Abstract
Chronic hepatitis B (CHB) infection is rarely eradicated by current antiviral nucleos(t)ide analogues. We found that α2,6-biantennary sialoglycans of HBV surface antigen (HBsAg) bound human SIGLEC-3 (CD33) by IP and ELISA, and the binding affinity between SIGLEC-3 and α2,6-biantennary sialoglycans was determined by biolayer interferometry (equilibrium dissociation constant [KD]: 1.95 × 10–10 ± 0.21 × 10–10 M). Moreover, HBV activated SIGLEC-3 on myeloid cells and induced immunosuppression by stimulating immunoreceptor tyrosine-based inhibitory motif phosphorylation and SHP-1/-2 recruitment via α2,6-biantennary sialoglycans on HBsAg. An antagonistic anti–SIGLEC-3 mAb reversed this effect and enhanced cytokine production in response to TLR-7 agonist GS-9620 in PBMCs from CHB patients. Moreover, anti–SIGLEC-3 mAb alone was able to upregulate the expression of molecules involved in antigen presentation, such as CD80, CD86, CD40, MHC-I, MHC-II, and PD-L1 in CD14+ cells. Furthermore, SIGLEC-3 SNP rs12459419 C, which expressed a higher amount of SIGLEC-3, was associated with increased risk of hepatocellular carcinoma (HCC) in CHB patients (HR: 1.256, 95% CI: 1.027–1.535, P = 0.0266). Thus, blockade of SIGLEC-3 is a promising strategy to reactivate host immunity to HBV and lower the incidence of HCC in the CHB patient population.
Keywords: Immunology, Infectious disease
Keywords: Glycobiology, Hepatitis, Immunotherapy
Introduction
HBV infection is a worldwide health problem, and chronic HBV infection frequently results in liver cirrhosis and hepatocellular carcinoma (HCC). In HBV-infected hepatocytes, viral DNA enters the nucleus and becomes covalently closed circular DNA (cccDNA), thereby producing a large number of viral proteins, including HBV surface antigen (HBsAg), early antigen (HBeAg), core antigen (HBcAg), and X protein (HBx). Chronic HBV (CHB) infection can be divided into 4 phases (1–3). The immune tolerance phase does occur in young CHB patients, although recent data suggest it is uncommon even in this age group (4). In the immune clearance phase, activation of host immunity causes liver inflammation, liver fibrosis with repeated episodes of liver injury, and detectable HBcAg-specific antibody. In the residual inactive phase (or low replicative phase), HBeAg seroconversion and decreased viral load are observed. Reactivation of HBV replication (viral DNA greater than 2 log10 IU/mL) occurs after remission. Severe liver inflammation is observed in the HBeAg-negative chronic hepatitis phase (or high replicative phase), even though patients are able to produce antibodies against HBcAg and HBeAg. It is still a mystery why CHB patients are unable to produce anti-HBsAg antibody during all 4 phases.
The persistent presence of serum HBsAg and viral DNA produced by HBV cccDNA in infected hepatocytes is the prominent feature of CHB infection (5). The ideal goals for CHB therapy include loss of HBsAg, induction of anti-HBsAg antibody, and eradication of cccDNA from the liver (5), but current antiviral treatments do not induce sufficient anti-HBsAg antibodies to achieve serum HBsAg loss. Present nucleotide analogue treatment results in a reduction in HBsAg in only 5%–8% of patients who are HBeAg positive and less than 1% of patients who are HBeAg negative (6–8). In the Checkmate 040 study, only 3 of 51 (6%) CHB patients with HCC had a limited decline of serum HBsAg level after anti–PD-1 mAb (nivolumab) treatment (9). Another extensive study showed 1 in 10 CHB patients had an HBsAg decline when treated with a combination of nivolumab (anti–PD-1 mAb) and GS-4774 (therapeutic HBV vaccine) (10). Blockade of the PD-1/PD-L1 pathway results in reactivation of HBV infection in 6 of 114 (5.3%) cancer patients with CHB infection (11), indicating that the efficacy of activating nonspecific T cell–mediated immunity for CHB treatment is still far from satisfactory.
It has been reported that HBsAg is able to attenuate IL-12 and IFN-α secretion in DCs stimulated with TLR-3 ligands (12, 13). Moreover, HBV has been shown to suppress poly (I:C) + IFN-γ–induced IL-12 as well as TLR-2 ligand–induced TNF-α expression in DCs (14). However, the underlying molecular mechanism is still unknown. Furthermore, TLR-7 agonist GS-9620 has been shown to induce IFN-α and reduce the HBsAg serum level in HBV-infected chimpanzees (15), but GS-9620 is unable to eliminate cccDNA in human hepatic cells (16) or to reduce the HBsAg serum level in clinical trials (17). These observations suggest that an alternative approach is needed to develop a better strategy to treat CHB patients.
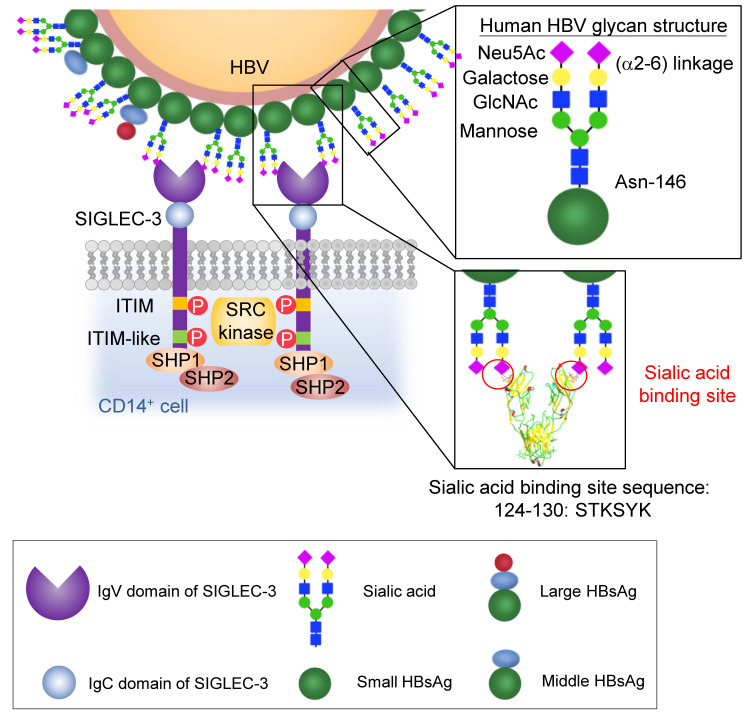
HBV has been reported to contain a biantennary α2–6 linked sialoglycan [Neu5Ac(α2-6)-Gal(β1-4)-GlcNAc(β1-2)-Man], which is identical to human biantennary α2–6 linked sialoglycans (18, 19), at Asn-146 of HBsAg (20, 21). Sialoglycans can modulate host immune responses via binding to the sialic acid–binding Ig-like lectins (SIGLECs) (22). SIGLECs comprise 2–17 extracellular Ig-like domains and a ligand-binding N-terminal V set domain. Except for SIGLEC-14, -15, and -16, the majority of human SIGLECs contain a cytoplasmic immunoreceptor tyrosine-based inhibitory motif (ITIM), which recruits the protein tyrosine phosphatase 1/2 (SHP-1/-2) to attenuate cell activation (23, 24). The downstream signaling of myeloid inhibitory SIGLECs (such as SIGLEC-3, -7, and -9) is similar to that of the ITIM-containing immune checkpoint receptor programmed cell death protein-1 (PD-1) and cytotoxic T lymphocyte–associated antigen 4 (CTLA4) (25–27), which are successfully targeted by antagonistic mAbs for cancer immunotherapy (28–31). Moreover, correlations between SNPs in the genes encoding SIGLECs and various diseases have been reported, such as the SIGLEC-3 (CD33) SNP associated with Alzheimer disease (32), the SIGLEC-9 SNP associated with chronic obstructive pulmonary disease (COPD) (33), and the SIGLEC-8 SNP associated with bronchial asthma (34). These observations suggest that SIGLECs may play critical roles in various human diseases.
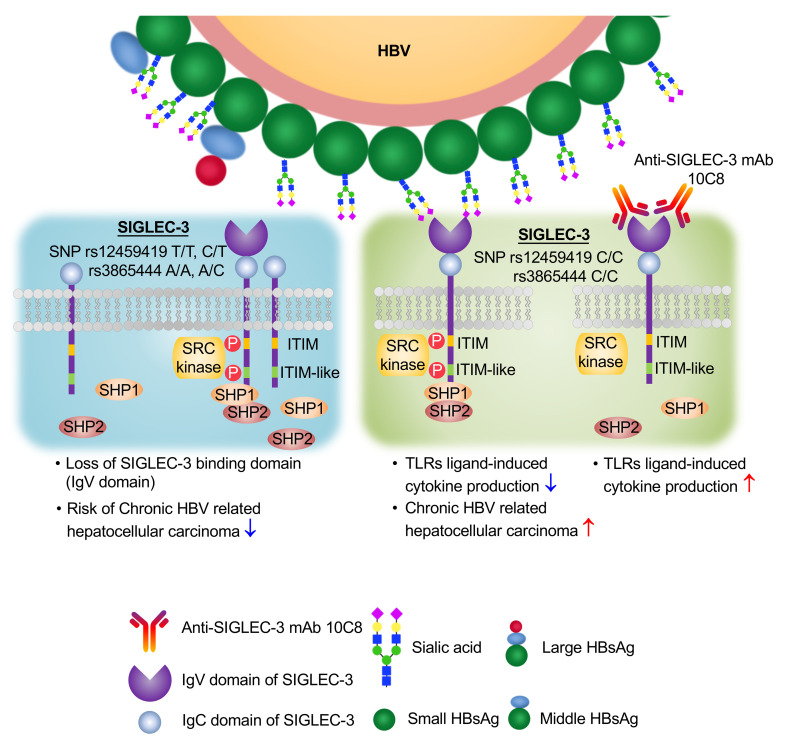
Because activation of SIGLECs downregulates TLR-induced cytokine release in DCs (35, 36), we sought to identify SIGLECs that interact with HBV sialoglycan and are responsible for HBV-induced immunosuppression. In this study, we demonstrated that HBV interacted with human SIGLEC-3 via α2–6 linked sialoglycan and was able to recruit SHP-1/-2 to attenuate TLR ligand–induced cytokine secretion. Interestingly, an antagonistic, neutralizing anti–SIGLEC-3 mAb (10C8) that did not cause SHP-1/-2 recruitment was able to reverse HBsAg-induced immunosuppression. Moreover, 10C8 blocked HBsAg-induced SHP-1/-2 recruitment, thereby upregulating molecules involved in antigen presentation. Furthermore, 10C8 had a synergistic effect with GS-9620 to upregulate cytokine production. It is interesting to note that SIGLEC-3 SNPs rs3865444 C/rs12459419 C are independent risk factors of HCC incidence in CHB patients. These observations suggest that SIGLEC-3 plays an important role in the progression of HBV infection, and blockade of SIGLEC-3 is a promising approach to enhance the GS-9620-mediated immunomodulatory effect. This speculation is in accord with the observation that Mylotarg, an antibody-drug conjugate targeting SIGLEC-3 in acute myeloid leukemia (AML), is more effective in patients with the rs12459419 C allele (37). Thus, a combination of anti–SIGLEC-3 mAb with GS-9620 may be able to reactivate host immunity to produce anti-HBsAg antibody to clear HBsAg and reduce HCC incidence in CHB patients.
Results
Glycan structure on HBsAg.
