Abstract
Keywords: Cell Biology, Oncology
Keywords: Cancer, Mouse models, Tumor suppressors
Introduction
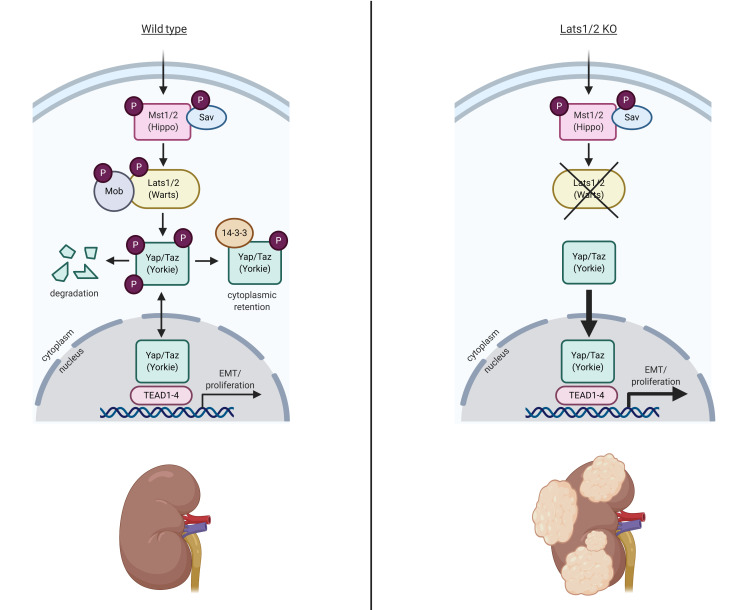
The Hippo/Warts (Mst/Lats) pathway is one of the major signaling pathways regulating organ size and tissue growth during embryonic development (1). The pathway controls the activity of several genes involved in cell differentiation, proliferation, and survival via a kinase-signaling cascade. Briefly, kinases Mst1 and Mst2 (orthologs of Drosophila Hippo) and Sav1 form a complex to activate Lats1 and Lats2 (Drosophila Warts) via phosphorylation. Lats1/2 kinases in turn phosphorylate the transcriptional coactivators Yap and Taz (orthologs of Drosophila Yorkie), leading to their retention in the cytoplasm, inhibiting these two major downstream effectors. When dephosphorylated, Yap/Taz can translocate into the nucleus and interact with cofactors to induce expression of genes that inhibit apoptosis and promote proliferation (1). The activity of the Hippo pathway is regulated at multiple levels, e.g., cell density, cell polarity, and cell-cell contact as well as signaling by atypical cadherins such as Fat and Dachsous (1, 2). In the absence of an upstream activator, Lats1 and Lats2 are ubiquitinated and targeted for proteasomal degradation. Merlin, a FERM domain–containing protein encoded by the NF2 gene, represses the ubiquitin ligase that targets Lats1/2 for degradation (3). Loss-of-function mutations in the upstream activators, kinases, or NF2 would all result in the inappropriate accumulation of Yap and Taz in the nucleus and activation of target genes.
Congruent with its important role in the regulation of tissue growth, the Hippo/Warts pathway has been implicated in the development of several cancers (4). Perturbation of Hippo pathway components has been shown to cause tumor formation in various organ systems in mice, and mutation and altered expression of a subset of Hippo pathway genes have been observed in a variety of human cancers, including liver, colon, prostate, ovarian, and breast (4). Several recent studies suggest that the Hippo/Warts pathway is misregulated in various cancers of the urogenital system. For instance, loss-of-function mutations in FAT4 were identified in 5% of screened human urothelial carcinoma (UC) samples (5), and multiple truncating mutations of Lats2 and NF2 have been identified in a subset of clear cell renal cell carcinoma (ccRCC) (6–8). In contrast, the YAP gene was amplified in 4% of 131 urothelial bladder carcinomas (8), and Liu et al. detected elevated levels of YAP in 53.1% (113/213) of bladder UC biopsies (9). These data are in agreement with the predicted roles of Fat4, Lats, and Merlin as tumor suppressors and Yap as an oncogene. Further, they indicate that the Hippo/Warts pathway is deregulated in human cancers of the urogenital system. However, direct evidence that activation of the pathway can promote urogenital tumorigenesis has not been provided. In fact, it has recently been reported that inactivation of Hippo (Mst1/2) in the kidney epithelia results in fibrosis and chronic kidney disease, in part through promotion of nuclear Yap (10). In addition, several studies have reported that nuclear YAP is observed in the epithelia of human polycystic kidney disease patients and transgenic activation of Yap in mouse kidney epithelium causes cyst formation, raising questions as to the precise role (if any) of the pathway in renal tumor formation (11, 12).
Here, we show that co-inactivation of the Warts kinases Lats1 and Lats2 in collecting duct or proximal tubules of adult mice is sufficient to induce the formation of metastatic, sarcomatoid renal tumors as well as UC of the renal pelvis. Tumor formation in Lats mutants requires functional Yap/Taz as demonstrated by coablation of Lats1/2 and Yap/Taz or treatment with the drug verteporfin. However, we find that expression of a mutated form of Yap that cannot be phosphorylated (Yap5SA) in adult kidney epithelia does not induce renal cell carcinoma (RCC) formation, suggesting that activated Yap is necessary but not sufficient for tumor formation.
Comparison of the transcriptomes of mouse tumors and human RCC tumors revealed a relatively small subset of human RCCs that significantly overlapped Lats mutant tumors. Further analysis will be required to determine whether Warts pathway mutants represent a unique RCC subtype. Overall, our findings provide valuable insight into the role of Hippo/Warts signaling in urogenital cancers and suggest that a subset of RCC patients may be amenable to treatment with Yap-inhibitory drugs. This model provides a new tool for the study of the cell types and mechanisms involved in renal cancer progression and metastasis and for investigation of potential therapeutics for human renal cancers with mutations in the Hippo/Warts pathway.
Results
Ablation of Lats1/2 in epithelia of adult kidneys leads to sarcomatoid tumors.
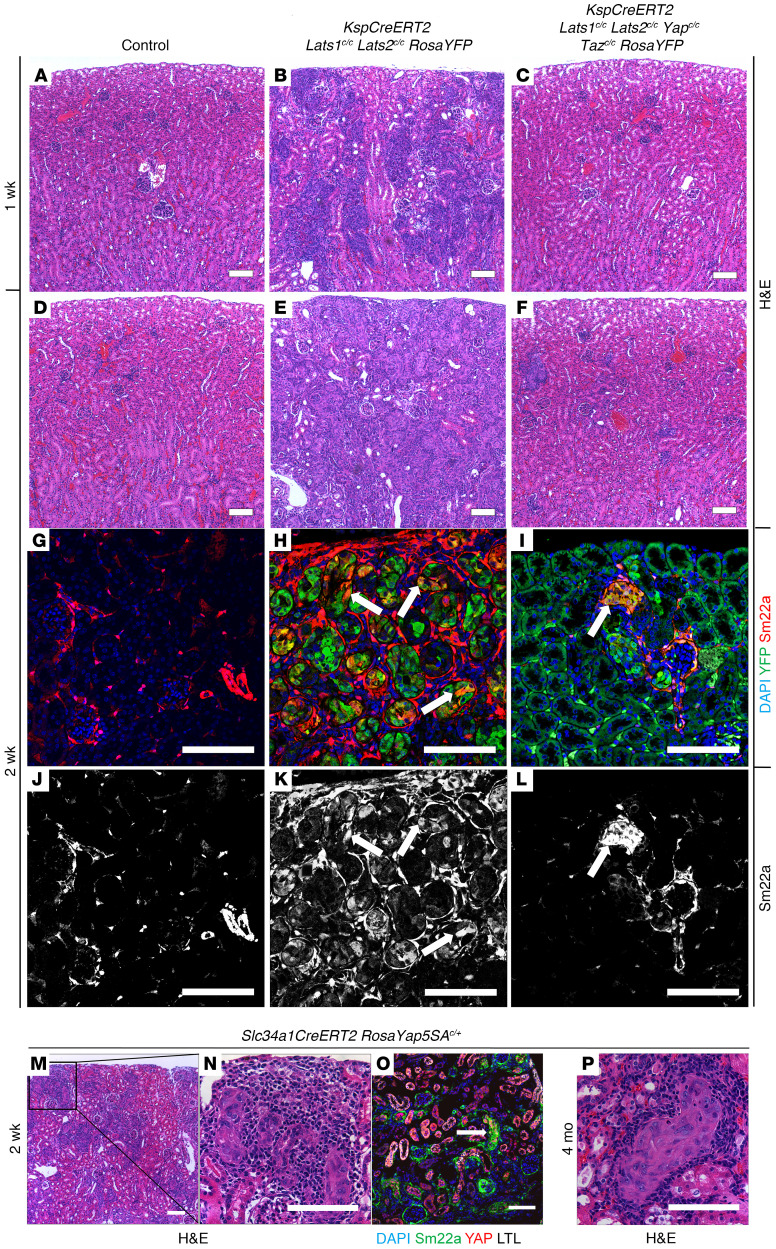
To investigate the role of the Hippo/Warts pathway mutation in the adult kidney, we ablated the serine/threonine kinases Mst1 and Mst2 (orthologs of Hippo) and Lats1 and Lats2 (orthologs of Warts) in adult kidney epithelia using a tamoxifen-inducible, kidney-specific Cre line (KspCreERT2). The Ksp promoter drives expression throughout the kidney epithelium with highest levels in the collecting ducts and more distal nephron segments (13). High-dose tamoxifen administration (5 mg/40 g body weight, 3 times) in 1-month-old KspCreERT2 Mst1fl/fl Mst2fl/fl mice had no discernible impact on their health. Histological examination of mutant kidneys 6 months after tamoxifen administration showed no gross abnormalities (data not shown). In contrast, high-dose tamoxifen administration to KspCreERT2 Lats1fl/fl Lats2fl/fl animals resulted in weight loss and premature lethality in mutants, with the majority of mice succumbing within 2 weeks of the final tamoxifen administration. Microscopic examination of H&E-stained sections revealed cellular transformation and neoplastic cell growth throughout the kidney. Mutant kidneys were enlarged with visibly disrupted patterning, and tubules were obstructed with luminal infiltration of cells (Figure 1, B, E, and F) compared with WT (Figure 1, A, C, and D).
