Abstract
Protein–protein interactions are fundamental to various aspects of cell biology with many protein complexes participating in numerous fundamental biological processes such as transcription, translation and cell cycle. MS-based proteomics techniques are routinely applied for characterising the interactome, such as affinity purification coupled to mass spectrometry that has been used to selectively enrich and identify interacting partners of a bait protein. In recent years, many orthogonal MS-based techniques and approaches have surfaced including proximity-dependent labelling of neighbouring proteins, chemical cross-linking of two interacting proteins, as well as inferring PPIs from the co-behaviour of proteins such as the co-fractionating profiles and the thermal solubility profiles of proteins. This review discusses the underlying principles, advantages, limitations and experimental considerations of these emerging techniques. In addition, a brief account on how MS-based techniques are used to investigate the structural and functional properties of protein complexes, including their topology, stoichiometry, copy number and dynamics, are discussed.
Keywords: Affinity purification coupled to mass spectrometry (AP-MS), Proximity-dependent biotinylation coupled to MS (PDB-MS), Cross-linking mass spectrometry (XL-MS), Co-fractionation mass spectrometry (coFrac-MS), Thermal proximity coaggregation (TPCA)
Introduction
The functions of proteins are primarily dictated by their higher-order structures and their propensity to form a protein network. Mathematical simulations have imposed an upper bound of ~ 650,000 protein–protein interactions (PPIs) among human proteins [1]. Although it has been demonstrated that artificial intelligence (AI) can predict the 3D structures and the folding of proteins with exceptional accuracy, such advances have not been extended to the quaternary structures of proteins [2]. Hence, large-scale investigations of PPIs are mainly performed with two broad categories of experimental techniques.
The first category of methods, which includes yeast two-hybrid (Y2H) assay and protein complementation assay (PCA), comprises assays that monitor binary interactions of proteins, whereby the physical interaction of a preselected bait and a prey protein is evaluated in a pairwise manner. Each of the pair is genetically fused with different portions of another split protein and subsequently co-expressed. When the bait–prey protein pair interacts, the two split protein tags resume their assembly and functions, resulting in gene expression, enzymatic activity or fluorescence that serve as readouts for reporting the direct interaction of the selected protein pair [3]. In contrast, the second category adopts the affinity purification coupled to the mass spectrometry (AP-MS) technique or its close variants. In such methods, a bait-specific antibody or affinity reagent is used to capture a bait protein from cell lysates, with simultaneous purification of its preys in bulk [4]. Titeca et al. named these two respective approaches as binary and co-complex technologies [3]. Since copurified proteins in AP-MS are not known a priori, a subsequent protein identification step is performed using MS. Thus, the AP-MS technique has an advantage over binary technologies because it enables identifying previously unknown interaction partners, besides offering sensitive, high-throughput and hypothesis-free assays.
This review discusses recent developments in co-complex methodologies, with a description of the techniques in the figures provided. Apart from deciphering the exact composition of a protein complex or protein network, we believe that it is equally essential to disentangle other properties of interacting proteins, such as (i) the topology that relates how each protein subunit is interconnected to contribute to the overall shape and relative spatial arrangements of a protein complex; (ii) the stoichiometry, or the ratio of each constituent protein subunit; (iii) the copy number which refers to the absolute number of each constituent subunit and (iv) the dynamics, which pertains to the alterations in the composition, topology and stoichiometry or copy number over time, upon external perturbations, or as a result of changes in cellular functions [2]. These, too, will be reviewed here (Table 1).
Table 1.
A table listing the pros and cons of MS-based methodologies for identifying and quantifying PPIs
| Methods | Pros | Cons |
|---|---|---|
| AP-MS | Co-immunoprecipitation can be performed without tagged baits expressed at physiological levels to identify endogenous PPIs | Co-IP with untagged baits is limited by the availability of antibodies, and the low expression levels of baits |
| Epitope tagging provides an alternative for purifying proteins lacking suitable antibodies | Epitope tags may interfere with the functions and solubility of the baits | |
| Transient transfection of tagged baits enhance their expression, thus improving the efficiency and throughput of the pulldowns | Ectopic expression of tagged baits may promote misfolding and mislocalization of the baits, promoting background contamination and spurious interactions | |
| PDB-MS | Allows detection of PPIs among both soluble and membrane proteins, as well as enriching for PPIs that are transient, weak, low abundance or have high turnover [11, 28, 29] | May react with biotin-phenol and H2O2 to produce reactive radicals resulting in cellular toxicity (APEX) [30] |
| Avoids post-lysis artefacts [11] | The accessibility and labelling efficiency of the biotinylating enzyme are locality-dependent, as its orientation and topology within the protein complex may impede its performance | |
| The affinity of biotin to streptavidin is robust yet reversible. Hence, highly stringent conditions for sample denaturing, solubilization, capture, wash and extraction of biotinylated proteins can be employed to maximize the recovery of hydrophobic proteins while minimizing nonspecific background contaminants | The high affinity of the streptavidin–biotin interaction may hinder the recovery of highly biotinylated proteins. PDB-MS suffers from false positives in the forms of high-abundance background proteins or artefacts from endogenous biotinylation | |
| The labelling time for different enzyme varies from 1 min to 24 h [12, 13, 16] | ||
| XL-MS | Crosslinking reagents can covalently connect two or more non-covalently interacting proteins, regardless of the duration and strength of the interaction. As such, even transient and weak PPIs can be preserved [45, 46] | The low efficiency (~ 1–5%) of crosslinking reagents, which often results in marginal crosslinks, where only the top 20–30% of proteins are detected |
| When used in combination with X-ray crystallography, CryoEM, NMR and native MS, the spatial constraint data from XL-MS can guide molecular modelling, construct a connectivity map for determining subunit topology, and map the dynamic behaviour of the protein complex [49–51 ] | The crosslinking reaction time may be relatively long (~ 30 min). Excessively long reaction time may result in large, crosslinked protein aggregates | |
| To expand the number and coverage of crosslinks, alternative modes of crosslinking can be employed, such as carboxyl-targeting reagents [40–42] | A crosslinker covalently links two linear peptides, giving rise to a hybrid dipeptide that can dramatically expand the search space during spectra matching, giving rise to the 'n-square problem' [68, 69] | |
| Co-Frac-MS | CoFrac-MS has high throughput, and it provides global identification and quantification of native protein complexes in one setting | False positives constitute a significant problem in the form of chance co-elution |
| It can be operated without genetic manipulation and overexpression, thereby inferring endogenous, physiologically relevant interactome [3] | ||
| CoFrac-MS combined with quantitative proteomics can delineate the relative distribution of a protein in multiple co-elution features. Thus, the stoichiometries and dynamics of a target protein within different co-isolated complexes can be simultaneously elucidated [85] | ||
| TPCA | TPCA permits system-wide profiling of protein complex dynamics, and it requires neither antibodies nor epitope tagging [87] | The current version of TPCA is limited to studying the dynamics of known or predicted protein complexes across cellular state and physiological conditions. Need to incorporate existing interaction data with graph/network clustering algorithms to identify novel protein complexes [87] |
| Little preparation time is required. It allows most of the study of protein complexes in situ and in vivo | ||
| TPCA profiling can be rapidly deployed to unravel the assembly state of protein complexes across cellular state, cell type, tissue and physiological conditions to provide insight into their functions in normal and diseased cells |
Affinity purification coupled to mass spectrometry (AP-MS)
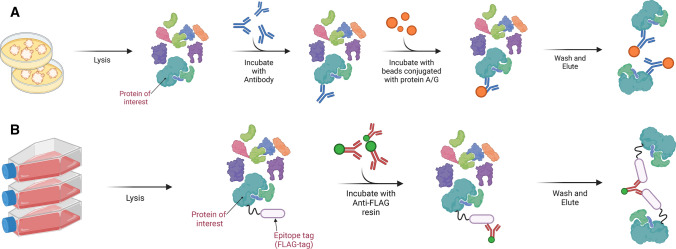
AP-MS is the most widely used high-throughput method for PPI study. In AP-MS, a bait protein is selectively purified with specific antibodies or other affinity reagents along with its potential interacting partners (preys) from a cell or tissue lysate. This step is followed by identifying and quantifying these purified proteins by MS. AP-MS experiments are then repeated with different baits. The combination of bait-prey pairs from these AP-MS experiments is then statistically computed to infer the protein network. An AP-MS assay typically involves several steps comprising (i) incubation of precleared protein lysate with beads conjugated with the bait or epitope tag-specific antibodies, (ii) washing procedures to minimize nonspecific binding, (iii) elution of the purified complexes, and (iv) identification of the eluted proteins with MS (Fig. 1). Whereas an ideal co-immunoprecipitation experiment characterizes endogenous PPIs using untagged bait protein expressed at physiological levels, it is usually limited by the repertoire of antibodies and the low expression levels of bait proteins. As an alternative, a bait protein can be created by genetically fusing a gene of interest to an epitope tag followed by its expression in a chosen cell line for optimal biological context. Such tags may comprise short peptides or proteins that are uniquely recognizable by readily available antibodies. Some examples of these tags are FLAG, c-myc, HA, polyHis and streptavidin. A comprehensive list of epitope tags has been documented by Vandemoortele et al. [5]. These tags can be fused in single or multiple copies, as well as in tandem with different tags for multiple rounds of purifications, namely tandem affinity purification (TAP).
