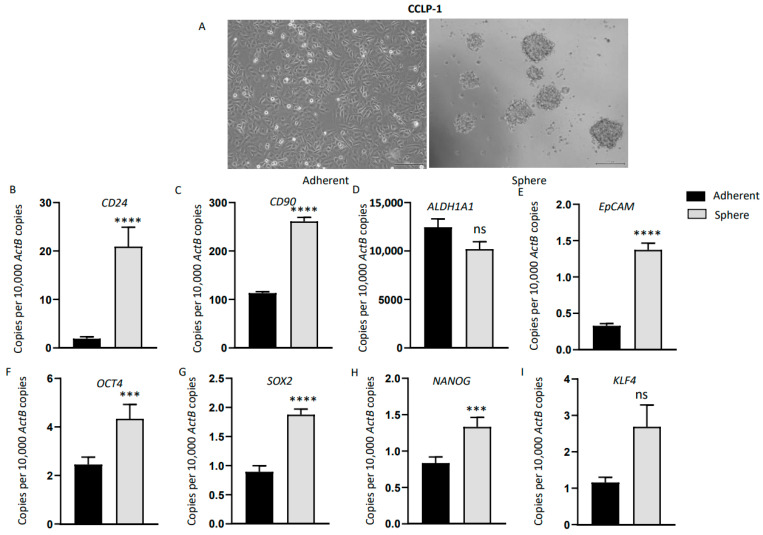
To assess the relationship between immune checkpoint modulators and CSCs, three-dimensional spheres were generated using human CCA cell lines HuCCT-1, CCLP-1 and EGI-1. By day 7, anchorage-independent spheres were enriched in all CCA cell lines cultured in serum-free stem cell medium (Figure 4A, Figure 5A and Figure S2A). We confirmed the stemness of CCA-derived spheres by examining the expression of stemness genes essential for the proliferation, self-renewal and differentiation of stem cells, including CD13, CD24, CD44, CD90, CD133, ALDH1A1, epithelial cell adhesion molecule (EpCAM), Krüppel-like factor 4 (Klf4) and octamer-binding transcription factor 4 (OCT4). These CSC markers have been widely used individually or in combination to characterize CSCs in CCA [18,31,32]. As controls, CCA cell lines were grown as adherent monolayers at the same density as the spheres. Three-dimensional sphere cultures and adherent cultures were subjected to RNA extraction on day seven. HuCCT-1 spheres showed markedly elevated expression of stemness markers CD13, CD24, CD133, ALDH1A1 and EpCAM compared with parental adherent cells (Figure 4B–F). In comparison with parental adherent cells, HuCCT-1 spheres also showed upregulation in the expression of the embryonic stem-cell-associated genes OCT4, Sox2, Nanog and Klf4 (Figure 4G–J). Likewise, CCLP-1-derived spheres showed increased expression of stemness markers CD24, CD90 and EpCAM compared with adherent parental cells (Figure 5B–E). In comparison with the adherent parental cells, CCLP-1 spheres showed higher mRNA levels of Oct4, Sox2, Nanog and Klf4 (Figure 5F–I). EGI-1-derived spheres expressed higher levels of CD24, CD44, ALDH1A1, EpCAM and Klf4 (Supplementary Materials Figure S2A–E). These findings indicate that the human CCA stem-like cells can be selectively enriched with stem cell serum-free medium.
Figure 4.
Anchorage-independent three-dimensional spheroid culture enriches HuCCT-1 CSCs. (A) Monolayer culture and 3-D culture of HuCCT-1 cells (scale bar = 200 µm). (B–J) qRT-PCR analysis demonstrated increased expression of embryonic stemness and surface CSC markers in HuCCT-1 spheres compared with HuCCT-1 adherent monolayer culture. Values are mean ± SD of three experiments in triplicate (* p < 0.05, ** p < 0.01, *** p < 0.005, **** p < 0.001, ns: not significant). ActB: β-Actin.
Figure 5.
Anchorage-independent three-dimensional spheroid culture of CCLP-1 cells enriches CSCs. (A) Monolayer culture and 3-D culture of CCLP-1 cells (scale bar = 200 µm). (B–I) Enhanced expression of embryonic stemness and cell surface CSC markers were observed in CCLP-1 spheres compared with CCLP-1 adherent monolayer culture by qRT-PCR. Values are mean ± SD of three experiments in triplicate (*** p < 0.005, **** p < 0.001, ns: not significant). ActB: β-Actin.
