Abstract
To meet the testing demands and overcome supply chain issues during the SARS-CoV-2 pandemic, many clinical laboratories validated multiple SARS-CoV-2 molecular testing platforms. Here, we compare three different molecular assays for SARS-CoV-2 that received emergency use authorization (EUA) from the U.S. Food and Drug Administration. In order to determine the agreement among Roche cobas® SARS-CoV-2 Test (Cobas), Abbott RealTime SARS-CoV-2 assay (ART), and Mayo Clinic Laboratory SARS-CoV-2 Molecular Detection Assay (Mayo LDT), 100 each of anterior nares (AN), nasopharyngeal (NP), oropharyngeal (OP), and NP+OP swabs were tested on each platform. The consensus result was defined as agreement by 2 or more methods. Furthermore, 30 positive NP swabs from each molecular platform (n = 90 total) were tested on the three platforms to determine the PPA among positive samples. ART platform called more specimens positive than the other two platforms. All three assays performed with greater than 90% agreement for NP specimens throughout the study.
Keywords: COVID-19, Molecular diagnostics, PCR, SARS-CoV-2
1. Background
As the global COVID-19 pandemic continues, clinical microbiology laboratories face continued demand for increased testing capacity amidst supply chain issues, forcing clinical laboratories to implement multiple assays for molecular detection of SARS-CoV-2. Such measures often led to the questions on which tests have the “best” clinical sensitivities and specificities. However, determining the clinical sensitivity and specificity is challenging when there is no gold standard clinical definition of COVID-19.
To determine how test results correlate among the different high-throughput molecular assays and to help guide patient care, we conducted a study to determine the agreement of results obtained on upper respiratory tract specimens tested with 3 molecular assays that have received emergency use authorization (EUA) from the U.S. Food and Drug Administration (FDA): Abbott RealTime SARS-CoV-2 assay (ART; Abbott Molecular, Inc., Des Plaines, IL), cobas® SARS-CoV-2 assay for use with the cobas® 6800 / 8800 systems (Cobas; Roche Molecular Systems, Inc., Pleasanton, CA), and Mayo Clinic Laboratory SARS-CoV-2 Molecular Detection Assay (Mayo LDT; a laboratory-developed assay with EUA).
2. Study design
Consecutive upper respiratory tract swab specimens leftover from routine SARS-CoV-2 detection testing at Mayo Clinic Laboratories during April and May of 2020 were used in this study. One hundred consecutive specimens from each of the following sources were obtained from persons under investigation (PUI) for COVID-19, and placed in 3 mL of viral transport media (VTM) or phosphate-buffered saline (PBS): anterior nares (AN), nasopharynx (NP), oropharynx (OP), or combined NP+OP. The combined NP+OP was an NP swab collected first, followed by collection of an OP swab from the same PUI, with both swabs placed into a 3- mL tube of transport media. For each specimen source, only one specimen submitted for routine testing on each PUI was used in this study, and the four specimen sources are not matched for the study subjects. Due to shortages of assay reagents and consumable supplies, all commercially available brands of VTM, PBS, and saline were acceptable for testing per FDA guidance in deeming them as equivalent (U.S. Food and Drug Administration 2021). All specimens were tested initially with Cobas, stored at 2°C to 8°C for up to 7 days before testing with the other 2 assays. The consensus result was defined as the result of at least 2 out of 3 testing platforms that were found to be in agreement. Testing with all three assays was performed per manufacturers’ instructions for use. For data analysis in this study, indeterminate results generated were grouped in the positive result category, as patients with indeterminate test results are managed clinically as having probable COVID-19. Specimens with this result type did not have sufficient volume for retesting in this study.
In addition, 30 known-positive NP specimens previously tested with one of the 3 assays (total of 90 specimens) were selected for determination of positive agreement among the 3 assays. These specimens were selected based on target CT values of the initial assay and classified as high-, medium-, or low-positives: CT of <12 high, 12 to 20 medium, and >20 low for ART (37 total cycles); <20 for high, 20 to 30 medium, and >30 low for Cobas (50 total cycles) and Mayo LDT (45 total cycles total), respectively. Although both target CT for each the Cobas and Mayo LDT were within the ranges, only the CT values of the ORF1ab target for Cobas and N target for Mayo LDT were used for data analysis in this study. After testing positive by the initial assay, each of these specimens was tested by the remaining 2 assays.
