Abstract
Soluble forms of aggregated tau misfolded protein, generally termed oligomers, are considered to be the most toxic species of the different assembly states that are the pathological components of neurodegenerative disorders. Therefore, a critical biomedical need exists for imaging probes that can identify and quantify them. We have designed and synthesized a novel fluorescent probe, pTP-TFE for which binding and selectivity profiles towards aggregated tau and Aβ proteins were assessed. Our results have shown pTP-TFE to be selective for early forms of soluble tau aggregates, with high affinity of dissociation constants (Kd) = 66 nM, and tenfold selectivity over mature tau fibrils. Furthermore, we found that pTP-TFE is selective for tau over Aβ aggregates and had good cell permeability. This selectivity of pTP-TFE towards early forms of aggregated tau protein ex vivo was also supported with studies on human brain tissue containing tau and Aβ pathology. To the best of our knowledge, this is the first fluorescent molecule to be reported to have this form of selectivity profile, which suggests that pTP-TFE is a unique probe candidate for imaging-based detection of early stages of Alzheimer's disease and other tauopathies.
pTP-TFE imaging probe can distinguish soluble tau aggregated proteins from other aggregated proteins enabling earlier detection of neurodegenerative diseases.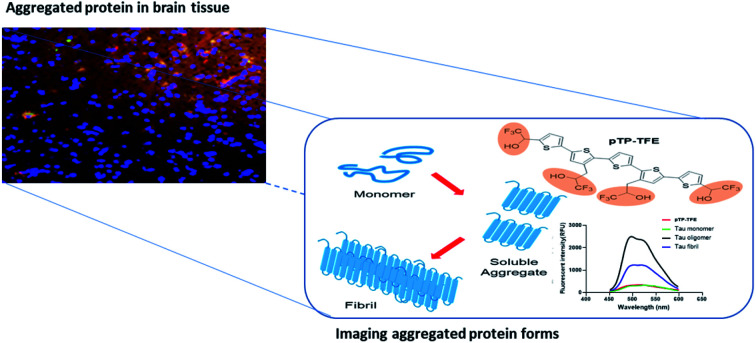
Introduction
The accumulation of misfolded protein aggregates is a pathological hallmark of neurodegenerative diseases.1 In the case of Alzheimer's disease (AD), the most prevalent pathology consists of extracellular β-amyloid protein (Aβ) plaques and intracellular tau protein-containing neurofibrillary tangles.2 Protein aggregates have high beta-sheet structural characteristic,3 which led to the identification of small molecules with specific binding affinity for them,4 several of which have been used successfully for in vivo imaging with positron emission tomography (PET).5 However, it is becoming increasingly apparent that proteins associated with neurodegenerative diseases can adopt different assembly states, which can impact disease onset and their progression to different extent.6 Research on tau protein has shown that the earlier forming smaller soluble aggregates, generally termed oligomers, are the most toxic species and their in vivo levels are likely to be more correlated with disease progression than the levels of mature fibrils.7 Given that early diagnosis of neurodegenerative disease has become an important objective, a critical need exists for the development of novel imaging probes that can identify and quantify soluble aggregated protein species in vivo. Moreover, such probes could greatly enhance our understanding of disease progression and be used for assessing the efficacy of new therapeutic candidates.
Efforts to design a facile imaging agent that can be used for investigation of soluble protein aggregates have thus far focused on Aβ-protein, leading to the development of a monoclonal antibody based probe ([125I]8D3-F(ab′)2-h158)8 and a modest number of small molecules, most notably BD-Oligo based on the boron-dipyrromethene (BODIPY) fluorescence structure9 and AN-SP based on a spiropyran scaffold.10 In addition, luminescent conjugated oligothiophenes (LCOs) have been shown to bind to soluble aggregates of both Aβ and tau protein.11 Despite the success in recent years of developing PET probes for imaging tau neurofibrillary tangles12 presently there are no probes that have selective binding toward tau soluble aggregated species. With tau protein pathology being a constituent feature of several neurodegenerative disorders, collectively termed tauopathies, the development of such probes, represents a major biomedical need.
The scaffold diversity of recently reported tau binding compounds highlights the fact that, despite recent progress, there is currently no clear structure activity relationship consensus. Nonetheless, flat π-conjugated scaffolds with length limited to 13 Å and polarizable ligands are thought to be essential design elements for tau probes.13 Previously reported LCO, pFTAA (Scheme 1), fulfils these requirements by including a pentathiophene backbone and four carboxylic acid (CA) functional groups and has shown affinity for the soluble aggregates of tau and Aβ. However, with no apparent selectivity between protein types (i.e. tau or Aβ) or their various forms (i.e. soluble and fibril).14–16
Scheme 1. Synthesis of pTP-TFE; synthesis of pFTAA (analogues to previously report).14.
Towards developing molecular imaging probes with selectivity for soluble aggregates we noted an important structural feature of mature tau fibrils is a positively charged flexible polyelectrolyte brush, also known as a fuzzy coat.17 On the basis that the positive charges of the coat may interact strongly with the negative charges present in anionic dyes such as pFTAA, we reasoned that we may enhance selectivity toward soluble aggregate forms by reducing this chemical property.18 To impart selectivity but retain the CA binding properties toward aggregated proteins, we chose to replace the CAs with bioisosteric 2,2,2-trifluoroethan-1-ol (TFE) functional groups, to yield the compound pTP-TFE (pentathiophene-trifluoroethanol – Scheme 1). TFE functional groups have been shown to act as excellent replacements for CA groups, while imparting superior pharmacokinetic properties on molecules.19 Furthermore, the inclusion of fluorine into the molecule introduces several other advantages; (i) provides a natural position for labelling with fluorine-18 radioisotope so that a PET probe can be developed;20 (ii) allows 19F magnetic resonance imaging (MRI) and magnetic resonance spectroscopy (MRS) measurements; and (iii) may enhance in vivo pharmacokinetic properties, such as metabolic instability and reduce blood plasma retention.