3.6. Enhanced Expression of Immune Modulators in CCA Derived Stem-Like Cells
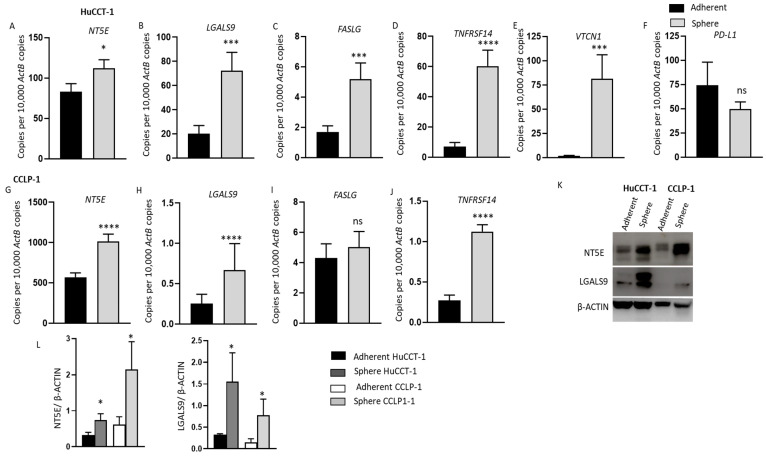
The expression of immune checkpoint regulators linked with poor outcome in patients with CCA was evaluated in CCA-derived CSCs. We compared the expression of immune checkpoints in spheres and adherent parental CCA cell lines using qRT-PCR. In HuCCT-1-derived spheres, we noted markedly increased expression of NT5E, LGALS9, FASLG, TNFRSF14 and VTCN1 compared with adherent cells (Figure 6A–E). However, the spheres showed lower PD-L1 expression levels compared with adherent cells (Figure 6F). Similarly, elevated expression of NT5E, LGALS9, FASLG and TNFRSF14 was detected in CCLP-1-derived spheres compared with parental adherent cells (Figure 6G–J). CCLP-1 cells did not express PD-L1 and VTCN1. IDO1, HAVCR2 and CD80 expression was undetectable in all cell lines. Western blot assay showed increased NT5E and LGALS9 protein expression in CCA-derived CSCs from HuCCT-1 and CCLP-1 compared with adherent cells (Figure 6K,L). EGI-1-derived spheres showed elevated expression of NT5E (Supplementary Materials Figure S2F). No significant difference in expression of other immune checkpoint molecules was noted in EGI-1-derived CSCs and adherent cells (Supplementary Materials Figure S2G–H). Given the high expression of immune checkpoints in CCA-derived CSCs, ICI-based immunotherapy may be used to target the CSC population in CCA to obtain effective and durable treatment outcomes.
Figure 6.
HuCCT-1 and CCLP-1 CSCs have elevated expression of immune modulators. (A–F) qRT-PCR analysis showed upregulation of immune modulators NT5E, LGALS9, FASLG, TNFRSF14 and VTCN1 in HuCCT-1 spheres compared with the parental adherent HuCCT-1 cells. (G–J) Increased expression of immune modulators NT5E, LGALS9, FASLG and TNFRSF14 was detected in mRNA from CCLP-1 spheres versus the parental adherent cells. (K) Western blots demonstrated the upregulation of NT5E and LGALS9 in both HuCCT-1 and CCLP-1 spheres versus the adherent parental cells. (L) Graphs represent quantification of Western blot analyses. Values are mean ± SD of three experiments in triplicate (* p < 0.05, *** p < 0.005, **** p < 0.001, ns: not significant). ActB: β-Actin.
3.7. Induction of Immune Checkpoint Modulator Expression during TGF-β1- and TNF-α-Mediated EMT in Human CCA Cells
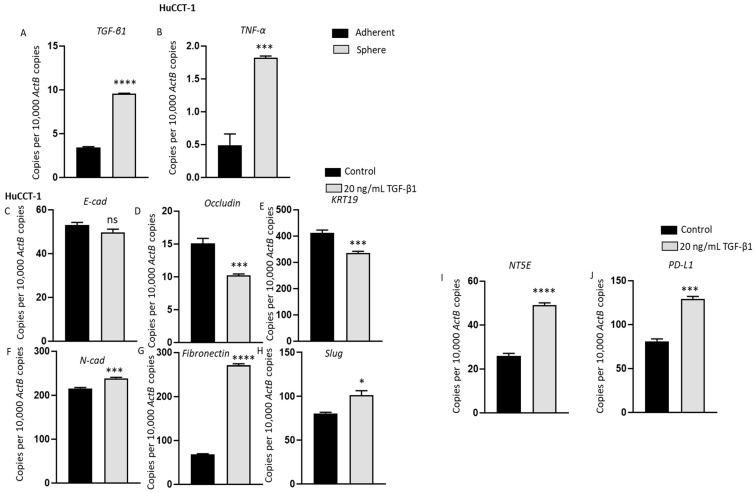
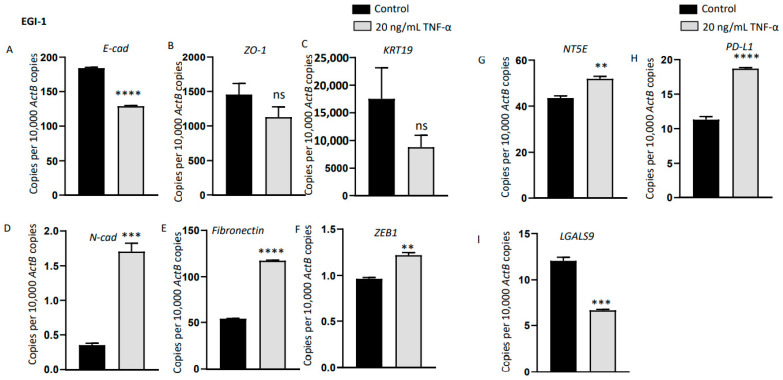
Emerging research has demonstrated a direct association between EMT and acquisition of stem-cell-like features [28,33]. We propose that the elevated expression of immune checkpoints in CSCs is regulated by EMT via TGF-β1 and/or TNF-α. EMT and CSC in turn enhance the invasion, metastasis and recurrence and may contribute to poor prognosis of CCA patients. Therefore, we investigated whether EMT inducers are expressed by CCA stem cells and if EMT can modulate immune checkpoint expression. For the subsequent study, we used cell lines that can undergo EMT upon induction with cytokines. Thus, we chose to study HuCCT-1 and EGI-1 that have an epithelial phenotype and excluded CCLP-1 cells with a mesenchymal phenotype. HuCCT-1 spheres showed upregulation in the expression of potent EMT drivers TGF-β and TNF-α compared with parent adherent cells (Figure 7A,B). Previously, EMT inducers have been shown to induce the expression of PD-L1 and other ICs in HCC [29]. In order to evaluate whether EMT is closely associated with modulation of ICs in CCA, HuCCT-1 cells were treated with 20ng/mL of TGF-β1. After 3 days, HuCCT-1 cells underwent EMT as evidenced by the decrease in epithelial markers E-Cad, Occludin and KRT19 and elevation in mesenchymal markers N-cad, Fibronectin and Slug (Figure 7C–H). HuCCT-1 did not respond to TNF-α stimulation. Elevated expressions of ICs, NT5E and PD-L1 were observed during TGF-β1-induced EMT in HuCCT-1 cells (Figure 7I,L). EGI-1 cells, on the other hand, were more responsive to TNF-α-driven EMT than TGF-β1. After stimulation of EGI-1 cells with 20 ng/mL TNF-α for 3 days, EGI-1 cells underwent EMT with a reduction in the expression of epithelial markers E-Cad, ZO-1 and KRT19 and upregulation in the expression of mesenchymal markers N-cad, Fibronectin and Zeb1 (Figure 8A–F). TNF-α stimulation also induced NT5E and PD-L1 expression (Figure 8G,H). These findings suggest that the expression of ICs in aggressive phenotypes such as cells undergoing EMT and stemness are closely linked.
