Abstract
Charcot-Marie-Tooth disease type 4J (CMT4J) is caused by recessive, loss-of-function mutations in FIG4, encoding a phosphoinositol(3,5)P2-phosphatase. CMT4J patients have both neuron loss and demyelination in the peripheral nervous system, with vacuolization indicative of endosome/lysosome trafficking defects. Although the disease is highly variable, the onset is often in childhood and FIG4 mutations can dramatically shorten life span. There is currently no treatment for CMT4J. Here, we present the results of preclinical studies testing a gene-therapy approach to restoring FIG4 expression. A mouse model of CMT4J, the Fig4–pale tremor (plt) allele, was dosed with a single-stranded adeno-associated virus serotype 9 (AAV9) to deliver a codon-optimized human FIG4 sequence. Untreated, Fig4plt/plt mice have a median survival of approximately 5 weeks. When treated with the AAV9-FIG4 vector at P1 or P4, mice survived at least 1 year, with largely normal gross motor performance and little sign of neuropathy by neurophysiological or histopathological evaluation. When mice were treated at P7 or P11, life span was still significantly prolonged and peripheral nerve function was improved, but rescue was less complete. No unanticipated adverse effects were observed. Therefore, AAV9-mediated delivery of FIG4 is a well-tolerated and efficacious strategy in a mouse model of CMT4J.
Keywords: Genetics, Neuroscience
Keywords: Gene therapy, Neurological disorders
Introduction
Charcot-Marie-Tooth disease (CMT) is a collection of inherited motor and sensory neuropathies. Mutations in over 80 genes can lead to CMT, and it is highly heterogeneous (1). Loss of peripheral motor and sensory axon function through demyelination, axon degeneration, or a combination of these mechanisms is a disease hallmark (2).
Recessive mutations in FIG4 underlie a rare form of the disease, CMT type 4J (CMT4J) (3). CMT4J usually involves the recurring I41T allele of FIG4 in a compound heterozygous state with a truncating allele, such as a frame shift (3–5). CMT4J patients are highly variable in their presentation and severity, with cases ascertained from childhood through the sixth decade of life, though many present at the younger end of this spectrum. Nerve biopsy and neurophysiological examinations indicate axon loss, but also demyelination (3–5). Primary cells isolated from CMT4J patients show vacuoles that are LAMP2 positive, suggesting defects in endosome/lysosome trafficking (6). Mutations in FIG4 also lead to other neurological and developmental disorders, depending on the severity of the loss of function caused by the mutations. Inheritance of 2 null alleles of FIG4 leads to Yunis-Varon syndrome, which includes skeletal defects and severe neurological symptoms, with vacuolization and neuronal loss, typically resulting in death in infancy (7). Homozygosity for the D783V allele of FIG4 results in bilateral temporooccipital polymicrogyria, which includes seizures (8). Thus, recessive mutations in FIG4 lead to a variety of neurological conditions that are generally severe and currently untreatable. Variants in FIG4 have also been associated with adult-onset amyotrophic lateral sclerosis and primary lateral sclerosis, although the pathogenicity of these alleles has not been firmly established (9).
The FIG4 protein is an phosphoinositol(3,5)P2-phosphatase. Importantly, it is in a complex with the enzymes that generate PI(3,5)P2, which are PIKFYVE and VAC14 (10, 11). PI(3,5)P2 levels are very dynamic and important for multivesicular body formation and endosome/lysosome trafficking. Loss of FIG4 destabilizes this complex, resulting in the mislocalization of all 3 proteins and a reduction of PI(3,5)P2. Mutations in VAC14 also cause neurological disease in both mice and humans (12, 13). Therefore, some of the FIG4 phenotype may be due to other components of this complex, such as VAC14. Consistent with this, Fig4 mutations in mice can be partially rescued with a catalytically inactive Fig4 transgene (C486S) that may serve to restore protein interactions with VAC14 and PIKFYVE (12). However, the phenotype is more effectively rescued by a WT Fig4 gene (14). Importantly, neuronal expression of Fig4 also rescues myelination, suggesting a non–cell autonomous effect (15). No adverse effects from Fig4 transgenic overexpression were observed. Taken together, these results indicate CMT4J-FIG4 mutations are excellent candidates for gene therapy to restore FIG4 expression in peripheral neurons.
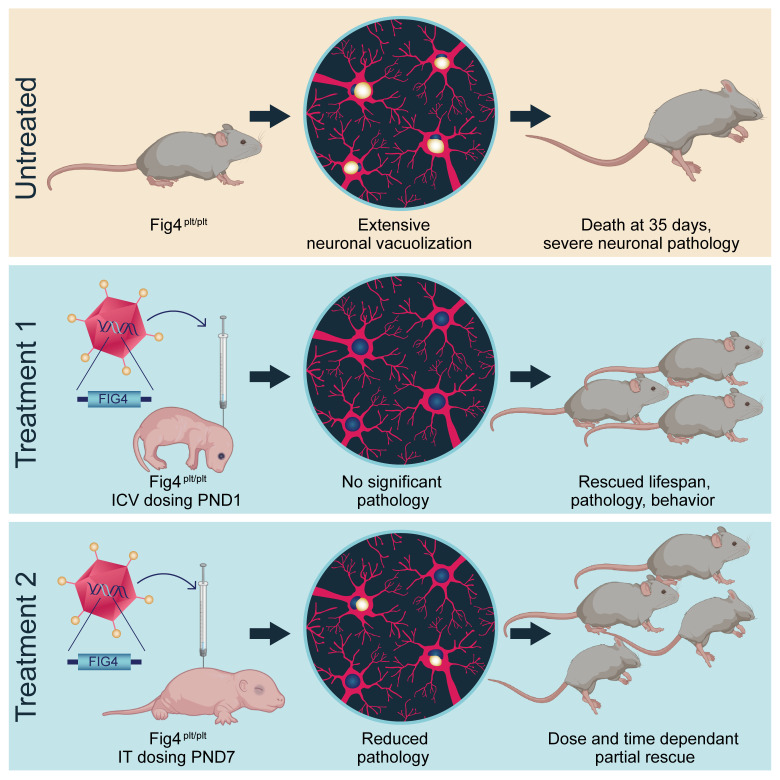
In the present study, we have performed preclinical testing of an adeno-associated virus serotype 9 (AAV9) vector to deliver a codon-optimized WT human FIG4 gene to neuronal and nonneuronal cells in a mouse model of CMT4J. The pale tremor (plt) mouse carries a transposon insertion in intron 18 of the mouse Fig4 gene, resulting in a loss-of-function allele. The cloning of this mouse mutation led to the identification of FIG4 as a human disease gene underlying CMT4J (3). The mice have a very similar phenotype, with demyelination, loss of motor neurons, and vacuolization in many regions of the nervous system, leading to a median life span of approximately 35 days when experiments are performed in a C57BL/6.C3H F1 genetic background (either inbred background alone survives only a few days). Restoration of FIG4 expression using the gene-therapy vector promoted survival to over 1 year when mice were treated at birth or at P4 and substantially prolonged survival. Gross motor performance, peripheral axon function, and histopathological phenotypes were all improved. Thus, AAV9-mediated delivery of FIG4 is efficacious in a mouse model of CMT4J.
Results
AAV9-FIG4 gene therapy improves survival of Fig4-deficient mice.
