Abstract
Novel Coronavirus with its highly transmittable characteristics is rapidly spreading, endangering millions of human lives and the global economy. To expel the chain of alteration and subversive expansion, early and effective diagnosis of infected patients is immensely important. Unfortunately, there is a lack of testing equipment in many countries as compared with the number of infected patients. It would be desirable to have a swift diagnosis with identification of COVID-19 from disease genes or from CT or X-Ray images. COVID-19 causes flus, cough, pneumonia, and lung infection in patients, wherein massive alveolar damage and progressive respiratory failure can lead to death. This paper proposes two different detection methods – the first is a Gene-based screening method to detect Corona diseases (Middle East respiratory syndrome-related coronavirus, Severe acute respiratory syndrome coronavirus 2, and Human coronavirus HKU1) and differentiate it from Pneumonia. This novel approach to healthcare utilizes disease genes to build functional semantic similarity among genes. Different machine learning algorithms - eXtreme Gradient Boosting, Naïve Bayes, Regularized Random Forest, Random Forest Rule-Based Model, Random Ferns, C5.0 and Multi-Layer Perceptron, are trained and tested on the semantic similarities to classify Corona and Pneumonia diseases. The best performing models are then ensembled, yielding an accuracy of nearly 93%. The second diagnosis technique proposed herein is an automated COVID-19 diagnostic method which uses chest X-ray images to classify Normal versus COVID-19 and Pneumonia versus COVID-19 images using the deep-CNN technique, achieving 99.87% and 99.48% test accuracy. Thus, this research can be an assistance for providing better treatment against COVID-19.
Keywords: COVID-19, Pneumonia, Gene-based screening, Functional semantic similarity matrix, Machine learning, CNN
1. Introduction
COVID-19 caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is a highly contagious disease. The Coronaviruses were thought to infect only animals until the world witnessed a severe acute respiratory syndrome (SARS) outbreak caused by SARS-CoV, 2002 in Guangdong, China [1]. In 2005, the human coronavirus HKU1 was first discovered. Almost a decade later, another endemic coronavirus known as Middle East respiratory syndrome coronavirus (MERS CoV) appeared in Middle Eastern countries. We have now seen the onset of COVID-19. Emerging from Wuhan, China in December 2019, COVID-19 spread rapidly around the world, affecting the people of approximately 215 countries. On 12 February 2020, the WHO warned that due to COVID-19, millions would die if it remained uncontrolled, and declared it as a Pandemic on March 11, 2020 [2]. According to the Worldometers data, over 21 million people have been infected, with over 0.76M deaths [3]. This pandemic has become a grim figure as the new cases have increased exponentially. Social distancing and contact tracing are two effective techniques proposed by the World Health Organization (WHO) to control the spread of this viral infection [4]. Thus, to avoid fast transmission of the virus, most countries made lockdown compulsory, which disrupted daily life and socio-economic conditions. Still, the situation is not entirely controlled.
Effective screening of infected patients helps them so as to become isolated and receive immediate treatment and care to mitigate the spread of the virus [5]. The reverse-transcription polymerase chain reaction (RT-PCR) is the accepted standard diagnostic method of COVID-19 [6]. However, because the number of RT-PCR testing kits, testing reagents, proper lab environment, PPE, and expertise is inadequate to meet demand, contaminated rates are rapidly increasing. Hence, researchers are trying to develop alternative detection techniques. Currently, Machine Learning and Deep Learning are used as successful AI techniques for effective diagnosis of diseases. The X-ray radiography method is easier and more cost-effective than CT scan images. Therefore, most researchers prefer the use of X-ray images rather than CT images.
Almost all of the Corona diseases start with cold-like symptoms and then advance to Pneumonia. COVID-19 Symptoms can be mild to severe, including fever, cough, and dyspnea to pneumonia, severe acute respiratory syndrome, septic shock, multi-organ failure and death in more serious cases [7]. From a report of [3] among the active cases, 2% of patients are critical and 98% are mild. Studies have found that the symptoms are changing gradually as the virus slightly changes its genetic makeup. In some current cases, corona-positive patients are found without any symptoms. For these reasons, the gene-based COVID-19 detection method can be a great alternative to the other methods. To mitigate the risk of developing certain diseases and to detect these diseases at an earlier stage, the knowledge of an individual's genetic make-up can be used [8]. Given a set of disease genes associated with a disease, they can be used to find further candidate genes for the disease [9] and also to detect and distinguish it from other diseases.
This study aims to mitigate the limitations of the traditional COVID-19 diagnostic method by demonstrating two fast and effective diagnostic techniques, a Gene-based Corona disease detection method and an automated computer-aided diagnosis (CAD) tool for the diagnosis of COVID-19, to differentiate it from pneumonia and healthy via from chest X-ray imagery. Genes and chest X-ray images associated with the diseases are first collected and pre-processed. Several techniques are applied to the disease genes to calculate the functional similarity measure matrices among them. The National Center for Biotechnology Information (NCBI) and Gene Ontology (GO) online databases are used for these purposes. Afterward, different machine learning algorithms are applied to the matrices for successful prediction. The X-ray images are incorporated by using a pre-trained CheXNet deep convolutional neural network (CNN) with model weights [10] to diagnose COVID-19 from Pneumonia versus Normal healthy images.
The main contributions of this paper are:
-
➢
A new approach to diagnose Corona diseases from disease genes with excellent performance.
-
➢
An extensive experimentation was done to select the best performing machine learning (ML) model on the best gene functional similarity measures.
-
➢
An ensemble technique of ML models was utilized to increase classification accuracy.
-
➢
An automated computer-aided method of COVID-19 detection was developed to be used with from Chest X-ray imagery with a transfer learned deep CNN model.
-
➢
A relative study was done of different image preprocessing techniques to achieve the best classification accuracy.
-
➢
An empirical study on different image augmentation techniques was employed to work with limited datasets and resolve the CNN model's overfitting problem.
The remainder of the paper is organized as follows: Related Works demonstrating the literature review can be found in section 2. Section 3 captures the proposed materials and methods of this study. Section 4 describes and discusses the results. Finally, the conclusion and future work of this research can be found in section 5.
2. Related works
Modern technology has made diagnosis and treatment easier and more convenient than ever before. The availability of large datasets and the success of deep learning have made the results of diagnostics tasks more accurate. This section highlights the studies and works done by other groups related to this research.
As of this writing, Coronavirus is still spreading, which causes danger to millions of people. To control the spread of the COVID-19, screening large numbers of suspected cases for appropriate quarantine and treatment measures is a priority. Yet, the RT-PCR testing process is time-consuming and also sometimes shows false-negative results, so researchers are trying to develop alternative detection techniques.
Paper [11] uses gene functional similarity to identify disease genes. Jianpeng Zhang et al. developed a deep learning model to detect COVID-19 from Chest X-ray images with high sensitivity for active cases [6]. In article [12], different online Chest X-ray datasets are combined, rearranged and then transfer learning methods are used for this disease detection. Paper [7,13] also developed an automated deep CNN model for detecting and distinguishing COVID-19 from Pneumonia using X-rays. Lin Li et al. proposes a deep learning neural network model called COVNet for COVID-19 detection and differentiates it from pneumonia and other lung diseases using CT scan images [14].