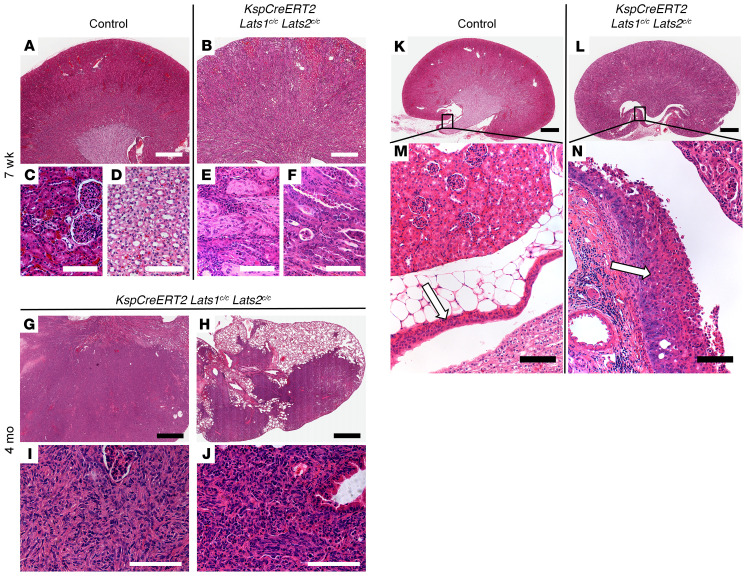
Figure 1. Ablation of Lats1/2 in epithelia of adult kidneys leads to sarcomatoid tumors and TCC.
H&E-stained sections of control kidney (A, C, D, K, and M) or KspCreERT2 Lats-KO kidney (B, E–G, I, L, and N) or lung (H and J) collected 7 weeks (A–F; n = 3 for control and mutant) or 4 months (G–J; n = 4) after the first treatment with high-dose (B, E, and F) or low-dose (G–J) tamoxifen. C–D, E–F, I, and J are high-magnification images of A, B, G, and H, respectively. C and E were taken in the cortex, while D and F are images of the papillae. (K–N) H&E-stained sections of control (K and M) or KspCreERT2 Lats1/2c/c (L and N) kidneys 7 weeks after tamoxifen. Arrows indicate control (M) or mutant (N) ureteric epithelia. Box in K corresponds to M, and box in L corresponds to N. Scale bars: (A, B, G, and H) 900 μm, (C–F, I, J, M, N) 100 μm, (K and L) 2 mm.
In an attempt to allow the transformed cells to progress, we sought to preserve some normal renal function by ablating Lats1/2 mosaically using a low dose of tamoxifen (2.5 mg/40 g body weight, once). The majority of KspCreERT2 Lats1/2 mutant mice given low-dose tamoxifen were viable beyond 2 weeks. Analysis of mutant kidneys collected from mice 4 months after low-dose tamoxifen administration revealed significant enlargement of the organ. Histological analysis revealed large areas where the normal parenchyma was completely replaced by spindle-shaped cells showing sarcomatoid features (Figure 1, G and I). Although structures resembling vascularized glomeruli were present, epithelial features were largely absent. The majority of mutants whose lungs were analyzed also had tumors within the lung (Figure 1, H and J). Like the renal tumors, lung tumors were completely composed of spindle-shaped cells with sarcomatoid features. To determine whether these were primary lung tumors or metastases from the kidney, we analyzed WT and mutant mice carrying the RosaYFP reporter allele 4 months after tamoxifen administration. YFP was not visible in KspCreERT2 RosaYFP Lats1fl/+ Lats2fl/+ lungs (Supplemental Figure 1C; supplemental material available online with this article; https://doi.org/10.1172/JCI144108DS1), although it was readily visible in the kidneys (Supplemental Figure 1A). In contrast, in KspCreERT2 RosaYFP Lats1fl/fl Lats2fl/fl animals, YFP was broadly visible in the kidney (Supplemental Figure 1B) and lungs, although YFP-positive cells in the lung appeared to be restricted to tumors (Supplemental Figure 1D). This and previous studies showing that the KspCre promoter is not active in the lung, and the fact that the lung tumors are multiple, variably sized (cannonball pattern), and found immediately adjacent to large blood vessels, suggest that the lung tumors metastasized from the kidney. A subset (~20%) of KspCreERT2 Lats1fl/fl Lats2fl/fl kidneys also displayed high-grade in situ urothelial carcinoma (UC) of the renal pelvis (Figure 1, K–N). It is important to note that the only times when tumors were not observed in tamoxifen-treated mutants, YFP was also not observable, suggesting that recombination had failed. Interestingly, we occasionally observed transformed cells within tumors that lacked lineage tracer. To test whether this observation was due to a failure to recombine at the Rosa26 locus, the incorporation of WT cells into tumors, or the silencing of the Rosa26 locus, we bred the Rosa26mT/mG reporter onto the KspCreERT2 Lats1/2 mutant background. The mT/mG mouse expresses the Tomato reporter ubiquitously and the GFP reporter after excision of a translational stop codon. Large patches of cells within tumors were observed that lacked both Tomato and GFP expression, indicating that in some mutant cells, transcription from the Rosa26 locus was absent, possibly because the cells were dying (Supplemental Figure 1, I–L).
Sarcomatoid renal cell carcinoma (sRCC) is a particularly aggressive form of renal cancer. sRCC is observed in all subclasses of RCC and is believed to represent a stage of the disease after multiple secondary mutations have accumulated, leading to a more dedifferentiated, terminal state (14). To determine whether Lats mutant cells gave rise to a recognizable class of RCC at earlier time points, we sacrificed mutant mice 4, 7, 14, and 28 days after high-dose tamoxifen treatment. Histological examination showed that the first cellular abnormalities occurred as early as 4 days after Cre induction, while at 7 days, large regions within the cortex and medulla of mutant kidneys showed neoplastic changes in epithelial cells and signs of intertubular fibrosis (Supplemental Figure 1, E and F). Over the next 7 days the area of transformed cells expanded so that, by 14 days after tamoxifen administration, the vast majority of lineage-traced cells showed signs of transformation. Immune infiltrate was also observed histologically at this point (Supplemental Figure 1G). By 28 days, most of the kidney was filled with transformed cells that were still not identifiable as a specific type of RCC (Supplemental Figure 1H). Starting at 3 months, we observed signs of sarcomatoid transformation (Figure 1 and not shown). At no time did mutant cells show histopathological features indicative of any of the accepted histological classes of RCC other than sarcomatoid.
RCC is thought to primarily arise from proximal tubules. As KspCre is most highly active in the distal nephron and collecting ducts, we next sought to determine whether Lats1/2 deletion in collecting duct specifically was sufficient to induce transformation. Previous studies showed that Lats1 and Lats2 are necessary for proper differentiation and branching morphogenesis of the embryonic collecting ducts (15). Therefore, we first attempted to delete Lats1/2 after branching morphogenesis had ceased using Pkhd1Cre, a line active in the early postnatal collecting ducts (16). One-week-old Pkdh1Cre Lats1/2fl/fl mouse kidneys were of relatively normal size, suggesting that no significant defect in ureteric bud branching had occurred. However, histological examination showed luminal infiltration of cells within the medullary collecting ducts and ureter (Supplemental Figure 2, B and D). Although these cells showed signs of transformation, all mutants died within 6 weeks of birth with obstructed ureters and hydronephrosis but without signs of fulminant tumor formation. To determine whether the mutant collecting duct cells could progress to tumors, we generated Lats1/2-null cell lines from collecting duct cells and implanted them under the capsule of a single kidney in immunocompromised (NOD/SCID) mice. WT (Lats1/2fl/fl) cells were implanted as controls. Mice carrying the null cells showed signs of cachexia (significant weight loss, hunched back) within 4 weeks of implantation and had to be sacrificed by 6 weeks according to the guidelines of our IACUC. Visual examination showed that mice implanted with Lats1/2 mutant cells had very large masses on their kidneys (Supplemental Figure 2F). Histological examination revealed large tumors composed entirely of spindle-shaped cells with sarcomatoid features growing on the host kidney (Supplemental Figure 2H). Kidneys receiving implants of control cells did not show signs of tumor growth (Supplemental Figure 2E). Instead, the control cells formed poorly differentiated epithelial tubules with little or no proliferative expansion (Supplemental Figure 2G). Interestingly, mice engrafted with the Lats1/2 mutant cell line showed not only lung metastases but also liver metastases, something never observed in KspCreERT2 mutants (data not shown). It is important to note that pelvic UCs were not observed in grafted kidneys, suggesting that the UCs present in KspCreERT2 mutants were most likely not the result of metastasis.
KspCre shows little or no activity in the proximal tubules. To determine the impact of ablating Lats1/2 in the proximal tubules, we crossed the Lats1/2-floxed alleles to mice carrying Slc34a1CreERT2, a Cre line that is active only in the S1 and S2 segments of the proximal tubule (17). One month after high-dose tamoxifen administration, Slc34a1CreERT2 Lats1/2fl/fl mice showed obvious lesions in the cortex of the kidney (Figure 2B) compared with control (Figure 2A). From 4 months after high-dose tamoxifen onward, Slc34a1CreERT2 Lats1/2fl/fl mice had large sarcomatoid tumors with little normal parenchyma remaining (Figure 2, C and D). Additionally, these mice developed metastases in the lung with sarcomatoid morphology as determined by pathological analysis of H&E-stained sections (Figure 2F). An H&E-stained section of control lung is shown in Figure 2E. No UCs were observed, nor were liver metastases. Similar to what was observed with the KspCreERT2 model, Slc34a1CreERT2-derived mutant cells never showed histopathology matching other RCC subtypes. Instead, they appeared to rapidly progress toward the sarcomatoid phenotype.