Fig. 1.
The AP-MS workflow. A A specific antibody can be used to selectively capture an untagged protein of interest (POI) that is expressed at physiological levels from the protein lysate. This untagged POI binds to other protein interactors directly or indirectly. Subsequently, beads conjugated with protein A/G are added to the protein mixture to capture the antibodies together with the protein assemblies. This is then followed by the washing and elution step to release the POI and its interactors for LC–MS/MS analysis. B For bait proteins lacking suitable antibodies, the POI can be genetically fused with an epitope tag, such as FLAG-tag or HA-tag. This bait-tag fusion construct can then be transfected transiently or stably into selected cell lines. Subsequently, resins conjugated to anti-epitope tag antibodies are added so that the POI and its interactors can be selectively enriched
For proteins lacking suitable antibodies, epitope tagging provides a general approach for purifying protein complexes; but with the downside that such tags may interfere with the functions and solubility of the bait protein. Besides, transient transfection can enhance the expression of the tagged baits, hence improving the efficiency and throughput of the pulldown experiments, but with the caveat that such ectopic expression may promote misfolding and mis-localization of the baits, thereby exacerbating background contamination and spurious interactions. A major challenge in AP-MS is the copurification of high-abundance, nonspecific-interacting proteins. Therefore, incorporating appropriate controls that discriminate bona fide interactors apart from nonspecific binding has become indispensable in AP-MS. Such controls may constitute the expression of empty vectors for pulldown experiments or the use of antibody isotypes, knockdown and knockout of the endogenous baits for co-IPs. Besides, TAP-tagging, which allows multiple washing and elution steps, can be used to minimize nonspecific interactions, albeit at the expense of losing weak and transient PPIs. It is also noteworthy that quantitative MS and dedicated bioinformatics algorithms such as SAINT, CRAPome and BioPlex can help differentiate background contamination by identifying significant differences in protein abundance between the experiment and the negative controls [6–8].
An interesting application for AP-MS was recently demonstrated by Gordon et al. for elucidating the PPIs for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) that causes the COVID-19 pandemic [9]. In this work, the authors cloned and expressed 26 of the 29 SARS-CoV-2 proteins carrying 2 × Strep tags in HEK293T cells. This allowed them to identify 332 high-confidence PPIs between human proteins and SARS-CoV-2 proteins. In their subsequent work, Gordon et al. exploited the AP-MS methodology for comparative viral-human PPIs for SARS-CoV-2, SARS-CoV-1 and Middle East respiratory syndrome coronavirus (MERS-CoV) [10]. Subsequently, the authors identified host proteins that could affect coronavirus proliferation, such as Tom70, a mitochondrial chaperone protein that interacts with ORF9b-coded protein from SARS-CoV-1 and SARS- CoV-2.
Proximity-based labelling coupled to mass spectrometry (PDB-MS)
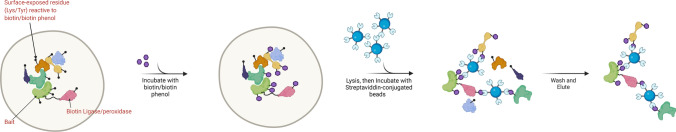
Proximity-dependent biotinylation coupled to MS (PDB-MS) involves expressing a bait protein that is genetically fused to a biotin ligase (BioID), a horseradish peroxidase (HRP), or a peroxidase (APEX) [11–13]. The fused enzymes are capable of catalysing externally added biotins or phenolic biotins into reactive biotin intermediates that subsequently diffuse out to biotinylate proteins in the vicinity of the bait. After biotin-labelling, cells are lysed, and pulldown is performed using streptavidin or neutravidin, followed by identification and quantification with MS [12, 13]. The detailed methodology is described in Fig. 2.
Fig. 2.
The PDB-MS workflow. In PDB, a biotin ligase (BioID), a horseradish peroxidase (HRP) or a peroxidase (APEX) is genetically fused to a selected bait protein and expressed in a chosen cell line. In vivo labelling is achieved by adding biotins (BioID) or biotin phenols (APEX) to the cells, whereby these molecules are converted to reactive biotin intermediates. These reactive intermediates then diffuse away from the enzyme in a distance-dependent manner to covalently modify lysine (BioID) or tyrosine (APEX) residues located in close proximity. After performing cell lysis in harsh, denaturing conditions, biotinylated proteins are enriched using resin conjugated with streptavidin or neutravidin for subsequent quantitative proteomics analysis
Central to PDB-MS is promiscuous biotinylation, a covalent modification process dependent on the random diffusion of reactive biotin intermediates. However, promiscuous biotinylation is constrained by distance, as proteins in close proximity to the bait/enzyme fusion are preferentially biotinylated, but the labelling strength dwindles with increasing distance. Therefore, it is noteworthy that PDB-MS defines the neighbourhood surrounding the bait within an “effective labelling radius” of the enzyme. These neighbouring proteins may constitute the actual physical contacts of the bait itself, or other proteins that happen to be present in the vicinity of the bait/enzyme fusion. The effective labelling radii for BirA*, a mutant biotin ligase used in BioID, and APEX, an ascorbate peroxidase, have been estimated to be ~ 10 nm and ~ 20 nm, respectively [14, 15].
Classic BioID was developed using E. coli-derived 35 kDa BirA* that harbours an R118G mutation that destabilizes the catalytic domain [12]. BirA* catalyzes the conversion of biotins to form the highly reactive biotinoyl-AMP intermediates, which dissociate prematurely, diffuse out and react with the neighbouring lysine residues in a promiscuous manner [12]. Meanwhile, BioID2 was developed from A. aeolicus-derived biotin ligase carrying an R40G mutation, rendering the ligase smaller in size (27 kDa) and catalytically more active. This resulted in more efficient biotinylation and minimal mis-localization of the bait [16]. Nevertheless, both BioIDs require an incubation period of 12–24 h. To improve the labelling speed, TurboID and MiniTurbo were adapted from BirA* ligase, with extensive engineering at the reactive biotin-5′-AMP binding motif (RBAM) [17]. Both mutants have enhanced efficiency and speed of biotinylation in 10 min. By introducing three mutations to RBAM of B. subtilis BirA*, Ramanathan et al. created a 28-kDa ligase named “BASU” with over 1000-fold faster kinetics and over 30-fold increased signal-to-noise ratio compared to BirA* [18].
Horseradish peroxidase (HRP) can also convert a substrate into free radicals in the presence of H2O2, thus covalently label neighbouring proteins on electron-rich amino acids [19]. However, HRP is mainly used for proximity labelling in oxidizing environments, such as the extracellular surface, due to its low reactivity in the reducing environment [20–22]. Notably, in the enzyme-mediated activation of radical source (EMARS) method, HRP is fused to a protein located on the cell surface or an antibody that can recognize this target protein. At the same time, the substrate constitutes an aryl azide group that has been conjugated with biotin and fluorescein tags [23]. Upon the addition of H2O2, the aryl azide group is activated by HRP to form a nitrene radical that can attack neighbouring cell surface proteins. At the same time, the biotin or fluorescein tags allow affinity purification of the labelled proteins with streptavidin- or antibody-immobilized beads for subsequent MS analysis. In another HRP-based proximity labelling method, which is named the “selective proteomic proximity labelling assay using tyramide” (SPPLAT), the substrate used is a biotin-tyramide or biotin phenol [24, 25].
APEX is a 27-kDa monomeric ascorbate peroxidase derived from pea and is active in the reducing environment. APEX was adapted to catalyse the oxidation of biotin-phenol to short-lived (< 1 ms) biotin-phenoxyl radicals in the presence of H2O2, [13]. These radicals can biotinylate tyrosine, tryptophan, cysteine and histidine residues. In an APEX experiment, cells expressing bait/APEX fusion are incubated with biotin-phenol for 30 min, followed by a 1-min exposure to H2O2 to induce biotinylation. APEX is an efficient enzyme that can generate sufficient signal-to-noise within a short period (1 min of labelling time versus 10 min for TurboID; and 18–24 h for BioID). As such, it allows a “time-lapse” analysis of a dynamic interactome at a superior temporal resolution, rather than a single “long-exposure” image lasting several hours. Nevertheless, APEX is limited by its low catalytic activity and sensitivity, as biotinylation often goes undetected when APEX is expressed at the physiological level. To address this, Ting's lab employed yeast display evolution to develop APEX2, a soybean-derived peroxidase that harbours an extra A134P mutation [26]. APEX2 possesses enhanced labelling efficiency and sensitivity. The stability and activity of APEX2 were further improved by Huang et al. by introducing a version of cysteine-free APEX2 with C32S mutation [27]. The directed evolution of proximity labelling components is discussed in detail by Bosch et al. [19].