Figure 7.
TGF-β1-mediated EMT induced upregulation of immune modulators in HuCCT-1 cells. qRT-PCR analysis revealed the upregulation in the expression of (A) TGF-β and (B) TNF-α in HuCCT-1 spheres compared with parent adherent cells. (C–H) qRT-PCR showed epithelial markers E-Cad, Occludin and KRT19 were decreased, and mesenchymal markers N-cad, Fibronectin and Slug were increased after 72 h of TGF-β1 treatment in HuCCT-1 cells. (I–J) qRT-PCR demonstrated elevated expression of immune modulators NT5E and PD-L1 upon 72 h of TGF-β1 treatment in HuCCT-1 cells. Values are mean ± SD of three experiments in triplicate (* p < 0.05, *** p < 0.005, **** p < 0.001, ns: not significant). ActB: β-Actin.
Figure 8.
TNF-α-mediated EMT induced upregulation of immune modulators in CCLP-1 cells. (A–F) qRT-PCR showed reduction in the expression of epithelial markers E-Cad, ZO-1 and KRT19, and elevation in the expression of mesenchymal markers N-cad, Fibronectin and ZEB1 after 72 h of TNF-α treatment in CCLP-1 cells. (G–I) qRT-PCR demonstrated elevated expression of immune modulators NT5E and PD-L1 and decreased level of LGALS9 upon 72 h of TNF-α treatment in CCLP-1 cells. Values are mean ± SD of three experiments in triplicate (** p < 0.01, *** p < 0.005, **** p < 0.001, ns: not significant). ActB: β-Actin.
3.8. Coordinate Expression of Immune Modulatory Genes, EMT Markers and Stemness Marker in CCA Patients
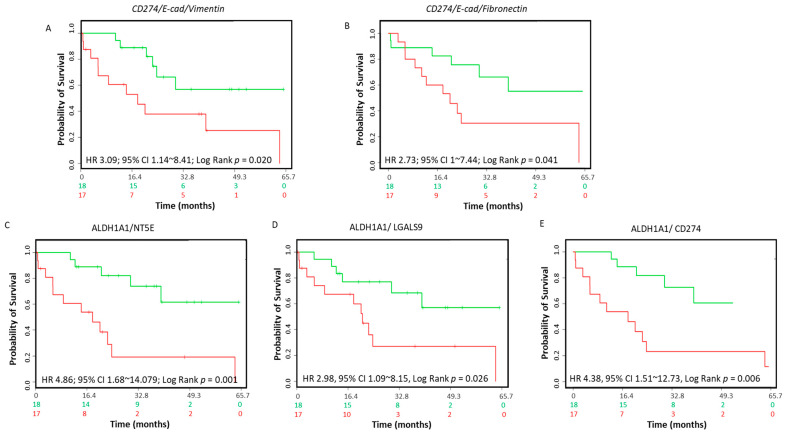
Elevated expression of PD-L1 has been recently reported in mesenchymal cells within CCA tumors [22]. We examined the association between PD-L1 with an EMT phenotype in a CCA patient cohort. Although expression of PD-L1 alone was not significantly associated with poor prognosis in the CCA patient dataset, coordinate downregulation of E-Cad (CDH1) expression and upregulation of VIM expression revealed overall worse survival (HR: 3.09, CI: 1.14~8.41, log-rank equal curves p = 0.020) in combination with PD-L1 (Figure 9A). Coordinate expression of PD-L1 with E-Cad and Fibronectin also showed poor overall survival in these patients (HR: 2.73, CI: 1~7.44, log-rank equal curves p = 0.041) (Figure 9B). We next evaluated the relationship between immune checkpoint genes and stemness genes in CCA patients. Expression of stemness marker ALDH1A1 showed poor overall survival when combined with NT5E (HR: 4.86, CI: 1.68~14.079, log-rank equal curves p = 0.001), LASGALS9 (HR: 2.98, CI: 1.09~8.15, log-rank equal curves p = 0.026) or PD-L1 (HR: 4.38, CI: 1.51~12.73, log-rank equal curves p = 0.006) (Figure 9C–E). Coordinate expression of other stemness genes including CD13, CD24, CD133, EpCAM, OCT4, SOX2, Nanog and KLF4 with immune checkpoints evaluated in this study did not show an association with overall survival in CCA patients (Supplementary Materials Table S2). There was no significant association of overall survival of CCA patients showing coordinate expressions of CSC markers and immune checkpoint genes FASLG, TNRSF14, VTCN1 and PD-L1 (Supplementary Materials Table S2). These findings reveal that elevated PD-L1 expression in CCA patients is closely linked with EMT status, and high co-expression of PD-L1, NT5E or LGALS9 with stemness marker ALDH1A1 is related to poor prognosis in CCA patients.