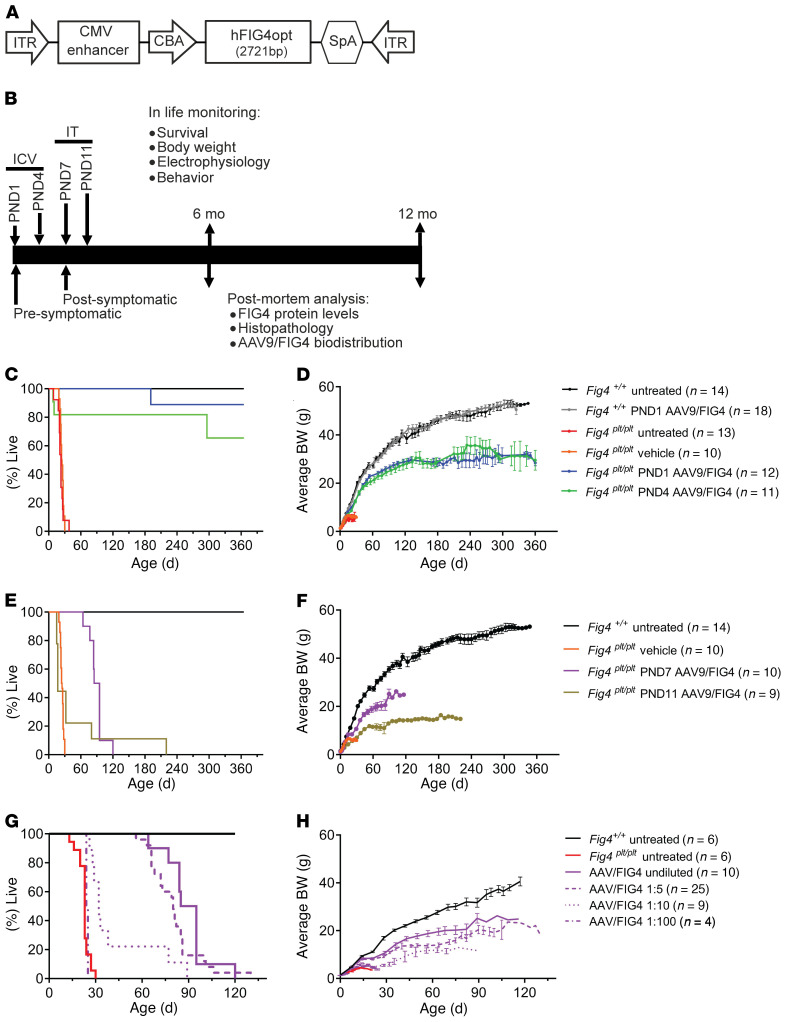
The Fig4-deficient mouse, or plt mouse, presents a severe phenotype characterized by reduced life span, small body size, tremors, peripheral neuropathy, loss of motor neurons, and hydrocephalus. The cloning of the spontaneous plt mutation in mice led to the identification of FIG4 as the gene causing CMT4J in humans (3). Therefore, we used the Fig4plt mice as a proof-of-concept preclinical model for a gene-replacement therapy to treat CMT4J using an AAV9-FIG4 vector (Figure 1A). The vector is a single-stranded AAV9 carrying a codon-optimized WT human FIG4 cDNA driven by a ubiquitously expressed chicken β-actin (CBA) promoter with a CMV enhancer. To first demonstrate efficacy of the virus and gene-replacement strategy, mice were treated at P1 and P4 with the highest feasible dose of AAV9-FIG4 (5.4 × 1011 vg) via i.c.v. injection (Figure 1B). All mice were genotyped at birth (P0), randomized among the different experimental groups, and injected blinded for genotype and treatment. All cohorts were monitored daily for survival and body weight. As was previously reported (3), Fig4plt/plt mice that were untreated or vehicle treated had a short life span, with a median survival age of 23 days (Figure 1C). After treatment with AAV9-FIG4, 90% of Fig4plt/plt mice injected at P1 survived more than 12 months (Figure 1C). Although Fig4plt/plt mice treated at P1 did gain weight and grow, they still showed a significantly lower body weight throughout their life spans when compared with Fig4+/+ littermates (Figure 1D). Both Fig4+/+ treated at P1 with AAV9-FIG4 and untreated WT mice had 100% survival after a year (Figure 1C), with no differences in growth (Figure 1D), suggesting the AAV9-FIG4 vector was well tolerated in control mice.
Figure 1. Neonatal delivery of AAV9-FIG4 vector improves survival of Fig4plt/plt mice.
(A) Representation of the AAV9-FIG4 vector, with a full-length, codon-optimized human FIG4 cDNA (hFIG4opt) under control of the CBA promoter, including a CMV-derived enhancer and a synthetic poly(A) tail (SpA). This expression cassette was flanked by an ITR sequence for packing into AAV9. (B) Outline of preclinical study design. Fig4plt/plt mice were treated by i.c.v. injection at P1 or P4 with AAV9-FIG4, representing the presymptomatic intervention and proof-of-concept group for efficacy testing. Fig4 WT littermates were dosed i.c.v. at P1, representing the safety group for AAV9-FIG4 toxicity evaluation. Both Fig4plt/plt and WT mice treated with vehicle or untreated were included as controls. For postsymptomatic treatments, Fig4plt/plt mice were treated at P7 or P11 via i.t. injection. A minimum of n = 5 mice per sex per genotype were enrolled in each group. All mice were monitored daily for body weight and survival. Electrophysiology and behavioral assessments were conducted at various time points after AAV9-FIG4 treatment. Some of the i.c.v. injected group were analyzed at 6 to 8 months after treatment and the rest at the 12- to 13-month end point. (C) Survival and (D) average body weights (BW) for mice injected with AAV9-FIG4 via i.c.v. (5.4 × 1011 vg) at P1 and P4. For postsymptomatic intervention, Fig4plt/plt mice were treated by i.t. injection (AAV9-FIG4, 1.35 × 1012 vg) at P7 and P11. (E) Survival and (F) average body weights are shown. Dose-response analysis was conducted by injecting Fig4plt/plt mice i.t. with AAV9-FIG4 diluted 1:5 (2.7 × 1011 vg), 1:10 (1.35 × 1011 vg), and 1:100 (1.35 × 1010 vg). (G) Survival and (H) average body weights are shown. The log-rank (Mantel-Cox) test was applied for survival curve comparisons. Growth curves show average body weight ± SEM.
We tested treatment efficacy by injecting at various later time points to approximate postonset treatments that may be necessary clinically: at P4 via i.c.v. injection and at P7 and P11 via lumbar intrathecal (i.t.) injections. As indicated in Figure 1C, 65% of the mice injected at P4 survived more than 1 year, and half of those that did not survive died on the same time course as untreated mice, possibly indicating failed injections. P4 Fig4plt/plt mice treated with AAV9-FIG4 followed a trajectory of growth and weight gain similar to that of P1-treated Fig4plt/plt mice (Figure 1D).
To test the efficacy of AAV9-FIG4 at later ages with a more advanced disease, we treated Fig4plt/plt mice at P7 and P11. Delivery i.t. of 1.35 × 1012 vg at P7 resulted in an increased life span with a median survival age of 90 days (Figure 1E), whereas mice injected i.t. with the same dose at P11 had a lower benefit with a median survival age of 17 days, not significantly different from that of vehicle-treated Fig4plt/plt mice (Figure 1E). All mice injected i.t. at P7 or P11 did grow and gain weight, but this varied with injection age, with P7-treated mice showing larger effects than the P11-treated cohort (Figure 1F).
These results indicate that treatment with AAV9-FIG4 is able to improve survival and growth of Fig4plt/plt mice. The success of the treatment was dependent on the age of injection, with greater benefit at early ages (P1 and P4), although i.t. treatment at P7 also significantly improved survival (P7-treated vs. untreated Fig4plt/plt mice, P < 0.0001) and growth.
As these experiments used the maximum feasible dose and produced a benefit in Fig4plt/plt mice both by i.c.v. injection and i.t. delivery, we next performed a dose-response experiment to determine the minimum dose able to produce a significant benefit with respect to body weight and survival. Since postonset treatment and i.t. delivery are likely to reflect translational applications in clinical trials, we performed the dose-response experiment in P7 animals treated via i.t. delivery. Both 1:5 (2.7 × 1011 vg, treated vs. untreated; P < 0.0001) and 1:10 dilutions (1.35 × 1011 vg, treated vs. untreated; P < 0.0001) still significantly extended life span in Fig4plt/plt animals, while a 1:100 dilution (1.35 × 1010 vg, treated vs. untreated; P = 0.3842) was not effective (Figure 1G). Similar results were observed in body weight, with Fig4plt/plt mice injected at 1:10 and 1:100 dilution gaining less weight compared with 1:5 and the maximum dose of AAV9-FIG4 (Figure 1H).