Hemdan et al. [15] proposed COVIDX-Net with seven different CNN models- VGG19, DenseNet201, ResNetV2, InceptionV3, InceptionResNetV2, Xception, MobileNetV2 to diagnose COVID-19 using 25 COVID-19 positive and 25 normal X-ray images. The model obtained the best accuracy of 90% for VGG19 and DenseNet201. A deep CNN model called DeCoVNet is proposed by Zheng et al. [16] to detect COVID-19 from non-COVID-19 normal CT images with 90.1% accuracy. The model of Sarhan A.M. et al. [17] proposed a fusion of Wavelet and SVM model to differentiate COVID-19 from Normal X-ray image acquiring 94.5% accuracy. Nasrin et al. [7] performed a binary classification of 50 COVID-19 vs 50 Normal images and obtained the highest classification accuracy of 98% using ResNet50. DarkCovidNet proposed by Ozturk et al. [18] obtained an accuracy of 98.08% for binary and 87.02% for three classes.
Wang et al. [19] uses CT images to classify COVID-19 from Pneumonia using the modified Inception (M-Inception) deep model. The model achieved a classification accuracy of 82.9%. Ying et al. performed two binary classification tasks- COVID-19 vs Pneumonia achieving 86% accuracy and COVID-19 vs Normal obtaining 94% accuracy using CT images with the DRE-Net model [20]. Sethy and Behera [21] proposed a model which is the fusion of ResNet50 features with an SVM classifier and achieved 95.38% accuracy for COVID-19 vs Pneumonia classification. They extracted the features of the pre-trained CNN model and then used an SVM classifier as the final layer.
Some other models are also employed for the purpose of multi-class classification. Xu et al. [22] achieved an 86.7% performance accuracy in detecting COVID-19 from Influenza-A viral pneumonia (IAVP) and healthy cases using pulmonary CT images. CovidAID by Mangal et al. [23] uses pre-trained CheXNet model and achieves a performance accuracy of 90.5%. COVID-Net, a deep CNN model by Wang and Wong [5] achieved a 93.3% accuracy for classifying COVID-19, Non-COVID-19 and Normal images. Asif et al. [24] used pre-trained Inception V3 model to diagnose COVID-19 from three-class classifications with 96% test accuracy from chest X-rays. Kumar R. [25] et al. proposed a model that classifies X-ray images to COVID-19, Normal and Pneumonia images with 97.7% accuracy.
Pneumonia is a contagious lung disease that creates breathing difficulty and severe respiratory problems with inflammation in lung alveoli. One of the major symptoms of COVID-19 is Pneumonia. Hence, it is very difficult to differentiate between Pneumonia and COVID-19. Identifying COVID-19 disease genes from Pneumonia disease genes appropriately, in turn means identifying COVID-19 from Pneumonia. ML classifiers trained on gene semantic similarity scores can differentiate disease genes by inferring hidden semantic similarities among genes. As AI and ML tools show efficient performance in diagnosing Pneumonia, they can be also applied to diagnose COVID-19 successfully.
To suppress the rapid transmission of the coronavirus, it is necessary to screen all suspected cases, quarantine them and provide immediate treatment. This study proposed a new diagnostic technique for Corona Disease that uses disease genes and performs well in distinguishing Corona disease from Pneumonia. Also, a fine-tuned CheXNet CNN model is proposed here that is pre-trained on the Pneumonia dataset [26] for the diagnosis of COVID-19 from X-ray images that serves two classifications tasks-COVID-19 vs Pneumonia and COVID-19 vs Normal images.
3. Proposed material and methods
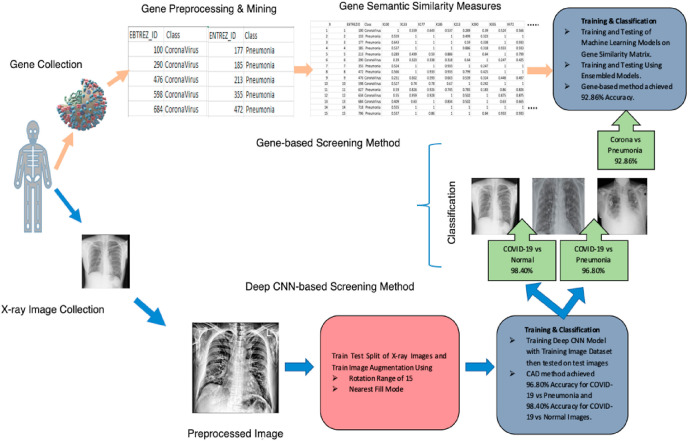
The proposed method demonstrated different steps from data collection to Corona detection using the Gene-based method and CAD method. The diagram of Fig. 1 displays the schematic representation of the proposed methodology.
Fig. 1.
Schematic representation of corona detection methods.
3.1. Data source
Two different screening methods have been developed here for the diagnosing of Corona diseases and COVID-19 from diseases genes and chest X-ray images. For this purpose, associated genes for all types of Coronavirus and Pneumonia for Homo sapiens are collected from NCBI Gene databases. As COVID-19 is a new term, there are only a few genes available online. So, Gene-based diagnostic study focuses on three types of Coronavirus diseases such as- Middle East respiratory syndrome-related coronavirus, Severe acute respiratory syndrome coronavirus 2 and human coronavirus HKU1. The National Center for Biotechnology Information (NCBI) [27,28] is a branch of the National Institutes of Health (NIH) and is part of the United States National Library of Medicine (NLM) containing a series of databases. The NCBI Gene database is a freely accessible online database with a huge collection of known and predicted genes.
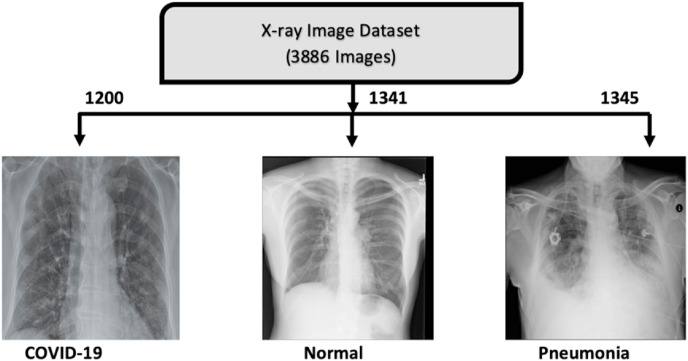
The online ‘COVID-19 Radiography Dataset’ [29] by Tawsifur Rahman, which is the ‘Winner of COVID-19 Dataset Award by Kaggle’ is used here for the CNN-based CAD method. This dataset collects COVID-19 images from Cohen JP [30] and different publications and Pneumonia and Normal images from the Kaggle pneumonia dataset of Paul M [31]. The dataset consists of 1200 COVID-19 images, 1341 Normal images and 1345 viral Pneumonia images. Fig. 2 summarizes the X-ray dataset.
Fig. 2.
X-ray dataset for COVID-19 detection.