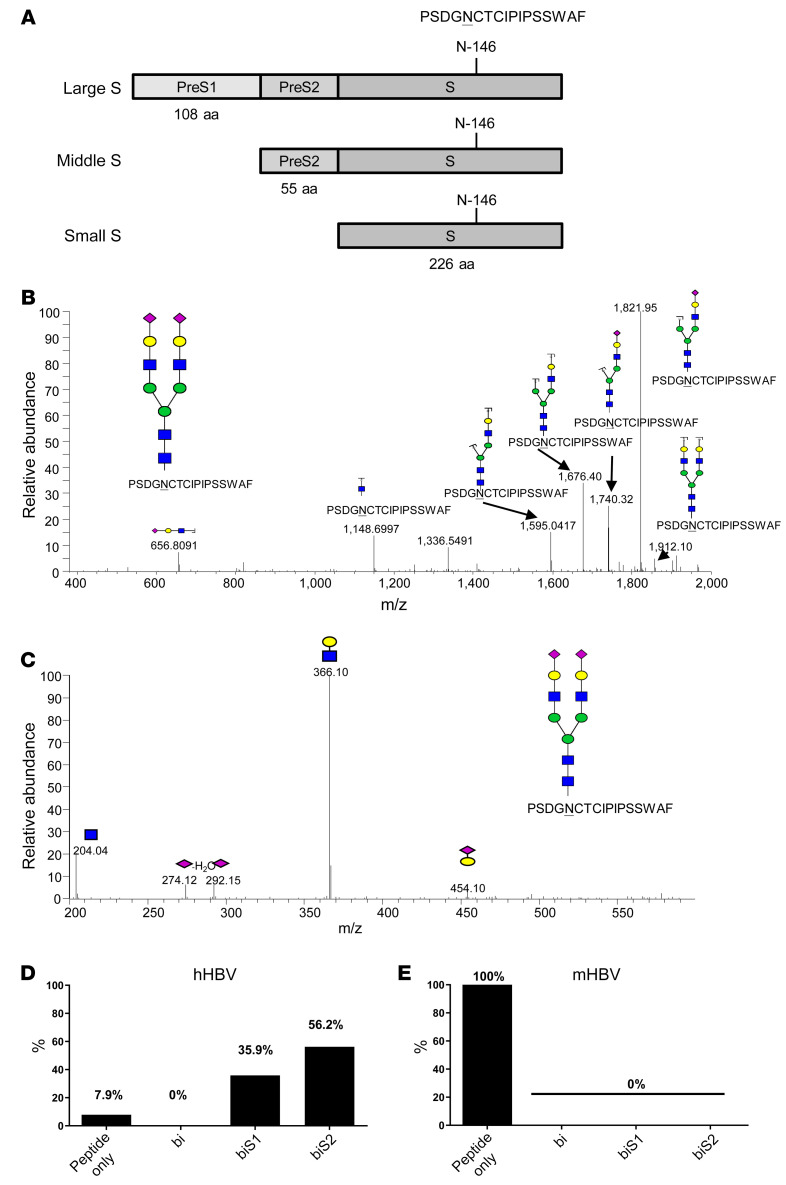
Since glycosylation is species specific, we compared the sialoglycans in HBV from CHB patients (hHBV) and transgenic mice (mHBV). The hHBV (genotype C) and mHBV (genotype D) were harvested from human and mouse sera, respectively, by CsCl2 ultracentrifugation followed by nano-flow LC-MS/MS analysis to determine glycan structures. We found that the P142SDGN146CTCIPIPSSWAF158 peptide fragment of human HBsAg (hHBsAg) contained the Neu5Ac(α2-6)-Gal(β1-4)-GlcNAc conjugated to Asn-146 (Figure 1, A and B). We further determined the linkage of the terminal sialic acid to galactose by pseudo-MS (Figure 1C), where the low m/z 274.12 and m/z 292.15 suggests the presence of Neu5Ac(α2-6)-Gal(β1-4)-GlcNAc (38). Approximately 92.1% of peptides contained biantennary N-glycans with either 2 terminal sialic acids (biS2, 56.2%) or 1 terminal sialic acid (biS1, 35.9%) (Figure 1D). In contrast, peptide P142SDGN146CTCIPIPSSWAF158 of mHBV did not contain an N-glycan conjugated to Asn-146 (Figure 1E). Thus, we conclude that the biantennary Neu5Ac(α2-6)-Gal(β1-4)-GlcNAc(β1-2)-Man conjugated to Asn-146 in HBsAg was only observed in hHBV but not mHBV.
Figure 1. Glycan structure of HBsAg.
(A) N-linked glycosylation site on hHBsAg (human HBsAg). (B) Glycan structure at Asn-146 of peptide P142SDGN146CTCIPIPSSWAF158 derived from hHBsAg (genotype C) by tandem mass spectrometry analysis. Neu5Ac, galactose; GlcNAc, mannose. (C) Determination of Neu5Ac linkage to galactose by pseudo-MS3 spectra. Pseudo-MS3 is an approach for the use of in-source fragmentation with electrospray ionization followed by product ion scan in a triple quadrupole mass spectrometer system. It is the combination of nonselective fragmentation followed by true MS/MS in a MALDI QqTOF mass spectrometer. (D and E) Percentage of biantennary sialoglycans in peptide P142SDGN146CTCIPIPSSWAF158 from hHBsAg (genotype C) and mHBsAg (genotype D). Bi (biantennary glycan with no terminal Neu5Ac), biS1 (single Neu5Ac), biS2 (2 Neu5Ac). Experiments were performed and repeated 3 times with the same results.
Interactions between HBV Asn-146 sialoglycan and human SIGLEC-3.
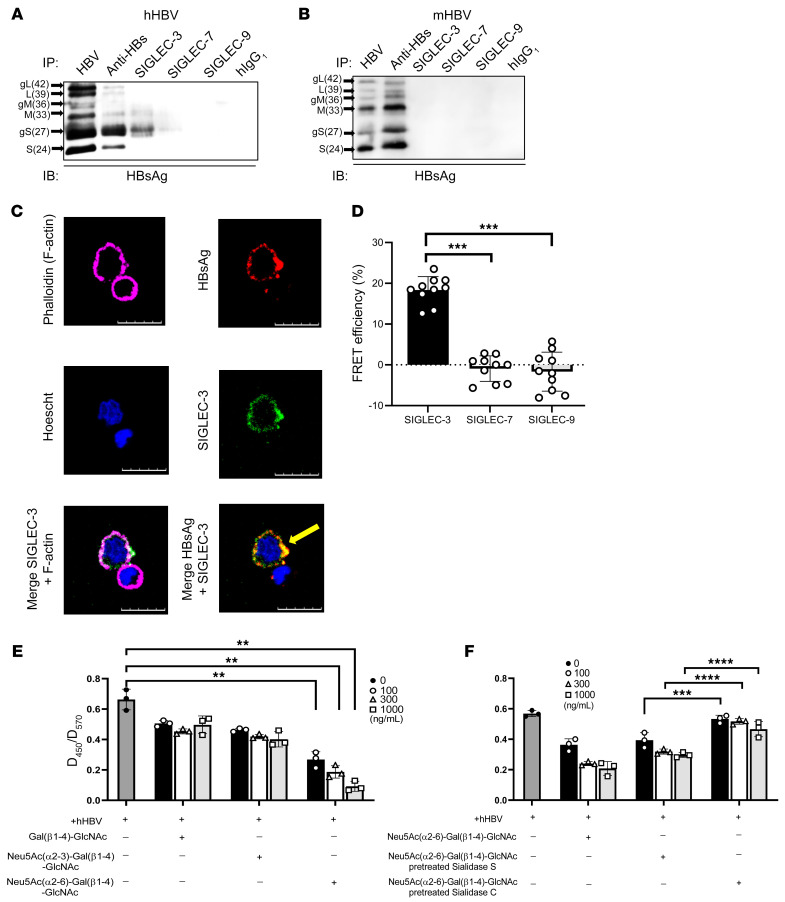
Because the biantennary sialoglycan on HBsAg is similar to the ligands of human SIGLEC-3, -7, and -9 (22), we asked whether the HBV virion interacts with any of these 3 SIGLECs. To address this question, recombinant SIGLEC-ECD.Fc fusion proteins (comprising the extracellular domain of SIGLEC-3, -7, and -9 and human IgG1 Fc portion) were used to pull down HBV virions, which were purified from human sera (genotype C) (Figure 2A) and mouse sera (genotype D) (Figure 2B) followed by Western blot analysis using HRP-conjugated anti-HBsAg antibodies (see complete unedited blots in the supplemental material). We found that HRP-conjugated anti-HBsAg antibody could detect 6 bands corresponding to glycosylated and unglycosylated HBsAg (large, middle, and small) of hHBV and mHBV (Figure 2, A and B). Although most of the HBsAgs pulled down from hHBV virions were glycosylated small HBsAg, a smaller amount of unglycosylated HBsAg was also observed (Figure 2A). In contrast, anti-HBsAg antibody pulled down unglycosylated middle HBsAg as well as glycosylated and unglycosylated small HBsAg from mHBV virion (Figure 2B). These observations suggest that most of HBsAg in hHBV virion was glycosylated small HBsAg, whereas HBsAg in mHBV virion was dominated by unglycosylated middle and small HBsAg. Among the 3 SIGLECs tested, only human SIGLEC-3 pulled down significant glycosylated small HBsAg from human CHB sera (Figure 2A). In contrast, none of the 3 SIGLECs pulled down HBsAg from mHBV virion under the same condition (Figure 2B). The specific interactions between SIGLEC-3 and HBsAg were confirmed by ELISA (Supplemental Figure 1A; supplemental material available online with this article; https://doi.org/10.1172/JCI141965DS1) and immunofluorescence staining (Supplemental Figure 1B). To further confirm the specific interactions between hHBV and human SIGLEC-3 ex vivo, we isolated CHB patients’ PBMCs and stained them with mouse anti–SIGLEC-3 mAb and goat anti-HBsAg mAb followed by incubation with Alexa Fluor 488–conjugated (green) goat anti-mouse mAb and Alexa Fluor 546–conjugated (red) donkey anti-goat mAb simultaneously; cell contour was marked by phalloidin (pink). Among the phalloidin-marked cells (Figure 2C), HBsAg (red) was only detected in SIGLEC-3–positive cells (green). Colocalization of HBsAg and SIGLEC-3 was also noted in SIGLEC-3–positive cells (yellow arrowhead, Figure 2C). We further quantified the frequency of cells expressing SIGLEC-3 and HBsAg in CHB patients’ PBMCs by using Metamorph software, and the expression profiling is as follows: SIGLEC-3+ (13.8% ± 1.4%), HBsAg+ (8.7% ± 0.7%), SIGLEC-3+HBsAg+ (7.8% ± 0.8%), SIGLEC-3–HBsAg– (86.1% ± 1.4%) (Supplemental Figure 1C). Thus, more than half of SIGLEC-3+ cells (56.5%) bound to HBsAg, and almost all the HBsAg+ cells were SIGLEC-3+ cells. It has been reported that CD14+ cells interact with HBsAg (39); thus, we performed multiple-color staining to identify cells that can interact with HBsAg (Supplemental Figure 2). Based on the gating strategy described in a previous study (40), SIGLEC-3 was expressed in all the CD14+ monocytes (R1) comprising CD14+ HLA-DR+ and CD14+-HLA-DR–/lo populations (R3). In contrast, SIGLEC-3 was not expressed in CD3+ T cells, CD19+ B cells, or CD3–CD56+ NK cells (R2) (Supplemental Figure 2). We further sorted out the CD14+ SIGLEC-3+ HLA-DR+ population and stained it with anti-HBsAg antibody (Supplemental Figure 3). We found that HBsAg was preferentially interacting with the CD14+ SIGLEC-3+HLA-DRhi population rather than the CD14+ SIGLEC-3+HLA-DRlo population (Supplemental Figure 3, A and B).
Figure 2. Interaction of HBV with SIGLEC-3.
(A and B) IP of hHBV and mHBV by SIGLEC-ECD.Fc fusion proteins. Markers identify S (small), M (middle), and L (large) HBsAg; g: glycosylated isoforms. (C) Colocalization of SIGLEC-3 and hHBV in CHB patients’ PBMCs under confocal microscopy (green: SIGLEC-3, pink: phalloidin, red: HBsAg, blue: Hoechst 33342) Scale bars: 10 μm. Experiments were performed and repeated 3 times with similar results. (D) FRET efficiency between hHBV and SIGLEC-3, -7, or -9 in freshly isolated PBMCs (2 × 105) from CHB patients. (n = 10) by 2-way ANOVA. (E and F) Competition assay of hHBV and SIGLEC-3 interaction by sialoglycan. (n = 3) (E) and sialidase C/sialidase S-treated sialoglycan; (n = 3) (F). Data are expressed as mean ± SD. **P < 0.01, ***P < 0.001, ****P < 0.0001 by 2-way ANOVA (E and F).