PDB-MS permits the detection of PPIs among both soluble and membrane proteins, apart from enriching for interactions that are transient, weak, of low abundance or have high turnover [11, 28, 29]. As PDB-MS biotinylates proteins in cells, it allows the labelling of fragile complexes or interactions in addition to avoiding post-lysis artefacts. Finally, the affinity of biotin to streptavidin is probably the strongest yet reversible biological interaction known. Consequently, highly stringent conditions for sample denaturing, solubilization, capture, wash and extraction of biotinylated proteins can be employed to maximize the recovery of hydrophobic proteins while minimizing nonspecific background contaminants. Notwithstanding, PDB-MS has several caveats. For instance, APEX may react with biotin-phenol and H2O2 to produce reactive radicals that result in cellular toxicity [30]. Furthermore, the accessibility and labelling efficiency of the biotinylating enzyme are locality-dependent, as its orientation and topology within the protein complex may impede its performance. The high affinity of the streptavidin–biotin interaction may also hinder the recovery of highly biotinylated proteins. Like AP-MS, PDB-MS suffers from false positives in the forms of high-abundance background proteins or artefacts from endogenous biotinylation. Hence, similar strategies applied in AP-MS to discriminate background contaminants, such as expression of biotinylating enzyme alone or fusing the bait to an irrelevant polypeptide, for instance, Green Fluorescent Protein (GFP), have been proposed [11].
Recently, Ke et al. designed and evaluated 12 different biotin-phenol analogues as proximity labelling probes for APEX2 [31]. Among these probes, the BP5 and BN2 were found to generate free radicals and conjugates to tyrosine residues with high efficiency and selectivity. These two probes were used to profile the spatiotemporal interactome of the EGFR signalling component STS1 with a minute timescale. As a result, they identified endosome markers, such as HGS, STAM and STAM2, at 10 min of EGF stimulation. This observation is consistent with the discovery that the endosome contained the highest number of STS1-interacting proteins during the internalization of EGFR induced by EGF [32]. In a separate study, Zhang et al. evaluated TurboID, BioID and BioID2 on their ability to identify the proteome that is proximal to N, which is a nucleotide-binding leucine-rich repeat (NLR) immune receptors that confer resistance to Tobacco mosaic virus (TMV) in plants [33]. Consequently, TurboID was found to produce the most efficient levels of biotinylation and that a putative E3 ubiquitin ligase, UBR7, was discovered to directly interacts with the TIR domain of N. Many more variants of proximity labelling methods have been published, such as NEDDylation, PUP-IT, photoactivable proximity labelling and sortase-mediated ligation [34–37]. However, due to limitation in space, they will not be discussed here.
Cross-linking mass spectrometry (XL-MS)
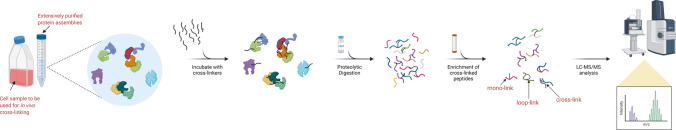
Crosslinking mass spectrometry (XL-MS) lies at the interface of interaction proteomics and structural biology [38, 39]. In- XL-MS, a selected protein or protein complex in their native states is first chemically crosslinked with reagents that can covalently tether amino acid residues that are spatially proximal. Crosslinked proteins are then proteolyzed, and the resulting peptide mixtures are separated and analyzed with LC–MS/MS. Subsequent database searching of the MS data elucidates the sequence of the crosslinked peptides, in addition to the crosslinked sites (Fig. 3).
Fig. 3.
The XL-MS workflow. Chemical crosslinking can be performed in vitro using extensively purified protein assemblies or in vivo using intact cells. The first step of chemical crosslinking involves adding a selected crosslinker to the protein mixture or cells. After chemical crosslinking, crosslinked proteins are digested to yield peptides. Typically, three types of cross-linked peptides are produced, i.e., the mono-linked peptides, the loop-linked peptides and the cross-linked peptides, among the many unlabelled peptides and unreacted crosslinkers. Due to the heterogeneity, the total pool of proteolyzed peptides is subjected to fractionation to enrich cross-linked peptides, subsequently mass-analysed by LC–MS/MS
Key to XL-MS experiments is the crosslinking reagents, typically small bifunctional molecules carrying two reactive groups separated by a carbon-chain spacer. Such bifunctional molecules can react with the respective side chains of two amino acids and covalently linking them together. Depending on the reactive groups, these crosslinkers can be classified into (i) amine-reactive (lysine-targeting), (ii) sulfhydryl-reactive (cysteine-targeting), (iii) carboxyl-reactive (targeting acidic amino acids) and (iv) photo-reactive categories, as comprehensively compiled by Steigenberger et al. [40]. On the other hand, crosslinkers can also be classified according to the length of the spacers or the number of functional groups that they carry. For example, some crosslinkers can carry zero-length spacers, while homobifunctional crosslinkers harbour two identical functional groups; heterobifunctional crosslinkers carry two different functional groups and trifunctional crosslinkers have three functional groups. For the latter, the third functional group (for example, biotin or phosphonic acid) is usually added as an affinity handle for enriching crosslinked peptides [41, 42]. Besides, a labile moiety can be incorporated in the spacer region, rendering crosslinked peptides cleavable by gas-phase fragmentation [43, 44]. MS-induced cleavage helps uncouple crosslinked peptides in MS2 so that the resulting pair of linear peptides can be individually sequenced in MS3, facilitating spectrum matching.
XL-MS has several favourable attributes. First, crosslinking reagents can covalently connect two or more non-covalently interacting proteins, regardless of the duration and strength of the interaction. As such, even transient and weak PPIs can be preserved [45, 46]. MS analysis would subsequently confirm the physical proximity and interaction of the two crosslinked proteins. XL-MS also helps pinpoint the localities of crosslinked amino acid side chains, thereby restricting physical interaction sites to certain structural regions [47]. Given that a crosslinker interconnects two amino acid residues, a value indicating the distance constraint, i.e. the sum of the crosslinker spacer arm length and the side chain lengths of the two linked residues, can be calculated to impart an upper bound of the physical distance [48]. When used in combination with X-ray crystallography, CryoEM and native MS, such spatial constraint data can guide molecular modelling, construct connectivity map for determining subunit topology and map the dynamic behaviour of the protein complex [49–51].
Chemical crosslinking has primarily been performed on highly purified, overexpressed protein complexes to overcome the low efficiency of crosslinking and minimize the search space during spectrum matching [52–54]. Due to the increasing sensitivity of MS, it is now possible to crosslink protein complexes before (in vivo XL) or after (on-beads XL) affinity purification. Better still, with only endogenous expression, the native structures and physiological interactions of protein complexes can be preserved, as exemplified by the protein phosphatase 2A (PP2A) study [49, 55]. Recently, chemical crosslinking has also been employed in a proteome-wide manner for cell lysates, intact cells or organelles, to simultaneously monitoring PPIs and their spatial information for the whole proteome [56–58].
Considerable advances have been made in XL-MS with respect to crosslinking chemistry, sample preparation, crosslink enrichment, MS technology and tools for data analysis and visualization [47]. These advances mainly address sample and data complexity [59]. A major limitation of XL-MS pertains to the low efficiency (~ 1–5%) of crosslinking reagents, which often results in marginal crosslinks, where only the top 20–30% of proteins are detected [54, 60, 61]. It should also be noted that the crosslinking reaction time may be relatively long (~ 30 min). Excessively long reaction time may result in large, crosslinked protein aggregates. Crosslinking reactions tend to produce four heterogeneous classes of crosslinks comprising (i) unreacted peptides, (ii) mono-links or dead-end links, (iii) loop-links and (iv) crosslinks [61, 62]. Only crosslinked and mono-linked peptides provide useful spatial information but are also the lowest in abundance. Strong cation exchange (SCX) or size exclusion chromatography (SEC) is often used to enrich these low-abundant crosslinks [63, 64]. Apart from that, affinity chromatography can be used to enrich crosslinked peptides harbouring trifunctional, affinity-tagged crosslinkers [41, 42, 65]. One way to expand the number and coverage of crosslinks is by applying alternative modes of crosslinking, for instance, by using carboxyl-targeting reagents. Such crosslinkers provide complementary spatial information to those obtained from the more commonly adopted lysine targeting chemistry [54]. Further, since a crosslinker covalently connects two linear peptides, this gives rise to a hybrid dipeptide that can dramatically expand the search space during spectra matching [66, 67]. This is because all theoretically possible peptide pairs in the protein database would need to be considered. One solution to this 'n-square problem' is to apply MS-cleavable crosslinkers in an MS2–MS3 strategy, whereby interpretation of mass spectra can be substantially simplified due to the availability of linear peptides and characteristic peaks. However, this gain inevitably comes at the expense of duty cycles and identification rates.
