Abstract
Recombinant proteins are an essential milestone for a plethora of different applications ranging from pharmaceutical to clinical, and mammalian cell lines are among the currently preferred systems to obtain large amounts of proteins of interest due to their high level of post-translational modification and manageable large-scale production. In this regard, human embryonic kidney 293 (HEK293) cells constitute one of the main standard lab-scale mammalian hosts for recombinant protein production since these cells are relatively easy to handle, scale-up, and transfect. Here, we present a detailed protocol for the cost-effective, reproducible, and scalable implementation of HEK293 cell cultures in suspension (suitable for commercially available HEK293 cells, HEK293-F) for high-quantity recombinant production of secreted soluble multi-domain proteins. In addition, the protocol is optimized for a Monday-to-Friday maintenance schedule, thus simplifying and streamlining the work of operators responsible for cell culture maintenance.
Graphic abstract:
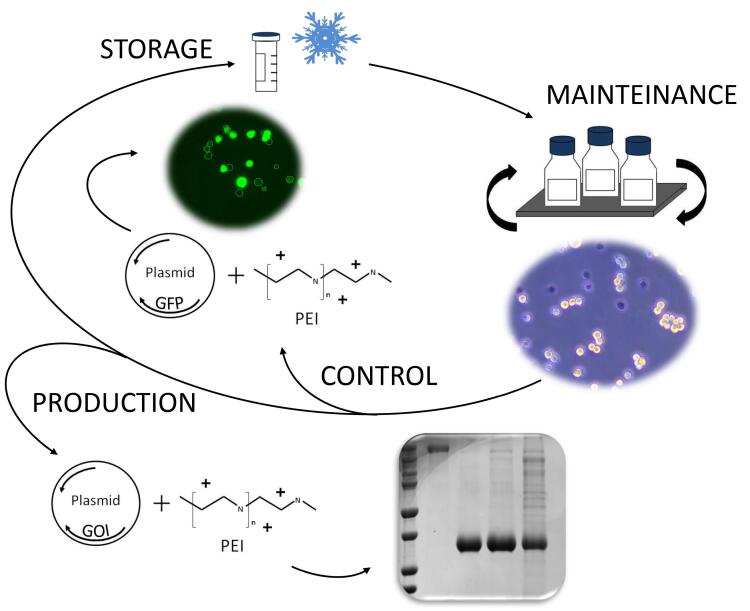
Schematic overview of the workflow described in this protocol
Keywords: Recombinant protein production, HEK293 cells, Suspension cultures, Extracellular proteins, Cell transfection, Polyethyleneimine
Background
Large amounts of high quality recombinant proteins are essential reagents for a variety of fields ranging from in vitro biochemical investigations to pharmaceutical and clinical applications ( Reeves et al., 2002 ; Olsen et al., 2003 ; Swiech et al., 2012 ; Forneris et al., 2017 ; Amanat et al., 2020 ). Mammalian cells are particularly preferred in this context due to their ability to perform complex post-translational modifications (PTMs) of proteins, and presently large-volume cultures of mammalian cells constitute a well-established methodology to obtain milligram-to-gram quantities of recombinant proteins (Wurm, 2004; Durocher et al., 2007 ; Chaudhary et al., 2011 ; Dyson, 2016; Arena et al., 2018 ). Among the most used mammalian cell lines, human embryonic kidney 293 (HEK293) cells have become the standard choice for research-scale production due to their relative ease of manipulation and high transfection efficiency (Thomas and Smart, 2005; Nettleship et al., 2015 ; Arena et al., 2018 ). The HEK293 cell line was derived in 1973 from HEK cells grown in tissue culture by the introduction of a sheared adenovirus 5 DNA into wild type cells, resulting in a partially dysregulated cell cycle and subsequent immortalization of the cell line ( Graham et al., 1977 ).
Although the literature offers many protocols to efficiently obtain high quantities of proteins of interest using mammalian cells as a recombinant host ( Aricescu et al., 2006 ; Durocher et al., 2007 ; Aricescu and Owens, 2013; Portolano et al., 2014 ; Arena et al., 2018 ; L’ Abbé et al., 2018 ), the difficulties associated with establishing scalable and reproducible setups in a cost-effective manner remain evident. Here, we present the optimized protocols, currently in use in our lab, for the maintenance and transfection of HEK293-F cells, a commercially available HEK293 cell line suitable for high-density culture and transient transfection in suspension ( Nettleship et al., 2015 ). This protocol allows the successful production of a variety of challenging recombinant protein targets, as described in our previous publications ( Banushi et al., 2016 ; Scietti et al., 2018 ; Angiolini et al., 2019 ; Chiapparino et al., 2020 ). Our protocol offers a comprehensive description of all the procedures needed to establish and operate a recombinant protein production facility using these cells in an affordable but highly reproducible lab-scale setting. The proposed optimized Monday-to-Friday schedule further facilitates the lab procedures without the need for monitoring cells during the weekend. Each section describes the step-by-step procedures for cell handling, in addition to a series of technical notes to facilitate troubleshooting and improve reproducibility.