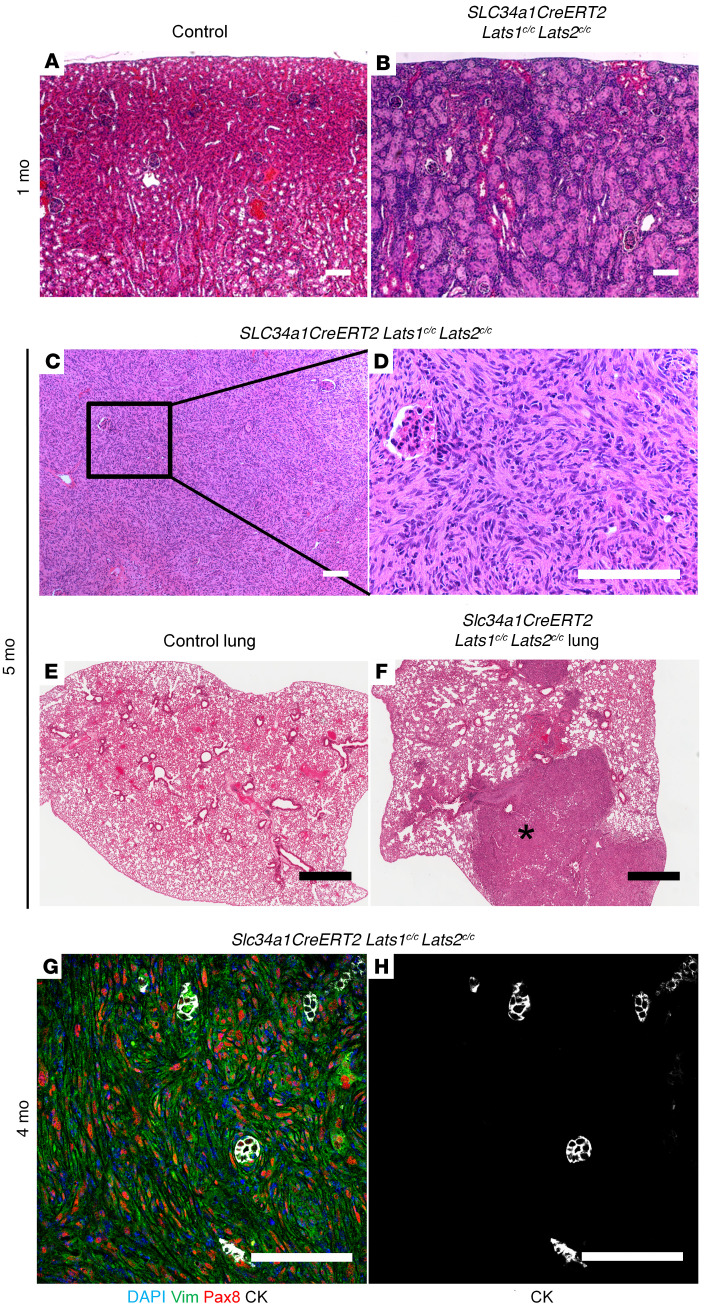
Figure 2. Ablation of Lats in the proximal tubules leads to metastatic sRCC.
(A–F) H&E-stained sections of control kidney (A), control lung (E), and Slc34a1CreERT2 Lats1/2c/c kidney (B–D) and lung (F). (B) Mutant kidney 1 month after tamoxifen (n = 3). (C, D, and F) Low-magnification kidney (C), high-magnification kidney (D), and high-magnification lung (F) from Slc34a1CreERT2 Lats1/2c/c mouse 5 months after tamoxifen (n = 3). D corresponds with the box in C. Asterisk in F shows a metastasis. (G and H) Immunofluorescence staining on sections of Slc34a1CreERT2 Lats1/2c/c kidney 4 months after tamoxifen (n = 3). (G) Blue, DAPI; green, vimentin; red, Pax8; white, cytokeratin. H is a single-channel image of the image in G showing only cytokeratin. Scale bars: (A–D, G, and H) 100 μm, (E and F) 900 μm.
We next sought to better classify the Slc34a1CreERT2 Lats mutant model. sRCCs can be identified according to their Pax8, cytokeratin, vimentin triple-positive status (18, 19). To assess the cellular identity of Lats mutant tumors, kidneys of Slc34a1CreERT2 Lats1/2fl/fl mice 4 months after tamoxifen were stained with antibodies to Pax8, cytokeratin, and vimentin. Although tumors showed high vimentin and Pax8 levels, we were unable to detect cytokeratin expression outside of normal (unrecombined) collecting ducts (Figure 2, G and H). This result implies that the tumors do not exhibit epithelial markers typical of sRCC, instead showing a fully mesenchymal phenotype.
In sum, our findings indicate that ablation of Lats1/2 in disparate epithelial cell types within the adult kidney is sufficient to rapidly induce the formation of metastatic renal tumors with sarcomatoid features with nearly 100% penetrance. To our knowledge, this is the first example of a genetically engineered model of metastatic kidney cancer.
Activation of Yap/Taz is necessary for tumor formation.
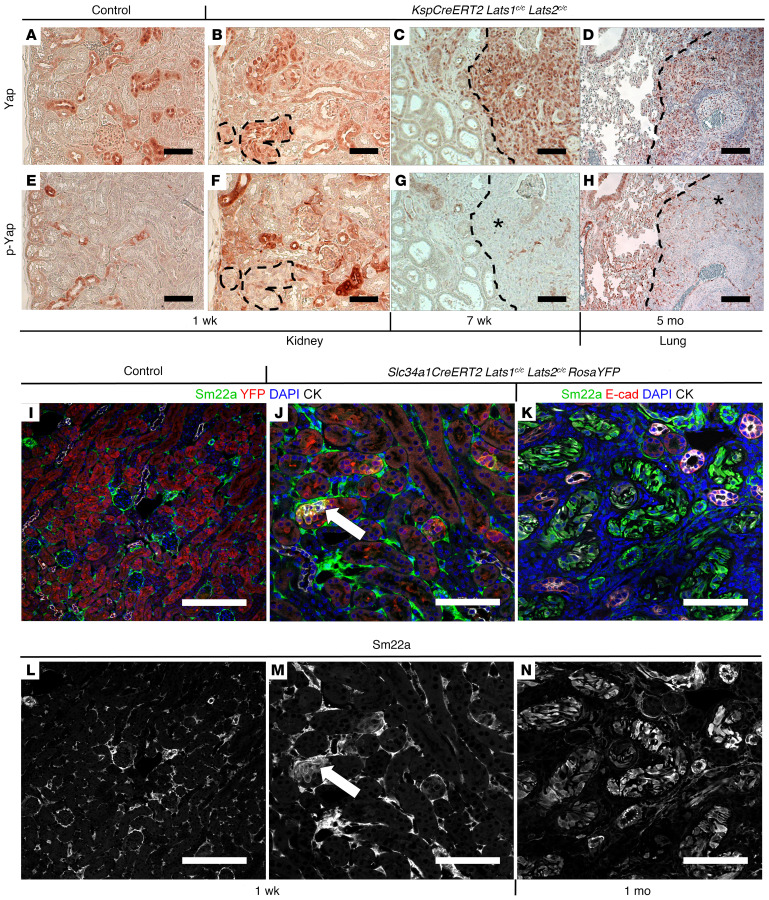
Kidney epithelial activation of Yap/Taz has been observed in multiple pathological but non-neoplastic contexts (10, 12, 20). While Lats1 and Lats2 have been shown to be key players in the Yap/Taz activity, there is some evidence for noncanonical roles for Lats kinases in tumor formation (21). To determine the role of Yap/Taz in the Lats1/2 mutant phenotype, we first examined Yap phosphorylation status and localization in Lats1/2 mutant kidney tumors and lung metastases. Immunohistochemical staining of mutant and control kidneys and mutant lung metastases revealed that loss of Lats1/2 resulted in absence of phosphorylated Yap from the cytoplasm, while total Yap was strongly enriched in the nuclei of mutant cells (Figure 3, A–H). To determine whether nuclear Yap was active, we examined the expression of a known target of Yap, Sm22a (22). Sm22a, also known as transgelin, is an actin-binding protein that is an early marker of smooth muscle cell differentiation (23). In the normal kidney, Sm22a is only found in vascular smooth muscle cells (Figure 3, I and L). In contrast, ectopic Sm22a expression was observed in Lats1/2 mutant epithelial cells as early as 7 days after tamoxifen, prior to overt signs of cellular transformation (Figure 3, J and M). This expression was maintained throughout late tumor stages, with the majority of sarcomatoid cells exhibiting high Sm22a (Figure 3, K and N).
Figure 3. Molecular analysis of Lats mutants.
(A–H) Immunohistochemical staining against Yap (A–D) or p-Yap (E–H) on sections of control kidney (A and E; n = 3), KspCreERT2 Lats1c/c Lats2c/c kidney 1 week (B and F; n = 3) or 7 weeks after tamoxifen (C and G; n = 3), and KspCreERT2 Lats1c/c Lats2c/c lung 5 months after tamoxifen (D and H; n = 3). (I–N) Immunofluorescence staining on control (I and L) or Slc34a1CreERT2 Lats1/2c/c RosaYFP kidneys (J, K, M, and N) 1 week (J and M; n = 3 control, n = 5 mutant) or 1 month (K and N; n = 4) after tamoxifen. (I and J) Green, Sm22a; red, YFP; blue, DAPI; white, cytokeratin. (K) Green, Sm22a; red, E-cadherin; blue, DAPI; white, cytokeratin. L–N are single-channel images of Sm22a from I–K, respectively. All scale bars: 100 μm.
To determine whether Yap and Taz activity is necessary for tumor formation, we bred conditionally inactive alleles of Yap and Taz into the KspCreERT2 Lats1/2 mutant background. WT, KspCreERT2 Lats1/2 mutant, and KspCreERT2 Lats1/2 Yap/Taz mutant mice were given high-dose tamoxifen (5 mg/40 g body weight, 3 times) at 1 month of age and sacrificed 7 or 14 days after the first dose. At 7 days after tamoxifen, H&E-stained Lats1/2 mutant kidneys showed multiple lesions in the cortex and medulla, while WT and Lats1/2 Yap/Taz mutant kidneys appeared phenotypically normal (Figure 4, A–C). After 14 days, compared with control (FIgure 4D), Lats1/2 mutant kidneys were filled with transformed cells as seen on H&E-stained sections (Figure 4E). In contrast, histological examination of Lats1/2 Yap/Taz mutant kidneys appeared relatively normal, although some small foci of transformed cells could be identified (Figure 4F). Additionally, epithelial and intertubular Sm22a expression was markedly decreased in Lats1/2 Yap/Taz mutant kidneys compared with Lats1/2 mutants (Figure 4, H–L). Control kidney Sm22a expression is shown in Figure 4, G and J. Staining of the few remaining lesions found in Lats1/2 Yap/Taz mutants revealed the presence of nuclear Yap, suggesting that these had not undergone complete recombination at the Yap locus or that they had acquired a mutation that allowed a functional Yap protein to be made (Supplemental Figure 3, A–C). These observations suggest that Yap and Taz are necessary for transformation of Lats1/2 mutant cells.
Figure 4. Yap and Taz are necessary for sRCC formation.
(A–F) H&E-stained sections of control (A and D), KspCreERT2 Lats1/2c/c RosaYFP (B and E), and KspCreERT2 Lats1/2c/c Yapc/c Tazc/c RosaYFP (C and F) kidneys 1 week (A–C; n = 8 control, n = 11 mutant, n = 3 rescue) or 2 weeks (D–F; n = 9 control, n = 8 mutant, n = 8 rescue) after tamoxifen. (G–L) Immunofluorescence staining on sections of control (G and J; n = 3), KspCreERT2 Lats1/2c/c RosaYFP (H and K; n = 3), and KspCreERT2 Lats1/2c/c Yapc/c Tazc/c RosaYFP (I and L; n = 3) kidneys. (G–I) Blue, DAPI; green, YFP; red, Sm22a. (J–L) Single-channel (Sm22a) corresponding with G–I, respectively. (M–O) H&E-stained (M and N) or immunofluorescence-stained (O) sections of Slc34a1CreERT2 RosaYap5SAc/+ kidney 2 weeks after tamoxifen (n = 3). N is a high-magnification image corresponding with the box in M. (O) Lesions in Slc34a1CreERT2 RosaYap5SAc/+ kidneys express high nuclear Yap and Sm22a. Blue, DAPI; green, Sm22a; red, Yap; white, Lotus Tetragonolobus Lectin (LTL). (P) H&E-stained section of Slc34a1CreERT2 RosaYap5SAc/+ kidney 4 months after tamoxifen (n = 3). All scale bars: 100 μm.