Figure 9.
Association between immune modulators, EMT markers, CSC markers and survival in CCA patients. The Kaplan–Meier survival curves in SurvExpress CCA patients for the gene expression of (A) CD274/E-Cad/Vimentin, (B) CD274/E-Cad/Fibronectin, (C) ALDH1A1/NT5E, (D) ALDH1A1/LGALS9, (E) ALDH1A1/CD274 in CCA patients. Low-risk groups are indicated in green and high-risk groups are indicated in red. The x-axis presents the study time in months. HR, CI and p-values are shown in the insert.
4. Discussion
In the present pilot study, the expression of immune modulators IDO1, NT5E and FASLG was related to poor prognosis in CCA patients. Furthermore, a combination of various ICs with putative immune modulators PD-1, PD-L1 and CTLA-4 was associated with poor CCA patient prognosis. Moreover, EMT and CSC were closely associated with the modulation of immune checkpoint molecules in CCA. PD-L1 and NT5E expression was closely associated with EMT, while coordinate expression of NT5E and LSGAL9 with CSC marker ALDH1A1 was linked with poor overall survival in CCA patients.
Indoleamine 2,3-dioxygenase 1 (IDO1) is an intracellular heme-containing enzyme that contributes to the immune escape of tumors [34]. Our observation of the association of high IDO1 with poor overall survival in CCA patients is consistent with observations in colorectal, non-small-cell lung and prostate cancers [35]. In contrast, high IDO1 expression levels in HCC patients have been correlated with better survival outcomes, indicating that IDO1 may not have immunosuppressive functions in this cancer [36,37]. Elevated IDO1 expression in cancers has been correlated with single-agent ICI therapy resistance [38]. Our findings suggest that combining IDO1 inhibitors with other ICIs may represent a promising strategy to expand CCA patient populations for immunotherapies. FASLG, a transmembrane protein of the tumor necrosis factor superfamily triggers apoptosis of T-cells [39]. Ecto-5′-nucleotidase (CD73 or NT5E) is a glycophosphatidylinositol-anchored receptor enzyme that blocks activation of T-cell when adenosine binds to its receptor [40]. A study found that CCA cell lines that expressed FASLG, induced cell death when cocultured with T-cells, indicative of the immune evasive function of FASLG/FAS axis in CCA [39]. We and others have previously found that NT5E expression in cancers was associated with poor prognosis [10,41]. We found that IDO1 expression was associated with clinical parameters, such as a presence of risk factors for HCC and tumor stage II and III, while the expression of FASLG in CCA patients was associated with the lymph node stage. IDO1, FASLG and NT5E showed no association with clinical parameters including vascular invasion, gender and tumor stage T0–T3.
This pilot study revealed that IDO1, FASLG, CD80, HAVCR2, CD73, CTLA-4, LGALS9, VTCN1 and TNFRSF14 in combination with PD-L1 is linked with poor outcome in CCA patients. The Cluster of differentiation 80 (CD80) regulates T-cell activation by binding to CTLA4. A study on biliary tract cancers including CCA reported that strong CD80 expression in tumor tissue was closely associated with resistance to adjuvant chemotherapy [42]. The T-cell immunoglobulin and mucin-domain 3 (TIM3 or HAVCR2) receptor limits T-cell responses by interacting with its ligand Galectin-9 (LGALS9 or Gal-9) [43]. In animal models, combining anti-HAVCR2 and anti-PD-1 has shown to suppress tumor growth [44].
V-set domain-containing T-cell activation inhibitor 1, VTCN1, (also named B7-H4, B7S1 or B7x) belongs to the B7 family and regulates T-cell-mediated antitumor responses. Studies in CCA patients showed high levels of VTCN1 expression were significantly related to poor prognosis [45,46]. LGALS9 is a tandem-repeat-type galectin that promotes antitumor immune responses by exerting antiproliferative effects on CAA cells [47]. T-cell immunoglobulin and ITIM domain (TIGIT) is an inhibitory immune checkpoint of the poliovirus receptor (PVR)/desmin family [48]. Inhibition of TIGIT alone or with PD-1 has shown to restore tumor-suppressive effects [49]. The tumor stroma of CCA patients showed infiltration of lymphocytes expressing ICs, including PD1 and TIGIT [50]. The function of tumor necrosis factor receptor superfamily member 14 (TNFRSF14) is not known in CCA.
We and others have noted PD-L1 expression was closely linked with EMT status [22]. This is the first study to examine the relationship between TGF-β1- and TNF-α-induced EMT and upregulation of PD-L1 and NT5E expression. Furthermore, we and others have reported that PD-L1 expression negatively impacts CCA patient prognosis [51]. In contrast, other studies show that CCA patients with low PD-L1 expression had a poorer prognosis [24,52].
A study showed that CCA-derived, CD133-positive CSCs displayed high levels of TGF-β1 and activation of the TGF-β1–pSmad2–EMT axis [53]. Similarly, we found CCA-derived CSCs showed a high expression of TGF-β1 along with TNF-α. A study reported that the PD-L1 low cell fraction isolated from HuCCT-1 cells was enriched with CSC-related characteristics compared with the PD-L1 high cell fraction [24]. This observation is consistent with our results demonstrating the downregulation of PD-L1 in CSCs enriched by HuCCT-1 cells. This study is the first to examine the relationship between CSC phenotype and other novel ICs including NT5E, LGALS9, TNFRSF14, FASLG and VTCN1. Our data suggest that CCA patient tumors with mesenchymal and CSC phenotypes might be targeted using immune checkpoint blockades.