By combining PDB-MS (with an effective labelling radius of 10–20 nm) and XL-MS (with a spatial constraint of ~ 1 nm), Liu et al. recently demonstrated that it is not only possible to define the neighbouring proteins of a single bait protein located at the human nuclear envelope interactome, but also to identify crosslinked peptides which originated from 109 literature-curated physical PPIs of 14 nuclear envelope proteins [68]. In another study, Courouble et al., by combining hydrogen–deuterium exchange MS (HDX-MS) with XL-MS, elucidated the structural dynamics of the SARS-CoV-2 full-length nsp7:nsp8 complex [69]. These complementary techniques validate the interaction surfaces from the published three-dimensional heterotetrameric crystal structure of the nsp7:nsp8 complex and suggest that the nsp7:nsp8 heterotetramer can dissociate into a stable dimeric unit.
Co-Fractionation coupled to mass spectrometry (coFrac-MS)
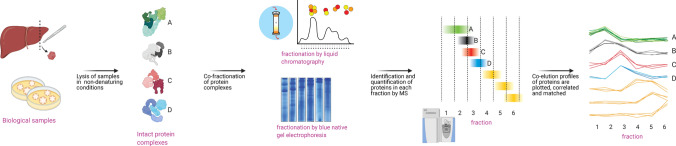
Spatiotemporal co-behaviour of biomolecules, such as co-expression or co-localization, has been proposed to imply functional or physical interactions [70]. Likewise, polypeptide constituents from the same assembly tend to co-migrate in the same analytical column under native conditions. Hence, proteins sharing similar co-fractionation profiles may suggest apparent co-localization [71]. This correlating relationship was initially exploited for organellar proteomics using density gradient centrifugation for biochemical fractionation, but this concept was extrapolated to interaction proteomics, giving rise to CoFractionation-MS (coFrac-MS) [72–76]. In CoFrac-MS, protein complexes in cell lysates are extensively fractionated under non-denaturing conditions with chromatographic or electrophoretic techniques. Each fraction is then proteolyzed, analyzed with LC–MS/MS, followed by identifying and quantifying its proteome composition. Subsequently, the fractionation profiles of individual protein complex subunits can be constructed. Since subunits of intact complexes tend to co-fractionate, protein complexes can be bioinformatically predicted from these data using the correlations between fractionation profiles as a feature of central importance.
As preserving the intactness of protein complex is vital, coFrac-MS workflows typically start with rapid cells/tissue lysis under refrigerated, native conditions, with minimal dilution [77]. This is followed by extensive biochemical/ biophysical separation of the protein complexes in native, non-denaturing states, whereby each fraction is subsequently subject to quantitative MS analysis. The abundance for each identified protein can then be captured from MS1 intensities, spectral counts or reporter ion intensities and computed to construct a co-elution profile reflecting the abundance of individual proteins across fractions. Finally, the co-elution profiles for co-fractionating proteins are correlated, matched and scored to detect and build the network for binary PPIs (Fig. 4).
Fig. 4.
The coFrac-MS workflow. Samples are lysed in mild conditions to preserve the integrity of protein complexes, separated under native or near-native conditions using column chromatography or native gel electrophoresis into fractions. Each fraction is then individually subjected to quantitative, bottom-up LC–MS/MS analysis. With the assistance of dedicated computational algorithms, the abundance of each protein is then plotted as co-migration profiles across fractions to construct an interactome network
One defining feature of coFrac-MS is the biochemical/ biophysical separation schemes used for resolving protein complexes in native or near-native conditions. Size-exclusion chromatography (SEC), ion-exchange (IEX) and hydrophobic interaction chromatography (HIC) are commonly used for co-fractionating soluble protein complexes according to their sizes, charges and hydrophobicity [75, 76, 78, 79]. With SEC, the separation of complexes is performed at near-native conditions, i.e., at neutral pH and physiological salt concentration, but is limited by its resolution [80]. Meanwhile, IEX (ion exchange) separation relies on ionic interaction. A variety of IEX materials, including SCX (strong cationic exchange), WCX (weak cationic exchange), SAX (strong anionic exchange) and WAX (weak anionic exchange) with differing charge properties, resolution and strength are commercially available. However, the presence of salt in the IEX mobile phases may disrupt native PPIs [81]. Conversely, high salt content is used to enhance the adsorption of hydrophobic protein surfaces to the solid support in HIC, and complexes are eluted upon decreasing salt gradient [82]. Apart from soluble complexes, it has been demonstrated that with mild or non-denaturing detergents, it is possible to co-fractionate membrane-bound complexes, for instance, the mitochondrial membrane-bound complexes using BN-PAGE [83, 84]. Since coFrac-MS potentially identifies thousands of PPIs in one experiment, the roles of dedicated algorithms are equally critical for delineating all possible combination of binary protein matrixes based on co-migration profiles. As reviewed in detail by Salas et al., such algorithms apply a variety of mathematical approaches comprising correlational metrics, co-apex measures; mutual information; Jaccard index and Euclidean distance [81].
The merits of coFrac-MS lie in its high throughput and its ability to provide global identification and quantification of native protein complexes in one setting. Furthermore, it can be operated without genetic manipulation and overexpression, thereby inferring endogenous, physiologically relevant interactome [3]. Besides, coFrac-MS combined with quantitative proteomics can delineate the relative distribution of a protein in multiple co-elution features. Thus, the stoichiometries and dynamics of a target protein within different co-isolated complexes can be simultaneously elucidated [83]. Nevertheless, there are caveats that we must consider in experimental design. Similar to AP-MS, false positives constitute a significant problem in the form of chance co-elution. This can be minimized by adopting high-resolution separation methods or combining multiple orthogonal separations, apart from more rigorous bioinformatic analyses.
In an interesting application, Mallam et al. applied CoFrac-MS in the form of SEC separation to analyze two equivalent cell culture lysates that served as a control and an RNase A-treated sample [85]. Upon fractionation, proteins in each fraction are identified with MS to build a proteome-wide protein co-elution profile for each condition. Following that, the authors evaluated the profiles from both samples to detect the elution shift of proteins upon RNase A treatment, which implies RNA–protein association. These elution shifts are then cross-referenced with known protein complexes to identify RNP complexes. As a result, co-Frac-MS allowed Mallam et al. to identify 1428 protein complexes that associate with RNA. Meanwhile, using SEC- or IEC-based separation combined with MS, Moutaoufik et al. generated mitochondrial interaction maps of human pluripotent embryonal carcinoma stem cells (ECSCs) and differentiated neuronal-like cells (DNLCs) [86]. The resulting PPI networks contain 6,442 interactions from ~ 600 mitochondrial proteins, revealing the dynamics of mitochondrial interactions during neuronal differentiation. Furthermore, they also demonstrated that C20orf24 is a respirasome assembly factor important for respiratory chain activity.
Thermal proximity coaggregation (TPCA)
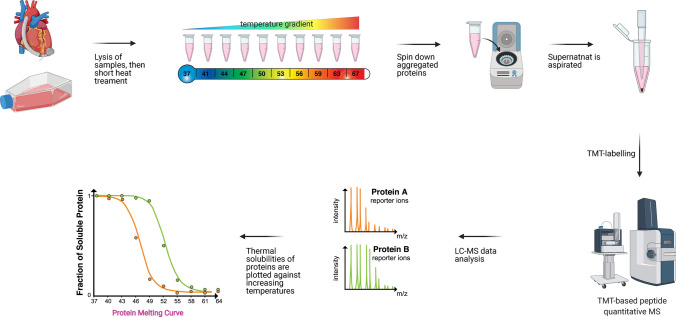
Thermal Proximity Coaggregation (TPCA) is a relatively recent and unconventional approach for proteome-wide profiling of protein complex dynamics [87]. It exploits the phenomenon that interacting proteins co-aggregate after heat-induced denaturation and co-precipitate. As a result, they have a high similarity in their thermal solubility compared to non-interacting proteins. The assembly state of known protein complexes can be inferred from the similarity or changes in protein thermal solubility to identify those modulated across cellular states or physiological conditions. To simultaneously monitor the dynamics for hundreds to thousands of protein complexes, proteome-wide quantification of protein thermal solubility is determined using quantitative MS, similar to that of thermal proteome profiling [88], which employs isobaric TMT (tandem mass tag) reagents to simultaneously quantify protein solubility across ten different temperatures from CETSA (Cellular Thermal Shift Assay) experiments [89] (Fig. 5).
Fig. 5.
The TPCA workflow. TPCA can be performed on intact cells or cell lysate. Lysed samples are first divided into an equal amount of aliquots and subjected to heat treatment with an increasing temperature gradient. Heat treatment induces denaturation and coaggregation of interacting proteins, which then co-precipitate. Upon centrifugation, the supernatant consisting of soluble proteins from different temperature treatment is retrieved for isobaric TMT-labelling and quantitative LC–MS/MS analysis. The abundance of each soluble proteins identified and quantified is then plotted against the temperatures to generate the “protein melting curve”
Current implementation of TPCA utilizes the CETSA protocol [90] to denature proteins and extract the soluble fraction, followed by TPP for proteome-wide quantification of protein solubility [91]. When the thermal solubilities of proteins are plotted against increasing temperatures, the so-called melting curve of proteins can be constructed to visualize TPCA signature across cell types or conditions. The similarity in protein thermal solubility between pairs of proteins across multiple temperatures can be quantified using measures like Euclidean distance [87] and Pearson's correlation [92]. Statistical significance of observed similarities and changes in thermal solubility between pairs of proteins are estimated through a bootstrapping approach using random pairs of proteins to establish random background distribution [87].