The direct hHBV-SIGLEC-3 interaction was further revealed by fluorescence resonance energy transfer (FRET). We found that high energy transfer was only observed between hHBV and SIGLEC-3 but not between hHBV and SIGLEC-7 or SIGLEC-9 in CHB patients’ PBMCs (Figure 2D). Addition of Neu5Ac(α2-6)-Gal(β1-4)-GlcNAc inhibited hHBV-SIGLEC-3 interaction in a dose-dependent manner (Figure 2E), whereas Neu5Ac(α2,3)-Gal(β1,4)-GlcNAc and Gal(β1,4)-GlcNAc did not inhibit hHBV–SIGLEC-3 interaction under the same condition (Figure 2E). To further confirm this observation, sialidase S (cleave α2–3 linked sialic acid) and sialidase C (cleave α2–3 and α2–6 linked sialic acids) were used to release terminal sialic acid from these sialoglycans. We found that sialidase C–treated but not sialidase S–treated sialoglycans were unable to inhibit hHBV–SIGLEC-3 interaction (Figure 2F). Thus, we conclude that the interactions between hHBV and SIGLEC-3 were via terminal α2–6 sialic acid of HBsAg-associated sialoglycans.
Determination of binding affinity between hHBV and human SIGLEC-3.
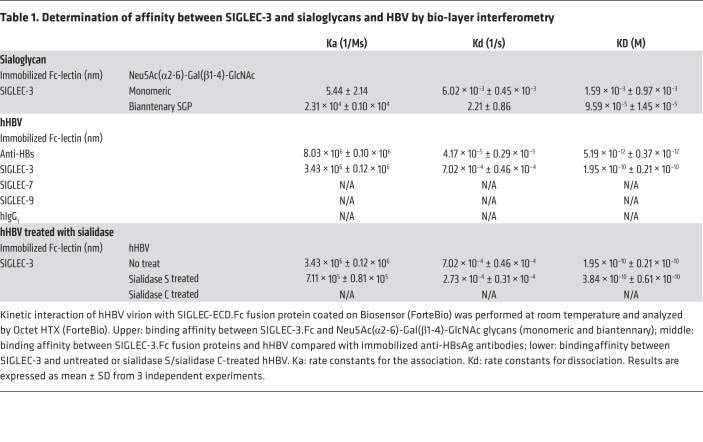
We further determined the binding affinity between hHBV and human SIGLEC-3. The equilibrium dissociation constant (KD) between recombinant bivalent SIGLEC-3.Fc (SIGLEC-3 is a dimeric protein based on crystal structure) and monomeric Neu5Ac(α2-6)-Gal(β1-4)-GlcNAc is 1.59 × 10–3 ± 0.97 × 10–3; the KD between bivalent SIGLEC-3.Fc and biantennary Neu5Ac(α2-6)-Gal(β1-4)-GlcNAc is 9.59 × 10–5 ± 1.45 × 10–5 (Table 1, upper panel). It is interesting to note that the KD between recombinant SIGLEC-3.Fc and hHBV increased to 1.95 × 10–10 ± 0.21 × 10–10 (Table 1, middle panel), suggesting the presence of multivalence binding between bivalent SIGLEC-3.Fc and hHBV. In contrast, mHBV did not bind to human SIGLEC-3 under the same condition (Supplemental Table 1). Furthermore, removal of α2–6 linked Neu5Ac from hHBV by sialidase C abolished SIGLEC-3 binding to hHBV (Table 1, lower panel). Thus, we conclude that hHBV bound to SIGLEC-3 via the terminal α2–6 sialic acid of the biantennary Neu5Ac(α2-6)-Gal(β1-4)-GlcNAc in HBsAg. By using the model proposed by Gargano et al. (41), the valence (N) between HBV and the recombinant SIGLEC-3.Fc is ‘2’, suggesting each monomeric SIGLEC-3 binds to 1 biantennary Neu5Ac(α2-6)-Gal(β1-4)-GlcNAc. The modeled interactions between hHBV and SIGLEC-3 and subsequent receptor crosslinking are shown in Figure 3.
Table 1. Determination of affinity between SIGLEC-3 and sialoglycans and HBV by bio-layer interferometry.
Figure 3. Schematic of hHBV-SIGLEC-3 interaction.
Human SIGLEC-3 interacts with Asn-146 sialoglycans (Neu5Ac(α2-6)-Gal(β1-4)-GlcNAc(β1-2)-Man) located on the small HBsAg. Our data demonstrated that 1 SIGLEC monomer binds to 2 terminal sialic acids on neighboring Asn-146 sialoglycans, thus allowing multiple-valence binding between HBV virion and myeloid cells, cross-linking SIGLEC-3, and triggering downstream signaling. Based on computer modeling (RCSB Protein Data Bank number 5J0B), the S124TKYSYK130 of human SIGLEC-3 is responsible for interaction with terminal sialic acids on Asn-146 sialoglycans.
SIGLEC-3–mediated hHBV immunosuppressive effect on monocyte-derived DCs.
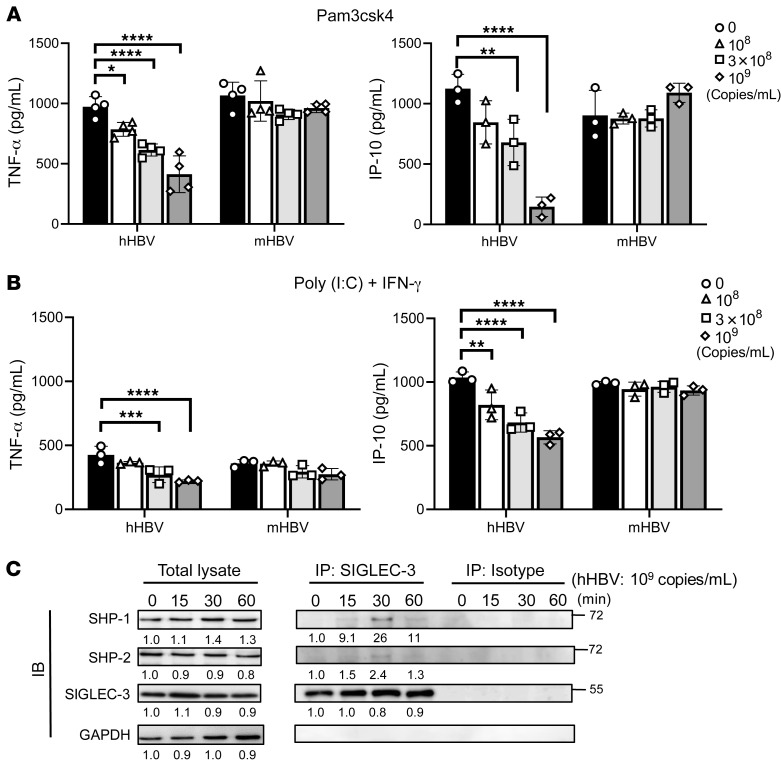
We further investigated the role of SIGLEC-3 in cytokine secretion from human CD14+ monocyte-derived DCs (moDCs) incubated with hHBV and TLR ligands. To address this question, moDCs from healthy donors were preincubated with hHBV or mHBV for 24 hours followed by incubation with Pam3csk4 (TLR-2 ligand) (Figure 4A) or poly (I:C) (TLR-3 ligand) + IFN-γ (Figure 4B). We found that hHBV suppressed the production of TNF-α and IP-10 (Figure 4, A and B) but not IL-1 β, IL-6, or IFN-γ from activated moDCs (Supplemental Figure 4). In contrast, mHBV had no inhibitory effect on cytokine secretion (Figure 4 and Supplemental Figure 4). We could not detect the expression of IFN-α, IL-12, IL-23, or TGF-β under the same condition. To further confirm that hHBV can induce inhibitory signals, hHBV was incubated with moDCs for up to 1 hour followed by IP using anti–SIGLEC-3 Ab (MAB1137, R&D Systems). As shown in Figure 4C, hHBV recruited SHP-1 and SHP-2 to SIGLEC-3 at 15 minutes, and the signal reached the peak at 30 minutes after incubation (Figure 4C; see complete unedited blots in the supplemental material). This observation suggests that hHBV is able to recruit SHP-1 and SHP-2 to SIGLEC-3.
Figure 4. SIGLEC-3 mediates the hHBV immunosuppressive effect.
(A and B) hHBV virion inhibited cytokine secretion in TLR ligand–stimulated moDCs. MoDCs (6 × 104 per well) pretreated with hHBV or mHBV in different doses for 24 hours before incubation with Pam3csk4 or poly (I:C) and IFN-γ for another 24 hours. Cytokine levels in the supernatant were determined by ELISA. Data are expressed as mean ± SD. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001 (2-way ANOVA) (n = 3–4). (C) Recruitment of SHP-1 and SHP-2 by hHBV is time dependent. MoDCs were treated with hHBV (109 copies/mL) to observe recruitment of SHP-1 and SHP-2 in moDCs by Western blotting. Signals were quantified by densitometry using ImageJ software. The values represent the fold-change normalized to the starting point. Experiments were performed and repeated 3 times with similar results.
Blockade of SIGLEC-3 reverses CD33 ligand and the HBV-induced suppressive effect.
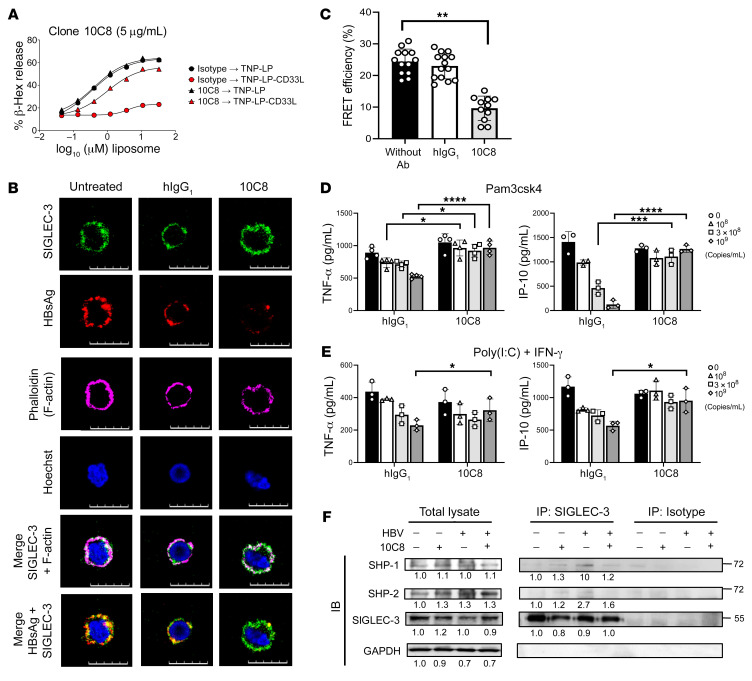
To further confirm the role of SIGLEC-3 in hHBV-induced immunosuppression, we generated an antagonistic mAb against human SIGLEC-3 (10C8) by using phage-displayed combinatorial libraries (42) and tested its effect in the TriNitroPhenol-conjugated liposome-induced (TNP-LP–induced) degranulation in mast cells (43). We found that TNP-LP was able to induce mast cell degranulation (Figure 5A), and incorporation of a synthetic SIGLEC-3 ligand containing liposome (TNP-LP-CD33 ligand) inhibited TNP-LP–induced degranulation (red circle, Figure 5A). Interestingly, 10C8 was able to reverse TNP-LP-CD33 ligand–mediated suppression (red triangle, Figure 5A), whereas isotype control antibody had no effect under the same condition (red circle, Figure 5A). We further asked whether 10C8 can inhibit HBV binding to SIGLEC-3 in CHB patients’ PBMCs. To address this question, PBMCs were incubated with commercially available anti–SIGLEC-3 mAb and anti-HBsAg mAb in the presence of 10C8 mAb or isotype mAb. In freshly isolated PBMCs, intense SIGLEC-3 (green) and HBsAg (red) were observed and colocalized (yellow) (left and middle columns, Figure 5B). Interestingly, the HBsAg (red) signal was very weak in the presence of 10C8, whereas the SIGLEC-3 (green) signal was not affected under the same condition (right column, Figure 5B). This observation suggests that 10C8 competed with detached HBsAg for reattachment to SIGLEC-3. Although 10C8 did not compete with Alexa Fluor 488–conjugated anti–SIGLEC-3 antibody used in this imaging (Figure 5B), it was able to inhibit FRET between SIGLEC-3 and HBsAg (Figure 5C). Furthermore, 10C8 was able to reverse the hHBV-mediated suppressive effect on cytokine secretion from Pam3csk4-activated (Figure 5D) or poly (I:C) + IFN-γ–activated (Figure 5E) moDCs, as well as to inhibit hHBV-induced recruitment of SHP-1 and SHP-2 to SIGLEC-3 (Figure 5F; see complete unedited blots in the supplemental material). These observations suggest that blockade of SIGLEC-3 was able to reverse hHBV-mediated immunosuppression in vitro.