This study is limited by a lack of CCA patient samples who have undergone treatment with immune checkpoint therapies. This pilot study, comprising a small number of CCA patients, may not provide adequate information and conclusions. Thus, further validation of the potential of immune checkpoint regulators as prognostic markers in larger cohorts of CCA patients will be more informative. Another limitation of this study is the lack of available clinical data for the SurvExpress CCA patients. Furthermore, the prognosis of CCA may be affected by the location of the tumor. In this study, we were unable to evaluate the independent association between distal, perihilar or intrahepatic CCA with the expression of immune checkpoints and patient prognosis. We were unable to assess the association between immune checkpoint expression and cancer-specific mortality. Further studies are needed to evaluate the correlation between immune checkpoint expression with parameters such as tumor location and cancer-specific mortality. While this study focused on the expression ICs in CCA tumor cells, comprehensive investigation is needed to validate the role of each individual IC in in vitro and in vivo CCA models. Studies particularly focusing on molecular mechanisms, functional assays, including motility and drug testing, and evaluation of ICs in animal models will be required to gain a better understanding of their function in CCA tissues. Further studies are needed to assess the expression of these molecules on tumor-infiltrating T-cells, which will also be helpful in predicting ICI responses. Additionally, studies on the co-expression of these immune checkpoints in CCA, as well as on how they influence or act with each other, will need to be assessed to enable effective and durable treatment.
5. Conclusions
CCA has a dismal prognosis with very limited therapeutic options. Accordingly, novel treatment modalities that are both effective and associated with durable responses are needed for the treatment of CCA. Immunotherapy in the form of ICIs is anticipated to be used as an effective treatment modality for CCA. Immune checkpoint molecules IDO1, FASLG, CD80, HAVCR2, CD73, CTLA-4, LGALS9, VTCN1, TNFRSF14 and PD-L1 may be useful as potential biomarkers for the treatment and prognosis of CCA patients. Additionally, ICI-based immunotherapy may be used to target EMT and CSC populations in CCA to achieve lasting and durable treatment outcomes. This study is the first attempt to examine the association between immune checkpoint modulator expression, CSCs and EMT status in CCA, providing a better understanding of the molecular events contributing to prognosis prediction. Further investigations of the underlying above-detailed mechanisms leading to overexpression of immune checkpoints in CCA microenvironment will provide better strategies for ICI therapy.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/jcm10102191/s1, Figure S1: Link between ICs in combination and survival in CCA patients. The Kaplan–Meier survival curves generated with the SurvExpress CCA patient dataset for the gene expression of (A) PD-1/TIGIT, (B) PD-1/TNFRSF14, (C) PD-1/CTLA4, (D) PD-1/FASLG, (E) CTLA4/IDO1 and (F) CTLA4/CD274. Green indicates low-risk group. Red indicates high-risk group. The x-axis shows the study time in months. HR, CI and P values are shown in the insert. Figure S2: Enrichment of CSCs with anchorage-independent three-dimensional spheroid culture in EGI-1 cells. (A) Monolayer culture and 3-D culture of EGI-1 cells (scale bar = 200 µm). (B-F) Increased expression of embryonic stemness and cell surface CSC markers was detected in EGI-1 spheres compared with adherent monolayer EGI-1 cells. (G-I) qRT-PCR detected higher expression of NT5E. Values are mean ± SD of three experiments in triplicate (* p < 0.05, ** p < 0.01, **** p < 0.001, ns: not significant). ActB: β-Actin. Table S1: SurvExpress-based overall survival of 35 CCA patients showing co-ordinate expression of PD-L1 and other ICs. Table S2: SurvExpress-based overall survival of 35 CCA patients showing co-ordinate expression of ICIs and CSC markers.
Author Contributions
L.C., P.P., D.H.C. and A.J. designed the study. L.C., P.P., R.S. (Ritu Shrestha), M.A., R.S. (Revati Sharma) and A.J. performed data acquisition. L.C., L.C., P.P., R.S. (Ritu Shrestha), M.A., R.S. (Revati Sharma) and A.J. analyzed and interpreted the data and drafted the manuscript. Critical manuscript revision was performed by G.K., M.A., K.R.B. and D.H.C. All authors have read and agreed to the published version of the manuscript.
Funding
The Gallipoli Medical Research Foundation and Reginald Ferguson Research Fellowship supported the present study.
Institutional Review Board Statement
Not applicable. The patient data utilized in this study are from publicly available datasets cBioPortal and SurvExpress, which store de-identified clinical data, such as gender, age, tumor type, tumor grade, treatment regimen, overall and progression-free survival data [26,27,54].
Informed Consent Statement
Not applicable. No patient samples were handled in this study. The patient data utilized in this study are from publicly available datasets CBioPortal and SurvExpress, which store de-identified clinical data, such as gender, age, tumor type, tumor grade, treatment regimen, overall and progression-free survival data [26,27,54].
