To the Editor
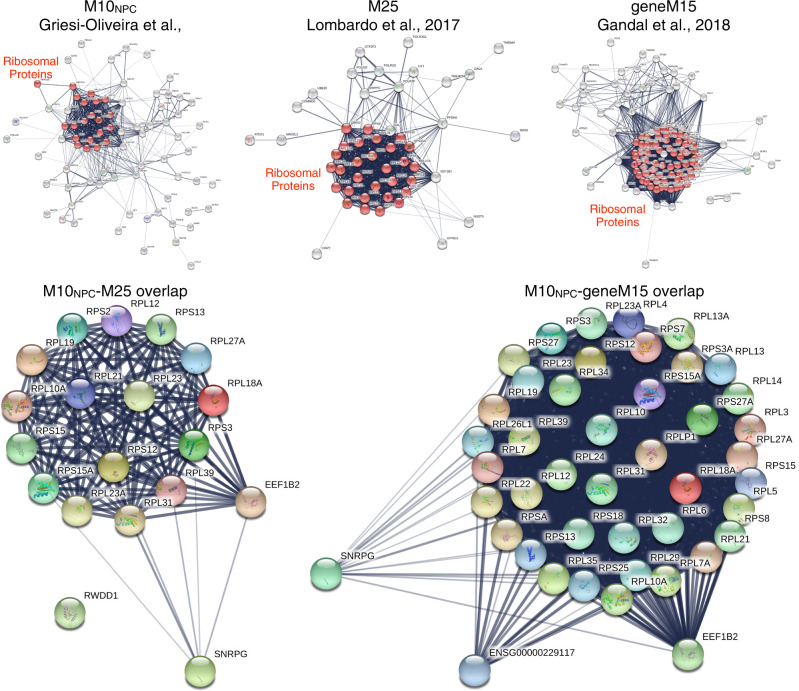
In a recent paper, Griesi-Oliveira et al. [1] identified a gene co-expression module in induced pluripotent stem cell (iPSC)-derived neural progenitor cells (NPCs) that is upregulated in expression in autism. The authors also show proteomic evidence to suggest that these earlier disruptions in translation in NPCs may lead to dysregulation of gene expression relevant to synaptic processes in neurons. These are valuable insights to contribute to the literature, particularly because iPSC models can be a useful model of early prenatal periods of development. Griesi-Oliveira et al. [1] also attempted to assess how similar this kind of transcriptomic dysregulation is to previously reported studies using gene expression data in post-mortem cortical tissue. As the authors rightly note, the papers they utilized for this analysis [2, 3] did not themselves report any co-expression modules with differential module eigengene expression in modules enriched for translation processes. However, in our recent paper [4], we re-analyzed data from these studies [2, 3] and indeed identified two consensus co-expression modules enriched in translation processes (M1 and M25) that were replicably upregulated in autism in both datasets. When we examine whether MNPC10-blue heavily overlaps with these translation-enriched modules, we indeed find high overlap with the translation-enriched M25 co-expression module (odds ratio (OR) = 15.52, p = 1.24e − 14). Evidence from a later study by Gandal et al. [5] also showed the presence of a co-expression module (geneM15) that is localized to the ribosome and enriched in excitatory neuronal cell types. This autism-upregulated geneM15 module from post-mortem cortical tissue highly overlaps with MNPC10-blue (OR = 19.10, p = 3.26e − 34) and the M25 module from our study (OR = 140.27, p = 1.09e − 54). Nearly all of the overlapping genes in MNPC10-blue and Lombardo et al.’s [4] M25 and Gandal et al.’s [5] geneM15 are ribosomal protein genes (e.g., RPL and RPS). See Fig. 1 for a graphical depiction of these gene sets and the overlap. The data and code that reproduces these results can be found here: https://github.com/mvlombardo/ipsc_translation. Thus, rather than there being no prior evidence of upregulated translation co-expression modules in post-mortem cortical tissue, there is such evidence from multiple studies [4, 5], as well as evidence to support that many of the same genes in MNPC10-blue are also those found within those translation-enriched co-expression modules from post-mortem cortical tissue.
