Abstract
With increasing age, individuals are more vulnerable to viral infections such as with influenza or the SARS-CoV-2 virus. One age-associated defect in human T cells is the reduced expression of miR-181a. miR-181ab1 deficiency in peripheral murine T cells causes delayed viral clearance after infection, resembling human immune aging. Here we show that naive T cells from older individuals as well as miR-181ab1–deficient murine T cells develop excessive replication stress after activation, due to reduced histone expression and delayed S-phase cell cycle progression. Reduced histone expression was caused by the miR-181a target SIRT1 that directly repressed transcription of histone genes by binding to their promoters and reducing histone acetylation. Inhibition of SIRT1 activity or SIRT1 silencing increased histone expression, restored cell cycle progression, diminished the replication-stress response, and reduced the production of inflammatory mediators in replicating T cells from old individuals. Correspondingly, treatment with SIRT1 inhibitors improved viral clearance in mice with miR-181a–deficient T cells after LCMV infection. In conclusion, SIRT1 inhibition may be beneficial to treat systemic viral infection in older individuals by targeting antigen-specific T cells that develop replication stress due to miR-181a deficiency.
Keywords: Aging
Keywords: Adaptive immunity, Cell cycle, Cellular senescence
Introduction
Life expectancy has dramatically increased over the last decades, with vaccinations preventing infections being a major contributing factor to this success of modern medicine. However, immune competence declines with aging, as evidenced by the increased susceptibility of old individuals to previously encountered as well as new pathogens (1). Moreover, in the elderly, the benefits of vaccination to prevent infectious disease are limited (2). In spite of annual vaccinations, a large number of deaths is associated with influenza infection. Ninety percent of influenza-associated deaths from respiratory or cardiovascular complications occur in persons 65 years of age or older (3). The current SARS-CoV-2 pandemic is a global health threat in particular for the elderly population (4). With the changing age demographics of the population, healthy immune aging is therefore of paramount importance.
The aging immune system has to cope with a decreased ability to produce new T cells in the thymus as well as with T cell–intrinsic changes that develop as a consequence of homeostatic T cell proliferation and stimulation by external cues. Homeostatic proliferation is mostly able to maintain a naive CD4+ T cell compartment that, in spite of moderate repertoire contraction, is sufficiently large and diverse (5), while naive CD8+ T cells are disproportionately lost and the CD8+ T cell compartment is shifted toward clonally expanded effector populations (6, 7). T cell–intrinsic changes are of particular interest because they offer the possibility of identifying means to compensate for the age-associated defects in adaptive immunity (8). Many of the age-associated T cell–intrinsic alterations are related to changes in microRNA expression. As one example, aged T cells are characterized by a lower expression of miR-181a and an increased expression of miR-21, a pattern that is also seen with T cell differentiation, suggesting that T cell aging at least in part involves regular differentiation pathways (9, 10). The increased expression of miR-21 induces the inhibition of negative feedback loops involving several signaling pathways, thereby causing degradation of FOXO1 and sustained mTORC1 signaling (10, 11). As a consequence, aged T cells preferentially differentiate into short-lived effector T cells, while development of T follicular helper and long-lived memory T cells is impaired. miR-181a is a rheostat of T cell receptor (TCR) activation by controlling the expression of several phosphatases, including PTPN22, SHP2, DUSP5, and DUSP6 (12). Due to increased DUSP6, naive CD4+ T cells from the elderly have reduced extracellular signal–related kinase (ERK) phosphorylation upon TCR stimulation (9). Mice with conditional deletion of miR-181ab1 in peripheral T cells exhibit features of human immune aging, including impaired expansion of antigen-specific T cells in antiviral responses and delayed viral clearance (13). Improving TCR signaling through DUSP6 silencing was not sufficient to repair T cell responses (13), suggesting additional miR-181a–mediated mechanisms.
Here, we found that miR-181a deficiency in proliferating T cells from old individuals as well as from conditional knockout mice reduced histone upregulation in the early S phase and thereby induced replication stress. Reduced histone expression was due to the miR-181a target SIRT1 that is overexpressed in miR-181a–deficient murine T cells and in aged human T cells and recruited to histone gene promoters. Inhibition of SIRT1 activity or reducing SIRT1 expression diminished the replication-stress response in in vitro studies of T cells from old adults as well as viral clearance in vivo in mice with miR-181ab1–deficient T cells.
Results
miR-181a deficiency impairs histone expression and cell cycle progression in murine antiviral responses.
In previous studies, we have developed a mouse model of conditionally deleted miR-181ab1 in peripheral T cells that exhibited defective antiviral T cell responses as they occur in humans with T cell aging (13). When cotransferring congenically marked wild-type (WT) (distal Lck-Cre+ Rosa26YFP miR-181ab1+/+) and miR-181a–/– (distal Lck-Cre+ Rosa26YFP miR-181ab1fl/fl) SMARTA CD4+ T cells (expressing a TCR specific for the LCMV glycoprotein 61–80 epitope) into C57BL/6J (B6) recipient mice, we found a reduced expansion of miR-181a–deficient SMARTA cells in response to LCMV infection (ref. 13 and Figure 1A). Apoptotic rates of SMARTA cells on day 7 after infection were similar in both mouse strains (Supplemental Figure 1; supplemental material available online with this article; https://doi.org/10.1172/JCI143632DS1). To identify the molecular basis for the defective T cell expansion, we performed RNA sequencing (RNA-seq) on WT and miR-181a–/– SMARTA T cells on day 7 after infection and found differential expression of 725 genes, 344 of which were more highly expressed in WT cells (adjusted P < 0.05). Gene set enrichment analysis (GSEA) of differentially expressed genes showed a significant enrichment of the cell cycle gene signature in WT SMARTA cells (Figure 1B). Notably, miR-181a deficiency globally reduced core histone gene expression in LCMV-responding SMARTA cells (Figure 1C). Reduced histone H2A, H2B, H3, and H4 protein levels in miR-181a–/– SMARTA cells were confirmed by Western blotting (Figure 1D). Reduced histone expression was only seen for activated proliferating T cells, while no difference was observed for unstimulated cells (Supplemental Figure 2).
Figure 1. miR-181a deficiency impairs histone expression and cell cycle progression in murine antiviral responses.
(A–E) Equal numbers of congenically marked WT and miR-181a–/– YFP+ SMARTA cells were cotransferred into B6 mice before LCMV infection. (A) Experimental scheme (left), representative flow plots of WT and miR-181a–/– SMARTA cells (middle), and SMARTA frequencies (right, mean ± SEM). (B and C) RNA-seq of WT and miR-181a–/– SMARTA CD4+ T cells on day 7 after LCMV infection. (B) GSEA of cell cycle gene signature in WT relative to miR-181a–/– SMARTA cells. (C) Volcano plot of core histone genes (red indicates adjusted P < 0.05). (D) Immunoblotting for histones in WT and miR-181a–/– SMARTA cells on day 7 after LCMV infection. (E) On day 5 after LCMV infection, recipient mice received BrdU for 1 hour prior to spleen harvest. Representative flow plots of BrdU incorporation and DNA content and summarized frequencies (mean ± SEM). (F and G) WT and miR-181a–/– mice infected with LCMV were injected with BrdU. (F) Number of Db LCMV GP33-tetramer+ CD8+ T cells (mean ± SEM). (G) Representative flow plots of BrdU incorporation and DNA content in tetramer+ CD8+ T cells (left) and summary of frequencies (right, mean ± SEM). (H) GSEA of the p53-induced gene set in WT relative to miR-181a–/– SMARTA cells. (I) Immunoblots for p53 on day 7 SMARTA cells and summary data of mean normalized intensities. (J) Immunoblots of SMARTA cells on day 7 after LCMV infection. Data are representative of 3 experiments with 3 to 5 mice per group (A and D), 1 experiment with 4 mice per group (B, C, and H), or representative of 2 experiments with 2 to 6 mice per group (E–G, I, and J). Comparison by 2-tailed, paired (A, E, and I) or unpaired Student’s t test (F and G). *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. NS, not significant.
Histone gene expression is cell cycle dependent and upregulated during DNA replication in the S phase (14). Reduced histone expression in miR-181a–deficient T cells may therefore reflect low numbers of S-phase cycling cells. Conversely, reduced histone expression slows cell cycle progression through the S phase (15, 16). To examine these possibilities, LCMV-infected mice that had received WT and miR-181a–/– SMARTA cells were pulsed with the nucleoside analog 5-bromo-2′-deoxyuridine (BrdU) for 1 hour. Cell cycle analysis determined by BrdU incorporation and DNA content revealed that miR-181a–deficient SMARTA cells significantly accumulated in the early S phase of the cell cycle, when compared with WT SMARTA cells (Figure 1E). Proliferation of miR-181a–/– CD8+ T cells in response to LCMV infection was also impaired as compared with WT CD8+ T cells. On day 7 after infection, activated CD8+ T cells expressing CD44 were reduced (Supplemental Figure 3A), as were CD8+ T cells recognizing the GP33 epitope of LCMV (Figure 1F). Also, LCMV-specific miR-181a–/– CD8+ T cells accumulated in the early S phase of the cell cycle, similar to the observation for CD4+ T cells (Supplemental Figure 3B and Figure 1G). Proliferative rates were different for LCMV-specific CD4+ and CD8+ T cells and depended on the day after infection; however, accumulation in the early S phase was consistent.
Further analysis of the transcriptome showed that the gene set induced by p53 was enriched in miR-181a–/– compared with WT SMARTA cells (Figure 1H), indicating a DNA damage response. Indeed, miR-181a–/– SMARTA cells had higher levels of p53 (Figure 1I), the DNA damage marker phosphorylated H2aX (γH2aX), and higher expression of the cell cycle inhibitor p21 (Figure 1J). Increased phosphorylation of CHK1 and RPA32 indicated increased ATR (ataxia telangiectasia and Rad3-related) activity, consistent with replication stress elicited by the stalled cell cycle progression and resulting in the activation of the p53 pathway.