To determine whether ablation of Yap/Taz would prolong the lifespan of the Lats1/2 mutants, we followed mutants over an extended time period. While only 2 of 23 Lats1/2 mutants (8.7%) were still alive 6 months after tamoxifen administration, 10 of 12 Lats1/2 Yap/Taz mutants (83.3%) were still alive at this time, indicating that ablation of Yap and Taz is sufficient to rescue Lats1/2 deletion–induced lethality (Supplemental Figure 3D).
We next sought to determine whether overexpression of active Yap was sufficient to drive transformation. To accomplish this, we used the Rosa26-lox-stop-lox-Yap5SA allele (RosaYap5SA). Upon Cre-mediated activation, this mutant expresses a form of Yap that cannot be phosphorylated because of mutation of 5 serine residues that have been shown to be targets of Lats and thus should phenocopy Lats1/2 loss (24). This mouse was bred to the Slc34a1CreERT2 line, and mice were given a high dose of tamoxifen at 1 month of age. Two weeks after the first tamoxifen dose, the kidneys of Slc34a1CreERT2 RosaYap5SA heterozygotes showed many small transformed lesions, morphologically similar to those found in Lats1/2 mutant kidneys. As expected, recombined cells displayed strong nuclear Yap accumulation and high Sm22a expression (Figure 4, M–O). To determine whether these cells would eventually form tumors, mice were also given a single, low dose of tamoxifen and collected 4 months later. Somewhat unexpectedly, mice carrying the activated allele of Yap in their proximal tubules showed signs of transformation and fibrosis but did not progress to RCC (Figure 4P). These data suggest that activation of Yap in the adult kidney epithelia is sufficient to transform cells but not to promote fulminant tumor formation.
Phenotyping and RNA sequencing give insights into molecular characteristics of Lats1/2 knockout–induced RCC.
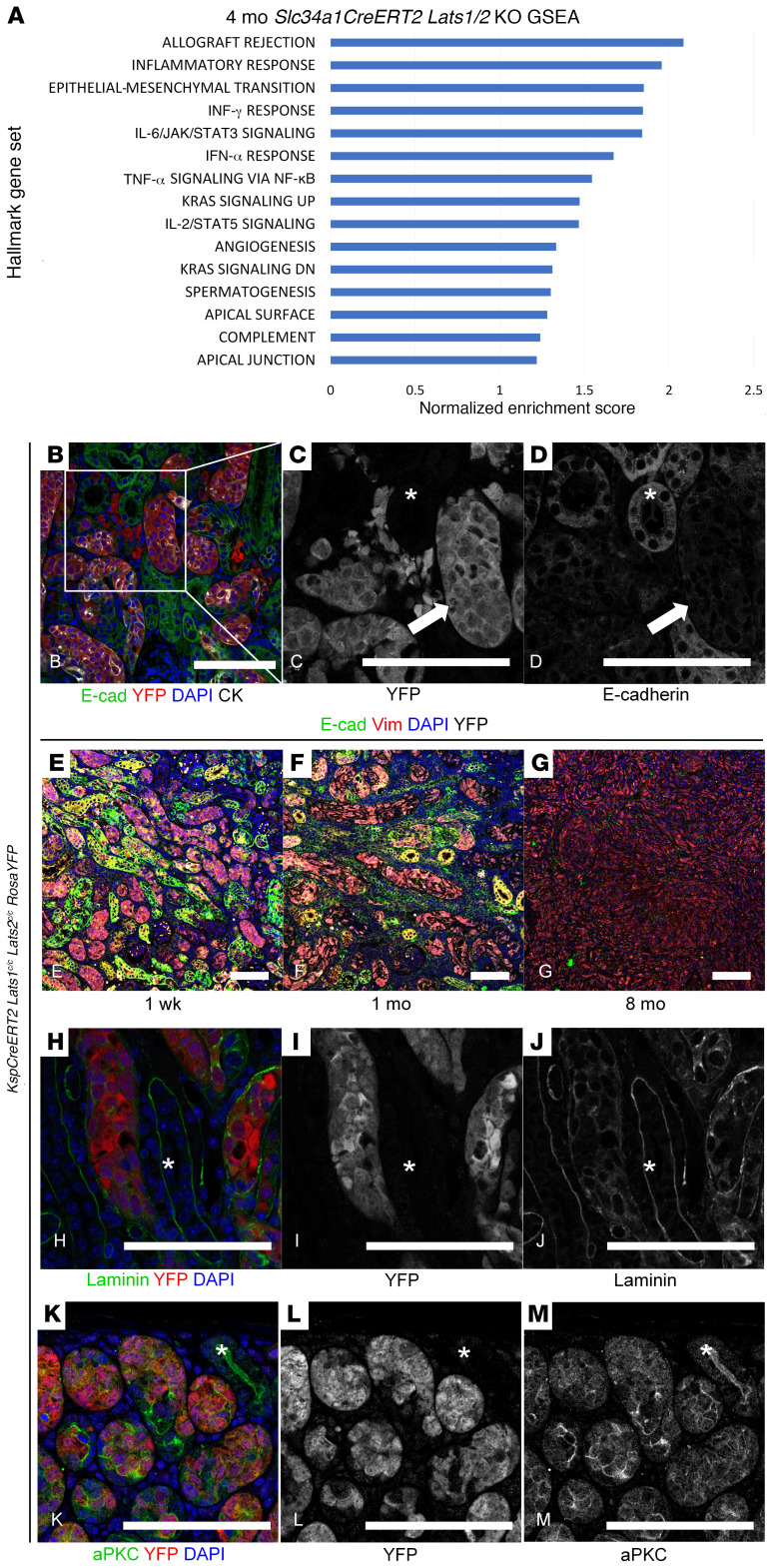
As the transcription factors Yap and Taz are necessary to transform Lats1/2 mutant renal epithelia, we next sought to identify the transcriptional targets promoting transformation by performing RNA sequencing of mutant cells isolated from Slc34a1CreERT2 Lats1/2 mutant kidney tumors and lung metastases. WT (Slc34a1CreERT2 RosaYFP) FACS-sorted proximal tubule cells were used as a control. Examination of the RNA-Seq data suggested that a number of canonical Yap/Taz targets, including Tagln (the gene encoding Sm22a), Ankrd1, and Cyr61, were upregulated in the Lats mutant cancers (22). In addition to the Yap pathway, gene set enrichment analysis of RNA-Seq data showed perturbation of genes involving various cellular processes including activation of epithelial-mesenchymal transition (EMT), inflammatory response, and IL-6/JAK/STAT3 signaling (Figure 5A).
Figure 5. Phenotypic analysis of Lats mutants.
(A) Gene set enrichment analysis (GSEA) on Slc34a1CreERT2 Lats1/2c/c kidneys 4 months after tamoxifen. (B–D) Immunofluorescence staining on section of KspCreERT2 Lats1c/c Lats2c/c RosaYFP kidney 1 week after tamoxifen. (B) Green, E-cadherin; red, YFP; blue, DAPI; white, cytokeratin. Mutant, YFP-positive cells display loss of E-cadherin (C and D, arrows), while WT, YFP-negative tubules express E-cadherin (C and D, asterisks). C and D are single-channel images of YFP and E-cadherin, respectively, corresponding to the box in B. (E–G) Immunofluorescence staining on sections of KspCreERT2 Lats1c/c Lats2c/c RosaYFP kidneys 1 week, 1 month, and 8 months after tamoxifen. Green, E-cadherin; red, vimentin; blue, DAPI; white, YFP. (H–M) Immunofluorescence staining on section of KspCreERT2 Lats1c/c Lats2c/c RosaYFP kidney 1 week after tamoxifen. (H–J) Green, laminin; red, YFP; blue, DAPI. WT, YFP-negative tubules display intact basal lamina (asterisks), while surrounding mutant, YFP-positive cells exhibit disorganized basal lamina. I and J are single-channel images of H showing YFP and laminin, respectively. (K–M) Green, aPKC; red, YFP; blue, DAPI. WT, YFP-negative tubules show apical localization of aPKC (K–M, asterisks), which is not maintained in surrounding mutant, YFP-positive tubules. L and M are single-channel images of G showing YFP and aPKC, respectively. Experiments shown in B–M were conducted on 3 different mutants. Scale bars: (B–M) 100 μm.
We next sought to validate some of the findings from the RNA-Seq data using immunofluorescence staining. By 1 week after tamoxifen administration, Lats1/2 mutant cells had lost expression of E-cadherin (Figure 5, B–D) and gained expression of the mesenchymal marker vimentin (Figure 5, E–G) (25). We next examined markers of a polarized epithelium, including genes expressed at the apical surface or in the basement membrane, as deficiencies in this process are also associated with EMT and loss of the basement membrane is required for metastasis (26, 27). Mutant cells showed greatly reduced expression of basolateral laminin (Figure 5, H–J), as well as a loss of apical localization of atypical protein kinase C (aPKC) (Figure 5, K–M). These findings, together with the Yap phosphorylation status and subcellular localization data described above, suggest that activated Yap contributes to tumorigenesis and that Lats1/2 mutant cells rapidly and directly undergo EMT several weeks before tumor formation.
Lats mutant tumors model human RCC.