FIG4 levels are restored in CNS after treatment with AAV9-FIG4.
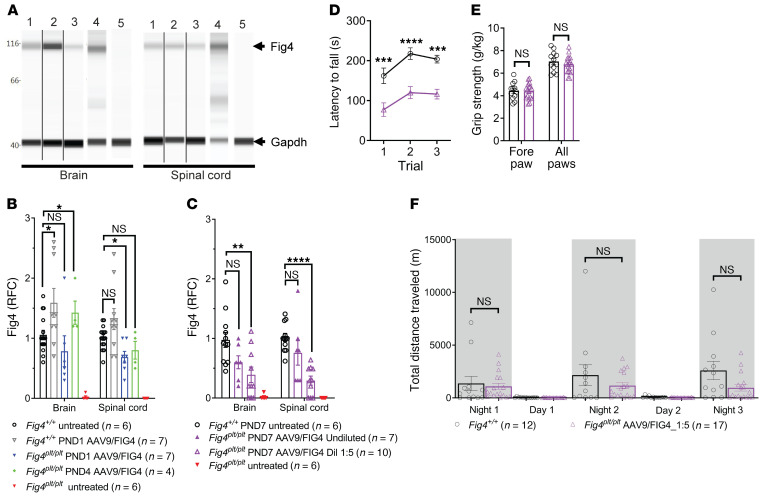
Although a truncated Fig4 mRNA has been reported in Fig4plt/plt mice, there was a complete absence of FIG4 protein by Western blot (3). We assessed the effectiveness of AAV9-FIG4 to induce human FIG4 expression and improve FIG4 protein levels in CNS tissues of treated mice. The Western blot analysis of whole brain or spinal cord showed absence of FIG4 in untreated Fig4plt/plt mice (Figure 2A, lane 5), whereas in mice injected at P1 or P4, FIG4 was detected at the same or slightly higher levels than in untreated Fig4+/+ mice (Figure 2, A and B). In mice treated at P7 with undiluted AAV9-FIG4, FIG4 protein was restored to slightly lower levels than in untreated Fig4+/+ mice in both brain and spinal cord (Figure 2C). As anticipated, FIG4 protein levels were dose dependent, with lower levels in brain and spinal cord when mice were injected with a 1:5 dilution of the maximum dose (Figure 2C); however, the low overall FIG4 levels still produced a significant improvement in survival of Fig4plt/plt mice (Figure 1G).
Figure 2. AAV9-FIG4 therapy restores FIG4 protein levels and behavioral outcomes of Fig4plt/plt mice.
FIG4 protein levels were assessed in whole-protein lysates from brain and spinal cord by Western blot. Mice treated at P1 or P4 by i.c.v. injection of the maximum AAV9-FIG4 dose were analyzed at 6 months of age. (A) Representative Western blot showing detection of FIG4 as a single band at 112 kDa, present in Fig4+/+ untreated mice (lane 1), Fig4+/+ treated at P1 (lane 2), Fig4plt/plt treated at P1 (lane 3), Fig4plt/plt treated at P4 (lane 4), and not present in Fig4plt/plt untreated mice (lane 5). (B) FIG4 levels were normalized to GAPDH as a loading control, and RFC calculated using Fig4+/+ untreated mice as reference. (C) Average RFC at 100–120 days of age for i.t. treated mice at P7 with maximum or 1:5 diluted AAV9-FIG4. (D–F) Fig4plt/plt mice treated by i.t. delivery at P7 with AAV9-FIG4 (1:5 dilution, 2.7 × 1011 vg) were assessed 35 days after treatment for motor coordination performance, grip strength, and general activity. (D) Rotarod analysis is reported as the latency to fall during 3 trials. (E) Average grip strength normalized to body weight in the forepaws and all paws is shown. (F) General activity was assessed by monitoring voluntary wheel running; the average total distance traveled at each day time cycle is reported. Data are represented as average ± SEM of combined males and females, since no significant differences were observed by sex. Groups were compared by 2-way ANOVA with Dunnett’s correction for multiple comparisons (B and C) and Mann-Whitney U test (D–F). *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
AAV9-FIG4 gene therapy rescues muscle strength, motor coordination, and overall activity in Fig4plt/plt mice.
An important clinical feature of CMT4J patients is the gradual loss of motor and neuromuscular function. To assess the benefits of the treatment on muscle strength, AAV9-FIG4–treated Fig4plt/plt mice were tested for grip strength and with rotarod and wheel running and compared with untreated Fig4 WT littermate mice to determine what fraction of strength could be recovered with treatment. Since untreated Fig4plt/plt mice do not survive long enough to perform these tests, they were not included. Mice treated with AAV9-FIG4 at P1 or P4 by i.c.v. delivery performed indistinguishably from WT mice at 6 months of age, indicating full rescue of rotarod, grip-strength, and wheel-running performance (Supplemental Figure 1, A–F; supplemental material available online with this article; https://doi.org/10.1172/JCI137159DS1). Importantly, there were also no behavioral changes in treated Fig4+/+ mice, supporting the safety of AAV9-FIG4. Dose-response experiments showed a good rescue of Fig4plt/plt mice treated i.t. with 1:5 diluted AAV9-FIG4 (2.7 × 1011 vg). Thus, we also assessed behavioral outcomes in Fig4plt/plt mice treated by i.t. injection at P7 with a lower dose of AAV9-FIG4 to test the capacity of the treatment to provide benefit in a postsymptomatic disease setting beyond the improvements in survival and growth already described (Figure 1, E and F). Rotarod analysis at 35 days of age revealed that the treated Fig4plt/plt mice had a shorter latency to fall when compared with Fig4 WT mice (Figure 2D), indicating the presence of some motor coordination deficits. However, in assessment of muscle strength, Fig4plt/plt treated mice showed similar grip strength when compared with WT mice (Figure 2E). Overall activity assessed by spontaneous wheel running showed no differences between treated Fig4plt/plt and WT controls, although a trend toward lower activity in Fig4plt/plt treated mice can be observed (Figure 2F).
Nerve conduction velocity is improved by AAV9-FIG4.
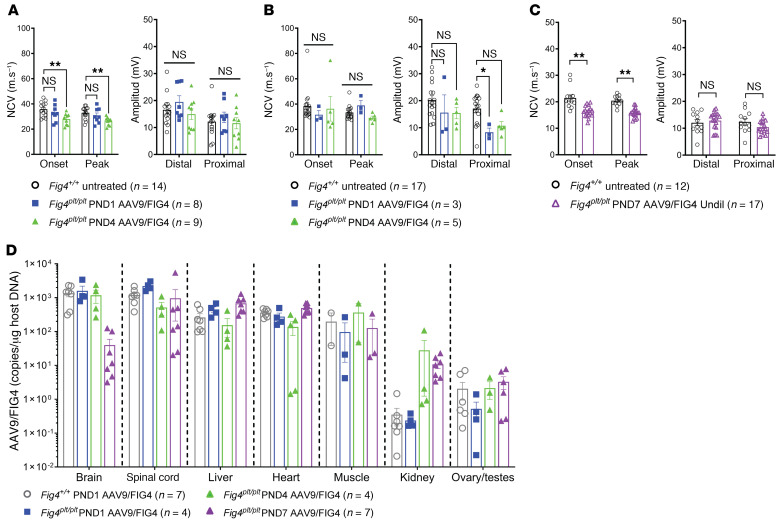
Fig4plt/plt mice develop hypomyelination features and reduced nerve conduction velocity (NCV) and compound muscle action potential (CMAP) amplitudes (3). When Fig4plt/plt mice were treated by i.c.v. injection with AAV9-FIG4 at P1, NCV was indistinguishable when compared with that of untreated Fig4+/+ mice at 6 months of age, while P4-injected mice showed a significant decrease in NCV (Figure 3A). However, when the remainder of the i.c.v. injection–treated cohort was assessed at 1 year after treatment, the AAV9-FIG4–treated mice showed NCV values indistinguishable from those of WT controls (Figure 3B). When AAV9-FIG4 was delivered by i.t. injection at P7, NCVs at 35 days of age were reduced in treated Fig4plt/plt mice compared with WT untreated controls (Figure 3C). Despite these differences in NCV outcomes, the CMAP amplitude in all treated Fig4plt/plt mice was similar to that of WT controls (Figure 3, A–C). Thus, the degree of rescue of nerve function depended on the age and route of dosing, with better outcomes observed in the mice treated at P1 (Figure 3, A and B).