All three molecular assays used in this study received EUA from the FDA and all are based on real-time RT-PCR method amplifying different viral target sequences. ART amplifies the RNA-dependent-RNA polymerase gene (RdRp) and the nucleocapsid gene (N), with a limit of detection (LoD) of 100 copies/mL (Mostafa et al., 2020, Degli-Angeli et al., 2020, Abbott Molecular, Inc 2020). Cobas amplifies the open reading frame 1ab (ORF1ab) and the envelope gene (E) with an LoD of 46 copies/mL (Mostafa et al., 2020, Roche Molecular Systems, Inc 2020). Mayo LDT detects the ORF1ab and N gene with an LoD of 156 copies/mL, using the NucliSENS® easyMag® (bioMérieux, Inc., Durham, NC) or eMag® (bioMérieux, Inc.) extraction systems, followed by amplification and detection with the LightCycler® 480 Real-Time PCR System (Roche Molecular Systems, Inc.) (Rodino et al., 2020, Mayo Clinic Laboratories, Inc 2020).
Percent positive agreement (PPA) and percent negative agreement (NPA) were calculated in reference to the consensus result, as defined above, which was defined as agreement by at least 2 of the 3 PCR tests. Correlation of CT values generated by 2 assays for all positive specimens was determined by Deming regression using MedCalc version 19.6 statistical program (MedCalc Software Ltd., MedCalc Software Ltd, Ostend, Belgium).
3. Results
Among the 400 total consecutive clinical swab specimens collected from the 4 anatomic sites (AN, NP, OP, NP+OP) and tested with the 3 molecular assays, ART had 100% PPA across all specimen types. Mayo LDT had 100% PPA for AN and NP+OP, while Cobas had 100% PPA for OP and NP+OP. A PPA of <95% was observed for AN and NP with Cobas and for NP and OP with Mayo LDT. NPA was >95% across all specimen types and combinations of assay. There were 16 (4%) total discrepancies among results of the 400 specimens (Table S1), and 12 of these discrepancies were weakly positive (CT >20) by ART but not detected by Cobas and/or Mayo LDT. The remaining 4 discrepancies were low positives (CT >30) or indeterminate results by Mayo LDT but not detected (TND) by ART and Cobas.
Among the 90 known positive specimens, ART showed 100% PPA, detecting all 30 specimens that were positive by Cobas and Mayo LDT ( Table 2 ). Only 4 discrepant results (4.4%; Table S2) were observed, with 3 of these discrepancies showing low positive results (CT >30) with ART but no detectable targets with the other 2 assays. The remaining discrepant result was a low positive by Cobas (CT >30 for both targets) but negative only by Mayo LDT.
Table 1.
Comparison of results of the ART, Cobas, and Mayo LDT assays among consecutive anterior nares, nasopharyngeal, and oropharyngeal swab specimens obtained from persons under investigation for COVID-19.
| Consensus resulta | ART |
Cobas |
Mayo LDT |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Detected | TND | PPA | NPA | Detected | TND | PPA | NPA | Detected | TND | PPA | NPA | |
| AN swab | ||||||||||||
| ?>Detected | 5 | 0 | 100% | 4 | 1 | 80.0% | 5 b | 0 | 100% | |||
| ?>TND | 2 | 93 | 97.9% | 0 | 95 | 100% | 0 | 95 | 100% | |||
| NP swab | ||||||||||||
| ?>Detected | 26 | 0 | 100% | 24 | 2 | 92.3% | 24 b | 2 | 92.3% | |||
| ?>TND | 2 | 72 | 97.3% | 0 | 74 | 100% | 0 | 74 | 100% | |||
| OP swab | ||||||||||||
| ?>Detected | 18 | 0 | 100% | 17 | 0 | 100% | 16 b | 1 | 94.1% | |||
| ?>TND | 1 | 81 | 98.8% | 0 | 83 | 100% | 0 | 83 | 100% | |||
| NP+OP swab | ||||||||||||
| ?>Detected | 7 | 0 | 100% | 7 | 0 | 100% | 7 b | 0 | 100% | |||
| ?>TND | 1 | 92 | 98.8% | 0 | 93 | 100% | 3 | 90 | 96.8% | |||
AN = anterior nares; D = detected; NP = nasopharyngeal; NPA = negative percent agreement; OP = oropharyngeal; PPA = positive percent agreement; TND = Target not detected.