Human proliferating naive CD4+ T cells of old adults have reduced histone expression and prolonged cell cycle S phase.
Because miR-181a expression declines with age in human naive CD4+ T cells (9), we examined whether reduced histone expression is also a feature of T cell proliferative responses of older individuals. Naive CD4+ T cells from healthy young (20–35 years) and old (65–85 years) adults were activated for 5 days with beads coated with anti-CD3 and anti-CD28 antibodies. Transcriptome analysis by RNA-seq revealed that activated old naive CD4+ T cells tended to have globally lower expression of core histone genes than young T cells (Figure 2A), recapitulating the pattern in miR-181a–deficient T cells responding to LCMV infection (Figure 1C). Quantification of histone transcripts (Figure 2B) and Western blot analysis of H2A, H2B, H3, and H4 (Figure 2C) in sorted cycling cells confirmed a significant reduction of core histones in cycling old T cells. In contrast, no difference in H3 and H4 protein expression was seen for unstimulated naive CD4+ T cells from young and old adults, suggesting that the difference was dependent on proliferation (Supplemental Figure 4). Flow cytometric analysis of histone proteins and DNA content showed the cell cycle–dependent change in histone expression, with histone H4 protein progressively increasing during S-phase progression and reaching the highest levels during G2/M phase (Figure 2D). In these flow cytometric studies, activated naive CD4+ T cells from old adults had lower histone protein levels in the S phase than those from young adults, while levels in the G2/M phase of the cell cycle were similar, demonstrating a selective defect in histone synthesis during DNA replication (Figure 2E). Moreover, day 5 activated naive CD4+ T cells showed an accumulation of aged T cells in the S phase, as compared with the young (Figure 2F). To examine cell cycle progression, naive CD4+ T cells from young and old individuals were activated for 5 days and pulsed with BrdU for 1 hour before analysis. Costaining of BrdU incorporation and DNA content revealed that cycling old T cells accumulated in the early S phase (Figure 2G). Consistent with the murine data, apoptotic rates were not different (Supplemental Figure 5A). However, in contrast, this increase in early-S-phase cells in proliferative T cell responses from old adults was not associated with decreased cell recovery, suggesting additional differences in cell cycle regulation (Supplemental Figure 5B). Indeed, we have previously described increased mTORC1 activation and increased c-MYC expression in T cell responses of older adults that may accelerate G1/S progression (10, 11). Taken together, these results show that aged human naive CD4+ T cells proliferating in vitro exhibit the same phenotype as miR-181a–deficient murine T cells responding in vivo to LCMV infection.
Figure 2. Human proliferating naive CD4+ T cells of old adults have reduced histone expression and prolonged cell cycle S phases.
Naive CD4+ T cells from healthy young (20- to 35-year-old) and old (65- to 85-year-old) individuals were activated with anti-CD3/anti-CD28 beads for 5 days. (A) Volcano plot of core histone gene expression from RNA-seq data (SRA: SRP158502). Red dots indicate significance (P < 0.05). (B) Live S- and G2/M-phase cycling cells were sorted based on DNA content. Core histone transcripts were measured by qRT-PCR. Results are presented relative to cycling young cells (n = 8, mean ± SEM). Expression levels of all histone genes are significantly different (P < 0.05), except that of HIST1H3A (P = 0.13) and HIST1H4B (P = 0.05), after correction for multiple comparisons. (C) Immunoblotting of histones in cycling young (Y) and old (O) cells (n = 4). (D) Gating strategy of cell cycle stages based on histone H4 and DNA content. (E) Histograms of histone H4 gated on the S- or G2/M-phase cells (left) and summary graphs from 8 experiments with 1 young and 1 old individual each (right). (F) Representative histograms (left) and summary graphs (right) of frequencies of G0/G1-, S-, and G2/M-phase cells from 10 young and 9 old individuals (mean ± SEM). (G) Representative flow plots of BrdU incorporation after 1-hour pulsing and of DNA content (left) and summary data of early and late S-phase cell frequencies (right; n = 8–9, mean ± SEM). (H–J) Day 5 activated naive CD4+ T cells were pulsed with EdU for 2 hours, followed by BrdU for 1 hour. (H) Representative flow plots of BrdU and EdU incorporation, (I) summary graphs of frequencies, and (J) percentages of BrdU–EdU+ S-exit cells among EdU+ cells (n = 9–10, mean ± SEM). Comparisons by 2-tailed, unpaired (B, F, G, I, and J) or paired (E) Student’s t test. **P < 0.01, ***P < 0.001. NS, not significant.
In primary T cell responses, cell cycle progression cannot be synchronized by removal of growth factors. To at least in part circumvent this limitation in cell cycle analysis, we used a sequential labeling approach; activated cells were first incubated with the nucleoside analog 5-ethynyl-2′-deoxyuridine (EdU) for 2 hours, followed by BrdU for 1 hour. Given that BrdU outcompetes EdU in DNA incorporation (17), cells just entering S phase are labeled as BrdU+EdU–, cells still in S phase are BrdU+EdU+, and S-exit cells are BrdU–EdU+ (refs. 18, 19, and Figure 2H). Cycling old T cells showed an accelerated entry into the S phase (Figure 2, H and I), corresponding to our previous finding of a sustained activation of AKT/mTORC1 signaling in activated old compared with young naive CD4+ T cells (10). As with miR-181a–deficient T cells in the conditional knockout mice, cycling old human T cells had a prolonged S phase and a significant delay in progression into G2/M phase compared with young cells, as shown by their significant accumulation of cells still in S phase (Figure 2I) and a reduced percentage of EdU-labeled cells that had progressed to the G2/M phase (Figure 2J).
Reduced histone expression promotes replication stress.
To determine whether reducing histone expression in young T cells reproduces the alteration in cell cycle progression seen with T cells from older adults, we targeted the transcription factor NPAT that controls core histone gene expression (20). Lentiviral transduction of shRNA targeting NPAT inhibited expression of NPAT and consequently downregulated histone expression at the transcript and protein level (Figure 3, A–C). Similar results were obtained with a second shNPAT construct (Supplemental Figure 6A). Although complete knockout of NPAT has been shown to arrest cells in the G1/S phase (21), reduced histone expression to the degree observed in old proliferating T cells did not affect the S-phase entry, but arrested T cells in early S phase, thereby extending the S phase (Figure 3, D and E). To determine whether the delay in cell cycle progression increased replication stress, we examined the activation of the ATR pathway. Indeed, reducing histone expression in activated young CD4+ T cells via NPAT silencing increased phosphorylation of RPA32 and CHK1. Downstream events, including the levels of the DNA damage marker γH2aX and the expression of the cell cycle inhibitor p21, were upregulated (Figure 3F and Supplemental Figure 6B).
Figure 3. Reduced histone expression promotes replication stress.
Naive CD4+ T cells from young adults were activated with anti-CD3/anti-CD28 beads and transduced with control (shCtrl) or NPAT (shNPAT) shRNA lentivirus for 6 days. (A) Immunoblots for NPAT in lentivirally transduced GFP+ cells and summary data of normalized intensities from 4 young adults (mean ± SEM). (B) Expression of indicated histone genes in sorted shRNA+ cells. Results, normalized to ACTB, are presented relative to shCtrl+ cells from 6 young adults (mean ± SEM). Differences in expression of all histone genes are statistically significant (P < 0.01), except for HIST1H1B (P = 0.03), HIST1H3B (P = 0.06), and HIST1H3D (P = 0.03), after correction for multiple comparisons using Holm’s step-down adjustment. (C) Immunoblots for histone H3 and H4 from shRNA+ cells and summary data of normalized intensities from 6 young adults (mean ± SEM). (D) Activated cells were pulsed with BrdU for 1 hour. Representative flow plots of BrdU incorporation and DNA content and summary data of frequencies (n = 5, mean). (E) Activated cells were pulsed with EdU for 2 hours, followed by BrdU for 1 hour. Percentage of BrdU–EdU+ S-exit cells among EdU+ cells from 3 young adults (mean). (F) Immunoblotting for p-RPA32 (S8), p-CHK1 (S345), γH2aX (S139), and p21 on sorted shRNA+ cells (left) and summary graphs of mean normalized intensities from 6 young adults (right, mean ± SEM). Comparisons by 2-tailed, paired Student’s t test (A–F). *P < 0.05, **P < 0.01.
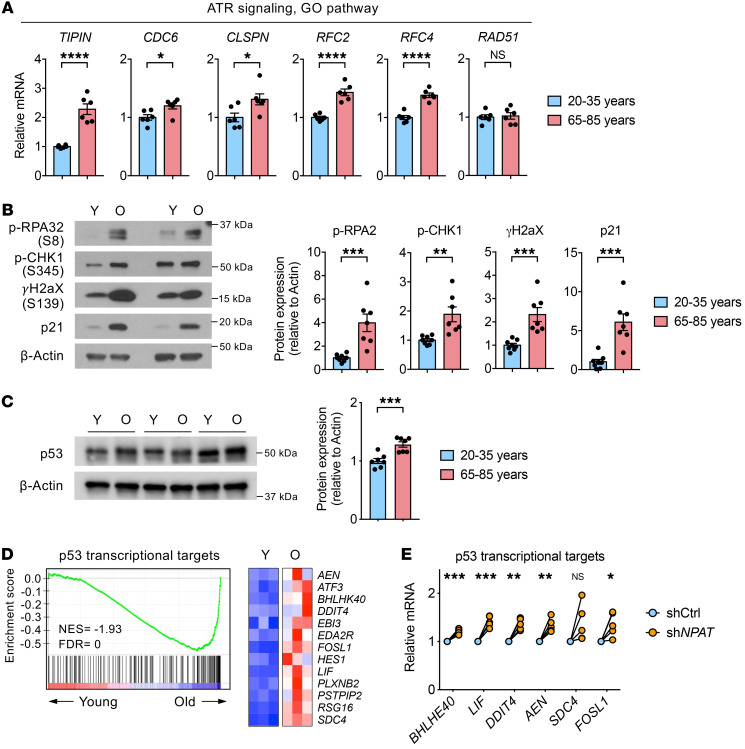
These data prompted us to examine whether cycling CD4+ T cells from old individuals have increased replication stress. Indeed, genes involved in the ATR signaling pathway were upregulated in cycling old compared with young T cells (Figure 4A); moreover, RPA32 and CHK1 were more phosphorylated (Figure 4B). Consistent with CHK1 activation, we also observed increases in γH2aX and p21 (Figure 4B). Taken together, these data demonstrated that reduced histone expression in cycling old T cells stalls cell cycle S-phase progression and thereby causes a replication-stress response.
