Abstract
Since the emergence of their primitive strains, the complexity surrounding their pathogenesis, constant genetic mutation and translation are contributing factors to the scarcity of a successful vaccine for coronaviruses till moment. Although, the recent announcement of vaccine breakthrough for COVID-19 renews the hope, however, there remains a major challenge of accessibility to urgently match the rapid global therapeutic demand for curtailing the pandemic, thereby creating an impetus for further search. The reassessment of results from a stream of experiments is of enormous importance in identifying bona fide lead-like candidates to fulfil this quest. This review comprehensively highlights the common pathomechanisms and pharmacological targets of HCoV-OC43, SARS-CoV-1, MERS-CoV and SARS-CoV-2, and potent therapeutic potentials from basic and clinical experimental investigations. The implicated targets for the prevention and treatment include the viral proteases (Mpro, PLpro, 3CLpro), viral structural proteins (S- and N-proteins), non-structural proteins (nsp 3, 8, 10, 14, 16), accessory protein (ns12.9), viroporins (3a, E, 8a), enzymes (RdRp, TMPRSS2, ADP-ribosyltransferase, MTase, 2’-O-MTase, TATase, furin, cathepsin, deamidated human triosephosphate isomerase), kinases (MAPK, ERK, PI3K, mTOR, AKT, Abl2), interleukin-6 receptor (IL-6R) and the human host receptor, ACE2. Notably among the 109 overviewed inhibitors include quercetin, eriodictyol, baicalin, luteolin, melatonin, resveratrol and berberine from natural products, GC373, NP164 and HR2P-M2 from peptides, 5F9, m336 and MERS-GD27 from specific human antibodies, imatinib, remdesivir, ivermectin, chloroquine, hydroxychloroquine, nafamostat, interferon-β and HCQ from repurposing libraries, some iron chelators and traditional medicines. This review represents a model for further translational studies for effective anti-CoV therapeutic designs.
Keywords: Coronaviridae, Pathophysiology, Drug targets, Multi-target pharmacology, Biochemical analysis, Therapeutic design
1. Introduction
The entire humanity has been experiencing a sudden outbreak of viral infectious diseases indefatigably. The emergence of new strains or re-emergence of mutated pathogenic types has always been creating a public health emergency while the development of effective vaccines lasts years or decades even with sophisticated resources and accelerated procedures. Notably among the recently observed global viral-mediated pandemics include the Spanish flu, Zika virus, Ebola virus and the current coronavirus (Dallmeier and Neyts, 2016; Giulietti et al., 2018). The coronaviridae family consists of seven species infectious to human cells, largely grouped into the alpha- and beta-coronaviruses (Cabezón and Arechaga, 2020). The members of the alpha- group include the human coronaviruses (HCoVs), HCoV-NL6E, HCoV-229E and feline infectious peritonitis (FIP) virus while HCoV-OC43, HCoV-HKU1, SARS-CoV-1, MERS-CoV and SARS-CoV-2 constitute the beta family with host reservoirs mostly in bats, rodents, cattle, palm civets and humans (Corman et al., 2018; Pillaiyar et al., 2020). Among the beta types, HCoV-OC43 was the first human coronavirus genotypic strains to be identified in the mid-1960s with a relative innocuous identification as a source of common cold until 2003, when a global epidemic outbreak of the severe acute respiratory syndrome (SARS) was reported with primary origin through exposures to a health care worker in Guangdong province, China. The Middle East respiratory syndrome coronavirus (MERS-CoV) was firstly discovered in Saudi Arabia and spread worldwide around 2012 with a higher mortality rate (≈40%) (Chen et al., 2017; Galante et al., 2016; Yamamoto et al., 2016). According to virologists, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is a β-CoV family member of coronaviridae of the order Nidovirales, similar to SARS-CoV-1 and MERS-CoV (Elfiky, 2020; Xia et al., 2020a).
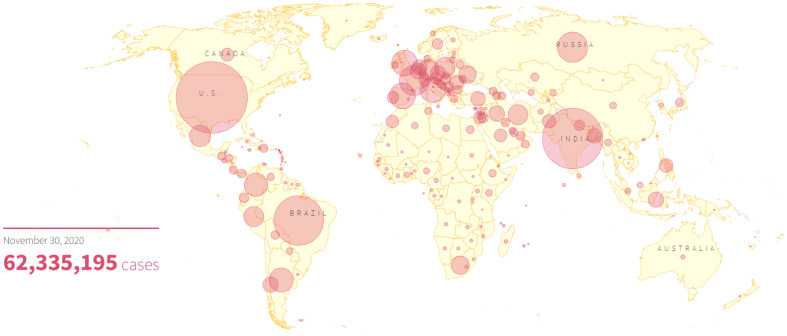
Coronavirus disease 2019 (COVID-19) is a newly emerged viral infection of SARS-CoV-2, firstly observed in a suburb city of China towards the end of the year 2019 and had spread across almost all continents of the world. It was later pronounced as COVID-19 pandemic by the World Health Organization (WHO) (Zhang et al., 2020a), the situation which severely threatens the global health, economy and population through the daily loss of lives, leading to its further declaration as public health emergency of international concern by WHO in January 2020 (Zhang et al., 2020a; Atri et al., 2020). The death rate of COVID-19 as of 30th November 2020 stands at 2.30% with 1,455,620 mortality of the global population out of the confirmed cases of 62,335,195, and up to 40,123,407 (64.40%) of infected patients have recovered while the estimated number of active cases remains 20,756,168 (33.30%). Around 220 countries and territories of the world are currently battling with the disease out of which about 196 have reported fatalities, prominently US, Brazil, India, Mexico, Italy and Russia (Fig. 1 ) (R. Graphics, 2020).
Fig. 1.
World map illustrating the global spread of coronavirus as of November 30, 2020. The number of confirmed cases is represented by the size of the circles with the US, Brazil, India and Mexico currently on top. (Reproduced from R. Graphics (2020), free access.)
1.1. Aetiology and symptoms of coronavirus infection
Although, almost all HCoV infections are primarily zoonotic with presumed origin from bats and have dynamic nature favouring the feasibility of a cross-species transmission (Corman et al., 2018; Kim et al., 2012). The human-to-human cross-infection occurs predominantly through respiratory droplets, during coughing, sneezing and sometimes through excretory products. These have been earmarked as factors greatly contributing to the exponential spread of these viral diseases, and sometimes in association with the lower respiratory tract infections. To further argue the hypothesis of common pathogenesis, the overview of chest radiographs and imaging outcomes reportedly indicates the organization of pneumonia-like arrangements by fibrocellular intra-alveolar structuring with bronchiolitis obliterans in the infections induced by MERS-CoV and SARS-CoV (van den Brand et al., 2015). The common symptoms of the diseases vary from mild such as fever, fatigue, nostril blockage, dry cough, flu-like to acute respiratory distress syndrome (ARDS), acute lung injury (ALI), pneumonia, cardiac arrest, dyspnea, liver damage and death, although asymptomatic cases have been frequently reported (Gupta et al., 2020; Wu et al., 2020; Zhang et al., 2020b). Oftentimes, COVID-19-related mortalities are reported with several comorbidities, predominantly, cardiovascular diseases, diabetes and renal failure (Albini et al., 2020). Thus, effective preventive measures are essential for the control of the pandemic, as such, the WHO recommended several preventive ethics and the use of hygienic products including povidone‑iodine (PVP-I) mouth wash (Anderson et al., 2020).
The global therapeutic demand to mitigate the impact of the deadly virus on humans is increasingly high while the new strains emerge at unpredictable rates. Despite the continuous sheer amount of concerted scientific efforts towards identifying efficacious and safe therapeutic options against the CoVs since the emergence of their primitive strains, the complexity surrounding their aetiology, constant genetic mutation and translation are contributing factors to the scarcity of a successful vaccine till the moment. Secondly, the available drugs in clinical trials display unpleasant side effects. Prominently, remdesivir was reported to cause some adverse events in 13 out of 35 (22.8%) patients from intensive care unit (ICU) and infectious disease ward (IDW) enrolled for clinical studies in Italy (Antinori et al., 2020). Although, the recent announcement of vaccine breakthrough for COVID-19 renews the hope, however, there remains a major challenge of accessibility to match the rapid global demand for the highly pathogenic disease. Thus, there is a need for sustained innovative, preventive and treatment countermeasures to upturn the effects of the unwanted phenomenon. The reassessment of results from a stream of experiments are of enormous importance in identifying bona fide lead-like candidates to fulfil this quest. Taking advantage of their similar pathology and pathomechanisms, a large range of prospective candidates are being experimented against the old and newly emerged strains. Many of these agents demonstrate strong inhibitory potentials across the strains at various experimental levels, a propensity for effective therapeutic development for the current and emerging CoVs. Although, some reports have been recently documented with a specific overview on either the therapeutic target or the class of inhibitors (Lim et al., 2016; Verma et al., 2020; Xian et al., 2020). However, an all-inclusive analysis of homologous human CoV strains from features through infection to treatment remains sparse. This review comprehensively highlights the common pathomechanisms and pharmacological targets of four beta-CoVs infectious to human, HCoV-OC43, SARS-CoV-1, MERS-CoV and SARS-CoV-2, and potential therapeutic candidates from the recently reported basic and clinical experimental investigations.
1.2. Structural features, pathophysiology of coronavirus infection and therapeutic targets
The understanding of the structural characteristics is essential for speedy approaches towards therapeutic development including structure-assisted drug design, high throughput screening (HTS), virtual screening (VS), basic and clinical experimental studies. Members of the pleomorphic viral family including the CoVs possess common structural features such as the positive-sense RNA molecule of around 30 kb, open reading frames (ORFs), structural proteins: envelope (E), membrane (M), spike (S) glycoprotein for viral infusion into the host cells through the viral-host receptor, angiotensin-converting enzyme 2 (ACE2) with an expression on the surface of the pulmonary, intestinal and renal cells inclusively (Fig. 2 ) (Albini et al., 2020; Huang et al., 2017). Similarly, a clinical case study indicates the complications of SARS-CoV-2-related syndrome with ARDs and diagnosed cardiac injury expressed as myocarditis, stress cardiomyopathy and demand ischemia, thereby inclusively implicating the interleukin-6 and aldose reductase as pathophysiological targets (Coyle et al., 2020). Due to these complications, the applications of the ACE-2 inhibitors, renin-angiotensin-aldosterone system (RAAS) inhibitors and/or angiotensin receptor blockers (ARBs) in the experimental trials of SARS-CoV prevention and treatment creates a scientific argument (Mourad and Levy, 2020). Some researchers suggest the discontinuation of ACE inhibitors and ARBs in the therapeutic trials based on the hypothesis that they facilitate SARS-CoV-2 diffusion into cells, thereby contributing to the severity, others claim there exist no basic or clinical experimental shreds of evidence to support the argument, as such they are recommended for continuous applications (Alexandre et al., 2020; Kai and Kai, 2020; Sriram and Insel, 2020; Verdecchia et al., 2020). In addition to the ACE2, sometimes the FURIN gene, a paired basic amino acid cleaving enzyme located on chromosome 15, performs relevant pathophysiological functions. Thus, its repressors/silencers are putatively identified as markers for mitigating the viral entry (Glinsky, 2020). Furthermore, the viral main proteases (Mpro) and chymotrypsin-like proteases (3C/3CLpro) and papain-like proteases (PLpro) are important enzymes which play pivotal roles in mediating replication and transcription, usually through the cleavage of viral polyprotein into intermediate or fully developed new virus proteins, thus constitute essential therapeutic targets (Kim et al., 2012; Jin et al., 2020). Other structural features include the nucleocapsid (N) phosphoprotein, the structural entities essentials for the formation of the RNP-enclosed virion particles for viral infection (Bhowmik et al., 2020). This protein encodes the viral genome into a helical structure, ribonucleocapsid protein (RNP) and plays essential parts in the viral multifarious activities including self-assembly.
Fig. 2.
The expression of ACE2 on the gut, lung, epithelial, endothelial, intestinal and testis cells. (Reproduced from Albini et al. (2020), Licenced under CC BY 4.0.)
There are only 600–1500 chemotherapeutic targets originally encoded by the human genome for small molecule drugs, but posttranslational modifications make the druggable targets considerably larger (Enriquez-Flores et al., 2020). However, CoVs exceptionally to other RNA viruses, possess a large genome associated with low rates of mutation. All CoVs contain a conserved macrodomain non-structural protein 3 (nsp 3) which enzymatically detaches the covalently bonded adenosine diphosphate (ADP)-ribose from protein targets, thereby reversing the ADP-ribosyltransferase activity and immune innate response. This indicates the possibility of ADP-ribosylation in the pathogenic CoV virulence factor and the potential of the enzyme as a target for vaccine development (Fehr et al., 2016). The CoVs also code for nsp14, a bifunctional enzyme with RNA cap guanine N-7-methyltransferase (MTase) and 3′-5′ exoribonucleic (ExoN) activities, thereby making it a potential therapeutic target. The translation and stability of the RNA are greatly influenced by the 5’cap structures of the eukaryotic messenger RNA (mRNA) in most viruses with replication occurring in the cytoplasm of eukaryotes which releases 2’-O-MTase for mRNA modification through the m7GppNm 5′-cap formation. The mechanisms enhance the replication and an escape to innate immune recognition within the host cells. However, these resistance-inclined processes require the stimulation of the 2’-O-MTase activity of SARS-CoV nsp16 by nsp10. Unarguably, the nsp16/nsp10 2’-O-MTase activation is thus, an important therapeutic target for overcoming viral replication and innate resistance. The expression was demonstrated in vitro using a biochemical assay for MTase activity, while a peptide, TP29 derived from the nsp16/nsp10 domain shows great experimental inhibition of the pathway (Atri et al., 2020; Wang et al., 2015). Similar to these are the ns12.9 accessory proteins which function as a viroporin and are reportedly involved in the pathogenesis and morphogenesis of virion in HCoV-OC43 (Zhang et al., 2015). Also reported is the nsp8 which participates in diverse non-canonical template-dependent polymerase activities including the metal ion-dependent RNA 3′-terminal adenylyl transferase (TATase) activity for viral replication in HCoV-229E (Tvarogová et al., 2019).
The viroporins are viral proteins associated with ion channel activity, stimulation of viral release from infected cells, and are essential for virulence pathogenesis and replication. The SARS-CoVs encode 3a, E and 8a viroporins. Whilst the 3a and E are actively involved in the control of various cell functions including replication and pathogenesis due to the presence of PDZ-binding motif (PBM), a potential for binding over 400 cellular proteins with PDZ domain, the 8E contributes less to these viability pathways (Castano-Rodriguez et al., 2018). Thus, the proteins are relevant targets for therapy and mutations for vaccine development.
Upon gaining entrance into the host respiratory tract, SARS-CoV-2 infuses the host cell in a similar way to SARS-CoV-1 and MERS-CoV via the membrane-bound ACE2 and employs the transmembrane protease serine 2 (TMPRSS2) for S protein activation, the progression of which the viral fusion and replication occur (Hoffmann et al., 2020; Xia et al., 2020b). The down-regulation of TMPRSS2 expression has been reported as a pharmacological pathway towards the inhibition of influenza A virus (Shen et al., 2020), a similar pathogenic organism to CoVs. Other host proteases such as the cathepsin and FURIN also undergo some proteolysis, facilitating the infusion of viral S-protein into the host cells (Yamamoto et al., 2016). Although, the possibility of interactions between the viral S-protein and some other human cell adhesion factors such as the dipeptidyl peptidase, cyclophilins and ezrin cannot be overruled (Vankadari and Wilce, 2020). Also, sex-dependent epidemiology is possibly observed in the current COVID-19 due to the encoding of ACE2 gene expression on X chromosomes (Atri et al., 2020), supporting the fact that the signal pathway of the human ACE2 is imperative in the control of COVID-19 infection in addition to the viral S-protein and the nucleocapsid NTD (Kang et al., 2020a; Wrapp et al., 2020; Wu and Yang, 2020).