Herein we report the synthesis of the novel fluorescent probe pTP-TFE as well as the characterization of its binding to tau and Aβ aggregated proteins in comparison to pFTAA. This includes the application of a sucrose gradient method to obtain known sizes of aggregate proteins and thereby quantify binding affinities of pTP-TFE towards specific soluble aggregated species. Finally, we assessed if pTP-TFE can penetrate intact live cell membranes and evaluated its binding to tau pathology in human brain tissue.
Results and discussion
Synthesis of pTP-TFE
The strategy for the synthesis of pTP-TFE relied on the trifluoromethylation of individual thiophene building blocks and their subsequent assembly to the pentameric target compound through Suzuki cross-coupling reactions (Scheme 1). Commercially available starting material (1) was brominated to yield 2 and subsequently treated with (trifluoromethyl)trimethylsilane and caesium fluoride to yield silyl protected compound 3, which was purified, deprotected (4) and reduced to give 2,2,2-trifluoroethan-1-ol bearing thiophene 5. 5 was cross-coupled to commercially available 2,5-bis-thiopheneboronic acid pinacol ester to yield 6, which was brominated to give 7. The terminal 2,2,2-trifluoroethan-1-ol bearing thiophene (10) was synthesized from commercially available starting material 2-bromo-5-trifluoroacetylthiophene (8), which was reduced (9) and subsequently boronated using bis(pinacolato)diboron (10). Finally, pTP-TFE was synthesized by Suzuki cross-coupling of substrates 7 and 10. Purification of pTP-TFE by preparative thin-layer chromatography (TLC) afforded the compound in acceptable purity for in vitro evaluation. The overall yield for the synthesis of pTP-TFE from starting material (1) is 0.7%. pFTAA was synthesized from starting material (1), analogues to the previously reported methodology.14 The fluorescence properties of pTP-TFE were obtained (Fig. S1.†pTP-TFE) including its quantum yield value, which at 0.27 is similar to pFTAA.14
Binding to Aβ and tau proteins in aggregation assay
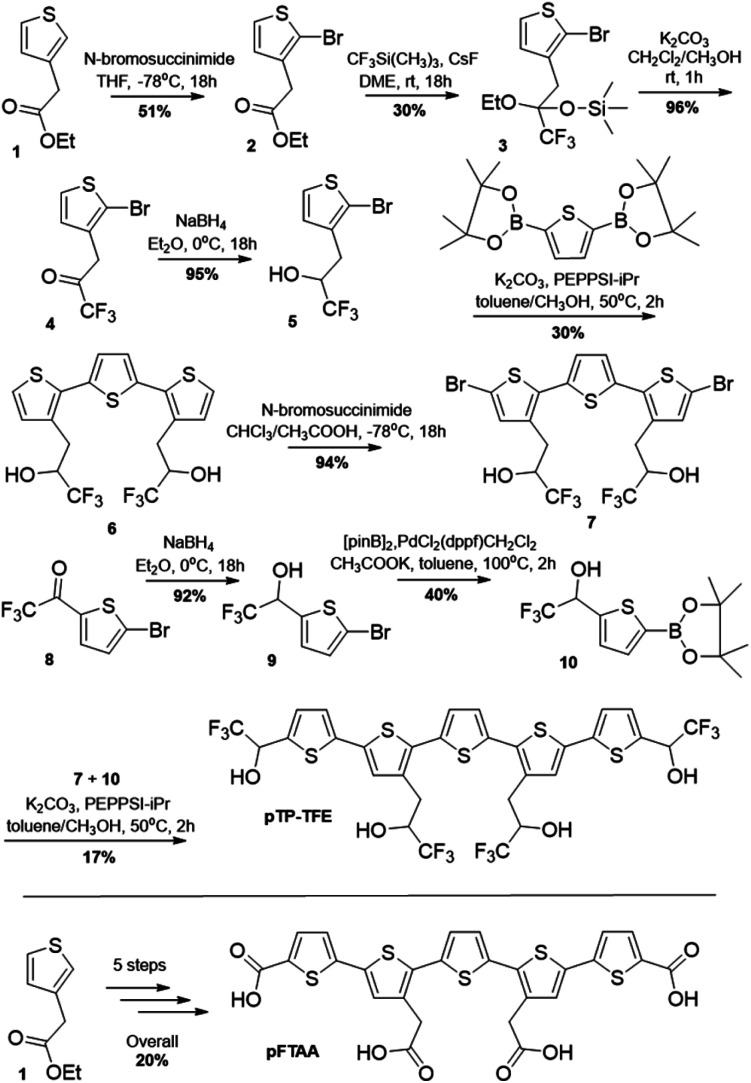
We started our investigation into pTP-TFE's binding and selectivity properties by performing an Aβ protein aggregation assay. In this assay monomeric Aβ40 is aggregated in the presence of pTP-TFE or pFTAA and the fluorescence intensities of the compounds monitored over time. An increase in fluorescence intensity is driven by realignment of π-orbitals due to conformation changes in the thiophene backbone and it is indicative of binding to protein aggregates.14 Initial results were encouraging as pTP-TFE showed an earlier response to Aβ40 aggregates in comparison to pFTAA (Fig. 1A). Transmission electron microscopy (TEM) images of Aβ40 aggregates at 90 min confirm the presence of small aggregate forms (Fig. 1B) and mature fibrils at later time points (700 min – Fig. 1D). To quantitatively assess the binding affinity of pTP-TFE and pFTAA to different types of aggregates, Aβ40 protein was collected from an aggregation assay (not containing compounds) at time points of 90 min, 200 min and 24 h. These aggregates were treated with different concentrations of the probes to determine binding constants (Kd) (Fig. 1E and F) by measuring their fluorescence intensity. These experiments confirmed that pTP-TFE displays a higher binding affinity to the earlier species (90 min) of aggregates (Kd = 7.58 μM), compared to pFTAA which showed no binding affinity for aggregates at this stage of maturation. Inversely, pFTAA showed stronger binding affinity to later aggregates (Kd = 0.93 μM {200 min}, Kd = 0.65 μM {24 h}) compared to pTP-TFE (Kd = 3.27 μM {200 min}, Kd = 3.79 μM {24 h}). In addition, the binding profile of pTP-TFE to a mixture of Aβ40 : Aβ42 (9 : 1 ratio), which is considered a more accurate representation of human cerebrospinal fluid Aβ aggregates in vivo, was also determined. On the basis of the analysis of fluorescence intensities during the assays (Fig. S6A†) pTP-TFE interacted with Aβ aggregates at an earlier time compared to pFTAA and ThT. Thus, we observe a similar relative interaction profile as with just Aβ40 (Fig. 1A). The binding affinities of pTP-TFE to Aβ40 : Aβ42 (9 : 1) mixture studied were Kd = 2.1 μM at 200 min and Kd = 0.75 μM at 24 h, therefore a higher binding affinity than just with Aβ40. This is consistent with higher affinity binding to Aβ42 as previously observed.21
Fig. 1. (A) Normalized fluorescence intensities of pTP-TFE, pFTAA and ThT in an Aβ aggregation assay starting with Aβ40 monomers (n = 3). (B–D) Representative TEM image of Aβ aggregates 90 min (B), 200 min (C) and 700 min (D) (white bar = 200 nm). (E and F) Fluorescence intensity based binding curve of pTP-TFE (E) and pFTAA (F) with Aβ aggregates collected at 90 min (red), 200 min (blue) and 24 hours (black) of the aggregation assay.
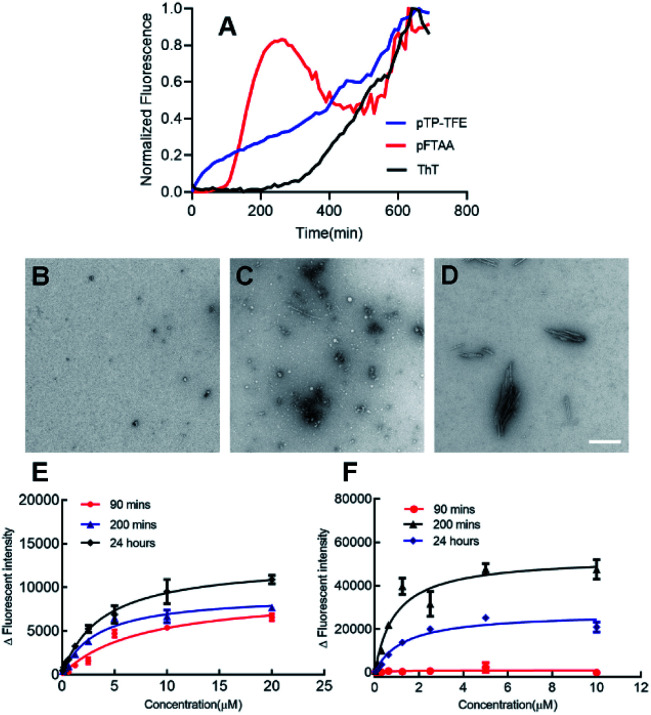
We then investigated the binding profile of pTP-TFE to tau protein. To this end, tau monomers were subjected to fibril-forming conditions using heparin and aliquots removed at 1, 5, 24, 48, 80, 96, 168 and 240 h. Treatment of these aliquots with pTP-TFE showed a gradual increase of fluorescence intensity, which reached a maximum after 96 h and declined thereafter until the end point of the experiment (Fig. 2A). Experiments with pFTAA showed a curve of similar shape and characteristic trajectory.16 TEM images of tau aggregates at different time points confirmed that initially small tau aggregates were detectable as early as 24 h (Fig. 2B), which became more apparent at 96 h (Fig. 2C) and then mature fibrils were found in the 240 h samples (Fig. 2D). These results suggest preferential binding of pTP-TFE to early soluble tau aggregates in comparison to pFTAA.
Fig. 2. (A) Normalized fluorescence intensities of pTP-TFE (n = 3), pFTAA (n = 3) and ThT (n = 3) in an aggregation assay starting with tau monomers. (B–D) Representative TEM image of tau aggregates 24 h (B), 96 h (C) and 240 h (D) (white bar = 200 nm). (E and F) Binding affinity of pTP-TFE to tau aggregates collected at 96 hours (E) and 240 hours (F) of the aggregation assay.
Interestingly, we observed a shift in the emission spectrum of pTP-TFE over the course of the experiment. Emission spectra recorded with tau aggregates at 24, 96 and 240 h show distinct shapes (Fig. S4A†). An analysis of the ratio of normalized fluorescence intensities at 495 nm and 560 nm revealed a gradual shift towards the blue until 96 h (ratio = 2.2 ± 0.20), followed by a shift back towards the red (ratio = 1.5 ± 0.74) (Fig. S4B†). These results suggest the pTP-TFE can bind to distinct oligomers of tau that are generated along the aggregation pathway of the protein. No emission spectrum shifts were observed in experiments with pFTAA (Fig. S5B†).