Figure 5. Anti–SIGLEC-3 mAb (10C8) blocks hHBV-sialolglycan-SIGLEC-3 interaction.
(A) 10C8 reverse TNP-LP-CD33 ligand–mediated suppression without TNP-LP–induced degranulation (n = 3). (B) PBMCs isolated from CHB patients were treated with 10C8 at 4°C and further incubated with Alexa Fluor 488–conjugated anti–SIGLEC-3 mAb (Abcam, green), Alexa Fluor 546–conjugated anti-HBsAg mAb (red), and phalloidin (pink) at 4°C. Samples were counterstained with Hoechst 33342 (blue) followed by observation under a confocal microscopy (Zeiss). (C) FRET efficiency between SIGLEC-3 and hHBV was attenuated by 10C8 in PBMCs from CHB patients. Data are expressed as mean ± SD. (n = 11–14) cells per condition. (D and E) Reversal of hHBV suppressed cytokine secretion from TLR ligand–stimulated moDCs by 10C8. MoDCs from healthy donors (6 × 104 per well) were pretreated for 1 hour with 10C8 (3 μg/mL) followed by incubation with indicated amounts of hHBV in the presence of Pam3csk4 (n = 3–4) (D) or poly (I:C) + IFN-γ (n = 3) (E) for 24 hours. Cytokine levels were determined by ELISA. (F) Inhibition of hHBV-mediated recruitment of SHP-1 and SHP-2 by 10C8. MoDCs from healthy donors (6 × 104 per well) were pretreated for 1 hour with 10C8 (3 μg/mL), which was followed by incubation with hHBV (109 copies/mL) for 30 minutes and then pulldown with anti–SIGLEC-3 mAb. The immunoprecipitates were fractionated on SDS-PAGE before blotting followed by probing with anti–SHP-1 and anti–SHP-2 mAb. Quantification was determined by densitometry using ImageJ software and the number represented the fold of each value normalized to the value of the untreated control. Data are expressed as mean ± SD. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001 (2-way ANOVA) (C–E). (B and F) Experiments were performed and repeated 3 times with similar results.
Blockade of SIGLEC-3 upregulates surface marker expression and cytokine production in GS-9620–activated PBMCs.
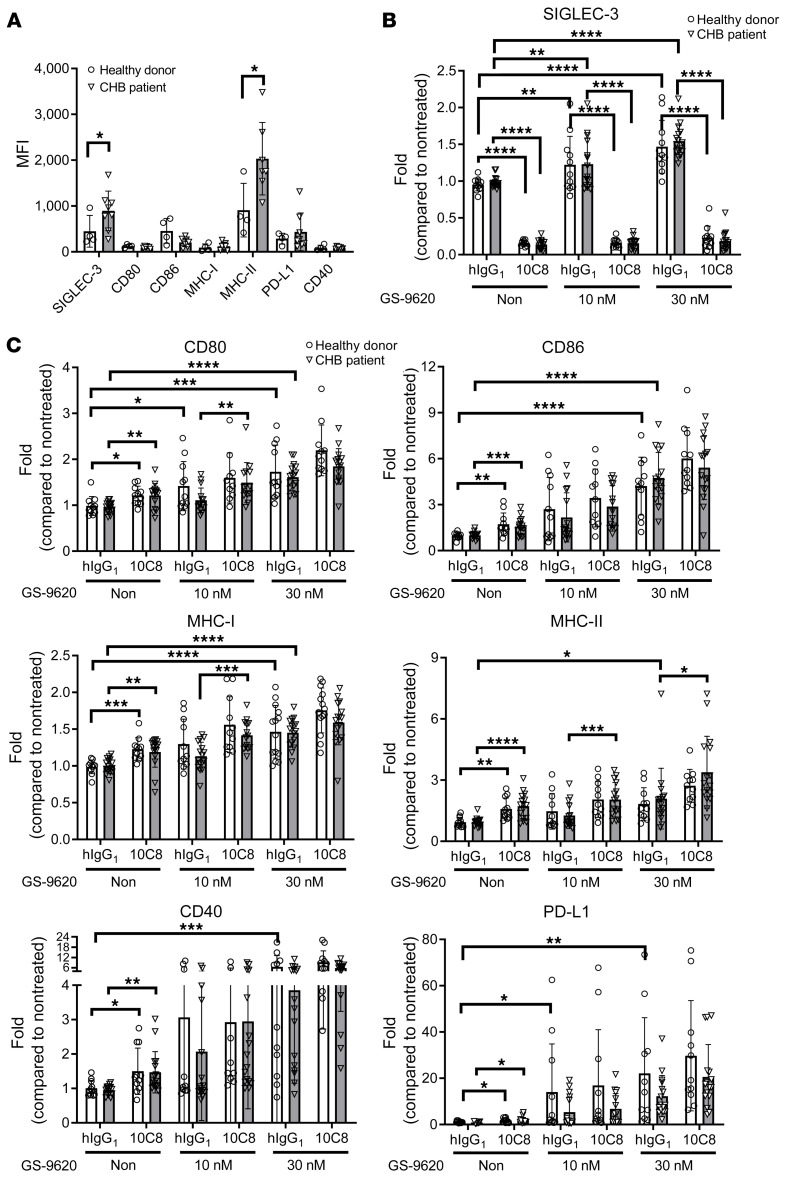
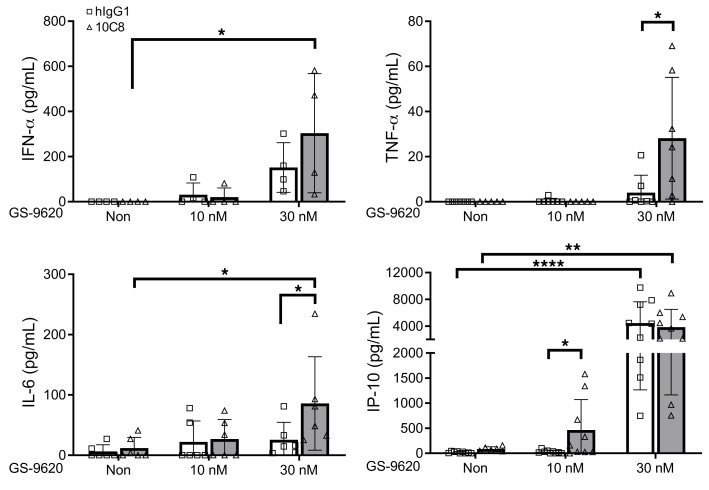
We further measured the expression of SIGLEC-3 and molecules involved in antigen presentation in CD14+ cells using CHB patients’ and healthy donors’ PBMCs. Compared with healthy donors, higher expression of SIGLEC-3 and MHC-II was observed in CHB patients’ CD14+ cells (Figure 6A). To understand whether a combination of 10C8 and TLR-7 agonist (GS-9620) can further enhance host immune responses, PBMCs were incubated with 10C8 in the presence or absence of GS-9620 to examine their effects in surface marker expression by flow cytometry. Based on the loss of fluorescence, 10C8 apparently downregulated the expression of SIGLEC-3 in CD14+ cells of healthy donors (83.4%) and CHB patients (85.9%) (Figure 6B). In contrast, GS-9620 had a modest effect on upregulation of SIGLEC-3 expression under the same condition (Figure 6B). Moreover, 10C8 alone was able to upregulate the expression of CD80 (23.1% vs. 24.6%), CD86 (75.9% vs. 57.5%), CD40 (49.3% vs. 52.2%), MHC-I (24.0% vs. 18.9%), MHC-II (67.7% vs. 82.1%), and PD-L1 (60.3% vs. 93.0%) in CD14+ cells from healthy donors and CHB patients, respectively (Figure 6C). In contrast to the weak effect of 10C8, GS-9620 was a potent inducer of PD-L1 in healthy donors’ and CHB patients’ CD14+ cells. At a low concentration of GS-9620 (10 nM), 10C8 further upregulated the expression of CD80, MHC-I, and MHC-II, and a higher concentration of GS-9620 (30 nM) alone was able to upregulate the expression of all the markers tested; in addition, 10C8 was still able to enhance the expression of MHC-II (61.7%) (Figure 6C). We further investigated the effect of 10C8 and GS-9620 in cytokine secretion by ELISA (Figure 7). It is interesting to note that 10C8 was able to enhance GS-9620–induced (30 nM) production of TNF-α (4.0→28.1 pg/mL) and IL-6 (25.5→85.8 pg/mL). Even though GS-9620 alone was a potent inducer of IP-10, 10C8 was still able to enhance IP-10 production (28.7→520.4 pg/mL) induced by a lower concentration of GS-9620 (10 nM) from CHB patients’ PBMCs. These observations suggest that 10C8 and GS-9620 had synergistic effects to upregulate antigen presentation and cytokine production in CHB patients.
Figure 6. 10C8 modulates surface marker expression in PBMCs from CHB patients.
(A) MFI of cell surface markers on CD14 + cells. PBMCs from healthy donors (n = 4) or CHB patients (n = 7–8) were incubated with 10C8 at 37°C for 1 hour followed by incubation with indicated antibody and measured by flow cytometry. (B) Fold change of SIGLEC-3 and (C) CD80, CD86, MHC-I, MHC-II, CD40, PD-L1 in CD14+ cells of PBMCs from CHB patients or healthy donors treated with or without GS-9620. PBMCs (1 × 106) from patients with CHB (n = 14–16) or healthy donors (n = 9–11) were pretreated for 1 hour with 10C8 (3 μg/mL) followed by GS-9620 treatment (10 nM and 30 nM) for another 24 hours. Cell surface markers were measured by flow cytometry. All data are expressed as mean ± SD. *P < 0.05, **P < 0.01, ***P < 0.001 ****P < 0.0001 (1-way ANOVA).
Figure 7. 10C8 modulates GS-9620–induced cytokine secretion in PBMCs from CHB patients.
PBMCs (1 × 106) from CHB patients (n = 4–9) were pretreated for 1 hour with 10C8 (3 μg/mL) followed by GS-9620 treatment at the indicated concentrations for another 24 hours. The supernatant was collected and cytokines were measured by ELISA. All data are expressed as mean ± SD. *P < 0.05, **P < 0.01, ***P < 0.001 ****P < 0.0001 (1-way ANOVA).
Association between SNPs in the SIGLEC-3 gene and clinical outcomes of CHB patients.
Two human SIGLEC-3 cosegregating SNPs, rs3865444 and rs12459419, have been shown to associate with the incidence of Alzheimer disease. While rs3865444 is located in the promoter region and is associated with modest changes in SIGLEC-3 expression (44), rs12459419 is located in intron 2 and is associated with increased exon-2 skipping, resulting in a truncated SIGLEC-3 and loss of the sialoglycan-binding V2 domain (Supplemental Figure 5A). It has been shown that rs12459419 C is the major allele and a risk factor for Alzheimer disease, and rs12459419 T (the minor allele) is protective in Alzheimer disease (heterozygous OR of 0.89 and homozygous OR of 0.81) (32). Thus, we asked whether these 2 SNPs are associated with disease progression in CHB patients. We retrospectively enrolled 3560 treatment-naive patients with CHB in the REVEAL-HBV (Risk Evaluation of Viral Load Elevation and Associated Liver Disease/Cancer-Hepatitis B Virus) cohort study. SNP rs3865444 was detected in 3555 cases, and SNP rs12459419 was detected in 3554 cases (Supplemental Table 4). The allele frequency of SNP rs3865444 in CHB patients was 3.89% (A/A homozygous minor), 27.66% (C/A, heterozygous), and 68.45% (C/C, homozygous major). The allele frequency of SNP rs12459419 in CHB patients was 3.97% (T/T, homozygous minor), 28.14% (C/T, heterozygous), and 67.90% (C/C, homozygous major). The cohort included 412 patients who progressed to liver cirrhosis and 377 patients who progressed to HCC. Compared with the rs12459419 T/C and T/T alleles (truncated SIGLEC-3), rs12459419 C/C major allele (full-length SIGLEC-3) (45) had a higher incidence of HCC (HR: 1.256, 95% CI: 1.027–1.535, P = 0.0266) in CHB patients under multivariate Cox proportional hazard regression analysis (Supplemental Table 2). The rs3685444 C/C allele had a similar incidence of HCC as rs12459419 C/C (Supplemental Table 2). In contrast, rs3865444 and rs12459419 SNPs did not associate with the incidence of liver cirrhosis (Supplemental Table 3). There was no difference between age, gender, genotype, HBeAg (positive vs. negative), baseline serum level of HBV DNA, baseline serum level of HBsAg, baseline serum level of alanine transaminase (ALT), baseline serum level of aspartate transaminase (AST), or age of diagnosis of liver cirrhosis or HCC between the alleles (Supplemental Table 4). These observations suggest that SIGLEC-3 polymorphism is associated with the progression to HCC in CHB patients. Thus, blockade of SIGLEC-3 is a promising approach to reactivate host immunity against HBV-mediated immunosuppression and may be able to reduce the risk of developing HCC in CHB patients.