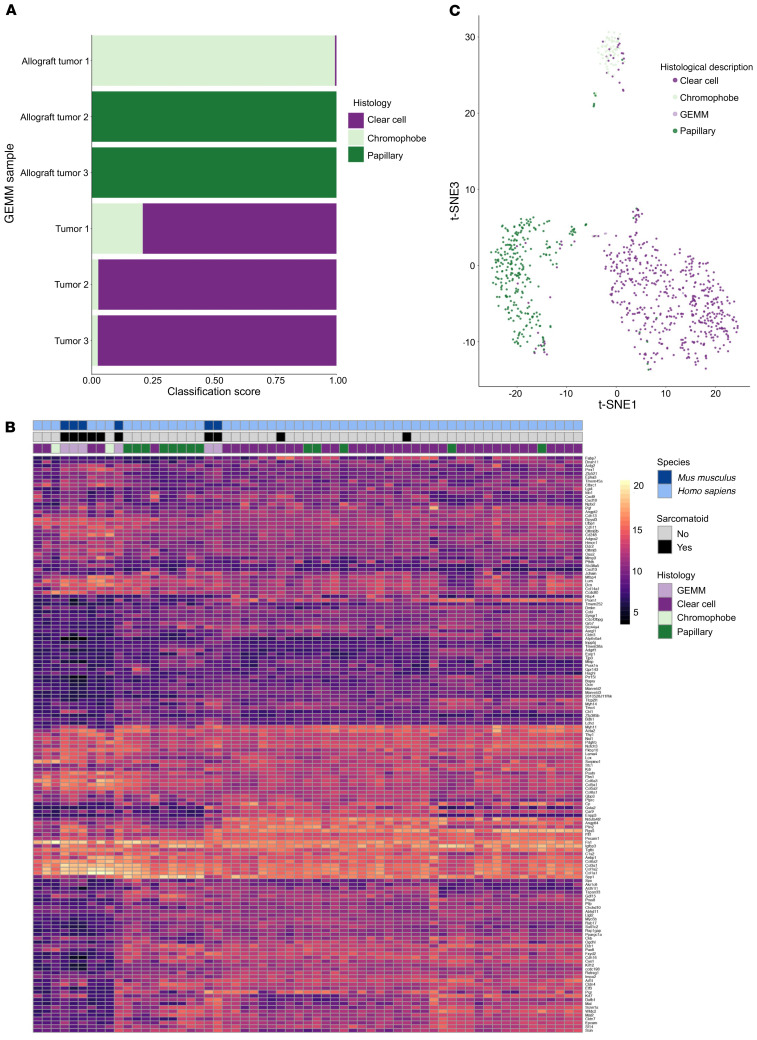
Genetic and molecular perturbations of the Salvador/Warts/Hippo pathway have been reported in numerous types of RCC as well as other, less common renal cancers. As mentioned, sarcomatoid features are usually observed in association with other subclasses of RCC. As we did not observe features reminiscent of a specific subclass of RCC, we sought to determine whether our genetically engineered mouse models resembled a specific tumor type at the molecular level. To address this question, we compared the transcriptomes of Slc34a1CreERT2 Lats1/2fl/fl (proximal tubule–derived) and Lats1/2 mutant inner medullary collecting duct–derived allograft tumors with the transcriptomes of clear cell, papillary, and chromophobe RCC specimens curated in The Cancer Genome Atlas Program (TCGA) database. A neural network was trained to classify the expression profiles of the human tumors. Then, the expression profiles for three Slc34a1CreERT2 Lats1/2fl/fl tumors and three tumors obtained from the above allograft procedure were submitted as input to the classifier. All three Slc34a1CreERT2 Lats1/2fl/fl tumors were classified as most similar to clear cell, even though they did not show a clear cell histology nor did they stain positive (other than in necrotic regions) for CAIX, a diagnostic marker of ccRCC (27) (Supplemental Figure 4). Interestingly, 2 allograft tumors were classified as most similar to papillary and one to chromophobe (Figure 6A). These classifications most likely represent the cell type of origin of the different tumors rather than overlapping pathological features as will be discussed below.
Figure 6. Bioinformatic comparison of Lats mutant tumors and human RCC.
(A) Neural network classification of Lats1/2 mutant tumors. (B) A gene expression heatmap of Lats1/2 mutant tumors and human RCC showing only our tumors and the human RCCs that cluster with them. (C) Lats1/2 mutant tumor and human RCC t-SNE. GEMM, genetically engineered mouse model.
To attempt to identify pathological overlap (vs. lineage overlap), we generated a list of genes most differentially expressed between RCCs with sarcomatoid features and those without and compared it with the mouse data set. As depicted in Supplemental Figure 4G, a heatmap of selected genes suggests that while the transcriptome of the genetically engineered mouse models (GEMMs) shows significant overlap with the majority of tumors from each of the subtypes of RCC, they also show significant differences that are shared by a small subset of human RCCs (Figure 6B). This is more robustly illustrated in the t-distributed stochastic neighbor embedding (t-SNE) presented in Figure 6C. These data suggest that although Lats mutant tumors share transcriptional properties with several types of RCC (including sarcomatoid and non-sarcomatoid histologies), they have the most overlap with the transcriptome of a small, undefined cluster of RCCs.
Nuclear YAP identifies a subset of RCCs.
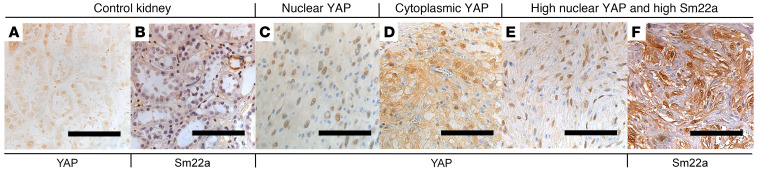
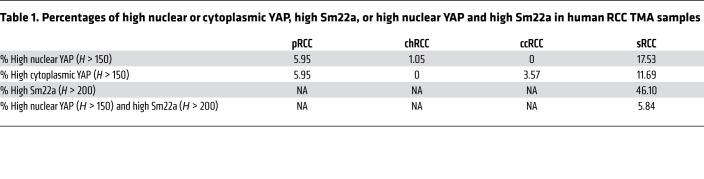
Activation of YAP has previously been documented in human RCC and other kidney malignancies. However, the relationship of YAP activation and sarcomatoid features has not been well documented. To assess YAP status in human RCC and sRCC, we performed immunohistochemical staining on tissue microarrays (TMAs) of human RCCs (Figure 7). TMAs contained samples of the following RCC types: sarcomatoid (sRCC), clear cell (ccRCC), papillary (pRCC), and chromophobe/renal oncocytoma (chRCC/RO). Two TMAs of each RCC subtype, including a total of 468 punch biopsies from 199 patients, were stained for total YAP. The YAP staining was quantified by percentage of intensity score (0 to 3), which was converted to an H-score of 0–300 for both cytoplasmic and nuclear localization. We designated an H-score greater than 150 as “high.” The tumor that most frequently scored as high for nuclear YAP was sRCC, with 17.53% of samples scoring 150 or higher. A low but significant number of pRCC and chRCC samples also showed high nuclear YAP (5.95% and 1.05%, respectively). No ccRCC samples scored in the high nuclear category. In contrast, assessment of cytoplasmic YAP revealed that sRCC had the highest percentage (11.69%), followed by pRCC (5.95%) and ccRCC (3.57%). No chRCC showed high cytoplasmic YAP (Table 1).
Figure 7. Human RCC TMAs reveal heterogeneity in YAP and YAP target expression.
Examples of immunohistochemical staining for YAP (A and C–E) or Sm22a (B and F) on sections of control kidney (A and B; n = 3 for YAP and Sm22a) or RCC TMA samples (C–F). C–E are representative images from staining on 468 biopsies from 199 patients; F is representative from staining on 173 biopsies from 51 patients. Scale bars: 100 μm.
Table 1. Percentages of high nuclear or cytoplasmic YAP, high Sm22a, or high nuclear YAP and high Sm22a in human RCC TMA samples.
To determine whether YAP nuclear accumulation correlated with activation of validated YAP target genes, we also stained the TMAs that contained only RCCs with sarcomatoid features with an antibody against Sm22a and correlated Sm22a levels with the presence of high nuclear YAP (H > 150) on the same patient samples. High Sm22a expression was defined as an H-score greater than 200. In contrast to the 17.53% of sRCC samples that showed high nuclear YAP, 46.1% showed high Sm22a. But high nuclear YAP along with high Sm22a was only observed in 5.84% of samples (Table 1). Thus, although high levels of nuclear YAP are most common in sRCC, such tumors represent only a fraction of sRCC cases, and coexpression with the YAP target Sm22a is observed in an even smaller subset, supporting the notion of molecular heterogeneity within RCC. We hypothesize that this small subset of sRCCs may represent a distinct subclass of RCC.
Yap represents a potential therapeutic target in RCC formation.
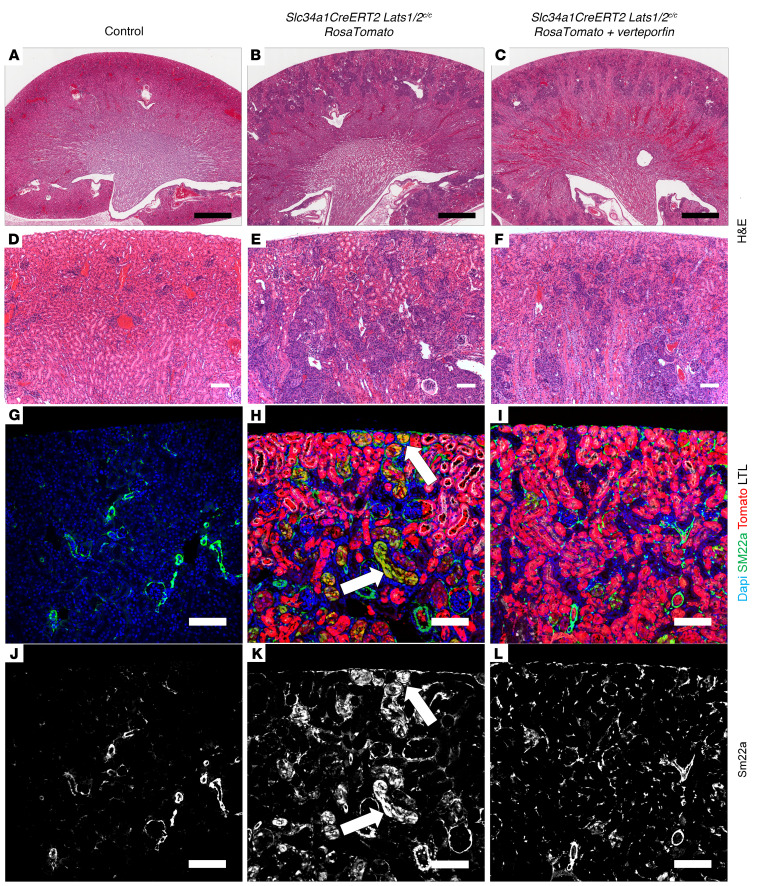
Our data suggest that activation of Yap in kidney epithelia is necessary to cause aggressive renal tumor formation in Lats1/2 mutants and that active YAP is found in a significant proportion of human RCCs. These findings indicate that YAP could represent a therapeutic target in a subset of RCCs. Thus, we sought to test the efficacy of 2 small-molecule antagonists of Yap, verteporfin and dobutamine, as therapeutics in our Lats1/2 mutant models (28, 29). Slc34a1CreERT2 Lats1/2fl/fl RosaTomato mice were given 3 doses of 5 mg/40 g body weight tamoxifen every other day. Mice were also injected with vehicle, 100 mg/kg verteporfin, or 5 mg/kg dobutamine on alternating days for 14 days. Mice were harvested 14 days after the first drug dose. We were unable to continue treatments beyond 14 days, as longer verteporfin treatment resulted in death of the treated animals (WT and mutants). H&E-stained sections showed that Lats1/2 mutant mice treated with verteporfin exhibited a marked decrease in transformed cells compared with those treated with vehicle, particularly in the most cortical regions of the kidney (Figure 8, B, C, E, and F). An H&E-stained control kidney section is shown in Figure 8, A and D. Immunofluorescence staining on these kidneys revealed a reduction in epithelial and intertubular Sm22a in the verteporfin-treated animals (Figure 8, H, I, K, and L). Control kidney Sm22a expression is shown in Figure 8, G and J. Dobutamine, however, had no effect on lesion size, with some dobutamine kidneys appearing to have increased levels of transformed cell relative to vehicle treated control, although this was not quantified (Supplemental Figure 5, B and D). Dobutamine-treated kidneys also maintained strong Sm22a expression (Supplemental Figure 5, F and H). These results suggest that Yap inhibitors may hold potential as therapeutics in RCCs showing misregulation of the Hippo/Warts pathway.