The binding affinity for pTP-TFE towards aggregates collected from the aggregation assay (not containing compounds) at 96 h and 240 h were determined by treatment with different concentrations of probe (Fig. 2E and F). pTP-TFE displayed exceptional affinity for small aggregates (96 h) with a Kd of 38 nM and, encouragingly, showed no detectable affinity towards the 240 h tau fibril sample (Kd < 10 μM). These results indicate good selectivity towards soluble aggregated species.
Binding to fractionated aggregates of Aβ and tau proteins
It has been well documented that protein samples collected at different time points of an aggregation assay contain heterogeneous mixtures of different aggregated species, from small soluble forms to fibrils. Therefore, we recognized the need to purify these samples to obtain homogenous protein aggregate fractions of tau in order to confirm the binding profile discussed above. To achieve this, we employed a sucrose gradient ultracentrifugation technique by means of which protein aggregates of different sizes can be separated into homogenous fractions22 and then used these samples for affinity measurement (Fig. 3).
Fig. 3. (A) Tau (0N4R) protein were collected in 100 μL aliquots every 24 hours from the aggregation reaction and then stored at −80 °C after flash freezing. After 240 hours of incubation 1 mL of tau aggregates was collected and loaded on the top of the ultracentrifuge. (B–G) TEM images of tau aggregates structures at fractions from 50% to 5% sucrose gradient after 4 hours of ultracentrifugation (white bar = 500 nm).
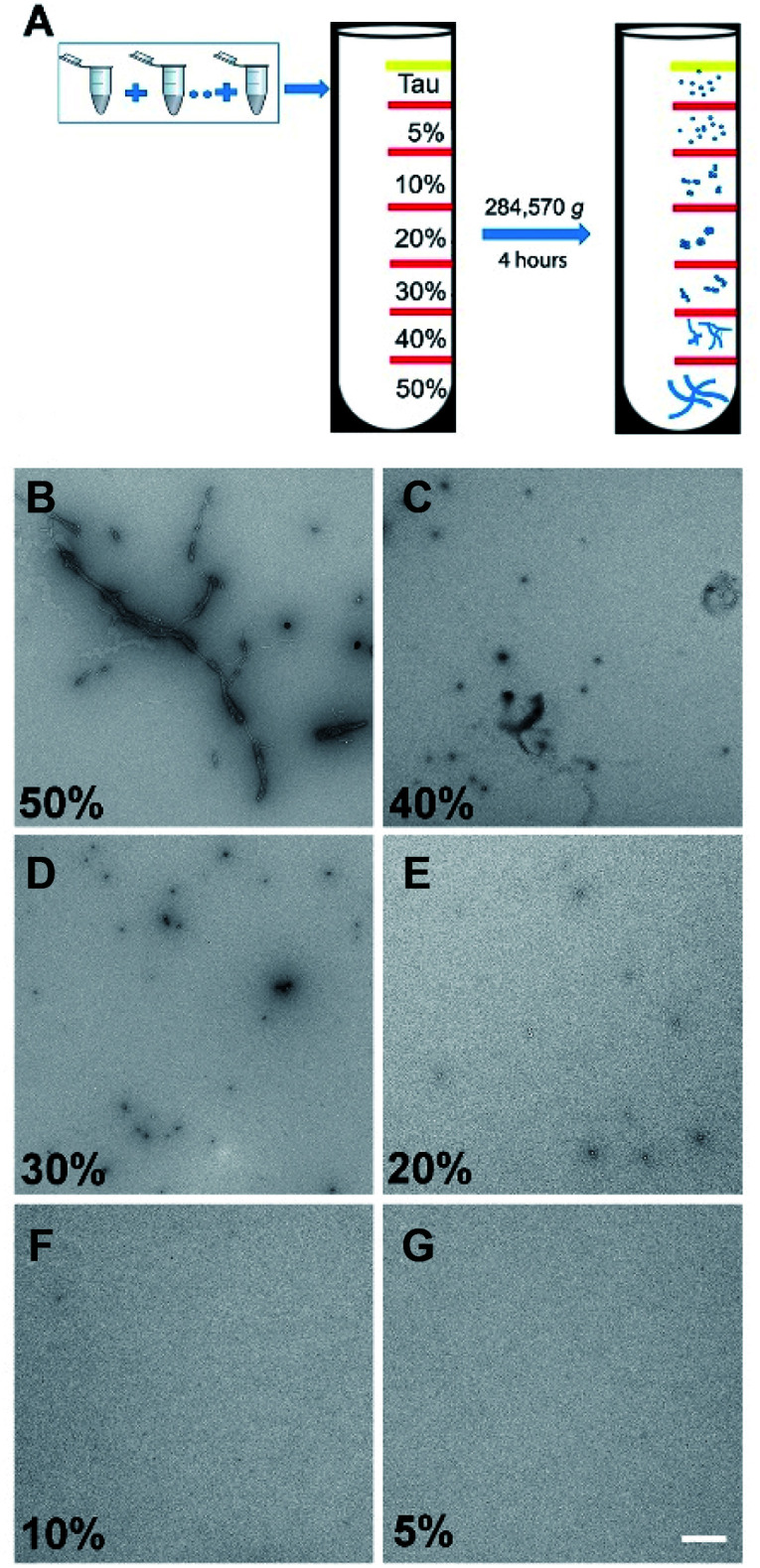
The binding affinities of pTP-TFE and pFTAA with 10% to 50% sucrose gradient tau aggregates were then tested by a fluorescent binding assay (Table 1). The data obtained confirms results from the previous experiments (Fig. S7†) indicating pTP-TFE binds most strongly to the 20% sucrose gradient fraction (Kd = 66 nM), which consist of soluble aggregated tau as confirmed by TEM (Fig. 3). Furthermore, selectivity of pTP-TFE for small aggregates over fibrils was confirmed by a difference in Kd for 40% and 50% sucrose gradient tau aggregates of approximately one order of magnitude. Comparisons of pTP-TFE to tau binding affinity data with that to Aβ aggregates at 200 min (Table 1) show a 50-fold selectivity in favour of tau (Kd(Aβ40) = 3.3 μM vs. Kd(tau) = 66 nM). This indicates that pTP-TFE is also selective for soluble tau aggregates over soluble Aβ aggregates. pFTAA, by comparison, displays highest affinity to tau aggregates in the 50% sucrose gradient fraction (Kd = 0.20 μM) and Aβ fibril aggregates (24 h; Kd = 0.65 μM; Table 1). These results support our hypothesis that the removal of negatively charged functional groups imparts higher selectivity for early aggregates of tau over mature fibrils. Encouragingly, these results also show that the use of TFE bioisostere endows the compound with selectivity for soluble aggregates of tau over soluble aggregates of Aβ.