Data Availability Statement
The data presented in this study are available in this article and supplementary material.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Rizvi S., Borad M.J., Patel T., Gores G.J. Cholangiocarcinoma: Molecular pathways and therapeutic opportunities. Semin. Liver Dis. 2014;34:456–464. doi: 10.1055/s-0034-1394144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bergquist A., von Seth E. Epidemiology of cholangiocarcinoma. Best Pract. Res. Clin. Gastroenterol. 2015;29:221–232. doi: 10.1016/j.bpg.2015.02.003. [DOI] [PubMed] [Google Scholar]
- 3.Bridgewater J., Galle P.R., Khan S.A., Llovet J.M., Park J.W., Patel T., Pawlik T.M., Gores G.J. Guidelines for the diagnosis and management of intrahepatic cholangiocarcinoma. J. Hepatol. 2014;60:1268–1289. doi: 10.1016/j.jhep.2014.01.021. [DOI] [PubMed] [Google Scholar]
- 4.Rizvi S., Gores G.J. Pathogenesis, diagnosis, and management of cholangiocarcinoma. Gastroenterology. 2013;145:1215–1229. doi: 10.1053/j.gastro.2013.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Valle J., Wasan H., Palmer D.H., Cunningham D., Anthoney A., Maraveyas A., Madhusudan S., Iveson T., Hughes S., Pereira S.P., et al. Cisplatin plus gemcitabine versus gemcitabine for biliary tract cancer. N. Engl. J. Med. 2010;362:1273–1281. doi: 10.1056/NEJMoa0908721. [DOI] [PubMed] [Google Scholar]
- 6.Topalian S.L., Drake C.G., Pardoll D.M. Immune checkpoint blockade: A common denominator approach to cancer therapy. Cancer Cell. 2015;27:450–461. doi: 10.1016/j.ccell.2015.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mertens J.C., Rizvi S., Gores G.J. Targeting cholangiocarcinoma. Biochim. Biophys. Acta Mol. Basis Dis. 2018;1864:1454–1460. doi: 10.1016/j.bbadis.2017.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pardoll D.M. The blockade of immune checkpoints in cancer immunotherapy. Nat. Rev. Cancer. 2012;12:252–264. doi: 10.1038/nrc3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kudo M. A New Era of Systemic Therapy for Hepatocellular Carcinoma with Regorafenib and Lenvatinib. Liver Cancer. 2017;6:177–184. doi: 10.1159/000462153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shrestha R., Prithviraj P., Anaka M., Bridle K.R., Crawford D.H., Dhungel B., Steel J.C., Jayachandran A. Monitoring Immune Checkpoint Regulators as Predictive Biomarkers in Hepatocellular Carcinoma. Front. Oncol. 2018;8:269. doi: 10.3389/fonc.2018.00269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eso Y., Seno H. Current status of treatment with immune checkpoint inhibitors for gastrointestinal, hepatobiliary, and pancreatic cancers. Therap. Adv. Gastroenterol. 2020;13 doi: 10.1177/1756284820948773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shrestha R., Bridle K.R., Crawford D.H., Jayachandran A. Immune checkpoint blockade therapies for HCC: Current status and future implications. Hepatoma Res. 2019;5:32. doi: 10.20517/2394-5079.2019.24. [DOI] [Google Scholar]
- 13.Iwai Y., Hamanishi J., Chamoto K., Honjo T. Cancer immunotherapies targeting the PD-1 signaling pathway. J. Biomed. Sci. 2017;24:26. doi: 10.1186/s12929-017-0329-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ueno M., Ikeda M., Morizane C., Kobayashi S., Ohno I., Kondo S., Okano N., Kimura K., Asada S., Namba Y., et al. Nivolumab alone or in combination with cisplatin plus gemcitabine in Japanese patients with unresectable or recurrent biliary tract cancer: A non-randomised, multicentre, open-label, phase 1 study. Lancet Gastroenterol. Hepatol. 2019;4:611–621. doi: 10.1016/S2468-1253(19)30086-X. [DOI] [PubMed] [Google Scholar]
- 15.Ioka T., Ueno M., Oh D.-Y., Fujiwara Y., Chen J.-S., Doki Y., Mizuno N., Park K., Asagi A., Hayama M., et al. Evaluation of safety and tolerability of durvalumab (D) with or without tremelimumab (T) in patients (pts) with biliary tract cancer (BTC) J. Clin. Oncol. 2019;37:387–387. doi: 10.1200/JCO.2019.37.4_suppl.387. [DOI] [Google Scholar]
- 16.Gibney G.T., Weiner L.M., Atkins M.B. Predictive biomarkers for checkpoint inhibitor-based immunotherapy. Lancet Oncol. 2016;17:e542–e551. doi: 10.1016/S1470-2045(16)30406-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lou Y., Diao L., Cuentas E.R., Denning W.L., Chen L., Fan Y.H., Byers L.A., Wang J., Papadimitrakopoulou V.A., Behrens C., et al. Epithelial-Mesenchymal Transition Is Associated with a Distinct Tumor Microenvironment Including Elevation of Inflammatory Signals and Multiple Immune Checkpoints in Lung Adenocarcinoma. Clin. Cancer Res. 2016;22:3630–3642. doi: 10.1158/1078-0432.CCR-15-1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Raggi C., Invernizzi P., Andersen J.B. Impact of microenvironment and stem-like plasticity in cholangiocarcinoma: Molecular networks and biological concepts. J. Hepatol. 2015;62:198–207. doi: 10.1016/j.jhep.2014.09.007. [DOI] [PubMed] [Google Scholar]
- 19.Kalluri R., Weinberg R.A. The basics of epithelial-mesenchymal transition. J. Clin. Investig. 2009;119:1420–1428. doi: 10.1172/JCI39104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kudo-Saito C., Shirako H., Takeuchi T., Kawakami Y. Cancer metastasis is accelerated through immunosuppression during Snail-induced EMT of cancer cells. Cancer Cell. 2009;15:195–206. doi: 10.1016/j.ccr.2009.01.023. [DOI] [PubMed] [Google Scholar]