Discussion
Although patient-derived HBsAg (genotype B and C) is sialylated at Asn-146, mouse-derived HBsAg (genotype D) is not sialylated at the same position. This observation is in accord with the absence of the inhibitory effect of mHBV in TLR-stimulated moDCs (Figure 4, A and B). A previous study showed that HBsAg produced from Chinese hamster ovary (CHO) cells contains glycosylated peptides, whereas HBsAg produced from yeast Hansenula polymorpha is nonglycosylated (46). Moreover, HBV produced from CHO cells is unable to attenuate cytokine secretion (IFN-α and IL-12) in plasmacytoid and myeloid DCs stimulated by TLR ligand [poly (I:C) or CpG-A] (12, 13). Because of the absence of α2–6-sialyltransferase (St6gal1) expression in CHO cells, the terminal sialic acid in the sialoglycan is in an α2–3 linkage (47). These observations suggest that α2–6 sialoglycans are crucial in HBV-mediated immunosuppression.
There are 10 genotypes of HBV (genotype A–J), and the genotypes with major global prevalence are genotypes A to D; the major genotypes in Taiwan are genotypes B and C (48–50). We found that the P142SDGN146CTCIPIPSSWAF158 peptide fragment of HBsAg, which contains the Neu5Ac(α2-6)-Gal(β1-4)-GlcNAc at Asn-146 (Figure 1, A and B), was identical in genotypes C–J. Moreover, Schmitt et al. have reported that Asn-146 glycosylation is present in genotypes A–D (20, 21). Furthermore, the corresponding region of HBsAg in genotypes A and B is P142TDGN146CTIPIPSSWAF158, which is almost identical to that of genotypes C and D, except S→T at amino acid residue 143 (51). We also found the genotype B of HBsAg contains the same Neu5Ac(α2-6)-Gal(β1-4)-GlcNAc glycan located at Asn-146 of P142TDGN146CTIPIPSSWAF158 (Supplemental Figure 6). These observations suggest that Asn-146 sialoglycans are conserved in all 10 genotypes.
Moreover, we found that HBV interacted with human SIGLEC-3 via the biantennary α2–6 linked sialoglycan located at Asn-146 of HBsAg and suppressed TLR ligand–induced cytokine production. We further produced an antagonistic, neutralizing anti–SIGLEC-3 mAb (10C8) to reverse HBV-mediated immune suppression and upregulate surface molecules critical in antigen presentation. Moreover, 10C8 had a synergistic effect with GS-9620 to upregulate cytokine production and the expression levels of MHC-I, MHC-II, and costimulatory signals (Figure 6). Anti–SIGLEC-3 mAb (clone WM53, BioLegend) was still able to detect SIGLEC-3 in cells pretreated with 10C8 at 4°C (Supplemental Figure 7A), and 10C8 was able to induce SIGLEC-3 internalization at 37°C (Supplemental Figure 7B). Therefore, we conclude that 10C8 was able to induce SIGLEC-3 internalization to downregulate its expression on the cell surface in the presence or absence of GS-9620 (Figure 6B). In contrast, we did not observe a synergistic effect between GS-9620 and anti–PD-L1 mAb (atezolizumab) under the same condition (Supplemental Figure 8). These observations suggest GS-9620 and 10C8 may have a synergistic effect to enhance antigen presentation to T cells to generate reactive host immunity against HBV in CHB patients. Because GS-9620 alone was unable to induce seroconversion or reduce HBsAg expression in a phase II clinical trial (52), a combination of GS-9620 and 10C8 seems a promising strategy to boost T cell immunity to clear HBV infection and prevent progression to HCC via enhancing antigen presentation and cytokine production.
Although chronic liver inflammation leads to liver cirrhosis and HCC (53), a recent study suggested that the occurrence of HCC but not liver cirrhosis correlates with immunosuppression and immune exhaustion (54). We demonstrated that SNPs of SIGLEC-3 (rs3865444 C and rs12459419 C) were associated with an increased risk of HCC (Supplemental Table 2) but not liver cirrhosis in CHB patients (Supplemental Table 3). These results are consistent with the argument that HBV can activate SIGLEC-3 to induce immunosuppression, thereby increasing the incidence of HCC but not liver cirrhosis. All these observations suggest that SIGLEC-3 plays a critical role in the pathogenesis of CHB and progression to HCC.
We were unable to find CHB patients with rs3865444 T/T and rs12459419 A/A given the low frequency of these alleles in Taiwanese people. Instead, we found 2 CHB patients with rs3865444 C/T and rs12459419 C/A alleles. Compared with patients with rs3865444 C/C and rs12459419 C/C, lower expression levels of SIGLEC-3 were observed in these 2 patients (Supplemental Figure 5B). However, the modulatory effects of 10C8 (with and without GS-9620) in surface marker expression (Supplemental Table 5) and cytokine production (Supplemental Table 6) were similar between patients with rs3865444 C/C and rs12459419 C/C (major alleles) or rs3865444 C/T and rs12459419 C/A. These findings suggest that 10C8 is still effective in patients with rs3865444 C/T and rs12459419 C/A.
Given that NIH and our funding agencies no longer support biomedical research in chimpanzees, the woodchuck is an alternative animal model to test the therapeutic effect of anti-human SIGLEC-3 mAb in vivo. However, none of our current antibodies can cross-react to woodchuck SIGLEC-3; therefore, we were unable to test the potential therapeutic effect of anti–SIGLEC-3 mAb in the woodchuck. The other potential animal model is the genetically modified mouse expressing human SIGLEC-3 without endogenous mouse Siglec-3 (Mcpt5-Cre+/–R26-SIGLEC-3+ mice, developed by James Paulson’s laboratory) (43). These Cre+/–R26-SIGLEC-3+ mice can be subjected to hydrodynamic injection with replication-competent HBV DNA to establish a chronic HBV infection model (55), thereby allowing us to test the potential therapeutic effect of 10C8 in vivo.
There are several potential limitations of this study regarding the potential therapeutic effect of anti–SIGLEC-3 mAb. First, all of the CHB patients represented in our collected data were treatment naive, and we were unable to show the response of CHB patients’ PBMCs incubated with HBV-specific peptides. This may be attributed to the low percentage of HBV-specific T cells in patients’ PBMCs (less than 1% of total CD3+ T cells after HBV-specific peptide stimulation) as determined by HBV tetramer. Second, SIGLEC-3 contains an ITIM motif similar to PD-1 and CTLA-4; thus, we cannot rule out the possibility that blockade of SIGLEC-3 by 10C8 may cause undesirable autoimmune side effects as observed in anti–PD-1 and anti–CTLA-4 mAb treatment, even though Siglec-3 (CD33) knockout mice do not develop autoimmunity (56). Unlike blockade of PD-1 and PD-L1 interaction (cell-cell interaction), 10C8 not only blocks SIGLEC-3–HBV interaction but may also inhibit interaction between SIGLEC-3 and endogenous sialoglycans. Furthermore, whether 10C8 can enhance HBsAg presentation and reactivate T cell response to HBV needs to be further examined in vivo. Third, liver cirrhosis was diagnosed by abdominal ultrasonography in the REVEAL-HBV cohort study. Given the lack of information regarding the status of hepatic encephalopathy and ascites in these patients, we were unable to evaluate the Child-Pugh score (a scoring system for evaluating the severity of liver cirrhosis). Fourth, the incidence of HCC is based on the record in the Taiwan Health Insurance database using international classification of disease (ICD) codes (ICD-9-CM diagnosis code 155.0 and ICD-10-CM diagnosis code C22.0), as well as by computerized data linkage with the National Cancer Registry and National Death Certification databases. However, neither the ICD code nor the computerized linkage to databases has HCC staging information. Therefore, we were unable to correlate the severity of liver cirrhosis with clinical staging of HCC in the REVEAL-HBV cohort study. Fifth, the recruited SHIP-1/2 in freshly isolated heathy donors’ PBMCs was only exposed to HBV for 24 hours; thus, the long-term effect of HBV-mediated immunosuppression needs be further investigated. Sixth, we incubated HBV virion with moDC derived from healthy donors and demonstrated that sialoglycan on HBsAg was able to modulate host immunity. However, because of the complicated components of the HBV virion, we cannot rule out that other viral components, such as pre-core antigen, may also modulate host immunity against HBV. Further study using humanized SIGLEC-3 transgenic mice will be able to reveal the beneficial versus unwanted side effects of anti–SIGLEC-3 mAb in the treatment of CHB.
Methods
Blood donors.
Naive untreated HBeAg-positive CHB patients (HBV DNA > 105 IU/mL) admitted to China Medical University Hospital were enrolled in this study to obtain PBMCs. All of the patients were negative for other viral infections, including hepatitis C virus, HIV, CMV, and EBV. Negative CMV and EBV infections were defined as negative for CMV IgM and EB-VCA IgM. Healthy donors were enrolled from the Taiwan Blood Services Foundation (Taipei, Taiwan).
REVEAL-HBV cohort study.
We enrolled 3560 treatment-naive patients with CHB from the REVEAL-HBV cohort study (57). In summary, a total of 23,820 male and female residents in Taiwan from age 30 to 65 years were enrolled in the REVEAL-HBV cohort study with informed consent during 1991–1992. Patients with coinfection by other etiologies such as hepatitis C virus, hepatitis D virus, or HIV, were excluded. Serum samples collected and frozen at cohort entry and follow-up examinations were tested. Regular health examinations using abdominal ultrasonography and serological tests occurred until 31 December 2005. HCC incidence was followed up to 31 December 2018 by using the Taiwan Health Insurance database with ICD codes (ICD-9-CM diagnosis code 155.0 and ICD-10-CM diagnosis code C22.0) as well as by computerized data linkage with the National Cancer Registry and National Death Certification databases. We defined the cutoffs for ALT and AST based on normal values.
Plasmids and fusion proteins.
The DNA fragments of the human SIGLEC-3/-7 ECD were amplified by reverse transcription PCR (RT-PCR) from human macrophages and subcloned into the pSecTag2-hIgG vector to generate the recombinant SIGLEC-3/-7-ECD.Fc fusion proteins. The SIGLEC-9-ECD.Fc cDNA was a gift from Takashi Angata (Institute of Biological Chemistry, Academia Sinica). The recombinant SIGLEC-3/-7/-9-ECD.Fc fusion proteins were overexpressed by the FreeStyle 293 Expression System (Invitrogen) according to the manufacturer’s instructions. The culture supernatants were harvested at day 3 and day 5 after transfection, and the recombinant fusion proteins were purified by protein A column (GE Healthcare).
Antibodies and reagents.
Antibodies used for Western blot, IP, confocal microscopy, and FACS are listed in Supplemental Table 7. Human GM-CSF and recombinant IL-4 were purchased from R&D Systems. Hoechst 33342 solution (62249), phalloidin, Dylight 488 (21833), and Pierce protein-free (TBS) blocking buffer (37570) were purchased from Thermo Fisher Scientific. Recombinant TLR-2 ligands (Pam3csk4, catalog: tlrl-pms) and TLR-3 [poly (I:C) HMW, catalog: tlrl-pic] were purchased from InvivoGen. GS-9620 (CAS 1228585-88-3) was purchased from Cayman Chemical. Glycans containing Neu5Ac(α2-6)-Gal(β1-4)-GlcNAc, Neu5Ac(α2-3)-Gal(β1-4)-GlcNAc, Gal(β1-4)-GlcNAc, and α2–6 linked sialylglycopeptide (58) were provided in-house. Protein A Sepharose 4 Fast Flow beads (17-5280-01) were from GE Healthcare. Anti-CD14 microbeads (130-050-201) were from Miltenyi Biotec GmbH. Sialidase S (GK80021) and sialidase C (GK80030) were from Prozyme.