3.2. Data preparation
Data preprocessing is one of the vital steps to enhance the quality of data and transform the raw data into a more suitable and efficient format. Collected Coronavirus and Pneumonia genes are rearranged according to their weight values as high-weighted genes are on top and sorted accordingly. Genes are collected in a summary format containing extensive information. The Genes are mined by removing irrelevant information and a dataframe combining genes from both classes with only gene id and disease class column is created. There are 108 genes found for Coronavirus and 252 for Pneumonia cases. Among the collected genes, 24 are common for both diseases. These common 24 genes are removed from both of the disease genes, resulting in 84 genes for Corona Disease and 228 genes for Pneumonia. As there exist more Pneumonia genes than Corona Disease genes, ML models can be slightly biased towards Pneumonia. So, to avoid the model's biasness, 84 top-weighted genes for both of the diseases are used for further processing. Table 1 shows the number of genes before and after preprocessing and gene mining.
Table 1.
Gene dataset summary.
| Disease Class | No. of Collected Genes | Common Genes | Genes after ignoring the Common Genes | No. of Genes after Preprocessing and Mining |
|---|---|---|---|---|
| Corona Disease | 108 | 24 | 84 | 84 |
| Pneumonia | 252 | 228 | 84 |
The X-ray images are rescaled to 0–1 ranges to make the training faster. Different image preprocessing and enhancement techniques such as- Histogram Equalization (HE), Adaptive Histogram Equalization (AHE), Gabor Filtering (GF), Histograms of Oriented Gradients (HOGs) and Local Binary Patterns (LBPs) are applied to the collected X-ray images. Among the techniques, AHE as an image contrast enhancement technique performs well. The images are then resized to 224 × 224 as the proposed CNN model accepts images of size 224 × 224 only.
3.3. Gene functional similarity matrix calculation
Gene functional similarity covers a wide area of biological and bioinformatics research including gene clustering, disease gene prediction, protein-protein interaction etc. Functional similarity between and among genes is the quantitative measure of the semantic relationships of their terms. It conveys more information about gene functions and semantic relationships and can be stored as a matrix. Gene expression profiles, Protein-Protein Interactions (PPI) networks or Gene Ontology (GO) can be used to identify the functional similarity of genes and their products. This study determines functional similarities based on GO annotations. Gene Ontology (GO) is the most frequently used vocabulary for representing gene functions with a well-defined structure and manual curation [32]. The Gene Ontology (GO) terms are structured as a hierarchical Directed Acyclic Graph (DAG). Biological Process (BP), Molecular Function (MF), and Cellular Component (CC) are the three orthogonal ontologies provided by GO. All of the five semantic similarity measures Resnik [33], Jiang [34], Lin [35], Schlicker [36] and Wang [37] methods are employed here with Maximum (max) and Best Match Average Strategy (BMA) combining strategy as a quantitative measure of gene functional similarities. The Resnik, Jiang, Lin and Schlicker measures are information content (IC) based methods and Wang is a Graph-based method. IC-based methods compute a semantic score between two GO terms based on the IC of their Most Informative Common Ancestor (MICA) term [11] and can be defined as- IC(t) = -log(p(t)); where, p(t) be the probability of usage of GO term t being used in a given GO corpus. Whereas, the Wang semantic similarity measure uses hierarchical DAG structure to estimate semantic similarity between genes.
The above semantic similarity measures can be represented as [38]-
Resnik method
| (1) |
Lin method
| (2) |
Rel method.
The Relevance method, which was proposed by Schlicker, combines Resnik's and Lin's method and can be defined as:
| (3) |
Jiang method.
The Jiang and Conrath's method can be defined as:
| (4) |
Wang method.
Given two GO terms A and B, the semantic similarity between these two terms can be defined as:
| (5) |
where SA(t) is the S-value of GO term t related to term A and SB(t) is the S-value of GO term t related to term B. SV(A) and SV(B) are the semantic values of GO terms A and B.
Assume that g1 and g2 are two distinct genes annotated by the GO terms sets GO1 = {go11, go12⋯go1m} and GO2 = {go21, go22⋯go2n}. To find the semantic similarity score of two genes g1 and g2, semantic similarity scores of their GO terms sets GO1 and GO2 are used. There are four methods-max, avg, rcmax, and BMA to combine semantic similarity scores of multiple GO terms implemented by GOSemSim package of R. The max semantic similarity combining technique calculates the maximum semantic similarity score over all possible pairs of GO terms {go11, go12⋯go1m} and {go21, go22⋯go2n} between these two GO term sets, GO1 and GO2. For example, it finds the semantic similarity between all pairs (go11, go21), (go11, go22), (go11, … go2n), (go12, go21), (go12, go22), (go12, … go2n), … … (go1m, … …. go2n) and then selects the max value as the semantic similarity value between gene g1 and g2 as like the equation below-
| (6) |
The BMA method also finds the pair wise semantic similarity values and computes the average of all maximum similarities on each row and column and is defined as:
| (7) |
3.4. Machine learning models construction and evaluation
Supervised machine learning methods are capable of training hidden gene-relationships from a given dataset and then using that learned knowledge to discriminate disease genes from non-disease genes. The gene functional similarity returns 169 × 172 matrices for all of the Res, Lin, Rel, Jiang and Wang measures. For representing biological concepts, more than 40 thousand GO terms are used. But, still GO semantic similarity returns null values for some genes that lack GO information. As semantic similarity only returns a value between 0 and 1, the null values need to be removed instead of replacing with 0. For Res, Lin, Rel and Jiang measures, 32 genes and for Wang measure 31 genes found with null values resulting in 137 × 140 and 138 × 141 matrixes respectively. Thus, the remaining final genes are 138 for the Wang measure with 140 feature columns and 1 label column and 137 genes with 139 features and 1 label column for all of the other measures. The gene dataset is then split into training and testing datasets using 80% of data as training data and 20% as test data. The following Table 2 shows the total number of available genes for each measure.
Table 2.
Available gene data.
| Semantic Similarity Measures | Total Genes = |
80% Training Gene Data = |
20% Test Gene Data = |
|---|---|---|---|
| Corona Genes + Pneumonia Genes | |||
| Res | 137 = 54 + 83 | 109 = 44 + 65 | 28 = 10 + 18 |
| Lin | 137 = 54 + 83 | 109 = 44 + 65 | 28 = 10 + 18 |
| Rel | 137 = 54 + 83 | 109 = 44 + 65 | 28 = 10 + 18 |
| Jiang | 137 = 54 + 83 | 109 = 44 + 65 | 28 = 10 + 18 |
| Wang | 138 = 54 + 83 | 110 = 44 + 66 | 28 = 11 + 17 |
The machine learning algorithms-eXtreme Gradient Boosting (xgbLinear), Naïve Bayes (NB), Regularized Random Forest (RRF), Random Forest Rule-Based Model (rfRules), Random Ferns (rFerns), C5.0 (C5) and Multi-Layer Perceptron (MLP) are then trained on the training gene dataset with five-fold cross-validation and tested on the remaining 20% test dataset. Among the models, xgbLinear and RRF perform best on the Wang measures with max combining technique and achieve 82.14% test accuracy. Contrarily, MLP gives the best performance on the Rel measure with BMA combining technique of about 89.29%.
xgbLinear is a method of eXtreme Gradient Boosting that can be used for both classification and regression using the xgboost library. To find the best tree model, it uses a specific Gradient Boosting method using more accurate and successful approximations.
NB is a supervised classification algorithm based on the Bayes’ Theorem. It predicts the best class in a way like the Bayes Theorem finds the best hypothesis from given prior knowledge.