Consensus result for each specimen type is defined as the agreement result obtained from ≥2 of the 3 assays.
Data included 1, 2, 4, and 2 indeterminate results of Mayo LDT for AN, NP, OP, and NP+OP swab specimens, respectively.
Table 2.
Comparison of SARS-CoV-2 detection results among 90 positive nasopharyngeal swab specimens tested with ART, Cobas, and Mayo LDT.
| Comparator assay with “Detected” results | Reference assay method |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| ART |
Cobas |
Mayo LDT |
|||||||
| Detected | TND | PPA | Detected | TND | PPA | Detected | TND | PPA | |
| ART | 30 | 0 | 100% | 30 | 0 | 100% | |||
| Cobas | 27 a | 3 | 90.0% | 30 | 0 | 100% | |||
| Mayo LDT | 27 b | 3 | 90.0% | 29 c | 1 | 96.7% | |||
PPA, positive percent agreement; TND, target not detected.
Data included 1 indeterminate result for Cobas.
Data included 1 indeterminate result for Mayo LDT.
Data include 2 indeterminate results for Mayo LDT.
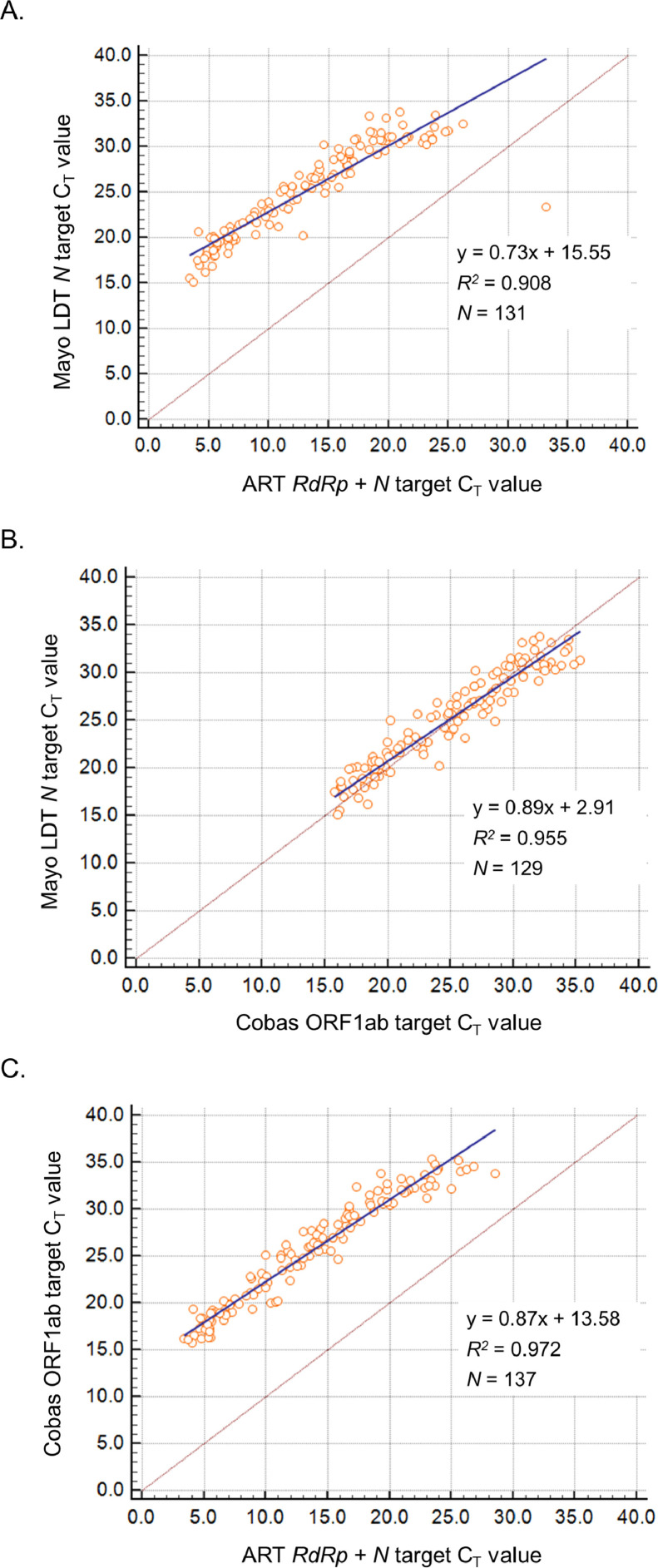
Correlation of the CT values of all positive specimen types by both assays compared was determined by Deming regression analysis for the 3 assays (Fig. 1 ). For ART, a single CT was generated for both amplified RdRp + N targets with the use of the same fluorophore for both probes, and first 10 amplification cycles were not counted by the signal analysis software for this assay. Cobas and Mayo LDT have two targets each that are measured in separate fluorescent channels, but only the CT values for ORF1ab and N targets were analyzed for Cobas and Mayo LDT, respectively. For Cobas, correlation of the E target CT values with those of ART and Mayo LDT (data not shown) are similar to the correlation observed for the Cobas ORF1ab target CT values, as the ORF1ab and E target CT values are very similar in all positive specimens tested with Cobas. Similar correlation patterns were observed for the Mayo LDT ORF1ab target (data not shown).