Figure 3. AAV9-FIG4 treatment provides a wide biodistribution and improves NCV outcomes in an age of injection– and dose-dependent manner.
Sciatic NCV of motor axons and CMAP was assessed at 6 months (A) and 12 months (B) after i.c.v. treatment of Fig4plt/plt mice and WT controls. Fig4plt/plt mice i.t. injected with AAV9-FIG4 1:5 dilution (2.7 × 1011 vg) were assessed at 35 days after treatment (C). Tissues were collected from mice treated with AAV9-FIG4 at 6 months (i.c.v. treatment) and 4 months (i.t. treatment) after treatment. The vector copy number was assessed by qPCR as copies/μg of genomic DNA. (D). Data are represented as the average ± SEM of combined males and females, since no significant differences were observed by sex. Groups were compared by 2-way ANOVA with correction for multiple comparisons using Dunnett’s test (A and B) or Bonferroni’s test (C). *P < 0.05; **P < 0.01.
Wide biodistribution upon i.t. delivery of AAV9-FIG4.
We assessed the viral copy number of AAV9-FIG4 in mice that were i.c.v. injected at P1–P4 (assessed 6 months after injection) and in mice that were i.t. injected at P7 (assessed approximately 100–120 days after injection). The AAV9-FIG4 vector was detected at high levels across multiple tissues (Figure 3D). Copy number was decreased in the brain of mice treated by i.t. injection compared with i.c.v. treated mice, which is likely a consequence of the different route of injection.
CNS and PNS pathology is improved in Fig4plt/plt AAV9-FIG4–treated mice.
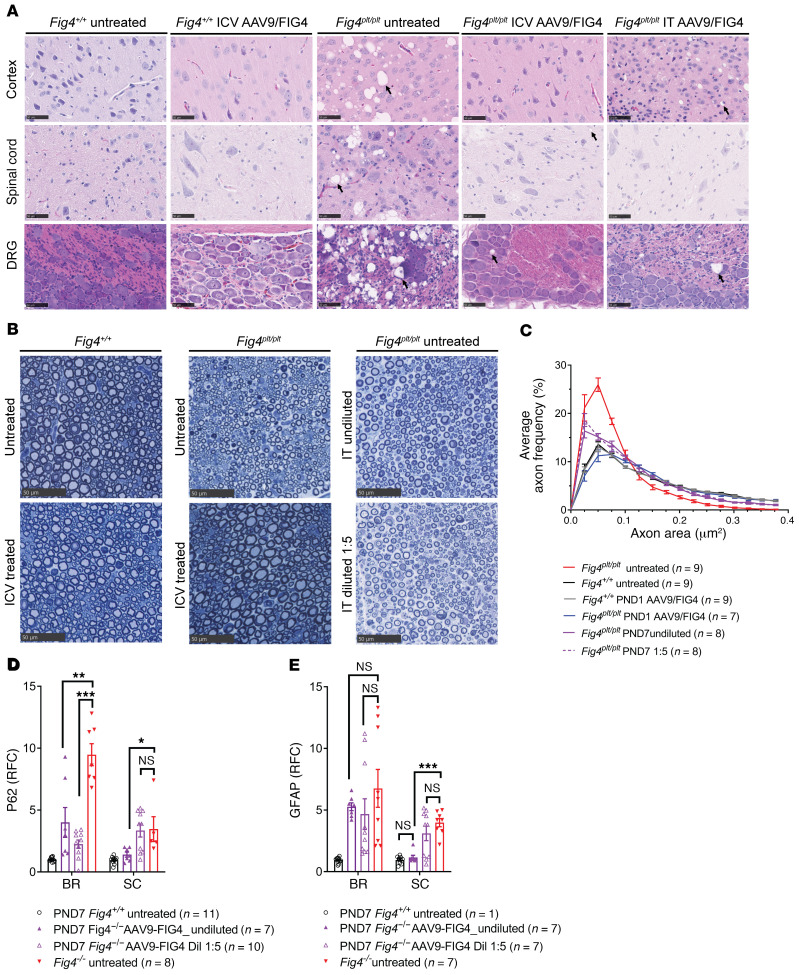
The Fig4plt/plt mice had an accumulation of autophagosomes in multiple areas of the brain (cortex, hippocampus, cerebellum and brain stem), spinal cord, and dorsal root ganglion (DRG), producing vacuolization in these tissues (3). We performed a histopathological analysis in Fig4plt/plt and Fig4+/+ treated mice with and without AAV9-FIG4 treatment. Untreated Fig4plt/plt mice showed pronounced vacuolization in DRG, spinal cord, and cerebral cortex at 23 days of age (Figure 4A). In contrast, 6 months after i.c.v. delivery of AAV9-FIG4 at P1, the degree of vacuole accumulation in Fig4plt/plt mice was almost completely rescued in DRG, spinal cord, and brain (Figure 4A). While the later delivery of AAV9-FIG4 by i.t. injection was generally less effective than the i.c.v. delivery at P1 or P4, Fig4plt/plt mice treated by i.t. delivery at P7 showed a similar reduction of vacuole accumulation in spinal cord and DRG, although the brain was less well rescued. No vacuoles were detected in Fig4+/+ mice when untreated or treated by i.c.v. injection with AAV9-FIG4, supporting the safety of the treatment (Figure 4A).
Figure 4. CNS pathology is reduced in AAV9-FIG4–treated Fig4plt/plt mice.
Histopathology changes were assessed in i.c.v. and i.t. AAV9-FIG4–treated and untreated Fig4plt/plt mice and WT controls. (A) Representative H&E images of motor cortex, thoracic spinal cord (ventral horn), and DRGs dissected from the indicated groups. Scale bars: 50 μm. Black arrows indicate cytoplasmic vacuoles. (B) Representative semithin sections of toluidine blue–stained sciatic nerve were analyzed for axon size distribution. Cross sections of sciatic nerve were scanned at ×40, and axon caliber for each axon in the section was determined using ImageJ software. (C) Distribution of the average axon frequency by size (axon area, 50 μm). (D and E) The autophagosome marker SQSTM1 (p62) and the astrocyte marker GFAP were analyzed by quantitative Western blot in protein lysates of brain (BR) and spinal cord (SC). p62 and GFAP signal were normalized to GAPDH loading control. RFC was calculated using Fig4+/+ untreated tissue as reference. Fig4plt/plt–derived protein extracts were prepared from 23-day-old untreated mice. Bars represent the average ± SEM. Statistical significance was calculated by 1-way ANOVA with Dunnett’s test correction for multiple comparisons. *P < 0.05; **P < 0.01; ***P < 0.001.
The histopathological analysis of sciatic nerves indicated that untreated Fig4plt/plt have an increased frequency of small caliber axons. The treatment with AAV9-FIG4 in Fig4plt/plt was able to produce an increase in the frequency of large caliber axons (Figure 4, B and C), which can be associated with the improvements in NCV reported by electrophysiology analysis (Figure 3). The mice injected i.t. at P7 with AAV9-FIG4 still presented a significantly elevated frequency of small caliber axons (Figure 4B).
AAV9-FIG4 gene therapy improves autophagosome pathology in CNS of Fig4plt/plt mice.