RRF implements a regularized random forest algorithm that can be used for both classification and regression. It applies the tree regularization framework to RF and can select a compact feature subset [39] of relevant and non-redundant features.
rfRules acts as both a classification and regression model. It generates a series of “if-then” rules to effectively classify classes.
rFerns is a machine learning classification algorithm that extends the Naïve Bayes algorithm. It can be considered as a constrained decision tree where at each level of the tree the same binary test is performed.
C5.0 is a classification algorithm that is well-known for producing decision trees. It can be used for both small and large datasets and its decision trees are relatively easy to understand and deploy.
MLP is a supervised classification and regression algorithm which is widely used in image and speech recognition. It is a multilayer feedforward artificial neural network generating a set of outputs from a set of inputs. MLP uses backpropagation.
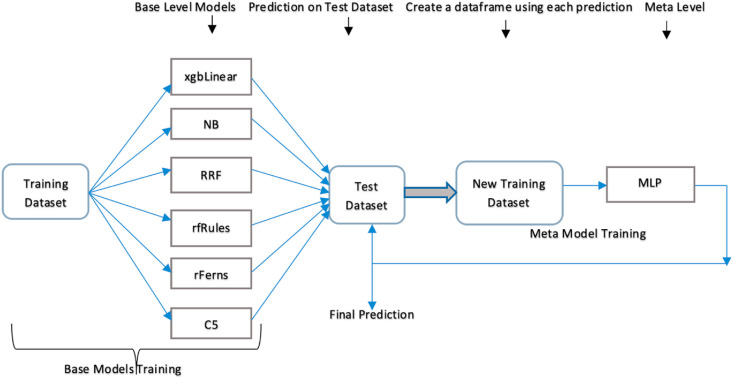
To obtain more accurate classification results, the machine learning models are ensembled using the stacking ensembled technique. Stacking ensemble is a technique that works with two levels of models. The base level or bottom level ensembles all of the base models using the original dataset as input and the meta level or top level contains a model that uses the base level's outputs as inputs. In this research, xgbLinear, NB, RRF, rfRules, rFerns, C5 models are used as base models and MLP is used as a top-level model. The top-level model makes a prediction on the diseases, whether it is corona disease or pneumonia. Wang measures with the max combining technique were chosen for the ensemble because they produced the best classification result for all of the seven classifiers. Fig. 3 displays the schematic representation of the stacking ensemble model.
Fig. 3.
Stacking ensemble model.
3.5. Image augmentation
Two binary classification tasks- COVID-19 vs Normal and COVID-19 vs Pneumonia are performed in the CNN-based CAD method. After preprocessing of chest X-ray images, they are labeled as 0 and 1 where 1 represents COVID-19 image and 0 represents the other class image- Normal or Pneumonia for both of the classification tasks. The images are then divided into training and testing datasets using a 7:3 ratio. Thus, there are 836 COVID-19, 942 Normal training images and 364 COVID-19, 399 Normal test images for COVID-19 vs Normal classification. And for COVID-19 vs Pneumonia image classification, there are 836 COVID-19 and 945 Pneumonia training images and 364 COVID-19, 400 Pneumonia test images. Training a deep CNN model with a limited number of data may cause the model to overfit. Thus, the model may perform well on training data but fails to generalize. So, to artificially increase the amount of training data and overcome the overfitting problem, augmentation is applied to the training dataset only. The training images are augmented using a rotation range of 15° and the nearest fill_mode.
3.6. CNN model construction
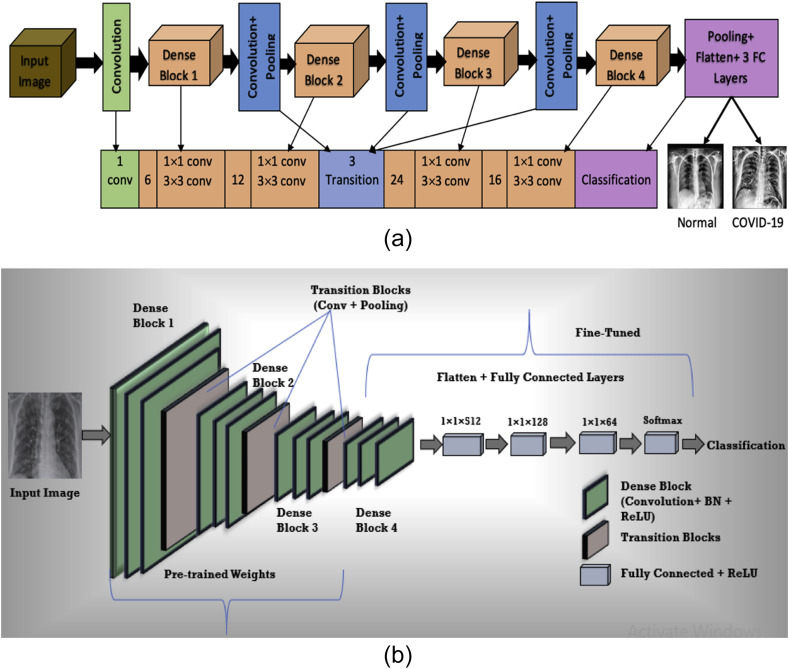
This research uses the CheXNet CNN model, which is the fine-tuned and transfer learned CheXNet model that was previously used by Ref. [10] & [45] for Pneumonia Detection. The original CheXNet model was proposed by the researchers from Stanford University [40] is a 121-layer DenseNet architecture. CheXNet was at first pre-trained on the ImageNet dataset and then trained on the CXR dataset of [41]. In our previous Pneumonia detection research, the fine-tuned CheXNet model was trained on [26] dataset. Here, the transfer learned CheXNet model is used with Softmax activation function at the final layer for binary classification and ReLU activation function for all other activation layers. Fig. 4 (a) represents the CheXNet model architecture and Fig. 4(b) shows the proposed CheXNet model architecture with pre-trained weights and fine tuning. The model uses Adam optimizer and binary cross entropy loss function and is trained end-to-end with a mini batch size of 32. During fine-tuning, the 1st to 409th layers kept freezing while the remaining 410th to 437th layers are trained. Before flattening, Max Pooling with 20% dropout is used for COVID-19 vs Normal classifier and Global Average Pooling with 20% dropout is used for COVID-19 vs Pneumonia classifier. The fully connected dense layers consist of 512, 128 and 64 neurons with a 10% dropout rate before the final layer. After training the CNN model with a training dataset several times with 40 epochs, the testing dataset is tested on the model and a prediction is made.
Fig. 4.
(a). Architectural design of CheXNet model.
(b). Proposed fine-tuned CheXNet model with pre-trained weights.
4. Results and discussion
This section provides the result and discusses the output of each step of the proposed methods and materials of the current research project. The results are described in the following subsections.