Materials and Reagents
Pipette tips (Sarstedt, catalog numbers: 70.1130 [10 ml], 70.760.002 [200 ml], 70.762 [1,000 ml]), to be autoclaved before use
Serological pipettes (Sarstedt, catalog numbers: 86.1685.001 [25 ml], 86.1254.001 [10 ml], 86.1253.001 [5 ml], 86.1252.001 [2 ml])
Polycarbonate square-bottomed bottles (Triforest Labware, catalog numbers: BCP0250 [250 ml], BCP0500 [500 ml], BCP2000 [2 L])
6-well polystyrene plates (Sarstedt, catalog number: 83.3920)
Filtropur 0.2 μm sterile filters (Sarstedt, catalog number: 83.1826.00)
1.6 ml CryoPure tubes (Sartstedt, catalog number: 72.380.992)
50 ml syringe without needle, luer-lock (VWR, catalog number: TERUSS-50L1)
10 ml syringe without needle, luer-lock (VWR, catalog number: 613-0973)
HEK293-F cells (Gibco, catalog number: R79007)
Freestyle 293 Expression Medium (Gibco, catalog number: 12338-026)
Penicillin-Streptomycin (Merck, catalog number: P0781)
Trypan Blue (Merck, catalog number: T8154; starting concentration: 0.4%, dilute 1:1 with PBS to reach a final working concentration of 0.2%)
Dimethylsulfoxide (Sigma-Aldrich, catalog number: D8414)
PEI MAX polyethyleneimine 40K powder (Polysciences, catalog number: 24765-1)
Primatone RL (Sigma Aldrich, catalog number: P4963)
OptiMEM I (1×) + Glutamax-1 (Gibco, catalog number: 51985-034)
pTT3-eGFP (Addgene, catalog number: 154346)
Equipment
Microvolume pipettes (Gilson, models: Pipetman P1000, P200, P100, P20)
Automated pipettor for serological pipettes (VWR, model: Pipetboy pro Integra)
BSL-2 laminar flow hood (Thermo Fisher, model: MSC Advantage)
Large-volume benchtop centrifuge with swing-out rotor (Beckman Coulter, model: Allegra X-15R with SX4750 rotor)
CO2 incubator with embedded shaker (Eppendorf, model: New Brunswick S41i, catalog number: S41I230011)
–80°C ultrafreezer (Thermo Fisher, model: TDE 300FV)
CoolCell™ controlled rate freezing apparatus (VWR, catalog number: 479-0492)
Water bath (Exacta Optech, model: WNB22)
Neubauer chamber (Merck, catalog number: BR717820)
Automated cell counter (DeNovix Inc., model: Celldrop)
Liquid nitrogen storage tank (Taylor-Wharton, model: VHC-35)
Procedure
All procedures described herein are performed with HEK293-F cells, which are maintained in Freestyle 293 Expression Medium, i.e., a chemically defined, protein-free medium optimized for growth and recombinant protein production in HEK293-F cells kept in suspension culture. Maintenance medium is supplemented with penicillin-streptomycin at a final concentration of 0.1% (v/v). All cell handling procedures are carried out under sterile conditions in a standard BSL-2 laminar flow hood. Cells are cultured in suspension inside polycarbonate square-bottomed bottles of different volumes or in multi-well polystyrene plates. When required, cells are centrifuged using a benchtop centrifuge at 1,000 × g for 10 min at room temperature. Scrupulous monitoring of the incubation settings is required since small changes will strongly affect both cell growth and transfection efficiency. CO2 supply, shaking orbit, and shaking speed are the most critical parameters. In particular, the latter is adjusted to prevent aggregation but at the same time avoid cell death caused by excess speed or accumulation of cell debris at the air–medium interface. In our setting, HEK293-F cells are maintained in New Brunswick S41i shaking incubators set at 37°C with 5% CO2. These incubators have a shaking orbit of 2.5 cm: after extensive benchmarking for identification of the best compromise between cell viability and aggregation, we noticed that a shaking speed between 120 and 130 rpm was optimal for HEK293-F cell growth in both bottles and flat-bottomed multi-well plates. The various sections describe methods that, in our opinion, may also be used for specific maintenance/transfection of other mammalian cell lines; however, this has not been tested thoroughly as with HEK293-F cells.