Binding affinity of pTP-TFE and pFTAA with 10% to 50% tau fraction (n = 3–6) and Aβ40 aggregates at 90 min, 200 min and 24 hours (n = 3) after ultracentrifugation, Kd values were fitted using GraphPad Prism5.
| Aggregates | pTP-TFE, Kd (μM) | pTP-TFE, binding ratioa | pFTAA, Kd (μM) | pFTAA, binding ratiob |
|---|---|---|---|---|
| 10% Sucrose tau | 0.96 ± 0.20 | 14 | 29.57 ± 2.10 | 148 |
| 20% Sucrose tau | 0.07 ± 0.03 | 1 | 2.75 ± 0.57 | 14 |
| 30% Sucrose tau | 3.04 ± 0.71 | 49 | 5.27 ± 1.25 | 26 |
| 40% Sucrose tau | 0.59 ± 0.20 | 9 | 1.29 ± 0.20 | 6 |
| 50% Sucrose tau | 1.08 ± 0.17 | 15 | 0.20 ± 0.01 | 1 |
| 90 Minute, Aβ | 7.58 ± 2.47 | 108 | No affinity | N/A |
| 200 Minute, Aβ | 3.27 ± 0.64 | 47 | 0.93 ± 0.30 | 5 |
| 24 Hour, Aβ | 3.79 ± 0.34 | 54 | 0.65 ± 0.19 | 3 |
Binding ratio is based on comparison of Kd values with the 20% sucrose tau Kd.
Binding ratio is based on comparison of Kd values with the 50% sucrose tau Kd.
Cellular uptake of pTP-TFE into primary human fetal neurons
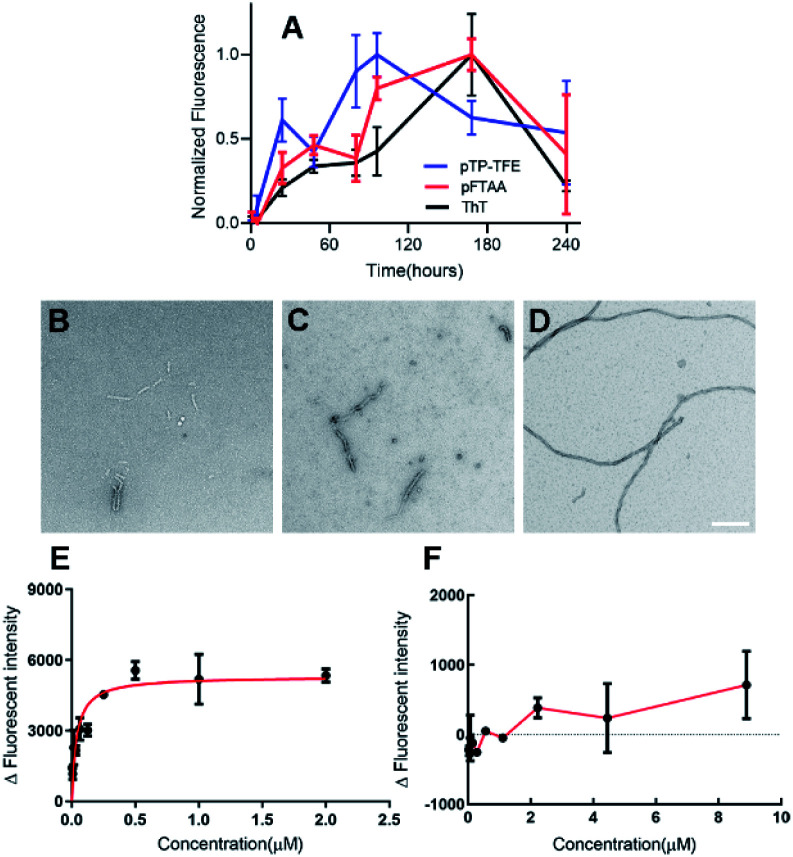
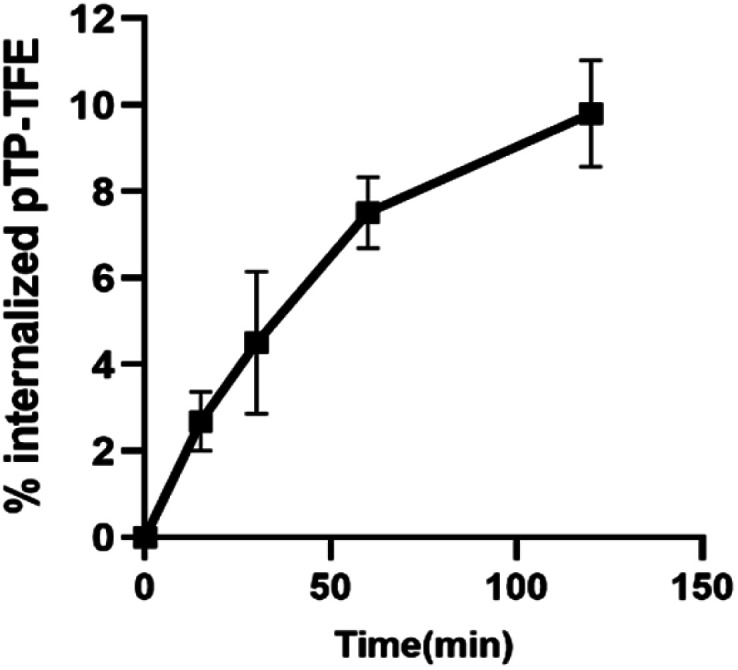
To determine whether pTP-TFE can penetrate live intact cell membranes we investigated the cellular uptake of pTP-TFE in live primary human fetal neurons. This was performed by adding pTP-TFE to a final concentration of 4 μM in a culture medium, and incubating for 15 min, 30 min, 60 min and 120 min. pTP-TFE uptake in neurons at each time point was determined by acetonitrile extraction of PBS-washed cultures, and quantified using an LC-MS/MS assay23 (Fig. S9†). This showed 5% of the total applied pTP-TFE in the neuronal cells within 30 min and 10% in 2 hours (Fig. 4 and Table S1†), which indicates the compound can cross the cell membrane with rapid uptake. The quantity and kinetics of this uptake are comparable to other compounds, including established PET probes in oncology24 and agents delivered into neuronal cells.25
Fig. 4. Kinetics of pTP-TFE uptake in primary human fetal neurons quantified by measuring the concentration of pTP-TFE in live neurons using LC-MS/MS; data presented as % of total administered amount. Y values calculated from Table S1† (n = 3).
Ex vivo imaging of early tau aggregates in AD and PSP human brain slices
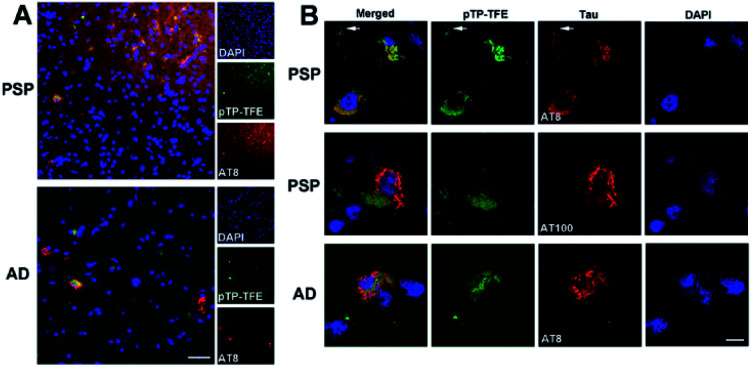