- 21.Mak M.P., Tong P., Diao L., Cardnell R.J., Gibbons D.L., William W.N., Skoulidis F., Parra E.R., Rodriguez-Canales J., Wistuba I.I., et al. A Patient-Derived, Pan-Cancer EMT Signature Identifies Global Molecular Alterations and Immune Target Enrichment Following Epithelial-to-Mesenchymal Transition. Clin. Cancer Res. 2016;22:609–620. doi: 10.1158/1078-0432.CCR-15-0876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ueno T., Tsuchikawa T., Hatanaka K.C., Hatanaka Y., Mitsuhashi T., Nakanishi Y., Noji T., Nakamura T., Okamura K., Matsuno Y., et al. Prognostic impact of programmed cell death ligand 1 (PD-L1) expression and its association with epithelial-mesenchymal transition in extrahepatic cholangiocarcinoma. Oncotarget. 2018;9:20034–20047. doi: 10.18632/oncotarget.25050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jayachandran A., Dhungel B., Steel J.C. Epithelial-to-mesenchymal plasticity of cancer stem cells: Therapeutic targets in hepatocellular carcinoma. J. Hematol. Oncol. 2016;9:74. doi: 10.1186/s13045-016-0307-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tamai K., Nakamura M., Mizuma M., Mochizuki M., Yokoyama M., Endo H., Yamaguchi K., Nakagawa T., Shiina M., Unno M., et al. Suppressive expression of CD274 increases tumorigenesis and cancer stem cell phenotypes in cholangiocarcinoma. Cancer Sci. 2014;105:667–674. doi: 10.1111/cas.12406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu Y., Chen M., Wu P., Chen C., Xu Z.P., Gu W. Increased PD-L1 expression in breast and colon cancer stem cells. Clin. Exp. Pharmacol. Physiol. 2017;44:602–604. doi: 10.1111/1440-1681.12732. [DOI] [PubMed] [Google Scholar]
- 26.Aguirre-Gamboa R., Gomez-Rueda H., Martínez-Ledesma E., Martínez-Torteya A., Chacolla-Huaringa R., Rodriguez-Barrientos A., Tamez-Peña J.G., Treviño V. SurvExpress: An online biomarker validation tool and database for cancer gene expression data using survival analysis. PLoS ONE. 2013;8:e74250. doi: 10.1371/journal.pone.0074250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gao J., Aksoy B.A., Dogrusoz U., Dresdner G., Gross B., Sumer S.O., Sun Y., Jacobsen A., Sinha R., Larsson E., et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jayachandran A., Shrestha R., Dhungel B., Huang I.T., Vasconcelos M.Y.K., Morrison B.J., Ramlogan-Steel C.A., Steel J.C. Murine hepatocellular carcinoma derived stem cells reveal epithelial-to-mesenchymal plasticity. World J. Stem Cells. 2017;9:159–168. doi: 10.4252/wjsc.v9.i9.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shrestha R., Bridle K.R., Crawford D.H., Jayachandran A. TNF-α-mediated epithelial-to-mesenchymal transition regulates expression of immune checkpoint molecules in hepatocellular carcinoma. Mol. Med. Rep. 2020;21:1849–1860. doi: 10.3892/mmr.2020.10991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Awan F.M., Naz A., Obaid A., Ali A., Ahmad J., Anjum S., Janjua H.A. Identification of Circulating Biomarker Candidates for Hepatocellular Carcinoma (HCC): An Integrated Prioritization Approach. PLoS ONE. 2015;10:e0138913. doi: 10.1371/journal.pone.0138913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cardinale V., Renzi A., Carpino G., Torrice A., Bragazzi M.C., Giuliante F., DeRose A.M., Fraveto A., Onori P., Napoletano C., et al. Profiles of cancer stem cell subpopulations in cholangiocarcinomas. Am. J. Pathol. 2015;185:1724–1739. doi: 10.1016/j.ajpath.2015.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu H.J., Chu P.Y. Role of Cancer Stem Cells in Cholangiocarcinoma and Therapeutic Implications. Int. J. Mol. Sci. 2019;20 doi: 10.3390/ijms20174154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Celià-Terrassa T., Jolly M.K. Cancer Stem Cells and Epithelial-to-Mesenchymal Transition in Cancer Metastasis. Cold Spring Harb. Perspect. Med. 2020;10 doi: 10.1101/cshperspect.a036905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cheong J.E., Sun L. Targeting the IDO1/TDO2-KYN-AhR Pathway for Cancer Immunotherapy—Challenges and Opportunities. Trends Pharmacol. Sci. 2018;39:307–325. doi: 10.1016/j.tips.2017.11.007. [DOI] [PubMed] [Google Scholar]
- 35.Ferdinande L., Decaestecker C., Verset L., Mathieu A., Moles Lopez X., Negulescu A.M., Van Maerken T., Salmon I., Cuvelier C.A., Demetter P. Clinicopathological significance of indoleamine 2,3-dioxygenase 1 expression in colorectal cancer. Br. J. Cancer. 2012;106:141–147. doi: 10.1038/bjc.2011.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li S., Han X., Lyu N., Xie Q., Deng H., Mu L., Pan T., Huang X., Wang X., Shi Y., et al. Mechanism and prognostic value of indoleamine 2,3-dioxygenase 1 expressed in hepatocellular carcinoma. Cancer Sci. 2018;109:3726–3736. doi: 10.1111/cas.13811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xu D., Liu X., Wang Y., Zhou K., Wu J., Chen J.C., Chen C., Chen L., Zheng J. Identification of immune subtypes and prognosis of hepatocellular carcinoma based on immune checkpoint gene expression profile. Biomed. Pharmacother. 2020;126:109903. doi: 10.1016/j.biopha.2020.109903. [DOI] [PubMed] [Google Scholar]