Figure 8. Treatment with verteporfin, a Yap inhibitor, partially rescues Lats1/2 deletion phenotype.
(A–F) H&E-stained sections of control (A and D; n = 3) or Slc34a1CreERT2 Lats1/2c/c RosaTomato kidneys 2 weeks after tamoxifen and vehicle (B and E; n = 4) or tamoxifen and verteporfin (C and F; n = 2 survived to 14 days). D–F are high-magnification images of the sections in A–C, respectively. (G–L) Immunofluorescence staining on sections of control (G and J) or Slc34a1CreERT2 Lats1/2c/c RosaTomato kidneys 2 weeks after tamoxifen and vehicle (H and K) or tamoxifen and verteporfin (I and L). (G–I) Blue, DAPI; green, Sm22a; red, Tomato; white, LTL. (J–L) Single-channel (Sm22a) corresponding with G–I, respectively. Lats mutant lesions express both Tomato and Sm22a (H and K, arrows), and such lesions are markedly reduced in the cortexes of mutant kidneys treated with verteporfin (I and L). Scale bars: (A–C) 900 μm, (D–L) 100 μm.
Discussion
Although there has been extensive speculation that the Hippo/Warts pathway plays a critical role in kidney cancer, direct evidence has been lacking. Although activation (nuclear localization) of the transcriptional regulator Yap has been observed in various renal tumors, it has also been observed in nonmalignant contexts including polycystic kidney disease and renal fibrosis (11, 30). Mutations in Hippo pathway components have been seen in both low-grade and high-grade renal neoplasms (31, 32). In addition, although loss-of-function mutations in upstream components of the pathway including NF2, Lats1, Lats2, and Fat4 are frequently observed in RCC tumors, the vast majority of mutations are heterozygous, which does not cause transformation in animal models. Further, transgenic overexpression of Yap in kidney epithelia has been shown to cause cyst formation, not cancer (12). Based on these findings, one would predict that nuclear localization of Yap or Taz alone is not sufficient to drive tumor formation. Here we show that inactivation of the Warts paralogs Lats1 and Lats2 in the epithelia of adult mouse kidneys drives nuclear accumulation of Yap and leads to metastatic RCC. While at least 1 allele of Yap or its paralog Taz is necessary for tumor formation in Lats mutants, activation of a mutant form of Yap that cannot be phosphorylated (Yap5SA) is not sufficient for tumor formation.
Multiple studies have observed different phenotypes associated with enriched nuclear Yap in mouse kidneys. There are a number of possible explanations for the disparity, some of which are more consistent with our data than others. One is that nuclear Yap may result in different phenotypes depending on the cell type in which it is activated (nuclear). Although we think there is certainly some credence to this hypothesis when comparing very disparate cell types such as epithelia and fibroblasts, we have shown that Lats1/2 inactivation in the proximal tubules has very different effects compared with Yap activation (via expression of Yap5SA) within the same cell type. In contrast, Lats1/2 inactivation in collecting ducts results in sarcomatoid tumors that are indistinguishable from those arising after inactivation in the proximal tubules. Thus, we think it is unlikely that the different phenotypes observed in different models are based on the identity of the cell type manipulated. Another possibility is that cytoplasmic (phosphorylated) Yap represses RCC formation and that this protein pool is lost in Lats mutant cells but not in other models tested (such as transgenic overexpression of Yap). Indeed, cytoplasmic Yap has been shown to sequester various proteins, and there were huge differences in the ratio of cytoplasmic to nuclear Yap observed in human renal cancers. A third possibility is that although Yap is necessary for RCC formation in Lats mutants, other substrates of Lats also contribute to and are necessary for tumor formation. These other substrates may be directly involved in tumorigenesis or indirectly promote chromosome instability, increasing the mutational burden of recombined cells. The speed at which Lats mutant cells progress to transformation as well as the penetrance and uniformity of response suggest it is unlikely that additional mutations are required. However, it is interesting to note that we did occasionally identify mutant cells in late-stage tumors of Slc34a1CreERT2 Lats1/2fl/fl mice that did not express a reporter allele from the Rosa26 locus (Supplemental Figure 1I and data not shown ). This is of interest because the Rosa26 locus lies in proximity to the Vhl locus on mouse chromosome 6 and Vhl is frequently mutated in RCC. Whether this observation was due to chromosome 6 loss, transcriptional silencing, or mutant cells on the verge of death (prenecrotic) is not known, but this was not simply the effect of failed recombination at the Rosa26 locus, as mutant cells did not express either myristoylated Tomato or myristoylated GFP in some cells when the dual-reporter Rosa26mT/mG line was crossed in. However, RNA-Seq data did not detect significant differences in mRNA expression of genes surrounding the Rosa26 locus, including Vhl, nor did we observe expression of CAIX in mutant tumors, suggesting that loss of Vhl activity and/or silencing of the surrounding region did not occur in the majority of mutant cells. A final possibility is that the differences observed in various models are caused by differences in the absolute levels of Yap/Taz in the nucleus. This is an attractive hypothesis as this has been proposed for other transcription factors such as β-catenin (33, 34). However, it is interesting to note that rescue of the Lats1/2 mutant phenotype required ablation of all 4 Yap/Taz alleles. We did not notice a partial rescue or reversion to a cystic or fibrotic phenotype in kidneys lacking 3 of the 4 alleles. Distinguishing which of these possibilities is the case will require additional investigation.
As mentioned, numerous studies have shown increased nuclear YAP in various kidney pathologies. Our analysis revealed that high nuclear YAP was most frequently observed in RCCs with sarcomatoid features (17.53%). However, high expression of Sm22a, a direct target of YAP in our Lats mutants, along with high nuclear YAP, was only observed in 5.84% of all sarcomatoid tumors examined. These observations raise a number of important points. First, sRCC is a molecularly (and most likely genetically) heterogeneous disease. Second, the observations suggest that the presence of YAP in the nucleus does not necessarily correlate with activation of downstream targets. Whether this is caused by alterations in the epigenetic landscape, presence or absence of cofactors, or the relative levels of YAP protein is unclear. It will be interesting to determine whether there is any sort of correlation between YAP and YAP target expression and mutational state, tumor progression, survival, and/or response to treatment.
Classifying the Lats mutant tumors has been somewhat complicated. Although the tumors have a clearly sarcomatoid phenotype, histological features of other “stages” or subclasses of RCC are not observed. Our analysis suggests that Lats mutants undergo EMT very quickly following tamoxifen administration, progressing to aggressive tumors with homogenous, undifferentiated morphology without first exhibiting morphological characteristics of lower-grade carcinoma (i.e., ccRCC). Thus, the tumors do not fit the WHO classification for sRCC (35). Bioinformatics analysis comparing transcriptomes of Lats1/2 mutant tumors with those of human RCC suggests that our GEMMs give rise to a renal tumor that shares characteristics with multiple human RCC subtypes, depending on the cell type in which Lats1 and Lats2 were inactivated. Deletion in the proximal tubules resulted in tumors most closely resembling ccRCC, while mutant collecting duct–derived allograft tumors identify most closely with either pRCC or chRCC. But we propose that this is a result of the tumors being grouped by similarities in gene expression arising from their cell type of origin rather than their mutant transcriptomes. The classifiers used for this analysis included only ccRCC, pRCC, and chRCC, so the tumors were placed into whichever of these categories was most similar, even if the gene expression did not have a large percentage of overlap. By analysis of the most differentially expressed genes between sarcomatoid and non-sarcomatoid RCC, Lats1/2 mutants cluster with a small group of human tumors that are molecularly distinct from other RCC subtypes. It is possible that this subset represents a novel, rare RCC subtype. It is important to note that there exists a small percentage of RCC cases (~5%) that do not fit classification criteria for the major RCC subtypes or other rare kidney neoplasms; these cases are known as unclassified renal cell carcinoma (uRCC). uRCC shows a high rate of metastasis, as in the Lats mutants, and is correlated with worse clinical outcome (36). The WHO recommends that RCCs with purely sarcomatoid features should be designated as uRCC (35). Interestingly, a recent study found that a significant proportion of uRCC cases harbor mutations in the upstream Hippo pathway component NF2 along with a corresponding disruption of Hippo signaling (32). With the alterations in Hippo signaling, highly metastatic behavior, and lack of alignment with major RCC subtypes, it is possible that our Lats1/2 mutant tumors most closely resemble this NF2-deficient subset of uRCC, potentially providing a mouse model of a heretofore understudied RCC subtype. However, we cannot rule out the possibility that the mouse model does not represent any identified human pathology. Identification of Lats mutant biomarkers along with transcriptomic analysis of NF2-deficient uRCCs will be required to clarify this issue.
Methods
Mice.
Lats1fl/fl, Lats2fl/fl, Yapfl/fl, Tazfl/fl, RosaYFP, RosaTomato, RosamTmG, Slc34a1CreERT2, KspCreERT2, RosaYap5SA, and Pkhd1Cre mice have all been previously described (13, 16, 17, 24, 37–42). Pkhd1Cre mice were acquired from the University of Texas Southwestern (UTSW) George M. O’Brien Kidney Research Core Center (NIH P30DK079328). NOD/SCID mice were purchased from Charles River Laboratories. All other mice were purchased from The Jackson Laboratory. Tamoxifen was administered by oral gavage to 6- to 8-week-old mice. Subcapsular injections were performed in 6- to 8-week old NOD/SCID mice (adapted from the protocol of Pavía-Jiménez et al., ref. 43).