Figure 4. Proliferating old CD4+ T cells have increased replication stress.
(A–C) Naive CD4+ T cells from young and old individuals were activated for 5 days. Cycling cells were sorted based on DNA content. (A) Quantitative RT-PCR of indicated transcripts associated with ATR signaling. Results, normalized to ACTB, are presented for old relative to cycling young cells (n = 6, mean ± SEM). (B) Immunoblot for p-RPA32 (S8), p-CHK1 (S345), γH2aX (S139), and p21 and summary graphs of normalized intensities from 8 young (Y) and 7 old (O) individuals (mean ± SEM). (C) Immunoblot for p53 and summary graph (n = 7, mean ± SEM). (D) GSEA of p53 transcriptional targets in activated old naive CD4+ T cells relative to their expression in young cells (left) and heatmap of selected genes from RNA-seq data (right) (SRA: SRP158502). (E) Naive CD4+ T cells from young adults were activated and transduced with shCtrl or shNPAT lentivirus for 6 days. Expression of indicated p53 target genes was determined in transduced cells by quantitative RT-PCR. Results, normalized to ACTB, are presented relative to shCtrl+ cells (n = 5). Comparisons by 2-tailed, unpaired (A–C) or paired Student’s t test (E), with correction for multiple comparisons using Holm’s step-down adjustment in E. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. NS, not significant.
Under replication stress, the tumor suppressor p53 is phosphorylated and activated by several kinases including ATR and CHK1 (22, 23), leading to transcriptional activation of genes involved in cell cycle control, apoptosis, and cellular senescence. Moreover, recent studies have implicated p53 directly in replication-stress responses. p53 has been shown to bind to stalled replication forks and is required to restart replication (24). We found that the expression of p53 was increased in cycling naive CD4+ T cells from old compared with young adults (Figure 4C). Consistent with p53 activation in replication stress, GSEA of genes differentially expressed on day 5 activated naive CD4+ T cells from young and old adults showed a significant enrichment of p53 transcriptional targets in old CD4+ T cells (Figure 4D). To determine whether reduced histone expression induces activation of the p53 pathway, we quantified transcripts of 6 p53 target genes in proliferating young naive CD4+ T cells that had been partially silenced for NPAT expression (Figure 4E). Compared with control-transduced CD4+ T cells, all of these transcripts were upregulated, suggesting that reduced histone expression at least in part accounts for the p53 signature in old proliferating T cells.
Collectively, these data indicate that reduced histone expression in activated old naive CD4+ T cells results in a prolongation of the S phase and an accumulation of cells in the early S phase, which promotes increased replication stress and activation of the p53 pathway.
Age-related increase in SIRT1 expression represses histone gene transcription.
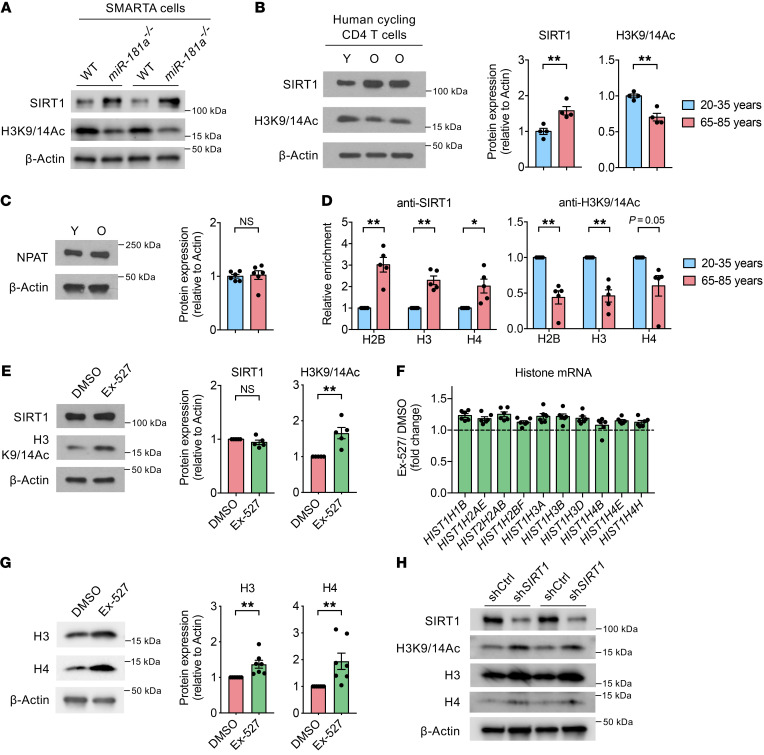
To identify the mechanism underlying reduced histone expression in proliferating old CD4+ T cells, we focused on miR-181a target genes, given that murine miR-181a–deficient T cells responding to LCMV infection recapitulated the findings in humans. SIRT1, which is an NAD+-dependent histone deacetylase and one of the direct targets of miR-181a (25), is known to promote transcriptional repression by deacetylating histone H3K9/14Ac and H4K16Ac (26). miR-181a–deficient SMARTA CD4+ T cells responding to LCMV had higher SIRT1 expression and lower amounts of histone H3K9/14Ac than WT cells (Figure 5A). Consistent with the mouse model of miR-181a deficiency, expression of SIRT1 is increased in resting naive CD4+ T cells from older individuals (27) and continues to be higher after T cell activation (Figure 5B). Accordingly, cycling old CD4+ T cells had globally lower amounts of acetylated histone H3K9/14Ac, indicating that overexpressed SIRT1 is functionally active (Figure 5B). Previous studies have shown an NPAT-dependent recruitment of SIRT1 to the histone gene promoters (28, 29). Expression of NPAT did not differ between young and old T cells (Figure 5C). However, CUT&RUN assays showed increased binding of SIRT1 and a concomitant decrease in H3K9/14Ac levels at histone gene promoters (Figure 5D). Pharmacological inhibition of SIRT1 activity with Ex-527 increased H3K9/14Ac levels without altering SIRT1 expression (Figure 5E) and thereby upregulated both histone transcripts and protein expression in T cells from old adults (Figure 5, F and G). A similar effect was seen with SIRT1 silencing (Figure 5H). A significant effect of SIRT1 inhibition was not seen in T cells from young adults (Supplemental Figure 7A). These data indicate that SIRT1 overexpression directly repressed histone gene expression in cycling old CD4+ T cells.
Figure 5. miR-181a–dependent increase in SIRT1 expression represses histone gene transcription.
(A) Equal numbers of congenically marked WT and miR-181a–/– YFP+ SMARTA cells were cotransferred into B6 mice before infection with LCMV. Immunoblots for SIRT1 and histone H3-K9/14 acetylation (H3K9/14Ac) of splenic SMARTA cells on day 7, representative of 3 experiments with 2 to 3 mice per group. (B–D) Human young (Y) and old (O) naive CD4+ T cells were activated for 5 days. Immunoblots for SIRT1 and histone H3K9/14Ac (B) and NPAT (C) in cycling cells (left); summary graphs of normalized intensities from 4–6 young and 4–6 old individuals (right, mean ± SEM). (D) CUT&RUN assay of SIRT1 binding and histone H3K9/14Ac enrichment at indicated histone gene promoters from 5 experiments with 1 young and 1 old individual each. Results are presented relative to cycling young cells (mean ± SEM). (E–G) Naive CD4+ T cells from old individuals were activated for 5 days. DMSO or Ex-527 was added on day 2. (E) Immunoblots of SIRT1 and histone H3K9/14Ac in cycling cells and summary data from 5 old adults (mean ± SEM). (F) Expression of indicated histone genes in cycling cells was determined by quantitative RT-PCR. Results are presented relative to DMSO-treated cells (n = 6, mean ± SEM). Differences in expression of all histone genes are statistically significant (P < 0.01), except for HIST1H4B (P = 0.25), after correction for multiple comparisons using Holm’s step-down adjustment. (G) Immunoblots of histones in cycling cells and summary data from 7 old adults (mean ± SEM). (H) Naive CD4+ T cells from old adults were activated with anti-CD3/anti-CD28 beads and transduced with control (shCtrl) or SIRT1 (shSIRT1) shRNA lentivirus for 6 days. Immunoblots for indicated proteins in shRNA+ cells (n = 2). Comparisons by 2-tailed, unpaired (B and C) or paired Student’s t test (D–G). *P < 0.05, **P < 0.01. NS, not significant.
SIRT1 inhibition in replicating old human T cells restores cell cycle progression and diminishes the replication-stress response.
We next examined whether SIRT1 inhibition restores the naive CD4+ T cell response of old individuals. Enhancing histone expression through SIRT1 inhibition increased the BrdU–EdU+ S-exit cell population in activated old CD4+ T cells, indicating accelerated cell cycle progression through the S phase (Figure 6A). With normal cell cycle kinetics restored, replication stress was diminished, as documented by reduced phosphorylated RPA32 and CHK1 levels and less extensive upregulation of γH2aX and p21 expression in T cells from old adults than in untreated cells (Figure 6B). In contrast to T cells from old adults, markers of replication stress were generally low in young activated T cells and did not show a consistent pattern upon treatment with the SIRT1 inhibitor (Figure 4B and Supplemental Figure 7B). Similar to pharmacological SIRT1 inhibition, partial silencing of SIRT1 expression reduced replication stress in T cell responses of old adults (Figure 6C). Also, SIRT1 inhibition in proliferating old naive CD4+ T cells reduced expression of p53 and its transcriptional targets (Figure 6, D and E). SIRT1 inhibition did not change apoptotic rates (Supplemental Figure 8), but ultimately improved the recovery rate of CD4+ T cells from old adults in the culture system (Figure 6F). As expected, inhibition of ATR in T cell responses from old adults accelerated S-phase exit and increased S to G2/M transition (Supplemental Figure 9A), but at the expense of vastly increased apoptotic rates (Supplemental Figure 9B).