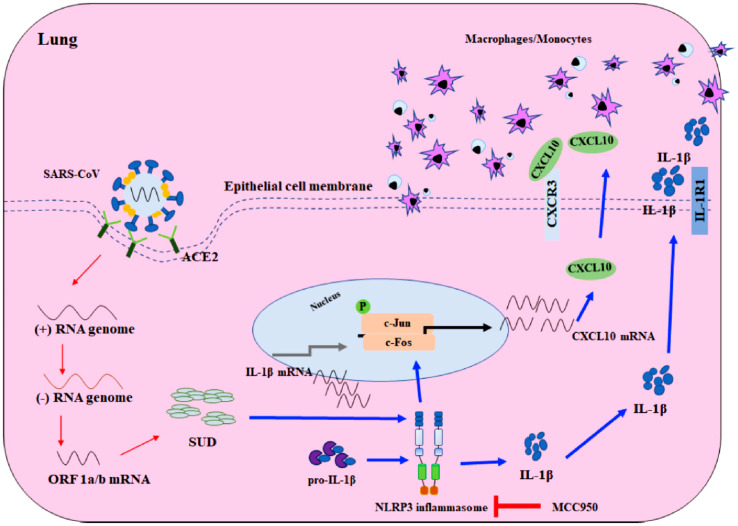
Some analyses such as the temporal kinome analysis (TKA), functional network analysis (FNA) and the overrepresentation analysis (ORA) on human MERS-CoV-infected hepatocytes with peptide kinome arrays indicate that the signalling responses on mitogen-activated protein kinase (MAPK)/extracellular signal-regulated kinase (ERK) and phosphoinositol 3-kinase (PI3K)/mammalian target of rapamycin (mTOR)/serine-threonine kinase (AKT) are modulated specifically in MERS-CoV infection (Kindrachuk et al., 2015). Similarly, the Abelson tyrosine-protein kinase 2 (Abl2) has also been implicated in the pathways of SARS-CoV-1, MERS-CoV and SARS-CoV-2 infection and replication (Coleman et al., 2016; Nabavi et al., 2020). SARS-CoVs are known to induce lung injury mediated by cytokine/chemokine storm, thus, SARS-CoV unique domain (SUD) consisting of the C, M and N macrodomains are possible targets for G-quadruplet binding activity. As experimental evidence, chemokine analysis indicates that the SUD actuated the nucleotide-binding oligomerisation domain, leucine-rich-repeat and pyrin domain-containing protein 3 (NLRP3) inflammasome-dependent C-X-C motif chemokine ligand 10 (CXCL10)-mediated pulmonary inflammation, confirmed by NLRP3 inflammasome inhibitor in a mouse model (Fig. 3 ) (Chang et al., 2020). The deamidated human triosephosphate isomerase, a glycolytic enzyme was demonstrated as another promising druggable target for the prevention/treatment of SARS-CoV-2 infection using in silico and in vitro experimental assays (Enriquez-Flores et al., 2020).
Fig. 3.
Schematic diagram of SARS-CoV SUD induced CXCL10-mediated pulmonary inflammation through the NLRP3 inflammasome pathway. Red arrow; viral transcription and translation, blue arrow; the presumed pathway induced by SARS-CoV SUD. (Reproduced from Chang et al. (2020), Licenced under CC BY 4.0.)
1.3. Homology of active therapeutic targets of coronavirus strains
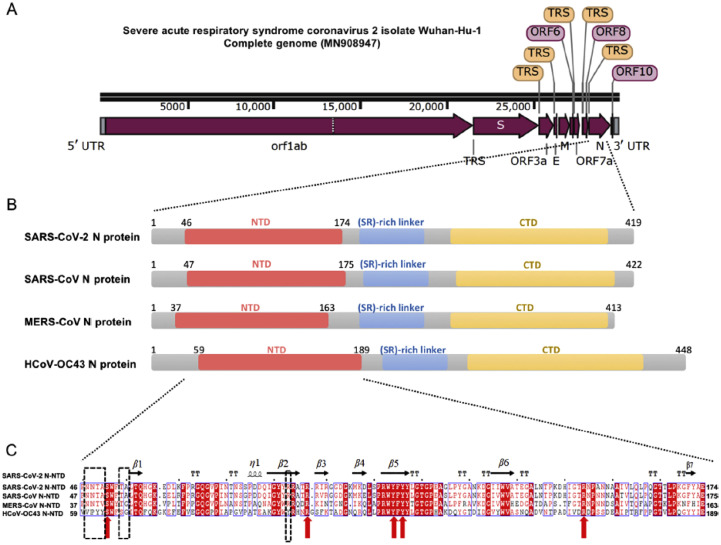
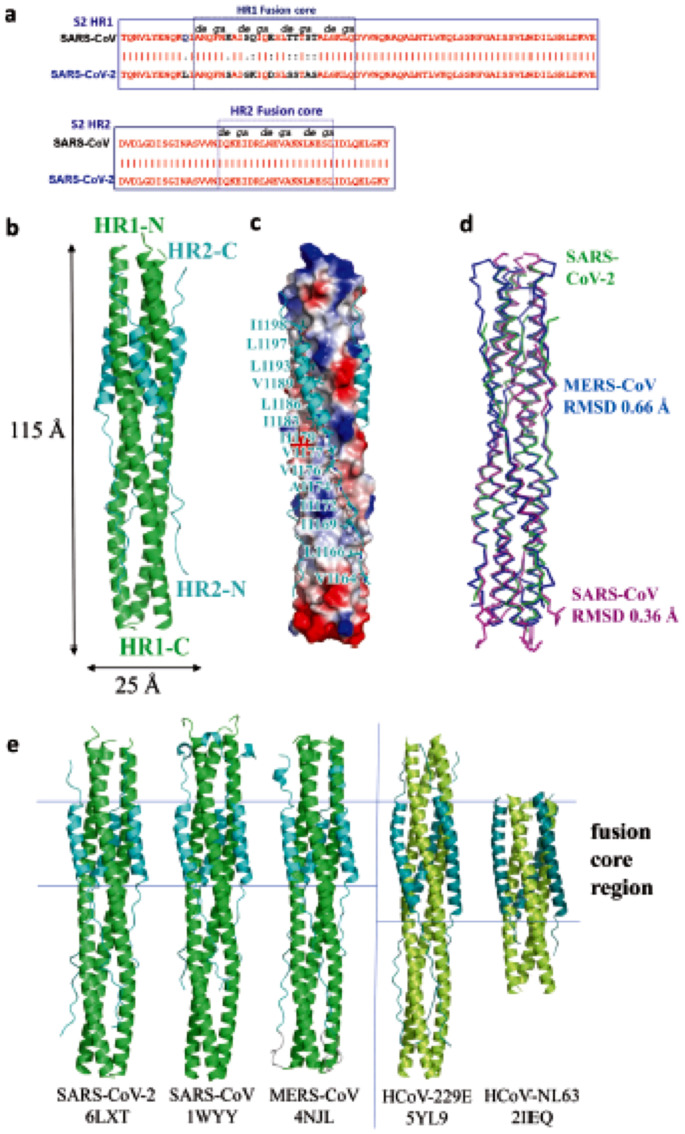
Possibly arising from their evolutional differentiation, the four strains of coronaviruses under focus, HCoV-OC43, SARS-CoV-1, MERS-CoV and SARS-CoV-2 have been resolved with genomic and structural homology especially at the active sites of their main therapeutic targets, the NTD of N-protein (Fig. 4 ) (Kang et al., 2020a), the S-protein at the 6-HB fusion core (Fig. 5 ) (Xia et al., 2020b; Xia et al., 2020c), 3CLpro (ul Qamar et al., 2020) and the PLpro (Fischer et al., 2020). This structural homology further unveils the underlying similarity in their molecular mechanisms of infection, pharmacological pathways and therapeutic hypotheses. Further analyses suggest the inhibitory mechanisms against the essential proteases for proteolytic activities of viral polyproteins as a common therapeutic pathway against the CoVs (Fischer et al., 2020).
Fig. 4.
Sequence features of SARS-CoV-2 nucleocapsid protein. (A) The complete genomic features of SARS-CoV-2 Wuhan-Hu-1 isolate (Gene bank: MN908947). UTR = untranslated region; TRS = transcriptional regulatory sequence; orf/ORF = open reading frame; E = envelope protein-encoding region; S = spike glycoprotein encoding region; N = nucleocapsid protein-encoding region; M = membrane protein-encoding region illustrated by Snap Gene Viewer. (B) Domain architectures of CoV N-protein. NTD = N-terminal RNA-binding domain; CTD = C-terminal dimerization domain. (C) Multiple sequence alignment of N-NTD of SARS-CoV-2 with that SARS-CoV (UniProtKB P59595), MERS-CoV (UniProtKB R9UM87) and HCoV-OC43 (UniProtKB P33469). Red arrows indicate conserved residues for ribonucleotide binding sites and dash boxes indicate residues variably in structural comparison. (Reproduced from Kang et al. (2020a), Licenced under CC BY-NC-ND 4.0.)
Fig. 5.
Overall structure of post-fusion 6-HB in SARS-CoV-2. (a) Sequence alignment of HR1 and HR2 domain in SARS-CoV and SARS-CoV-2. (b) Structure of SARS-CoV-2 6-HB shown in cartoon representation with HR1 (green) and HR2 (cyan) and structural dimensions are indicated in Å. (c) Electrostatic surface of HR1 trimer of SARS-CoV-2 6-HB and cartoon representation of HR2 domain, the important binding residues of which are shown in sticks and labelled. (d) The superposition of the 6-HB structure of SARS-CoV (PDB 1WYY), MERS-CoV (PDB 4NJL) and SARS-CoV-2 (PDB 6LXT) shown in the ribbon. The RMSD between the structures is indicated. The sequence comparison of the 6-HB structure of different HCoVs is shown in cartoon representation with different colours for HR1 and HR2. The helical fusion core regions are indicated. (Reproduced from Xia et al. (2020b), Licenced under CC BY 4.0.)
1.4. Medical hypotheses and randomised trials
The emergencies usually associated with CoV outbreaks warrant reasonable therapeutic suggestions and randomised trials by experts based on previous relevant experimental experiences. The complication of CoV infection with other ailments oftentimes regulates the treatment options. For instance, the Royal College of Paediatrics and Child Health reported complications of Kawasaki disease (KD) syndromes in some children infected with SARS-CoV-2, and this led to the suggestion of some KD suppressors such as the serum interferon-gamma (IFNγ) and interleukin 6 (IL-6) with diminishing inflammatory cytokine mechanisms as adjuvant treatments (Bar-Or et al., 2020). Impressive antiviral activities of some phytochemicals such as saikosaponins which include the inhibition of influenza virus and several inflammatory mediators such as reactive oxygen species (ROS), cyclooxygenase-2 (COX-2) and the interleukins (IL-6, IL-8, IL-10) mainly prompted their suggestive trials during the first wave of COVID-19 (Bahbah et al., 2020). In a commentary reported by Bleasel and Peterson, emetine, an alkaloid from ipecacuanha root demonstrates stronger inhibitory potentials against MERS-CoV and SARS-CoV-1 in 500–2000-folds of the effect against Entamoeba histolytica, its main target as anti-amoebiasis. Although the treatment is associated with emetic complications in patients, it is manageable with 5-HT3 antagonists such as ondansetron. It is therefore suggested to deliver emergent therapeutic demand against COVID-19 (Bleasel and Peterson, 2020). Clinical impediments of neuroinvasive and neurotropic effects have been reportedly observed in association with COVID-19 in forms of neurological symptoms such as hyposmia, anosmia and dysgeusia, indicating the applicability of doxycycline in treatment studies (Bonzano et al., 2020). Chloroquine (CQ) and its derivative, hydroxychloroquine (HCQ) are among the most popular inhibitors of enveloped viruses such as dengue, Nipah and Hendra paramyxoviruses, human immunodeficiency virus (HIV) and the CoVs in vitro through the hijacking of the endocytic pathway, the process in which the endosomal lipid, bis(monoacylglycero)phosphate reportedly plays a vital role. Other suggested mechanisms of chemotherapeutic actions of these weak bases include the increment in the lysosomal pH through the alkylation of phagolysosome, inhibition of glycosylation, interference with the virus-receptor binding and proliferation, and inhibition of the acidic proteases that are responsible for the maturation of viral fusion proteins (Carriere et al., 2020; Colson et al., 2020; Monti and Montecucco, 2020). Although, with limited experiments, the in vivo efficacy is strongly debated still, however, it is earmarked as early therapeutic targets for SARS-CoV treatment (Carriere et al., 2020). Other therapeutic hypotheses for clinical investigation include teicoplanin, a glycopeptide antibiotic for treating several bacterial infections (Baron et al., 2020), heparin, an anti-inflammation agent (Belen-Apak and Sarialioglu, 2020), mevalonate pathway inhibitors and statins owing to their anti-inflammatory and immunomodulatory effects which benefit patients suffering from viral pathology such as influenza (Bifulco and Gazzerro, 2020). From the reported activities, these bioactive agents are hypothesized to possess potentials for improving prognosis in coronavirus pathologies.
2. Potential inhibitors from experimental studies
Since the similarities in pathophysiologies and the protein residual sequencing (especially at the active sites) of the existing and emerging CoV strains support the hypothesis of similar pathomechanisms of infection, similar approaches could be rationally employed for their prevention and treatment (Xia et al., 2020b), which have been virtually demonstrated (Xia et al., 2020b; Mercurio et al., 2021; Yadav et al., 2020). From experimental studies, the inhibition of the 3CLpro and the PLpro have been documented for lopinavir, although experimental arguments favour its inhibition of HIV protease more (Cattaneo et al., 2020). The S-protein and ACE2 by arbidol, viral RNA-dependent RNA polymerase (RdRp) by remdesivir and ribavirin, TMPRSS2 by camostat mesylate and ACE2 glycosylation by CQ analogues (Liu et al., 2020a). Other conventional medications known for the mitigation of ARDS, influenza, acute lung injury (ALI), inflammation, hypertension, immunomodulation have also been evaluated against pandemics, including losartan, statins and ARBs (Atri et al., 2020; Phadke and Saunik, 2020). Improved immune responses in SARS-CoV and MERS-CoV infected patients have also been investigated upon administration of chemokine and cytokine modulators such as tocilizumab and the human mesenchymal cells (Bennardo et al., 2020; Ceribelli et al., 2020; Shetty, 2020).
Among the herbal medicines screened, respiratory detox shot and Shuanghuanglian oral liquid, traditional Chinese medicines with known antiviral history experimented in early prevention and control of COVID-19 through inhibition of SARS-CoV-2's 3CLpro, PLpro, spike protein etc. by theoretical binding and in vitro studies (Su et al., 2020; Yang et al., 2020). The Xeubijing injection was also demonstrated with the active expression of the inflammatory response and lung injury, indicated by the improvement on lymphocyte count, partial pressure of CO2 in the artery and oxygenation index in patients with critical complicated conditions of COVID-19 (Ma et al., 2020a). Similarly, the Liu Shen capsule, Lianhuaqingwen and Gene-Eden-Vir/Novirin have been reported with inhibitory potential against SARS-CoV-2 through the regulation of innate immune response, inhibition of cell entry and replication, inhibition of viral proteases and reduction in pro-inflammatory cytokines (Ma et al., 2020b; Polansky and Lori, 2020; Runfeng et al., 2020).
The natural product, melatonin is a protective agent against some pathogenesis associated with viral infections and ARDS has also been speculated as an adjuvant in the treatment of some chronic ailments induced by COVID-19 including ARDS, ALI and pneumonia (Zhang et al., 2020b). Other phytochemicals such as theaflavin, quercetin, baicalin, berberine, eriodictyol, lycorine etc. with experimental antiviral potency have also been reviewed among the currently available options for further studies (Boukhatem and Setzer, 2020).
Sometimes the inhibitors are in the form of antibodies which could mediate through prophylactic therapeutic development and monoclonal immunotherapies. For instance, the monoclonal antibody (mAb), 5F9 binds to the NTD of MERS-CoV at the S1 subunit as a neutralizing epitope, thereby mapping the S-protein for viral infusion (Chen et al., 2017), and the murine antisera which reportedly neutralised the MERS-CoV activity in vitro and in vivo basic and clinical experimental models (New et al., 2019).
Although scientists are still in search of an effective vaccine against CoV infections, some candidates are already undergoing randomised clinical trials while others are still in the basic and pre-clinical experimental stages. Their sources range from traditional, scriptural, herbal and conventional medicines most of which exhibit multi-targeting and networking pharmacology, a propensity for overcoming drug resistance associated with the pandemic. The overview of these important therapeutic innovations is comprehensively addressed according to the CoV strain targets as follows.
2.1. Inhibitors of HCoV-OC43
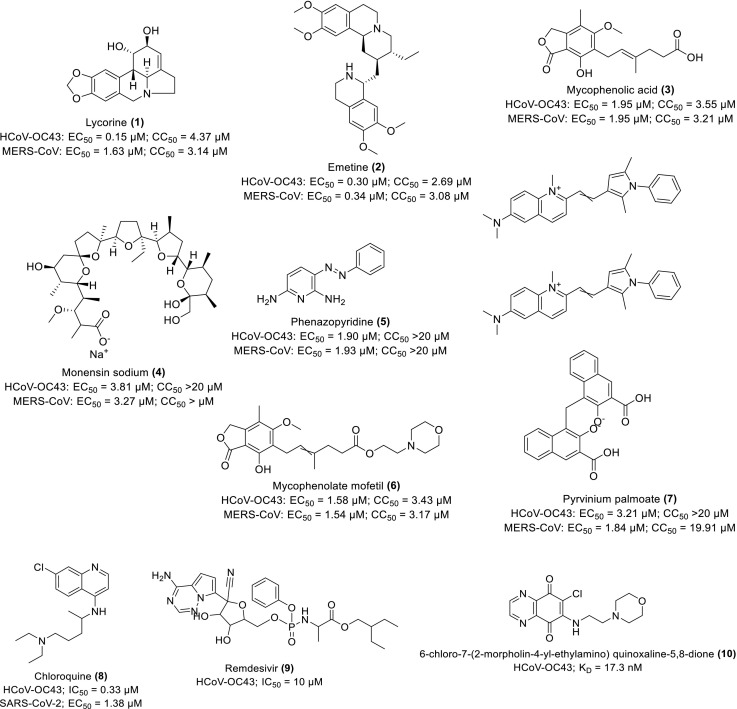
Shen and co-researchers performed an HTS assay on 2000 approved drugs out of which 56 hits were retrieved. Upon further biochemical analysis against the wild type CoV strains, seven repurposing drugs, 1–7 (Scheme 1 ) were identified with strong inhibition against the viral replications of HCoV-OC43 and MERS-CoV in vitro at low EC50 and CC50 values of <5 and >20 μM respectively. The further evaluation shows that 1 protected the BALB/c mice from the lethality of HCoV-OC43 as observed using novel RT bioluminescence imaging for monitoring HCoV-OC43 distribution in vivo mice while 2 blocked the entry of MERS-CoV (Shen et al., 2019). These compounds especially, 1 and 2 are therefore recommended as potential therapeutics against CoV entry and replication upon further study.
