Abstract
Mendelian and early-onset severe psychiatric phenotypes often involve genetic variants having a large effect, offering opportunities for genetic discoveries and early therapeutic interventions. Here, the index case is an 18-year-old boy, who at 14 years of age had a decline in cognitive functioning over the course of a year and subsequently presented with catatonia, auditory and visual hallucinations, paranoia, aggression, mood dysregulation, and disorganized thoughts. Exome sequencing revealed a stop-gain mutation in RCL1 (NM_005772.4:c.370 C > T, p.Gln124Ter), encoding an RNA 3′-terminal phosphate cyclase-like protein that is highly conserved across eukaryotic species. Subsequent investigations across two academic medical centers identified eleven additional cases of RCL1 copy number variations (CNVs) with varying neurodevelopmental or psychiatric phenotypes. These findings suggest that dosage variation of RCL1 contributes to a range of neurological and clinical phenotypes.
Subject terms: Genetics, Psychiatric disorders
Introduction
Large-scale studies of common variants provide insight into the genetic architecture of psychiatric disorders such as schizophrenia and bipolar disorder [1, 2]. But even though these studies have involved hundreds of thousands of participants, they have typically explained only a small fraction of the genetic contribution to these focal illnesses, and have identified scores of loci that, with follow up, have pointed to putative risk genes [3–5]. Exome sequencing (ES) of unrelated cases with idiopathic schizophrenia (and controls) have identified gene sets involved in illness risk, but have not, with the possible exception of SETD1A and UNC13B, clearly identified specific risk genes [6–8]. And while common variant and ES studies with even larger samples will likely provide important insights into the genetic nature of psychiatric disorders, the advantages of other experimental designs should also be considered. For example, analyses of rare Mendelian forms of common diseases (e.g., extreme phenotypes) is an effective strategy to discover genes that influence idiopathic forms of illness [9, 10]. Among these, some may bear mutations that have a profound effect on the phenotype (‘large effect’ genes). Similarly, ‘genetics first’ approaches that focus on rare genetic disorders or recurrent copy number variants can provide candidate gene sets [9, 11, 12]. However, these approaches often provide several candidate genes within CNVs, and determining the most relevant gene for follow-up can be a daunting task [13, 14].
The increasing use of ES and genome sequencing (GS) in medical genetics clinics has spurred a dramatic increase in the number of individual cases reported in which highly penetrant genetic mutations are now associated with specific phenotypes [9, 14]. Collecting such data from sites around the world is now becoming more feasible and is enabling the systematic study of specific rare genotype-phenotype associations followed by subsequent biological experimentation.
However, there are a number of methodological issues that must be considered when analyzing rare genetic mutations in genes associated with severe Mendelian phenotypes.
Here, we utilize a clinical pipeline for identifying causal genes in Mendelian phenotypes, identifying a premature stop-gain in RCL1 in a patient with very early onset psychosis (VEOP). While RCL1 has not yet been reported in the context of psychotic symptoms, a rare missense variant in RCL1 has been linked to an increased risk of depression [15]. Querying of two large academic medical centers for RCL1 associated copy number variation (CNV) cases further identified 13 additional RCL1 deletion and duplication patients with varying neurological and psychiatric phenotypes. We then performed human brain immunohistochemistry and transcriptome expression analyses across tissue types to demonstrate a cell type basis for RCL1 expression in the brain. We conclude that dosage variation in RCL1 may confer large effects on the brain and behavior.
Methods (short methods)
We utilized a strategy for discovering genes of large effect in Mendelian forms of severe mental illness (see Fig. 1) that begins with next-generation sequencing and identification of rare variants or copy number changes across the genome, with an emphasis on known genes with neuropsychiatric involvement according to the published literature and internal databases, using well-defined filters and protocols [16, 17]. The biological relevance of the candidate variants in affected intervals was queried with help of the Allen Brain Atlas (https://portal.brain-map.org/) and other publicly available bioinformatics databases such as ClinGen (https://clinicalgenome.org/) and String (http://string-db.com/); model organism databases such as zfin (https://zfin.org/) were also used to obtain evidence for or against the variant/gene/interval’s hypothesized involvement in the phenotype. We also queried whether any family-based studies show segregation of genetic variants with phenotypes of interest and whether the gene or locus is involved in related phenotypes. GWAS data were examined to determine if the gene is a candidate for this phenotype or related phenotypes—because this process can be labor-intensive, we used aggregation tools such as https://www.ebi.ac.uk/gwas/home. For example, the overlap between psychiatric and neurodevelopmental phenotypes such as autism spectrum disorder (ASD), psychosis, and schizophrenia requires a broad literature search for a comprehensive investigation of whether the gene or locus might be involved in one of these neurological disorders. Finally, if there was evidence of possible involvement, we identified additional patients to support the relationship between gene/interval involvement by querying large academic medical centers that have databases of next-generation sequencing data and phenotypes or by using the Matchmaker exchange platform (https://www.matchmakerexchange.org/) [18].
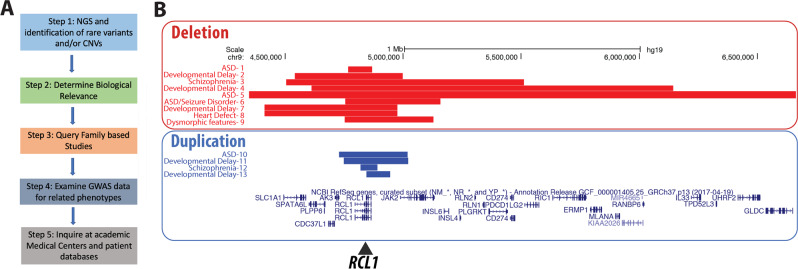
Fig. 1. Framework and additional cases.
a Study framework to verify candidate mutations resulting in severe Mendelian RCL1 phenotypes. b RCL1 copy number deletions (red) and duplications (blue) identified in academic medical centers in this study. Phenotypes consist of a range of neuropsychiatric features, individuals numbered 1–13. Note that individuals 8 and 9 were too young to display psychiatric symptoms. The genomic alignment panel includes genes surrounding RCL1 on chromosome 9 (9p24.1), which consists of 11 exons encompassing 68 kb, and additional RCL1 isoforms (color figure online).
Human subjects and samples
Research performed at Boston Children’s Hospital on human samples was conducted according to protocols approved by the institutional review board of Boston Children’s Hospital. Subjects were evaluated and identified in the Boston Children’s Hospital Developmental Neuropsychiatry clinic, and written as well as verbal consent/assent was obtained under the Gene Discovery Core protocol of the Manton Center for Orphan Disease Research. Patient data has been deposited into DECIPHER (https://decipher.sanger.ac.uk/patient/421796/). De-identified querying of samples at two other large academic medical centers (Baylor College of Medicine/Baylor Genetics and The Hospital for Sick Children, Toronto) was performed as per institutionally approved methods.
Proband family next-generation sequencing and chromosomal microarray analysis (CMA)
The patient underwent phlebotomy for CMA of peripheral blood lymphocytes. Genomic DNA was examined by array-based comparative genomic hybridization (aCGH) using the ClariView Array (Claritas Genomics, Cambridge, MA). The array contains DNA oligonucleotide probes in or flanking most exons of the evaluated genes and is designed to detect most single-exon deletions and duplications. Probe sequences and locations are based on Genome Reference Consortium build 37 (GRCh37)/UCSC hg19. Data analysis was performed with Agilent CytoGenomics 5.0.0.38 software (Cambridge, MA).
The proband and both parents underwent ES at the Yale Center for Mendelian Genomics. Whole-exome libraries were prepared using the KAPA Hyper kit, and the products were enriched with IDT’s xGen Exome Research Panel v1.0 using multiplexed capture of 16 samples. Exomes were sequenced on an Illumina HiSeq 4000 using paired-end chemistry at a read length of 100 bp. FASTQs were aligned by Codified Genomics (proprietary algorithm, Houston, TX). De novo, homozygous, heterozygous, and rare variants were examined if they passed quality criteria adapted from Yuen et al. [16]: (1) read depth in the proband and parents ≥10X; (2) allele frequencies <1% in all population databases; (3) residual variation intolerance score (RVIS) percentile <70 and loss-of-function observed/expected upper bound fraction (LOEUF) <0.6 [19]; and (4) read ratio ≥25% of the alternate allele in the proband of the trio.