Fig4plt/plt mice show an accumulation of autophagosomes with increased levels of the autophagosome marker SQSTM1 (p62), usually followed by increased numbers of reactive astrocytes (11). Western blot analysis in brain and spinal cord of Fig4plt/plt mice treated at P7 by i.t. delivery revealed a reduction in the abundance of p62 in both brain and spinal cord tissue when AAV9-FIG4 was injected at the maximum dose, but a significant p62 reduction was observed only in brain in mice treated with diluted virus (Figure 4D). The gliosis marker GFAP was significantly decreased in spinal cord of Fig4plt/plt mice treated at P7 with the maximum dose of AAV9-FIG4; however, the diluted virus did not produce a change (Figure 4E). Thus, the improvement in these cellular markers was dose dependent, and Fig4plt/plt mice treated by i.t. injection with a lower dose showed less improvement. These findings demonstrate that AAV9-FIG4 directly corrects histopathological features in relevant neuronal cells types in the brain and spinal cord (Figure 4A).
Histopathology analysis and safety of AAV9-FIG4 treatment.
To evaluate the safety of AAV9-FIG4 gene-therapy treatment, we performed histopathological analysis of multiple tissues dissected at 6 or 12 months after treatment. The analysis at 6 months across multiple tissues in WT mice treated with AAV9-FIG4 at P1 by i.c.v. delivery and untreated mice showed no major pathology findings as a consequence of the treatment (Supplemental Table 1). Most notable findings in liver were micro- and macrovesicular lipidosis and glycogen accumulation. We hypothesize this may be a consequence of long-term exposure to a diet gel rich in carbohydrates (DietGel 76A). However, 12 months after treatment, hepatocellular carcinomas (HCCs) were found in Fig4+/+ mice in both untreated and i.c.v. AAV9-FIG4–treated groups, and also in some of the Fig4plt/plt i.c.v. treated mice. As indicated in Supplemental Table 2, we found an increased incidence of HCCs in i.c.v. treated Fig4+/+ mice (Fisher’s exact test; P = 0.0152). Liver tumor masses were also identified in Fig4plt/plt i.c.v. treated mice (P1 and P4); however, their incidence was not significantly increased compared with in untreated mice (Fisher’s exact test, P = 0.5962).
HCC has been previously reported in mice treated with AAV vectors using strong promoters. Much of the tumorigenesis is attributed to integration of the vectors at the Rian locus, a rodent-specific sequence (16). To assess the possible integration of AAV9-FIG4 in the mouse genome and whether the tumorigenic activity was related to insertion at the Rian locus as previously reported, 1 solid mass was dissected from each liver sample, and whole-genome sequencing was performed. The sequencing analysis detected unique integration events of the viral vector in multiple genomic regions (Supplemental Figure 2A), with the Rian locus having the highest number of insertions. In 80% of all HCCs analyzed, it was possible to identify integrations at the Rian locus (P = 0.02; Supplemental Figure 2B). An elevated number of unique insertion events was also mapped to 2 genes, Cps1 and the Atrn-UTR3 (Supplemental Figure 2A); however, these events were not associated with HCC samples (Supplemental Figure 2, C and D). All vector integrations identified at the Rian locus included the inverted terminal repeat (ITR) and promoter regions of AAV9-FIG4.
We also performed a clinical blood chemistry analysis in untreated Fig4plt/plt mice at 23 days old and AAV9-FIG4–treated mice at 2, 7, and 12 months of age (Supplemental Figure 3). No significant differences were observed between AAV9-FIG4–treated and untreated cohorts for electrolytes (Na+, K+, Cl–), glucose, creatinine, cholesterol, and triglycerides (results not shown). However, liver enzymes were elevated in plasma of untreated Fig4plt/plt mice (Supplemental Figure 3). A noticeable decrease in liver enzymes was observed in both i.c.v. and i.t. treated mice at 2 to 8 months. However, i.c.v. treated mice at the 12 to 13 month end point showed a significant increase in liver enzymes, particularly the treated Fig4+/+ mice, which is likely a consequence of the hepatic carcinomas detected in several of these mice (Supplemental Figure 3).
Discussion
Our studies demonstrate the efficacy and tolerability of AAV9-delivered FIG4 as a gene-therapy treatment for CMT4J. The plt mouse is a valid disease model for CMT4J, although perhaps more analogous to the more severe clinical designation of Yunis-Varon syndrome rather than the relatively more mild designation of CMT4J. Indeed, the identification of Fig4 as the mutated gene in plt mice led to the identification of FIG4 as the gene underlying CMT4J. Previous genetic studies in the plt mice established that the mutation is a recessive loss of function and that restoring Fig4 in the nervous system by transgenic expression was largely sufficient to rescue the phenotype (15). Our gene-therapy strategy used AAV9 delivery of a codon-optimized human FIG4 gene to accomplish much the same thing in a translationally relevant manner. Delivery of this vector at P1 or P4 extended the life span of the plt mice from a few weeks to at least a year. Those mice had nearly normal behavioral, neurophysiological, and histopathological phenotypes, though they were still smaller than WT mice. Delaying treatment to P7 or P11 still provided benefit, but the rescue was less complete than at the earlier time points. The improvement we observed in nerve conduction in peripheral motor axons and the reduced vacuolization in the ventral spinal cord and dorsal root ganglia indicate that the vector directly targets cell populations clinically relevant to CMT4J. The only adverse effect noted, the development of HCCs, is anticipated from the literature (16–19), and we confirmed that the incidence of these tumors can be explained by a peculiarity of the neonatal mouse genome. Thus, our results provide strong preclinical support for pursuing an AAV9-mediated gene-replacement strategy with FIG4 as a treatment for CMT4J.
In human genetic studies, CMT4J is consistent with straightforward, recessive loss-of-function alleles of FIG4. Typically, CMT4J patients are compound heterozygous for a presumed null allele (such as a frameshift or premature stop codon) and a nonsynonymous variant, most frequently the I41T substitution (5). The plt mouse harbors a null allele of Fig4 and thus likely represents a more severe genetic perturbation than is found in most CMT4J patients. Consistent with this, the mice have an aggressive, early onset phenotype and show vacuolization in not just lower motor neurons, but also in sensory neurons and in various regions of the brain. Delivery of the AAV9-FIG4 vector directly to the nervous system by i.c.v. was able to largely prevent these phenotypes, even rescuing the pathophysiology of neuronal populations in the brain that may not be directly affected in CMT4J patients. Conversely, CMT4J patients and the plt mice had signs of demyelinating neuropathy, and the Schwann cell–specific deletion of Fig4 in mice resulted in a CMT phenotype, though not as severe as the ubiquitous deletion, suggesting FIG4 expression is required in Schwann cells as well (5). However, in peripheral nerves of mice treated with AAV9-FIG4 at P1 or P7, the g ratio (axon diameter/total fiber diameter) was not different than in WT control mice, suggesting that any peripheral myelination defects were corrected by treatment (Supplemental Figure 4A). Furthermore, mice treated by i.c.v. delivery at P1 had normal nerve-conduction velocities, normal motor performance, and normal nerve histology. To assess whether these benefits were from transduction of neurons or Schwann cells, we used in situ hybridization to detect huFIG4 expression in combination with Schwann cell and motor neuron markers (Supplemental Figure 4B). In the sciatic nerve, 2% of all cells (DAPI nuclei) were transduced, and 7% of Pmp22-positive Schwann cells were also huFIG4 positive. In contrast, in spinal cord sections, approximately 80% of Chat-positive motor neurons in the ventral horn were also huFIG4 positive. Previous transgenic mouse studies indicate that neuronal expression of Fig4 was sufficient to rescue many aspects of the phenotype, including demyelination, suggesting a non–cell autonomous role for FIG4 (15). Our results using AAV9 are consistent with this interpretation, and the effects with this vector were profound when delivered at early postnatal ages. The low level of transduction in Schwann cells may account for some of the benefit observed, but presently, vectors and dosing regimens that efficiently transduce Schwann cells are not well established. In contrast, given the high efficiency transduction of neurons we observed, we feel this accounts for most of the prolonged survival and improved motor performance and motor neuron physiology in the plt mice.