Scheme 1.
Chemical structures of some repurposing drugs and their activity against HCoV-OC43 and MERS-CoV.
The HCoV-OC43 on emergence was pinpointed as the major cause of common cold and upper respiratory tract infections with resistance to several available drugs (Shen et al., 2016). Owing to this speculation, Shen and co-workers developed four recombinants HCoVs-OC43 expressing the Renilla luciferase (Rluc) receptor gene. They demonstrated that the HCoV-OC43 replication was strongly inhibited by 8 in higher terms than 9 in vitro with an inhibitory concentration (IC50) value of 0.33 μM and 10 μM respectively (Scheme 1). The adopted assays include Rluc activity, dual-luciferase reporter, small interfering RNA screening (siRNA), a double-stranded, non-coding RNA molecule with a length comprising of 20–25 base pairs, Western blot and antiviral assays. Other experimental protocols include cell viability assay using BHK-21, Huh and HEK-293 T cell lines, and in vivo mice model (Shen et al., 2016).
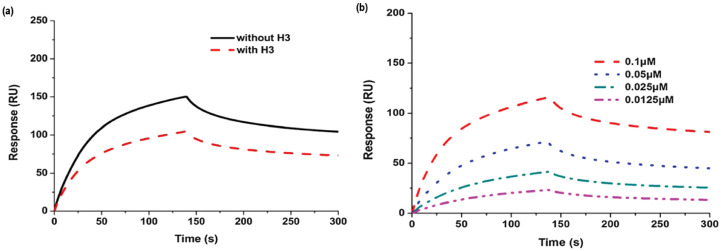
A biophysical Surface Plasmon Resonance (SPR) analysis shows a significant decrease (>20%) in the RNA binding capacity of the N-protein of HCoV-OC43 induced by 10. Further analyses by X-ray crystallography and fluorescence titration indicate that the mechanism of this action includes the antagonistic binding interaction between the compound and the NTD of the protein to prevent RNA-HCoV-OC43 interaction in a concentration- and time-dependent manner (Fig. 6 ) (Chang et al., 2016).
Fig. 6.
Effects of H3 on the RNA binding activity of NP (a) Sensorgram of the interaction between the immobilized single-stranded RNA and full-length HCoV-OC43 NPs in the presence of H3 at 2 μM; (b) Sensorgram of the interaction between the immobilized single-stranded RNA and full-length HCoV-OC43 NPs at different concentrations in the presence of H3. (Reproduced from Chang et al. (2016) with permission, copyright Royal Society of Chemistry 2016.)
Among the reported peptides for the inhibitory activity against the CoV S-protein, 11 with amino acid sequence SLDQINVTFLDEYEMKKLEEAIKKLEEGSGSGPEG4 display interesting activity against SARS-CoV-2 S-protein-mediated membrane fusion and pseudoinfection quantitatively with IC50 of 1.30 and 15.8 nM respectively. The application of 0.5 mg/kg of 11 intranasally in new-born mice pre and post-infection with HCoV-OC43 at TCID50 of 100 protected the mice from the viral infection. The mouse (100%) recovered from a significant reduction in body weight and 100% mortality upon HCoV-OC43 infection, indicating the promising prophylactic and therapeutic effects of the peptide for further studies (Xia et al., 2020b).
2.2. Inhibitors of SARS-CoV-1
In similitude to the report of Shen et al. (2016), and based on their expression of genetic regulation and RNA interference (RNAi) effects, the siRNAs have been designed into bioactive agents against virulent diseases such as cancer, Zika virus and SARS-CoV-1 where effective therapeutic options remain challenging (Giulietti et al., 2018; Ghosh et al., 2020). These agents reportedly demonstrate interesting inhibitory activities indicated by a 50–95% reduction in viral loads in vitro against SARS-CoV-1 and MERS-CoV, through various mechanisms, targeting the encoding sequence for the viral RdRp, proteolytic enzymes, helicase and N-protein. Enlisted among them include CN1548054 (12), CN101173275 (13), CN101085986 (14), CN101113158 (15), WO2005019410 (16) and US8653252 (17). Some of these potent anti-CoV agents have been developed into nanoparticles for enhanced delivery. Thus, these siRNA-based therapeutics are later suggested by Ghosh and co-workers for trials in SARS-CoV-2 pharmacological pathways by specifically targeting the viral nsp5 and RdRp (Ghosh et al., 2020).
The 3CLpro of the coronaviruses including the SARS-CoV-1 contains the chymotrypsin-like folds and a catalytic dyad/triad with a conserved cysteine residue as an active site. Thus, potent inhibitors implicated with the inhibitory activity against the viral replication through the 3CLpro pathomechanisms have been documented. Among them are the dipeptidyl residues with various active arrowheads. They demonstrate effective antiviral activities through the inhibition of 3CLpro of SARS-CoV-1 as observed using fluorescence resonance energy transfer (FRET) protease assay. The peptide derivatives, 18–20 with aldehyde, α-ketonamide and bisulfite adduct arrowheads possess IC50 values of 3.48 ± 1.59, 4.66 ± 0.19 and 4.35 ± 0.10 μM compared to a standard antiviral drug, rupintrivir with IC50 value >50 μM. The potential 3CLpro inhibitors, 18 and 20 were further investigated against FIP using TCID50, Western blot and nonspecific cytotoxicity assays, where they show synergistic/additive efficacy in combination with cathepsin B inhibitor against the virulent infection (Kim et al., 2012; Kim et al., 2013). Based on the experimental results, the compounds are suggested as potential 3CLpro-targeted anti-SARS-CoV-1 worthy of further therapeutic advancement.
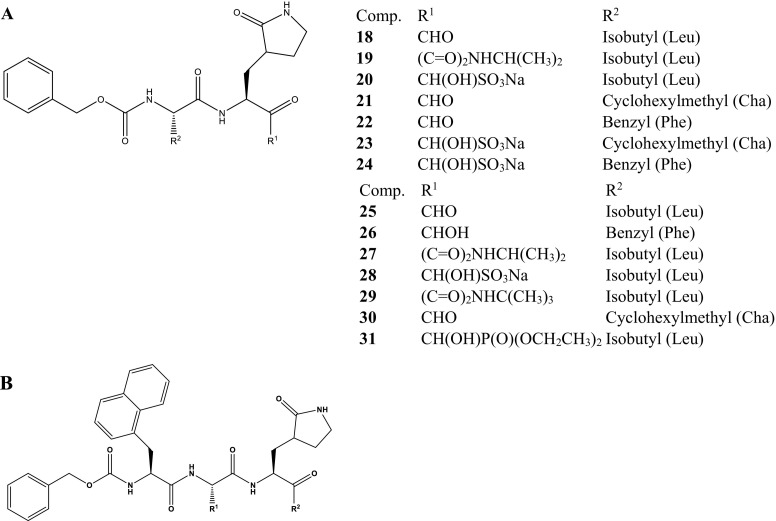
In 2016, Kim et al. reported new series of the dipeptidyl, 18–24 and tripeptidyl, 25–31 potent inhibitors of feline coronavirus (FCoV) 3CLpro (Scheme 2 ) with interesting activities (Table 1 ) (Kim et al., 2015). These are considered relevant since the FCoV shares common pathogenesis with SARS-CoV-1 in respect to immune response and depletion of T cells along the 3CLpro pathway as defense mechanisms. The anti-CoV activities were investigated by the addition of each compound to the Crandell-Rees feline kidney (CRFK) cells which are immediately infected with FCoV. The viral titres and effective concentrations at 50% (EC50) values were determined by the TCID50 method after 24 h of incubation as well as the half-cytotoxic concentration (CC50) of each compound. Thus, through extrapolation, the results indicate the promising potential of the peptidyl derivatives to inhibit CoV replication by targeting the 3CLpro for further development, especially 18, 20, 23, 25, 28 and 30.
Scheme 2.
Chemical structures of the peptidyl inhibitors of FCoV/SARS-CoV-1 3CLpro (A) Dipeptidyl derivatives; (B) Tripeptidyl derivatives (Kim et al., 2015).
Table 1.
Anti-FCoV activities of the dipeptidyl and tripeptidyl derivatives.
| Dipeptidyl |
Tripeptidyl |
||||
|---|---|---|---|---|---|
| Compound | FCoV (EC50 μM) | CC50 (μM) | Compound | FCoV (EC50 μM) | CC50 (μM) |
| 18 | 0.02 ± 0.01 | >150 | 25 | 0.02 ± 0.01 | 70.29 ± 5.6 |
| 20 | 0.04 ± 0.04 | >150 | 26 | 0.86 ± 0.72 | 40.57 ± 10 |
| 21 | 0.10 ± 0.03 | >150 | 27 | 0.54 ± 0.28 | 32.34 ± 1.9 |
| 22 | 0.43 ± 0.31 | >150 | 28 | 0.04 ± 0.03 | 61.91 ± 0.2 |
| 23 | 0.06 ± 0.05 | >150 | 29 | 0.18 ± 0.12 | 32.01 ± 1.3 |
| 24 | 0.12 ± 0.05 | >150 | 30 | 0.06 ± 0.06 | 21.96 ± 5.1 |
| 31 | 0.06 ± 0.001 | >150 | |||
Measured as the mean and standard error of the means (SEM).
The dipeptidyl, 20 were further investigated for safety, pharmacokinetics and 3CLpro-targeted efficacy in vivo FIP virus-infected cats. The experimental fatality of the cats was 100% once the clinical indications become obvious after the infection. Upon treatment with 20, the cats fully recover from the associated ailments such as fever, weight loss, lymphopenia, ascites, jaundice (mucous membrane/plasma) and other signatory illnesses, then regain normal healthy lives within 20 days of treatment (Table 2 ) (Kim et al., 2016). Although, the increase in the rate of recovery with the treatment duration supports the continuous administration for effective recovery, however, the compound interestingly worth further translational study as inhibitory therapeutics against CoV infections targeting the 3CLpro pathways.
Table 2.
Clinical and laboratory observations in cats infected with FIP virus before and during treatment.
| Clinical and laboratory observations before and during treatment | |||||||
|---|---|---|---|---|---|---|---|
| Cat | Fever | Weight loss | Jaundice | Ascites | Lymphopenia (<676/μL) | Treatment duration (dpi) | Outcome |
| P07 | + | + | + | NA | + | 20 days (15–34) | Recovered |
| P10 | + | + | + | NA | + | 20 days (15–34) | Recovered |
| P02 | + | + | + | Mild | + | 15 days (20–34) | Recovered |
| P03 | + | + | + | Mild | + | 16 days (19–34) | Recovered |
| P17 | + | + | + | Profound | + | 14 days (21–34) | Recovered |
| P24 | + | + | + | Profound | + | 14 days (21–34) | Recovered |
| P15 | + | + | + | Profound | + | 4 days (18–21) | Euthanised |
| P16 | + | + | + | Profound | + | 7 days (18–24) | Euthanised |
NA = Not apparent; dpi = days post infection. (Reproduced from Kim et al. (2016), Licenced under CC BY 4.0.)
Mucroporin-M1 (32), a highly penetrating defensive peptide from the scorpion venom with amino sequence LRFLIKSLIKRLVSAFK reportedly exhibit remarkable virucidal effects against SARS-CoV-1, thus suggested for further modification as an anti-SARS-CoV agent targeting the viral RNA (Abd El-Baky et al., 2018). Additionally, a β-defensin-based peptide, P9 (33) with sequence NGAICWGPCPTAFRQIGNCGHFKVRCCKIR reportedly exhibits potent antiviral activity against several respiratory viruses including SARS-CoV-1 and MERS-CoV, enhanced by its high binding affinity to the viral glycoproteins and their respective active amino acid residues. The proposed mechanisms of its activities include the effective prevention of endosomal acidification and, blockage of the viral-host membrane fusion and RNA release from late endosomes with IC50 and TC50 of 1.2 and 380 μg/mL respectively (Zhao et al., 2016).
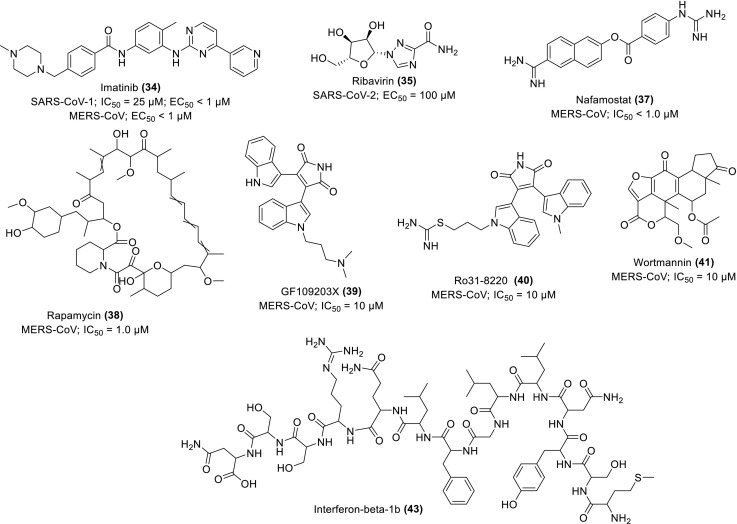
The dose-dependent inhibition of viral virion along the endosomal membrane of Abl2 has been experimentally demonstrated as a prophylactic and therapeutic mechanism of imatinib (34) (Scheme 3 ), an anticancer and Abl kinase inhibitor in the onset treatment of SARS-CoV-1 infection. The application of the 25 μM of the drug for 4 h causes the inhibition of genomic RNA production in 1000-folds, indicating its potency to prevent viral replication. The data obtained suggest the effective inhibition of the virus occurring before the RNA production without affecting the intracellular trafficking of the pseudo-type viruses, similar to its effect on MERS-CoV (Coleman et al., 2016). The viral genetic stability becomes disrupted due to the nucleotide mismatch induced in vitro by the ExoN at the RNA 3′-end and its inactivation in vivo. Thus, Ferron et al. demonstrated the RNA synthesis and proofreading pathway for SARS-CoV-1 through the association of nsp14 with nsp12 (low-fidelity) viral RNA polymerase using 35 (Scheme 3). Although, the drug is significantly incorporated, however, it readily escaped from the RNA, signalling rationale for limited activity in vivo (Ferron et al., 2018).
Scheme 3.
Chemical structures of some repurposing drugs and their activity against MERS-CoV and SARS-CoVs.
2.3. Inhibitors of MERS-CoV
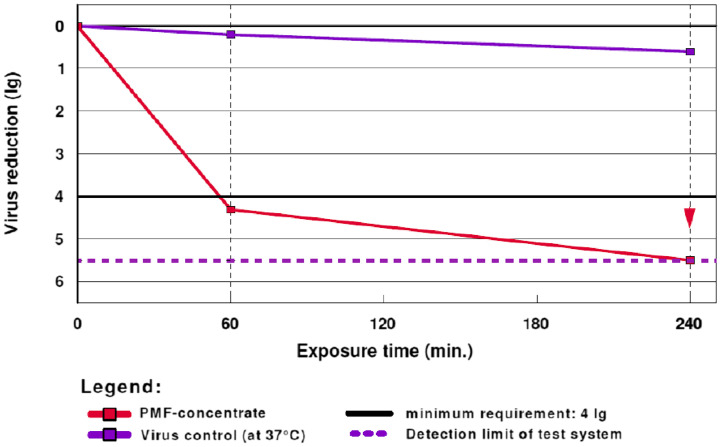
A traditional Prophetic Medicine fraction (PMF/PM 701) (36) extracted from camel's urine, popular among the Arabs for treating many diseases were investigated in vitro for activity on TGEV, a model for MERS-CoV. Other assays employed to study its antiviral/virostatic potentials include endpoint dilution titration, infective dose calculation and quantitative virucidal activity. At a concentration of 20.34% of PMF, 99.50% of TGEV became activated with viral reduction factor (RF) of 4.31 ± 0.48 after 60 min and the value increased to >5.50 ± 0.45 (99.99%) after 240 min (Fig. 7 ). The presence of zinc and copper ions in the regimen was suggested as a contributing factor to their interesting virucidal activity, although further studies are recommended to identify the major chemical compositions and the mechanisms of antiviral actions of 36 (Al Attas, 2019). Thus, this traditional medicine could be further evaluated for applicability in the fight against viral infection.