Purification and quantification of HBV.
HBV was purified from blood samples of patients with CHB and HBV-transgenic mice (generated in-house; ref. 59). The protocol for HBV purification was followed as described previously (60–62). HBV viral load was determined by RT-PCR according to a standard HBV plasmid provided in-house. HBsAg level was quantified by the HBsAg ELISA kit (General Biological).
Identification of glycan structure on HBsAg by nano-LC-MS/MS.
The analysis of glycan structure on HBsAg was performed as described previously (63, 64). In brief, purified HBV from patients with CHB and transgenic mice was fractionated on 12% SDS-PAGE, and the gel was cut into pieces to separate various forms of HBsAg followed by trypsin and chymotrypsin digestion. High-resolution and high-mass accuracy nano-flow LC-MS/MS experiments were performed on an LTQFT Ultra (linear quadrupole ion trap Fourier transform ion cyclotron resonance) mass spectrometer (Thermo Electron) equipped with a nano-electrospray ion source (New Objective, Inc.), an Agilent 1100 Series binary HPLC pump (Agilent Technologies), and a Famos autosampler (LC Packings). The digestion solution was injected (6 μL) at 10 μL/min flow rate onto a self-packed precolumn (150 μm inner diameter [I.D.] × 20 mm, 5 μm, 100 Å). Chromatographic separation was performed on a self-packed reversed-phase C18 nano-column (75 μm I.D. × 300 mm, 5 μm, 100 Å) using 0.1% formic acid in water as mobile phase A and 0.1% formic acid in 80% acetonitrile as mobile phase B operated at 300 nL/min flow rate. The survey full-scan MS condition was mass range m/z 320–2000 and resolution 50,000 at m/z 400.The 5 most intense ions were sequentially isolated for MS2 by LTQ. Electrospray voltage was maintained at 1.8 kV and capillary temperature was set at 200°C.
Removal of sialic acid from HBV and sialoglycan by sialidase.
Sialidase S and sialidase C were used to remove terminal sialic acid from HBV or sialoglycans according to the manufacturers’ instructions. In brief, HBV virions (quantified by BSA method) were incubated with sialidase S or sialidase C under an appropriate dose at 37°C for 1 hour in reaction buffer according to the manufacturer’s instructions. The sialidase-treated glycan substrates were used for the HBV–SIGLEC-3 binding assay and competition assay.
Interactions between HBV and SIGLEC.Fc fusion proteins.
HBV virion (109 copies/mL) was incubated with recombinant SIGLEC-3/-7/-9-ECD.Fc for 16 hours at 4°C followed by incubation with Sepharose-conjugated protein A beads for IP. The samples were fractionated on 12% SDS-PAGE before blotting to PVDF membrane and incubated in blocking buffer (0.05% Tween 20 in TBS containing 5% BSA) at 4°C overnight. Blots were incubated with HRP-conjugated goat-anti-HBsAg Ab (BS-1557G-HRP) for 1 hour at room temperature (RT) followed by incubation with streptavidin-HRP (SIGMA 1:10,000) for another hour at RT, and signals were developed by ECL.
Competition assay.
SIGLEC-ECD.Fc fusion proteins (5 μg) were coated on microtiter plates followed by the addition of hHBV or mHBV in the presence of various concentrations of the following glycans: Neu5Ac(α2-6)-Gal(β1-4)-GlcNAc, Neu5Ac(α2-3)-Gal(β1-4)-GlcNAc, and Gal(β1-4)-GlcNAc at 4°C for 12 hours with protein-free blocking buffer at RT. The sialidase-treated and untreated sialoglycans were used for the competition assay as described previously (65). After washing with PBST, HBV associated with SIGLEC-ECD.Fc was detected by addition of rabbit anti-HBsAg antibody (RT, 2 hours) and HRP-conjugated anti-rabbit antibody (RT, 2 hours) followed by coloration reaction using tetramethylbenzidine (TMB) (BD Pharmingen) as substrate and 2N H2SO4 as stop solution. The result was determined by using a microplate reader set to 450 nm.
Determination of receptor-virus interaction by bio-layer interferometry.
The interactions between human SIGLEC.Fc fusion protein and HBV were determined by bio-layer interferometry (ForteBio) (66). In brief, the biosensor (ForteBio) was immobilized with the SIGLEC.Fc, and the binding between the SIGLEC.Fc and HBV was performed with running buffer (10 mM Tris·HCl, 150 mM NaCl, 2 mM CaCl2, 2 mM MgCl2, pH 7.4) at RT. The valence between HBV and human SIGLEC-3 was determined by the models proposed by Gargano et al. (41) and Liang et al. (67), which correlate the strength of multivalent association with that of monovalent association as shown in the following: KA, surf = F (S × 10–2)n–1× (KA, mono)n, where KA, surf is the reciprocal of KD (HBV and SIGLEC-3); KA, mono is the reciprocal of KD (biantennary α2–6 linked sialoglycopeptide and SIGLEC-3); n is the number of valency; F is the statistic factor in the system; and S is equal to 40(Å), which was estimated by Chwan-Deng Hsiao according to the crystal structure of SIGLEC-3 published on Research Collaboratory for Structural Bioinformatics (RCSB) Protein Data Bank (number: 5J0B). We used 3 individual bio-layer interferometry results to calculate the n (number of valency).
Generation and characterization of anti–SIGLEC-3 mAbs and selection of antagonistic antibody.
Anti–SIGLEC-3 mAbs were produced from phage-displayed synthetic antibody libraries as described previously (68). The generic human antibody was constructed based on combining the functional single-chain variable fragment (scFv) variants with human IgG (provided by Tse-Wen Chang, Genomics Research Center, Academia Sinica) and produced by the FreeStyle 293 Expression System. The effect of anti–SIGLEC-3 mAb in mast cells by the TNP-LP–induced degranulation method was performed according to the previous method (43).
Cell culture.
PBMCs were isolated from the whole blood of healthy human donors and CHB patients by standard density-gradient centrifugation with Ficoll-Paque (Amersham Biosciences). In brief, whole blood from donors supplied with anticoagulants (EDTA or acid citrate dextrose (ACD) was diluted with PBS and carefully transferred (25 mL) into a 50 mL conical tube with 12.5 mL Ficoll-Paque. After centrifugation in a swinging bucket without a break (760 g, 20 minutes), the mononuclear cell layer was harvested and washed with PBS.
The culture of moDCs was carried out as described previously (69, 70). In brief, CD14+ cells were purified from PBMCs by high-gradient magnetic sorting using the VarioMACS technique with anti-CD14 microbeads (Miltenyi Biotec GmbH). Cells were then incubated in complete RPMI 1640 medium with 10% FCS (Hyclone) supplemented with human GM-CSF/IL-4 for 6–7 days to generate moDCs. The efficiency of moDC maturation was more than 95% (Supplemental Figure 9).
To test the inhibitory effect of HBV, moDCs (6 × 104 per well) were incubated with HBV virion in different concentrations for 24 hours before the addition of Pam3csk4 (0.05 μg/mL) or poly (I:C) (100 μg/mL) and IFN-γ (100 ng/mL) for another 24 hours. Supernatants were collected to determine cytokine levels by ELISA kits (R&D Systems and Thermo Fisher Scientific) according to the manufacturers’ instructions. To identify the antagonistic anti–SIGLEC-3 mAbs, healthy donor moDCs (6 × 104 per well) and CHB patient PBMCs (6 × 104 per well) were incubated with anti–SIGLEC-3 mAb for 1 hour followed by adding HBV virion for 24 hours before the addition of Pam3csk4 (0.05 μg/mL) or poly (I:C) (100 μg/mL) and IFN-γ (100 ng/mL) for another 24 hours. Supernatants were collected to determine cytokine levels by ELISA kits (R&D Systems and Thermo Fisher Scientific) according to the manufacturers’ instructions. In brief, the capture antibody was coated on microtiter plates for 12 hours at RT followed by the addition of supernatant in reagent diluent (RT, 2 hours). Then an addition of detection antibody (RT, 2 hours) followed by streptavidin-HRP (RT, 20 minutes) and coloration reaction using TMB (BD Pharmingen) as substrate and 2N H2SO4 as stop solution. The result was determined by using a microplate reader set to 450 nm.
Endotoxin removal from antibodies using Triton X-114.
The endotoxin removal protocol was based on a previous protocol with some modifications (71). Triton X-114 (Sigma-Aldrich, 93422) was added to the protein solution to a final Triton X-114 concentration of 1% v/v. After incubating at 4°C for 30 minutes, the sample was transferred to a water bath set at 37°C and incubated for 5 minutes followed by centrifugation at 13,000 g for 10 minutes at 37°C. The upper part containing the protein was collected, and the extraction procedure was repeated 1 time. To remove Triton X-114 in the protein sample, Bio-Beads SM-2 (Bio-Rad, 1528920) were added to the collected supernatant and incubated for 1 hour at 4°C with constant stirring. Using sedimentation, the Bio-Beads were removed from the protein. The endotoxin level in protein was determined by the LAL test (Thermo Fisher Scientific, 88282).
Detection of cell surface marker by flow cytometry.
PBMCs (1 × 106) from CHB patients and healthy donors were treated with LPS-free anti–SIGLEC-3 mAb (10C8), anti-PD-L1 (atezolizumab, Roche), and hIgG1 control (3 μg/mL) for 1 hour followed by stimulation with GS-9620 (10 nM or 30 nM). The supernatant and cells were collected at 24 hours. Cell surface markers were measured by flow cytometry, and cytokine production levels were measured by ELISA kits (R&D Systems and Thermo Fisher Scientific). To detect SIGLEC-3, human PBMCs from healthy donors or CHB patients were isolated by Ficoll gradient following the manufacturer’s instructions (Amersham). Cells were stained with antibodies listed in Supplemental Table 7. All flow cytometry data were acquired using a FACSVerse flow cytometer and analyzed using FlowJo software (Tree Star).
Confocal microscopy.
PBMCs (2 × 105 cells) isolated from CHB patients were incubated with or without anti–SIGLEC-3 mAb (3 μg/mL, 10C8) on ice. Cells were washed and fixed at 4°C for 1 hour followed by incubating in permeability buffer (0.5% Triton X-100 in PBS) for 15 hours and blocking buffer (3% BSA) at RT for 1 hour before addition of primary antibodies. After incubation at 4°C for 24 hours, cells were incubated with secondary antibodies at RT for another hour followed by the addition of phalloidin (1 unit/μL) and Hoechst 33342 for 10 minutes before observation under a Leica (Wetzlar) SP5 confocal microscope.
FRET and fluorescence lifetime imaging.
The processes of seeding, fixation, permeabilization, and blocking of PBMCs isolated from patients with CHB are described above in Confocal Microscopy. SIGLEC-3 (specified as “donor”) was incubated with primary antibody (NBP2-32819) at 4°C for 24 hours followed by incubation with donkey anti-mouse (H+L) Alexa Fluor 647–conjugated secondary antibody (ab150107) at RT for 1 hour. HBsAg (specified as “acceptor”) was incubated with primary antibody 100 μL of 10 μg/mL (BS-1557G) at 4°C for 24 hours followed by the addition of donkey anti-goat (H+L) Alexa Fluor 546–conjugated secondary antibody (A11056) at RT for 1 hour. After staining, cells were washed with PBS and resuspended in mounting solution and covered by coverslips. For fluorescence lifetime imaging (FLIM) and FRET analysis, FLIM was recorded on a Leica SP5 confocal microscope. Fluorescence lifetimes were measured in cells expressing only the FRET donor and cells expressing the combination of FRET donor and acceptor. Samples were pulsed with a laser (488 nm) and emitted wavelength (500 to 550 nm) was collected; 10,000 photons were recorded for each sample. FRET efficiency (E) was calculated according to the equation: E = 1–(τDA/τD), where τDA is the mean fluorescence lifetime of cells coexpressing FRET donor and acceptor and τD is the mean fluorescence lifetime of cells expressing FRET donor only (72).