4.1. Data collection and preparation
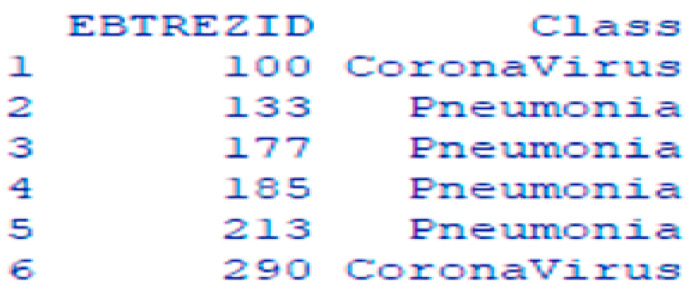
The collected gene data from NCBI Gene repositories are two summary type text files, one for Corona disease and the other for Pneumonia. There are 24 common genes found between Corona and Pneumonia diseases. They are removed from both files and 84 top-weighted genes from both classes are selected to maintain a balanced, unbiased dataset. After preprocessing and mining genes from the collected gene data files, a dataframe is created containing ENTREZID and Class column. The head of the dataframe is shown in Fig. 5 below-
Fig. 5.
Head of the gene input dataframe.
All processes in the Gene-based screening method are carried out in the R programming language in windows 10, 64-bit environment.
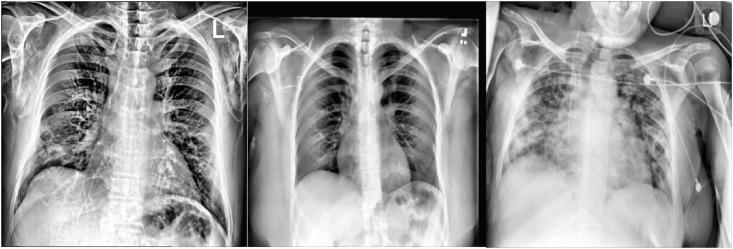
In the CNN-based CAD method, collected X-ray images are preprocessed using the AHE contrast enhancement technique. The enhanced COVID-19, Normal and Pneumonia images are shown in Fig. 6 .
Fig. 6.
Enhanced COVID-19, normal, pneumonia image.
70% of images are now used as training images and augmentation is applied on them to artificially increase the number of training images. The remaining 30% of images are used for testing the CNN model.
4.2. Gene functional semantic similarity measures calculation
Semantic Similarity can be used to measure the functional closeness of Gene Ontology (GO). The R packages org.Hs.eg.db [42] and GOSemSim [43] are used for the gene semantic similarity matrix estimation. As COVID-19 is a new term, some of the gene information of Corona disease and very few of Pneumonia lack GO information which returns null semantic similarity scores. Semantic similarity only returns a value between 0 and 1, thus null values need to be removed. Then the training and testing datasets are constructed using an 8:2 ratio. Among the semantic similarity measure methods (Resnik, Lin, Rel, Jiang and Wang), the Wang method achieves the best results with the max combining technique.
4.3. Classification using gene-based screening
To determine the hidden functional similarities between Corona disease genes and Pneumonia genes, xgbLinear, NB, RRF, rfRules, rFerns, C5 and MLP machine learning classifiers are trained and tested on Corona and Pneumonia gene functional similarities. The performance of any machine learning algorithm depends highly on the amount of data available. Huge data can make the algorithm more accurate than limited data. This is the main shortcoming of our study. Because of the unavailability of a large amount of gene data for coronavirus and updated GO information, the Machine Learning (ML) model accuracy got negatively affected. The above seven ML models are trained using a 5-fold cross-validation technique on each of the five-similarity matrices of training dataset with two combining techniques resulting in a total of 70 classifiers and make a prediction on the test dataset. Sensitivity and Specificity are also calculated for each of the ML models. Table 3, Table 4 show the performance of various machine learning classifiers built over functional similarities scores using the max and BMA combining techniques, respectively.
Table 3.
Average accuracy of xgbLinear, NB, RRF, rfRules, rFerns, C5 and MLP models on identifying Corona disease on Resnik, Lin, Rel, Jiang and Wang measures using max technique.
| ML Models | Average Accuracy from |
||||
|---|---|---|---|---|---|
| Resnik | Rel | Lin | Jiang | Wang | |
| xgbLinear | 64.29% | 64.29% | 53.57% | 46.43% | 82.14% |
| NB | 64.29% | 60.71% | 57.14% | 57.14% | 78.57% |
| RRF | 64.29% | 53.57% | 53.57% | 53.57% | 82.14% |
| rfRules | 57.14% | 57.14% | 53.57% | 53.57% | 71.43% |
| rFerns | 60.71% | 60.71% | 64.29% | 64.29% | 75% |
| C5 | 53.57% | 64.29% | 71.43% | 75% | 75% |
| MLP | 60.71% | 64.29% | 60.71% | 57.14% | 75% |
Table 4.
Average accuracy of xgbLinear, NB, RRF, rfRules, rFerns, C5 and MLP models on identifying Corona disease on Resnik, Lin, Rel, Jiang and Wang measures using BMA technique.
| ML Models | Average Accuracy from |
||||
|---|---|---|---|---|---|
| Resnik | Rel | Lin | Jiang | Wang | |
| xgbLinear | 71.43% | 57.14% | 64.29% | 64.29% | 64.29% |
| NB | 64.29% | 53.57% | 53.57% | 64.29% | 75% |
| RRF | 67.86% | 57.14% | 53.57% | 57.14% | 67.86% |
| rfRules | 60.71% | 53.57% | 57.14% | 67.86% | 71.43% |
| rFerns | 50% | 53.57% | 53.57% | 64.29% | 71.43% |
| C5 | 71.43% | 60.71% | 67.86% | 67.86% | 71.43% |
| MLP | 78.57% | 89.29% | 71.43% | 75% | 75% |
Table 3, Table 4 show that among the xgbLinear, NB, RRF, rfRules, rFerns, C5 and MLP models with Resnik, Lin, Rel, Jiang and Wang measures and max and BMA technique, MLP with Rel provides the best classification results of 89.29% for the BMA technique. In comparison, the other models performed poorly on the Rel measure. Moreover, all of the models perform well on the Wang measure for max technique. Hence, the Wang measure with max combining technique was selected for further processing. Fig. 7 represents the confusion matrix for all of the seven ML models for Wang measure with max technique.
Fig. 7.
Confusion matrix of (a) xgbLinear (b) NB (c) RRF (d) rfRules (e) rFerns (f) C5 and (g) MLP models on the Wang measure with max combining technique.
Some other ML models, i.e., random forest, cforest, gamboost, bstsm, bstTree, xgbTree, SVM, C5.0Cost were also applied to the gene functional similarity measures. But these models were ignored because of their lower performances during ensemble. To achieve more accurate performance from the ML models, the models (xgbLinear, NB, RRF, rfRules, rFerns, C5 and MLP) are ensembled using the stacking ensemble technique. MLP acts as a top layer model on the stack and predicts based on all the other base models’ responses. The stacking ensemble method improved the classification accuracy from 82.14% to 92.86%.
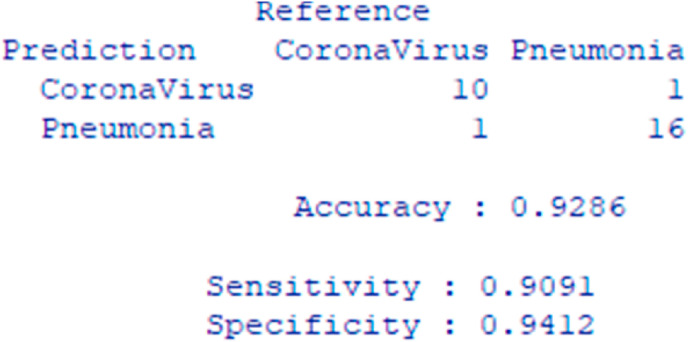
Fig. 8 shows the confusion matrix of the ensembled model with sensitivity and specificity score. The model achieves 90.91% sensitivity and 94.12% specificity.