Fig. 7.
Increase in virus reduction (lg) after the administration of PMF. The RF drastically increases to 4.31 after 60 min and subsequently >5.50 after 240 min. (Reproduced from Al Attas (2019), © ALÖKI Kft, Budapest, Hungary.)
The S-protein of MERS-CoV recognises a dipeptidyl peptidase-4 (DPP4)/cluster of differentiation 26 (CD26) as a host receptor, as such, the virus may gain entry through membrane fusion at the plasma membrane or via the endosomes after endocytosis. Either of the processes serves as additional proteolysis which favours the activation of the viral S-protein for fusion. A serine protease inhibitor, nafamostat (37) (Scheme 3) reportedly inhibits the S-mediated membrane fusion and demonstrates a potent blockage mechanism against MERS-CoV infection in vitro when cell lines expressing Rluc-based split reporter protein were used to assay a cell-based TMPRSS2-dependent fusion of S protein, with DMSO as a control. The cell lines, Calu3 and 293FT-derived effector cells were also used to express the MERS-CoV-S-protein fusion membrane in dual split protein assay. The strongest inhibition of the membrane is observed upon administration of 37 at concentrations of 1.0 and 10 μM where the relative cell fusion reduces to <10% and almost 0% respectively, although the fusion was better blocked in Calu3 than in 293FT cells (Yamamoto et al., 2016). Thus, nafamostat deserves further studies for therapeutic development.
Kindrachuk and co-workers demonstrate the experimental inhibition of MERS-CoV infection by some licenced kinase inhibitors in vitro using TKA, FNA and ORA analyses. The pharmacological pathways/mechanisms of action were suggested as the inhibitory signalling expression on the MAPK/ERK and PI3K/mTOR/AKT. The most potent inhibitors were shown as rapamycin (38), GF109203X (39), Ro31-8220 (40) and Wortmannin (41) (Scheme 3) with percentage inhibition estimated as 60, 58, 46 and 28% respectively each at a concentration of 10 μM. Among the four drugs, rapamycin exhibited the highest inhibitory potential even at a lower concentration of 1 μM, further supporting the implication of mTOR as a therapeutic target in the effective control of MERS-CoV infection (Kindrachuk et al., 2015), thus amenable for consideration in vaccine development.
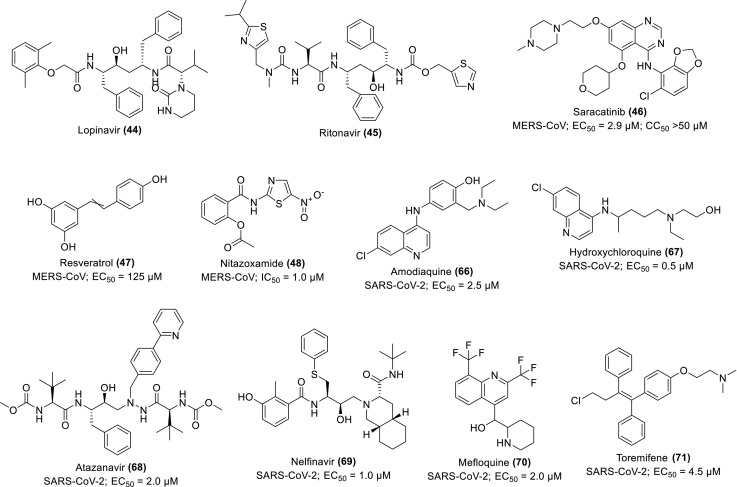
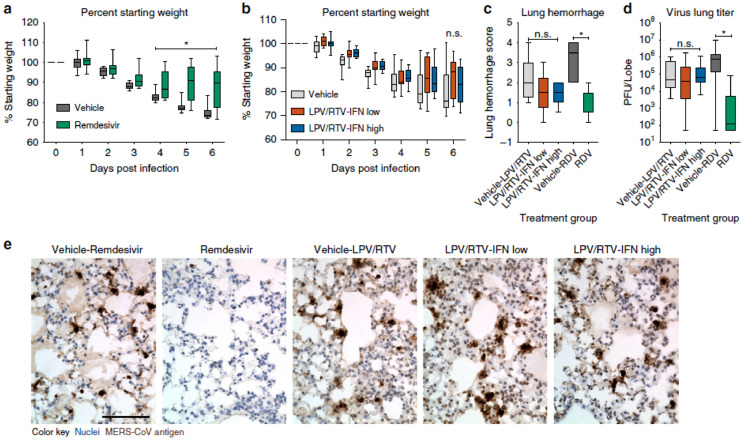
Oftentimes, combination therapy enhances anti-CoV activities in clinical trials. For instance, improved clinical outcome was observed when lopinavir/ritonavir (42) and interferon-β1b (43) (Scheme 3) were separately assessed in common marmosets infected with severe MERS-equivalent disease. From the clinical pathology and radiological findings, the mean clinical scores were observed between 50.9 and 95.0% and a decreased weight loss compared to the set treated with mycophenolate mofetil (MMF) and the untreated set. Other improvements are mild bronchointerstitial pneumonia and minimal pulmonary infiltrates in association with mean viral loads in the necropsied lung which changed from 0.59–1.06 log10. The mortality rate for the set treated with 42 and 43 stands between 0 and 33% respectively after 36 h postinoculation compared to the untreated and MMF-treated sets with 67% recorded mortality respectively. This strongly supports further evaluation of 42 as well as 43 in clinical trials of MERS-treatment (Chan et al., 2015). Similarly, 9 and 43 reportedly demonstrate superior antiviral activities against MERS-CoV in vitro compared to interferon-β1b (43), lopinavir (44) and ritonavir (45) (Scheme 4 ) individually or in combination. Upon further investigation in vivo mice, the drug shows promising potential for prophylaxis and treatment by improving the pulmonary function and reducing replication (viral loads), and other severe lung pathology (Fig. 8 ) (Sheahan et al., 2020). The study provides supporting evidence of the therapeutic potential of 9 against MERS-CoV.
Scheme 4.
Chemical structures of some repurposing drugs and their activity against MERS-CoV and SARS-CoV-2.
Fig. 8.
Therapeutic remdesivir reduces replication and pathology. Percent starting weight of 10–12-week-old female Ces1c-l-hDPP4 mice infected with 5E+04 pfu MERS M35C4 and treated with (a) subcutaneous vehicle for remdesivir (N = 13) or remdesivir (25 mg/kg, N = 14) B1D beginning 1 dpi or (b) vehicle for lopinavir/ritonavir-interferon-β (N = 15), lopinavir/ritonavir-interferon-β low (N = 16) or lopinavir/ritonavir-interferon-β high (N = 16) B1D. The oral vehicle of lopinavir/ritonavir (160/40/mg/kg) was administered orally once daily. Interferon-β low (1× human equivalent dose of 1.6 MIU/kg) and high (2× human equivalent dose of 40 MIU/kg) or PBS vehicles were administered via subcutaneous injection every other day. Asterisks indicate statistical difference by two-way ANOVA with Turkey's multiple comparison test. (c) Lung hemorrhage 6 dpi for all animals in (a), (b) scored on a scale of 0–4, where 0 is a normal pink healthy lung and 4 is a diffusely discoloured dark red lung. (d) MERS-CoV lung titer 6 dpi in mice as described in (a), (b). Asterisks indicate statistical significance (N group described in (a) and (b), p < 0.05) by one-way ANOVA with Kruskal-Wallis test for (c), (d). Data for (a-d) are compiled from two independent experiments. For the box and whisker plots, the boxes encompass the 25th to 75th percentile, the line is at the median, while the whiskers represent the range. (e) Representative photomicrographs of MERS-CoV antigen (brown) and hematoxylin stained nuclei (blue) in mouse lung tissue sections from 6 dpi. The black bar is 100 μM. (Reproduced from Sheahan et al. (2020), Licenced under CC BY 4.0.)
An Abl kinase inhibitor and anticancer drug, 34 was observed with good inhibitory potentials against the entry virion of MERS-CoV when its administration at 10 μM for 4 h inhibits the production of MERS-CoV RNA in 50-folds (Coleman et al., 2016). In a similar pattern, saracatinib (46), a renowned inhibitor of the Scr-family of tyrosine kinases (SFK) selectively inhibits MERS-CoV replication in Huh7 cells with EC50 and CC50 values 2.9 μM and >50 μM respectively. The proposed pharmacological mechanism is the suppression of SFK signalling pathways. Its synergistic effect was also observed in combination with gemcitabine, a potent inhibitor of RNA viruses (Shin et al., 2018).
The inhibitory potentials of resveratrol (stilbenol) (47), a natural antioxidant agent in grape seeds and skin as well as in red wine was evaluated in vitro against the MERS-infected Vero cells using MTT and neutral red uptake (NRU) assays. Other experimental procedures adopted for the assessment of the antiviral effects of the phytochemical on viral RNA and protein expression include quantitative PCR, immunofluorescence, Western blotting and plaque assays. The compound significantly inhibits MERS-CoV, indicated by the reduction in MERS-induced apoptosis through the suppression of the viral replication by its N-protein at 125 μM with an increase in the effects upon elongation of the treatment duration (Lin et al., 2017). Similarly, experimental demonstration of the anti-MERS-CoV effect was reported for nitazoxamide (48), an antiviral drug with a broad spectrum of activity. The drug inhibits the viral N-protein expression, suppresses the production of IL-6 and pro-inflammatory cytokines in vivo mice model (Rossignol, 2016). The compounds are therefore suggested for further investigation as potential therapeutics against the CoV infections, especially resveratrol whose potentials in combination with indomethacin were proposed as adjuncts for the treatment of SARS-CoV-2 (Marinella, 2020).
As an enveloped, positively-sensed virus with single strands of RNA, MERS-CoV employs S-protein S1 (SS1) subunit to interactively bind to its target receptor, DPP4/CD6 and becomes fused to the plasma/endosomal membrane through a fusion peptide located at N-terminal of S2 subunit. In the S2 site, the heptad repeats 1 (HR1) and the heptad repeats 2 (HR2) bind together to form a 6-helix bundle (6-HB) core for viral-cell membrane fusion. Peptides derived from the HR2 (HR2P) (49) can actively interact with the binding site of HR1, block the formation of the 6-HB fusion core, thereby inhibiting viral S-protein-mediated cell-cell fusion. Through the mechanism, the viral infection by pseudoviruses is effectively prevented with or without mutation in the HR1 region. Both 49 and its derivative, HR2P-M2 (50) strongly inhibit the MERS-CoV infection through the formation of 6-HB core in vitro in dose-dependent with an IC50 value of 0.97 and 0.55 μM respectively. When administered intranasally, 50 effectively protected the adenovirus serotype-5-human DPP4-transduced mice from MERS-CoV infection in vivo as assessed through fluorescence N-PAGE, circular dichroism spectroscopy, native polyacrylamide gel electrophoresis and Vero 81 cells for lung viral titer. Quantitatively, the viral titer reduction in the lung was >1000-folds and the effect became enhanced upon the combination with interferon-β (Channappanavar et al., 2015). The interesting results pose the regimen, 50/interferon-β (51) as an effective preventive option against MERS-CoV infection, thus, deserves further studies.
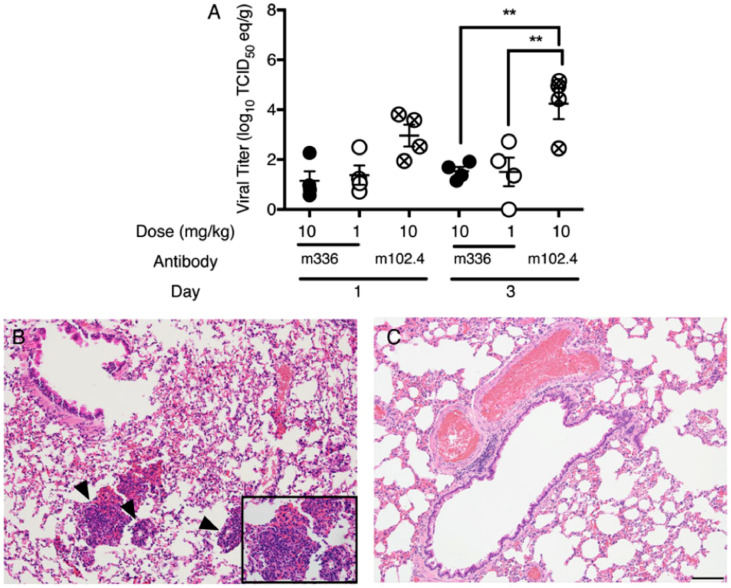
The propensity of some antibody as an effective countermeasure for prevention CoV infection was further demonstrated by Houser and co-workers when they investigated the potential of human mAb (m336) (52) to prevent rabbits from MERS-CoV infection. The antibody, 52 was administered in the volume range of 0.2–2.0 mL at 1 mL/kg and 10 mL/kg intravenously and intranasally respectively before the infection with 1 mL of 105 TCID50 of MERS-CoV isolate (HCoV-EMC/2012) propagated in Vero81 cells with OPti-MEM 5% FBS. The prophylaxis with m336 shows a very promising protective potential with a reduction in the pulmonary RNA titers of the virus by 40–9000-folds when compared with the irrelevant controls, hmAb and m102.4 (specifically for Henda virus) which show no effect (Fig. 9 ) (Houser et al., 2016). The study affiliated with the US government, strongly suggests the trials of a specific human antibody, 52 as an effective preventive strategy against CoV infections.
Fig. 9.
Viral titers in rabbit lungs after intravenous prophylaxis with neutralizing monoclonal antibody, m336. Rabbits received MERS-CoV-relevant m336 or irrelevant hmAb/m102.4 as control intravenously 24 h before the intranasal administration of 105 TCID50 of MERS-CoV isolate (HCoV-EMC/2012) at infective doses. (A) Lungs were analysed on days 1 and 3 post-infection, and viral titers were analysed using quantitative real-time-polymerase chain reaction (RT-PCR). (B) and (C) are mages of hematoxylin-eosin-stained sections from a rabbit treated with 10 mg/kg of m336 or the control (data from 4 rabbits/group). Arrowheads and insets indicate the areas of intense perivascular inflammation. All images were taken on day 3 post-infection. ⁎⁎p < 0.005 and ⁎⁎⁎p < 0.001 by 1-way variance analysis with the Turkey multiple comparison test. (Reproduced from Houser et al. (2016), Licence under US Government work.)
Niu and co-researchers reported the neutralizing activities of 13 series of monoclonal/polyclonal antibodies against MERS-CoV. The Abs were isolated from the human Ab libraries, transchromosomic bovines, transgenic humanised mice and naïve B cells of an infected person. Nine (9) of the Abs labelled Ab28 (53), Ab33-2 (54), Ab38 (55), Ab39-1 (56), Ab39-2 (57), Ab40 (58), Ab42 (59), Ab43 (60) and Ab45 (61) display strong binding against the viral S-protein in >30-folds while the weakly-bound Ab8 (62) is in <15-fold of H7, a mAb for hemaglutination of influenza virus, used as a control. The most promising two, Ab27 (MERS-GD27) (63) and Ab33-1 (MERS-GD33) (64) recovered from infected patient demonstrate the highest potent inhibitory effect against the live MERS-CoV in vitro with respective IC50 values of 0.0010 and 0.0013 μg/mL while their strong affinity for S-protein of MERS-CoV (KD) are evaluated as 0.775 and 0.575 nM respectively. Their combination shows additive/synergistic effects against the pseudotyped MERS-CoV (Niu et al., 2018). Thus, they are recommended for further translational development into effective therapeutics and vaccines for MERS-CoV infection.
An Amb (5F9) (65) located at the epitope of S1 subunit of MERS-CoV NTD efficiently neutralised the activity of wild-type strain of MERS-CoV, EMC/2012 with quantitative effects in term of IC50 as 0.2 μg/mL (Chen et al., 2017). This supports the scientific argument that antibodies are naturally effective as prophylactic agents and immunotherapy against the onset of CoV infection. For further study, the antibody is available at the National Library of Medicine (https://www.nlm.nih.gov/copyright.html).