Fig. 1. Graphs showing protein–protein interactions from STRING (https://string-db.org) for genes within co-expression modules MNPC10-blue from Griesi-Oliveira et al. [1] (top left), M25 from Lombardo et al. [4] (top middle), and geneM15 from Gandal et al. [5] (top right).
Ribosomal proteins are highlighted in red. In the bottom left are genes that overlap between MNPC10-blue and M25, while on the bottom right are genes that overlap between MNPC10-blue and geneM15.
A second point of interest is that our prior paper makes a statement about how the cortical transcriptome is hierarchically disorganized in autism [4]. Evidence to support this comes from the result that eigengenes from upregulated and downregulated co-expression modules are highly correlated, with increases in the expression of translation modules being associated with decreased expression of synaptic-enriched modules. In addition, dysregulated co-expression modules show a high degree of physical interactions at the protein level—that is, translation-enriched modules like M25 show a high level of protein–protein interactions with proteins from genes from synaptic-enriched co-expression modules. Thus, the statement that upregulated MNPC10-blue might be linked to dysregulation of neuronal co-expression modules in this iPSC dataset is indeed foreshadowed and predicted by our observations of this same kind of emergent disorganization across co-expression modules observed in post-mortem cortical tissue.
Finally, it is worth discussing the potential importance of these upregulated co-expression modules enriched in translation processes with respect to evidence already in the literature. The authors have cited that protein synthesis is a cellular process of interest in autism. Indeed, translational control has been of broad long-standing interest in autism research as many of the earliest known single gene mutations with high penetrance for autism are those that affect translational control (e.g., FMR1, TSC2, PTEN) [6]. These genes, along with others (e.g., EIF4E, UBE3A, NF1, MECP2, SHANK3), all affect AKT-PI3K-mTOR signaling pathways [7–11] and these pathways are known to be affected in idiopathic cases of autism [12, 13]. While these are important prior starting points for talking about protein synthesis in autism, there is a missed opportunity here to expand beyond this evidence. For example, the MNPC10-blue co-expression module has a large number of genes coding for 40S and 60S ribosomal subunits (RPL and RPS genes) and these genes are key members of our previously reported M25-upregulated translation-enriched co-expression module [4] as well as the autism-upregulated geneM15 module from Gandal et al. [5]. These genes as well as the ribosome cellular component itself are largely not focused on in the literature. The evidence from our paper [4] and others [14], as well as this new evidence from Griesi-Oliveira et al. [1] should underscore the idea that future work should focus more heavily on this type of translation-relevant biology. Furthermore, such upregulation of translation-relevant biology (e.g., genes for ribosomal proteins) are known to be similarly upregulated in maternal immune activation [15] and in non-neural cell types such as blood leukocytes from patients with autism [16, 17]. Finally, similar co-expression modules identified in blood leukocytes are associated with functional neural phenotypes relevant to receptive speech processing in autism with poor language outcome [18].