Human tissue brain preparation and immunohistochemistry
Control fetal (21 weeks gestation), flash froze neonatal (270 days post-birth), and adult cerebral cortex samples were prepared as previously described in Smith et al. [20]. Briefly, samples were sectioned at 20–30 μm thickness (Leica Cryostat) and mounted immediately onto warm charged SuperFrost Plus slides (Fisher). Whole tissue imaging was performed on a Zeiss Axio Observer or LSM710 confocal with image tiling at 20X magnification. Images were uniformly corrected by Zen Blue Software during stitching together. Following fixation (4% PFA), primary antibodies included mouse anti-RBFOX3 (NeuN, Millipore) and rabbit anti-RCL1 (Sigma), and secondary antibodies included AlexaFluor (1:500; 488 nm, 563 nm) donkey anti-mouse and donkey anti-rabbit (Invitrogen). See Supplemental Table 1 for details.
Table 1.
Variants of interest in the index patient.
| Gene | Genomic location (hg19) | Variant type | HBVS protein | Allele count in gnomAD | Zygosity | Parent of origin | Variant interpretation (ACMG/AMP guidelines via InterVar) [43] |
|---|---|---|---|---|---|---|---|
| CAPN1 | 11:64975716 A > G, ENST00000527323.1:c.1712A > G | Cryptic splice (Donor) | p.Asn571Ser |
4/ 210744 |
Het | Pat | VUS |
| CAPN1 |
11:64977855 C > T, ENST00000527323.1:c.1991C > T |
Nonsynonymous | p.Ser664Leu |
3/ 247776 |
Het | Mat | VUS |
| RCL1 | 9:4827019 C > T, ENST00000381750.4:c.370 C > T | Stop-gain | p.Gln124Ter | 0 | Het | Pat | Pathogenic |
Human RNA expression analysis
The Allen Human Brain Atlas maintains an online database of transcriptional expression profiling across cortical brain regions from age 8 weeks post-conception to adult [21]. BrainSpan data analysis of (ENST00000381750.9) at chr9:4792944-4861066 was referenced on November 2, 2019. RNA-seq expression measured in RPKM (reads per kilobase exon per million mapped reads) was obtained from the BrainSpan project data and summarized to Gencode v10 exons for all annotated neocortex tissues aged 8 weeks post-conception to 38 years. See Supplemental Table 1 for details.
Results
Clinical findings in the proband and affected father
The proband is currently an 18-year-old man who at 14 years of age experienced neuropsychiatric symptoms over the course of a year that accelerated 4 months prior to his presentation with catatonia, auditory and visual hallucinations, paranoia, aggression, mood dysregulation, and disorganized thoughts. An extensive medical evaluation was performed due to the accelerated nature of the patient’s decline just prior to presentation with catatonia. The patient had a relatively unremarkable development before his psychiatric hospitalization at age 14. He was born full-term after an uncomplicated pregnancy. He had some articulation problems that were corrected with speech therapy. He required tympanostomy tubes as an infant. All other developmental milestones were achieved on time. The patient began experiencing an increase in anxiety starting at age 13. At age 13 years and 9 months, he had a syncopal episode that was evaluated with an electroencephalogram (EEG) which showed intermittent left temporal slowing. However, brain magentic resonance imaging (MRI) obtained at this time was normal. After two additional near syncopal episodes, he was evaluated by a neurologist who concluded that the episodes were not likely seizures. He had previously experienced a head injury without loss of consciousness while playing sports. Prior to his decline in functioning, he was an above-average student and varsity athlete with no history of a learning disability, attention-deficit/hyperactive disorder (ADHD), or special education services.
As the patient’s anxiety increased during the year leading up to his first clearly psychotic episode, his grades declined, and he lost interest in activities that he had once enjoyed. Shortly thereafter, the patient presented with psychosis and catatonia and was hospitalized at age 14. Due to catatonia, an evaluation for autoimmune and other neurologic causes of psychosis was performed. His cerebrospinal fluid (CSF) was found to have elevated protein of 80 mg/dl (normal range: 15–45 mg/dl), which was consistently elevated on four serial repeat lumbar punctures. An F-18-fluoro-2-deoxyglucose positron emission tomography scan showed mild to moderate midbrain and frontal hypometabolism that was regarded as abnormal but nonspecific. The rest of these evaluations, including repeat EEG and MRI as well as serum and CSF autoantibody screens and metabolic studies, were normal.
The elevated CSF protein prompted concern for an autoimmune cause for the patient’s condition. He was treated with methylprednisone and intravenous immunoglobulin but proceeded to symptomatically worsen. Before immune or antipsychotic therapy, he responded to benzodiazepines with some improvement in global function from approximately 20% of baseline to 65% of baseline. However, the patient continued responding to internal stimuli and hallucinations. He had an inadequate response to several antipsychotics and achieved his best response to combined treatment with clozapine and paliperidone. While the patient’s hallucinations and delusions are controlled on this treatment, his thoughts have remained somewhat disorganized. He has had difficulty with urinary frequency and incontinence possibly attributable to a side effect of clozapine. Repeat EEG at age 17 showed mild generalized background slowing and absence of a posterior dominant rhythm consistent with a mild encephalopathy, possibly attributable to his underlying condition and/or effect of clozapine.
Neuropsychological evaluations were completed 4, 12, 29, and 44 months after his functional regression began at age 14 years with his latest evaluation completed at age 17 years 10 months. These evaluations found that intellectual functioning was in the low average range and remained consistent between testing episodes. His full-scale IQ across the four evaluations was 82/87/83/78, respectively, with the differences not considered significant. Comparing the initial evaluations at 4 and 12 months to the final evaluation, performance on formal measures of attention, graphomotor output speed, auditory verbal learning, and memory improved. Word retrieval and graphomotor output speed were within age expectations at the last evaluation. Scores on a measure of working memory were stable. Comparing the final two evaluations, he had much more difficulty focusing, sustaining attention, and encoding/learning at the final evaluation. However, at the final evaluation, he was able to hold on to the information he learned and had better scores on recognition items. His neuropsychologist concluded that his scores on measures of verbal learning/memory did not support decay in memory, but rather an impairment in encoding/learning likely secondary to attention and executive function deficits as well as a slow rate of information processing.
His catatonia has remitted with clonazepam. He meets DSM-5 criteria for schizophrenia, with a history of unspecified depressive disorder and catatonia as confirmed by the structured clinical interview for DSM-5 research version (SCID-5-RV) [22]. Repeated neuropsychological examinations undertaken after his functional decline found low average intelligence with deficits in sustained attention, processing speed, fine motor speed, dexterity in both hands, verbal memory encoding, and verbal fluency.
The patient’s family history is notable on his father’s side for early cognitive decline, multiple sclerosis (MS), anxiety, depression, alcoholism, obsessive-compulsive disorder, bipolar disorder, and suicidal ideation. His father is reported to have emotional regulation and anger control difficulties that worsened through mid-adulthood though he has not been clinically diagnosed. The patient’s paternal grandfather has had an early cognitive decline that was initially noted after a stroke at age 61 years and has continued to progress. His paternal aunt has had multiple psychiatric hospitalizations, and another paternal aunt has MS. On his mother’s side, there is a psychiatric family history of anxiety and dementia in the maternal great grandmother (the patient’s mother’s paternal grandmother) that started in her 70 s and also affected her siblings in their 70 s.
Identification of RCL1 stop-gain paternally inherited variant and additional variants
ES showed heterozygosity for a paternally inherited early stop codon in the RCL1 gene ([OMIM # 611405] NC_000009.11:g.4827019 C > T, NM_005772.4:.370 C > T, NP_005763.3:p.Gln124Ter). This variant has not been recorded in any database. However, a private Gln124His variant was identified in an internal cohort of patients with interstitial cystitis. No structural variants involving RCL1 were identified in the human gene mutation database and no gold-standard variants involving RCL1 were listed in the database of genomic variants (DGV) (September 18, 2020).