Our best rescue of the plt/CMT4J phenotype was obtained by treating as early as possible, at P1. We faced 2 experimental limitations in this regard. First, the plt mice have an early and aggressive phenotype, reducing the feasibility of true postonset trials in this mouse model. In addition, given the size of the FIG4 gene and promoter sequences, we were restricted to using a single-stranded AAV9 that is likely to take weeks to achieve full expression levels in transduced cells (20–22). With this in mind, it is likely a combination of factors that contribute to later treatment being less effective at P7 or P11: (a) increased degenerative disease state with irreversible damage, (b) expression that may be too slow to overcome the aggressive phenotype, and (c) decreased brain vector biodistribution in i.t. P7/P11 injections versus P1/P4 injections. Extrapolating these mouse time points to patients is challenging. Patients are likely to provide a window of treatment from months to years, not days, but we still fully anticipate that the earlier treatment can be provided, the greater the anticipated benefit. Our findings of reduced vacuolization in the DRG provide a hopeful result. This phenotype is evident even at perinatal time points (3), and the fact that it is largely rescued with postnatal delivery of a single-stranded AAV9 vector suggests there may be reversal of this cellular pathology, not just prevention or arrested progression. While AAV9-mediated replacement of FIG4 is clearly not going to restore lost neurons, this result in mouse DRGs does suggest it may be able to reverse some hallmarks of neuronal dysfunction. The route of delivery is also a consideration. Our results suggest that transduction of neurons was responsible for the bulk of the benefit observed, supporting a targeted delivery to the CNS, such as i.t. injection. In addition, i.t. delivery has practical benefits, such as smaller doses, and thus smaller scale vector production; it also incurs less exposure of other tissues, such as the liver and heart, that are efficiently transduced by AAV9 when delivered systemically (23, 24).
The appearance of HCCs with delivery of AAV9-FIG4 is not unanticipated based on literature, and we confirmed that our findings are consistent with these previous reports (16, 17, 19). In the neonatal mouse genome, AAV9 vectors are prone to integration in the Rian locus (16). Most of the integrations at Rian target a microRNA, Mir341. Only the integration of strong promoter sequences in this location was reported to upregulate the expression of neighboring genes that may contribute to the tumorogenecity of these events (16). Integrations in Rian were the most abundant genomic abnormality found in HCCs analyzed after AAV9 transduction in our study and in previous analyses (16, 18). Interestingly, the frequency of HCCs in mice goes down rapidly with AAV9 transduction at older ages, suggesting the neonatal genome is more prone to these integrations, probably due to chromatin structure. Also, the Rian locus is not conserved in nonrodent species, and HCC has not been reported as a toxicological complication in AAV9 studies in humans or nonhuman primates. Given that we found HCC in 10 of 15 of our AAV9-FIG4–treated mice and 2 of 9 of our untreated mice at a year of age and that we found integrations of the AAV9 vector and promoter sequence at the Rian locus in 8 of 10 of the treated tumor samples, we feel that the incidence of HCCs in these mice can be accounted for by the proclivity of AAV9 vectors with strong promoters to integrate at the Rian locus in neonatal mice and promote tumor formation.
There is currently no treatment for CMT4J. The biology of PI(3,5)P2 is well studied, but also very complicated. It is not immediately obvious how one would design a pharmacological strategy to treat this disease. In addition, since FIG4 is in a complex with the kinase PIKFYVE, PI(3,5)P2 levels are lower in the plt mice in contrast to what one might expect from the loss of the phosphatase. In contrast, the genetics of CMT4J seem to be relatively simple, with recessive loss-of-function mutations causing disease, although there are still some unresolved issues regarding cell autonomy and the variability of clinical severity despite the apparent consistency of genotypes (5). The latter may be due to more complex genetic background effects, which are suggested by the mouse model as well. However, our preclinical studies with AAV9 delivery of FIG4 were largely successful. It is worth exploring whether this approach could be further optimized, by also transducing Schwann cells, for example, but considering the often severe and debilitating symptoms of CMT4J patients and the lack of other treatment options, our results indicate that this gene-therapy approach warrants further investigation.
Methods
Animals and treatments
Fig4-deficient mice carrying the plt allele were maintained under specific pathogen–free conditions at The Jackson Laboratory. Mice homozygous for the plt allele are neonatal lethal on a C57BL/6J background (B6.Cg-Fig4plt1/MmJ, catalog 17800), and they can survive up to 2 weeks on a C3HeB/FeJ background (C3Fe.Cg-Fig4plt1/MmJ, catalog 17801). However, in an F1 hybrid background, B6;C3H mice homozygous for the plt allele can survive 3 to 4 weeks (25). In this study, homozygous mice for the plt allele were generated by crossing heterozygous mice of both congenic stocks, generating an F1 hybrid B6;C3Fe-Fig4plt1/MmJ. For efficacy and safety testing, 10 to 18 homozygous male and female mice, Fig4plt/plt, and WT littermates, Fig4+/+, were randomly assigned to treatment groups at P1 or P4 and dosed by i.c.v. injection with 2 μl of either AAV9-FIG4 vector (5.4 × 1011 vg) or vehicle or left untreated. For postsymptomatic efficacy testing, 10 to 14 homozygous male and female mice, Fig4plt/plt, were treated at P7 or P11 by i.t. injection with 5 μl of either AAV9-FIG4 vector (13.5 × 1011 vg) or vehicle or left untreated. Lower dose i.t. injections were conducted with the same volume and vehicle composition. All injections were performed blinded for compound and genotype. All mice were monitored daily for body weight, body conditioning, and survival. Mice were euthanized when a body weight loss greater than 15% or a body conditioning score lower than 2 (on a scale 1–3) was detected.
AAV9-FIG4 production
An AAV9 vector was engineered to encode the codon-optimized human FIG4 gene for expression of the active human FIG4 enzyme. Three plasmids were used for manufacturing the AAV9-FIG4 vector: the packaging/helper plasmids pGSK2/9 and pXX6-80 (which provide the AAV and adenoviral helper functions needed for AAV vector production in mammalian cell culture) and the vector plasmid (pTR-CBA-hFIG4opt-spA). The vector plasmid contains the new 2721 bp optimized DNA-coding sequence (hFIG4opt; ATUM) cloned between a 1612 bp CBA promoter with a CMV enhancer and a 48 bp synthetic polyadenylation (SpA) signal. The transgene expression cassette was flanked by WT AAV2 ITRs (Figure 1A). The viral vector was produced by a triple-transfection method in HEK293 cells at the University of North Carolina Vector Core, as described (26). The final vector solution was formulated in PBS containing 350 mM NaCl (final concentration) and 5% sorbitol.
Western blot
Whole-tissue lysates were prepared in RIPA buffer (Thermo Fisher Scientific) containing Halt Protease Inhibitor Cocktail (Thermo Fisher Scientific). Tissue was homogenized using FastPrep Z tubes (MP Biomedicals). Total soluble protein content was quantified by DC protein assay (Bio-Rad) and adjusted to 4 mg/ml in RIPA buffer. Protein lysates were prepared for automated Western blot using the Simple Wes system following vendor protocol (ProteinSimple). Anti-FIG4/Sac3 (clone N202/7, Antibodies Inc.) was used at a 1:50 dilution, anti-P62/SQSTM1 (clone ab56416, Abcam) was used at a 1:2000 dilution, and anti-GFAP (clone NB300-141, Novus Biologicals) was used at 1:2000 dilution. Anti-GAPDH (mouse monoclonal MAB5718, R&D Systems, or rabbit polyclonal NB300-322, Novus Biologicals) was used as loading control at a 1:5000–1:10000 dilution. Chemiluminescent signal was captured, and the resulting intensities were analyzed using the Compass for Simple Western software package (ProteinSimple). The target protein peak area was normalized to the loading control peak area following ProteinSimple recommendations and standard guidelines (27, 28). The relative fold change (RFC) was calculated using Fig4+/+ mice as reference.