2.4. Inhibitors of SARS-CoV-2
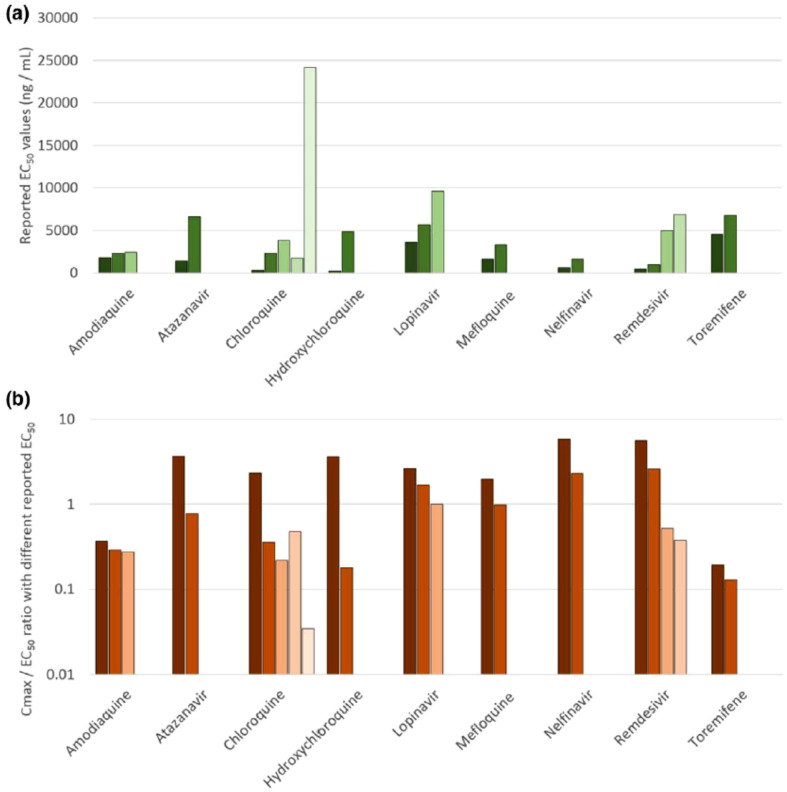
Considering the achievable chemotherapeutic concentrations in the targeted human compartments relevant to prognosis and pharmacology of COVID-19 including lung and plasma, Arshad and co-researchers predicted the accumulation of various repurposed drugs in the onset of the viral prevention and treatment within lung tissues using combined physicochemical and pharmacological parameters for modelling. The maximal effective concentrations at 90% (EC90) of the drugs were re-evaluated from in vitro activity records against SARS-CoV-2 expressed in terms of the achievable maximum plasma concentration (Cmax) at an effective dose (Cmax/EC90 ratio). Based on the pharmacokinetic parameters within the human plasma and lung, the drug molecules with the highest tendency to exert chemotherapeutic viral suspension were conveniently screened. Those predicted to achieve lung concentrations between 1.5 and 10-folds higher than the reported EC50 include amodiaquine (66), HCQ (67), atazanavir (68), 44, 8, nelfinavir (69), 9, mefloquine (70) and toremifene (71) with EC50 of 0.5–4.5 μM (Fig. 10 ; Scheme 4). The researchers suggested more focus on the EC90 evaluation mode and the contextual prioritisation of achievable potentials within the human target compartment such as the lung in further COVID-19 therapeutic studies (Arshad et al., 2020). Although, the application of some repurposed antibiotics such as azithromycin in this quest has generated some concerns of microorganism resistance and associated macrolides (Kow and Hasan, 2020).
Fig. 10.
(a) Variations in reported EC50 values for SARS-CoV-2 across the drugs with more than one reported value. (b) The implications of the variability in reported EC50 in terms of Cmax/EC50 ratio. Amodiaquine and toremifene were estimated to exhibit subtherapeutic pharmacokinetics irrespective of the EC50 value used. Similarly, nelfinavir was estimated to have higher Cmax than EC50 values irrespective of the EC50 used in the analysis. The interpretation for other drugs is strongly dependent upon the EC50 used which is a major concern. (Reproduced from Arshad et al. (2020), Licenced under CC BY-NC-ND 4.0.)
From an in vitro experimental evaluation, ivermectin (72) (Scheme 4), an FDA-approved anti-parasite demonstrates a strong antiviral effect against SARS-CoV-2. After single addition to Vero-hSLAM cells, 2-h post-infection with SARS-CoV-2, TaqMan real-time-polymerase chain reaction (RT-PCR) assay shows a significant reduction of 93% in the viral RNA after 24 h, which subsequently increased to 99.8% and observed in ≈5000-folds at 48 h, indicating the total loss of the virions. The IC50 against the supernatant RdRp gene was recorded as 2.2 μM, supporting its eligibility for further studies as a potent inhibitor of SARS-CoV-2 (Caly et al., 2020).
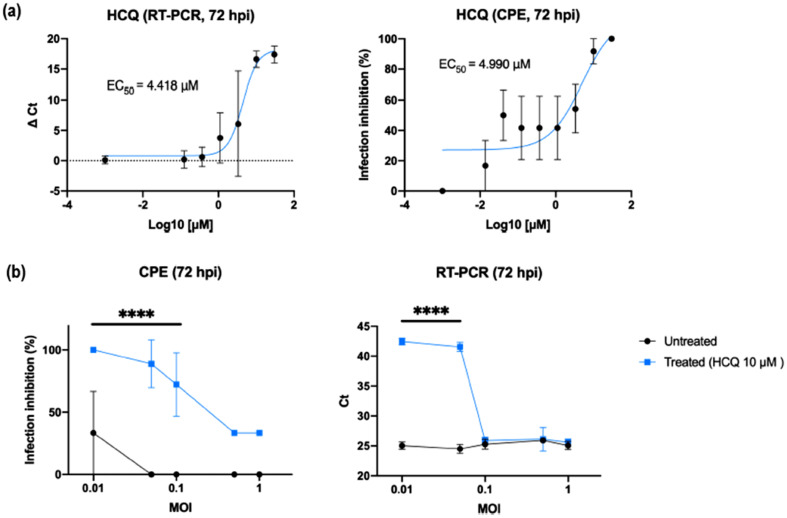
The in vitro activity of effective dosage of 67 was also reported to be maximally achieved when administered before and after infection of Caco-2 and Vero E6 cells. This suggests its potency for the inhibition of SARS-CoV-2 infection through combined prophylactic and therapeutic modes. The theory was experimentally validated by its EC50 of 4.418 and 4.990 μM for cytopathic effect (CPE) and RT-PCR analysis respectively using various multiplicities of infection (MOI) on both treated and untreated cells (Fig. 11 ) (Clementi et al., 2020). Similarly, Liu and co-workers also demonstrated experimentally the availability, safety and efficacy of 67 against SARS-CoV-2 in comparison with 8 and 9 (Liu et al., 2020b). This supports the worthiness of its further study, although, some researchers debate its activity for protection against SARS-CoV-2 infection even in combination with antibiotics such as azithromycin (Maisonnasse et al., 2020).
Fig. 11.
Effects of HCQ as combined prophylactic and therapeutic agent against SARS-CoV-2. (a) HCQ dose-dependent assessment. HCQ EC50 against SARS-CoV-2 was obtained by both CPE and RT-PCR analysis on results from a full-time experimental setting on Vero E6 cells. Mean values and SD (for RT-PCR values) and SEM (for CPE values) are reported for all experimental replicates. All conditions were tested in quadruplicate and tested in duplicate in RT-PCR. (b) HCQ antiviral effect using different MOI. CPE analysis resulted in a statistical difference between treated and untreated cells (⁎⁎⁎⁎P < 0.0001) when using 0.1 MOI or less, RT-PCR only from 0.05 MOI to decrease (⁎⁎⁎⁎P < 0.0001). (Reproduced from Clementi et al. (2020), Licenced under CC BY 4.0.)
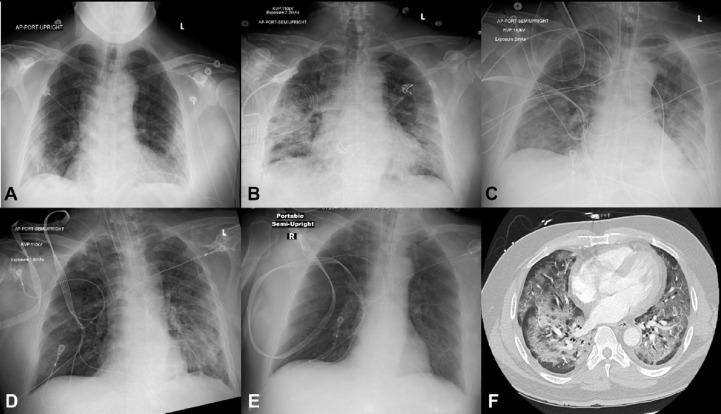
A clinical case was reported in respect of a 57-year old male patient suffering from SARS-CoV-2 infection associated with ARD complications as myocarditis, stress cardiomyopathy, acute coronary syndrome and demand ischemia and treated with tocilizumab (73), an inhibitor of interleukin-6 receptor (IL-6R) and AT-100 (74), an inhibitor of aldose reductase. One of the main pathomechanisms of SARS-CoV-2 infection is the invasion of the pneumocytes and myocytes where it induces critical illnesses including ARDs and cardiac injury through the generation of inflammation cascade, necessitating the administration of methylprednisolone, a corticosteroid against myocarditis. With other medical supports, the patient became fully recovered after 8 days on admission as shown by the observable normal lung parenchyma using a chest radiograph (Fig. 12 ) and was discharged on day 19 (Coyle et al., 2020). This clinical trial demonstrates the efficacy of some repurposed drugs such as 73, 74 and methylprednisolone (75) in the treatment of onset SARS-CoV-2 complicated with ARDs and myocarditis.
Fig. 12.
Serial chest radiograph and computed tomography of a patient diagnosed with complicated SARS-CoV-2 infection and treated to recovery. (A) Initial chest radiograph with bibasilar patchy interstitial opacities. (B) Chest radiograph on day 3 with worsening bilateral opacities. (C) Chest radiograph on day 5 following intubation. (D) Chest radiograph on day 6 with marked improvement in aeration. (E) Chest radiograph on day 8 showing normal lung parenchyma. (F) Computed tomography (CT) of the chest on day 3 revealed extensive diffuse bilateral airspace consolidations and ground-glass opacities most pronounced in the right upper lobe and bilateral lower lobes, with areas of subpleural sparing. Trace bilateral pleural effusions and minimal coronary artery calcification were noted. AP = anteroposterior; L = left; R = right. (Reproduced from Coyle et al. (2020), Licenced under CC BY-NC-ND 4.0.)
The attenuation of ARD complications in SARS-CoV-2 infections was demonstrated by iron chelators, deferiprone (76), deferoxamine (77) and deferasirox (78) commonly applied to treat iron overload. The multidentate ligands suppress the endothelial inflammation through inhibition of IL-6 synthesis by decreasing the signalling pathway of the nuclear factor, NF-kB. Other pathophysiological mechanisms of anti-SARS-CoV-2-induced ARDS exhibited by these agents include inhibition of viral replication, prevention of pulmonary fibrosis, decrease in iron availability and upregulation of lymphocytes (B cells) to improve lymphopenia expression (Dalamaga et al., 2020). These agents are generally identified with antiviral, antioxidants and immunomodulatory profiles, thus, reasonably recommended for further studies in the search for effective treatment against SARS-CoV-2.
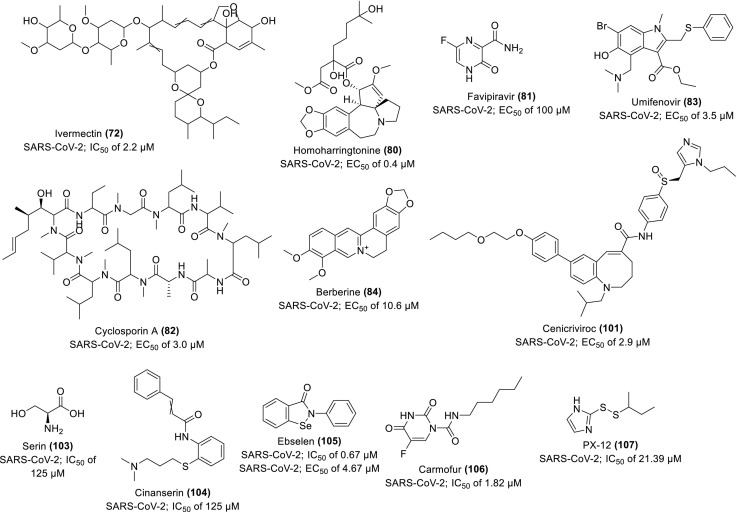
To further evaluate the synergistic antiviral efficacy of combinatorial therapy of some repurposed drugs, Choy and co-workers experimented with the anti-SARS-CoV-2 activity of some drugs (individually and in combination) in vitro Vero E6 cells. They observed 64.9% viral inhibition when 0.195/6.25 μM of combined drugs, 2/9 (79) compared to the 50% inhibition recorded when 23.15, 26.63, 2.55 and 0.40 μM of 9, 44, 2 and homoharringtonine (80) respectively were tested individually. The positive controls of 35 and favipiravir (81) show no inhibition each at 100 μM. They suggested the clinical trial of the combinatorial approach owing to the synergistic effects observed in 79 combined therapy (Choy et al., 2020). In a similar report, 9 displays the strongest inhibitory potential against SARS-CoV-2, indicated by the lowest EC50 value of 0.99 μM in vitro infected Vero E6 cells compared to other repurposable drugs, 8, cyclosporine A (82), umifenovir (83), 44 and berberine (84) (Scheme 5 )whose EC50 values are 1.38, 3.00, 3.50, 5.20 and 10.60 μM respectively (Pizzorno et al., 2020). Its synergistic activity was also observed in combination with diltiazem (85), a host-directed drug. It demonstrates good pharmacokinetics, safety and tolerability (Humeniuk et al., 2020), thus recommended for further pathophysiological investigation.
Scheme 5.
Chemical structures of some repurposing drugs and their activity against SARS-CoV-2.
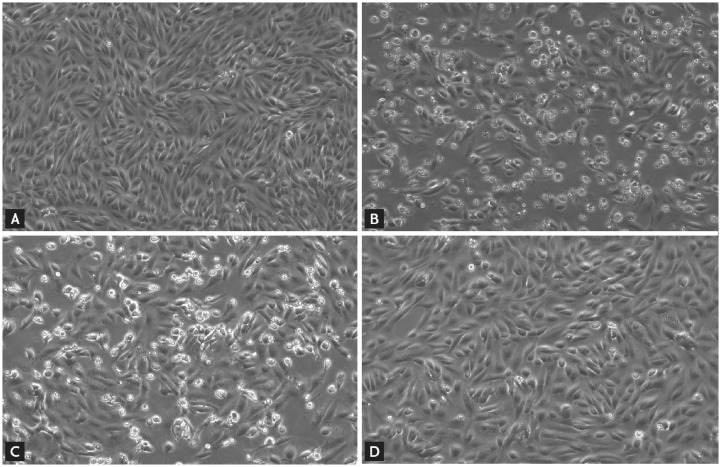
The administration of combined 44/45 (86) (7:1.75 μg/mL) and 67 (1–2 μg/mL) to SARS-CoV-2-inoculated Vero cells individually or in combination reportedly induce CPE of virus and reduced viral loads at various degrees after 48 h, studied using qRT-PCR analysis. In this in vitro experiment, 86 at normal plasma steady-state concentration (PSC) demonstrates a higher anti-SARS-CoV-2 potential with lower CPE and reduced viral loads while 67 exhibits lesser potential all in comparison with the control (Fig. 13 ) (Kang et al., 2020b), supporting the anti-SARS-CoV-2 potential of lopinavir/ritonavir for further development.
Fig. 13.
Antiviral activity by the cytopathic effect of Vero cells after 48 h of inoculation with SARS-CoV-2. (A) Positive control; (B) Negative control; (C) 2 μg/mL HCQ, equivalent to PSC of 800 mg once daily; (D) 7:1.75 μg/mL lopinavir/ritonavir, equivalent to PSC of 400/100 mg twice daily. (Reproduced from Kang et al. (2020b), Licenced under CC By 4.0.)
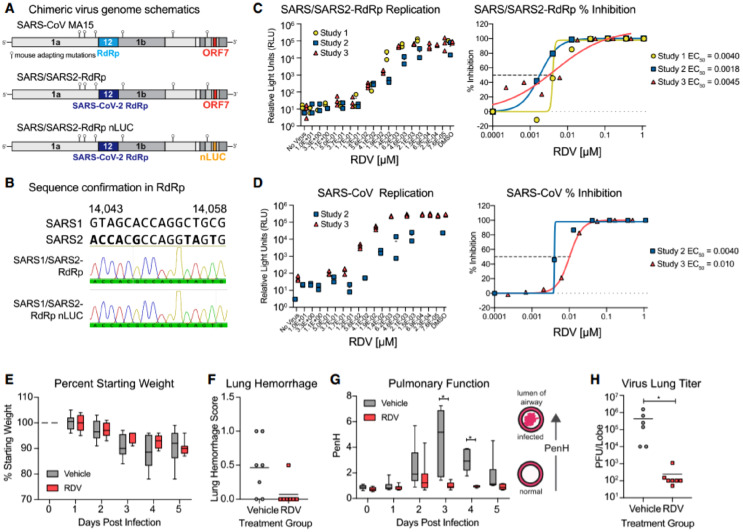
Pruijssers and co-workers also demonstrate an experiment supporting the anti-SARS-CoV-2 efficacy of 9 in vitro and non-clinical in vivo models using a SARS-CoV-1 encoding the SARS-CoV-2 RdRp (SARS/SARS2-RdRp). The drug strongly inhibits the viral replication in cultured human lung and primary airway epithelial cells through the RdRp with an EC50 value of 0.10 μM. It improved the pulmonary functions and reduced the viral loads in SARS-CoV-2-infected mice (Fig. 14 ) (Pruijssers et al., 2020).