The patient was also compound heterozygous for two variants in the CAPN1 gene: NC_000011.9:g.64977855 C > T, NM_001198868.1:c.1991C > T, NP_001185797.1:p.Ser664Leu (Sift = 0.0, Polyphen2 = P, MutationTaster = disease-causing), and NC_000011.9:g.64975716 A > G, NM_001198868.1:c.1712A > G, NP_001185797.1:p.Asn571Ser (Sift = 0.04, Polyphen2 = D, MutationTaster = disease-causing). All variants were confirmed by Sanger sequencing in the trio (see Table 1).
The patient’s compound heterozygous variants in CAPN1 were classified as variants of uncertain significance (Table 1). CAPN1 (OMIM # 114220) encodes calpain-1, a large subunit of μ-calpain, a calcium-activated cysteine protease widely present in the central nervous system. Mutations in CAPN1 have been linked to hereditary spastic paraplegia type 76, which is characterized by adult-onset, chronically progressive corticospinal tract dysfunction (SPG76) with variable cerebellar dysfunction, peripheral neuropathy, and urinary symptoms including incontinence [23–25]. At the most recent follow-up, the patient has not manifested any signs of pyramidal tract dysfunction (spasticity, hyperreflexia, abnormal plantar response), cerebellar dysfunction/ataxia, or peripheral neuropathy, although we cannot exclude that these may later develop with age. Thus, we are not able to rule out that the compound heterozygosity of CAPN1 mutations is contributing in some way to the patient’s presentation (e.g., urinary symptoms).
The patient’s clinical microarray (CMA) revealed a paternally inherited copy number loss in 2q13 (Min/Max coordinates [hg19]: 110862474-110983457; 110833650-111406694; Min/Max Size: 120 kb/573 kb, containing genes MALL, NPHP1, LIMS3, RGPD6, and BUB1). Mutations of NPHP1 (OMIM #607100) are associated with autosomal recessive nephronophthisis. A sequence variant in the other allele was not seen on ES. There are also overlapping deletions listed in the DECIPHER database (e.g., cases 337636, 271516, 258446, 259866, 259955, 308018, and 283832). Associated features include ASD, intellectual disability, cognitive delay, global developmental delay, delayed speech, central hypoventilation, and dysphagia. A copy number loss encompassing a similar interval (including MALL, NPHP1, LIMS3, RGPD6, and BUB1) has been reported in ClinVar as a variant of uncertain significance with the phenotype of developmental delay and facial dysmorphisms (SCV000709783.2). A similar interval to the one reported in our proband with the exception of BUB1 was present in DGV gold-standard variants (defined as curated variants from a selected number of studies in DGV) with a frequency of 0.07% (gssvL65158, phenotype unknown). 2q13 deletions encompassing NPHP1 are present in 0.571% of controls in the UK Biobank (865 in 151,619 individuals) and 0.571% of other controls (152 in 26,628 individuals) [26].
The premature stop codon in the RCL1 index case and the 2q13 copy number loss are paternally inherited. There is a paternal family history of psychiatric hospitalizations and difficulties with emotional regulation in the father and grandfather.
Blood phenotypes
Common variants in RCL1 affect red blood cell and platelet indices, including significantly decreasing or increasing mean corpuscular volume (MCV), mean corpuscular hemoglobin (MCH) [27], and red blood cell count (RBC), affecting platelet count (decreasing [28, 29] and increasing [30, 31]) and increasing platelet crit [30] (defined as platelet count × mean platelet volume [MPV]/10,000). Therefore, blood panel values were abstracted from the proband’s medical record, including 20 assessments over the course of over 3 years. While MCV was in the normal range, MCH was low at 27.2 ± 0.37 picograms/cell (normal range 28.2–30.5), and his RBC was elevated at 5.2 ± 0.22 million cells per µL (normal range 3.7–4.9). In addition, MPV was elevated with a mean of 11.3 ± 0.55 fL (normal range 7.5–8.3). In agreement with previous RCL1 blood phenotypes, the proband’s RCL1 variant is likely contributing to the shifted MCV, RBC, and MPV phenotypes.
Robust RCL1 transcriptional expression during human development
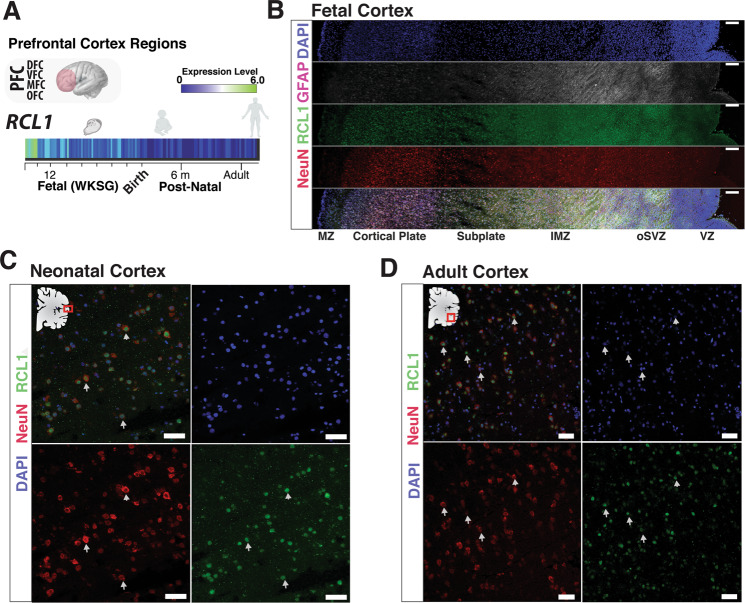
RCL1 demonstrates robust RNA expression in the prefrontal cortical regions during early human development which extends into the neonatal period and subsequently decreases into adulthood (Fig. 2a). To evaluate RCL1 localization in the developing human cortex, we performed immunohistochemical analyses of RCL1 in the mid-gestation fetal cortex (20 weeks gestation) which demonstrate that RCL1 is ubiquitious across the cortical column (ventrical zone to cortical plate), with robust expression in deep-layer CTP2 + neurons (cortical layer 5/6) and among cells in the ventricular zone, including GFAP + progenitors (Fig. S1). However, RCL1 did not colocalize with GFAP + fibers located in more superficial cortical regions (Fig. S2c). Immunohistochemical analyses of the human neonate cortex (270 days post-birth) and adult cortex (37 year old) revealed the highest RCL1 signal colocalized with a neuronal marker (NeuN, Fig. 1) with the limited signal in the white matter (Fig. S1a). In agreement with IHC data, analysis of adult human neocortex single cell RNA sequencing data demonstrates that RCL1 is broadly expressed across several neuronal types, including enrichment within specific excitatory and inhibitory neuron clades (Fig. S2).
Fig. 2. RCL1 expression in human brain development.
a Bulk transcriptome analysis of prefrontal cortical regions demonstrates RCL1 transcripts enriched during gestational weeks (WKSG) and decrease postnatally, presented as log2 RPKM (reads per kilobase per million) values. DFC dorsolateral prefrontal cortex, VFC ventrolateral prefrontal cortex, MFC medial frontal cortex, OFC orbital frontal cortex. Transcriptome data from Allen Institute for Brain Science Atlas. b Confocal fluorescence image of a mid-gestation human coronal fetal cortex tissue with RCL1 antibody co-labeling with markers, glial fibrillary acidic protein (GFAP), a neuronal marker (NeuN), and nuclei marker (DAPI). RCL1 is lowly, yet uniformly present across developing cortex layers, with an abundant signal within the cortical plate. Scale bar 100 µm. MZ marginal zone, IMZ intermediate (fiber) zone, oSVZ outer sub-ventricular zone, VZ ventricular zone. RCL1 and cell type specific antibody staining with corresponding confocal fluorescence imaging of (c) 9-month-old human cortex (neonatal) and (d) adult cortex (37-year-old). Neuronal marker (NeuN) and global nuclei marker (DAPI) show RCL1 present in both neurons and non-neuronal cell types. Arrows indicate cells colocalized for NeuN and RCL1. Scale bar, 50 µm.