Detection of SIGLEC-3–associated SHP-1 and SHP-2 in HBV-treated moDCs.
MoDCs were incubated with HBV with or without the addition of 10C8 followed by resuspension in lysis buffer before incubation with anti–SIGLEC-3 antibody. Samples were immunoprecipitated by using protein A–conjugated Sepharose, and the immunoprecipitates were fractionated on SDS–PAGE before transfer to PVDF membrane and probed with anti-SHP-1 and SHP-2 antibody. Immunoblots were developed by incubating with HRP-conjugated anti-rabbit IgG antiserum and enhanced chemiluminescence detection reagents (Amersham). To detect the total amount of SIGLEC-3, the blot was stripped with Re-Blot Plus Strong solution (Chemicon, 2504) before being probed with mouse anti–SIGLEC-3 antibody.
Genotyping of SIGLEC-3.
Serum samples of CHB patients (200 μL) were applied for DNA extraction using the QIAamp DNA blood mini kit (QIAGEN). DNA concentration was determined using a Nanodrop 2000 spectrophotometer (Thermo Fisher Scientific). About 10–20 ng of DNA was subject to genotyping analysis. Genotyping for rs12459419 and rs3865444 of SIGLEC-3 was carried out by TaqMan technology (Thermo Fisher Scientific) using a predesign-specific probe (assay ID C_1487394_10 and C_1487395_40, respectively) and TaqMan Genotyping Master Mix following standard protocol. Amplification and fluorescent signal detection were performed using ABI 7500HT or QuantStudio 5 (Thermo Fisher Scientific) in a 384-plate format with a condition of 95°C for 10 minutes, 50 cycles at 95°C for 15 seconds, 60°C for 1 minute. Genotyping results were analyzed using TaqMan Genotyper Software (Thermo Fisher Scientific, version 1.5.0).
Statistics.
Experimental values are expressed as mean ± SD. All experiments were repeated at least 3 times, and the results were evaluated by the paired sample t test, 1-way ANOVA, and 2-way ANOVA using the Prism software package (GraphPad version 5.00). A P value of less than 0.05 was regarded as significant. Continuous variables were compared between 2 groups using the ANOVA test, labeled as “A” in Supplemental Table 4, and presented as the median ± IQR. Categorical variables were analyzed using the χ2 test, labeled as “C” in Supplemental Table 4. Cox proportional hazard regression analysis was used to identify factors associated with HCC incidence. SAS version 9.4 was used for statistical analyses.
Study approval.
This study was conducted under the Helsinki Declaration of 1975. All patients provided written informed consent before enrollment, and the study was approved by the Research Ethics Committee of China Medical University Hospital, Taichung (CMUH104-REC2-159), and the Research Ethics Committee of Academia Sinica, Taipei (AS-IRB01-15055, AS-IRB-BM-15017).
Author contributions
TYT, as first author, designed and performed experiments (HBV viral purification, glycosylation of HBV by MS, ELISA and Western blot setup for the HBV-SIGLEC-3 binding assay, anti–SIGLEC-3 mAb production and functional testing, experiments of HBV interactions with moDCs under TLR stimulation and antibody treatment, confocal and FRET analysis, patient collection and SNP analysis), and wrote the manuscript. MTH, as co–first author, performed experiments (analysis of cell surface marker and cytokine secretion in PBMC under antibody treatment and stratified by SNP, confocal imaging of SIGLEC and HBV, characterization of SIGLEC-3 expression in PBMCs and its binding with HBV), analyzed data, and checked the manuscript. PSS performed experiments and data analysis. CYP, CDH, and YHY analyzed data. MHT provided HBV transgenic mice antibodies. WCC, HIY, CYB, CJH, and MHP performed SIGLEC-3 SNP analysis. ASY and CMY produced anti-SIGLEC mAbs. YPL performed glycan structure analysis of hepatitis B surface antigen. CYW did synthesis of sialoglycans. SD and JP characterized anti–SIGLEC-3 mAbs. SLH designed experiments, did data interpretation, and wrote the manuscript.
Supplementary Material
Acknowledgments
This study was supported by Academic Sinica. We thank Tse-Wen Chang for providing the human IgG backbone to generate anti-SIGLEC-3 mAbs, and Kuo-Yao Tseng for bio-layer interferometry analysis. We are grateful to GRC Mass Core Facility for help with MS analysis and the High-throughput Screening Facility for kinetic study using the Octet HTX system. We thank Eileen Press and Karl Normington for review of the manuscript and SIGLEC-3 genotype discussions. We would like to thank Pei-Jer Chen for inspirational discussion and suggestions for this work. We are also grateful to the deceased Ding-Shinn Chen who kindly supported all aspects of this work. This work was supported by Academia Sinica (107-2101-01-18-03, AS-IA-109-L02), Translational Medical Research Program (AS-TM-108-02-10), and Biotechnology Research Park Translational Project (AS-BRPT-110-02).
Version 1. 06/01/2021
Electronic publication
Funding Statement
By both China Medical University and Academia Sinica
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2021, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2021;131(11):e141965.https://doi.org/10.1172/JCI141965.
Contributor Information
Tsung-Yu Tsai, Email: d18101@mail.cmuh.org.tw.
Ming-Ting Huang, Email: cyanobacteria08@gmail.com.
Pei-Shan Sung, Email: pei_jon77120@hotmail.com.
Cheng-Yuan Peng, Email: cypeng@mail.cmuh.org.tw.
Mi-Hua Tao, Email: bmtao@ibms.sinica.edu.tw.
Hwai-I Yang, Email: hiyang@gate.sinica.edu.tw.
Wei-Chiao Chang, Email: wcc@tmu.edu.tw.
An-Suei Yang, Email: yangas@gate.sinica.edu.tw.
Chung-Ming Yu, Email: yucmbuu@gate.sinica.edu.tw.
Ya-Ping Lin, Email: yaping65@gate.sinica.edu.tw.
Ching-Yu Bau, Email: u100011304@gmail.com.
Chih-Jen Huang, Email: tw.cj.huang@gmail.com.
Mei-Hung Pan, Email: panmaegan@gmail.com.
Chung-Yi Wu, Email: cyiwu@gate.sinica.edu.tw.
Chwan-Deng Hsiao, Email: hsiao@gate.sinica.edu.tw.
Yi-Hung Yeh, Email: yyh4625@gate.sinica.edu.tw.
Shiteng Duan, Email: andrew_dst@hotmail.com.
Shie-Liang Hsieh, Email: slhsieh@gate.sinica.edu.tw.
References
- 1.Rehermann B, Nascimbeni M. Immunology of hepatitis B virus and hepatitis C virus infection. Nat Rev Immunol. 2005;5(3):215–229. doi: 10.1038/nri1573. [DOI] [PubMed] [Google Scholar]
- 2.Liaw YF, Chu CM. Hepatitis B virus infection. Lancet. 2009;373(9663):582–592. doi: 10.1016/S0140-6736(09)60207-5. [DOI] [PubMed] [Google Scholar]
- 3.Liaw YF. Natural history of chronic hepatitis B virus infection and long-term outcome under treatment. Liver Int. 2009;29(Suppl 1):100–107. doi: 10.1111/j.1478-3231.2008.01941.x. [DOI] [PubMed] [Google Scholar]
- 4.Schwarz KB, et al. Phenotypes of chronic hepatitis B in children from a large North American cohort. J Pediatr Gastroenterol Nutr. 2019;69(5):588–594. doi: 10.1097/MPG.0000000000002467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liang TJ. Hepatitis B: the virus and disease. Hepatology. 2009;49(5 suppl):S13–S21. doi: 10.1002/hep.22881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marcellin P, et al. Tenofovir disoproxil fumarate versus adefovir dipivoxil for chronic hepatitis B. N Engl J Med. 2008;359(23):2442–2455. doi: 10.1056/NEJMoa0802878. [DOI] [PubMed] [Google Scholar]
- 7.Chang TT, et al. A comparison of entecavir and lamivudine for HBeAg-positive chronic hepatitis B. N Engl J Med. 2006;354(10):1001–1010. doi: 10.1056/NEJMoa051285. [DOI] [PubMed] [Google Scholar]
- 8.Lai CL, et al. Entecavir versus lamivudine for patients with HBeAg-negative chronic hepatitis B. N Engl J Med. 2006;354(10):1011–1020. doi: 10.1056/NEJMoa051287. [DOI] [PubMed] [Google Scholar]
- 9.El-Khoueiry AB, et al. Nivolumab in patients with advanced hepatocellular carcinoma (CheckMate 040): an open-label, non-comparative, phase 1/2 dose escalation and expansion trial. Lancet. 2017;389(10088):2492–2502. doi: 10.1016/S0140-6736(17)31046-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gane E, et al. Anti-PD-1 blockade with nivolumab with and without therapeutic vaccination for virally suppressed chronic hepatitis B: A pilot study. J Hepatol. 2019;71(5):900–907. doi: 10.1016/j.jhep.2019.06.028. [DOI] [PubMed] [Google Scholar]
- 11.Zhang X, et al. Hepatitis B virus reactivation in cancer patients with positive Hepatitis B surface antigen undergoing PD-1 inhibition. J Immunother Cancer. 2019;7(1):322. doi: 10.1186/s40425-019-0808-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shi B, et al. HBsAg inhibits IFN-α production in plasmacytoid dendritic cells through TNF-α and IL-10 induction in monocytes. PLoS One. 2012;7(9):e44900. doi: 10.1371/journal.pone.0044900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Op den Brouw ML, et al. Hepatitis B virus surface antigen impairs myeloid dendritic cell function: a possible immune escape mechanism of hepatitis B virus. Immunology. 2009;126(2):280–289. doi: 10.1111/j.1365-2567.2008.02896.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Visvanathan K, et al. Regulation of Toll-like receptor-2 expression in chronic hepatitis B by the precore protein. Hepatology. 2007;45(1):102–110. doi: 10.1002/hep.21482. [DOI] [PubMed] [Google Scholar]
- 15.Lanford RE, et al. GS-9620, an oral agonist of Toll-like receptor-7, induces prolonged suppression of hepatitis B virus in chronically infected chimpanzees. Gastroenterology. 2013;144(7):1508–1517. doi: 10.1053/j.gastro.2013.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Niu C, et al. Toll-like receptor 7 agonist GS-9620 induces prolonged inhibition of HBV via a type I interferon-dependent mechanism. J Hepatol. 2018;68(5):922–931. doi: 10.1016/j.jhep.2017.12.007. [DOI] [PubMed] [Google Scholar]
- 17.Agarwal K, et al. Safety and efficacy of vesatolimod (GS-9620) in patients with chronic hepatitis B who are not currently on antiviral treatment. J Viral Hepat. 2018;25(11):1331–1340. doi: 10.1111/jvh.12942. [DOI] [PubMed] [Google Scholar]
- 18.Varki A. Colloquium paper: uniquely human evolution of sialic acid genetics and biology. Proc Natl Acad Sci U S A. 2010;107(Suppl 2):8939–8946. doi: 10.1073/pnas.0914634107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Appenheimer MM, et al. Biologic contribution of P1 promoter-mediated expression of ST6Gal I sialyltransferase. Glycobiology. 2003;13(8):591–600. doi: 10.1093/glycob/cwg066. [DOI] [PubMed] [Google Scholar]
- 20.Schmitt S, et al. Structure of pre-S2 N- and O-linked glycans in surface proteins from different genotypes of hepatitis B virus. J Gen Virol. 2004;85(pt 7):2045–2053. doi: 10.1099/vir.0.79932-0. [DOI] [PubMed] [Google Scholar]
- 21.Schmitt S, et al. Analysis of the pre-S2 N- and O-linked glycans of the M surface protein from human hepatitis B virus. J Biol Chem. 1999;274(17):11945–11957. doi: 10.1074/jbc.274.17.11945. [DOI] [PubMed] [Google Scholar]
- 22.Macauley MS, et al. Siglec-mediated regulation of immune cell function in disease. Nat Rev Immunol. 2014;14(10):653–666. doi: 10.1038/nri3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Taylor VC, et al. The myeloid-specific sialic acid-binding receptor, CD33, associates with the protein-tyrosine phosphatases, SHP-1 and SHP-2. J Biol Chem. 1999;274(17):11505–11512. doi: 10.1074/jbc.274.17.11505. [DOI] [PubMed] [Google Scholar]
- 24.Crocker PR, et al. Siglecs and their roles in the immune system. Nat Rev Immunol. 2007;7(4):255–266. doi: 10.1038/nri2056. [DOI] [PubMed] [Google Scholar]