Fig. 14.
Anti-SARS-CoV-2 RdRp activity of remdesivir. (A) Schematic of the recombinant SARS-CoV mouse-adapted MA15 strain chimeric virus genomes. SARS/SARS2-RdRp and SARS/SARS2-RdRp-nanoluciferase (nLUC) constructed by exchanging the SARS-CoV-1 MA15 RdRp with the SARS-CoV-2 RdRp. ORF7 is replaced by nLUC in SARS2-RdRp-nLUC. (B) The presence of the SARS-CoV-2 RdRp confirmed by Sanger sequencing in stocks of both recombinant chimeric viruses. The alignment indicates that SARS-CoV-2 RdRp-specific nucleotides are present in both chimeric viruses as shown by the Sanger sequencing and histogram. (C) SARS/SARS2-RdRp-nLUC replication in Huh7 cells in the presence of remdesivir (left) and associated percentage inhibition (right). (D) SARS-CoV-1 replication in Huh7 cells in the presence of remdesivir (left) and associated percentage inhibition (right). (E) Percent starting weight of 17-week-old female Ces1c-l- mice infected intranasally with 1 × 103 PFU of SARS/SARS2-RdRp and treated subcutaneously with 25 mg/kg remdesivir or vehicle 1-day post-infection (dpi) and twice daily thereafter. (F) Lung hemorrhage at 5 dpi. (G) Pulmonary function by WBP. The PenH metric shown is a surrogate marker of pulmonary obstruction. p < 0.0001 as determined by two-way ANOVA with Sidak's multiple comparisons test. (H) Lung titer at 5 dpi as measured by plaque assay. p = 0.0012 by Mann-Whitney test. In (E) and (G), boxes encompass the 25th–75th percentile, a line is drawn at the median and whiskers represent the range. (Reproduced from Pruijssers et al. (2020), Licenced under CC BY-NC-ND 4.0.)
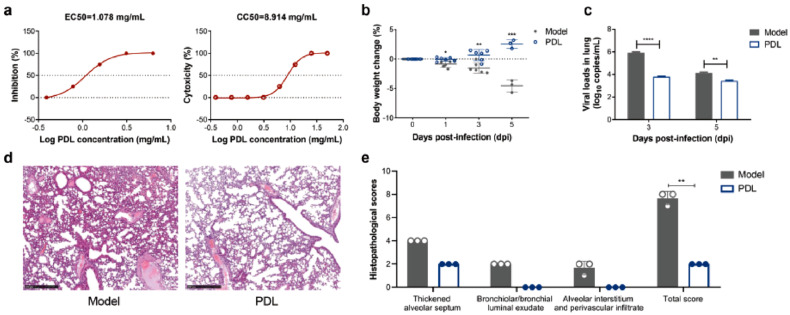
Among the traditional medicines, Pudilan Xioyan liquid (PDL) (87), a Chinese herbal mixture from Isatis indigotica, Taxacum mongolicum, Corydalis bungeana and Scutellaria baicalensis was evaluated with strong anti-SARS-CoV-2 activity with EC50, CC50 and selectivity index of 1.078 mg/mL, 8.914 mg/mL and 8.27 respectively in vitro model. When 4 mL/kg was administered 1-h post inoculation once daily in hACE2 transgenic SARS-CoV-2-infected mice, a significant increase in body weight and change in viral loads within the lung tissues were noticed at 1-day post-infection, quantitatively with histopathological scores, dpi (p = 0.0120), 3dpi (p = 0.0020) and 5dpi (p = 0.0006) compared to the control group (Fig. 15 ), suggesting the potent efficacy of the natural herbal regimen (Deng et al., 2020).
Fig. 15.
Therapeutic efficacy of PDL against SARS-CoV-2 in vitro and in vivo. (a) Cytotoxic effect and antiviral activity of PDL in Vero E6 cells by a reduction in cytopathic effect. (b) Weight loss in SARS-CoV-2-infected hACE2 transgenic mice with(out) PDL treatment. (c) Changes in viral loads from lung tissues indicated by quantitative RT-PCR at 3dpi and 5dpi. (d) Histopathological analysis of lung tissues in SARS-CoV-2-infected hACE2 transgenic mice with(out) PDL treatment (Bar = 500 μm). (e) Histopathological scores of PDL-treated group and untreated group (control), including the lung samples at 5dpi. The scoring index is based on the severity of the lung histopathology. (Reproduced from Deng et al. (2020), Licenced under CC BY 4.0.)
The suppressors of the FURIN gene such as the combined quercetin (88)/vitamin D (89)/estradiol (90) [91] putatively demonstrates mitigating activities against COVID-19 in gene silencing and overexpression, and relevant transgenic mouse models. Using molecular docking screening and gene set enrichment analysis (GSEA) of expression profiling experiments (EPEs), 88, 89 and combined 88/89 shows interference of 85, 70 and 93% respectively with functions of SARS-CoV-2 proteins in human cells (Glinsky, 2020). The scaffolds of the natural-based bioactive agents, 88 and 90 are therefore suggested for the development of effective therapeutic candidates for the prevention and treatment of COVID-19. Russo and co-researchers also investigated the anti-SARS-CoV-2 potentials of some natural products of the polyphenol/flavonoid family including 88, luteolin (91), baicalin (92), hesperetin (93), epicatechin gallate (94), gallocatechin gallate (95), papyriflavonol (96), amentoflavone (97) and scutellarein (98). They demonstrate promising inhibitory activity against some key targets implicated in SARS-CoV-2 infection and replication such as the NTPase/helicase, PLpro and 3CLpro with a fair expression for systemic toxicity, therefore, suggested for further evaluation (Russo et al., 2020).
The innate immune-inclined mast cells (MCs) are implicated in the pathogenesis of SARS-CoV-2 as targets for mediating inflammation. The activation of these MCs by microbes including SARS-CoV-2 through the TLRs occurs by the release of pro-inflammatory chemicals and cytokines. In a mouse experimental model, an MC stabilizer, sodium chromo-glycate (SCG) (99) retards the release of the pathogenic molecules, thereby improving the pathological changes, suppressing inflammation and enhancing the mouse survival during the viral infection. Similarly, palmitoylethanolamide (PEA) (100) exhibits protective and inhibitory activity on the respiratory tract against the viral infection (Gigante et al., 2020), thus, both are hypothesized as a potent therapy for the SARS-CoV-2-induced inflammation and MC activation.
A potent anti-inflammatory and immunomodulatory antagonist of the C—C chemokine receptor type 5 (CCR5), cenicriviroc (101) (Scheme 5) reportedly inhibits the replication of SARS-CoV-2 in vitro. The anti-HIV-1 drug demonstrates a strong preventive activity against the SARS-CoV-2-induced cell damage and inhibitory potential against the viral RNA with EC50 values of 19.0 and 2.9 μM respectively, thus, suggested for further consideration in the COVID-19 therapeutic design (Okamoto et al., 2020).
The Michael acceptor inhibitor (N3) (102) reportedly inhibits the Mpro of SARS-CoV-1, MERS-CoV and some other virulent viral infections. When evaluated against SARS-CoV-2 using some computer-aided structural and in vitro assays, the drug exhibits a very strong inhibitory potency, such that the corresponding dissociation/inactivation constants (ki/k3) could not be feasibly measured. Among the potential inhibitors with similar interactions to 102 in respect to amino acid residues in the active substrate-binding pocket of the SARS-CoV-2 Mpro, serin (103) and cinanserine (104) (renown serotonergic antagonist) (Scheme 5) demonstrate optimization potential with an IC50 value of 125 μM and CC50 > 200 μM respectively. The HTS and tandem mass spectrometry results show that among the screened drug‑leads, ebselen (105), carmofur (106) and PX-12 (107) (Scheme 5) bind covalently to the catalytic cysteine dyad (C145) of SARS-CoV-2 Mpro, thereby modifying the enzyme completely or partially. The drug 105 demonstrates the strongest inhibitory potentials with IC50 of 0.67 ± 0.09 while 106 and 107 have 1.82 ± 0.06 and 21.39 ± 7.06 μM respectively. Further evaluation of the enzymatic inhibition using RT-PCR cell-based in vitro assay supports the strongest antiviral potential of 105 in good competition with 102, a co-crystallized inhibitor of SARS-CoV-2 Mpro with an EC50 value of 4.67 and 16.77 μM respectively (Jin et al., 2020). Thus, 105, an organoselenium compound with known antioxidant, anti-inflammatory and cytoprotective activity shows a promise for effective inhibition of SARS-CoV-2 as a potential for therapeutic development.
In a similar fashion to the inhibitory activity against the MERS-CoV infection, two specific human mAbs labelled CA1 (108) and CB6 (109) from COVID-19-infected patients show a promising neutralizing potential specifically against SARS-CoV-2 in vitro. Further experiment favours the inhibition of SARS-CoV-2 infection by 109 as both prophylactic and a treatment agent in the rhesus monkey. The Ab induced great reduction in viral loads and alleviation of SARS-CoV-2 infection-related lung damage. The structural study suggestively supports the mechanism of action as the interference at the SARS-CoV-2-ACE2 binding site, thereby preventing the viral interactive infusion into the host receptor (Shi et al., 2020), thus, indicating the propensity of 109 as a promising therapeutic candidate upon further translational study.
3. Conclusion and perspectives
The scientific theory of the similarity in pathomechanisms of human coronavirus infections, pathophysiology and therapeutic targets has aided the hypotheses of effective pharmacological approaches towards their prevention and treatment. These phenomena have been verified through basic, pre-clinical and clinical, experimental in vitro and in vivo models through which several therapeutic targets and bioactive agents with potentials for both prophylaxis and treatment of CoV infections have been identified from various sources. This review comprehensively highlights the common underlying pathomechanisms and pharmacological targets of HCoV-OC43, SARS-CoV-1, MERS-CoV and SARS-CoV-2 infections, and proven therapeutic potentials from basic and clinical experimental investigations. The reported pathomechanisms involve the human-to-human cross-infection predominantly through respiratory droplets, during coughing, sneezing and sometimes through excretory products.
The earmarked therapeutic targets and their potent inhibitors from experimental studies for the prophylaxis and treatment include the viral proteases, Mpro, PLproand 3CLpro by anti-CoV agents 18–31, 88, 91–98 and 102–107, viral structural proteins, S-glycoprotein by 11, 37, 42, 43, 49, 50, 53–65 and 79, N-proteins by 1–7, 10, 47 and 48, non-structural proteins, nsp 16/nsp 10 2’-OMTase by TP29, nsp 5 by 12–17, nsp 12/nsp 14 by 35. Others include enzymes such as the RdRp inhibited by 9, 12–17, 35 and 72, TMPRSS2 by 8, 9, 44, 66–71, FURIN by 91, kinases such as MAPK/ERK and PI3K/mTOR/AKT reportedly interact with 38–41, Abl2 by 34, IL-receptors by 48, 73–78 and 87, TLR by 99, 100 and the human host ACE2 interfered by 91. The active inhibitors are identified from various sources for easier accessibility. Notably among them include the traditional Chinese and Prophetic medicines, repurposing libraries, peptides, antibodies, iron chelators, siRNAs and combined therapy. Interestingly, most of the highlighted inhibitors exhibit a multi-targeting and networking pharmacology amenable for overcoming drug resistance and mitigating the comorbidities associated with CoV infections.
Although, more robust experimental studies especially clinical and non-clinical in vivo animal/human models are required to reveal details of underlying molecular and cellular mechanisms surrounding the promising efficacy of the agents for the current and emerging CoV strains. Secondly, the study of pharmacodynamics, pharmacokinetics and safety of these candidates are inadequately reported, as such more scientific effort is required especially on the yet-to-be-approved agents. Importantly, this review has not proclaimed the approval of any reported candidate for medical application. However, it reveals the essential therapeutic targets and potent inhibitors amenable for further translational studies into effective pharmaceuticals for the current and emerging CoV infections.
Conflicts of interest
The authors declare no conflict in this study.
Acknowledgments
Acknowledgement
This work was supported by Tetfund Nigeria, Ministry of Higher Education of Malaysia, HICoE (grant no. 311/CDADAH/4401009) and Universiti Sains Malaysia GA Scheme (grant no. 308.AIPS.415401).