Identification of additional patients with copy number variants including RCL1
To determine if the observed RCL1 variant could cause the proband’s presentation, we queried cases undergoing clinical CMA testing at two major medical centers for individuals with CNVs impacting RCL1. The Hospital for Sick Children’s population (N = 44,113 total individuals) is from various published and unpublished sets including MSSNG, the database of clinical variants, and microarrays. The Baylor genetics cohort consists of ~58,000 total individuals, containing probands of diverse phenotypes and parental samples. Thirteen patients were identified with CNVs encompassing RCL1; nine with copy number losses and four with copy number gains (Tables 2 and 3 and Fig. 1).
Table 2.
Additional RCL1 copy number losses: cases from academic medical centers.
| Case # | Inheritance | Start (hg19) | End (hg19) | Size | Copy number change | Age | Phenotype |
|---|---|---|---|---|---|---|---|
| Toronto SickKids* | |||||||
| 1 | Paternal | 4,767,677 | 4,869,801 | 102 kb | Loss | Not available | Autism spectrum disorder (ASD), inherited from father |
| 2 | Unknown, seen in sibling (unknown phenotype) | 4,542,988 | 4,999,498 | 457 kb | Loss | Not available | Developmental delay, 2q13 duplication |
| 3 | Unknown | 4,504,448 | 5,510,644 | 1006 kb | Loss | Not available | Schizophrenia |
| 4 | Unknown | 203,861 | 6,648,114 | 6,444,253 | Loss | Not available | Developmental delay |
| 5 | Unknown | 4,611,869 | 6,144,065 | 1,532,196 | Loss | Not available | Developmental delay, ASD |
| Baylor genetics | |||||||
| 6 | Paternal | 4,744,770 | 5,037,925 | 293 kb | Loss | 19 years | ASD, seizure disorder |
| 7 | Paternal | 4,428,574 | 4,979,623 | 551 kb | Loss | 6 years | Moderate developmental delay, intellectual disability, dysmorphic features |
| 8 | Unknown | 4,428,574 | 4,979,623 | 551 kb | Loss | 0 months | Congenital heart defect, small deletion of PABPC4L |
| 9 | Unknown | 4,781,753 | 5,015,889 | 234 kb | Loss | 4 months | Dysmorphic features, cleft lip and palate |
Table 3.
Copy number gains in RCL1 from Academic Medical Center 1 (Toronto SickKids*).
| Case # | Inheritance | Start (hg19) | End (hg19) | Size | Copy number change | Age | Phenotype |
|---|---|---|---|---|---|---|---|
| 10 | Paternal | 4,725,823 | 5,012,037 | 286 kb | Gain (×3) | Not available | ASD |
| 11 | Unknown | 4,744,779 | 5,015,759 | 271 kb | Gain (×3) | Not available |
Developmental delay and ADHD; pathogenic 16p11.2 copy number loss |
| 12 | Unknown | 4,814,948 | 4,886,266 | 71 kb | Gain (×3) | Not available | Schizophrenia |
| 13 | Unknown | 4,841,229 | 4,938,706 | 97 kb | Gain (×3) | Not available | Developmental delay |
Discussion
We present an index case of an 18-year-old male whose decline in functioning starting at age 14 led to a DSM-5 diagnosis of schizophrenia with a history of catatonia and unspecified depressive disorder. ES revealed a paternally inherited premature stop codon in RCL1 p.(Gln124Ter), a gene with CNVs identified in 13 additional individuals, 11 of whom have neuropsychiatric phenotypes.
Biological evidence and analysis of RCL1 in human tissue
RCL1 is a ubiquitously expressed, highly conserved eukaryotic gene located on chromosome 9 (9p24.1) involved in ribosome biogenesis [32–37]. RCL1 is expressed globally across tissues (ENSG00000120158.11; https://gtexportal.org/home/gene/RCL1), with the highest expression levels in the liver, adipose, and arterial tissues. Defects in ribosomal biogenesis have been reported to result in a range of phenotypes, including embryonic lethality, growth delays, craniofacial defects, sterility, skin and skeletal abnormalities, anemia, cirrhosis, and cognitive impairment [38, 39]. A review of data in gnomAD (v2.1.1, accessed 04/03/2020) identified only six presumptive loss-of-function mutations in RCL1 (20.6 expected), resulting in a LOEUF of 0.57 [19]. The presence of only six individuals heterozygous for RCL1 loss-of-function mutations among the 141,456 supposedly healthy individuals in the gnomAD cohort is consistent with the pathogenicity of RCL1 haploinsufficiency under a model accounting for later onset and variable expressivity of a psychiatric condition.
Archived Zfin data of the RCL1 zebrafish orthologue rcl1 (transgenic insertion rcl1 hi2452Tg zebrafish, ZFIN ID: ZDB-GENE-040930-11) lists phenotype “day 2: slightly smaller head and eye; day 5: small head and eyes, underdeveloped liver/gut, a little pericardial edema” [40]. While no behavioral phenotype or seizure activity is listed, it is interesting that smaller head size is a documented phenotype of zebrafish rcl1 disruption [41], and rcl1 was one of 315 genes identified in the screen as being essential for zebrafish development (Dr. Adam Amsterdam, personal communication). Similar to human data presented here, the zebrafish rcl1 gene is highly expressed in utero (See Fig. 2a) consistent with the hypothesis that a significant early neurodevelopmental component carries the risk for schizophrenia spectrum disorders, deriving from processes operating in prenatal development [42].
RCL1 expression is enriched to neurons in the neonatal and adult human brain
Our postnatal human neocortex IHC experiments indicate that RCL1 is primarily expressed in nuclei of neurons, supporting its role as a RNA 3′-terminal phosphate cyclase-like protein involved in ribosome biogenesis, with some expression in the cytoplasm as well. However, we did not observe RCL1 colocalized with layer 1 interlaminar astrocyte processes as previously reported in the adult cortex [15], nor did we find significant expression within the adult white matter layer or within GFAP + fibers. Taken together, our RNA expression analysis and IHC data demonstrate that RCL1 is likely enriched in neurons during the neonatal and adolescence period, offering a pathological basis for early life psychosis. Further exploration of RCL1 expression in the adult cortex at the single-cell transcriptome level shows RCL1 expression in both excitatory and inhibitory neuronal subtypes, without expression in non-neuronal cells, suggesting that RCL1 dysfunction could disrupt cell types differentially.
Family-based studies
A recent GS study focused on the identification of rare variants in families from an isolated population reported a missense p.Leu372Phe variant (rs115482041) in RCL1 segregating with depression in a multi-generation pedigree [15]. The variant showed significant association with depressive symptoms (N = 2393, βT-allele = 2.33, P-value = 1 × 10−4) and explained 2.9% of the estimated genetic variance of depressive symptoms (22%) in the genetically isolated population. Both of the homozygous carriers exhibited high scores on the depressive symptom rating scale and were also diagnosed with major depressive disorder requiring a combination of psychotherapy and antidepressant treatment. Despite the variant being twice as rare in a neighboring outbred population (MAF < 0.5%), the same variant showed a similar effect and significant association with depressive symptoms (N = 1604, βT-allele = 3.60, P-value = 3 × 10−2) [15].
This missense change in Rs115482041of Leu to Phe is predicted to be Likely Benign using ACMG criteria [43]. This is in contrast to the variant discussed in our proband, which results in a premature truncation of the protein and is thus scored as Pathogenic using ACMG criteria. In accordance with Mort et al., which predicted that genes important in binding have an abundance of missense changes [44] (n = 214.7 expected in gnomAD for RCL1, and 258 observed) and a paucity of nonsense changes (20.6 expected, 6 observed), we conclude that RCL1 binds partners during ribosomal biogenesis [45]. The evidence that the amino acid change in Rs115482041 confers a risk of a psychiatric phenotype supports the hypothesis that a nonsense mutation in RCL1 could also increase risk of neuropsychiatric phenotypes.
Investigation of GWAS studies
RCL1 variants have been reported to influence red blood cell volume [27, 46], platelet count [31, 47], and platelet crit [30] in several genome-wide association studies, suggesting a role for RCL1 in disorders of the blood. Our patient shows an elevation of MPV, providing supporting evidence that the RCL1 variant is physiologically active and producing a known associated phenotype.