These insights raise questions for how future work might expand on the relevance of upregulated ribosomal protein genes in brain development in autism? Here I suggest a couple of routes that could be examined. First, the upregulated expression of ribosomal protein genes could suggest that ribosome biogenesis is enhanced, and thus protein synthesis is also enhanced. Therefore, examination of ribosome biogenesis could be an important avenue of future work in autism, particularly if enhanced ribosome biogenesis affects specific cell types (e.g., excitatory neurons, radial glia). Second, enhanced protein synthesis—be it through enhanced ribosome biogenesis or other known autism-associated mechanisms (e.g., FMR1, TSC2, PTEN, mTOR signaling)—could lead to enhanced cell proliferation. It is also known that cell proliferation and protein synthesis are tightly linked [19], and in cancer research, many mechanisms relevant to suppressing or enhancing cell growth and proliferation affect ribosome biogenesis and protein synthesis [20]. This route may be relevant for examining molecular mechanisms that could underlie accelerated early brain overgrowth in autism [11, 21] and/or subtypes of patients with much larger brains [22]. Given the insights from Griesi-Oliveira et al. [1] for showing this process is already upregulated in NPCs, this avenue could be interesting from the standpoint that specific types of progenitor cells (e.g., ventricular or outer radial glia) in very early fetal brain development are implicated in the expansion of cortical surface area [23, 24]. Third, ribosomal proteins can have non-ribosomal functions, such as involvement in the immune system [20]. These functions can emerge because steps in ribosome biogenesis can be disturbed (i.e. ribosomal stress) and lead to an accumulation of ribosome-free ribosomal proteins in the cell. Whether these extra-ribosome functions are a factor in autism is unknown, but this could be another route for how enhanced ribosomal proteins could potentially affect autistic patients.
In conclusion, the discoveries of Griesi-Oliveria et al. [1] are important and highlight the idea that upregulated translation processes and their interaction with other important biological processes taking place at the synapse be prioritized as a fruitful area for future discoveries. While translational control is itself an area of much interest in autism [6–10], there is likely more to this topic than currently understood and more emphasis could also be placed on the importance of ribosomal proteins.
Compliance with ethical standards
Conflict of interest
The author declares no conflict of interest.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Griesi-Oliveira K, Fogo MS, Pinto BGG, Alves AY, Suzuki AM, Morales AG, et al. Transcriptome of iPSC-derived neuronal cells reveals a module of co-expressed genes consistently associated with autism spectrum disorder. Mol Psychiatry. 2020. 10.1038/s41380-020-0669-9. [DOI] [PMC free article] [PubMed]
- 2.Voineagu I, Wang X, Johnston P, Lowe JK, Tian Y, Horvath S, et al. Transcriptomic analysis of autistic brain reveals convergent molecular pathology. Nature. 2011;474:380–4. doi: 10.1038/nature10110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gupta S, Ellis SE, Ashar FN, Moes A, Bader JS, Zhan J, et al. Transcriptome analysis reveals dysregulation of innate immune response genes and neuronal activity-dependent genes in autism. Nat Commun. 2014;5:1–8. doi: 10.1038/ncomms6748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lombardo MV, Courchesne E, Lewis NE, Pramparo T. Hierarchical cortical transcriptome disorganization in autism. Mol Autism. 2017;8:29. doi: 10.1186/s13229-017-0147-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gandal MJ, Zhang P, Hadjimichael E, Walker RL, Chen C, Liu S, et al. Transcriptome-wide isoform-level dysregulation in ASD, schizophrenia, and bipolar disorder. Science. 2018;362:eaat8127. doi: 10.1126/science.aat8127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kelleher RJ, Bear MF. The autistic neuron: troubled translation? Cell. 2008;135:401–6. doi: 10.1016/j.cell.2008.10.017. [DOI] [PubMed] [Google Scholar]