Finally, we investigated whether the selectivity demonstrated in in vitro assays could be replicated in ex vivo human brain tissue. To this end we evaluated the binding of pTP-TFE to tau pathology in human brain tissue of progressive supranuclear palsy (PSP), a pure tauopathy, and Alzheimer's disease, which contains both Aβ and tau pathology. Low-magnification epifluorescence microscopy showed that pTP-TFE signals can be found colocalizing with hyperphosphorylated, AT8-positive tau pathology in both PSP and AD brains (Fig. 5A). Closer inspection using confocal microscopy revealed that pTP-TFE signals were predominantly observed within the cell soma, co-localizing with AT8-positive tau pathology, although the presence of pTP-TFE signals could also be found in the axonal/dendritic compartments (Fig. 5B, arrows). Conversely, co-localization with tau fibril antibody AT100 is less well established; AT100-positive immunoreactivity has been shown to selectively label filamentous tau pathology,26 this substantiates our in vitro binding data and suggests that pTP-TFE demonstrates preferential reactivity to early stage soluble tau aggregates.
Fig. 5. pTP-TFE staining with human PSP (n = 5) and AD brain slides (n = 4). (A) Epifluorescence images of pTP-TFE in AD and PSP human brain slides with scale bar = 50 μm. (B) Confocal images of pTP-TFE in AD and PSP with scale bar = 10 μm. The arrows indicate an axonal/dendritic compartment.
Conclusions
We have designed and synthesized a novel fluorescent probe, pTP-TFE for which binding and selectivity profiles towards aggregated tau and Aβ proteins were assessed. This included use of a sucrose gradient ultracentrifugation method for more accurate quantification of binding affinities to specific size tau aggregates. Our results show pTP-TFE to be selective for soluble tau aggregates, with a high affinity of Kd = 66 nM, and ten-fold selectivity over mature fibrils. Furthermore, we found that pTP-TFE is tau selective over Aβ, the other major misfolded protein aggregate of neurodegenerative disorders. To the best of our knowledge, pTP-TFE is the first fluorescent molecule to have this form of selectivity for soluble tau aggregates over tau fibrils. In addition, we established that pTP-TFE could penetrate intact live cell membranes rapidly, while its selectivity towards early forms of aggregated tau protein was also supported by studies on human brain tissue containing tau and Aβ pathology.