-
Cell thawing and culture initiation
-
Pre-warm the Freestyle 293 medium supplemented with 0.1% penicillin-streptomycin (hereafter referred to as Medium) to 37°C in a water bath (for a 1 L bottle, this may take a couple of hours).
Note: Although the addition of antibiotic to the Medium is generally not recommended, penicillin-streptomycin concentrations up to 10 ml/L (equal to 1%) are compatible with cell cultures and, based on several tests carried out using different antibiotic concentrations (0 ml/L–10 ml/L), do not impact cell growth or transfection efficiency. According to these results, we recommend supplementing the cell cultures with 1 ml/L (0.1%) penicillin-streptomycin to minimize the risk of contamination.
Rapidly thaw an aliquot of frozen HEK293-F cells (each aliquot should contain approximately 10 × 106 cells/ml) in the 37°C water bath. This operation should take less than 1 min.
-
Directly dispense the cells into a 250 ml bottle containing 20 ml Medium to reach a final concentration of 0.5 × 106 cells/ml.
Note: HEK293-F cells are extremely sensitive and prone to contamination during the thawing step; therefore, the greatest care must be taken during this step. It is strongly advised to aspirate the entire cell volume from the frozen vial in a single pipetting operation and dispense the cells directly into the pre-warmed Medium in the culture bottle. Any further manipulation can increase the risk of contamination.
Incubate the cells at 37°C, 5% CO2 with 120 rpm shaking, taking care to loosen the bottle cap to allow gas exchange.
-
Check the cell growth rate daily and assess the viability using the Trypan Blue assay as follows:
Harvest approximately 1 ml cells from the bottle.
Dilute 15 µl cells 1:1 with 0.2% Trypan Blue.
-
Count live cells using a Neubauer chamber (or an automatic cell counter, Figure 1) and assess cell viability using the following formula:
Mean number of cells counted in 4 squares × 2 (dilution factor) × 104
Note: Under good culture conditions, suspension-cultured HEK293-F cells display an average doubling time of 24 h; however, 24 h after thawing, the cells typically do not show any signs of duplication. Starting from 48 h post-thawing, the cells should duplicate every 24 h, reaching a final concentration of approximately 2 × 106 cells/ml on day 3. At this timepoint, it is recommended to replace the Medium by centrifuging the cells at 1,000 × g for 10 min at room temperature and resuspending in fresh Medium.
72 h after thawing, the cells should have reached a final concentration of approximately 2 × 106 cells/ml: harvest the cells by centrifugation at 1,000 × g for 10 min at room temperature.
Discard the supernatant and gently resuspend the cell pellet in fresh, pre-warmed Medium using a volume sufficient to reach a final concentration of 0.5 × 106 cells/ml.
-
Culture the cells in the shaking incubator using the same settings as stated in Step A4, following the schedule for cell maintenance described in Procedure B.
Note: An effective weekly schedule suggestion includes thawing cells on a Monday (day 0), as this will allow the operator to monitor cell growth (as well as possible contamination) daily throughout the first week. Day 3 (72 h after thawing) should therefore correspond to a Thursday, on which cells should be split to a final concentration of 0.5 × 106 cells/ml. Splitting again the following day (Friday) will guarantee exponential growth and optimal cell density over the weekend without intervention from the operator.
-
-
Cell maintenance
Subculture the cells three times a week as follows:
-
Every Monday and Wednesday, count the cells and calculate the total cell number and viability as described in Step A5.
Note: Avoid daily cell splitting: in our experience, this will drastically slow down cell growth and eventually result in a significant reduction in transfection efficiency.
-
Split cells into a new bottle. Seed cells at a final concentration of 0.5 × 106 cells/ml in the required volume of Medium following the guidelines suggested in Table 1.
Notes:
In our experience, the optimal seeding concentration for HEK293-F cells is 2 × 106 cells/ml. It is detrimental to allow the cells to overgrow since they consume all the nutrients in the Medium and dead cell debris accumulates, posing a problem for subsequent transfection procedures. Cell debris can be easily visualized as the formation of a sticky ring around the walls of the culture bottle (Figure 2) .