- 25.Chemnitz JM, et al. SHP-1 and SHP-2 associate with immunoreceptor tyrosine-based switch motif of programmed death 1 upon primary human T cell stimulation, but only receptor ligation prevents T cell activation. J Immunol. 2004;173(2):945–954. doi: 10.4049/jimmunol.173.2.945. [DOI] [PubMed] [Google Scholar]
- 26.Marengere LE, et al. Regulation of T cell receptor signaling by tyrosine phosphatase SYP association with CTLA-4. Science. 1996;272(5265):1170–1173. doi: 10.1126/science.272.5265.1170. [DOI] [PubMed] [Google Scholar]
- 27.Adams OJ, et al. Targeting sialic acid-Siglec interactions to reverse immune suppression in cancer. Glycobiology. 2018;28(9):640–647. doi: 10.1093/glycob/cwx108. [DOI] [PubMed] [Google Scholar]
- 28.Callahan MK, Wolchok JD. At the bedside: CTLA-4- and PD-1-blocking antibodies in cancer immunotherapy. J Leukoc Biol. 2013;94(1):41–53. doi: 10.1189/jlb.1212631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hao C, et al. Efficacy and safety of anti-PD-1 and anti-PD-1 combined with anti-CTLA-4 immunotherapy to advanced melanoma: a systematic review and meta-analysis of randomized controlled trials. Medicine (Baltimore) 2017;96(26):e7325. doi: 10.1097/MD.0000000000007325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chae YK, et al. Current landscape and future of dual anti-CTLA4 and PD-1/PD-L1 blockade immunotherapy in cancer; lessons learned from clinical trials with melanoma and non-small cell lung cancer (NSCLC) J Immunother Cancer. 2018;(1):39. doi: 10.1186/s40425-018-0349-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tanvetyanon T, et al. PD-1 checkpoint blockade alone or combined PD-1 and CTLA-4 blockade as immunotherapy for lung cancer? Expert Opin Biol Ther. 2017;17(3):305–312. doi: 10.1080/14712598.2017.1280454. [DOI] [PubMed] [Google Scholar]
- 32.Estus S, et al. Evaluation of CD33 as a genetic risk factor for Alzheimer’s disease. Acta Neuropathol. 2019;138(2):187–199. doi: 10.1007/s00401-019-02000-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ishii T, et al. Influence of SIGLEC9 polymorphisms on COPD phenotypes including exacerbation frequency. Respirology. 2017;22(4):684–690. doi: 10.1111/resp.12952. [DOI] [PubMed] [Google Scholar]
- 34.Angata T. Associations of genetic polymorphisms of Siglecs with human diseases. Glycobiology. 2014;24(9):785–793. doi: 10.1093/glycob/cwu043. [DOI] [PubMed] [Google Scholar]
- 35.Chen GY, et al. Broad and direct interaction between TLR and Siglec families of pattern recognition receptors and its regulation by Neu1. Elife. 2014;3:e04066. doi: 10.7554/eLife.04066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pillai S, et al. Siglecs and immune regulation. Annu Rev Immunol. 2012;30:357–392. doi: 10.1146/annurev-immunol-020711-075018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lamba JK, et al. CD33 splicing polymorphism determines Gemtuzumab Ozogamicin response in de novo acute myeloid leukemia: report from randomized phase III children’s oncology group trial AAML0531. J Clin Oncol. 2017;35(23):2674–2682. doi: 10.1200/JCO.2016.71.2513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen C-H, et al. Rapid identification of terminal sialic acid linkage isomers by pseudo-MS3 mass spectrometry. Isar J Chem. 2015;55(3–4):412–422. doi: 10.1002/ijch.201400141. [DOI] [Google Scholar]
- 39.Vanlandschoot P, et al. LPS-binding protein and CD14-dependent attachment of hepatitis B surface antigen to monocytes is determined by the phospholipid moiety of the particles. J Gen Virol. 2002;83(Pt 9):2279–2289. doi: 10.1099/0022-1317-83-9-2279. [DOI] [PubMed] [Google Scholar]
- 40.Huang A, et al. Myeloid-derived suppressor cells regulate immune response in patients with chronic hepatitis B virus infection through PD-1-induced IL-10. J Immunol. 2014;193(11):5461–5469. doi: 10.4049/jimmunol.1400849. [DOI] [PubMed] [Google Scholar]
- 41.Gargano JM, et al. Multivalent inhibition of AB(5) toxins. J Am Chem Soc. 2001;123(51):12909–12910. doi: 10.1021/ja016305a. [DOI] [PubMed] [Google Scholar]
- 42.Hsu HJ, et al. Assessing computational amino acid beta-turn propensities with a phage-displayed combinatorial library and directed evolution. Structure. 2006;14(10):1499–1510. doi: 10.1016/j.str.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 43.Duan S, et al. CD33 recruitment inhibits IgE-mediated anaphylaxis and desensitizes mast cells to allergen. J Clin Invest. 2019;129(3):1387–1401. doi: 10.1172/JCI125456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Malik M, et al. CD33 Alzheimer’s risk-altering polymorphism, CD33 expression, and exon 2 splicing. J Neurosci. 2013;33(33):13320–13325. doi: 10.1523/JNEUROSCI.1224-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Raj T, et al. CD33: increased inclusion of exon 2 implicates the Ig V-set domain in Alzheimer’s disease susceptibility. Hum Mol Genet. 2014;23(10):2729–2736. doi: 10.1093/hmg/ddt666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Diminsky D, et al. Comparison between hepatitis B surface antigen (HBsAg) particles derived from mammalian cells (CHO) and yeast cells (Hansenula polymorpha): composition, structure and immunogenicity. Vaccine. 1997;15(6–7):637–647. doi: 10.1016/s0264-410x(96)00239-3. [DOI] [PubMed] [Google Scholar]
- 47.Lee EU, et al. Alteration of terminal glycosylation sequences on N-linked oligosaccharides of Chinese hamster ovary cells by expression of beta-galactoside alpha 2,6-sialyltransferase. J Biol Chem. 1989;264(23):13848–13855. doi: 10.1016/S0021-9258(18)80078-3. [DOI] [PubMed] [Google Scholar]
- 48.Kramvis A, et al. Hepatitis B virus genotypes. Vaccine. 2005;23(19):2409–2423. doi: 10.1016/j.vaccine.2004.10.045. [DOI] [PubMed] [Google Scholar]
- 49.Kao JH. Hepatitis B virus genotypes and hepatocellular carcinoma in Taiwan. Intervirology. 2003;46(6):400–407. doi: 10.1159/000074999. [DOI] [PubMed] [Google Scholar]
- 50.Lin CL, Kao JH. Hepatitis B virus genotypes and variants. Cold Spring Harb Perspect Med. 2015;5(5):a021436. doi: 10.1101/cshperspect.a021436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tatematsu K, et al. A genetic variant of hepatitis B virus divergent from known human and ape genotypes isolated from a Japanese patient and provisionally assigned to new genotype J. J Virol. 2009;83(20):10538–10547. doi: 10.1128/JVI.00462-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Janssen HLA, et al. Safety, efficacy and pharmacodynamics of vesatolimod (GS-9620) in virally suppressed patients with chronic hepatitis B. J Hepatol. 2018;68(3):431–440. doi: 10.1016/j.jhep.2017.10.027. [DOI] [PubMed] [Google Scholar]
- 53.Tu T, et al. Chronic viral hepatitis and its association with liver cancer. Biol Chem. 2017;398(8):817–837. doi: 10.1515/hsz-2017-0118. [DOI] [PubMed] [Google Scholar]
- 54.Lim CJ, et al. Multidimensional analyses reveal distinct immune microenvironment in hepatitis B virus-related hepatocellular carcinoma. Gut. 2019;68(5):916–927. doi: 10.1136/gutjnl-2018-316510. [DOI] [PubMed] [Google Scholar]
- 55.Huang LR, et al. An immunocompetent mouse model for the tolerance of human chronic hepatitis B virus infection. Proc Natl Acad Sci U S A. 2006;103(47):17862–17867. doi: 10.1073/pnas.0608578103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kim MY, et al. Genetic inactivation of CD33 in hematopoietic stem cells to enable CAR T cell immunotherapy for acute myeloid leukemia. Cell. 2018;173(6):1439–1453. doi: 10.1016/j.cell.2018.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen CJ, Yang HI. Natural history of chronic hepatitis B REVEALed. J Gastroenterol Hepatol. 2011;26(4):628–638. doi: 10.1111/j.1440-1746.2011.06695.x. [DOI] [PubMed] [Google Scholar]
- 58.Liu L, et al. Improved isolation and characterization procedure of sialylglycopeptide from egg yolk powder. Carbohydr Res. 2017;452:122–128. doi: 10.1016/j.carres.2017.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chen CC, et al. Long-term inhibition of hepatitis B virus in transgenic mice by double-stranded adeno-associated virus 8-delivered short hairpin RNA. Gene Ther. 2007;14(1):11–19. doi: 10.1038/sj.gt.3302846. [DOI] [PubMed] [Google Scholar]
- 60.Tsuge M, et al. Infection of human hepatocyte chimeric mouse with genetically engineered hepatitis B virus. Hepatology. 2005;42(5):1046–1054. doi: 10.1002/hep.20892. [DOI] [PubMed] [Google Scholar]
- 61.Kim CY, Tilles JG. Purification and biophysical characterization of hepatitis B antigen. J Clin Invest. 1973;52(5):1176–1186. doi: 10.1172/JCI107284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cruceanu A, et al. A simple procedure for purification of hepatitis B-antigen. Med Microbiol Immunol. 1973;159(1):83–88. doi: 10.1007/BF02122652. [DOI] [PubMed] [Google Scholar]
- 63.Yu CY, et al. Automated glycan sequencing from tandem mass spectra of N-linked glycopeptides. Anal Chem. 2016;88(11):5725–5732. doi: 10.1021/acs.analchem.5b04858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Madsen JA, et al. Concurrent automated sequencing of the glycan and peptide portions of O-linked glycopeptide anions by ultraviolet photodissociation mass spectrometry. Anal Chem. 2013;85(19):9253–9261. doi: 10.1021/ac4021177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Brinkman-Van der Linden EC, Varki A. New aspects of siglec binding specificities, including the significance of fucosylation and of the sialyl-Tn epitope. Sialic acid-binding immunoglobulin superfamily lectins. J Biol Chem. 2000;275(12):8625–8632. doi: 10.1074/jbc.275.12.8625. [DOI] [PubMed] [Google Scholar]
- 66.Lo YL, et al. Dengue virus infection is through a cooperative interaction between a mannose receptor and CLEC5A on macrophage as a multivalent hetero-complex. PLoS One. 2016;11(11):e0166474. doi: 10.1371/journal.pone.0166474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Liang PH, et al. Quantitative microarray analysis of intact glycolipid-CD1d interaction and correlation with cell-based cytokine production. J Am Chem Soc. 2008;130(37):12348–12354. doi: 10.1021/ja8012787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chen IC, et al. High throughput discovery of influenza virus neutralizing antibodies from phage-displayed synthetic antibody libraries. Sci Rep. 2017;7(1):14455. doi: 10.1038/s41598-017-14823-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.You RI, et al. Apoptosis of dendritic cells induced by decoy receptor 3 (DcR3) Blood. 2008;111(3):1480–1488. doi: 10.1182/blood-2007-09-114850. [DOI] [PubMed] [Google Scholar]
- 70.Hsu TL, et al. Modulation of dendritic cell differentiation and maturation by decoy receptor 3. J Immunol. 2002;168(10):4846–4853. doi: 10.4049/jimmunol.168.10.4846. [DOI] [PubMed] [Google Scholar]
- 71.Teodorowicz M, et al. Optimized Triton X-114 assisted lipopolysaccharide (LPS) removal method reveals the immunomodulatory effect of food proteins. PLoS One. 2017;12(3):e0173778. doi: 10.1371/journal.pone.0173778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wallrabe H, Periasamy A. Imaging protein molecules using FRET and FLIM microscopy. Curr Opin Biotechnol. 2005;16(1):19–27. doi: 10.1016/j.copbio.2004.12.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.