Behavior assessments
Rotarod.
An Ugo-Basile accelerating rotarod (model 47600) for mice was used for this test. Mice were acclimated to the testing room for 60 minutes. The test began by placing the mice on the rotating rod at 4 rpm, then accelerating the rod up to 40 rpm over the course of 300 seconds. Each mouse was subjected to 3 consecutive trials with a 45-second resting interval. The time in seconds at which the mouse fell from the rod was recorded.
Grip strength.
A commercially available grip-strength meter (Bioseb) was used to measure forelimb grip strength and/or combined forelimb/hind limb grasp strength. For all measurements, a wire grid coupled to a strain gauge that measures peak force (kg) was used. The mice were weighed and allowed to acclimate to the testing room for a minimum of 60 minutes before the test began. For forepaw testing, mice were held by their tails and lowered toward the grid to allow for visual placing and for the mouse to grip the grid with its forepaws. The animal was firmly pulled horizontally away from the grid for 6 consecutive trials with a brief intertrial rest period. The first 3 trials measured forepaw grip only, while the last 3 included all 4 paws. The force peak was normalized to mouse body weight.
Spontaneous wheel running.
Animals were singly housed in a new cage with a running wheel, with food and water ad libitum. Running wheels (Med Associates) were equipped with a wireless transponder that recorded activity on the running wheels (rpm) in sync with a computer that timestamped events. The mice were left undisturbed during the testing period of 3 nights and 2 days. Data were evaluated for total distance traveled during each time period.
Electrophysiology
Sciatic NCV was previously described (29). It was determined by measuring differences in CMAP latency in the muscles of the hind paw following sciatic nerve stimulation at 2 points (hip and ankle, NCV = distance between stimuli/difference in latency [hip – ankle]). The presence of conduction block was also evaluated by measuring the integrated CMAP in response to distal and proximal stimulation, where conduction block results in a smaller response to proximal stimulation, which must propagate a longer distance. Briefly, mice were anesthetized with isoflurane (1.5%–2 %) and placed on a thermostatically regulated heating pad to maintain normal body temperature. For recording, the active needle electrode was inserted in the center of the paw and a reference electrode was placed in the skin between the first and second digits. The distance between points of stimulation and the recorded latencies was used in the calculation of velocity.
Histopathology analysis
Half of the cohort treated at P1 and P4 were euthanized at 6 months of age, and brain, spinal cord, skeletal muscle, heart, liver, kidney, gonads, and mesenteric and cervical lymph nodes were dissected. Tissues were sectioned in half; one section was flash frozen for protein and DNA extraction and a second section was fixed in 10% NBF for histopathological assessments. Fixed brain, heart, liver, kidney, lymph nodes, and gonads were paraffin embedded, and 5 μm sections were stained with H&E. Lumbar spinal cord was decalcified with Immunocal (StatLab) overnight, and spinal cord and DRGs were dissected. DRGs were mounted in HistoGel (Thermo Fisher Scientific) for paraffin embedding, sectioning, and H&E staining. Histopathology analysis was conducted blinded for genotype and treatment by a board-certified veterinary pathologist at The Jackson Laboratory.
For histology nerve analysis, sciatic nerve was dissected and fixed overnight at 4°C in EM fixative (2 % PFA, 2 % glutaraldehyde, 0.1 M cacodylate buffer; pH = 7.2). Tissue was embedded in plastic blocks, and semithin sections (1 μm) were stained with toluidine blue. Slides were scanned at ×40 magnification and digital images analyzed with ImageJ (NIH) for axon area distribution.
AAV9-FIG4 ISH
Analysis of AAV9/huFIG4 expression was conducted by ISH using RNAScope probes (ACD, Biotechne) specific for huFIG4 (hFIG4-codon-No-XMm-C2, catalog 870051-C2), Pmp22 (Probe - Mm-Pmp22, catalog 816571), and Chat (Probe – Mm-Chat, catalog 408731). Mice injected i.c.v. with AAV9/huFIG4 or vehicle were euthanized at 4 weeks of age. Spinal cord and sciatic nerve were dissected and mounted longitudinally in OCT. Longitudinal cryosections of spinal cord and sciatic nerve were double stained for Pmp22/huFIG4 (sciatic nerve) and Chat/huFIG4 (spinal cord), following RNAScope protocols. Images were captured in a Zeiss AxioImager.Z2 and the number of positive cells by ROI manually counted using ImageJ software.
AAV9-FIG4 biodistribution
For analysis of vector biodistribution, mice treated by i.c.v. injection with AAV9-FIG4 at P1 or P4 were euthanized at 6 months of age, and mice treated by i.t. injection were euthanized at 3 months of age. Genomic DNA was extracted from brain, spinal cord, liver, heart, muscle, kidney, and gonads. Vector copy number was quantified by quantitative PCR (qPCR), as previously described (24), using a specific set of primers for hFIG4opt (forward: 5′-CGAGGACACCAACATGATCTAC-3′, reverse: 5′-CCACGTTCTGGAAGATTCTAAGG-3′), and Laminb2 (forward: 5′-GGACCCAAGGACTACCTCAAGGG-3′, reverse: 5′-AGGGCACCTCCATCTCGGAAAC-3′) to normalize and calculate the copies of viral genomes by μg of host DNA (vg/μg).
HCC whole-genome sequencing
One tumor mass was dissected from each mouse liver, together with a liver sample visually free of tumor, and both were flash frozen. gDNA was extracted following standard methods. For library preparation 1 μg by sample of gDNA was used as input material. Sequencing libraries were generated using the DNA Library Prep Kit (NEBNext) following the manufacturer’s recommendations. The resulting libraries were fed into Illumina sequencers (HiSeqX, Illumina).
For sequence analysis, 2 tools, Virus-Clip (30) and BatVI (31), were used to detect possible integrations of the viral reference sequence pTR-CBA-hFIG4opt-spA. Both tools rely on using split-reads to determine possible integration sites. A split-read is identified when one portion of a sequencing read maps to one genomic location and another portion of the same sequencing read maps to a different genomic location. The tools return the number of split-reads supporting each proposed integration event. The total number of reads (sequencing depth) at each integration site and the fraction of split-reads called by each tool at each integration site were calculated.
Statistics
Data were analyzed and graphed using GraphPad Prism 7 software. Mann-Whitney U test was used for comparisons between 2 groups. One-way or 2-way ANOVA test with Dunnett’s correction was used for multiple group comparisons. Statistical significance was set at P < 0.05 for all comparisons.
Study approval
All animal treatments and procedures were approved by the Animal Care and Use Committee at The Jackson Laboratory.
Author contributions
The AAV9-FIG4 vector was designed and produced by RMB and SJG. Mouse studies and subsequent molecular analyses were performed by MP, CD, LB, TM, JC, HW, and RW. Experimental design and interpretation were done by RWB, SJG and CL, with assistance from RMB, GML, and MP. The manuscript was written by MP, RWB, SJG, and CL, with input from all authors.
Supplementary Material
Acknowledgments
The authors are grateful for funding and support from the Cure CMT4J/Talia Duff Foundation (to SJG and CL). This work was supported by The Center for Precision Genetics at The Jackson Laboratory (NIH grants U54 OD020351 and U54 OD020351). The authors are grateful to Scientific Services at The Jackson Laboratory for behavioral phenotyping, histology, and genome sequencing. These services are supported by NIH grant CA034196. RWB was supported by NIH R37 NS054154. GML was supported by NIGMS R01 GM24872. The authors are also grateful to Jun Li (Wayne State University) for helpful discussions on the Fig4plt mice and CMT4J clinical features.
Version 1. 04/20/2021
In-Press Preview
Version 2. 06/01/2021
Electronic publication
Footnotes
Conflict of interest: SJG and RMB are inventors on the FIG4 vector design and have received royalty income from Neurogene LLC related to this invention.