Using TPP and CETSA protocols, data for TPCA analysis can be obtained from both cell lysate and intact cells. In the former, cells are first lysed before heat denaturation, while in the latter, intact cells are first heated before cell lysis. In the first proof-of-concept work demonstrating TPCA can be used to identify protein complexes modulated across cell types, cellular states and cellular conditions, protein complexes were observed to exhibit much stronger TPCA signature (i.e. co-aggregating) in data from intact cells than from cell lysate. As the first proof-of-concept experiment, TPCA was performed to identify protein complexes modulated across different cell types, cellular states and cellular conditions [87]. The final results showed that protein complexes obtained from intact cells exhibited a higher level of co-aggregation (stronger TPCA signature) than those originated from cell lysate [87]. This observation suggests the integrity of protein complexes might have been compromised after cell lysis. Notably, for many protein complexes that exhibit TPCA signature only in intact cells, they are often associated and likely dependent on subcellular scaffolds like chromatin and membrane for structural stability, which is probably absent in cell lysate. Taken together, these observations suggest TPCA will be valuable for studying protein complexes in situ, particularly for weak-binding protein complexes that easily dissociate after cell lysis. Importantly, TPCA can reveal the subcomplex organization of megacomplexes like the nuclear pore complex and the proteasome [87, 92]. Also, it has been reported that phosphorylation can affect the thermal solubility of protein through modulating PPIs, suggesting the ability to identify phosphorylation-dependent protein complexes [93]. Interestingly, similar to CETSA and TPP, it has also been shown that TPCA analysis can be extended to in vivo specimens such as tissues and blood samples [87, 94].
TPCA for system-wide profiling of protein complex dynamics has the advantages of requiring neither antibodies nor epitope tagging. It requires little preparation time compared to existing methods, and most importantly, permits the study of protein complexes in situ and in vivo. The current version of TPCA could be deployed to study the dynamics of known or predicted protein complexes across cellular states and physiological conditions efficiently, but need to incorporate existing interaction data with graph/network clustering algorithms to identify novel protein complexes. Nevertheless, Hashimoto et al. recently demonstrated novel protein–protein interactions could be inferred among the small set of viral proteins using only TPCA data [95]. Large-scale human interactome projects and integrative data analysis have uncovered many novel but functionally uncharacterized protein complexes. TPCA profiling can be rapidly deployed to unravel the assembly state of these protein complexes across cellular state, cell type, tissue and physiological conditions to provide insight into their functions in normal and diseased cells. The thermal protein solubility of proteins can be rapidly generated across species, and with data now available over 13 species ranging from human to archaea species. Thus, we envision that the TPCA analysis approach could be widely adopted to study protein complexes and protein interactions across the tree of life [96–98].
Current challenges
Despite being capable of wholesale copurification and detection of PPIs, the MS-based co-complex strategy is plagued with problems, particularly concerning the limited recovery of transient and low-affinity PPIs and false positives originating from high abundant proteins as backgrounds. Among these co-complex techniques, XL-MS can confirm the direct interaction of two interacting proteins due to the presence of inter-protein crosslinks. On the other hand, AP-MS can capture and identify direct and indirect binding partners of bait proteins. In comparison, both coFrac-MS and TPCA rely on correlation algorithms to infer PPIs from co-localization and coaggregation data of proteins. As such, they too, do not provide direct evidence of physical interactions. Therefore, PPI data derived from these methods should preferably be followed up meticulously using orthogonal methods, apart from validation with targeted MS or SWATH-MS.
To discriminate signal from noise, it is necessary for high-throughput PPI investigations to refer to the so-named "gold standards", databases containing curated and unequivocal interactions [8, 99]. However, it is noteworthy that gold standard databases are assembled from different experiments and techniques, each with a unique set of biases [100]. This is because PPIs can be context-specific and transient. Single datasets, which are typically generated by a single technique, can disagree with gold standards. These variabilities may reflect actual biological differences, or technical biases. Therefore, gold-standard databases may fail to support the subset of interactions that are missing due to experimental conditions or technical limitations.
Although a common aim of the abovementioned techniques is to tease apart qualitatively the exact composition of protein complexes, additional information gained from these experiments may further elucidate the structural and functional properties of identified PPIs. This additional information encompasses the topology and the quantitative measurement of the stoichiometry, copy number and dynamics of these identified protein complexes [101]. As of now, MS-based proteomics and structural biology have increasingly merged with MS-based methods progressively used to complement structural biology tools [102]. The topology of a protein complex relates how each protein subunit is interconnected to contribute to the overall shape and relative spatial arrangements of a complex. Currently, XL-MS and several dedicated structural MS techniques such as native MS, hydrogen/deuterium (H/D) exchange and hydroxy radical foot-printing have been employed to unravel protein complex topology. Notably, XL-MS can yield valuable data on the spatial constraint, subunit connectivity and direct PPIs at a proteome-wide scale. Meanwhile, the determination of stoichiometry and copy number within a protein complex with biological MS has been chiefly accomplished using native MS and absolute quantification using peptide-based MS. Notably, the determination of these two values using peptide-based MS measurement is highly dependent on knowing the concentrations of each constituent in the complex under study. This means that MS-based absolute quantification, which entails spiking in known and quantified reference peptides for external or internal calibration, is required to accurately determine the concentration of proteins [101].
Future perspectives
Contemporary development in chemical and synthetic biology has further enriched the toolbox to disentangle protein networks. One promising area is click chemistry, which possesses exceptional biorthogonality, efficiency and selectivity has been increasingly adopted in proteomics, particularly for probing new protein synthesis and post-translational modifications [103]. Meanwhile, a synthetic biology tool, named genetic code expansion, enables site-specific incorporation of unnatural amino acids (UAAs) into a protein of interest (POI) by exploiting amber codon suppression. A TAG stop codon is practically first introduced to the target gene at the target locale, followed by transient transfection of a tRNA complementary to this stop codon (tRNACUA), the UAA, and an orthogonal aminoacyl tRNA synthetase.
By combining both technologies, Smits et al. genetically encoded a UAA, i.e., p-azido-L-phenylalanine, a phenylalanine analogue containing a clickable azide group, which serves as a small handle for selectively enriching the POI using copper-free click chemistry [104]. Relative to the traditional epitope tags, this small handle is less likely to interfere with the localization, solubility and functions of the POI. Besides, the incorporation of UAA carrying photo-reactive groups such as aryl azides, benzophenones and diazirines has been reported for proximity-dependent labelling and stabilizing in vivo transient PPIs’ covalent capture [105]. Recently, bifunctional UAAs, such as DiZASeC, containing both clickable handles and photo-crosslinker side chains, have also been reported [106]. These bifunctional UAAs enable a POI and its physiological protein interactors to be “locked” in vivo via covalent crosslinking upon UV-irradiation, thus preventing their dissociation by subsequent cell lysis and proteolytic digest. The tryptic peptides are then reacted with click chemistry reagents so that only peptides harbouring the UAA are labelled and affinity-purified for MS analysis.
Conclusion
Essentially, the five widely used strategies that we have reviewed here elucidate PPIs based on the principles of copurification (AP-MS), proximity (XL-MS and PDB-MS) and the co-behaviour of physically interacting proteins (co-Frac-MS and TPCA), which may inevitably result in some distinct bias. However, these methods are not mutually exclusive; but instead, their complementarity should be exploited. A good example would be combining PDB-MS and XL-MS, or XL-MS with HDX-MS, as mentioned above. We also noted that the blurring of the boundary between PPI studies and structural biology. Notably, PDB-MS and XL-MS can refine structural data obtained from X-ray, NMR and cryoEM, apart from MS-based approaches such as native MS and HDX-MS. The elucidation of the composition of protein complexes and their interacting surfaces, topology, stoichiometry, copy number and dynamics would further enhance the utility of these tools for integrated structural biology in the future.
Acknowledgements
The authors would like to thank SCIEX Malaysia for technical and research support. They are also grateful to Ms. Shu Ning Low for producing the figures. All figures are created with BioRender.com.
Author contributions
TYL and CSHT contributed to the conception and design of the review article; TYL, MAM, SES, AV, NSAR, YFP and CSHT drafted the article and interpreting the relevant literature; TYL, YFP and CSHL revised it critically for important intellectual content. All authors reviewed and approved the final draft.