Fig. 8.
Confusion matrix of the stacked ensembled model.
Authors of [11] obtained 80% AUC values on the Gene-based screening method to identify ASD disease candidate genes. As this technique was not yet applied by other researchers for the Corona detection task, the proposed model could be an ideal supporting model for Corona Disease and Pneumonia Detection with approximately 93% classification accuracy.
4.4. Performances of CNN model
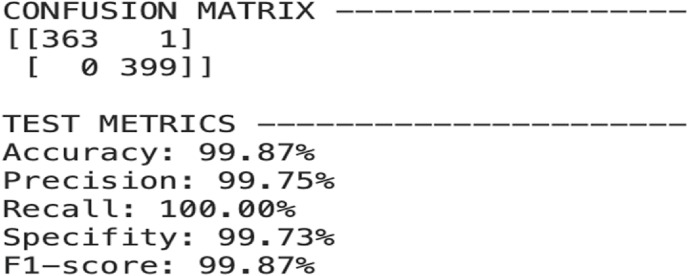
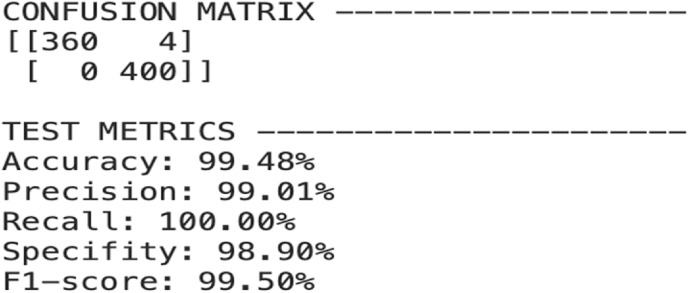
All tasks for the CNN-based CAD method, including training and testing were completed in python on a mac operating system with Google colab gpu and keras framework (using TensorFlow backend). The augmented training chest X-ray image dataset with a fixed image input size of 224 × 224 is fed to the pre-trained CheXNet model for training. The model is trained several times for 40 epochs and then tested on the test dataset. For the COVID-19 vs Normal image classification, the model achieves test accuracy of 99.87% and for the COVID-19 vs Pneumonia classification, the obtained accuracy is 99.48%. The following Fig. 9 and Fig. 10 below represent the confusion matrix with performance parameters- Accuracy, Precision, Recall, Sensitivity and F1-score for both of the classification tasks.
Fig. 9.
Confusion matrix with performance parameters for COVID-19 vs Normal classification.
Fig. 10.
Confusion matrix with performance parameters for COVID-19 vs Pneumonia classification.
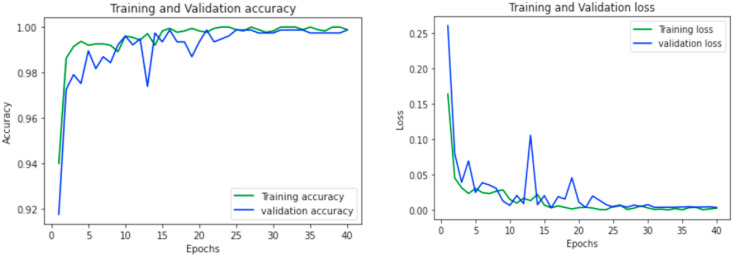
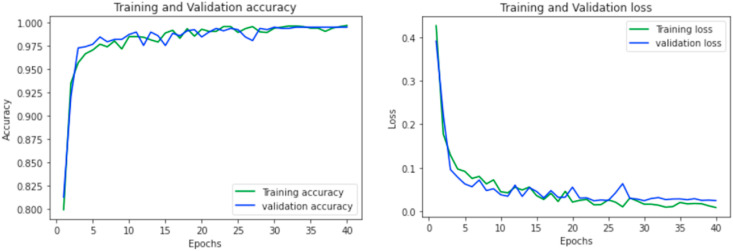
Learning curves are used to demonstrate the model performances during training over each epoch. The accuracy and loss learning curves for both of the proposed classifiers (COVID-19 vs Normal and COVID-19 vs Pneumonia) during different epochs are reported in Fig. 11 and Fig. 12 .
Fig. 11.
Accuracy and loss curve for COVID-19 vs Normal classification model.
Fig. 12.
Accuracy and loss curve for COVID-19 vs Pneumonia classification model.
4.5. Comparison with different models
This section is designed to compare the proposed CNN-based COVID-19 detection model with the existing models. The transfer learned, fine-tuned CheXNet model is used in this research as it shows better performances in classifying both COVID-19 vs Normal and COVID-19 vs Pneumonia images.
Six different pre-trained models are trained, validated and tested on the same COVID-19 Radiography Dataset [29] on the same mac operating system using Google colab gpu with keras framework (using TensorFlow backend) in python. The models VGG-16, VGG-19, Resnet50, EfficientNet, MobileNetV2 models are pre-trained on ImageNet dataset and the CheXNet model pre-trained on CXR dataset [41] are used here for comparison. Table 5, Table 6 depict the experimental results of these six models with the proposed CheXNet (pre-trained on pneumonia dataset) model for COVID-19 vs Normal image classification and COVID-19 vs Pneumonia image classification respectively.
Table 5.
Experimental results of different models on the same dataset for COVID-19 vs Normal classification.
| CNN Models | Accuracy | Precision | Recall | Specificity | F1-score | Confusion Matrix |
|---|---|---|---|---|---|---|
| CheXNet | 99.21% | 98.52% | 100% | 98.35% | 99.25% | 358 6 |
| 0 399 | ||||||
| Resnet50 | 99.34% | 98.76% | 100% | 98.63% | 99.38% | 359 5 |
| 0 399 | ||||||
| VGG-19 | 99.61% | 99.25% | 100% | 99.18% | 99.63% | 361 3 |
| 0 399 | ||||||
| MobileNetV2 | 99.61% | 99.25% | 100% | 99.17% | 99.62% | 361 3 |
| 0 399 | ||||||
| VGG-16 | 99.74% | 99.75% | 99.74% | 99.73% | 99.74% | 363 1 |
| 1 398 | ||||||
| EfficientNet | 99.74% | 99.50% | 100% | 99.45% | 99.75% | 362 2 |
| 0 399 | ||||||
| Proposed Model | 99.87% | 99.75% | 100% | 99.73% | 99.87% | 363 1 |
| 0 399 |
Table 6.