RCL1 may be involved in the response to antipsychotic treatment in schizophrenia [48], and a case-control study has shown that an intergenic SNP variant between the HNRNPA1P41 pseudogene and RCL1 is statistically significantly associated with Alzheimer’s disease (AD) and related dementias (rs9969783 chr9:4,888,441 [hg19], effect size: −0.2949 [unit decrease] std dev: 0.0615 p-value: 1.63E−06) [49]. Psychosis occurs in up to 50% of individuals with AD and is associated with significantly worse clinical outcomes [50, 51]. Atypical antipsychotics are sometimes used in AD, suggesting shared mechanisms [52], and similar neuropsychological deficits in processing speed and executive function have been observed in individuals with very late onset schizophrenia-like psychosis and AD + psychosis [53]. In addition, a recent study found that a schizophrenia polygenic risk score was associated with AD + psychosis [54]. These new findings point towards psychosis in AD sharing some genetic liability with schizophrenia and are consistent with the hypothesis that dysregulation of RCL1 is related to various brain-related illness phenotypes.
Additional patients with copy number variants including RCL1
Through collaborative data sharing with major academic medical centers, we were able to identify additional individuals with CNVs in our candidate gene, many of whom had neuropsychiatric phenotypes. Two small copy number losses have childhood-onset psychosis and ASD as their phenotypes. Two of the youngest patients with copy number losses in RCL1 did not have diagnoses of developmental delay or psychiatric phenotypes, though it was too early in their lives for those phenotypes to be recorded.
Nine additional unrelated probands who harbor CNVs involving the RCL1 gene were identified at Toronto SickKids (Tables 2 and 3). Seven probands in this group have been diagnosed with significant neurodevelopmental phenotypes and two with schizophrenia. The two probands diagnosed with schizophrenia have a duplication and deletion in the RCL1 gene, respectively. The copy number gain with the smallest interval (71,319 bp and duplicating exons 2–9, Patient #12) has a schizophrenia phenotype. However, this CNV’s impact on function is unknown, as it is overlapping one end of the gene and leaving one copy intact. One other patient with a developmental delay has a duplication interval overlapping one end of the gene spanning exons 6–9 (Patient #13), which may also leave a copy of the gene intact. Unfortunately, recontacting patients for a more detailed history or additional samples was not possible.
At Baylor genetics, four patients were identified with copy number loss of RCL1 smaller than one megabase. One patient (Patient #8, originally diagnosed with a cardiac defect and a carrier of an additional PABPC4L deletion), is now 10 years of age with no reported neuropsychiatric phenotype. Another patient (Patient #9) was too young at the time of testing to have shown a psychiatric phenotype (at 4 months) and an update was not available. The other two patients have ASD and seizures or moderate developmental delay, intellectual disability, and dysmorphic features, respectively.
Several of the academic medical center cases are also paternally inherited, and some harbor additional CNVs. The first (Patient #11) has a CNV in 16p11.2, which is known to have widely varying phenotypes, ranging from schizophrenia to developmental delay and intellectual disability to no remarkable phenotypes at all [55]. Another (Patient #2) has a reciprocal 2q13 duplication. While the index case has a 2q13 copy number loss, this patient has developmental delay and a 456,511 bp CNV loss encompassing RCL1 and an additional 2q13 duplication. It is possible that a second genetic mutation or CNV is required to unmask the predisposition to a neuropsychiatric phenotype, in accordance with Girirajan et al. [56].
ClinGen does not list any benign gains or losses encompassing RCL1 but lists several pathogenic gains and losses, with phenotypes including developmental delay, low-set ears, abnormal gait, facial abnormalities, ASD, and intellectual disability.
Loss of MALL, NPHP1, LIMS3, and BUB1
Chromosome 2q13 deletions and duplications are associated with developmental delay as well as psychiatric and behavioral disorders [57, 58]. The 2q13 copy number loss is associated with ASD, but in this patient, only the BUB1 gene overlaps in an autism-associated 2q13 interval described by Guivarch et al. [59]. Mutations in BUB1 can cause mosaic variegated aneuploidy and increase the risk of colorectal cancer at a young age [60]. A paper describing individuals with various overlapping intervals involving RGPD6 and BUB1 reported one individual as having normal neuropsychiatric development but described others with developmental delay/intellectual disability, ASD, and pervasive developmental disorder - not otherwise specificed (PDD-NOS) [61]. Moreover, given the above-average athletic and scholastic abilities of the proband before his first psychiatric episode, and the CNV’s frequency among control sequencing databases (0.571% in the UK Biobank), it is difficult to conceive that the 2q13 interval is causative to the patient’s psychosis phenotype. Lastly, the patient’s interval also does not include candidate genes for schizophrenia in the 2q13 region (ANAPC1, BCL2L11, or MERTK) mentioned in Costain et al. [62].
CAPN1
The proband also has compound heterozygous missense mutations in CAPN1, which may be resulting in an atypical version of the Mendelian disease spastic paraplegia-76 (SPG76), in particular since this subtype is associated with young adult spastic paraplegia onset. While SPG76 is not traditionally associated with early-onset psychosis, an SPG76 case report details psychosis in one patient at age 45, 20 years after the onset of lower limb stiffness and gait instability. This patient had a more deleterious homozygous nonsense CAPN1 mutation (p.Trp392*), and was wheelchair-bound at age 40 years [63]. The individual had chronic depression (with a positive family history), and at age 45 years manifested severe cognitive and behavioral disturbances mimicking the clinical and neuroimaging features of behavioral variants of Frontotemporal Dementia, which include moderate cortical atrophy [63]. She also experienced urinary incontinence and urgency, though the onset of the urinary symptoms was 13 years after the onset of gait instability and lower limb stiffness. Currently, there are over 80 genetic subtypes of SP resulting in a spectrum of associated phenotypes, including the SPAST gene in SPG4, a microtubule-associated gene, which is associated with an increased rate of schizophrenia and psychosis in the affected individuals [64].
For the proband in this study, the paternal CAPN1 variant (p.Asn571Ser) is extremely rare with an allele count of 4 in gnomAD (n = 210,744), with 2/4 of these CAPN1 carriers belonging to the gnomAD psychiatric cohort. Similarly, the maternal variant (p.Ser664Leu) is present in three individuals in gnomAD (n = 247776), one of whom belongs to the psychiatric cohort. While CAPN1 is relatively tolerant to missense changes (gnomAD, o/e ratio of 0.78), both of the proband’s missense CAPN1 variants are in the top 1% of deleteriousness as both have CADD scores above 24. CAPN1 is a recessive gene and previous pathogenic variants have both been homozygous and compound heterozygous variants. ClinVar lists 35 reports of CAPN1 mutations: 11/35 are pathogenic, 20/35 are (likely) benign and 4 are either VUS or have conflicting interpretations of pathogenicity. In terms of mutation type distribution in ClinVar, 8/11 of the pathogenic variants are loss of functions, two are missense and one is a non-essential splice site. Thus, the evidence for the compound heterozygous missense mutations in CAPN1 to cause SPG76 in this patient is strong but not as compelling as if one of the variants were a loss-of-function. However, we cannot rule out that the proband is pre-symptomatic or oligosymptomatic for SPG76, although he does not yet have the core feature of leg spasticity. It is possible these phenotypes will develop over time; reports of SPG76 in the literature state age of onset ranging from 15 to 45 years [24, 63, 65–68]. Electrophysiology tests such as EMG/NCS or lower limb SSEP’s have not yet been performed on this patient, given the normal motor exams in the past and the likely difficulty of tolerating such tests with his psychiatric phenotype.
Limitations
Our inability to recontact the additional cases prevented us from comprehensively phenotyping the patients, and we were unable to follow up on cardiac abnormalities and dysmorphic features that were noted in some patients and may have been present but unreported in others. Ideally, the patients should have been monitored longitudinally. In addition, consenting limitations prevented the querying of additional potentially causative mutations in CNV carriers.