Fig. 1.
Correlation of cycle threshold (CT) values observed in the ART, Cobas, and Mayo LDT EUA assays. Correlation was determined by Deming regression analysis of paired CT values between Mayo LDT and ART (A), Mayo LDT and Cobas (B), and Cobas and ART (C). Regression equations, coefficients of determinate (R2), and total number of eligible specimens (N) are shown for each graph. Note that CT values for ART are lower because the signal analysis software was not programmed to count the first 10 amplification cycles.
4. Discussion
Multiple assays with different nuances may cause challenges in result interpretation. Complicating matters, there is no gold standard diagnosis of COVID-19, and therefore, clinical sensitivity and clinical specificity cannot readily be calculated. Our study supports previous publications showing variability in sensitivity among different specimen types tested on multiple platforms (Lee et al., 2021, Eberle et al., 2021). While other studies highlighted the challenges of utilizing multiple platforms, they compare neither the performance between ART and Cobas nor performance of an LDT that is not modified from the original CDC assay. Our study also compares a relatively large sample size of 4 different specimen types, while several other published studies compared only one specimen type (Liotti et al., 2021, Zhen et al., 2020, Zhen et al., 2020, Lowe et al., 2020).
Our study has several limitations; all of the 400 prospectively collected clinical specimens were submitted from reference testing laboratory clients without information on the duration of symptoms in the subjects tested. Since the clinical specimens were collected in May 2020 when FDA EUA were granted to assays for testing only PUI, one can only assume that all clinical specimens submitted for testing in our laboratory were collected from PUI. Furthermore, since the 4 specimen types (AN, NP, OP, NP+OP) tested across the 3 different assays in our study did not come from the same 100 subjects, we cannot conclude which specimen type was best for detection of SARS-CoV-2 by each of these 3 assays. Another limitation is that we did not have sufficient volume to retest specimens that had discrepant results. The discrepant results may be explained by several different reasons including extraction, possible contamination, or less sensitive limit of detection (LoD) in the Cobas and/or Mayo LDT assays. The LoD explanation is plausible for 12 out of 16 discrepant results. However, the remaining 4 discrepant results do not appear to be due to the LoD and could be due to the individual assay design of each target. Additional studies are in progress to evaluate detection rates among matched clinical samples and saliva from individual PUIs, as there is increasing interest in using saliva for noninvasive testing.
Overall, ART, Cobas, and Mayo LDT performed similarly in generating positive or negative results. Our data suggest that ART is the most analytically sensitive of these three assays, and this high sensitivity can likely be attributed to the assay design in which the same fluorophore is used on both TaqMan probes hybridizing to the viral RdRp and N gene sequences amplified in the assay. In contrast, the Mayo LDT and Cobas use separate fluorophores for the probes targeting different viral target sequences (ORF1ab and E sequences for Cobas; ORF1ab and N sequences for Mayo LDT). Our study data also highlight the finding that not all CT values can be compared across different real-time PCR assays to gauge relative viral load in the same or different clinical specimens obtained from a given individual. As the CT value is dependent on assay design and signal analysis software application, the same specimen tested will likely generate different CT values by different PCR assays.