A fundamental parameter to consider when performing cell splitting is the air/Medium ratio used for cell culture. HEK293-F cells are extremely sensitive to this parameter: we recommend filling the culture bottles with Medium to no more than a quarter of their total volume capacity to ensure proper oxygen exchange and prevent excess aggregation. At the same time, too low a culture volume will cause cell death due to increased mechanical stress induced by shaking, which may be solved by reducing the shaking speed but may interfere with the need to standardize the incubator settings to enable multiple cultures at the same time. The suggested optimal culture volumes (Table 1) were selected after extensive testing based on the assessment of cell viability and accumulation of cell aggregates and debris using bottles of several sizes and shapes, in addition to multi-well plates, with the incubator settings as described in Step A4 .
When splitting cells, it is not necessary to perform washes with PBS or other buffers to remove cell debris. Such operations are typically avoided in this protocol to limit the risk of contamination and minimize stressful operations related to cell handling.
Grow the cells in the shaking incubator using the same settings as described in Step A4.
-
Every Friday, count the cells and calculate the total cell number and viability as described in Step A5.
Note: Although HEK293-F are immortalized cells, we observed a sort of senescence, with a drastic reduction in the transfection efficiency of the old batches as compared with the newer ones. This may negatively impact the yield of recombinant protein production, in particular for recombinant targets with production yields below 1 mg/L using “fresh” HEK293-F cells. For this reason, we encourage the initiation of a new cell culture batch by thawing a fresh aliquot of HEK293-F cells at passage 25–30 (i.e., every 11 weeks). This will ensure that the cells subject to transient transfection will never be older than passage 30–32.
-
Split cells into a new bottle, seeding them at a final concentration of 0.3 × 106 cells/ml in the required volume of Medium.
Note: It is strongly recommended to split the cells to concentrations no lower than 0.3 × 106 cells/ml, as we noticed that this may eventually lead to a drastic decrease in transfection efficiency even when the cells appear to grow normally.
Culture the cells in the shaking incubator using the same settings as described in Step A4.
-
-
Cell cryopreservation
HEK293-F cells should be stored in liquid nitrogen. Prepare new aliquots of HEK293-F cells for cryopreservation as follows:
Seed at least 100 ml cells from a maintenance stock as described in Procedure B at a final concentration of 0.5 × 106/ml and culture them as described in Step A4.
-
After 24 h, count the cells as indicated in Step A5 and calculate the total cell number and viability: the cell concentration should have reached 1 × 106/ml.
Note: HEK293-F cells are extremely sensitive to the concentration used for cryopreservation. We carried out extensive testing using multiple cell concentrations to assess the quality of the reconstituted HEK293-F cell batches after thawing. We noticed a remarkable increase in cell viability and transfection efficiency when the cells were frozen at a concentration of 10 × 106 cells/ml. When using higher cell concentrations in cryostocks, we did not observe changes in duplication rates but did see significantly lower transfection efficiencies. For this reason, we strongly recommend not to exceed this cell concentration.
Freshly prepare the freezing medium composed of Medium supplemented with 10% DMSO. Filter the freezing medium through a 0.2 µm filter.
Harvest the cells by centrifugation at 1,000 × g for 10 min at room temperature, discard the supernatant, and gently resuspend the cell pellet in the freezing medium to a final concentration of 10 × 106 cells/ml.
Rapidly aliquot 1 ml suspension into the cryovials and transfer to a pre-conditioned controlled-rate freezing apparatus to progressively lower the cell temperature by 1°C per min.
Transfer the cell aliquots to –80°C for 24 h.
The following day, transfer the cells to the liquid nitrogen tank for long-term storage.
-
Preparation of polyethyleneimine for cell transfection
Discovered in the early 1970s for its ability to precipitate DNA (Atkinson and Jack, 1973), polyethyleneimine (PEI) was shown 20 years later to be capable of delivering genetic material into mammalian cell lines ( Boussif et al., 1995 ; Goula et al., 1998 ; Pollard et al., 1998 ; Ringenbach et al., 1998 ; Longo et al., 2013 ). This polymer has a high density of positive charge that allows it to condense DNA molecules. The DNA–PEI complexes have a net positive charge and can bind to the cell membrane by interacting non-specifically with negatively charged glycoproteins, proteoglycans, and sulfated proteoglycans located on the cell surface ( Pham et al., 2006 ).
In our protocol, we use linear polyethyleneimine (PEI) due to its ease of preparation, long-term stability, low cost, high transfection efficiency, and very low cell toxicity.