Conclusions
VEOP is an extreme phenotype of the more common adult-onset psychosis, thus making VEOP a candidate for Mendelian inheritance patterns. Mendelian disorders are more likely to involve genes of large effect. Therefore, we began with an index case of VEOP and utilized our systematic approach to identifying candidate genes for further investigation. Having established the involvement of RCL1 in the brain and behavior of our index VEOP patient, we queried two academic medical centers and confirmed the involvement of RCL1 in 13 additional patients with neurological disorders. RCL1 deletions and duplications were associated with a range of neurological and psychiatric phenotypes. Copy number loss of RCL1 was associated with developmental delay, intellectual disability, ASD, seizures, and schizophrenia in two separate patient populations at the two medical centers. Copy number gain of RCL1 was associated with developmental delay, ASD, schizophrenia, and ADHD.
Our multistep study design had several advantages. Next-generation sequencing of rare Mendelian forms of complex disease increases the chances of discovering genes of large effect. Examination of model organism data on RCL1 supports the hypothesis that disruption of RCL1 results in a neurological phenotype. In addition, we used the Allen Brain Atlas database to screen RNA expression and validated these results through in-house functional studies to gain biological insights. Our screening included cell type RCL1 expression patterns from neonatal life to adulthood and offers a pathological transcriptional timeline that aligns with early developmental processes leading up to the psychosis period.
In conclusion, our findings identify RCL1 as an important candidate gene for a range of neurological and psychiatric phenotypes, and we highlight published evidence that RCL1 protein is, in fact, present in the developing human brain. Our findings will undoubtedly guide future investigations into the genetic and pathological bases of neuropsychiatric phenotypes observed in the RCL1 cases. Details of the etiology, neurobiological manifestations, and the role of other genetic loci or nongenetic effects in shaping phenotypes are yet to be elucidated.
Supplementary information
Acknowledgements
The authors are grateful for the participation of the families reported herein and Johnathan Hecht for assistance with human tissue. Human tissue was obtained from the NIH NeuroBioBank at the University of Maryland, Baltimore, MD. We acknowledge the referring clinicians for providing clinical samples and data used in this analysis. Support was provided by Stanley Center of the Broad Institute, Tommy Fuss Center for Neuropsychiatric Disease Research, Anne and Paul Marcus Family Foundation, Robin and Jonathan Klein Family, and NIH U54 HD090255 in support of the Boston Children’s Hospital IDDRC Molecular Genetics Core Facility. Additional work was supported by NIH F32 NS100033801 and K99 NS112604 to RS and R01 NS035129 to CAW. The authors thank the Centre for Applied Genomics and the University of Toronto McLaughlin Centre for technical support and funding. SWS holds the GlaxoSmithKline-CIHR Chair in Genome Sciences at The Hospital for Sick Children and the University of Toronto.
Author contributions
LHR, MH, EG, ED, EDA, DC, HMH, FP, MPG, AC, CAW, CAG, DCG, and JGH collected, interpreted, and/or evaluated clinical data; CAB, EG, JL, KC, JB, AR, WB, JH, MZ, PBA, RS, AWH, LHR, DJS, JAR, AHB, RAG, SWS, DCG, and JGH, analyzed human genetics data; RS, performed and analyzed cell-based experiments; CAB, RS, JAR, MZ, LHR, AR, JGH, DCG, and SWS, wrote the manuscript, all co-authors edited the manuscript.
Data availability
Schizophrenia sample IDs (from Psychiatric Genomic Consortium): Ed_PT-267W@cas_scz_edin_eur_A6.0*Ed_PT-267W. PT-BJMD@fam_scz_butr_eur_A6.0*1403_0_pca. Proband patient data has been deposited into DECPHER https://decipher.sanger.ac.uk/patient/421796.
Compliance with ethical standards
Conflict of interest
The Department of Molecular and Human Genetics at Baylor College of Medicine receives revenue from clinical genetic testing at Baylor Genetics Laboratories. JGH holds equity in and is founding head of the scientific advisory board for Mightier/Neuromotion Labs, a company making emotional regulation training video games, and has received consulting income from Alkermes, Inc., a pharmaceutical company. The remaining authors declare no competing financial interests.
Ethics
Informed consent was obtained for the proband and family whose results are reported in this study. The study was approved by the Institutional Review Board of Boston Children’s Hospital under protocol # 10-02-0053.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Catherine A Brownstein, Richard S. Smith
Supplementary information
The online version contains supplementary material available at 10.1038/s41380-021-01035-y.
References
- 1.Pardiñas AF, Holmans P, Pocklington AJ, Escott-Price V, Ripke S, Carrera N, et al. Common schizophrenia alleles are enriched in mutation-intolerant genes and in regions under strong background selection. Nat Genet. 2018;50:381–9. doi: 10.1038/s41588-018-0059-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Leonenko G, Di Florio A, Allardyce J, Forty L, Knott S, Jones L, et al. A data-driven investigation of relationships between bipolar psychotic symptoms and schizophrenia genome-wide significant genetic loci. Am J Med Genet. 2018;177:468–75. doi: 10.1002/ajmg.b.32635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sekar A, Bialas AR, de Rivera H, Davis A, Hammond TR, Kamitaki N, et al. Schizophrenia risk from complex variation of complement component 4. Nature. 2016;530:177–83. doi: 10.1038/nature16549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fromer M, Roussos P, Sieberts SK, Johnson JS, Kavanagh DH, Perumal TM, et al. Gene expression elucidates functional impact of polygenic risk for schizophrenia. Nat Neurosci. 2016;19:1442–53. doi: 10.1038/nn.4399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Consortium SWGotPG. Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511:421–7. doi: 10.1038/nature13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Purcell SM, Moran JL, Fromer M, Ruderfer D, Solovieff N, Roussos P, et al. A polygenic burden of rare disruptive mutations in schizophrenia. Nature. 2014;506:185–90. doi: 10.1038/nature12975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Singh T, Kurki MI, Curtis D, Purcell SM, Crooks L, McRae J, et al. Rare loss-of-function variants in SETD1A are associated with schizophrenia and developmental disorders. Nat Neurosci. 2016;19:571–7. doi: 10.1038/nn.4267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Egawa J, Hoya S, Watanabe Y, Nunokawa A, Shibuya M, Ikeda M, et al. Rare UNC13B variations and risk of schizophrenia: Whole-exome sequencing in a multiplex family and follow-up resequencing and a case-control study. Am J Med Genet. 2016;171:797–805. doi: 10.1002/ajmg.b.32444. [DOI] [PubMed] [Google Scholar]
- 9.Hansen AW, Murugan M, Li H, Khayat MM, Wang L, Rosenfeld J, et al. A genocentric approach to discovery of mendelian disorders. Am J Hum Genet. 2019;105:974–86. doi: 10.1016/j.ajhg.2019.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lesage S, Brice A. Parkinson’s disease: from monogenic forms to genetic susceptibility factors. Hum Mol Genet. 2009;18:R48–59. doi: 10.1093/hmg/ddp012. [DOI] [PubMed] [Google Scholar]
- 11.Sanders SJ, Sahin M, Hostyk J, Thurm A, Jacquemont S, Avillach P, et al. A framework for the investigation of rare genetic disorders in neuropsychiatry. Nat Med. 2019;25:1477–87. doi: 10.1038/s41591-019-0581-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stessman HA, Bernier R, Eichler EE. A genotype-first approach to defining the subtypes of a complex disease. Cell. 2014;156:872–7. doi: 10.1016/j.cell.2014.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alexander Arguello P, Addington A, Borja S, Brady L, Dutka T, Gitik M, et al. From genetics to biology: advancing mental health research in the genomics ERA. Mol Psychiatry. 2019;24:1576–82. doi: 10.1038/s41380-019-0445-x. [DOI] [PubMed] [Google Scholar]
- 14.Brownstein CA, Beggs AH, Homer N, Merriman B, Yu TW, Flannery KC, et al. An international effort towards developing standards for best practices in analysis, interpretation and reporting of clinical genome sequencing results in the CLARITY challenge. Genome Biol. 2014;15:R53. doi: 10.1186/gb-2014-15-3-r53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Amin N, de Vrij FMS, Baghdadi M, Brouwer RWW, van Rooij JGJ, Jovanova O, et al. A rare missense variant in RCL1 segregates with depression in extended families. Mol Psychiatry. 2018;23:1120–6. doi: 10.1038/mp.2017.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yuen RKC, Merico D, Bookman M, Howe JL, Thiruvahindrapuram B, Patel RV, et al. Whole genome sequencing resource identifies 18 new candidate genes for autism spectrum disorder. Nat Neurosci. 2017;20:602–11. doi: 10.1038/nn.4524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zarrei M, Burton CL, Engchuan W, Young EJ, Higginbotham EJ, MacDonald JR, et al. A large data resource of genomic copy number variation across neurodevelopmental disorders. NPJ Genom Med. 2019;4:26. doi: 10.1038/s41525-019-0098-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Philippakis AA, Azzariti DR, Beltran S, Brookes AJ, Brownstein CA, Brudno M, et al. The matchmaker exchange: a platform for rare disease gene discovery. Hum Mutat. 2015;36:915–21. doi: 10.1002/humu.22858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Karczewski KJ, Francioli LC, Tiao G, Cummings BB, Alföldi J, Wang Q, et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020;581:434–43. doi: 10.1038/s41586-020-2308-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smith RS, Kenny CJ, Ganesh V, Jang A, Borges-Monroy R, Partlow JN, et al. Sodium channel SCN3A (Na V 1.3) regulation of human cerebral cortical folding and oral motor development. Neuron. 2018;99:905–13.e7. doi: 10.1016/j.neuron.2018.07.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jones AR, Overly CC, Sunkin SM. The Allen Brain Atlas: 5 years and beyond. Nat Rev Neurosci. 2009;10:821–8. doi: 10.1038/nrn2722. [DOI] [PubMed] [Google Scholar]
- 22.First MB, Williams JBW, Karg RS, Spitzer RL. Structured clinical interview for DSM-5—research version (SCID-5 for DSM-5, research version; SCID-5-RV). Arlington, VA: American Psychiatric Association; 2015.