Experimental results of different models on the same dataset for COVID-19 vs Pneumonia classification.
| CNN Models | Accuracy | Precision | Recall | Specificity | F1-score | Confusion Matrix |
|---|---|---|---|---|---|---|
| CheXNet | 99.08% | 98.76% | 99.50% | 98.63% | 99.13% | 359 5 |
| 2 398 | ||||||
| Resnet50 | 98.43% | 97.32% | 99.75% | 96.98% | 98.52% | 353 11 |
| 1 399 | ||||||
| VGG-19 | 99.35% | 99.50% | 99.25% | 99.45% | 99.37% | 362 2 |
| 3 397 | ||||||
| MobileNetV2 | 99.08% | 98.28% | 100% | 98.08% | 99.13% | 357 7 |
| 0 400 | ||||||
| VGG-16 | 99.35% | 99.26% | 99.50% | 99.17% | 99.38% | 359 3 |
| 2 400 | ||||||
| EfficientNet | 99.35% | 98.77% | 100% | 98.63% | 99.38% | 359 5 |
| 0 400 | ||||||
| Proposed Model | 99.48% | 99.01% | 100% | 98.90% | 99.50% | 363 1 |
| 0 399 |
From Table 5, Table 6, it is clear that the proposed model provides better performances than the existing models on the same dataset of Tawsifur [29]. Tawsifur et al. also proposed a model classifying Normal and COVID-19 images having an accuracy of 99.7% [44]. Diagnosis of COVID-19 can be either from CT scan images or from chest X-ray images. Comparison results of the proposed model with other binary and multi-class classification models on the different datasets also prove that the proposed model outperforms other state-of-the-art models for the diagnosis of COVID-19. Table 7 summarizes the comparative performances of different models on different datasets to our proposed model performance.
Table 7.
Comparison of different models with proposed model.
| CNN Models | Image Type | Classification Type | Accuracy |
|---|---|---|---|
| Hemdan et al. [15] | X-ray | COVID-19 vs Normal | 90% |
| Zheng et al. [16] | CT | COVID-19 vs Normal | 90.1% |
| Ying et al. [20] | CT | COVID-19 vs Normal | 94% |
| Sarhan A.M et al. [17] | X-ray | COVID-19 vs Normal | 94.5% |
| Narin et al. [7] | X-ray | COVID-19 vs Normal | 98% |
| Ozturk et al. [18] | X-ray | COVID-19 vs Normal | 98.08% |
| Tawsifur R [44]. | X-ray | COVID-19 vs Normal | 99.7% |
| Wang et al. [19] | CT | COVID-19 vs Pneumonia | 82.9% |
| Ying et al. [20] | CT | COVID-19 vs Pneumonia | 86% |
| Sethy and Behera [21] | X-ray | COVID-19 vs Pneumonia | 95.38% |
| Xu et al. [22] | CT | COVID-19 vs IAVP vs Normal | 86.7% |
| Mangal et al. [23] | X-ray | COVID-19 vs Normal vs Pneumonia | 90.5% |
| Wang and Wong [5] | X-ray | COVID-19 vs Non-COVID-19 vs Normal | 93.3% |
| Asif et al. [24] | X-ray | COVID-19 vs Normal vs Pneumonia | 96% |
| Kumar R [25]. | X-ray | COVID-19 vs Normal vs Pneumonia | 97.7% |
| Tawsifur R [44]. | X-ray | COVID-19 vs Normal vs Pneumonia | 97.9% |
| Proposed Model | X-ray | COVID-19 vs Pneumonia | 99.48% |
| COVID-19 vs Normal | 99.87% |
The above comparison stated that the proposed binary COVID-19 diagnosis model performs superior to the compared binary and multi-class model. Thus, it could become a great supporting tool for fighting the COVID-19 pandemic.
5. Conclusion
As Pneumonia is a major symptom of COVID-19, it is very difficult to differentiate COVID-19 or Corona diseases from Pneumonia. In this study, two cost-effective, rapid, and automatic Corona disease diagnostic methods were demonstrated. Gene Ontology (GO) is the most frequently used term by researchers to calculate gene functional similarity. Genes with higher functional similarity may belong to the same hierarchical path of GO with higher semantic terms. The identification of disease from associated genes through GO-based gene similarity measures can open a new era in complex disease diagnosis. ML classifiers with a large gene dataset may help to obtain improved accuracy. In the gene-based detection method, ML classifiers are applied in identifying and predicting the Corona disease from gene functional similarities calculated using different semantic similarity measures. Stacking ensembles of different machine learning models improve performance accuracy. Chest X-ray imagery is readily available, and the cost-effective images conveys potential information to assist radiologists in diagnosis disease. The proposed CNN-based CAD method provides a simple model that demonstrates superior results in diagnosing COVID-19 from X-ray imagery. In the future, the authors will try to overcome the data shortage limitation and optimize the model to classify more diseases with an effective result.
Funding
This research did not receive any specific grants from funding agencies in the public, commercial, or not-for-profit sectors.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
The authors are grateful to the participants who contributed to this research. No funding to declare.
References
- 1.Shereen M.A., Khan S., Kazmi A., Bashir N., Siddique R. COVID-19 infection: origin, transmission, and characteristics of human coronaviruses. J Adv Res. 2020;98–91 doi: 10.1016/j.jare.2020.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.World Health Organization (WHO) https://www.who.int/emergencies/diseases/novel-coronavirus-2019
- 3.Worldometers.info https://www.worldometers.info/coronavirus/
- 4.Farooq M., Hafeez A. 2020. COVID-ResNet: a deep learning framework for screening of COVID19 from radiographs. arXiv:2003.14395. [Google Scholar]
- 5.Wang L., Wong A. 2020. COVID-net: a tailored deep convolutional neural network design for detection of COVID-19 cases from chest radiography images. arXiv preprint arXiv:2003.09871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang J., Xie Y., Li Y., Shen C., Xia Y. 2020. COVID-19 screening on chest X-ray images using deep learning based anomaly detection. arXiv. [Google Scholar]
- 7.Narin A., Kaya C., Pamuk Z. 2020. Automatic detection of coronavirus disease (COVID-19) using X-ray images and deep convolutional neural networks. arXiv preprint arXiv:2003.10849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hall W.D., Morley K.I., Lucke J.C. The prediction of disease risk in genomic medicine. EMBO Rep. 2004;S26-S22 doi: 10.1038/sj.embor.7400224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cáceres J.J., Paccanaro A. Disease gene prediction for molecularly uncharacterized diseases. PLoS Comput Biol. 2019;15:7. doi: 10.1371/journal.pcbi.1007078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Habib N., Hasan M.M., Rahman M.M. Fusion of deep convolutional neural network with PCA and logistic regression for diagnosis of pediatric pneumonia on chest X-rays. Network Biol. 2020;76–62 [Google Scholar]