Funding
TYL is financially supported by the Geran Universiti Penyelidikan (GUP-2020-078) scheme awarded by Universiti Kebangsaan Malaysia (UKM). This work is also supported by grants from National Natural Science Foundation of China (Grant 22074060) and Shenzhen Innovation of Science and Technology Commission Grant (Grant JCY20200109140814408) awarded to CSHT. MAM and TYL are also supported by the Dana Impak Perdana scheme (DIP-2018-011 and DIP-2020-002) awarded by Universiti Kebangsaan Malaysia (UKM). This work is also funded by the Malaysia Ministry of Higher Education Grant FRGS/1/2019/SKK08/UNIM/02/2 awarded to YFP.
Declarations
Conflict of interest
The authors declare that there are no conflicts of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Saiful Effendi Syafruddin and M. Aiman Mohtar contributed equally.
Contributor Information
Teck Yew Low, Email: lowteckyew@ppukm.ukm.edu.my.
Chris Soon Heng Tan, Email: christan@sustech.edu.cn.
References
- 1.Stumpf MPH, Thorne T, de Silva E, et al. Estimating the size of the human interactome. Proc Natl Acad Sci USA. 2008;105:6959–6964. doi: 10.1073/pnas.0708078105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Senior AW, Evans R, Jumper J, et al. Improved protein structure prediction using potentials from deep learning. Nature. 2020;577:706–710. doi: 10.1038/s41586-019-1923-7. [DOI] [PubMed] [Google Scholar]
- 3.Titeca K, Lemmens I, Tavernier J, Eyckerman S. Discovering cellular protein-protein interactions: technological strategies and opportunities. Mass Spectrom Rev. 2019;38:79–111. doi: 10.1002/mas.21574. [DOI] [PubMed] [Google Scholar]
- 4.Wu Z, Malty R, Moutaoufik MT, et al. Advances in experimental medicine and biology. New York LLC: Springer; 2019. A tag-based affinity purification mass spectrometry workflow for systematic isolation of the human mitochondrial protein complexes; pp. 83–100. [DOI] [PubMed] [Google Scholar]
- 5.Vandemoortele G, Eyckerman S, Gevaert K. Pick a tag and explore the functions of your pet protein. Trends Biotechnol. 2019;37:1078–1090. doi: 10.1016/j.tibtech.2019.03.016. [DOI] [PubMed] [Google Scholar]
- 6.Choi H, Larsen B, Lin ZY, et al. SAINT: probabilistic scoring of affinity purificationg-mass spectrometry data. Nat Methods. 2011;8:70–73. doi: 10.1038/nmeth.1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mellacheruvu D, Wright Z, Couzens AL, et al. The CRAPome: a contaminant repository for affinity purification-mass spectrometry data. Nat Methods. 2013;10:730–736. doi: 10.1038/nmeth.2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huttlin EL, Ting L, Bruckner RJ, et al. The BioPlex Network: A Systematic Exploration of the Human Interactome. Cell. 2015;162:425–440. doi: 10.1016/j.cell.2015.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gordon DE, Jang GM, Bouhaddou M, et al. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020;583:459–468. doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gordon DE, Hiatt J, Bouhaddou M, et al. Comparative host-coronavirus protein interaction networks reveal pan-viral disease mechanisms. Science (-80) 2020 doi: 10.1126/science.abe9403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gingras AC, Abe KT, Raught B. Getting to know the neighborhood: using proximity-dependent biotinylation to characterize protein complexes and map organelles. Curr Opin Chem Biol. 2019;48:44–54. doi: 10.1016/j.cbpa.2018.10.017. [DOI] [PubMed] [Google Scholar]
- 12.Roux KJ, Kim DI, Raida M, Burke B. A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. J Cell Biol. 2012;196:801–810. doi: 10.1083/jcb.201112098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rhee HW, Zou P, Udeshi ND, et al. Proteomic mapping of mitochondria in living cells via spatially restricted enzymatic tagging. Science (-80) 2013;339:1328–1331. doi: 10.1126/science.1230593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim DI, Birendra KC, Zhu W, et al. Probing nuclear pore complex architecture with proximity-dependent biotinylation. Proc Natl Acad Sci USA. 2014;111:E2453–E2461. doi: 10.1073/pnas.1406459111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martell JD, Deerinck TJ, Sancak Y, et al. Engineered ascorbate peroxidase as a genetically encoded reporter for electron microscopy. Nat Biotechnol. 2012;30:1143–1148. doi: 10.1038/nbt.2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Di K, Sc J, Ka N, et al. An improved smaller biotin ligase for BioID proximity labeling Dae. Mol Biol Cell. 2016;27:1188–1196. doi: 10.1091/mbc.E15-12-0844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Branon TC, Bosch JA, Sanchez AD, et al. Efficient proximity labeling in living cells and organisms with TurboID. Nat Biotechnol. 2018;36:880–898. doi: 10.1038/nbt.4201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ramanathan M, Majzoub K, Rao DS, et al. RN A-protein interaction detection in living cells. Nat Methods. 2018;15:207–212. doi: 10.1038/nmeth.4601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bosch JA, Chen CL, Perrimon N. Proximity-dependent labeling methods for proteomic profiling in living cells: an update. Wiley Interdiscip Rev Dev Biol. 2021;10:1–17. doi: 10.1002/wdev.392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hopkins C, Gibson A, Stinchcombe J, Futter C. Chimeric molecules employing horseradish peroxidase as reporter enzyme for protein localization in the electron microscope. Methods Enzymol. 2000;327:35–45. doi: 10.1016/S0076-6879(00)27265-0. [DOI] [PubMed] [Google Scholar]
- 21.Cijsouw T, Ramsey AM, Lam TKT, et al. Mapping the proteome of the synaptic cleft through proximity labeling reveals new cleft proteins. Proteomes. 2018 doi: 10.3390/proteomes6040048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li J, Han S, Li H, et al. Cell-surface proteomic profiling in the fly brain uncovers wiring regulators. Cell. 2020;180:373–386.e15. doi: 10.1016/j.cell.2019.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jiang S, Kotani N, Ohnishi T, et al. A proteomics approach to the cell-surface interactome using the enzyme-mediated activation of radical sources reaction. Proteomics. 2012;12:54–62. doi: 10.1002/pmic.201100551. [DOI] [PubMed] [Google Scholar]
- 24.Li XW, Rees JS, Xue P, et al. New insights into the DT40 B cell receptor cluster using a proteomic proximity labeling assay. J Biol Chem. 2014;289:14434–14447. doi: 10.1074/jbc.M113.529578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rees JS. Methods in molecular biology. Humana Press Inc; 2019. Proteomic proximity labeling to reveal interactions between biomolecules; pp. 13–28. [DOI] [PubMed] [Google Scholar]
- 26.Lam SS, Martell JD, Kamer KJ, et al. Directed evolution of APEX2 for electron microscopy and proximity labeling. Nat Methods. 2014;12:51–54. doi: 10.1038/nmeth.3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sen HM, Lin WC, Chang JH, et al. The cysteine-free single mutant C32S of APEX2 is a highly expressed and active fusion tag for proximity labeling applications. Protein Sci. 2019;28:1703–1712. doi: 10.1002/pro.3685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Trinkle-Mulcahy L. Recent advances in proximity-based labeling methods for interactome mapping. F1000Res. 2019 doi: 10.12688/f1000research.16903.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hung V, Lam SS, Udeshi ND, et al. Proteomic mapping of cytosol-facing outer mitochondrial and ER membranes in living human cells by proximity biotinylation. Elife. 2017 doi: 10.7554/eLife.24463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Che Y, Khavari PA. Research techniques made simple: emerging methods to elucidate protein interactions through spatial proximity. J Invest Dermatol. 2017;137:e197–e203. doi: 10.1016/j.jid.2017.09.028. [DOI] [PubMed] [Google Scholar]
- 31.Ke M, Yuan X, He A, et al. Spatiotemporal profiling of cytosolic signaling complexes in living cells by selective proximity proteomics. Nat Commun. 2021 doi: 10.1038/s41467-020-20367-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Orre LM, Vesterlund M, Pan Y, et al. SubCellBarCode: proteome-wide mapping of protein localization and relocalization. Mol Cell. 2019;73:166–182.e7. doi: 10.1016/j.molcel.2018.11.035. [DOI] [PubMed] [Google Scholar]
- 33.Zhang Y, Song G, Lal NK, et al. TurboID-based proximity labeling reveals that UBR7 is a regulator of N NLR immune receptor-mediated immunity. Nat Commun. 2019;10:3252. doi: 10.1038/s41467-019-11202-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hill ZB, Pollock SB, Zhuang M, Wells JA. Direct proximity tagging of small molecule protein targets using an engineered NEDD8 ligase. J Am Chem Soc. 2016;138:13123–13126. doi: 10.1021/jacs.6b06828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu Q, Zheng J, Sun W, et al. A proximity-tagging system to identify membrane protein–protein interactions. Nat Methods. 2018;15:715–722. doi: 10.1038/s41592-018-0100-5. [DOI] [PubMed] [Google Scholar]