- 38.Brown Z.J., Yu S.J., Heinrich B., Ma C., Fu Q., Sandhu M., Agdashian D., Zhang Q., Korangy F., Greten T.F. Indoleamine 2,3-dioxygenase provides adaptive resistance to immune checkpoint inhibitors in hepatocellular carcinoma. Cancer Immunol. Immunother. 2018;67:1305–1315. doi: 10.1007/s00262-018-2190-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li Z.Y., Zou S.Q. Fas counterattack in cholangiocarcinoma: A mechanism for immune evasion in human hilar cholangiocarcinomas. World J. Gastroenterol. 2001;7:860–863. doi: 10.3748/wjg.v7.i6.860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hay C.M., Sult E., Huang Q., Mulgrew K., Fuhrmann S.R., McGlinchey K.A., Hammond S.A., Rothstein R., Rios-Doria J., Poon E., et al. Targeting CD73 in the tumor microenvironment with MEDI9447. Oncoimmunology. 2016;5:e1208875. doi: 10.1080/2162402X.2016.1208875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Loi S., Pommey S., Haibe-Kains B., Beavis P.A., Darcy P.K., Smyth M.J., Stagg J. CD73 promotes anthracycline resistance and poor prognosis in triple negative breast cancer. Proc. Natl. Acad. Sci. USA. 2013;110:11091–11096. doi: 10.1073/pnas.1222251110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ghidini M., Pizzo C., Botticelli A., Hahne J.C., Passalacqua R., Tomasello G., Petrelli F. Biliary tract cancer: Current challenges and future prospects. Cancer Manag. Res. 2019;11:379–388. doi: 10.2147/CMAR.S157156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Anderson A.C. Tim-3: An emerging target in the cancer immunotherapy landscape. Cancer Immunol. Res. 2014;2:393–398. doi: 10.1158/2326-6066.CIR-14-0039. [DOI] [PubMed] [Google Scholar]
- 44.Ngiow S.F., von Scheidt B., Akiba H., Yagita H., Teng M.W., Smyth M.J. Anti-TIM3 antibody promotes T cell IFN-γ-mediated antitumor immunity and suppresses established tumors. Cancer Res. 2011;71:3540–3551. doi: 10.1158/0008-5472.CAN-11-0096. [DOI] [PubMed] [Google Scholar]
- 45.Xie N., Cai J.B., Zhang L., Zhang P.F., Shen Y.H., Yang X., Lu J.C., Gao D.M., Kang Q., Liu L.X., et al. Upregulation of B7-H4 promotes tumor progression of intrahepatic cholangiocarcinoma. Cell Death Dis. 2017;8:3205. doi: 10.1038/s41419-017-0015-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhao X., Guo F., Li Z., Jiang P., Deng X., Tian F., Li X., Wang S. Aberrant expression of B7-H4 correlates with poor prognosis and suppresses tumor-infiltration of CD8+ T lymphocytes in human cholangiocarcinoma. Oncol. Rep. 2016;36:419–427. doi: 10.3892/or.2016.4807. [DOI] [PubMed] [Google Scholar]
- 47.Kobayashi K., Morishita A., Iwama H., Fujita K., Okura R., Fujihara S., Yamashita T., Fujimori T., Kato K., Kamada H., et al. Galectin-9 suppresses cholangiocarcinoma cell proliferation by inducing apoptosis but not cell cycle arrest. Oncol. Rep. 2015;34:1761–1770. doi: 10.3892/or.2015.4197. [DOI] [PubMed] [Google Scholar]
- 48.Blessin N.C., Simon R., Kluth M., Fischer K., Hube-Magg C., Li W., Makrypidi-Fraune G., Wellge B., Mandelkow T., Debatin N.F., et al. Patterns of TIGIT Expression in Lymphatic Tissue, Inflammation, and Cancer. Dis. Markers. 2019:5160565. doi: 10.1155/2019/5160565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chauvin J.M., Pagliano O., Fourcade J., Sun Z., Wang H., Sander C., Kirkwood J.M., Chen T.H., Maurer M., Korman A.J., et al. TIGIT and PD-1 impair tumor antigen-specific CD8⁺ T cells in melanoma patients. J. Clin. Investig. 2015;125:2046–2058. doi: 10.1172/JCI80445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sato Y., Tanaka S., Kinoshita M., Takemura S., Shinkawa H., Kokudo T., Hasegawa K., Tanaka H., Yoshimoto H., Mori A., et al. Immunosuppressive tumor microenvironment in occupational cholangiocarcinoma: Supportive evidence for the efficacy of immune checkpoint inhibitor therapy. J. Hepato-Biliary-Pancreat Sci. 2020;27:860–869. doi: 10.1002/jhbp.788. [DOI] [PubMed] [Google Scholar]
- 51.Xu G., Sun L., Li Y., Xie F., Zhou X., Yang H., Du S., Xu H., Mao Y. The Clinicopathological and Prognostic Value of PD-L1 Expression in Cholangiocarcinoma: A Meta-Analysis. Front. Oncol. 2019;9:897. doi: 10.3389/fonc.2019.00897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhu Y., Wang X.Y., Zhang Y., Xu D., Dong J., Zhang Z., Yi C.H., Jia H.L., Yang X. Programmed death ligand 1 expression in human intrahepatic cholangiocarcinoma and its association with prognosis and CD8. Cancer Manag. Res. 2018;10:4113–4123. doi: 10.2147/CMAR.S172719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cai X., Li J., Yuan X., Xiao J., Dooley S., Wan X., Weng H., Lu L. CD133 expression in cancer cells predicts poor prognosis of non-mucin producing intrahepatic cholangiocarcinoma. J. Transl. Med. 2018;16:50. doi: 10.1186/s12967-018-1423-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cerami E., Gao J., Dogrusoz U., Gross B.E., Sumer S.O., Aksoy B.A., Jacobsen A., Byrne C.J., Heuer M.L., Larsson E., et al. The cBio Cancer Genomics Portal: An Open Platform for Exploring Multidimensional Cancer Genomics Data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data presented in this study are available in this article and supplementary material.