A 1 mg/ml PEI solution (1× stock) can be prepared as follows:
Dissolve 100 mg PEI MAX in ultrapure water in a 100 ml glass beaker by gently stirring.
-
Carefully add 1 M NaOH dropwise to the PEI solution until the final pH is between 6.90 and 7.10.
Note: Immediately after adding the water, the pH of the PEI MAX solution will be around 2. Due to the lack of a buffering component in the solution, titration with 1 M NaOH should be carried out very carefully to reach neutral conditions.
Transfer the PEI solution into a graduated cylinder and adjust the final volume to 100 ml to reach a final concentration of 1 mg/ml.
Sterile-filter the solution using a 0.2 µm filter.
-
Aliquot the solution into 500 µl stocks and store at 4°C.
Note: PEI MAX solutions are stable for roughly 6 months. We recommend checking the transfection efficiencies periodically and preparing fresh PEI MAX solution when required.
-
Preparation of protein hydrolysate supplement for cell transfection
Supplementation of peptones (protein hydrolysates) to the cell culture media after transfection is known to produce a significant increase in production yield, especially for secreted proteins ( Pham et al., 2003 ; Pham et al., 2005 ). In our setup, we found that supplementation of Primatone RL, a meat protein enzymatic hydrolysate, boosts the recombinant protein production by an average of 2.5-fold.
A 6% Primatone RL (10× stock) solution can be prepared as follows:
Prepare a 100 ml aliquot of Medium in the hood.
-
Take the Medium aliquot out of the hood and dissolve 6 g Primatone RL.
Note: During Primatone RL preparation, special care should be taken to avoid contamination. Since this compound consists of a very thin powder, it is strongly recommended not to pour the Medium directly on it from the stock, but instead to prepare a dedicated aliquot in a separate container and dissolve the Primatone powder outside the sterile environment.
Vigorously vortex the solution to completely dissolve the meat hydrolysate.
-
Place the Primatone solution in the hood, sterile-filter using a 0.2 µm filter, and store at 4°C.
Note: Use the solution within one month.
The final working concentration (0.6%) should be added to the cell cultures at least 4 h after transfection.
-
Cell transfection
The following protocol was standardized for basic transient cell transfection for the recombinant production of secreted extracellular protein targets:
24 h prior to cell transfection, count the cells 1:1 as indicated in Step A5 and calculate the total cell number and viability.
-
Seed the cells in the appropriate container at a final concentration of 0.5 × 106/ml, adjusting the volume according to Table 1.
Note: It is strongly recommended to seed the cells at 0.5 × 106/ml and further incubate for at least an additional 24 h prior to transient transfection. As discussed in (B), avoid two consecutive cell splits within 24 h as this will strongly impact transfection efficiency.
Incubate the cells as described in Step A4.
The following day, count the cells as indicated in Step A5, and calculate the total cell number and viability (cells should have doubled and their final concentration should have reached 1 × 106/ml).
-
Prepare the transfection mix as follows:
Use 1 µg pure plasmid DNA for every 1 × 106 cells.
Use a 1:5 DNA:PEI MAX ratio.
-
Calculate the final transfection volume (in µl) as follows:
Transfection volume = total µg DNA × 25 µl/µg
Note: We performed several experiments to elucidate the optimal volume for cell transfection. We established that 25 µl per µg DNA represents a good ratio for an efficient transfection mix.
Prepare two different sterile containers, each containing half the calculated transfection volume of OptiMEM I (1×) + Glutamax-1 medium.
Dispense the plasmid DNA into one container and PEI MAX into the other.
Transfer the DNA–OptiMEM solution dropwise into the PEI–OptiMEM solution to obtain the transfection mix.
Vortex the transfection mix for 5 s, three times.
-
Incubate the transfection mix at room temperature for 10 min.
Note: The incubation time of the DNA–PEI mix prior to transient transfection may also heavily impact protein production yield. We recommend testing different incubation times (5, 10, and 15 min) in a small-scale expression trial of the protein target of interest.
Add the transfection mix dropwise to the cell suspension while gently shaking the culture bottle in a circular motion.
-
4 h post-transfection, add Primatone RL, prepared as previously described, at a final concentration of 0.6%.
Note: Ensure that the transfection procedure is performed in the morning: this will allow the addition of Primatone RL (Step F9) during working hours.
Incubate the transfected cells as described in Step A4 for the appropriate number of days in accordance with the recombinant target protein of interest.