References
- Abd El-Baky N., Uversky V.N., Redwan E.M. Virucidal activity of cell-penetrating peptides of viral origin. J. Biomol. Struct. Dyn. 2018;36(7):1739–1746. doi: 10.1080/07391102.2017.1333459. [DOI] [PubMed] [Google Scholar]
- Al Attas S.A. Antiviral activity of extracted fraction from camel urine against corona and influenza a (H1n1) viruses. Appl. Ecol. Environ. Res. 2019;17(5) doi: 10.15666/aeer/1705_1102311031. [DOI] [Google Scholar]
- Albini A., Di Guardo G., Noonan D.M., Lombardo M. The SARS-CoV-2 receptor, ACE-2, is expressed on many different cell types: implications for ACE-inhibitor- and angiotensin II receptor blocker-based cardiovascular therapies. Intern. Emerg. Med. 2020;15(5):759–766. doi: 10.1007/s11739-020-02364-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandre J., Cracowski J.L., Richard V., Bouhanick B., C.-w.g.o.t.F.S.o.P.T. Drugs Renin-angiotensin-aldosterone system and COVID-19 infection. Ann. Endocrinol. (Paris) 2020;81(2–3):63–67. doi: 10.1016/j.ando.2020.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson D.E., Sivalingam V., Kang A.E.Z., Ananthanarayanan A., Arumugam H., Jenkins T.M., Hadjiat Y., Eggers M. Povidone-iodine demonstrates rapid in vitro virucidal activity against SARS-CoV-2, the virus causing COVID-19 disease. Infect. Dis. Ther. 2020;9(3):669–675. doi: 10.1007/s40121-020-00316-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antinori S., Cossu M.V., Ridolfo A.L., Rech R., Bonazzetti C., Pagani G., Gubertini G., Coen M., Magni C., Castelli A., Borghi B., Colombo R., Giorgi R., Angeli E., Mileto D., Milazzo L., Vimercati S., Pellicciotta M., Corbellino M., Torre A., Rusconi S., Oreni L., Gismondo M.R., Giacomelli A., Meroni L., Rizzardini G., Galli M. Compassionate remdesivir treatment of severe Covid-19 pneumonia in intensive care unit (ICU) and Non-ICU patients: Clinical outcome and differences in post-treatment hospitalisation status. Pharmacol. Res. 2020;158:104899. doi: 10.1016/j.phrs.2020.104899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arshad U., Pertinez H., Box H., Tatham L., Rajoli R.K.R., Curley P., Neary M., Sharp J., Liptrott N.J., Valentijn A., David C., Rannard S.P., O’Neill P.M., Aljayyoussi G., Pennington S.H., Ward S.A., Hill A., Back D.J., Khoo S.H., Bray P.G., Biagini G.A., Owen A. Prioritization of anti-SARS-Cov-2 drug repurposing opportunities based on plasma and target site concentrations derived from their established human pharmacokinetics. Clin. Pharmacol. Ther. 2020;108:775–790. doi: 10.1002/cpt.1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atri D., Siddiqi H.K., Lang J., Nauffal V., Morrow D.A., Bohula E.A. COVID-19 for the cardiologist: a current review of the virology, clinical epidemiology, cardiac and other clinical manifestations and potential therapeutic strategies. JACC: Basic Transl. Sci. 2020 doi: 10.1016/j.jacbts.2020.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahbah E.I., Negida A., Nabet M.S. Purposing Saikosaponins for the treatment of COVID-19. Med. Hypotheses. 2020;140:109782. doi: 10.1016/j.mehy.2020.109782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baron S.A., Devaux C., Colson P., Raoult D., Rolain J.M. Teicoplanin: an alternative drug for the treatment of COVID-19? Int. J. Antimicrob. Agents. 2020;55(4):105944. doi: 10.1016/j.ijantimicag.2020.105944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-Or D., Rael L.T., Brody E.N. Insights into pediatric multi-system inflammatory syndrome and COVID-19. Clin. Chim. Acta. 2020;510:121–122. doi: 10.1016/j.cca.2020.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belen-Apak F.B., Sarialioglu F. The old but new: can unfractioned heparin and low molecular weight heparins inhibit proteolytic activation and cellular internalization of SARS-CoV2 by inhibition of host cell proteases? Med. Hypotheses. 2020;142:109743. doi: 10.1016/j.mehy.2020.109743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennardo F., Buffone C., Giudice A. New therapeutic opportunities for COVID-19 patients with Tocilizumab: possible correlation of interleukin-6 receptor inhibitors with osteonecrosis of the jaws. Oral Oncol. 2020;106:104659–104660. doi: 10.1016/j.oraloncology.2020.104659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhowmik D., Nandi R., Jagadeesan R., Kumar N., Prakash A., Kumar D. Identification of potential inhibitors against SARS-CoV-2 by targeting proteins responsible for envelope formation and virion assembly using docking based virtual screening, and pharmacokinetics approaches. Infect. Genet. Evol. 2020;84:104451. doi: 10.1016/j.meegid.2020.104451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bifulco M., Gazzerro P. Statins in coronavirus outbreak: It’s time for experimental and clinical studies. Pharmacol. Res. 2020;156:104803. doi: 10.1016/j.phrs.2020.104803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleasel M.D., Peterson G.M. Emetine, ipecac, ipecac alkaloids and analogues as potential antiviral agents for coronaviruses. Pharmaceuticals (Basel) 2020;13(3) doi: 10.3390/ph13030051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonzano C., Borroni D., Lancia A., Bonzano E. Doxycycline: from ocular rosacea to COVID-19 anosmia. New insight into the coronavirus outbreak. Front Med. (Lausanne) 2020;7:200. doi: 10.3389/fmed.2020.00200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boukhatem M.N., Setzer W.N. Aromatic herbs, medicinal plant-derived essential oils, and phytochemical extracts as potential therapies for coronaviruses: future perspectives. Plants (Basel) 2020;9(6) doi: 10.3390/plants9060800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabezón E., Arechaga I. Drug weaponry to fight against SARS-CoV-2. Front. Mol. Biosci. 2020;7 doi: 10.3389/fmolb.2020.00204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caly L., Druce J.D., Catton M.G., Jans D.A., Wagstaff K.M. The FDA-approved drug ivermectin inhibits the replication of SARS-CoV-2 in vitro. Antivir. Res. 2020;178:104787. doi: 10.1016/j.antiviral.2020.104787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carriere F., Longhi S., Record M. The endosomal lipid bis(monoacylglycero) phosphate as a potential key player in the mechanism of action of chloroquine against SARS-COV-2 and other enveloped viruses hijacking the endocytic pathway. Biochimie. 2020 doi: 10.1016/j.biochi.2020.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castano-Rodriguez C., Honrubia J.M., Gutierrez-Alvarez J., DeDiego M.L., Nieto-Torres J.L., Jimenez-Guardeno J.M., Regla-Nava J.A., Fernandez-Delgado R., Verdia-Baguena C., Queralt-Martin M., Kochan G., Perlman S., Aguilella V.M., Sola I., Enjuanes L. Role of severe acute respiratory syndrome coronavirus viroporins E, 3a, and 8a in replication and pathogenesis. mBio. 2018;9(3) doi: 10.1128/mBio.02325-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cattaneo D., Cattaneo D., Gervasoni C., Corbellino M., Galli M., Riva A., Gervasoni C., Clementi E., Clementi E. Does lopinavir really inhibit SARS-CoV-2? Pharmacol. Res. 2020;158:104898. doi: 10.1016/j.phrs.2020.104898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceribelli A., Motta F., De Santis M., Ansari A.A., Ridgway W.M., Gershwin M.E., Selmi C. Recommendations for coronavirus infection in rheumatic diseases treated with biologic therapy. J. Autoimmun. 2020;102442 doi: 10.1016/j.jaut.2020.102442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J.F., Yao Y., Yeung M.L., Deng W., Bao L., Jia L., Li F., Xiao C., Gao H., Yu P., Cai J.P., Chu H., Zhou J., Chen H., Qin C., Yuen K.Y. Treatment with Lopinavir/ritonavir or interferon-beta1b improves outcome of MERS-CoV infection in a nonhuman primate model of common marmoset. J. Infect. Dis. 2015;212(12):1904–1913. doi: 10.1093/infdis/jiv392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C.K., Jeyachandran S., Hu N.J., Liu C.L., Lin S.Y., Wang Y.S., Chang Y.M., Hou M.H. Structure-based virtual screening and experimental validation of the discovery of inhibitors targeted towards the human coronavirus nucleocapsid protein. Mol. BioSyst. 2016;12(1):59–66. doi: 10.1039/c5mb00582e. [DOI] [PubMed] [Google Scholar]
- Chang Y.S., Ko B.H., Ju J.C., Chang H.H., Huang S.H., Lin C.W. SARS Unique Domain (SUD) of severe acute respiratory syndrome coronavirus induces NLRP3 inflammasome-dependent CXCL10-mediated pulmonary inflammation. Int. J. Mol. Sci. 2020;21(9) doi: 10.3390/ijms21093179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Channappanavar R., Lu L., Xia S., Du L., Meyerholz D.K., Perlman S., Jiang S. Protective effect of intranasal regimens containing peptidic middle east respiratory syndrome coronavirus fusion inhibitor against MERS-CoV infection. J. Infect. Dis. 2015;212(12):1894–1903. doi: 10.1093/infdis/jiv325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Lu S., Jia H., Deng Y., Zhou J., Huang B., Yu Y., Lan J., Wang W., Lou Y., Qin K., Tan W. A novel neutralizing monoclonal antibody targeting the N-terminal domain of the MERS-CoV spike protein. Emerg. Microbes Infect. 2017;6(5) doi: 10.1038/emi.2017.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choy K.T., Wong A.Y., Kaewpreedee P., Sia S.F., Chen D., Hui K.P.Y., Chu D.K.W., Chan M.C.W., Cheung P.P., Huang X., Peiris M., Yen H.L. Remdesivir, lopinavir, emetine, and homoharringtonine inhibit SARS-CoV-2 replication in vitro. Antivir. Res. 2020;178:104786. doi: 10.1016/j.antiviral.2020.104786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clementi N., Criscuolo E., Diotti R.A., Ferrarese R., Castelli M., Dagna L., Burioni R., Clementi M., Mancini N. Combined prophylactic and therapeutic use maximizes hydroxychloroquine anti-SARS-CoV-2 effects in vitro. Front. Microbiol. 2020;11:1704. doi: 10.3389/fmicb.2020.01704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman C.M., Sisk J.M., Mingo R.M., Nelson E.A., White J.M., Frieman M.B. Abelson kinase inhibitors are potent inhibitors of severe acute respiratory syndrome coronavirus and Middle East respiratory syndrome coronavirus fusion. J. Virol. 2016;90(19):8924–8933. doi: 10.1128/JVI.01429-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colson P., Rolain J.M., Raoult D. Chloroquine for the 2019 novel coronavirus SARS-CoV-2. Int. J. Antimicrob. Agents. 2020;55(3):105923. doi: 10.1016/j.ijantimicag.2020.105923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corman V.M., Muth D., Niemeyer D., Drosten C. Elsevier; 2018. Hosts and Sources of Endemic Human Coronaviruses, Advances in Virus Research; pp. 163–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyle J., Igbinomwanhia E., Sanchez-Nadales A., Danciu S., Chu C., Shah N. A recovered case of COVID-19 myocarditis and ARDS treated with corticosteroids, tocilizumab, and experimental AT-001. JACC Case Rep. 2020;2(9):1331–1336. doi: 10.1016/j.jaccas.2020.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalamaga M., Karampela I., Mantzoros C.S. Commentary: could iron chelators prove to be useful as an adjunct to COVID-19 treatment regimens? Metabolism. 2020;108:154260. doi: 10.1016/j.metabol.2020.154260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dallmeier K., Neyts J. Zika and other emerging viruses: aiming at the right target. Cell Host Microbe. 2016;20(4):420–422. doi: 10.1016/j.chom.2016.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng W., Xu Y., Kong Q., Xue J., Yu P., Liu J., Lv Q., Li F., Wei Q., Bao L. Therapeutic efficacy of Pudilan Xiaoyan Oral Liquid (PDL) for COVID-19 in vitro and in vivo. Signal Transduct. Target Ther. 2020;5(1):66. doi: 10.1038/s41392-020-0176-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elfiky A.A. Anti-HCV, nucleotide inhibitors, repurposing against COVID-19. Life Sci. 2020;117477 doi: 10.1016/j.lfs.2020.117477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enriquez-Flores S., Flores-Lopez L.A., Garcia-Torres I., de la Mora-de la Mora I., Cabrera N., Gutierrez-Castrellon P., Martinez-Perez Y., Lopez-Velazquez G. Deamidated human triosephosphate isomerase is a promising druggable target. Biomolecules. 2020;10(7) doi: 10.3390/biom10071050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehr A.R., Channappanavar R., Jankevicius G., Fett C., Zhao J., Athmer J., Meyerholz D.K., Ahel I., Perlman S. The conserved coronavirus macrodomain promotes virulence and suppresses the innate immune response during severe acute respiratory syndrome coronavirus infection. mBio. 2016;7(6) doi: 10.1128/mBio.01721-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferron F., Subissi L., Silveira De Morais A.T., Le N.T.T., Sevajol M., Gluais L., Decroly E., Vonrhein C., Bricogne G., Canard B., Imbert I. Structural and molecular basis of mismatch correction and ribavirin excision from coronavirus RNA. Proc. Natl. Acad. Sci. U. S. A. 2018;115(2):E162–E171. doi: 10.1073/pnas.1718806115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer A., Sellner M., Neranjan S., Smieško M., Lill M.A. Potential inhibitors for novel coronavirus protease identified by virtual screening of 606 million compounds. Int. J. Mol. Sci. 2020;21(10):3626. doi: 10.3390/ijms21103626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galante O., Avni Y.S., Fuchs L., Ferster O.A., Almog Y. Coronavirus NL63-induced adult respiratory distress syndrome. Am. J. Respir. Crit. Care Med. 2016;193(1):100–101. doi: 10.1164/rccm.201506-1239LE. [DOI] [PubMed] [Google Scholar]
- Ghosh S., Firdous S.M., Nath A. siRNA could be a potential therapy for COVID-19. EXCLI J. 2020;19:528–531. doi: 10.17179/excli2020-1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gigante A., Aquili A., Farinelli L., Caraffa A., Ronconi G., Enrica Gallenga C., Tete G., Kritas S.K., Conti P. Sodium chromo-glycate and palmitoylethanolamide: a possible strategy to treat mast cell-induced lung inflammation in COVID-19. Med. Hypotheses. 2020;143:109856. doi: 10.1016/j.mehy.2020.109856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giulietti M., Righetti A., Cianfruglia L., Sabanovic B., Armeni T., Principato G., Piva F. To accelerate the Zika beat: candidate design for RNA interference-based therapy. Virus Res. 2018;255:133–140. doi: 10.1016/j.virusres.2018.07.010. [DOI] [PubMed] [Google Scholar]
- Glinsky G.V. Tripartite combination of candidate pandemic mitigation agents: vitamin D, quercetin, and estradiol manifest properties of medicinal agents for targeted mitigation of the COVID-19 pandemic defined by genomics-guided tracing of SARS-CoV-2 targets in human cells. Biomedicines. 2020;8(5) doi: 10.3390/biomedicines8050129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta R., Ghosh A., Singh A.K., Misra A. Clinical considerations for patients with diabetes in times of COVID-19 epidemic. Diabetes Metab. Syndr. 2020;14(3):211. doi: 10.1016/j.dsx.2020.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S., Kruger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A., Muller M.A., Drosten C., Pohlmann S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181(2):271–280. doi: 10.1016/j.cell.2020.02.052. e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houser K.V., Gretebeck L., Ying T., Wang Y., Vogel L., Lamirande E.W., Bock K.W., Moore I.N., Dimitrov D.S., Subbarao K. Prophylaxis with a Middle East respiratory syndrome coronavirus (MERS-CoV)-specific human monoclonal antibody protects rabbits from MERS-CoV infection. J. Infect. Dis. 2016;213(10):1557–1561. doi: 10.1093/infdis/jiw080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S.H., Lee T.Y., Lin Y.J., Wan L., Lai C.H., Lin C.W. Phage display technique identifies the interaction of severe acute respiratory syndrome coronavirus open reading frame 6 protein with nuclear pore complex interacting protein NPIPB3 in modulating Type I interferon antagonism. J. Microbiol. Immunol. Infect. 2017;50(3):277–285. doi: 10.1016/j.jmii.2015.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humeniuk R., Mathias A., Cao H., Osinusi A., Shen G., Chng E., Ling J., Vu A., German P. Safety, tolerability, and pharmacokinetics of remdesivir, an antiviral for treatment of COVID-19, in healthy subjects. Clin. Transl. Sci. 2020 doi: 10.1111/cts.12840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Z., Du X., Xu Y., Deng Y., Liu M., Zhao Y., Zhang B., Li X., Zhang L., Peng C., Duan Y., Yu J., Wang L., Yang K., Liu F., Jiang R., Yang X., You T., Liu X., Yang X., Bai F., Liu H., Liu X., Guddat L.W., Xu W., Xiao G., Qin C., Shi Z., Jiang H., Rao Z., Yang H. Structure of M(pro) from SARS-CoV-2 and discovery of its inhibitors. Nature. 2020;582(7811):289–293. doi: 10.1038/s41586-020-2223-y. [DOI] [PubMed] [Google Scholar]