Copyright: © 2021, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2021;131(11):e137159.https://doi.org/10.1172/JCI137159.
See the related Commentary at Adeno-associated virus gene therapy to the rescue for Charcot-Marie-Tooth disease type 4J.
Contributor Information
Maximiliano Presa, Email: Maximiliano.Presa@jax.org.
Rachel M. Bailey, Email: Rachel.Bailey@UTSouthwestern.edu.
Crystal Davis, Email: crystal.davis@jax.org.
Tara Murphy, Email: Tara.murphy@jax.org.
Jenn Cook, Email: Jenn.cook@jax.org.
Randy Walls, Email: Randy.walls@jax.org.
Hannah Wilpan, Email: Hannah.knowlton@jax.org.
Laurent Bogdanik, Email: Laurent.bogdanik@jax.org.
Guy M. Lenk, Email: glenk@umich.edu.
Robert W. Burgess, Email: robert.burgess@jax.org.
Steven J. Gray, Email: steven.gray@utsouthwestern.edu.
Cathleen Lutz, Email: cat.lutz@jax.org.
References
- 1.Timmerman V, et al. Genetics of Charcot-Marie-Tooth (CMT) disease within the frame of the Human Genome Project success. Genes (Basel) 2014;5(1):13–32. doi: 10.3390/genes5010013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Saporta MA, Shy ME. Inherited peripheral neuropathies. Neurol Clin. 2013;31(2):597–619. doi: 10.1016/j.ncl.2013.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chow CY, et al. Mutation of FIG4 causes neurodegeneration in the pale tremor mouse and patients with CMT4J. Nature. 2007;448(7149):68–72. doi: 10.1038/nature05876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nicholson G, et al. Distinctive genetic and clinical features of CMT4J: a severe neuropathy caused by mutations in the PI(3,5)P2 phosphatase FIG4. Brain. 2011;134(pt 7):1959–1971. doi: 10.1093/brain/awr148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hu B, et al. Myelin abnormality in Charcot-Marie-Tooth type 4J recapitulates features of acquired demyelination. Ann Neurol. 2018;83(4):756–770. doi: 10.1002/ana.25198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang X, et al. Mutation of FIG4 causes a rapidly progressive, asymmetric neuronal degeneration. Brain. 2008;131(pt 8):1990–2001. doi: 10.1093/brain/awn114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Campeau PM, et al. Yunis-Varón syndrome is caused by mutations in FIG4, encoding a phosphoinositide phosphatase. Am J Hum Genet. 2013;92(5):781–791. doi: 10.1016/j.ajhg.2013.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baulac S, et al. Role of the phosphoinositide phosphatase FIG4 gene in familial epilepsy with polymicrogyria. Neurology. 2014;82(12):1068–1075. doi: 10.1212/WNL.0000000000000241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chow CY, et al. Deleterious variants of FIG4, a phosphoinositide phosphatase, in patients with ALS. Am J Hum Genet. 2009;84(1):85–88. doi: 10.1016/j.ajhg.2008.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jin N, et al. VAC14 nucleates a protein complex essential for the acute interconversion of PI3P and PI(3,5)P(2) in yeast and mouse. EMBO J. 2008;27(24):3221–3234. doi: 10.1038/emboj.2008.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lenk GM, et al. Pathogenic mechanism of the FIG4 mutation responsible for Charcot-Marie-Tooth disease CMT4J. PLoS Genet. 2011;7(6):e1002104. doi: 10.1371/journal.pgen.1002104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lenk GM, et al. Biallelic mutations of VAC14 in pediatric-onset neurological disease. Am J Hum Genet. 2016;99(1):188–194. doi: 10.1016/j.ajhg.2016.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang Y, et al. Loss of Vac14, a regulator of the signaling lipid phosphatidylinositol 3,5-bisphosphate, results in neurodegeneration in mice. Proc Natl Acad Sci U S A. 2007;104(44):17518–17523. doi: 10.1073/pnas.0702275104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ferguson CJ, et al. Neuronal expression of Fig4 is both necessary and sufficient to prevent spongiform neurodegeneration. Hum Mol Genet. 2012;21(16):3525–3534. doi: 10.1093/hmg/dds179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Winters JJ, et al. Congenital CNS hypomyelination in the Fig4 null mouse is rescued by neuronal expression of the PI(3,5)P(2) phosphatase Fig4. J Neurosci. 2011;31(48):17736–17751. doi: 10.1523/JNEUROSCI.1482-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chandler RJ, et al. Vector design influences hepatic genotoxicity after adeno-associated virus gene therapy. J Clin Invest. 2015;125(2):870–880. doi: 10.1172/JCI79213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Colella P, et al. Emerging issues in AAV-mediated in vivo gene therapy. Mol Ther Methods Clin Dev. 2018;8:87–104. doi: 10.1016/j.omtm.2017.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Donsante A, et al. AAV vector integration sites in mouse hepatocellular carcinoma. Science. 2007;317(5837):477. doi: 10.1126/science.1142658. [DOI] [PubMed] [Google Scholar]
- 19.Ferla R, et al. Low incidence of hepatocellular carcinoma in mice and cats treated with systemic adeno-associated viral vectors. Mol Ther Methods Clin Dev. 2021;20:247–257. doi: 10.1016/j.omtm.2020.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gray SJ, et al. Optimizing promoters for recombinant adeno-associated virus-mediated gene expression in the peripheral and central nervous system using self-complementary vectors. Hum Gene Ther. 2011;22(9):1143–1153. doi: 10.1089/hum.2010.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zincarelli C, et al. Analysis of AAV serotypes 1-9 mediated gene expression and tropism in mice after systemic injection. Mol Ther. 2008;16(6):1073–1080. doi: 10.1038/mt.2008.76. [DOI] [PubMed] [Google Scholar]
- 22.Malik AK, et al. Kinetics of recombinant adeno-associated virus-mediated gene transfer. J Virol. 2000;74(8):3555–3565. doi: 10.1128/JVI.74.8.3555-3565.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bailey RM, et al. Comparison of high-dose intracisterna magna and lumbar puncture intrathecal delivery of AAV9 in mice to treat neuropathies. Brain Res. 2020;1739:146832. doi: 10.1016/j.brainres.2020.146832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gray SJ, et al. Preclinical differences of intravascular AAV9 delivery to neurons and glia: a comparative study of adult mice and nonhuman primates. Mol Ther. 2011;19(6):1058–1069. doi: 10.1038/mt.2011.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lenk GM, Meisler MH. In: Conn PM ed. Methods in Enzymology. Academic Press; 2014:245–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grieger JC, et al. Production of recombinant adeno-associated virus vectors using suspension HEK293 cells and continuous harvest of vector from the culture media for GMP FIX and FLT1 clinical vector. Mol Ther. 2016;24(2):287–297. doi: 10.1038/mt.2015.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Taylor SC, Posch A. The design of a quantitative western blot experiment. Biomed Res Int. 2014;2014:8. doi: 10.1155/2014/361590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fosang AJ, Colbran RJ. Transparency is the key to quality. J Biol Chem. 2015;290(50):29692–29694. doi: 10.1074/jbc.E115.000002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bogdanik LP, et al. Loss of the E3 ubiquitin ligase LRSAM1 sensitizes peripheral axons to degeneration in a mouse model of Charcot-Marie-Tooth disease. Dis Model Mech. 2013;6(3):780–792. doi: 10.1242/dmm.010942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ho DWH, et al. Virus-Clip: a fast and memory-efficient viral integration site detection tool at single-base resolution with annotation capability. Oncotarget. 2015;6(25):20959–20963. doi: 10.18632/oncotarget.4187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tennakoon C, Sung WK. BATVI: Fast, sensitive and accurate detection of virus integrations. BMC Bioinformatics. 2017;18(Suppl 3):71. doi: 10.1186/s12859-017-1470-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.