-
Periodically check cell viability (if cell viability drops below 70%, this may indicate that the recombinant protein is toxic and may therefore require early harvesting).
Notes:
Proper cell harvesting time is of fundamental importance when producing recombinant proteins in HEK293-F cells. Our rule-of-thumb based on extensive empirical testing is that for secreted recombinant protein targets, the optimal harvesting time for maximum protein yield is 6–7 days post-transfection. Such an extended culture time is not recommended in the case of intracellular or transmembrane proteins, which are typically harvested between 48 h and 72 h post-transfection. Nevertheless, continuous monitoring of cell viability is mandatory, particularly for new targets, to prevent the accumulation of cell debris in the Medium caused by unexpected toxicity associated with recombinant protein production. We noticed that, depending on the plasmid used for transfection, HEK293-F cells can be particularly resilient to the presence of the plasmid and may still continue to grow after the transfection procedure.
A negative control experiment for the comparative assessment of cell viability can be performed by transfecting cells with a plasmid lacking a mammalian cell promoter, e.g., a “dummy” DNA cloning vector.
-
6 days after transfection, count the cells 1:1 as indicated in Step A5 and calculate the total cell number and viability.
Note: For an optimized Monday-to-Friday schedule, organize the transfection days for Tuesday and Thursday: this will allow both splitting the cells for transfection (Step F1) and harvesting at 6–7 days post-transfection (Step F12) on weekdays.
Transfer the cell culture to conical centrifuge tubes and harvest the culture media by centrifugation at 1,000 × g for 15 min at 10°C.
Keep the supernatant and proceed according to your protein purification protocol.
-
Evaluation of transfection efficiency using eGFP
As discussed in (B), HEK293-F cells become senescent after a number of passages; therefore, it is essential to periodically monitor the transfection performance. We recommend checking transfection efficiency with a positive control at least twice a month. Mock transfections using expression plasmids containing the gene for enhanced green fluorescent protein (eGFP, e.g., pTT3-eGFP) can be used for this purpose. Measurement of the transfection efficiency evaluates the number of cells that have successfully incorporated the recombinant DNA and are therefore capable of producing the recombinant protein(s) of interest. The ratio between the number of cells displaying green fluorescence and the total number of cells yields the transfection efficiency value (Figure 3).
To perform the control transfection with eGFP, follow the protocol steps (Steps F1–F8), then:
Incubate the transfected cells as described in Step A4 for 72 h.
-
Observe the cells under the microscope and collect images using both brightfield and green fluorescence.
Note: When using an automatic cell counter instead of manual counting with a Neubauer chamber, use an untransfected sample to set the threshold for GFP signal detection in the instrument. The advantage of the CellDrop™ automatic cell counter is that it does not require any consumables, such as slides, typically used by other systems.
-
Transfection efficiency is calculated by counting the green cells (blue light imaging) and the total cells (white light imaging), and applying the following formula:
Transfection efficiency % = ((total number of green cells)/(total cell number)) × 100
The expected efficiency should be between 65% and 80%. Lower transfection efficiencies may suggest problems with some of the reagents or indicate that the cell batch may need to be replaced.
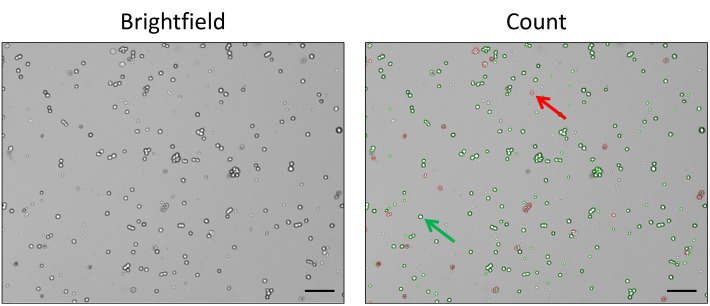
Figure 1. Example of automatic cell counting.
Comparison of cells after treatment with Trypan Blue counted using the CellDrop™ automatic cell counter (Denovix Inc.). The brightfield image (left) was processed by the DenovixEasyApps software for identification of live cells (green circles, as highlighted by the green arrow, total count in the image = 3.17 × 106/ml) and dead cells (red circles, as highlighted by the red arrow, total count in the image = 5.75 × 105/ml). Scale bar, 100 µm.
Table 1. Recommended cell culture volumes and associated containers.