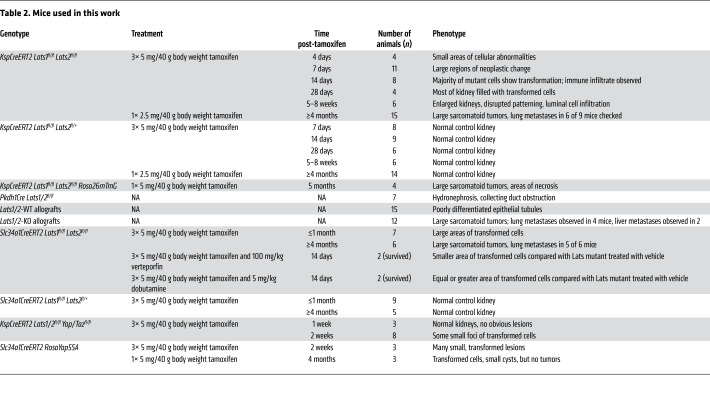
Table 2 shows all mice and genotypes used.
Table 2. Mice used in this work.
Histology, immunofluorescence, and immunohistochemistry on tissue sections.
For histological analysis, kidneys were fixed in 4% paraformaldehyde, embedded in paraffin, sectioned into 5 μm slices, and subjected to H&E staining. Immunofluorescence staining on paraffin-embedded tissue sections was performed as follows. Paraffin sections were dewaxed 3 times for 5 minutes in xylene, washed sequentially in 100%, 70%, 50%, and 30% EtOH for 5 minutes each, and rinsed in deionized water for 5 minutes. Slides were permeabilized in PBS with 0.1% Triton X (PBST) 3 times for 5 minutes, then boiled in 10 mM sodium citrate antigen retrieval buffer (pH 6). After rinsing in PBST, slides were blocked in 5% serum for 1 hour at room temperature. Slides were incubated in primary antibody diluted in 5% serum PBST overnight at 4°C. Primary antibodies were used as follows: E-cadherin (Invitrogen, catalog 131900; dilution 1:500), YFP (Aves Labs, catalog AB_2307313; dilution 1:500), pan-cytokeratin (Sigma-Aldrich, catalog C2562; dilution 1:500), phosphorylated Yap (p-Yap) (Cell Signaling Technology, catalog 13008; dilution 1:100), vimentin (Santa Cruz Biotechnology, catalog sc7557; dilution 1:100), laminin (Sigma-Aldrich, catalog L9393; dilution 1:500), Sm22a (Abcam, catalog ab14106; dilution 1:500), Lotus Tetragonolobus Lectin (LTL) (Vector Laboratories, catalog B1035, B1325; dilution 1:500), Tomato (MyBioSource, catalog MBS448092; dilution 1:200). Slides were washed 3 times for 5 minutes in PBST, incubated in fluorophore-conjugated secondary antibodies diluted 1:500 in 5% serum PBST for 1 hour at room temperature, then washed 3 times for 5 minutes in PBST. Finally, slides were incubated in DAPI (diluted 1:1000 in water) for 10 minutes, washed 3 times for 5 minutes in PBST, and mounted using VECTASHIELD Antifade Mounting Medium (Vector Laboratories, catalog H-1000).
Immunohistochemistry on paraffin-embedded tissue sections was performed as follows: Paraffin sections were dewaxed 3 times for 5 minutes in xylene, washed sequentially in 100% (twice), 70%, 50%, and 30% EtOH for 2 minutes each, and rinsed in PBS for 5 minutes. Slides were boiled in 10 mM sodium citrate antigen retrieval buffer (pH 6), then permeabilized in TBS with 0.025% Triton X (PBST) twice for 5 minutes. Next, slides were blocked in 1% BSA, 10% FBS in TBS for 1 hour at room temperature. Slides were incubated in primary antibody diluted in blocking solution overnight at 4°C. Primary antibodies were used as follows: Yap (Cell Signaling Technology, catalog 4912; dilution 1:100), p-Yap (Cell Signaling Technology, catalog 13008; dilution 1:100), Sm22a (Abcam, catalog ab14106; dilution 1:100), CAIX (Invitrogen, catalog PA1-16592; dilution 1:400). Slides were washed 3 times for 5 minutes in TBST, then subjected to peroxidase treatment (45 mL cold methanol, 750 μL 30% hydrogen peroxide) for 15 minutes. After a 5-minute wash in TBST, slides were incubated with HRP-conjugated secondary antibody for 1 hour at room temperature. Slides were washed 3 times for 5 minutes in TBST, then incubated in DAB solution (Vector Laboratories, catalog SK-4100) until desired stain darkness. After washing in deionized water, slides were either counterstained with hematoxylin or dehydrated. Dehydration was achieved with sequential 2-minute washes in 30%, 50%, 70%, and 100% EtOH; then slides were washed 3 times for 5 minutes in xylene. Finally, slides were mounted with Permount Mounting Medium (Fisher Scientific, catalog SP15-100).
All antibodies used in this study for immunofluorescence and immunohistochemistry are standardly used in the field and have been extensively tested and validated. Previously reported expression patterns were confirmed by the Carroll laboratory.
Isolation of adult mouse proximal tubule cells for FACS.
Adult (~6 months old), control (Slc34a1CreERT2 Lats1c/c Lats2c/+ RosaTomato), and mutant (Slc34a1CreERT2 Lats1c/c Lats2c/c RosaTomato) mice that had been given high-dose tamoxifen 4 months previous were sacrificed via CO2 inhalation and cervical dislocation. Kidneys were dissected out and put in ice-cold PBS, medullary regions were excised using a razor blade, and remaining kidney tissue was minced finely and placed into a 5 mL round-bottom tube containing 1 mL 1.5% collagenase A/PBS. Tubes were placed in a 37°C water bath for 15–20 minutes, with trituration after the first 2 minutes using a 1 mL pipette tip with the end cut off; 2.5 mL ice-cold 10% FBS/PBS was added to the tube to stop the reaction, with pipetting to mix well. Suspension was filtered with a 100 μm filter, pipetting on top and washing 3 times with 1 mL 10% FBS/PBS. Flow-through was collected in a 50 mL conical tube and centrifuged at 323g for 5 minutes at 4°C. Supernatant was removed, and the pellet was resuspended in 2 mL 1% FBS/PBS. Suspension was filtered with a 40 μm filter, pipetting on top and washing 3 times with 1% FBS/PBS. Flow-through was aliquoted into four to six 1.5 mL Eppendorf tubes and centrifuged at 323g for 5 minutes at 4°C. After supernatant was removed, the pellet was resuspended in 1 mL RBC lysis buffer. Suspension was then centrifuged at 323g for 5 minutes at 4°C, and supernatant was removed. (If needed, the RBC lysis step can be repeated.) The pellet was resuspended in 1 mL ice-cold 1% FBS/PBS and centrifuged at 323g for 5 minutes at 4°C twice in order to remove all RBC lysis buffer. Finally, suspension was filtered using a FACS tube with 35 μm filter top (Falcon, catalog 352235), rinsing twice with 1 mL 1% FBS/PBS.
Bioinformatics.
All genes with extant expression measurements in the TCGA database were mapped to mouse orthologs. Transcript abundance was estimated without alignment of reads using Salmon (44) against an index of coding sequences from the Ensembl GRCm38 assembly. Transcript-level abundance was imported and count and offset matrices generated using the tximport R/Bioconductor package (45). Differential expression analysis was performed using the DESeq2 R/Bioconductor package (46). RCC expression data were downloaded from the TCGA database using the TCGAbiolinks R/Bioconductor package (47). A variance-stabilizing transformation implemented in the vst function of DESeq2 was applied prior to further processing. Batch correction using the ComBat function from the sva package was applied to ensure comparability of mouse and human expression data. A sequential neural network with 2 hidden layers each containing 1024 nodes with dropout layers following each hidden layer was trained with the Adam optimizer using focal sparse categorical cross entropy loss (48) with the TensorFlow platform (TensorFlow White Papers, 2015). Graph regularization (49) was employed. Fivefold cross-validation was used during training.
The data discussed in this publication were deposited in the NCBI’s Gene Expression Omnibus database (50) and are accessible through GEO Series accession number GSE164879 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE164879).
Cell line generation.
Collecting duct cells were isolated from the tip of the renal papilla of 4-week-old Lats1fl/fl Lats2fl/fl mice and immortalized by infection with T antigen (51). Clonal lines were generated by serial dilution. Null cell lines were generated from floxed lines by infection with adenovirus carrying a ubiquitously expressed Cre transgene (CAG-Cre). Clonal lines were established and assayed for deletion of Lats1 and Lats2 using genomic PCR. Several independent lines were derived for each genotype. Cell lines were regularly tested for mycoplasma contamination.
Human tissues.
Human RCC TMAs were obtained from the UTSW kidney cancer SPORE CORE B Laboratory.
Statistics.
Survival curves of KspCreERT2 Lats1/2 mutant and KspCreERT2 Lats1/2 Yap/Taz mutant mice were compared via log-rank (Mantel-Cox) test. A P value less than 0.05 was considered significant.
Study approval.
All animals were housed, maintained, and used according to protocols approved by the Institutional Animal Care and Use Committees at the University of Texas Southwestern Medical Center and following the guidelines from the National Cancer Institute at Frederick (Frederick, Maryland, USA) Animal Care and Use Committee.
Author contributions
PC, US, CC, XP, and TJC designed the experiments. PC, US, BT, CC, XP, and JY acquired data. PC, US, CC, PK, and BE conducted data analysis. Resources were provided by GM, RZ, VM, JM, JLC, PK, and TJC. PC, US, CC, and TJC wrote the original draft of the manuscript. PC, CC, RZ, VM, PK, BE, and TJC reviewed and edited the manuscript. TJC acquired funding.
Supplementary Material
Acknowledgments
We thank Ondine Cleaver, Denise Marciano, and James Brugarolas and members of the UTSW Kidney Cancer Center and the Carroll, Cleaver, and Marciano laboratories for comments. We thank Eric Olson of the University of Texas Southwestern Medical Center for providing Yapfl/fl and Tazfl/fl mice, and Randy L. Johnson of The University of Texas MD Anderson Cancer Center for providing RosaYap5SA mice. This work was supported by grants from the Cancer Prevention and Research Institute of Texas (RP160340) and the NIH (R24DK106743, R01DK095057, P50CA196516) to TJC, from the NIH (P50CA196516) to PK, and from the NIH (R01DK069921) and the Department of Veterans Affairs, Veterans Health Administration, Office of Research and Development (1I01BX002196) to RZ. The graphical abstract was created with BioRender (Biorender.com).
Version 1. 06/01/2021
Electronic publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2021, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2021;131(11):e144108.https://doi.org/10.1172/JCI144108.
Contributor Information
Phoebe Carter, Email: phoebe.carter@utsouthwestern.edu.
Ulrike Schnell, Email: ulrikeschnell@googlemail.com.
Christopher Chaney, Email: Christopher.chaney@utsouthwestern.edu.
Betty Tong, Email: betty.tong@utsouthwestern.edu.
Xinchao Pan, Email: xinchao.pan@gmail.com.
Jianhua Ye, Email: yjh6scxh@cau.edu.cn.