Figure 6. SIRT1 inhibition in replicating old human T cells restores cell cycle progression and diminishes the replication-stress response.
(A and B) Naive CD4+ T cells from old individuals were activated for 5 days. DMSO or Ex-527 was added on day 2. (A) Day 5 activated cells were pulsed with EdU for 2 hours, followed by BrdU for 1 hour. Representative flow plots of BrdU and EdU incorporation (left) and summary graphs of BrdU–EdU+ cell frequencies and percentages of BrdU–EdU+ S-exit cells among EdU+ cells (right; n = 7, mean). (B) Immunoblots of indicated proteins in cycling cells from 6 old adults (mean ± SEM). (C) Naive CD4+ T cells from old adults were activated with anti-CD3/anti-CD28 beads and transduced with control (shCtrl) or SIRT1 (shSIRT1) shRNA lentivirus for 6 days. Immunoblots for indicated proteins in shRNA+ cells (n = 3). (D) Immunoblots for p53 in cycling cells in experiments with 3 old adults. (E) Expression of indicated p53 transcriptional target genes in cycling old cells was determined by quantitative RT-PCR. Results are presented relative to DMSO-treated cells (n = 5). (F) Number of activated naive CD4+ T cells from 9 old adults recovered on day 5 (mean). Comparisons by 2-tailed, paired Student’s t test (A, B, E, and F), with correction for multiple comparisons using Holm’s step-down adjustment in E. *P < 0.05, **P < 0.01, ***P < 0.001.
Replication stress accounts for the activation of proinflammatory pathways in CD4+ T cell responses in old adults.
The transcriptome of naive CD4+ T cells from old adults exhibited enrichment for the gene sets characteristic of cellular senescence and senescence-associated secretory phenotype (SASP) on day 5 after in vitro stimulation (Figure 7A). This gene signature was not observed in naive resting CD4+ T cells from old adults, and thus was dependent on the proliferative response (Supplemental Figure 10). To determine whether this inflammatory profile was related to the reduced histone expression and the associated replication stress, we examined the effect of NPAT silencing on the expression of selected SASP-related inflammatory mediators. Reducing histone expression in young activated CD4+ T cells increased the expression of IRF5, IRF7, CSF1, CSF2, and CCL3 (Figure 7B). Conversely, treatment of naive CD4+ T cell cultures from old adults with the SIRT1 inhibitor attenuated the transcription of these mediators (Figure 7C). However, further studies showed that more definitive markers of cellular senescence were lacking. While p21 was clearly upregulated (Figure 4B), p16 was very low in cultured cells and not different between young and old adults (Figure 7D), consistent with the observation that activated old CD4+ T cells were not in irreversible growth arrest that is typical of cellular senescence. The only human T cells that have appreciable amounts of p16 are growth-arrested terminally differentiated Temra cells (Figure 7D). β-Galactosidase activity was increased in activated old compared with young CD4+ T cells (Figure 7E). However, this increase could not be reproduced by NPAT silencing (Figure 7F) nor could it be repaired by SIRT1 inhibition (Figure 7G). Taken together, these results show that replication stress induces an inflammatory signature, but the associated DNA damage responses are not sufficient to cause cellular senescence.
Figure 7. Replication stress accounts for the activation of proinflammatory pathways in CD4+ T cell responses in old adults.
(A) GSEA of gene signatures of cellular senescence and SASP in activated old (O) naive CD4+ T cells relative to their expression in young (Y) cells (left) and heatmaps of selected genes from RNA-seq data (right; SRA: SRP158502). (B and C) Expression of indicated genes associated with inflammatory mediators was determined by quantitative RT-PCR in shCtrl+ or shNPAT+ cells (B) and in DMSO- or Ex-527–treated cells (C). Results, normalized to ACTB, are presented relative to shCtrl+ or DMSO-treated cells, respectively (n = 5). (D) Immunoblots for p16 in activated, proliferating naive CD4+ T cells from 3 young and 3 old adults. CD45RA+CCR7– terminally differentiated effector memory cells (CD8+ Temra) are included as positive control for p16 expression. (E) Naive CD4+ T cells from young and old individuals were activated for 8 days. Histograms of green fluorescence, indicative of senescence-associated β-galactosidase activity (left), and summary graph of geometric MFI (right) from 7 young and 7 old individuals (mean ± SEM). The filled gray histogram represents no substrate. (F) Histograms of senescence-associated β-galactosidase activity on shCtrl+ or shNPAT+ cells from 1 young adult. (G) Histograms of senescence-associated β-galactosidase activity in DMSO- or Ex-527–treated cells and summary graph from 3 old adults (mean). Comparisons by 2-tailed, paired (B, C, and G) or unpaired Student’s t test (E), with correction for multiple comparisons using Holm’s step-down adjustment in B and C. *P < 0.05, **P < 0.01, ***P < 0.001. NS, not significant.
SIRT1 inhibition augments in vivo antigen-specific T cell responses.
Given that SIRT1 inhibition restored alterations in proliferative T cell responses of older individuals, we next assessed the physiologic impact of SIRT1 inhibition in vivo. We transferred small numbers of WT or miR-181a–/– SMARTA CD4+ T cells into B6 recipient mice and infected them with LCMV 1 day later. Starting the day after the infection, infected mice were treated with daily Ex-527 or left untreated. Cell cycle analysis on day 5 after infection showed that SIRT1 inhibition resolved the accumulation of miR-181a–deficient SMARTA cells in the early S phase, while not changing the cell cycle kinetics of WT SMARTA cells (Figure 8A). Evidence of replication stress was low in WT cells irrespective of treatment. In contrast, miR-181a–deficient SMARTA cells had an increased replication-stress response that was improved with SIRT1 inhibition (Figure 8B). Consequently, SIRT1 inhibition improved proliferation of miR-181a–/– but not WT LCMV-specific CD4+ T cells (Figure 8C). Similar results were obtained for CD8+ T cell responses, as determined by tetramer staining of CD8+ T cells specific for the GP33 epitope. While no treatment effect was seen in WT mice, SIRT1 inhibition improved the expansion of GP33-specific miR-181a–/– CD8+ T cells (Figure 8D and Supplemental Figure 11). The restored cycling behavior was functionally important; viral clearance, impaired in mice with miR-181a–/– T cells compared with WT mice, was improved in the treated miR-181a–/– mice (Figure 8E). To exclude off-target effects of the SIRT1 inhibitor Ex-527, adoptive transfer experiments were performed with miR-181a–/– SMARTA cells transduced with shSirt1 constructs. Silencing resulted in an approximately 50% reduction in SIRT1 protein and increased H3K9/14Ac (Figure 8F). Sirt1 silencing reduced the percentage of miR-181a–deficient SMARTA cells in the early S phase (Figure 8G), reduced replication-stress responses (Figure 8H), and improved SMARTA cell recovery (Figure 8I).
Figure 8. SIRT1 inhibition augments antigen-specific T cell responses in vivo.
(A–C) WT or miR-181a–/– SMARTA cells were transferred into B6 hosts before infection with LCMV. DMSO or Ex-527 was given i.p. daily starting the day after the infection. On day 5 after infection, mice were injected with BrdU 1 hour prior to harvest. (A) Representative flow plots of BrdU incorporation and DNA content and summary data (mean ± SEM). (B) Immunoblots of SMARTA cells. (C) Number of SMARTA cells in the spleen (mean ± SEM). (D and E) WT and miR-181a–/– mice were infected with LCMV and given DMSO or Ex-527 daily. (D) Number of Db LCMV GP33-tetramer+ CD8+ T cells on day 7 (mean ± SEM). (E) Viral titers on day 6 after LCMV infection (mean ± SEM). (F–I) miR-181a–deficient SMARTA T cells were retrovirally transduced with either shCtrl or shSirt1. (F) Immunoblots of shRNA+ cells and summary graphs (n = 4, mean). (G) Sorted shCtrl+ or shSirt1+ miR-181a–deficient SMARTA cells were transferred into B6 hosts, followed by LCMV infection. On day 5, mice were injected with BrdU 1 hour prior to harvest. Representative flow plots of BrdU incorporation and DNA content and summary data (mean ± SEM). (H) Immunoblots of shRNA+ cells. (I) Number of shRNA+ SMARTA cells in the spleen (mean ± SEM). Data are pooled from 3 independent experiments with 6–10 mice per group (A and C), representative of 3 independent experiments with 2 mice per group (B), pooled from 2 experiments with 5–10 mice per group (D and E), or 1 experiment with 4–5 mice per group (F–I). Comparisons by 1-way ANOVA followed by Tukey’s post hoc test (A and C–E) and paired (F) or unpaired Student’s t test (G and I). *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. NS, not significant.
Discussion
The ability to cope with proliferative stress is at the core of a functional adaptive immune system that heavily depends on T cells expanding into effector and memory T cells upon the recognition of antigen. Here, we report that naive T cells from older individuals had excessive replication stress due to reduced histone expression in replicating cells and a delayed cell cycle progression through the S phase. Heightened replication-stress response activated the ATR pathway, thereby increasing p53 activity and inducing the production of inflammatory mediators in expanding T cells from older individuals. Reduced histone expression in the S phase of proliferating T cells was caused by the overexpression of the miR-181a target SIRT1 in miR-181a–deficient murine T cells and aged human T cells. SIRT1 directly repressed expression of histone genes by binding to their promoters and reducing histone acetylation. Inhibition of SIRT1 activity in replicating old human T cells increased histone expression, restored cell cycle progression, and diminished the replication-stress response. In an in vivo mouse model of T cell aging, SIRT1 inhibition increased expansion of miR-181a–deficient T cells in response to LCMV infection, improving viral clearance.