Overview of the minimal, maximal, and optimal cell culture volumes in different containers used for cell culture.
| Container type | Minimal volume | Maximal volume | Optimal volume |
| 250 ml bottle | 10 ml | 50 ml | 25 ml |
| 500 ml bottle | 50 ml | 100 ml | 80 ml |
| 2 L bottle | 100 ml | 500 ml | 250 ml |
| 6-well plate | 2 ml | 3 ml | 2 ml |
Figure 2. Sticky ring of dead cells.
The formation of a sticky ring on the lower part of the bottle constitutes the typical presentation of cell debris accumulation.
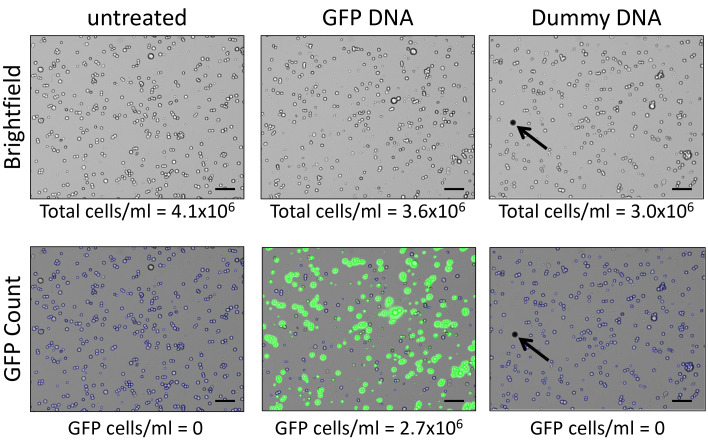
Figure 3. Evaluation of transfection efficiency.
Comparison between untreated cells (left) and those transfected with either GFP (center) or dummy DNA (right). For each sample, the comparative evaluation was performed using the CellDrop™ automatic cell counter (Denovix Inc.). The brightfield images (top) were subjected to GFP fluorescence counting (excitation = 470 nm ± 20 nm; emission = 525 nm ± 25 nm) using the DenovixEasyApps software. When unexpected objects different from cells are present in the counting chamber (e.g., air bubbles, as indicated by the black arrow in the right panel), the instrument automatically excludes them from the final cell count. Scale bar, 100 µm.
Acknowledgments
We thank Dr. F. Magnani (University of Pavia) and Dr. S. Wulhfard for useful discussions and suggestions, in addition to all members of the Armenise-Harvard laboratory of Structural Biology for continuous support, testing, and benchmarking of protein production conditions.
This project received funding from Fondazione Giovanni Armenise-Harvard (grant id. CDA2013 to FF), the Italian Association for Cancer Research (AIRC, “My First AIRC Grant” id. 20075 to FF), Fondazione Cariplo (grant ids. 2014-0881 and 2015-0768 to FF), the Mizutani Foundation for Glycoscience (grant id. 200039), the Italian Ministry of Education, University and Research (MIUR) (Rita Levi-Montalcini Award 2012 to FF), and Dipartimenti di Eccellenza Program (grant id. 2018-2022, to the Dept. of Biology and Biotechnology "L. Spallanzani", University of Pavia). AC2 was the recipient of a Marie Curie individual fellowship from the European Union’s Horizon 2020 research and innovation program (grant agreement COTETHERS - n. 745934). None of the funding sources played a role in study design, collection, analysis, and interpretation of data, writing of the report, or the decision to submit this article for publication.
Author Contributions: FF supervised the set-up and development of the cell culture infrastructure. MC established the initial cell culture setup, with support from MP and AC1. SF and AC2 optimized the culture setup and standardized the culture and transfection procedures. SF carried out the comparative analysis presented in this work and elaborated on the proposed Monday-to-Friday schedule. SF and FF organized the protocol sections and wrote the paper, with contributions from all authors.
Competing interests
The authors declare that they have no known competing financial interests or personal relationships that could have influenced the work reported in this paper.
Citation
Readers should cite both the Bio-protocol article and the original research article where this protocol was used.