- 23.Kocoglu C, Gundogdu A, Kocaman G, Kahraman-Koytak P, Uluc K, Kiziltan G, et al. Homozygous CAPN1 mutations causing a spastic-ataxia phenotype in 2 families. Neurol Genet. 2018;4:e218. doi: 10.1212/NXG.0000000000000218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gan-Or Z, Bouslam N, Birouk N, Lissouba A, Chambers DB, Vérièpe J, et al. Mutations in CAPN1 cause autosomal-recessive hereditary spastic paraplegia. Am J Hum Genet. 2016;98:1038–46. doi: 10.1016/j.ajhg.2016.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Su W, Zhou Q, Wang Y, Chishti A, Li QQ, Dayal S, et al. Deletion of the Capn1 gene results in alterations in signaling pathways related to Alzheimer’s disease, protein quality control and synaptic plasticity in mouse brain. Front Genet. 2020;11:334. doi: 10.3389/fgene.2020.00334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kendall KM, Rees E, Escott-Price V, Einon M, Thomas R, Hewitt J, et al. Cognitive performance among carriers of pathogenic copy number variants: analysis of 152,000 UK biobank subjects. Biol Psychiatry. 2017;82:103–10. doi: 10.1016/j.biopsych.2016.08.014. [DOI] [PubMed] [Google Scholar]
- 27.van der Harst P, Zhang W, Mateo Leach I, Rendon A, Verweij N, Sehmi J, et al. Seventy-five genetic loci influencing the human red blood cell. Nature. 2012;492:369–75. doi: 10.1038/nature11677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li J, Glessner JT, Zhang H, Hou C, Wei Z, Bradfield JP, et al. GWAS of blood cell traits identifies novel associated loci and epistatic interactions in Caucasian and African–American children. Hum Mol Genet. 2013;22:1457–64. doi: 10.1093/hmg/dds534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kanai M, Akiyama M, Takahashi A, Matoba N, Momozawa Y, Ikeda M, et al. Genetic analysis of quantitative traits in the Japanese population links cell types to complex human diseases. Nat Genet. 2018;50:390–400. doi: 10.1038/s41588-018-0047-6. [DOI] [PubMed] [Google Scholar]
- 30.Astle WJ, Elding H, Jiang T, Allen D, Ruklisa D, Mann AL, et al. The allelic landscape of human blood cell trait variation and links to common complex disease. Cell. 2016;167:1415–29. doi: 10.1016/j.cell.2016.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gieger C, Radhakrishnan A, Cvejic A, Tang W, Porcu E, Pistis G, et al. New gene functions in megakaryopoiesis and platelet formation. Nature. 2011;480:201–8. doi: 10.1038/nature10659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Horn DM, Mason SL, Karbstein K. Rcl1 protein, a novel nuclease for 18 S ribosomal RNA production. J Biol Chem. 2011;286:34082–7. doi: 10.1074/jbc.M111.268649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Delprato A, Al Kadri Y, Pérébaskine N, Monfoulet C, Henry Y, Henras AK, et al. Crucial role of the Rcl1p-Bms1p interaction for yeast pre-ribosomal RNA processing. Nucl Acids Res. 2014;42:10161–72. doi: 10.1093/nar/gku682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Henras AK, Plisson-Chastang C, O’Donohue MF, Chakraborty A, Gleizes PE. An overview of pre-ribosomal RNA processing in eukaryotes. Wiley Interdiscip Rev RNA. 2015;6:225–42. doi: 10.1002/wrna.1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Billy E, Wegierski T, Nasr F, Filipowicz W. Rcl1p, the yeast protein similar to the RNA 3′-phosphate cyclase, associates with U3 snoRNP and is required for 18S rRNA biogenesis. EMBO J. 2000;19:2115–26. doi: 10.1093/emboj/19.9.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Turner AJ, Knox AA, Prieto JL, McStay B, Watkins NJ. A novel small-subunit processome assembly intermediate that contains the U3 snoRNP, nucleolin, RRP5, and DBP4. Mol Cell Biol. 2009;29:3007–17. doi: 10.1128/MCB.00029-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Warner JR. The economics of ribosome biosynthesis in yeast. Trends Biochem Sci. 1999;24:437–40. doi: 10.1016/s0968-0004(99)01460-7. [DOI] [PubMed] [Google Scholar]
- 38.Freed EF, Bleichert F, Dutca LM, Baserga SJ. When ribosomes go bad: diseases of ribosome biogenesis. Mol Biosyst. 2010;6:481–93. doi: 10.1039/b919670f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Danilova N, Gazda HT. Ribosomopathies: how a common root can cause a tree of pathologies. Dis Model Mech. 2015;8:1013–26. doi: 10.1242/dmm.020529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thisse B, Thisse C. Fast release clones: a high throughput expression analysis. ZFIN Direct Data Submission, 2004.