A loss in histones has been implicated in accelerating aging in several model systems (30–35). Decreased core histone proteins increased genomic instability and induced nucleosome loss and inappropriate gene transcription in budding yeast (36). Conversely, increasing histone expression extended the yeast life span (31). In contrast with these global histone aberrations, the reduced histone expression observed in the studies described here was tightly linked to cell replication, and the consequences for cell function were therefore different. The linkage to cycling is not surprising given that histone transcription is highly upregulated during the early S phase for packaging the newly replicated DNA into chromatin (14). Reduced histone synthesis during DNA replication results in a slow replication fork progression and cell cycle arrest (15, 16). Consistent with this histone function, cycling old T cells exhibited delayed cell cycle progression through the S phase, with an accumulation of cells in the early S phase. Moreover, reducing histone expression in young T cells altered cell cycle progression, reproducing the findings in proliferating old T cells.
In proliferating aged human naive T cells expressing lower amounts of miR-181a as well as in miR-181a–deficient LCMV-specific murine T cells, reduced upregulation of histone transcription induced a replication-stress response. In rapidly proliferating cells, such as T cells undergoing clonal expansion after antigenic stimulation, DNA replication generates aberrant replication forks with accumulation of single-stranded DNA, which intrinsically triggers the replication-stress response (37). At stressed replication forks, replication protein A (RPA) binds to single-stranded DNA and recruits ATR to ensure accurate genomic duplication, leading to activation of downstream kinase CHK1 (38) and phosphorylation of histone variant H2AX (γH2aX, ref. 39). These downstream markers of replication stress, induced by reducing histone expression in proliferating young T cells, were upregulated in T cell responses from older adults.
Controlling replication stress is important in the generation and survival of memory T cells. To avoid replication stress, memory precursor T cells slow down the cell division rate at the peak response after a period of rapid expansion (40). Moreover, central memory precursors cycle more slowly early after infection than short-lived effector T cells (41). Increased replication stress in proliferating antigen-specific T cells may explain recent findings on the influence of age on T cell responses after vaccination. Impaired vaccine response after vaccination with the live varicella zoster virus (VZV) vaccine strain in older individuals resulted from accelerated attrition of effector T cells after the peak response and therefore reduced generation of long-lived memory cells (42). The greater attrition after day 14 after vaccination was correlated with gene expression signatures on day 8 and day 14 VZV-specific T cells. Specifically, gene expression modules involved in cell cycle regulation, DNA replication, and DNA repair pathways were predictive for attenuated vaccination-induced increases in VZV-specific memory T cells (42), raising the possibility that increased replication stress is detrimental to cell survival.
Replication stress has been implicated in inducing cellular senescence. Activation of ATR kinase alone without DNA damage induces cellular senescence (43), demonstrating the direct causal link between replication-stress response and senescence. Importantly, activation of p53 by DNA damage kinases is involved in this process (43, 44), in part by inducing downstream targets including p21 and thereby inhibiting the cell cycle (45). Consistent with this notion, we found significant enrichment of gene signatures of p53 activation and production of inflammatory mediators in proliferating old compared with young T cells. However, replication stress was not sufficient to induce the classical hallmarks of cellular senescence, such as p16 expression or increased β-galactosidase activity.
Reduced histone expression was shared between replicating T cells of older individuals and miR-181a–deficient murine T cells responding to LCMV infection; therefore, this was likely caused by a miR-181a target. Subsequent studies identified SIRT1 as this target. Previous studies have shown that SIRT1 binds to the transcription factor NPAT that regulates histone expression (28). Indeed, we could identify SIRT1 recruitment to the histone gene promoters and demonstrate local deacetylation of H3K9/14Ac in replicating CD4+ T cells from old adults.
The finding that increased expression of SIRT1 accounted for the age-associated T cell defect was unexpected, because SIRT1 is generally considered a longevity gene. In yeast and worms, Sir2, the nonmammalian homolog of SIRT1, promotes organismal life span extension (46, 47). In rodents, a decline in SIRT1 levels with age in various tissues has been hypothesized to contribute to the aging process (48, 49). Moreover, increasing SIRT1 activity with resveratrol (50, 51) or with the NAD+ precursor nicotinamide riboside (52) has been shown to be beneficial. In contrast with many other tissues, SIRT1 expression increases in naive and memory T cell subsets with age, mostly due to a decline in miR-181a. An exception are terminally differentiated effector memory T cells (Temra), in which SIRT1 expression is decreased and SIRT1 deficiency is important for the high glycolytic activity and the effector functions of these cells (53).
SIRT1 is present in both the nucleus and the cytoplasm; it is involved in the regulation of many core biological processes through deacetylating a range of proteins. Functional consequences of SIRT1 activity or the lack thereof therefore highly depends on the setting and the cell type. Complete knockout of SIRT1 impaired antiviral responses in animal models, mostly by impairing metabolic function and autophagy in antigen-presenting cells (54, 55). Compared with this complete SIRT1 deficiency, differences in SIRT1 expression or activity were small in our studies and we did not see any adverse effects on T cell survival. Of note, we did not see any beneficial or harmful effects of SIRT1 inhibition in WT mice or CD4+ T cells from young adults, suggesting that the influence of SIRT1 on histone expression was only seen with elevated concentrations. However, we consistently saw increased replication stress in miR-181a–deficient T cells or CD4+ T cells from old adults that improved with SIRT1 inhibition. In theory, SIRT1 should deacetylate p53 (56–58) and therefore counterregulate the p53 activation induced by the replication stress. This may explain the protective effects described for hepatitis C infection (59). However, if this feedback loop exists in the setting of miR-181a deficiency and age, it was clearly not sufficient to prevent a p53 activation signature in the CD4+ T cell population.
In addition to causing replication stress in proliferating T cells as described here, high expression of SIRT1 in T cell responses has a dampening effect, mainly through its ability to deacetylate transcription factors that are involved in different cellular processes (60). In the setting of exhausted T cells, such as in hepatitis C infection, increased SIRT1 expression appears to be beneficial by restoring functionality (59). However, in general, increased expression of SIRT1 induces T cell anergy, in part by deacetylating c-Jun and impairing AP1 activity (61). It is therefore possible that the beneficial effects of SIRT1 inhibition in our studies include improved TCR signaling in addition to restoring histone upregulation. However, we saw the beneficial effects on T cell expansion when the SIRT1 inhibitor was added after TCR stimulation in vitro. Similarly, SIRT1 inhibition was only started 1 day after LCMV infection. Taken together, these results show that the major benefits of SIRT1 inhibition appear to derive from reducing replication stress by increasing the transcription of histones in replicating T cells.
Delayed viral clearance due to reduced expansion of antigen-specific T cells is seen with increasing age and accounts for the increased morbidity and mortality of older individuals from viral infection, such as with the West Nile fever virus and probably SARS-CoV-2 (4, 62). Similarly, reduced generation of memory T cells contributes to the impaired protection of old adults from vaccination (63). Inhibition of SIRT1 in the early stages of a viral infection or in the days after vaccination to reduce replication stress may be a useful intervention to improve T cell immunity. So far, efforts have focused on identifying SIRT1 activators to improve longevity, and SIRT1 inhibitors have not been widely explored. Only Ex-527 (Selisistat) has been tested in phase I studies of Huntington disease (64). While not efficacious, it was well tolerated, suggesting that SIRT1 inhibitors could be developed that are safe and improve the T cell response to a viral infection.
Methods
Human study population and cell isolation.
Peripheral blood samples were obtained from 16 healthy individuals who did not have a history of autoimmune disease, diabetes mellitus, renal disease, cardiovascular disease, or cancer except skin cancer. In addition, buffy coats and leukoreduction system (LRS) chambers from 112 blood or platelet donors were purchased from the Stanford Blood Center. These samples were deidentified except for whether donors were younger than 35 years or older than 65 years. Untouched CD4+ T cells were purified from peripheral blood or LRS chambers of healthy volunteers with a Human CD4+ T Cell Enrichment Kit (STEMCELL Technologies), followed by density gradient centrifugation using Lymphoprep (STEMCELL Technologies). Naive CD4+ T cells were further isolated by negative selection with anti-CD45RO magnetic beads (Miltenyi Biotec) or by using a Human Naive CD4+ T Cell Enrichment Kit (STEMCELL Technologies).
Human primary T cell culture.
Isolated T cells were activated with Dynabeads Human T-Activator CD3/CD28 (Thermo Fisher Scientific) in RPMI 1640 (Sigma-Aldrich) supplemented with 10% FBS and 100 U/mL penicillin and streptomycin (Thermo Fisher Scientific). To inhibit SIRT1 activity, the SIRT1 inhibitor Ex-527 (5 μM, R&D Systems) or vehicle (DMSO, Sigma-Aldrich) was added to the culture on day 2 after T cell activation. VE-822 (5 μM, Selleckchem) was used to inhibit ATR activity. For BrdU labeling of replicating cells, activated T cells were treated with 10 μM BrdU (Sigma-Aldrich) for 1 hour prior to analysis. For concomitant EdU labeling, activated T cells were treated with 10 μM EdU (Thermo Fisher Scientific) for 2 hours, followed by 10 μM BrdU for 1 hour prior to analysis.
Lentivirus production and transduction of human T cells.
To knock down NPAT, we used a lentiviral vector expressing NPAT shRNA (Dharmacon). SIRT1 shRNA (Dharmacon) was used for SIRT1 knockdown. Lentivirus was produced by transfection of a lentiviral vector, along with psPAX2 (plasmid 12260, Addgene) and pMD2.G (plasmid 12259, Addgene) expression vectors into HEK293T cells (ATCC, CRL-11268) using FuGENE (Promega). Lentiviral particles were collected 48 and 72 hours after transfection, filtered through a 0.45-μm syringe filter (Millipore), concentrated using PEG-it solution (System Biosciences), and titered on HEK293T cells. For lentiviral transduction, naive CD4+ T cells were activated with anti-CD3/anti-CD28 beads and transduced with lentivirus at a multiplicity of infection of 10 in the presence of 8 μg/mL polybrene (Sigma-Aldrich) and 10 U/mL human IL-2 (Peprotech). After 36 hours, activated cells were washed and cultured with 2 μg/mL soluble anti-CD28 (CD28.2, BD Biosciences) and 10 U/mL human IL-2 (Peprotech) on plates coated with 1 μg/mL anti-CD3 (CD3-2, Mabtech).