The high affinity and selectivity for early soluble aggregates of tau and its cell permeability make pTP-TFE a best in class molecular tool for the study of tauopathy development and progression as well as a proof of concept chemotype for the development of small molecule therapeutics, with enhanced affinity and selectivity. We aim to utilize the potential displayed by pTP-TFEin vitro and ex vivo through in vivo optical, fluorine-19 MRI/MRS and fluorine-18 PET imaging modalities in the future.
Conflicts of interest
There are no conflicts to declare.
Supplementary Material
Acknowledgments
The authors thank Dr Gabriele Schierle and Dr Na Yu (Chemical Engineering Department, University of Cambridge), Dr Karin Muller (Cambridge Advanced Imaging Center) for technical support, Prof James Rowe (Department of Clinical Neurosciences, University of Cambridge), Dr David Williamson and Cambridge Brain Bank for the post-mortem brain samples, EPSRC Mass Spectrometry Service (University of Swansea). Cambridge Brain Bank is supported by the NIHR Cambridge Biomedical Research Centre. This study was funded by the National Institute for Health Research (NIHR) Cambridge Biomedical Research Centre: Dementia and Neurodegeneration theme, Medical Research Council grant (MR/K02308X/1), the Engineering and Physical Sciences Research Council (ST, EP/P008224/1), Royal Society (DK), and Amgen Foundation Scholarship (ER). WLK and BV are supported by the Medical Research Council (MR/S005528/1). GT was supported by the Hungarian Brain Research Program (2017-1.2.1-NKP-2017-00002).
Electronic supplementary information (ESI) available. See DOI: 10.1039/c9sc05620c
Notes and references
- Chiti F. Dobson C. M. Annu. Rev. Biochem. 2006;7:333. doi: 10.1146/annurev.biochem.75.101304.123901. [DOI] [PubMed] [Google Scholar]
- Ballatore C. Lee V. M. Trojanowski J. Q. Nat. Rev. Neurosci. 2007;8:663. doi: 10.1038/nrn2194. [DOI] [PubMed] [Google Scholar]; Ittner L. M. Götz J. Nat. Rev. Neurosci. 2011;12:67. doi: 10.1038/nrn2194. [DOI] [PubMed] [Google Scholar]; Ghetti B. Oblak A. L. Boeve B. F. Johnson K. A. Dickerson B. C. Goedert M. Neuropathol. Appl. Neurobiol. 2015;41:24. doi: 10.1038/nrn2194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eanes E. D. Glenner G. G. J. Histochem. Cytochem. 1968;16:673. doi: 10.1177/16.11.673. [DOI] [PubMed] [Google Scholar]; Fitzpatrick A. W. Falcon B. He S. Murzin A. G. Murshudov G. Garringer H. J. Crowther R. A. Ghetti B. Goedert M. Scheres S. H. Nature. 2017;547:185. doi: 10.1177/16.11.673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones M. R. Mathieu E. Dyrager C. Faissner S. Vaillancourt Z. Korshavn K. J. Lim M. H. Ramamoorthy A. Yong V. W. Tsutsui S. Stys P. K. Chem. Sci. 2017;8:5636. doi: 10.1039/C7SC01269A. [DOI] [PMC free article] [PubMed] [Google Scholar]; Li Y. Xu D. Sun A. Ho S. L. Poon C. Y. Chan H. N. Ng O. T. Yung K. K. Yan H. Li H. W. Wong M. S. Chem. Sci. 2017;8:8279. doi: 10.1039/C7SC01269A. [DOI] [PMC free article] [PubMed] [Google Scholar]; Nilsson K. P. FEBS Lett. 2009;583:2593. doi: 10.1039/C7SC01269A. [DOI] [PubMed] [Google Scholar]
- Liu Y. Yang Y. Sun M. Cui M. Fu Y. Lin Y. Li Z. Nie L. Chem. Sci. 2017;8:2710. doi: 10.1039/C6SC04798J. [DOI] [PMC free article] [PubMed] [Google Scholar]; Nordberg A. Rinne J. O. Kadir A. Långström B. Nat. Rev. Neurol. 2010;6:78. doi: 10.1039/C6SC04798J. [DOI] [PubMed] [Google Scholar]
- Eisenberg D. Jucker M. Cell. 2012;148:1188. doi: 10.1016/j.cell.2012.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]; Guo J. L. Covell D. J. Daniels J. P. Iba M. Stieber A. Zhang B. Riddle D. M. Kwong L. K. Xu Y. Trojanowski J. Q. Lee V. M. Cell. 2013;154:103. doi: 10.1016/j.cell.2012.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda S. Sahara N. Saito Y. Murayama S. Ikai A. Takashima A. Neurosci. Res. 2006;54:197. doi: 10.1016/j.neures.2005.11.009. [DOI] [PubMed] [Google Scholar]; Goedert M. Alzheimers Disease & Dementia. 2016;12:1040. doi: 10.1016/j.neures.2005.11.009. [DOI] [PubMed] [Google Scholar]