- 7.Santini E, Huynh TN, MacAskill AF, Carter AG, Pierre P, Ruggero D, et al. Exaggerated translation causes synaptic and behavioural aberrations associated with autism. Nature. 2013;493:411–5. doi: 10.1038/nature11782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gkogkas CG, Khoutorsky A, Ran I, Rampakakis E, Nevarko T, Weatherill DB, et al. Autism-related deficits via dysregulated eIF4E-dependent translational control. Nature. 2013;493:371–7. doi: 10.1038/nature11628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Santini E, Klann E. Reciprocal signaling between translational control pathways and synaptic proteins in autism spectrum disorders. Sci Signal. 2014;7:re10. doi: 10.1126/scisignal.2005832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Winden KD, Ebrahimi-Fakhari D, Sahin M. Abnormal mTOR activation in autism. Annu Rev Neurosci. 2018;41:1–23. doi: 10.1146/annurev-neuro-080317-061747. [DOI] [PubMed] [Google Scholar]
- 11.Courchesne E, Pramparo T, Gazestani VH, Lombardo MV, Pierce K, Lewis NE. The ASD living biology: from cell proliferation to clinical phenotype. Mol Psychiatry. 2019;24:88–107. doi: 10.1038/s41380-018-0056-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tang G, Gudsnuk K, Kuo S-H, Cotrina ML, Rosoklija G, Sosunov A, et al. Loss of mTOR-Dependent macroautophagy causes autistic-like synaptic pruning deficits. Neuron. 2014;83:1131–43. doi: 10.1016/j.neuron.2014.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gazestani VH, Pramparo T, Nalabolu S, Kellman BP, Murray S, Lopez L, et al. A perturbed gene network containing PI3K–AKT, RAS–ERK and WNT–β-catenin pathways in leukocytes is linked to ASD genetics and symptom severity. Nat Neurosci. 2019;22:1624–34. doi: 10.1038/s41593-019-0489-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hetman M, Slomnicki LP. Ribosomal biogenesis as an emerging target of neurodevelopmental pathologies. J Neurochem. 2019;148:325–47. doi: 10.1111/jnc.14576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lombardo MV, Moon HM, Su J, Palmer TD, Courchesne E, Pramparo T. Maternal immune activation dysregulation of the fetal brain transcriptome and relevance to the pathophysiology of autism spectrum disorder. Mol Psychiatry. 2018;23:1001–13. doi: 10.1038/mp.2017.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pramparo T, Pierce K, Lombardo MV, Barnes CC, Marinero S, Ahrens-Barbeau C, et al. Prediction of autism by translation and immune/inflammation coexpressed genes in toddlers from pediatric community practices. JAMA Psychiatry. 2015;72:386–94. doi: 10.1001/jamapsychiatry.2014.3008. [DOI] [PubMed] [Google Scholar]
- 17.Tylee DS, Hess JL, Quinn TP, Barve R, Huang H, Zhang-James Y, et al. Blood transcriptomic comparison of individuals with and without autism spectrum disorder: a combined-samples mega-analysis. Am J Med Genet B. 2017;174:181–201. doi: 10.1002/ajmg.b.32511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lombardo MV, Pramparo T, Gazestani V, Warrier V, Bethlehem RAI, Carter Barnes C, et al. Large-scale associations between the leukocyte transcriptome and BOLD responses to speech differ in autism early language outcome subtypes. Nat Neurosci. 2018;21:1680–8. doi: 10.1038/s41593-018-0281-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ruggero D, Pandolfi PP. Does the ribosome translate cancer? Nat Rev Cancer. 2003;3:179–92. doi: 10.1038/nrc1015. [DOI] [PubMed] [Google Scholar]
- 20.Zhou X, Liao W-J, Liao J-M, Liao P, Lu H. Ribosomal proteins: functions beyond the ribosome. J Mol Cell Biol. 2015;7:92–104. doi: 10.1093/jmcb/mjv014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hazlett HC, Gu H, Munsell BC, Kim SH, Styner M, Wolff JJ, et al. Early brain development in infants at high risk for autism spectrum disorder. Nature. 2017;542:348–51. doi: 10.1038/nature21369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Amaral DG, Li D, Libero L, Solomon M, Van de Water J, Mastergeorge A, et al. In pursuit of neurophenotypes: the consequences of having autism and a big brain: neurophenotypes of autism. Autism Res. 2017;10:711–22. doi: 10.1002/aur.1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rakic P. Specification of cerebral cortical areas. Science. 1988;241:170–6. doi: 10.1126/science.3291116. [DOI] [PubMed] [Google Scholar]
- 24.Cadwell CR, Bhaduri A, Mostajo-Radji MA, Keefe MG, Nowakowski TJ. Development and arealization of the cerebral cortex. Neuron. 2019;103:980–1004. doi: 10.1016/j.neuron.2019.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]