Mice, LCMV infection, retroviral transduction, and treatment.
Generation of miR-181ab1–deficient mice in peripheral T cells was described previously (13). Briefly, miR-181ab1fl/fl mice (gift from C.Z. Chen, Stanford University) (65) were crossed with Rosa26-YFP and distal Lck-Cre (dLck-Cre) mice (Jackson Laboratory) to generate conditional knockout of miR-181ab1 in mature T cells. SMARTA TCR-transgenic mice (gift from R. Ahmed, Emory University, Atlanta, Georgia, USA), with TCR specific for the LCMV glycoprotein 61–80 epitope presented by IAb, were crossed with dLck-Cre+ Rosa26YFP miR-181ab1fl/fl mice and fully backcrossed to C57BL/6J (B6, Jackson Laboratory) mice for at least 6 generations. dLck-Cre+ Rosa26YFP miR-181ab1+/+ mice were used as WT control. For adoptive cotransfer experiments of WT and miR-181a–/– SMARTA cells, CD4+ T cells were negatively isolated from the spleens of WT (CD45.1+) and miR-181a–/– (CD45.2+) SMARTA mice using a CD4 T Cell Isolation Kit (Miltenyi Biotec). Cells were then mixed at a 1:1 ratio, and a total of 1 × 104 YFP+ SMARTA CD4+ T cells were injected i.v. into B6 recipient mice 1 day prior to infection. The Armstrong strain of LCMV (gift from R. Ahmed) was grown in BHK cells and titered in Vero cells (R. Ahmed). Mice were infected i.p. at a dose of 2 × 105 plaque-forming units (PFU). Organs were homogenized, and LCMV titers were determined by plaque assay on Vero cells. All mice analyzed were infected at 8–10 weeks of age, and both sexes were included.
To silence SIRT1, Sirt1 shRNA (5′-TGCTGTTGACAGTGAGCGAAGGGTAATCAATACCTGTTTGTAGTGAAGCCACAGATGTACAAACAGGTATTGATTACCCTCTGCCTACTGCCTCGGA-3′) was cloned into an MSCV-based retroviral pLMPd-Ametrine vector for the expression of mir30-flanked shRNA (gift from Y.S. Choi, Seoul National University, Seoul, South Korea) (66). The pLMPd-Ametrine vector with mouse shCD19 (shCtrl) was used as a control. For retroviral transduction, virions were produced and collected after transfection of a vector into Plat-E cells (Cell Biolabs) and filtered through a 0.45-μm syringe filter (Millipore). miR-181a–deficient naive SMARTA CD4+ T cells were activated with anti-CD3 (145-2C11, Thermo Fisher Scientific) and anti-CD28 (37.51, Thermo Fisher Scientific) and transduced with retrovirus at 24 and 36 hours after stimulation. Retrovirally transduced miR-181a–deficient SMARTA CD4+ T cells (Ametrine+YFP+CD4+) were sorted, and 1 × 104 shRNA+ SMARTA cells were intravenously transferred into B6 hosts, followed by LCMV infection 3 days later (13).
To inhibit SIRT1 activity, mice were given SIRT1 inhibitor Ex-527 (10 mg/kg in DMSO, R&D Systems) i.p. daily starting the day after LCMV infection. For cell cycle analysis, LCMV-infected mice were injected with 2 mg BrdU in 200 μL PBS i.p. at 1 hour prior to tissue harvest. All mice analyzed were infected at 8 to 10 weeks of age, and both sexes were included.
Cell preparations and flow cytometry.
Single-cell suspensions were placed in RPMI 1640 media supplemented with 10% FBS and 100 U/mL penicillin and streptomycin. For cell surface staining, cells were incubated with fluorescently conjugated antibodies at 4°C in antibody staining buffer (PBS with 1% FBS). To stain LCMV-specific CD8+ T cells, splenocytes were incubated with Db GP33–41 (residues KAVYNFATC, herein referred to as GP33) tetramers (NIH Tetramer Core Facility) along with cell surface antibodies at 4°C for 30 minutes in antibody staining buffer. For staining of histone H4, activated cells were fixed with Cytofix Buffer (BD Biosciences) for 10 minutes at 37°C, followed by permeabilization with Perm Buffer III (BD Biosciences) for 30 minutes on ice. Cells were then incubated with anti–histone H4 antibody (ab7311, Abcam) for 1 hour at room temperature, followed by fluorescently conjugated secondary antibody against rabbit IgG (4414, Cell Signaling Technology). DNA content was analyzed by staining cells with propidium iodide in the presence of RNase A (Abcam) or 7-AAD (BD Biosciences). To sort live cycling cells, activated cells were stained with Vybrant DyeCycle Violet stain (Thermo Fisher Scientific) at 37°C for 30 minutes. S- and G2/M-phase cells were then sorted based on DNA content using a FACSAria cell sorter (BD Biosciences). BrdU-incorporating cells were identified using the BrdU Flow Kit (BD Biosciences) according to the manufacturer’s instructions. Briefly, cells were surface stained, fixed and permeabilized, treated with DNase I (Sigma-Aldrich), and stained with an anti-BrdU antibody. EdU-incorporating cells were analyzed using Click-iT EdU Flow Cytometry Assay Kits (Thermo Fisher Scientific) according to the manufacturer’s instructions, prior to BrdU staining (MoBU-1, Thermo Fisher Scientific). Senescence-associated β-galactosidase activity was assessed by using a CellEvent Senescence Green Flow Cytometry Assay Kit (Thermo Fisher Scientific) according to the manufacturer’s instructions. Briefly, activated cells were fixed with 4% paraformaldehyde for 10 minutes at room temperature and stained with fluorescent substrate in CellEvent Senescence buffer at 37°C for 2 hours. An Annexin V Apoptosis Detection Kit (BD Biosciences) was used to detect apoptotic cells. Dead cells were excluded from the analysis using LIVE/DEAD Fixable Aqua (Thermo Fisher Scientific) or LIVE/DEAD Fixable Yellow Dead Cell Stain Kit (Thermo Fisher Scientific). The following fluorochrome-conjugated antibodies were used for flow cytometry: anti-CD4 (RM4.5, BioLegend), anti-CD8 (53-6.7, BioLegend), anti-CD3 (145-2C11, Thermo Fisher Scientific), anti-CD28 (37.51, Thermo Fisher Scientific), anti-CD44 (IM7, BioLegend), anti-CD62L (MEL-14, BioLegend), anti-CD45.1 (A20, BioLegend), anti-CD45.2 (104, BioLegend), and anti-Vα2 (B20.1, BioLegend) for murine T cells; anti-CD3 (HIT3a, BD Biosciences), anti-CD4 (RPA-T4, BD Biosciences), anti-CD8 (RPA-T8, BD Biosciences), anti-CD25 (M-A251, BD Biosciences), anti-CD45RA (HI100, BD Biosciences), and anti-CCR7 (G043H7, BioLegend) for human T cells. Cells were analyzed on an LSRII or LSR Fortessa (BD Biosciences), and flow cytometry data were analyzed using FlowJo (TreeStar).
RNA-seq and data analysis.
Equal numbers of congenically marked WT and miR-181a–/– YFP+ SMARTA CD4+ T cells were cotransferred into B6 hosts that were infected with LCMV 1 day later. On day 7 after infection, WT and miR-181a–/– YFP+ SMARTA cells were sorted. Total RNA was prepared using the RNeasy Micro Kit (Qiagen), and RNA quality and quantity were examined by a 2100 Bioanalyzer (Agilent Technologies). cDNA synthesis and library preparation were performed with an Ovation RNA-Seq System (NuGEN). Libraries were pooled and sequenced on an Illumina NextSeq 500 at the Stanford Functional Genomics Facility. RNA-seq reads generated from the sequencing runs were analyzed using the nf-core pipeline (67) to determine read counts mapped to genes in the GRCm38 genome. The data were further analyzed using Bioconductor packages edgeR and CQN. The downstream analysis to identify differentially expressed genes was performed as described in Chen et al. (68) with addition of offsets from conditional quantile normalization CQN (69), followed by application of gene-wise negative binomial generalized linear models (70).
GSEA.
GSEA software from the Broad Institute (http://software.broadinstitute.org/gsea/index.jsp) was used to determine the enrichment of gene signatures in the transcriptome of WT and miR-181a–/– SMARTA CD4+ T cells on day 7 after LCMV infection (this study), of activated young and old naive CD4+ T cells (SRA: SRP158502), and of unstimulated young and old naive CD4+ T cells (SRA: PRJNA638216). We ranked the genes by log2(fold-change), and preranked GSEA was performed using the Molecular Signatures Database (http://www.gsea-msigdb.org/gsea/msigdb/index.jsp).
RNA isolation and quantitative RT-PCR.
Total RNA was isolated using the RNeasy Plus Micro Kit (Qiagen) and converted to cDNA using Maxima First Strand cDNA Synthesis Kits (Thermo Fisher Scientific). Quantitative RT-PCR was performed on the ABI 7900HT system (Applied Biosystems) using Power SYBR Green PCR Master Mix (Thermo Fisher Scientific), according to the manufacturers’ instructions. Oligonucleotide primer sets are listed in Supplemental Table 1. Expression levels were normalized to ACTB expression and are displayed as relative n-fold differences.
CUT&RUN assay.