- 36.Wang HH, Altun B, Nwe K, Tsourkas A. Proximity-based sortase-mediated ligation. Angew Chem-Int Ed. 2017;56:5349–5352. doi: 10.1002/anie.201701419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tamura T, Takato M, Shiono K, Hamachi I. Development of a photoactivatable proximity labeling method for the identification of nuclear proteins. Chem Lett. 2020;49:145–148. doi: 10.1246/cl.190804. [DOI] [Google Scholar]
- 38.Young MM, Tang N, Hempel JC, et al. High throughput protein fold identification by using experimental constraints derived from intramolecular cross-links and mass spectrometry. Proc Natl Acad Sci USA. 2000;97:5802–5806. doi: 10.1073/pnas.090099097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rappsilber J, Siniossoglou S, Hurt EC, Mann M. A generic strategy to analyze the spatial organization of multi-protein complexes by cross-linking and mass spectrometry. Anal Chem. 2000;72:267–275. doi: 10.1021/ac991081o. [DOI] [PubMed] [Google Scholar]
- 40.Steigenberger B, Albanese P, Heck AJR, Scheltema RA. To cleave or not to cleave in XL-MS? J Am Soc Mass Spectrom. 2020;31:196–206. doi: 10.1021/jasms.9b00085. [DOI] [PubMed] [Google Scholar]
- 41.Steigenberger B, Pieters RJ, Heck AJR, Scheltema RA. PhoX: an IMAC-enrichable cross-linking reagent. ACS Cent Sci. 2019;5:1514–1522. doi: 10.1021/acscentsci.9b00416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tan D, Li Q, Zhang MJ, et al. Trifunctional cross-linker for mapping protein-protein interaction networks and comparing protein conformational states. Elife. 2016 doi: 10.7554/eLife.12509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kao A, Chiu C, Vellucci D, et al. Development of a novel cross-linking strategy for fast and accurate identification of cross-linked peptides of protein complexes. Mol Cell Proteomics. 2011;10(M110):002212. doi: 10.1074/mcp.m110.002212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Müller MQ, Dreiocker F, Ihling CH, et al. Cleavable cross-linker for protein structure analysis: reliable identification of cross-linking products by tandem MS. Anal Chem. 2010;82:6958–6968. doi: 10.1021/ac101241t. [DOI] [PubMed] [Google Scholar]
- 45.De Jong L, De Koning EA, Roseboom W, et al. In-culture cross-linking of bacterial cells reveals large-scale dynamic protein-protein interactions at the peptide level. J Proteome Res. 2017;16:2457–2471. doi: 10.1021/acs.jproteome.7b00068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Walker-Gray R, Stengel F, Gold MG. Mechanisms for restraining cAMP-dependent protein kinase revealed by subunit quantitation and cross-linking approaches. Proc Natl Acad Sci USA. 2017;114:10414–10419. doi: 10.1073/pnas.1701782114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Leitner A, Faini M, Stengel F, Aebersold R. Crosslinking and mass spectrometry: an integrated technology to understand the structure and function of molecular machines. Trends Biochem Sci. 2016;41:20–32. doi: 10.1016/j.tibs.2015.10.008. [DOI] [PubMed] [Google Scholar]
- 48.Merkley ED, Rysavy S, Kahraman A, et al. Distance restraints from crosslinking mass spectrometry: mining a molecular dynamics simulation database to evaluate lysine-lysine distances. Protein Sci. 2014;23:747–759. doi: 10.1002/pro.2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu F, Heck AJR. Interrogating the architecture of protein assemblies and protein interaction networks by cross-linking mass spectrometry. Curr Opin Struct Biol. 2015;35:100–108. doi: 10.1016/j.sbi.2015.10.006. [DOI] [PubMed] [Google Scholar]
- 50.Yu C, Huang L. Cross-linking mass spectrometry: an emerging technology for interactomics and structural biology. Anal Chem. 2018;90:144–165. doi: 10.1021/acs.analchem.7b04431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tang X, Wippel HH, Chavez JD, Bruce JE. Crosslinking mass spectrometry: a link between structural biology and systems biology. Protein Sci. 2021;30:773–784. doi: 10.1002/pro.4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Leitner A, Joachimiak LA, Bracher A, et al. The molecular architecture of the eukaryotic chaperonin TRiC/CCT. Structure. 2012;20:814–825. doi: 10.1016/j.str.2012.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Staals RHJ, Zhu Y, Taylor DW, et al. RNA targeting by the type III-A CRISPR-Cas Csm complex of thermus thermophilus. Mol Cell. 2014;56:518–530. doi: 10.1016/j.molcel.2014.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Leitner A, Joachimiak LA, Unverdorben P, et al. Chemical cross-linking/mass spectrometry targeting acidic residues in proteins and protein complexes. Proc Natl Acad Sci USA. 2014;111:9455–9460. doi: 10.1073/pnas.1320298111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Herzog F, Kahraman A, Boehringer D, et al. Structural probing of a protein phosphatase 2A network by chemical cross-linking and mass spectrometry. Science (-80) 2012;337:1348–1352. doi: 10.1126/science.1221483. [DOI] [PubMed] [Google Scholar]
- 56.Liu F, Lössl P, Rabbitts BM, et al. The interactome of intact mitochondria by cross-linking mass spectrometry provides evidence for coexisting respiratory supercomplexes. Mol Cell Proteomics. 2018;17:216–232. doi: 10.1074/mcp.RA117.000470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kaake RM, Wang X, Burke A, et al. A new in vivo cross-linking mass spectrometry platform to define protein-protein interactions in living cells. Mol Cell Proteomics. 2014;13:3533–3543. doi: 10.1074/mcp.M114.042630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liu F, Rijkers DTS, Post H, Heck AJR. Proteome-wide profiling of protein assemblies by cross-linking mass spectrometry. Nat Methods. 2015;12:1179–1184. doi: 10.1038/nmeth.3603. [DOI] [PubMed] [Google Scholar]
- 59.Ngounou Wetie AG, Sokolowska I, Woods AG, et al. Investigation of stable and transient protein-protein interactions: past, present, and future. Proteomics. 2013;13:538–557. doi: 10.1002/pmic.201200328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Leitner A, Walzthoeni T, Kahraman A, et al. Probing native protein structures by chemical cross-linking, mass spectrometry, and bioinformatics. Mol Cell Proteom. 2010;9:1634–1649. doi: 10.1074/mcp.R000001-MCP201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Klykov O, Steigenberger B, Pekta S, et al. Efficient and robust proteome-wide approaches for cross-linking mass spectrometry. Nat Protoc. 2018;13:2964–2990. doi: 10.1038/s41596-018-0074-x. [DOI] [PubMed] [Google Scholar]
- 62.Schilling B, Row RH, Gibson BW, et al. MS2Assign, automated assignment and nomenclature of tandem mass spectra of chemically crosslinked peptides. J Am Soc Mass Spectrom. 2003;14:834–850. doi: 10.1016/S1044-0305(03)00327-1. [DOI] [PubMed] [Google Scholar]
- 63.Fritzsche R, Ihling CH, Götze M, Sinz A. Optimizing the enrichment of cross-linked products for mass spectrometric protein analysis. Rapid Commun Mass Spectrom. 2012;26:653–658. doi: 10.1002/rcm.6150. [DOI] [PubMed] [Google Scholar]
- 64.Leitner A, Reischl R, Walzthoeni T, et al. Expanding the chemical cross-linking toolbox by the use of multiple proteases and enrichment by size exclusion chromatography. Mol Cell Proteomics. 2012 doi: 10.1074/mcp.M111.014126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sinz A. Chemical cross-linking and mass spectrometry to map three-dimensional protein structures and protein-protein interactions. Mass Spectrom Rev. 2006;25:663–682. doi: 10.1002/mas.20082. [DOI] [PubMed] [Google Scholar]
- 66.Chen ZL, Meng JM, Cao Y, et al. A high-speed search engine pLink 2 with systematic evaluation for proteome-scale identification of cross-linked peptides. Nat Commun. 2019;10:1–12. doi: 10.1038/s41467-019-11337-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Barysz HM, Malmström J. Development of large-scale cross-linking mass spectrometry. Mol Cell Proteom. 2018;17:1055–1066. doi: 10.1074/mcp.R116.061663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Liu CH, Chien MJ, Chang YC, et al. Combining proximity labeling and cross-linking mass spectrometry for proteomic dissection of nuclear envelope interactome. J Proteome Res. 2020;19:1109–1118. doi: 10.1021/acs.jproteome.9b00609. [DOI] [PubMed] [Google Scholar]