- 11.Asif M., Martiniano H.F.M.C.M., Vicente A.M., Couto F.M. Identifying disease genes using machine learning and gene functional similarities, assessed through Gene Ontology. PloS One. 2018;13:12. doi: 10.1371/journal.pone.0208626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Apostolopoulos I.D., Bessiana T. 2020. COVID-19: automatic detection from X-ray images utilizing transfer learning with convolutional neural networks. arXiv:2003.11617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Afshara P., Heidarianb S., Naderkhania F., Oikonomouc A., Plataniotis K.N. 2020. COVID-CAPS: a capsule network-based framework for identification of COVID-19 cases from X-ray images. arXiv:2004.02696vol. 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li L., Qin L., Xu Z., Yin Y., Wang X., Kong B., Bai J., Lu Y., Fang Z., Song Q., Cao K., Liu D., Wang G., Xu Q., Fang X., Zhang S., Xia J., Xia J. Using artificial intelligence to detect COVID-19 and community-acquired pneumonia based on pulmonary CT: evaluation of the diagnostic accuracy. Radiology. 2020 doi: 10.1148/radiol.2020200905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hemdan E.E.D., Shouman M.A., Karar M.E. 2020. COVIDX-net: a framework of deep learning classifiers to diagnose COVID-19 in X-ray images. arXiv preprint arXiv:2003.11055. [Google Scholar]
- 16.Zheng C., Deng X., Fu Q., Zhou Q., Feng J., Ma H., Liu W., Wang X. Deep learning-based detection for COVID-19 from chest CT using weak label. medRxiv. 2020 doi: 10.1101/2020.03.12.20027185. [DOI] [Google Scholar]
- 17.Sarhan A.M. Detection of COVID-19 cases in chest X-ray images using wavelets and support vector machines. Biomed Eng. 2020 doi: 10.21203/rs.3.rs-37558/v1. [DOI] [Google Scholar]
- 18.Ozturk T., Talo M., Yildirim E.A., Baloglu U.B., Yildirim O., Acharya U.R. Automated detection of COVID-19 cases using deep neural networks with X-ray images. Comput Biol Med. 2020;121 doi: 10.1016/j.compbiomed.2020.103792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang S., Kang B., Ma J., Zeng X., Xiao M., Guo J., Cai M., Yang J., Li Y., Meng X., Xu B. medRxiv; 2020. A deep learning algorithm using CT images to screen for Corona Virus Disease (COVID-19) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Song Y., Zheng S., Li L., Zhang X., Zhang X., Huang Z., Chen J., Zhao H., Jie Y., Wang R., Chong Y., Shen J., Zha Y., Yang Y. Deep learning enables accurate diagnosis of novel coronavirus (COVID-19) with CT images. medRxiv. 2020 doi: 10.1101/2020.02.23.20026930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sethy P.K., Behera S.K. Detection of coronavirus disease (COVID-19) based on deep features. Preprints. 2020 doi: 10.20944/preprints202003.0300.v1. [DOI] [Google Scholar]
- 22.Xu X., Jiang X., Ma C., Du P., Li X., Lv S., Yu L., Chen Y., Su J., Lang G., Li Y., Zhao H., Xu K., Ruan L., Wu W. 2020. Deep learning system to screen coronavirus disease 2019 pneumonia. arXiv preprint arXiv:200209334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mangal A., Kalia S., Rajgopal H., Rangarajan K., Namboodiri V., Banerjee S., Arora C. 2020. CovidAID: COVID-19 detection using chest X-ray.http://arxiv.org/abs/2004.09803 arXiv:2004.09803. Apr. [Google Scholar]
- 24.Asif S., Wenhui Y., Jin H., Tao Y., Jinhai S. Classification of COVID-19 from chest X-ray images using deep convolutional neural networks. medRxiv. 2020 [Google Scholar]
- 25.Kumar R., Arora R., Bansal V., Sahayasheela V.J., Buckchash H., Imran J. Accurate prediction of COVID-19 using chest X-ray images through deep feature learning model with SMOTE and machine learning classifiers. medRxiv. 2020 doi: 10.1101/2020.04.13.20063461. [DOI] [Google Scholar]
- 26.Kermany D.S. Identifying medical diagnoses and treatable diseases by image-based deep learning. Cell. 2018 doi: 10.1016/j.cell.2018.02.010. 1131–1122. [DOI] [PubMed] [Google Scholar]
- 27.The human genome project. The New York times. https://archive.nytimes.com/www.nytimes.com/library/national/science/genome-index.html
- 28.Research Institute Posts Gene Data on Internet The New York times june 26. 1997. https://archive.nytimes.com/www.nytimes.com/library/cyber/week/062697gene.html
- 29.Tawsifur R., Amith K Dr.M.C. COVID-19 radiography dataset. 2021. https://www.kaggle.com/tawsifurrahman/covid19-radiography-database
- 30.Cohen J.P., Morrison P., Dao L. 2020. COVID-19 image data collection. arXiv:2003.11597. [Google Scholar]
- 31.Paul M. 2018. Chest X-ray images (pneumonia)https://www.kaggle.com/paultimothymooney/chest-xray-pneumonia [Google Scholar]
- 32.Wei Q., Khan I.K., Ding J., Yerneni S., Kihara D. NaviGO: interactive tool for visualization and functional similarity and coherence analysis with gene ontology. BMC Bioinf. 2017;18:177. doi: 10.1186/s12859-017-1600-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Philip, Resnik Semantic similarity in a taxonomy: an information-based measure and its application to problems of ambiguity in natural language. J Artif Intell Res. 1999:130–195. [Google Scholar]
- 34.Jiang Jay J., Conrath D.W. Proceedings of 10th international conference on research in computational linguistics. 1997. Semantic similarity based on corpus statistics and lexical taxonomy.http://www.citebase.org/abstract?id=oai:arXiv.org:cmp-lg/9709008 [Google Scholar]
- 35.Lin, Dekang . Proceedings of the 15th international conference on machine learning. 1998. An information-theoretic definition of similarity. 304–296. [DOI] [Google Scholar]
- 36.Schlicker Andreas, Domingues F.S., Rahnenführer J., Lengauer T. A new measure for functional similarity of gene products based on gene ontology. BMC Bioinf. 2006;7:302 doi: 10.1186/1471-2105-7-302. doi.org/1471-2105-7-302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang James Z., Du Z., Payattakool R., Yu P.S., Chen C.F. A new method to measure the semantic similarity of go terms. Bioinformatics. 2007:1274–1281. doi: 10.1093/bioinformatics/btm087. [DOI] [PubMed] [Google Scholar]
- 38.Yu G. Southern Medical University; 2020. School of basic medical sciences.https://bioconductor.org/packages/devel/bioc/vignettes/GOSemSim/inst/doc/GOSemSim.html#citation [Google Scholar]
- 39.Deng H., Runger G. Gene selection with guided regularized random forest. Pattern Recogn. 2013;3489–3483 [Google Scholar]
- 40.Rajpurkar P., Irvin J. 2017. Chexnet: radiologist-level pneumonia detection on chest X-rays with deep learning. arXiv preprint arXiv:1711.05225. [Google Scholar]
- 41.Wang X., Peng Y., Lu L., Lu Z., Bagheri M., Summers R.M. Proceedings of the IEEE conference on computer vision and pattern recognition. vols. 2106–2097. 2017. Chestx-ray8: hospital-scale chest x-ray database and benchmarks on weakly-supervised classification and localization of common thorax diseases. [DOI] [Google Scholar]
- 42.Carlson M. 2015. org.Hs.eg.db: genome wide annotation for Human. R Package version 312. [DOI] [Google Scholar]
- 43.Yu G. Gene ontology semantic similarity analysis using GOSemSim. In: Kidder B., editor. Stem cell transcriptional networks. Methods in molecular biology. Humana; New York, NY: 2020. 215-207. [DOI] [PubMed] [Google Scholar]
- 44.Muhammad E.H.C., Tawsifur R., Amith K. Can AI help in screening Viral and COVID-19 pneumonia? IEEE Access. 2020;8:132665–132676. [Google Scholar]
- 45.Habib N., Hasan M.M., Reza M.M. Ensemble of CheXNet and VGG-19 Feature Extractor with Random Forest Classifier for Pediatric Pneumonia Detection. SN COMPUT. SCI. 2020 doi: 10.1007/s42979-020-00373-y. [DOI] [PMC free article] [PubMed] [Google Scholar]