CUT&RUN was performed with a CUT&RUN Assay Kit (Cell Signaling Technology) according to the manufacturer’s instructions. Briefly, sorted cycling cells were bound to concanavalin A magnetic beads, permeabilized, and incubated with antibodies against SIRT1 (07-131, Millipore), acetyl-histone H3 (H3K9/14Ac; 06-599, Millipore), or control rabbit IgG (DA1E, Cell Signaling Technology) overnight at 4°C. Cells were then incubated with protein A–protein G–MNase (pAG-MNase), and released DNA fragments were extracted and used for quantitative RT-PCR. Spike-In DNA was added for normalization. Oligonucleotide primer sets for H2B, H3, and H4 gene promoters were described previously (29). Results were normalized to those obtained with IgG control or Spike-In DNA and are presented relative to those of cycling young cells.
Western blotting.
Cells were lysed in RIPA buffer containing PMSF and protease and phosphatase inhibitors (Santa Cruz Biotechnology) for 30 minutes on ice. Proteins were resolved in denaturing 4%–15% SDS-PAGE (Bio-Rad), transferred onto PVDF membranes (Millipore), and probed with antibodies against histone H3 (ab1791, Abcam), histone H4 (ab7311, Abcam), histone H2A (D6O3A, Cell Signaling Technology), histone H2B (D2H6, Cell Signaling Technology), p53 (DO-1, Santa Cruz Biotechnology), NPAT (A302-772A, Bethyl Laboratories), phospho-RPA32 (S8; E5A2F, Cell Signaling Technology), phospho-RPA32 (S4/S8; A300-245A, Bethyl Laboratories), phospho-CHK1 (S345; 133D3, Cell Signaling Technology), phospho-histone H2A.X (γH2aX) (S139; 20E3, Cell Signaling Technology), p21 Waf1/Cip1 (12D1, Cell Signaling Technology), p21 (EPR3993, Abcam), p16 INK4A (D7C1M, Cell Signaling Technology), SIRT1 (D1D7, Cell Signaling Technology), acetyl-histone H3 (H3K9/14Ac; 06-599, Millipore), and β-actin (13E5, Cell Signaling Technology). Membranes were developed using HRP-conjugated secondary antibodies and Pierce ECL Western blotting substrate (Thermo Fisher Scientific).
Data availability.
The accession number for the RNA-seq data of WT and miR-181a–/– SMARTA CD4+ T cells in this study is SRA SRP307046. The accession number for the RNA-seq data of day 5 activated naive CD4+ T cells from young and old adults is SRA SRP158502. The accession number for the RNA-seq data of resting naive CD4+ T cells from young and old adults is SRA PRJNA638216.
Statistics.
Statistical analysis was performed using Prism (GraphPad). Paired or unpaired 2-tailed Student’s t tests were used for comparing 2 groups. One-way ANOVA with Tukey’s post hoc test was used for multiple-group comparisons. To correct for multiple testing, we used Holm’s step-down method with a false discovery rate of 5%. P less than 0.05 was considered statistically significant.
Study approval.
Human studies were approved by the Stanford University Institutional Review Board, and all participants gave informed written consent. All animal experiments were approved by the Stanford University Institutional Animal Care and Use Committee.
Author contributions
CK, CMW, and JJG designed and analyzed the experiments. CK, JJ, ZY, and WC performed the experiments. CK, RRJ, CEG, and BH performed and analyzed the RNA-seq data. LT performed statistical analyses. CK and JJG wrote the manuscript, with all authors providing feedback.
Supplementary Material
Acknowledgments
We thank C.Z. Chen (Stanford University) for providing miR-181ab1fl/fl mice; R. Ahmed (Emory University) for providing SMARTA mice, LCMV-Armstrong, BHK cells, and Vero cells; the NIH Tetramer Core Facility (Atlanta, Georgia, USA) for providing tetramers; Y.S. Choi (Seoul National University) for the LMPd-Ametrine vector; and B. Carter of the Palo Alto Veterans Administration Flow Cytometry Core for assistance with flow cytometry and cell sorting. This work was supported by NIH grants R01 AR042527, R01 HL117913, R01 AI108906, R01 HL142068, and P01 HL129941 (to CMW); R01 AI108891, R01 AG045779, U19 AI057266, R01 AI129191 (to JJG); and with resources and the use of facilities at the Palo Alto Veterans Administration Healthcare System. Tetramers were provided by the NIH Tetramer Core Facility supported by contract HHSN272201300006C from the NIAID. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Version 1. 06/01/2021
Electronic publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2021, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2021;131(11):e143632.https://doi.org/10.1172/JCI143632.
Contributor Information
Chulwoo Kim, Email: cwkim@stanford.edu.
Jun Jin, Email: jinjun@stanford.edu.
Zhongde Ye, Email: zhongde@stanford.edu.
Rohit R. Jadhav, Email: jadhav@stanford.edu.
Bin Hu, Email: bhu616@stanford.edu.
Wenqiang Cao, Email: wenqcao@stanford.edu.
Lu Tian, Email: lutian@stanford.edu.
Cornelia M. Weyand, Email: cweyand@stanford.edu.
Jörg J. Goronzy, Email: jgoronzy@stanford.edu.
References
- 1.Del Giudice G, et al. Fighting against a protean enemy: immunosenescence, vaccines, and healthy aging. NPJ Aging Mech Dis. 2018;4:1. doi: 10.1038/s41514-017-0020-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gustafson CE, et al. Influence of immune aging on vaccine responses. J Allergy Clin Immunol. 2020;145(5):1309–1321. doi: 10.1016/j.jaci.2020.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thompson WW, et al. Mortality associated with influenza and respiratory syncytial virus in the United States. JAMA. 2003;289(2):179–186. doi: 10.1001/jama.289.2.179. [DOI] [PubMed] [Google Scholar]
- 4.Zhang JJ, et al. Clinical characteristics of 140 patients infected with SARS-CoV-2 in Wuhan, China. Allergy. 2020;75(7):1730–1741. doi: 10.1111/all.14238. [DOI] [PubMed] [Google Scholar]
- 5.Qi Q, et al. Diversity and clonal selection in the human T-cell repertoire. Proc Natl Acad Sci U S A. 2014;111(36):13139–13144. doi: 10.1073/pnas.1409155111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Czesnikiewicz-Guzik M, et al. T cell subset-specific susceptibility to aging. Clin Immunol. 2008;127(1):107–118. doi: 10.1016/j.clim.2007.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nikolich-Zugich J, et al. Age-related changes in CD8 T cell homeostasis and immunity to infection. Semin Immunol. 2012;24(5):356–364. doi: 10.1016/j.smim.2012.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goronzy JJ, Weyand CM. Mechanisms underlying T cell ageing. Nat Rev Immunol. 2019;19(9):573–583. doi: 10.1038/s41577-019-0180-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li G, et al. Decline in miR-181a expression with age impairs T cell receptor sensitivity by increasing DUSP6 activity. Nat Med. 2012;18(10):1518–1524. doi: 10.1038/nm.2963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim C, et al. Activation of miR-21-regulated pathways in immune aging selects against signatures characteristic of memory T cells. Cell Rep. 2018;25(8):2148–2162.e5. doi: 10.1016/j.celrep.2018.10.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jin J, et al. FOXO1 deficiency impairs proteostasis in aged T cells. Sci Adv. 2020;6(17):eaba1808. doi: 10.1126/sciadv.aba1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li QJ, et al. miR-181a is an intrinsic modulator of T cell sensitivity and selection. Cell. 2007;129(1):147–161. doi: 10.1016/j.cell.2007.03.008. [DOI] [PubMed] [Google Scholar]
- 13.Kim C, et al. Defects in antiviral T cell responses inflicted by aging-associated miR-181a deficiency. Cell Rep. 2019;29(8):2202–2216.e5. doi: 10.1016/j.celrep.2019.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marzluff WF, et al. Metabolism and regulation of canonical histone mRNAs: life without a poly(A) tail. Nat Rev Genet. 2008;9(11):843–854. doi: 10.1038/nrg2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mejlvang J, et al. New histone supply regulates replication fork speed and PCNA unloading. J Cell Biol. 2014;204(1):29–43. doi: 10.1083/jcb.201305017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barcaroli D, et al. FLASH is required for histone transcription and S-phase progression. Proc Natl Acad Sci U S A. 2006;103(40):14808–14812. doi: 10.1073/pnas.0604227103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bradford JA, Clarke ST. Dual-pulse labeling using 5-ethynyl-2’-deoxyuridine (EdU) and 5-bromo-2’-deoxyuridine (BrdU) in flow cytometry. Curr Protoc Cytom. 2011;Chapter 7:Unit 7.38. doi: 10.1002/0471142956.cy0738s55. [DOI] [PubMed] [Google Scholar]
- 18.Gitlin AD, et al. Humoral immunity. T cell help controls the speed of the cell cycle in germinal center B cells. Science. 2015;349(6248):643–646. doi: 10.1126/science.aac4919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Flach J, et al. Replication stress is a potent driver of functional decline in ageing haematopoietic stem cells. Nature. 2014;512(7513):198–202. doi: 10.1038/nature13619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao J, et al. NPAT links cyclin E-Cdk2 to the regulation of replication-dependent histone gene transcription. Genes Dev. 2000;14(18):2283–2297. doi: 10.1101/gad.827700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ye X, et al. The cyclin E/Cdk2 substrate p220(NPAT) is required for S-phase entry, histone gene expression, and Cajal body maintenance in human somatic cells. Mol Cell Biol. 2003;23(23):8586–8600. doi: 10.1128/MCB.23.23.8586-8600.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tibbetts RS, et al. A role for ATR in the DNA damage-induced phosphorylation of p53. Genes Dev. 1999;13(2):152–157. doi: 10.1101/gad.13.2.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lavin MF, Gueven N. The complexity of p53 stabilization and activation. Cell Death Differ. 2006;13(6):941–950. doi: 10.1038/sj.cdd.4401925. [DOI] [PubMed] [Google Scholar]
- 24.Roy S, et al. p53 orchestrates DNA replication restart homeostasis by suppressing mutagenic RAD52 and POLθ pathways. Elife. 2018;7:e31723. doi: 10.7554/eLife.31723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou B, et al. Downregulation of miR-181a upregulates sirtuin-1 (SIRT1) and improves hepatic insulin sensitivity. Diabetologia. 2012;55(7):2032–2043. doi: 10.1007/s00125-012-2539-8. [DOI] [PubMed] [Google Scholar]