Author contributions
JDY, JJG, MJB, and ARE contributed to the conception and design of the study. ACB, ECF, and JJG acquired the data. ARE and JDY analyzed the data. ARE and JDY drafted the article. All authors contributed to the revisions and gave final approval of the version submitted.
Acknowledgment
We thank the all staff of the laboratories in the Division of Clinical Microbiology and throughout the Department of Laboratory Medicine and Pathology for their continued dedication, especially during this pandemic.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.diagmicrobio.2021.115441.
Appendix. Supplementary materials
References
- U.S. Food and Drug Administration . 2021. COVID-19 Frequently Asked Questions.https://www.fda.gov/emergency-preparedness-and-response/coronavirus-disease-2019-covid-19/covid-19-frequently-asked-questions [10th May 2021] Available at: [Google Scholar]
- Mostafa HH, Hardick J, Morehead E, Miller JA, Gaydos CA, Manabe YC. Comparison of the analytical sensitivity of seven commonly used commercial SARS-CoV-2 automated molecular assays. J Clin Virol. 2020;130 doi: 10.1016/j.jcv.2020.104578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degli-Angeli E, Dragavon J, Huang ML, Lucic D, Cloherty G, Jerome KR. Validation and verification of the Abbott RealTime SARS-CoV-2 assay analytical and clinical performance. J Clin Virol. 2020;129 doi: 10.1016/j.jcv.2020.104474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abbott Molecular, Inc . 2020. Abbott RealTime SARS-CoV-2 assay instructions for use.https://www.fda.gov/media/136258/download [10th May 2021] Available at: [Google Scholar]
- Roche Molecular Systems, Inc . 2020. cobas® SARS-CoV-2: Qualitative assay for use on the cobas 6800/8800 Systems.https://www.fda.gov/media/136049/download [10th May 2021] Available at: [Google Scholar]
- Rodino KG, Espy MJ, Buckwalter SP, Walchak RC, Germer JJ, Fernholz E. Evaluation of saline, phosphate buffered saline and minimum essential medium as potential alternatives to viral transport media for SARS-CoV-2 testing. J Clin Microbiol. 2020;58(6) doi: 10.1128/JCM.00590-20. e00590-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayo Clinic Laboratories, Inc . 2020. SARS-CoV-2 Molecular Detection Assay EUA summary.https://www.fda.gov/media/137163/download [10th May 2021] Available at: [Google Scholar]
- Lee RA, Herigon JC, Benedetti A, Pollock NR, Denkinger CM. Performance of saliva, oropharyngeal swabs, and nasal swabs for SARS-CoV-2 molecular detection: a systematic review and meta-analysis. J Clin Microbiol. 2021;59(5) doi: 10.1128/JCM.02881-20. e02881-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberle U, Wimmer C, Huber I, Neubauer-Juric A, Valenza G, Ackermann N. Comparison of nine different commercially available molecular assays for detection of SARS-CoV-2 RNA. Eur J Clin Microbiol Infect Dis. 2021 doi: 10.1007/s10096-021-04159-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liotti FM, Menchinelli G, Marchetti S, Morandotti GA, Sanguinetti M, Posteraro B. Evaluation of three commercial assays for SARS-CoV-2 molecular detection in upper respiratory tract samples. Eur J Clin Microbiol Infect Dis. 2021;40:269–277. doi: 10.1007/s10096-020-04025-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhen W, Smith E, Manji R, Schron D, Berry GJ. Clinical evaluation of three sample-to-answer platforms for detection of SARS-CoV-2. J Clin Microbiol. 2020;58(8) doi: 10.1128/JCM.00783-20. e00783-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhen W, Manji R, Smith E, Berry GJ. Comparison of four molecular in vitro diagnostic assays for the detection of SARS-CoV-2 in nasopharyngeal specimens. J Clin Microbiol. 2020;58(8) doi: 10.1128/JCM.00743-20. e00743-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe CF, Matic N, Ritchie G, Lawson T, Stefanovic A, Champagne S. Detection of low levels of SARS-CoV-2 RNA from nasopharyngeal swabs using three commercial molecular assays. J Clin Virol. 2020;128 doi: 10.1016/j.jcv.2020.104387. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.