- Kai H., Kai M. Interactions of coronaviruses with ACE2, angiotensin II, and RAS inhibitors-lessons from available evidence and insights into COVID-19. Hypertens. Res. 2020;43(7):648–654. doi: 10.1038/s41440-020-0455-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang S., Yang M., Hong Z., Zhang L., Huang Z., Chen X., He S., Zhou Z., Zhou Z., Chen Q. Crystal structure of SARS-CoV-2 nucleocapsid protein RNA binding domain reveals potential unique drug targeting sites. Acta Pharm. Sin. B. 2020;10:1228–1238. doi: 10.1016/j.apsb.2020.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang C.K., Seong M.W., Choi S.J., Kim T.S., Choe P.G., Song S.H., Kim N.J., Park W.B., Oh M.D. In vitro activity of lopinavir/ritonavir and hydroxychloroquine against severe acute respiratory syndrome coronavirus 2 at concentrations achievable by usual doses. Korean J. Intern. Med. 2020;35(4):782–787. doi: 10.3904/kjim.2020.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y., Lovell S., Tiew K.-C., Mandadapu S.R., Alliston K.R., Battaile K.P., Groutas W.C., Chang K.-O. Broad-spectrum antivirals against 3C or 3C-like proteases of picornaviruses, noroviruses, and coronaviruses. J. Virol. 2012;86(21):11754–11762. doi: 10.1128/JVI.01348-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y., Mandadapu S.R., Groutas W.C., Chang K.-O. Potent inhibition of feline coronaviruses with peptidyl compounds targeting coronavirus 3C-like protease. Antivir. Res. 2013;97(2):161–168. doi: 10.1016/j.antiviral.2012.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y., Shivanna V., Narayanan S., Prior A.M., Weerasekara S., Hua D.H., Kankanamalage A.C., Groutas W.C., Chang K.O. Broad-spectrum inhibitors against 3C-like proteases of feline coronaviruses and feline caliciviruses. J. Virol. 2015;89(9):4942–4950. doi: 10.1128/JVI.03688-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y., Liu H., Galasiti Kankanamalage A.C., Weerasekara S., Hua D.H., Groutas W.C., Chang K.O., Pedersen N.C. Reversal of the progression of fatal coronavirus infection in cats by a broad-spectrum coronavirus protease inhibitor. PLoS Pathog. 2016;12(3) doi: 10.1371/journal.ppat.1005531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kindrachuk J., Ork B., Hart B.J., Mazur S., Holbrook M.R., Frieman M.B., Traynor D., Johnson R.F., Dyall J., Kuhn J.H., Olinger G.G., Hensley L.E., Jahrling P.B. Antiviral potential of ERK/MAPK and PI3K/AKT/mTOR signaling modulation for Middle East respiratory syndrome coronavirus infection as identified by temporal kinome analysis. Antimicrob. Agents Chemother. 2015;59(2):1088–1099. doi: 10.1128/AAC.03659-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kow C.S., Hasan S.S. Use of azithromycin in COVID-19: a cautionary tale. Clin. Drug Investig. 2020 doi: 10.1007/s40261-020-00961-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim Y.X., Ng Y.L., Tam J.P., Liu D.X. Human coronaviruses: a review of virus–host interactions. Diseases. 2016;4(3):26. doi: 10.3390/diseases4030026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S.C., Ho C.T., Chuo W.H., Li S., Wang T.T., Lin C.C. Effective inhibition of MERS-CoV infection by resveratrol. BMC Infect. Dis. 2017;17(1):144. doi: 10.1186/s12879-017-2253-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C., Zhou Q., Li Y., Garner L.V., Watkins S.P., Carter L.J., Smoot J., Gregg A.C., Daniels A.D., Jervey S. ACS Publications; 2020. Research and Development on Therapeutic Agents and Vaccines for COVID-19 and Related Human Coronavirus Diseases. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Cao R., Xu M., Wang X., Zhang H., Hu H., Li Y., Hu Z., Zhong W., Wang M. Hydroxychloroquine, a less toxic derivative of chloroquine, is effective in inhibiting SARS-CoV-2 infection in vitro. Cell Discov. 2020;6:16. doi: 10.1038/s41421-020-0156-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Q., Qiu M., Zhou H., Chen J., Yang X., Deng Z., Chen L., Zhou J., Liao Y., Chen Q., Zheng Q., Cai L., Shen L., Yang Z. The study on the treatment of Xuebijing injection (XBJ) in adults with severe or critical Corona Virus Disease 2019 and the inhibitory effect of XBJ against SARS-CoV-2. Pharmacol. Res. 2020;160:105073. doi: 10.1016/j.phrs.2020.105073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Q., Pan W., Li R., Liu B., Li C., Xie Y., Wang Z., Zhao J., Jiang H., Huang J., Shi Y., Dai J., Zheng K., Li X., Yang Z. Liu Shen capsule shows antiviral and anti-inflammatory abilities against novel coronavirus SARS-CoV-2 via suppression of NF-kappaB signaling pathway. Pharmacol. Res. 2020;158:104850. doi: 10.1016/j.phrs.2020.104850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maisonnasse P., Guedj J., Contreras V., Behillil S., Solas C., Marlin R., Naninck T., Pizzorno A., Lemaitre J., Goncalves A., Kahlaoui N., Terrier O., Fang R.H.T., Enouf V., Dereuddre-Bosquet N., Brisebarre A., Touret F., Chapon C., Hoen B., Lina B., Calatrava M.R., van der Werf S., de Lamballerie X., Le Grand R. Hydroxychloroquine use against SARS-CoV-2 infection in non-human primates. Nature. 2020 doi: 10.1038/s41586-020-2558-4. [DOI] [PubMed] [Google Scholar]
- Marinella M.A. Indomethacin and resveratrol as potential treatment adjuncts for SARS-CoV-2/COVID-19. Int. J. Clin. Pract. 2020;74(9) doi: 10.1111/ijcp.13535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercurio I., Tragni V., Busto F., De Grassi A., Pierri C.L. Protein structure analysis of the interactions between SARS-CoV-2 spike protein and the human ACE2 receptor: from conformational changes to novel neutralizing antibodies. bioRxiv. 2021;78:1501–1522. doi: 10.1007/s00018-020-03580-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monti S., Montecucco C. Can hydroxychloroquine protect patients with rheumatic diseases from COVID-19? Response to: ‘Does hydroxychloroquine prevent the transmission of COVID-19?’ by Heldwein and Calado and ‘SLE, hydroxychloroquine and no SLE patients with COVID-19: a comment’ by Joob and Wiwanitkit. Ann. Rheum. Dis. 2020;79(6) doi: 10.1136/annrheumdis-2020-217524. [DOI] [PubMed] [Google Scholar]
- Mourad J.J., Levy B.I. Interaction between RAAS inhibitors and ACE2 in the context of COVID-19. Nat. Rev. Cardiol. 2020;17(5):313. doi: 10.1038/s41569-020-0368-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabavi S.F., Habtemariam S., Clementi E., Berindan-Neagoe I., Cismaru C.A., Rasekhian M., Banach M., Izadi M., Bagheri M., Bagheri M.S., Nabavi S.M. Lessons learned from SARS-CoV and MERS-CoV: FDA-approved Abelson tyrosine-protein kinase 2 inhibitors may help us combat SARS-CoV-2. Arch. Med. Sci. 2020;16(3):519–521. doi: 10.5114/aoms.2020.94504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- New R.R.C., Moore B.D., Butcher W., Mahood R., Lever M.S., Smither S., O'Brien L., Weller S.A., Bayliss M., Gibson L.C.D., Macleod C., Bogus M., Harvey R., Almond N., Williamson E.D. Antibody-mediated protection against MERS-CoV in the murine model. Vaccine. 2019;37(30):4094–4102. doi: 10.1016/j.vaccine.2019.05.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu P., Zhang S., Zhou P., Huang B., Deng Y., Qin K., Wang P., Wang W., Wang X., Zhou J., Zhang L., Tan W. Ultrapotent human neutralizing antibody repertoires against Middle East respiratory syndrome coronavirus from a recovered patient. J. Infect. Dis. 2018;218(8):1249–1260. doi: 10.1093/infdis/jiy311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto M., Toyama M., Baba M. The chemokine receptor antagonist cenicriviroc inhibits the replication of SARS-CoV-2 in vitro. Antivir. Res. 2020;182:104902. doi: 10.1016/j.antiviral.2020.104902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phadke M., Saunik S. COVID-19 treatment by repurposing drugs until the vaccine is in sight. Drug Dev. Res. 2020;81:541–543. doi: 10.1002/ddr.21666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillaiyar T., Wendt L.L., Manickam M., Easwaran M. The recent outbreaks of human coronaviruses: a medicinal chemistry perspective. Med. Res. Rev. 2020;41:72–135. doi: 10.1002/med.21724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pizzorno A., Padey B., Dubois J., Julien T., Traversier A., Duliere V., Brun P., Lina B., Rosa-Calatrava M., Terrier O. In vitro evaluation of antiviral activity of single and combined repurposable drugs against SARS-CoV-2. Antivir. Res. 2020;181:104878. doi: 10.1016/j.antiviral.2020.104878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polansky H., Lori G. Coronavirus disease 2019 (COVID-19): first indication of efficacy of Gene-Eden-VIR/Novirin in SARS-CoV-2 infection. Int. J. Antimicrob. Agents. 2020;55(6):105971. doi: 10.1016/j.ijantimicag.2020.105971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruijssers A.J., George A.S., Schafer A., Leist S.R., Gralinksi L.E., Dinnon K.H., 3rd, Yount B.L., Agostini M.L., Stevens L.J., Chappell J.D., Lu X., Hughes T.M., Gully K., Martinez D.R., Brown A.J., Graham R.L., Perry J.K., Du Pont V., Pitts J., Ma B., Babusis D., Murakami E., Feng J.Y., Bilello J.P., Porter D.P., Cihlar T., Baric R.S., Denison M.R., Sheahan T.P. Remdesivir inhibits SARS-CoV-2 in human lung cells and chimeric SARS-CoV expressing the SARS-CoV-2 RNA polymerase in mice. Cell Rep. 2020;32(3) doi: 10.1016/j.celrep.2020.107940. 107940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R. Graphics Map and Charts Showing the Spread of Coronavirus. 2020. https://graphics.reuters.com/CHINA-HEALTH-MAP/0100B59S39E/index.html
- Rossignol J.F. Nitazoxanide, a new drug candidate for the treatment of Middle East respiratory syndrome coronavirus. J. Infect Public Health. 2016;9(3):227–230. doi: 10.1016/j.jiph.2016.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Runfeng L., Yunlong H., Jicheng H., Weiqi P., Qinhai M., Yongxia S., Chufang L., Jin Z., Zhenhua J., Haiming J., Kui Z., Shuxiang H., Jun D., Xiaobo L., Xiaotao H., Lin W., Nanshan Z., Zifeng Y. Lianhuaqingwen exerts anti-viral and anti-inflammatory activity against novel coronavirus (SARS-CoV-2) Pharmacol. Res. 2020;156:104761. doi: 10.1016/j.phrs.2020.104761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo M., Moccia S., Spagnuolo C., Tedesco I., Russo G.L. Roles of flavonoids against coronavirus infection. Chem. Biol. Interact. 2020;328:109211. doi: 10.1016/j.cbi.2020.109211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheahan T.P., Sims A.C., Leist S.R., Schafer A., Won J., Brown A.J., Montgomery S.A., Hogg A., Babusis D., Clarke M.O., Spahn J.E., Bauer L., Sellers S., Porter D., Feng J.Y., Cihlar T., Jordan R., Denison M.R., Baric R.S. Comparative therapeutic efficacy of remdesivir and combination lopinavir, ritonavir, and interferon beta against MERS-CoV. Nat. Commun. 2020;11(1):222. doi: 10.1038/s41467-019-13940-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen L., Yang Y., Ye F., Liu G., Desforges M., Talbot P.J., Tan W. Safe and sensitive antiviral screening platform based on recombinant human coronavirus OC43 expressing the luciferase reporter gene. Antimicrob. Agents Chemother. 2016;60(9):5492–5503. doi: 10.1128/AAC.00814-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen L., Niu J., Wang C., Huang B., Wang W., Zhu N., Deng Y., Wang H., Ye F., Cen S. High-throughput screening and identification of potent broad-spectrum inhibitors of coronaviruses. J. Virol. 2019;93(12) doi: 10.1128/JVI.00023-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen L.W., Qian M.Q., Yu K., Narva S., Yu F., Wu Y.L., Zhang W. Inhibition of Influenza A virus propagation by benzoselenoxanthenes stabilizing TMPRSS2 gene G-quadruplex and hence down-regulating TMPRSS2 expression. Sci. Rep. 2020;10(1):7635. doi: 10.1038/s41598-020-64368-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shetty A.K. Mesenchymal stem cell infusion shows promise for combating coronavirus (COVID-19)-induced pneumonia. Aging Dis. 2020;11(2):462. doi: 10.14336/AD.2020.0301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi R., Shan C., Duan X., Chen Z., Liu P., Song J., Song T., Bi X., Han C., Wu L., Gao G., Hu X., Zhang Y., Tong Z., Huang W., Liu W.J., Wu G., Zhang B., Wang L., Qi J., Feng H., Wang F.S., Wang Q., Gao G.F., Yuan Z., Yan J. A human neutralizing antibody targets the receptor-binding site of SARS-CoV-2. Nature. 2020;584(7819):120–124. doi: 10.1038/s41586-020-2381-y. [DOI] [PubMed] [Google Scholar]
- Shin J.S., Jung E., Kim M., Baric R.S., Go Y.Y. Saracatinib inhibits middle east respiratory syndrome-coronavirus replication in vitro. Viruses. 2018;10(6) doi: 10.3390/v10060283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sriram K., Insel P.A. Risks of ACE inhibitor and ARB usage in COVID-19: evaluating the evidence. Clin. Pharmacol. Ther. 2020;108(2):236–241. doi: 10.1002/cpt.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su H.X., Yao S., Zhao W.F., Li M.J., Liu J., Shang W.J., Xie H., Ke C.Q., Hu H.C., Gao M.N., Yu K.Q., Liu H., Shen J.S., Tang W., Zhang L.K., Xiao G.F., Ni L., Wang D.W., Zuo J.P., Jiang H.L., Bai F., Wu Y., Ye Y., Xu Y.C. Anti-SARS-CoV-2 activities in vitro of Shuanghuanglian preparations and bioactive ingredients. Acta Pharmacol. Sin. 2020;41(9):1167–1177. doi: 10.1038/s41401-020-0483-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tvarogová J., Madhugiri R., Bylapudi G., Ferguson L.J., Karl N., Ziebuhr J. Identification and characterization of a human coronavirus 229E nonstructural protein 8-associated RNA 3′-terminal adenylyltransferase activity. J. Virol. 2019;93(12) doi: 10.1128/JVI.00291-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ul Qamar M.T., Alqahtani S.M., Alamri M.A., Chen L.-L. Structural basis of SARS-CoV-2 3CLpro and anti-COVID-19 drug discovery from medicinal plants. J. Pharm. Anal. 2020;10:313–319. doi: 10.1016/j.jpha.2020.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Brand J.M., Smits S.L., Haagmans B.L. Pathogenesis of Middle East respiratory syndrome coronavirus. J. Pathol. 2015;235(2):175–184. doi: 10.1002/path.4458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vankadari N., Wilce J.A. Emerging COVID-19 coronavirus: glycan shield and structure prediction of spike glycoprotein and its interaction with human CD26. Emerg. Microbes Infect. 2020;9(1):601–604. doi: 10.1080/22221751.2020.1739565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdecchia P., Reboldi G., Cavallini C., Mazzotta G., Angeli F. ACE-inibitori, sartani e sindrome respiratoria acuta da coronavirus 2. G. Ital. Cardiol. 2020;21(5):321–327. doi: 10.1714/3343.33127. [DOI] [PubMed] [Google Scholar]
- Verma S., Twilley D., Esmear T., Oosthuizen C.B., Reid A.-M., Nel M., Lall N. Anti-SARS-CoV natural products with the potential to inhibit SARS-CoV-2 (COVID-19) Front. Pharmacol. 2020;11:1514. doi: 10.3389/fphar.2020.561334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Sun Y., Wu A., Xu S., Pan R., Zeng C., Jin X., Ge X., Shi Z., Ahola T. Coronavirus nsp10/nsp16 methyltransferase can be targeted by nsp10-derived peptide in vitro and in vivo to reduce replication and pathogenesis. J. Virol. 2015;89(16):8416–8427. doi: 10.1128/JVI.00948-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.-L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367(6483):1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D., Yang X.O. TH17 responses in cytokine storm of COVID-19: an emerging target of JAK2 inhibitor Fedratinib. J. Microbiol. Immunol. Infect. 2020;53(3):368–370. doi: 10.1016/j.jmii.2020.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C., Liu Y., Yang Y., Zhang P., Zhong W., Wang Y., Wang Q., Xu Y., Li M., Li X. Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods. Acta Pharm. Sin. B. 2020;10:766–788. doi: 10.1016/j.apsb.2020.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia S., Liu M., Wang C., Xu W., Lan Q., Feng S., Qi F., Bao L., Du L., Liu S. Inhibition of SARS-CoV-2 (previously 2019-nCoV) infection by a highly potent pan-coronavirus fusion inhibitor targeting its spike protein that harbors a high capacity to mediate membrane fusion. Cell Res. 2020;30(4):343–355. doi: 10.1038/s41422-020-0305-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia S., Liu M., Wang C., Xu W., Lan Q., Feng S., Qi F., Bao L., Du L., Liu S., Qin C., Sun F., Shi Z., Zhu Y., Jiang S., Lu L. Inhibition of SARS-CoV-2 (previously 2019-nCoV) infection by a highly potent pan-coronavirus fusion inhibitor targeting its spike protein that harbors a high capacity to mediate membrane fusion. Cell Res. 2020;30(4):343–355. doi: 10.1038/s41422-020-0305-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia S., Zhu Y., Liu M., Lan Q., Xu W., Wu Y., Ying T., Liu S., Shi Z., Jiang S. Fusion mechanism of 2019-nCoV and fusion inhibitors targeting HR1 domain in spike protein. Cell. Mol. Immunol. 2020;17(7):765–767. doi: 10.1038/s41423-020-0374-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xian Y., Zhang J., Bian Z., Zhou H., Zhang Z., Lin Z., Xu H. Bioactive natural compounds against human coronaviruses: a review and perspective. Acta Pharm. Sin. B. 2020;10:1163–1174. doi: 10.1016/j.apsb.2020.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav R., Imran M., Dhamija P., Suchal K., Handu S. Virtual screening and dynamics of potential inhibitors targeting RNA binding domain of nucleocapsid phosphoprotein from SARS-CoV-2. J. Biomol. Struct. Dyn. 2020:1–16. doi: 10.1080/07391102.2020.1778536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto M., Matsuyama S., Li X., Takeda M., Kawaguchi Y., Inoue J.I., Matsuda Z. Identification of nafamostat as a potent inhibitor of Middle East respiratory syndrome coronavirus S protein-mediated membrane fusion using the Split-protein-based cell-cell fusion assay. Antimicrob. Agents Chemother. 2016;60(11):6532–6539. doi: 10.1128/AAC.01043-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang R., Liu H., Bai C., Wang Y., Zhang X., Guo R., Wu S., Wang J., Leung E., Chang H., Li P., Liu T., Wang Y. Chemical composition and pharmacological mechanism of Qingfei Paidu Decoction and Ma Xing Shi Gan Decoction against Coronavirus Disease 2019 (COVID-19): in silico and experimental study. Pharmacol. Res. 2020;157:104820. doi: 10.1016/j.phrs.2020.104820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R., Wang K., Ping X., Yu W., Qian Z., Xiong S., Sun B. The ns12.9 accessory protein of human coronavirus OC43 is a viroporin involved in virion morphogenesis and pathogenesis. J. Virol. 2015;89(22):11383–11395. doi: 10.1128/JVI.01986-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z.-j., Wu W.-y., Hou J.-j., Zhang L.-l., Li F.-f., Gao L., Wu X.-d., Shi J.-y., Rong Zhang R., Long H.-l., Li M., Wu W.-y., Guo D.-a., Chen K.-x., Hofmann L.A., Ci Z.-h. Active constituents and mechanisms of Respiratory Detox Shot, a traditional Chinese medicine prescription, for COVID-19 control and prevention: network-molecular docking-LC-MS analysis. J. Integr. Med. 2020;18:229–241. doi: 10.1016/j.joim.2020.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R., Wang X., Ni L., Di X., Ma B., Niu S., Liu C., Reiter R.J. COVID-19: melatonin as a potential adjuvant treatment. Life Sci. 2020;117583 doi: 10.1016/j.lfs.2020.117583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H., Zhou J., Zhang K., Chu H., Liu D., Poon V.K., Chan C.C., Leung H.C., Fai N., Lin Y.P., Zhang A.J., Jin D.Y., Yuen K.Y., Zheng B.J. A novel peptide with potent and broad-spectrum antiviral activities against multiple respiratory viruses. Sci. Rep. 2016;6:22008. doi: 10.1038/srep22008. [DOI] [PMC free article] [PubMed] [Google Scholar]