- 26.Vaquero A, et al. Human SirT1 interacts with histone H1 and promotes formation of facultative heterochromatin. Mol Cell. 2004;16(1):93–105. doi: 10.1016/j.molcel.2004.08.031. [DOI] [PubMed] [Google Scholar]
- 27.Ye Z, et al. Regulation of miR-181a expression in T cell aging. Nat Commun. 2018;9(1):3060. doi: 10.1038/s41467-018-05552-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.He H, et al. CBP/p300 and SIRT1 are involved in transcriptional regulation of S-phase specific histone genes. PLoS One. 2011;6(7):e22088. doi: 10.1371/journal.pone.0022088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ma R, et al. Exogenous pyruvate represses histone gene expression and inhibits cancer cell proliferation via the NAMPT-NAD+-SIRT1 pathway. Nucleic Acids Res. 2019;47(21):11132–11150. doi: 10.1093/nar/gkz864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dang W, et al. Histone H4 lysine 16 acetylation regulates cellular lifespan. Nature. 2009;459(7248):802–807. doi: 10.1038/nature08085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Feser J, et al. Elevated histone expression promotes life span extension. Mol Cell. 2010;39(5):724–735. doi: 10.1016/j.molcel.2010.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.O’Sullivan RJ, et al. Reduced histone biosynthesis and chromatin changes arising from a damage signal at telomeres. Nat Struct Mol Biol. 2010;17(10):1218–1225. doi: 10.1038/nsmb.1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu L, et al. Chromatin modifications as determinants of muscle stem cell quiescence and chronological aging. Cell Rep. 2013;4(1):189–204. doi: 10.1016/j.celrep.2013.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pal S, Tyler JK. Epigenetics and aging. Sci Adv. 2016;2(7):e1600584. doi: 10.1126/sciadv.1600584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Goronzy JJ, et al. Epigenetics of T cell aging. J Leukoc Biol. 2018;104(4):691–699. doi: 10.1002/JLB.1RI0418-160R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hu Z, et al. Nucleosome loss leads to global transcriptional up-regulation and genomic instability during yeast aging. Genes Dev. 2014;28(4):396–408. doi: 10.1101/gad.233221.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Berti M, Vindigni A. Replication stress: getting back on track. Nat Struct Mol Biol. 2016;23(2):103–109. doi: 10.1038/nsmb.3163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zeman MK, Cimprich KA. Causes and consequences of replication stress. Nat Cell Biol. 2014;16(1):2–9. doi: 10.1038/ncb2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ward IM, Chen J. Histone H2AX is phosphorylated in an ATR-dependent manner in response to replicational stress. J Biol Chem. 2001;276(51):47759–47762. doi: 10.1074/jbc.C100569200. [DOI] [PubMed] [Google Scholar]
- 40.Kinjyo I, et al. Real-time tracking of cell cycle progression during CD8+ effector and memory T-cell differentiation. Nat Commun. 2015;6:6301. doi: 10.1038/ncomms7301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kretschmer L, et al. Differential expansion of T central memory precursor and effector subsets is regulated by division speed. Nat Commun. 2020;11(1):113. doi: 10.1038/s41467-019-13788-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Qi Q, et al. Defective T memory cell differentiation after varicella zoster vaccination in older individuals. PLoS Pathog. 2016;12(10):e1005892. doi: 10.1371/journal.ppat.1005892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Toledo LI, et al. ATR signaling can drive cells into senescence in the absence of DNA breaks. Genes Dev. 2008;22(3):297–302. doi: 10.1101/gad.452308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Di Micco R, et al. Oncogene-induced senescence is a DNA damage response triggered by DNA hyper-replication. Nature. 2006;444(7119):638–642. doi: 10.1038/nature05327. [DOI] [PubMed] [Google Scholar]
- 45.Rufini A, et al. Senescence and aging: the critical roles of p53. Oncogene. 2013;32(43):5129–5143. doi: 10.1038/onc.2012.640. [DOI] [PubMed] [Google Scholar]
- 46.Kaeberlein M, et al. The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes Dev. 1999;13(19):2570–2580. doi: 10.1101/gad.13.19.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tissenbaum HA, Guarente L. Increased dosage of a sir-2 gene extends lifespan in Caenorhabditis elegans. Nature. 2001;410(6825):227–230. doi: 10.1038/35065638. [DOI] [PubMed] [Google Scholar]
- 48.Gong H, et al. Age-dependent tissue expression patterns of Sirt1 in senescence-accelerated mice. Mol Med Rep. 2014;10(6):3296–3302. doi: 10.3892/mmr.2014.2648. [DOI] [PubMed] [Google Scholar]
- 49.Cho SH, et al. SIRT1 deficiency in microglia contributes to cognitive decline in aging and neurodegeneration via epigenetic regulation of IL-1β. J Neurosci. 2015;35(2):807–818. doi: 10.1523/JNEUROSCI.2939-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jin F, et al. Neuroprotective effect of resveratrol on 6-OHDA-induced Parkinson’s disease in rats. Eur J Pharmacol. 2008;600(1–3):78–82. doi: 10.1016/j.ejphar.2008.10.005. [DOI] [PubMed] [Google Scholar]
- 51.Tanno M, et al. Induction of manganese superoxide dismutase by nuclear translocation and activation of SIRT1 promotes cell survival in chronic heart failure. J Biol Chem. 2010;285(11):8375–8382. doi: 10.1074/jbc.M109.090266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Canto C, Auwerx J. Targeting sirtuin 1 to improve metabolism: all you need is NAD(+)? Pharmacol Rev. 2012;64(1):166–187. doi: 10.1124/pr.110.003905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jeng MY, et al. Metabolic reprogramming of human CD8+ memory T cells through loss of SIRT1. J Exp Med. 2018;215(1):51–62. doi: 10.1084/jem.20161066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Owczarczyk AB, et al. Sirtuin 1 regulates dendritic cell activation and autophagy during respiratory syncytial virus-induced immune responses. J Immunol. 2015;195(4):1637–1646. doi: 10.4049/jimmunol.1500326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Elesela S, et al. Sirtuin 1 regulates mitochondrial function and immune homeostasis in respiratory syncytial virus infected dendritic cells. PLoS Pathog. 2020;16(2):e1008319. doi: 10.1371/journal.ppat.1008319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Luo J, et al. Negative control of p53 by Sir2alpha promotes cell survival under stress. Cell. 2001;107(2):137–148. doi: 10.1016/S0092-8674(01)00524-4. [DOI] [PubMed] [Google Scholar]
- 57.Vaziri H, et al. hSIR2(SIRT1) functions as an NAD-dependent p53 deacetylase. Cell. 2001;107(2):149–159. doi: 10.1016/S0092-8674(01)00527-X. [DOI] [PubMed] [Google Scholar]
- 58.Langley E, et al. Human SIR2 deacetylates p53 and antagonizes PML/p53-induced cellular senescence. EMBO J. 2002;21(10):2383–2396. doi: 10.1093/emboj/21.10.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhou Y, et al. Protection of CD4+ T cells from hepatitis C virus infection-associated senescence via ΔNp63-miR-181a-Sirt1 pathway. J Leukoc Biol. 2016;100(5):1201–1211. doi: 10.1189/jlb.5A0316-119RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chang HC, Guarente L. SIRT1 and other sirtuins in metabolism. Trends Endocrinol Metab. 2014;25(3):138–145. doi: 10.1016/j.tem.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang J, et al. The type III histone deacetylase Sirt1 is essential for maintenance of T cell tolerance in mice. J Clin Invest. 2009;119(10):3048–3058. doi: 10.1172/JCI38902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Brien JD, et al. Key role of T cell defects in age-related vulnerability to West Nile virus. J Exp Med. 2009;206(12):2735–2745. doi: 10.1084/jem.20090222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Goronzy JJ, Weyand CM. Successful and maladaptive T cell aging. Immunity. 2017;46(3):364–378. doi: 10.1016/j.immuni.2017.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sussmuth SD, et al. An exploratory double-blind, randomized clinical trial with Selisistat, a SirT1 inhibitor, in patients with Huntington’s disease. Br J Clin Pharmacol. 2015;79(3):465–476. doi: 10.1111/bcp.12512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fragoso R, et al. Modulating the strength and threshold of NOTCH oncogenic signals by mir-181a-1/b-1. PLoS Genet. 2012;8(8):e1002855. doi: 10.1371/journal.pgen.1002855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Choi YS, Crotty S. Retroviral vector expression in TCR transgenic CD4+ T cells. Methods Mol Biol. 2015;1291:49–61. doi: 10.1007/978-1-4939-2498-1_5. [DOI] [PubMed] [Google Scholar]
- 67.Ewels PA, et al. The nf-core framework for community-curated bioinformatics pipelines. Nat Biotechnol. 2020;38(3):276–278. doi: 10.1038/s41587-020-0439-x. [DOI] [PubMed] [Google Scholar]
- 68.Chen Y, et al. From reads to genes to pathways: differential expression analysis of RNA-Seq experiments using Rsubread and the edgeR quasi-likelihood pipeline. F1000Res. 2016;5:1438. doi: 10.12688/f1000research.8987.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hansen KD, et al. Removing technical variability in RNA-seq data using conditional quantile normalization. Biostatistics. 2012;13(2):204–216. doi: 10.1093/biostatistics/kxr054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.McCarthy DJ, et al. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 2012;40(10):4288–4297. doi: 10.1093/nar/gks042. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The accession number for the RNA-seq data of WT and miR-181a–/– SMARTA CD4+ T cells in this study is SRA SRP307046. The accession number for the RNA-seq data of day 5 activated naive CD4+ T cells from young and old adults is SRA SRP158502. The accession number for the RNA-seq data of resting naive CD4+ T cells from young and old adults is SRA PRJNA638216.