Glenda Mernaugh, Email: merna3017@comcast.net.
Jennifer L. Cotton, Email: jennifer.l.cotton@gmail.com.
Vitaly Margulis, Email: Vitaly.Margulis@UTSouthwestern.edu.
Junhao Mao, Email: junhao.mao@umassmed.edu.
Roy Zent, Email: roy.zent@vumc.org.
Bret M. Evers, Email: bret.evers@utsouthwestern.edu.
Payal Kapur, Email: payal.kapur@utsouthwestern.edu.
Thomas J. Carroll, Email: thomas.carroll@utsouthwestern.edu.
References
- 1.Yu FX, Guan KL. The Hippo pathway: regulators and regulations. Genes Dev. 2013;27(4):355–371. doi: 10.1101/gad.210773.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Grusche FA, et al. Upstream regulation of the Hippo size control pathway. Curr Biol. 2010;20(13):R574–R582. doi: 10.1016/j.cub.2010.05.023. [DOI] [PubMed] [Google Scholar]
- 3.Li W, et al. Merlin/NF2 suppresses tumorigenesis by inhibiting the E3 ubiquitin ligase CRL4(DCAF1) in the nucleus. Cell. 2010;140(4):477–490. doi: 10.1016/j.cell.2010.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Harvey KF, et al. The Hippo pathway and human cancer. Nat Rev Cancer. 2013;13(4):246–257. doi: 10.1038/nrc3458. [DOI] [PubMed] [Google Scholar]
- 5.Guo G, et al. Whole-genome and whole-exome sequencing of bladder cancer identifies frequent alterations in genes involved in sister chromatid cohesion and segregation. Nat Genet. 2013;45(12):1459–1463. doi: 10.1038/ng.2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dalgliesh GL, et al. Systematic sequencing of renal carcinoma reveals inactivation of histone modifying genes. Nature. 2010;463(7279):360–363. doi: 10.1038/nature08672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cancer Genome Atlas Research Network Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature. 2013;499(7456):43–49. doi: 10.1038/nature12222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cancer Genome Atlas Research Network Comprehensive molecular characterization of urothelial bladder carcinoma. Nature. 2014;507(7492):315–322. doi: 10.1038/nature12965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu JY, et al. Overexpression of YAP 1 contributes to progressive features and poor prognosis of human urothelial carcinoma of the bladder. BMC Cancer. 2013;13:349. doi: 10.1186/1471-2407-13-349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu C, et al. Tubule-specific Mst1/2 deficiency induces CKD via YAP and non-YAP mechanisms. J Am Soc Nephrol. 2020;31(5):946–961. doi: 10.1681/ASN.2019101052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Happe H, et al. Altered Hippo signalling in polycystic kidney disease. J Pathol. 2011;224(1):133–142. doi: 10.1002/path.2856. [DOI] [PubMed] [Google Scholar]
- 12.Cai J, et al. A RhoA-YAP-c-Myc signaling axis promotes the development of polycystic kidney disease. Genes Dev. 2018;32(11–12):781–793. doi: 10.1101/gad.315127.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shao X, et al. Epithelial-specific Cre/lox recombination in the developing kidney and genitourinary tract. J Am Soc Nephrol. 2002;13(7):1837–1846. doi: 10.1097/01.ASN.0000016444.90348.50. [DOI] [PubMed] [Google Scholar]
- 14.Shuch B, et al. Sarcomatoid renal cell carcinoma: a comprehensive review of the biology and current treatment strategies. Oncologist. 2012;17(1):46–54. doi: 10.1634/theoncologist.2011-0227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reginensi A, et al. A critical role for NF2 and the Hippo pathway in branching morphogenesis. Nat Commun. 2016;7:12309. doi: 10.1038/ncomms12309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shibazaki S, et al. Cyst formation and activation of the extracellular regulated kinase pathway after kidney specific inactivation of Pkd1. Hum Mol Genet. 2008;17(11):1505–1516. doi: 10.1093/hmg/ddn039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kusaba T, et al. Differentiated kidney epithelial cells repair injured proximal tubule. Proc Natl Acad Sci U S A. 2014;111(4):1527–1532. doi: 10.1073/pnas.1310653110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.DeLong W, et al. Sarcomatoid renal cell carcinoma. An immunohistochemical study of 18 cases. Arch Pathol Lab Med. 1993;117(6):636–640. [PubMed] [Google Scholar]
- 19.Truong LD, Shen SS. Immunohistochemical diagnosis of renal neoplasms. Arch Pathol Lab Med. 2011;135(1):92–109. doi: 10.5858/2010-0478-RAR.1. [DOI] [PubMed] [Google Scholar]
- 20.Ma S, Guan KL. Polycystic kidney disease: a Hippo connection. Genes Dev. 2018;32(11–12):737–739. doi: 10.1101/gad.316570.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Britschgi A, et al. The Hippo kinases LATS1 and 2 control human breast cell fate via crosstalk with ERα. Nature. 2017;541(7638):541–545. doi: 10.1038/nature20829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhao B, et al. TEAD mediates YAP-dependent gene induction and growth control. Genes Dev. 2008;22(14):1962–1971. doi: 10.1101/gad.1664408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Owens GK, et al. Molecular regulation of vascular smooth muscle cell differentiation in development and disease. Physiol Rev. 2004;84(3):767–801. doi: 10.1152/physrev.00041.2003. [DOI] [PubMed] [Google Scholar]
- 24.Zhao B, et al. A coordinated phosphorylation by Lats and CK1 regulates YAP stability through SCF(beta-TRCP) Genes Dev. 2010;24(1):72–85. doi: 10.1101/gad.1843810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lamouille S, et al. Molecular mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell Biol. 2014;15(3):178–196. doi: 10.1038/nrm3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nakaya Y, et al. RhoA and microtubule dynamics control cell-basement membrane interaction in EMT during gastrulation. Nat Cell Biol. 2008;10(7):765–775. doi: 10.1038/ncb1739. [DOI] [PubMed] [Google Scholar]
- 27.Gunaratne A, Di Guglielmo GM. Par6 is phosphorylated by aPKC to facilitate EMT. Cell Adh Migr. 2013;7(4):357–361. doi: 10.4161/cam.25651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu-Chittenden Y, et al. Genetic and pharmacological disruption of the TEAD-YAP complex suppresses the oncogenic activity of YAP. Genes Dev. 2012;26(12):1300–1305. doi: 10.1101/gad.192856.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bao Y, et al. A cell-based assay to screen stimulators of the Hippo pathway reveals the inhibitory effect of dobutamine on the YAP-dependent gene transcription. J Biochem. 2011;150(2):199–208. doi: 10.1093/jb/mvr063. [DOI] [PubMed] [Google Scholar]
- 30.Szeto SG, et al. YAP/TAZ are mechanoregulators of TGF-β-smad signaling and renal fibrogenesis. J Am Soc Nephrol. 2016;27(10):3117–3128. doi: 10.1681/ASN.2015050499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Argani P, et al. Biphasic hyalinizing psammomatous renal cell carcinoma (BHP RCC): a distinctive neoplasm associated with somatic NF2 mutations. Am J Surg Pathol. 2020;44(7):901–916. doi: 10.1097/PAS.0000000000001467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen YB, et al. Molecular analysis of aggressive renal cell carcinoma with unclassified histology reveals distinct subsets. Nat Commun. 2016;7:13131. doi: 10.1038/ncomms13131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lowry WE, et al. Defining the impact of beta-catenin/Tcf transactivation on epithelial stem cells. Genes Dev. 2005;19(13):1596–1611. doi: 10.1101/gad.1324905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ramalingam H, et al. Disparate levels of beta-catenin activity determine nephron progenitor cell fate. Dev Biol. 2018;440(1):13–21. doi: 10.1016/j.ydbio.2018.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moch H, et al. The 2016 WHO classification of tumours of the urinary system and male genital organs-part a: renal, penile, and testicular tumours. Eur Urol. 2016;70(1):93–105. doi: 10.1016/j.eururo.2016.02.029. [DOI] [PubMed] [Google Scholar]
- 36.Zisman A, et al. Unclassified renal cell carcinoma: clinical features and prognostic impact of a new histological subtype. J Urol. 2002;168(3):950–955. doi: 10.1016/S0022-5347(05)64549-1. [DOI] [PubMed] [Google Scholar]
- 37.Heallen T, et al. Hippo pathway inhibits Wnt signaling to restrain cardiomyocyte proliferation and heart size. Science. 2011;332(6028):458–461. doi: 10.1126/science.1199010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xin M, et al. Regulation of insulin-like growth factor signaling by Yap governs cardiomyocyte proliferation and embryonic heart size. Sci Signal. 2011;4(196):ra70. doi: 10.1126/scisignal.2002278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xin M, et al. Hippo pathway effector Yap promotes cardiac regeneration. Proc Natl Acad Sci U S A. 2013;110(34):13839–13844. doi: 10.1073/pnas.1313192110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Srinivas S, et al. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev Biol. 2001;1:4. doi: 10.1186/1471-213X-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Muzumdar MD, et al. A global double-fluorescent Cre reporter mouse. Genesis. 2007;45(9):593–605. doi: 10.1002/dvg.20335. [DOI] [PubMed] [Google Scholar]
- 42.Madisen L, et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci. 2010;13(1):133–140. doi: 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pavía-Jiménez A, et al. Establishing a human renal cell carcinoma tumorgraft platform for preclinical drug testing. Nat Protoc. 2014;9(8):1848–1859. doi: 10.1038/nprot.2014.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Patro R, et al. Salmon provides fast and bias-aware quantification of transcript expression. Nat Methods. 2017;14(4):417–419. doi: 10.1038/nmeth.4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Soneson C, et al. Differential analyses for RNA-seq: transcript-level estimates improve gene-level inferences. F1000Res. 2015;4:1521. doi: 10.12688/f1000research.7563.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Love MI, et al. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Colaprico A, et al. TCGAbiolinks: an R/Bioconductor package for integrative analysis of TCGA data. Nucleic Acids Res. 2016;44(8):e71. doi: 10.1093/nar/gkv1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Lin T, et al. Focal loss for dense object detection. Paper presented at: 2017 IEEE International Conference on Computer Vision; October 22–29, 2017; Venice, Italy. https://www.computer.org/csdl/proceedings/iccv/2017/12OmNAXxXaK Accessed April 26, 2021.
- 49. Bui TD, et al. Neural graph machines: learning neural networks using graphs. https://arxiv.org/abs/1703.04818 Accessed April 26, 2021.
- 50.Edgar R, et al. Gene expression omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30(1):207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Husted RF, et al. Characteristics of papillary collecting duct cells in primary culture. Am J Physiol. 1988;255(6 pt 2):F1160–F1169. doi: 10.1152/ajprenal.1988.255.6.F1160. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.