- Sehlin D. Fang X. T. Cato L. Antoni G. Lannfelt L. Syvänen S. Nat. Commun. 2016;7:10759. doi: 10.1038/ncomms10759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teoh C. L. Su D. Sahu S. Yun S. W. Drummond E. Prelli F. Lim S. Cho S. Ham S. Wisniewski T. Chang Y. T. J. Am. Chem. Soc. 2015;137:13503. doi: 10.1039/C6CC08826K. [DOI] [PMC free article] [PubMed] [Google Scholar]; Jameson L. P. Dzyuba S. V. Bioorg. Med. Chem. Lett. 2013;23:1732. doi: 10.1039/C6CC08826K. [DOI] [PMC free article] [PubMed] [Google Scholar]; Lim S. Haque M. M. Su D. Kim D. Lee J. S. Chang Y. T. Kim Y. K. Chem. Commun. 2017;53:1607. doi: 10.1039/C6CC08826K. [DOI] [PubMed] [Google Scholar]
- Lv G. Sun A. Wei P. Zhang N. Lan H. Yi T. Chem. Commun. 2016;52:8865. doi: 10.1039/C6CC02741E. [DOI] [PubMed] [Google Scholar]
- Klingstedt T. Shirani H. Åslund K. A. Cairns N. J. Sigurdson C. J. Goedert M. Nilsson K. P. Chem.–Eur. J. 2015;19:10179. doi: 10.1002/chem.201301463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villemagne V. L. Doré V. Burnham S. C. Masters C. L. Rowe C. C. Nat. Rev. Neurol. 2018;14:225. doi: 10.1038/nrneurol.2018.9. [DOI] [PubMed] [Google Scholar]; Goedert M. Yamaguchi Y. Mishra S. K. Higuchi M. Sahara N. Frontiers in Neurology. 2018;15:70. doi: 10.1038/nrneurol.2018.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verwilst P. Kim H. S. Kim S. Kang C. Kim J. S. Chem. Soc. Rev. 2018;47:2249. doi: 10.1039/C7CS00706J. [DOI] [PubMed] [Google Scholar]
- Åslund A. Sigurdson C. J. Klingstedt T. Grathwohl S. Bolmont T. Dickstein D. L. Glimsdal E. Prokop S. Lindgren M. Konradsson P. Holtzman D. M. ACS Chem. Biol. 2009;4:673. doi: 10.1021/cb900112v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klingstedt T. Nilsson K. P. Biochem. Soc. Trans. 2012;40:704. doi: 10.1042/BST20120009. [DOI] [PubMed] [Google Scholar]
- Klingstedt T. Shirani H. Mahler J. Wegenast-Braun B. M. Nyström S. Goedert M. Jucker M. Nilsson K. P. Chem.–Eur. J. 2015;21:9072. doi: 10.1002/chem.201500556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerson J. E. Kayed R. Frontiers in Neurology. 2013;17:93. doi: 10.3389/fneur.2013.00093. [DOI] [PMC free article] [PubMed] [Google Scholar]; León L. D. Karla I. García-Gutiérrez P. Serratos I. N. Palomera-Cárdenas M. Figueroa-Corona M. D. Campos-Peña V. Meraz-Ríos M. A. J. Alzheimer's Dis. 2013;35:319. doi: 10.3233/JAD-121765. [DOI] [PubMed] [Google Scholar]
- Lasagna-Reeves C. A. Castillo-Carranza D. L. Sengupta U. Guerrero-Munoz M. J. Kiritoshi T. Neugebauer V. Jackson G. R. Kayed R. Sci. Rep. 2012;2:700. doi: 10.1038/srep00700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballatore C. Huryn D. M. Smith A. B. ChemMedChem. 2013;8:385. doi: 10.1002/cmdc.201200585. [DOI] [PMC free article] [PubMed] [Google Scholar]; Ducharme Y. Blouin M. Carrière M. C. Chateauneuf A. Côté B. Denis D. Frenette R. Greig G. Kargman S. Lamontagne S. Martins E. Bioorg. Med. Chem. Lett. 2005;15:1155. doi: 10.1002/cmdc.201200585. [DOI] [PubMed] [Google Scholar]
- Nordeman P. Johansson L. B. Bäck M. Estrada S. Hall H. Sjölander D. Westermark G. T. Westermark P. Nilsson L. Hammarström P. Nilsson K. P. ACS Med. Chem. Lett. 2016;7:368. doi: 10.1021/acsmedchemlett.5b00309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamin G. Teplow D. B. J. Neurochem. 2017;140:210. doi: 10.1111/jnc.13887. [DOI] [PMC free article] [PubMed] [Google Scholar]; Jan A. Gokce O. Luthi-Carter R. Lashuel H. A. J. Biol. Chem. 2008;283:28176. doi: 10.1111/jnc.13887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda S. Sahara N. Saito Y. Murayama M. Yoshiike Y. Kim H. Miyasaka T. Murayama S. Ikai A. Takashima A. Biochemistry. 2007;46:3856. doi: 10.1021/bi061359o. [DOI] [PubMed] [Google Scholar]
- Bhat J. Narayan A. Venkatraman J. Chatterji M. J. Microbiol. Methods. 2013;94:152. doi: 10.1016/j.mimet.2013.05.010. [DOI] [PubMed] [Google Scholar]; Richards K. H. Schanze N. Monk R. Rijntjes E. Rathmann D. Köhrle J. A. PLoS One. 2017;12:1. doi: 10.1016/j.mimet.2013.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacAskill M. G. Tavares A. S. Wu J. Lucatelli C. Mountford J. C. Baker A. H. Newby D. E. Hadoke P. W. Sci. Rep. 2017;7:44233. doi: 10.1038/srep44233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasadsri L. Kreuter J. Hattori H. Iwasaki T. George J. M. J. Biol. Chem. 2009;284:6972. doi: 10.1074/jbc.M805956200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clavaguera F. Akatsu H. Fraser G. Crowther R. A. Frank S. Hench J. Probst A. Winkler D. T. Reichwald J. Staufenbiel M. Ghetti B. Goedert M. Tolnay M. Proc. Natl. Acad. Sci. U. S. A. 2013;110:9535. doi: 10.1073/pnas.1301175110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.