- 41.Amsterdam A, Nissen RM, Sun Z, Swindell EC, Farrington S, Hopkins N. Identification of 315 genes essential for early zebrafish development. Proc Natl Acad Sci USA. 2004;101:12792–7. doi: 10.1073/pnas.0403929101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Clifton NE, Hannon E, Harwood JC, Di Florio A, Thomas KL, Holmans PA, et al. Dynamic expression of genes associated with schizophrenia and bipolar disorder across development. Transl Psychiatry. 2019;9:74. doi: 10.1038/s41398-019-0405-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li Q, Wang K. InterVar: clinical interpretation of genetic variants by the 2015 ACMG-AMP guidelines. Am J Hum Genet. 2017;100:267–80. doi: 10.1016/j.ajhg.2017.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mort M, Ivanov D, Cooper DN, Chuzhanova NA. A meta-analysis of nonsense mutations causing human genetic disease. Hum Mutat. 2008;29:1037–47. doi: 10.1002/humu.20763. [DOI] [PubMed] [Google Scholar]
- 45.Karbstein K, Doudna JA. GTP-dependent formation of a ribonucleoprotein subcomplex required for ribosome biogenesis. J Mol Biol. 2006;356:432–43. doi: 10.1016/j.jmb.2005.11.052. [DOI] [PubMed] [Google Scholar]
- 46.Ganesh SK, Zakai NA, van Rooij FJ, Soranzo N, Smith AV, Nalls MA, et al. Multiple loci influence erythrocyte phenotypes in the CHARGE Consortium. Nat Genet. 2009;41:1191–8. doi: 10.1038/ng.466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shameer K, Denny JC, Ding K, Jouni H, Crosslin DR, de Andrade M, et al. A genome- and phenome-wide association study to identify genetic variants influencing platelet count and volume and their pleiotropic effects. Hum Genet. 2014;133:95–109. doi: 10.1007/s00439-013-1355-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yu H, Yan H, Wang L, Li J, Tan L, Deng W, et al. Five novel loci associated with antipsychotic treatment response in patients with schizophrenia: a genome-wide association study. Lancet Psychiatry. 2018;5:327–38. doi: 10.1016/S2215-0366(18)30049-X. [DOI] [PubMed] [Google Scholar]
- 49.Beecham GW, Hamilton K, Naj AC, Martin ER, Huentelman M, Myers AJ, et al. Genome-wide association meta-analysis of neuropathologic features of Alzheimer’s disease and related dementias. PLoS Genet. 2014;10:e1004606. doi: 10.1371/journal.pgen.1004606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Murray PS, Kumar S, Demichele-Sweet MA, Sweet RA. Psychosis in Alzheimer’s disease. Biol Psychiatry. 2014;75:542–52. doi: 10.1016/j.biopsych.2013.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ropacki SA, Jeste DV. Epidemiology of and risk factors for psychosis of Alzheimer’s disease: a review of 55 studies published from 1990 to 2003. Am J Psychiatry. 2005;162:2022–30. doi: 10.1176/appi.ajp.162.11.2022. [DOI] [PubMed] [Google Scholar]
- 52.Ballard C, Howard R. Neuroleptic drugs in dementia: benefits and harm. Nat Rev Neurosci. 2006;7:492–500. doi: 10.1038/nrn1926. [DOI] [PubMed] [Google Scholar]
- 53.Van Assche L, Van Aubel E, Van de Ven L, Bouckaert F, Luyten P, Vandenbulcke M. The neuropsychological profile and phenomenology of late onset psychosis: a cross-sectional study on the differential diagnosis of very-late-onset schizophrenia-like psychosis, dementia with Lewy bodies and Alzheimer’s type dementia with psychosis. Arch Clin Neuropsychol. 2019;34:183–99. doi: 10.1093/arclin/acy034. [DOI] [PubMed] [Google Scholar]
- 54.Creese B, Vassos E, Bergh S, Athanasiu L, Johar I, Rongve A, et al. Examining the association between genetic liability for schizophrenia and psychotic symptoms in Alzheimer’s disease. Transl psychiatry. 2019;9:273. doi: 10.1038/s41398-019-0592-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Steinman KJ, Spence SJ, Ramocki MB, Proud MB, Kessler SK, Marco EJ, et al. 16p11.2 deletion and duplication: characterizing neurologic phenotypes in a large clinically ascertained cohort. Am J Med Genet. 2016;170:2943–55. doi: 10.1002/ajmg.a.37820. [DOI] [PubMed] [Google Scholar]
- 56.Girirajan S, Rosenfeld JA, Coe BP, Parikh S, Friedman N, Goldstein A, et al. Phenotypic heterogeneity of genomic disorders and rare copy-number variants. N Engl J Med. 2012;367:1321–31. doi: 10.1056/NEJMoa1200395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen CP, Lin SP, Lee CL, Chern SR, Wu PS, Chen YN, et al. Recurrent 2q13 microduplication encompassing MALL, NPHP1, RGPD6, and BUB1 associated with autism spectrum disorder, intellectual disability, and liver disorder. Taiwan J Obstet Gynecol. 2017;56:98–101. doi: 10.1016/j.tjog.2016.12.003. [DOI] [PubMed] [Google Scholar]
- 58.Wolfe K, McQuillin A, Alesi V, Boudry Labis E, Cutajar P, Dallapiccola B, et al. Delineating the psychiatric and behavioral phenotype of recurrent 2q13 deletions and duplications. Am J Med Genet. 2018;177:397–405. doi: 10.1002/ajmg.b.32627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Guivarch J, Chatel C, Mortreux J, Missirian C, Philip N, Poinso F. An atypical autistic phenotype associated with a 2q13 microdeletion: a case report. J Med Case Rep. 2018;12:79. doi: 10.1186/s13256-018-1620-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.de Voer RM, Geurts van Kessel A, Weren RD, Ligtenberg MJ, Smeets D, Fu L, et al. Germline mutations in the spindle assembly checkpoint genes BUB1 and BUB3 are risk factors for colorectal cancer. Gastroenterology. 2013;145:544–7. doi: 10.1053/j.gastro.2013.06.001. [DOI] [PubMed] [Google Scholar]
- 61.Yu HE, Hawash K, Picker J, Stoler J, Urion D, Wu BL, et al. A recurrent 1.71 Mb genomic imbalance at 2q13 increases the risk of developmental delay and dysmorphism. Clin Genet. 2012;81:257–64. doi: 10.1111/j.1399-0004.2011.01637.x. [DOI] [PubMed] [Google Scholar]
- 62.Costain G, Lionel AC, Merico D, Forsythe P, Russell K, Lowther C, et al. Pathogenic rare copy number variants in community-based schizophrenia suggest a potential role for clinical microarrays. Hum Mol Genet. 2013;22:4485–501. doi: 10.1093/hmg/ddt297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Souza PVS, Silva LHL, Badia BML, Farias IB, Pinto WBVR, Oliveira ASB. SPG76: an extremely rare hereditary spastic paraplegia with a new expanding complicated phenotype. Rev Neurol. 2019;175:572–4. doi: 10.1016/j.neurol.2019.01.397. [DOI] [PubMed] [Google Scholar]
- 64.McMonagle P, Hutchinson M, Lawlor B. Hereditary spastic paraparesis and psychosis. Eur J Neurol. 2006;13:874–9. doi: 10.1111/j.1468-1331.2006.01379.x. [DOI] [PubMed] [Google Scholar]
- 65.Kocoglu C, Gundogdu A, Kocaman G, Kahraman-Koytak P, Uluc K, Kiziltan G, et al. Homozygous. Neurol Genet. 2018;4:e218. doi: 10.1212/NXG.0000000000000218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang Y, Hersheson J, Lopez D, Hammer M, Liu Y, Lee KH, et al. Defects in the CAPN1 gene result in alterations in cerebellar development and cerebellar ataxia in mice and humans. Cell Rep. 2016;16:79–91. doi: 10.1016/j.celrep.2016.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tadic V, Klein C, Hinrichs F, Münchau A, Lohmann K, Brüggemann N. CAPN1 mutations are associated with a syndrome of combined spasticity and ataxia. J Neurol. 2017;264:1008–10. doi: 10.1007/s00415-017-8464-5. [DOI] [PubMed] [Google Scholar]
- 68.Travaglini L, Bellacchio E, Aiello C, Pro S, Bertini E, Nicita F. Expanding the clinical phenotype of CAPN1-associated mutations: a new case with congenital-onset pure spastic paraplegia. J Neurol Sci. 2017;378:210–2. doi: 10.1016/j.jns.2017.05.014. [DOI] [PubMed] [Google Scholar]
- 69.Marshall CR, Howrigan DP, Merico D, Thiruvahindrapuram B, Wu W, Greer DS, et al. Contribution of copy number variants to schizophrenia from a genome-wide study of 41,321 subjects. Nat Genet. 2017;49:27–35. doi: 10.1038/ng.3725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pinto D, Delaby E, Merico D, Barbosa M, Merikangas A, Klei L, et al. Convergence of genes and cellular pathways dysregulated in autism spectrum disorders. Am J Hum Genet. 2014;94:677–94. doi: 10.1016/j.ajhg.2014.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Schizophrenia sample IDs (from Psychiatric Genomic Consortium): Ed_PT-267W@cas_scz_edin_eur_A6.0*Ed_PT-267W. PT-BJMD@fam_scz_butr_eur_A6.0*1403_0_pca. Proband patient data has been deposited into DECPHER https://decipher.sanger.ac.uk/patient/421796.