- 69.Courouble VV, Dey SK, Yadav R, et al. Resolving the dynamic motions of SARS-CoV-2 nsp7 and nsp8 proteins using structural proteomics. bioRxiv Prepr Serv Biol. 2021 doi: 10.1101/2021.03.06.434214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Feiglin A, Ashkenazi S, Schlessinger A, et al. Co-expression and co-localization of hub proteins and their partners are encoded in protein sequence. Mol Biosyst. 2014;10:787–794. doi: 10.1039/c3mb70411d. [DOI] [PubMed] [Google Scholar]
- 71.Mann M. The origins of organellar mapping by protein correlation profiling. Proteomics. 2020 doi: 10.1002/pmic.201900330. [DOI] [PubMed] [Google Scholar]
- 72.Andersen JS, Wilkinson CJ, Mayor T, et al. Proteomic characterization of the human centrosome by protein correlation profiling. Nature. 2003;426:570–574. doi: 10.1038/nature02166. [DOI] [PubMed] [Google Scholar]
- 73.Dunkley TPJ, Watson R, Griffin JL, et al. Localization of organelle proteins by isotope tagging (LOPIT) Mol Cell Proteom. 2004;3:1128–1134. doi: 10.1074/mcp.T400009-MCP200. [DOI] [PubMed] [Google Scholar]
- 74.Dong M, Yang LL, Williams K, et al. A “tagless” strategy for identification of stable protein complexes genome-wide by multidimensional orthogonal chromatographic separation and iTRAQ reagent tracking. J Proteome Res. 2008;7:1836–1849. doi: 10.1021/pr700624e. [DOI] [PubMed] [Google Scholar]
- 75.Havugimana PC, Hart GT, Nepusz T, et al. A census of human soluble protein complexes. Cell. 2012;150:1068–1081. doi: 10.1016/j.cell.2012.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kristensen AR, Gsponer J, Foster LJ. A high-throughput approach for measuring temporal changes in the interactome. Nat Methods. 2012;9:907–909. doi: 10.1038/nmeth.2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bludau I, Heusel M, Frank M, et al. Complex-centric proteome profiling by SEC-SWATH-MS for the parallel detection of hundreds of protein complexes. Nat Protoc. 2020;15:2341–2386. doi: 10.1038/s41596-020-0332-6. [DOI] [PubMed] [Google Scholar]
- 78.Kristensen AR, Foster LJ. Protein correlation profiling-SILAC to study protein–protein interactions. Methods Mol Biol. 2014;1188:263–270. doi: 10.1007/978-1-4939-1142-4_18. [DOI] [PubMed] [Google Scholar]
- 79.Minic Z, Dahms TES, Babu M. Chromatographic separation strategies for precision mass spectrometry to study protein–protein interactions and protein phosphorylation. J Chromatogr B Anal Technol Biomed Life Sci. 2018;1102–1103:96–108. doi: 10.1016/j.jchromb.2018.10.022. [DOI] [PubMed] [Google Scholar]
- 80.Kunji ERS, Harding M, Butler PJG, Akamine P. Determination of the molecular mass and dimensions of membrane proteins by size exclusion chromatography. Methods. 2008;46:62–72. doi: 10.1016/j.ymeth.2008.10.020. [DOI] [PubMed] [Google Scholar]
- 81.Salas D, Stacey RG, Akinlaja M, Foster LJ. Next-generation interactomics: considerations for the use of co-elution to measure protein interaction networks. Mol Cell Proteom. 2019;19:mcp.R119.001803. doi: 10.1074/mcp.R119.001803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Rackiewicz M, Große-Hovest L, Alpert AJ, et al. Hydrophobic interaction chromatography for bottom-up proteomics analysis of single proteins and protein complexes. J Proteom Res. 2017;16(2318):2323. doi: 10.1021/acs.jproteome.7b00015. [DOI] [PubMed] [Google Scholar]
- 83.Scott NE, Rogers LD, Prudova A, et al. Interactome disassembly during apoptosis occurs independent of caspase cleavage. Mol Syst Biol. 2017;13:906. doi: 10.15252/msb.20167067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Heide H, Bleier L, Steger M, et al. Complexome profiling identifies TMEM126B as a component of the mitochondrial complex i assembly complex. Cell Metab. 2012;16:538–549. doi: 10.1016/j.cmet.2012.08.009. [DOI] [PubMed] [Google Scholar]
- 85.Mallam AL, Sae-Lee W, Schaub JM, et al. Systematic discovery of endogenous human ribonucleoprotein complexes. Cell Rep. 2019;29:1351–1368.e5. doi: 10.1016/j.celrep.2019.09.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Moutaoufik MT, Malty R, Amin S, et al. Rewiring of the human mitochondrial interactome during neuronal reprogramming reveals regulators of the respirasome and neurogenesis. iScience. 2019;19:1114–1132. doi: 10.1016/j.isci.2019.08.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Tan CSH, Go KD, Bisteau X, et al. Thermal proximity coaggregation for system-wide profiling of protein complex dynamics in cells. Science (-80) 2018;359:1170–1177. doi: 10.1126/science.aan0346. [DOI] [PubMed] [Google Scholar]
- 88.Savitski MM, Reinhard FBM, Franken H, et al. Tracking cancer drugs in living cells by thermal profiling of the proteome. Science (-80) 2014 doi: 10.1126/science.1255784. [DOI] [PubMed] [Google Scholar]
- 89.Molina DM, Jafari R, Ignatushchenko M, et al. Monitoring drug target engagement in cells and tissues using the cellular thermal shift assay. Science (-80) 2013;341:84–87. doi: 10.1126/science.1233606. [DOI] [PubMed] [Google Scholar]
- 90.Jafari R, Almqvist H, Axelsson H, et al. The cellular thermal shift assay for evaluating drug target interactions in cells. Nat Protoc. 2014;9:2100–2122. doi: 10.1038/nprot.2014.138. [DOI] [PubMed] [Google Scholar]
- 91.Franken H, Mathieson T, Childs D, et al. Thermal proteome profiling for unbiased identification of direct and indirect drug targets using multiplexed quantitative mass spectrometry. Nat Protoc. 2015;10:1567–1593. doi: 10.1038/nprot.2015.101. [DOI] [PubMed] [Google Scholar]
- 92.Becher I, Andrés-Pons A, Romanov N, et al. Pervasive protein thermal stability variation during the cell cycle. Cell. 2018;173:1495–1507.e18. doi: 10.1016/j.cell.2018.03.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Huang JX, Lee G, Cavanaugh KE, et al. High throughput discovery of functional protein modifications by hotspot thermal profiling. Nat Methods. 2019;16:894–901. doi: 10.1038/s41592-019-0499-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Perrin J, Werner T, Kurzawa N, et al. Identifying drug targets in tissues and whole blood with thermal-shift profiling. Nat Biotechnol. 2020;38:303–308. doi: 10.1038/s41587-019-0388-4. [DOI] [PubMed] [Google Scholar]
- 95.Hashimoto Y, Sheng X, Murray-Nerger LA, Cristea IM. Temporal dynamics of protein complex formation and dissociation during human cytomegalovirus infection. Nat Commun. 2020 doi: 10.1038/s41467-020-14586-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Jarzab A, Kurzawa N, Hopf T, et al. Meltome atlas—thermal proteome stability across the tree of life. Nat Methods. 2020;17:495–503. doi: 10.1038/s41592-020-0801-4. [DOI] [PubMed] [Google Scholar]
- 97.Volkening JD, Stecker KE, Sussman MR. Proteome-wide analysis of protein thermal stability in the model higher plant arabidopsis thaliana. Mol Cell Proteomics. 2019;18:308–319. doi: 10.1074/mcp.RA118.001124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Mateus A, Bobonis J, Kurzawa N, et al. Thermal proteome profiling in bacteria: probing protein state in vivo. Mol Syst Biol. 2018 doi: 10.15252/msb.20188242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wan C, Borgeson B, Phanse S, et al. Panorama of ancient metazoan macromolecular complexes. Nature. 2015;525:339–344. doi: 10.1038/nature14877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Stacey RG, Skinnider MA, Chik JHL, Foster LJ. Context-specific interactions in literature-curated protein interaction databases. BMC Genomics. 2018;19:758. doi: 10.1186/s12864-018-5139-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wohlgemuth I, Lenz C, Urlaub H. Studying macromolecular complex stoichiometries by peptide-based mass spectrometry. Proteomics. 2015;15:862–879. doi: 10.1002/pmic.201400466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Faini M, Stengel F, Aebersold R. The evolving contribution of mass spectrometry to integrative structural biology. J Am Soc Mass Spectrom. 2016;27:966–974. doi: 10.1007/s13361-016-1382-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Parker CG, Pratt MR. Click chemistry in proteomic investigations. Cell. 2020;180:605–632. doi: 10.1016/j.cell.2020.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Smits AH, Borrmann A, Roosjen M, et al. Click-MS: tagless protein enrichment using bioorthogonal chemistry for quantitative proteomics. ACS Chem Biol. 2016;11:3245–3250. doi: 10.1021/acschembio.6b00520. [DOI] [PubMed] [Google Scholar]
- 105.Nguyen TA, Cigler M, Lang K. Expanding the genetic code to study protein-protein interactions. Angew Chemi-Int Ed. 2018;57:14350–14361. doi: 10.1002/anie.201805869. [DOI] [PubMed] [Google Scholar]
- 106.He D, Xie X, Yang F, et al. Quantitative and comparative profiling of protease substrates through a genetically encoded multifunctional photocrosslinker. Angew Chem Int Ed. 2017;56:14521–14525. doi: 10.1002/anie.201708151. [DOI] [PubMed] [Google Scholar]