References
- 1. Amanat F., Stadlbauer D., Strohmeier S., Nguyen T. H. O., Chromikova V., McMahon M., Jiang K., Arunkumar G. A., Jurczyszak D., Polanco J., Bermudez-Gonzalez M., Kleiner G., Aydillo T., Miorin L., Fierer D. S., Lugo L. A., Kojic E. M., Stoever J., Liu S. T. H., Cunningham-Rundles C., Felgner P. L., Moran T., García-Sastre A., Caplivski D., Cheng A. C., Kedzierska K., Vapalahti O., Hepojoki J. M., Simon V. and Krammer F.(2020). A serological assay to detect SARS-CoV-2 seroconversion in humans. Nature Medicine 26(7): 1033-1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Angiolini F., Belloni E., Giordano M., Campioni M., Forneris F., Paronetto M. P., Lupia M., Brandas C., Pradella D., Di Matteo A., Giampietro C., Jodice G., Luise C., Bertalot G., Freddi S., Malinverno M., Irimia M., Moulton J. D., Summerton J., Chiapparino A., Ghilardi C., Giavazzi R., Nyqvist D., Gabellini D., Dejana E., Cavallaro U. and Ghigna C.(2019). A novel L1CAM isoform with angiogenic activity generated by NOVA2-mediated alternative splicing. Elife 8: e44305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Arena T. A., Harms P. D. and Wong A. W.(2018). High Throughput Transfection of HEK293 Cells for Transient Protein Production. Methods Mol Biol 1850: 179-187. [DOI] [PubMed] [Google Scholar]
- 4. Aricescu A. R., Lu W. and Jones E. Y.(2006). A time- and cost-efficient system for high-level protein production in mammalian cells. Acta Crystallogr D Biol Crystallogr 10): 1243-1250. [DOI] [PubMed] [Google Scholar]
- 5. Aricescu A. R. and Owens R. J.(2013). Expression of recombinant glycoproteins in mammalian cells: towards an integrative approach to structural biology. Curr Opin Struct Biol 23(3): 345-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Atkinson A. and Jack G. W.(1973). Precipitation of nucleic acids with polyethyleneimine and the chromatography of nucleic acids and proteins on immobilised polyethyleneimine. Biochim Biophys Acta 308(7): 41-52. [DOI] [PubMed] [Google Scholar]
- 7. Banushi B., Forneris F., Straatman-Iwanowska A., Strange A., Lyne A. M., Rogerson C., Burden J. J., Heywood W. E., Hanley J., Doykov I., Straatman K. R., Smith H., Bem D., Kriston-Vizi J., Ariceta G., Risteli M., Wang C., Ardill R. E., Zaniew M., Latka-Grot J., Waddington S. N., Howe S. J., Ferraro F., Gjinovci A., Lawrence S., Marsh M., Girolami M., Bozec L., Mills K. and Gissen P.(2016). Regulation of post-Golgi LH3 trafficking is essential for collagen homeostasis. Nat Commun 7: 12111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Boussif O., F. Lezoualc'h, Zanta M. A., Mergny M. D., Scherman D., Demeneix B. and Behr J. P.(1995). A versatile vector for gene and oligonucleotide transfer into cells in culture and invivo: polyethylenimine . Proc Natl Acad Sci U S A 92(16): 7297-7301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chaudhary S., Pak J. E., Pedersen B. P., Bang L. J., Zhang L. B., Ngaw S. M., Green R. G., Sharma V. and Stroud R. M.(2011). Efficient expression screening of human membrane proteins in transiently transfected Human Embryonic Kidney 293S cells. Methods 55(4): 273-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chiapparino A., De Giorgi F., Scietti L., Faravelli S., Roscioli T. and Forneris F.(2020). A cooperative network of molecular“hot spots” highlights the complexity of LH3 collagen glycosyltransferase activities. bioRxiv: 841486. [Google Scholar]
- 11. Durocher Y., Pham P. L., St-Laurent G., Jacob D., Cass B., Chahal P., Lau C. J., Nalbantoglu J. and Kamen A.(2007). Scalable serum-free production of recombinant adeno-associated virus type 2 by transfection of 293 suspension cells. J Virol Methods 144(1-2): 32-40. [DOI] [PubMed] [Google Scholar]
- 12. Dyson M. R.(2016). Fundamentals of Expression in Mammalian Cells. Adv Exp Med Biol 896: 217-224. [DOI] [PubMed] [Google Scholar]
- 13. Forneris F., Canciani A., Ciossani G., Palamini M., Chiapparino A., Gabrieli P., Guarino S. R., Campioni M., Nenci S. and Magnani F.(2017). Versatile medium-throughput strategies for recombinant expression screening in structural biology. Acta Crystallographica Section A 73(a2): C1276. [Google Scholar]
- 14. Goula D., Benoist C., Mantero S., Merlo G., Levi G. and Demeneix B. A.(1998). Polyethylenimine-based intravenous delivery of transgenes to mouse lung. Gene Ther 5(9): 1291-1295. [DOI] [PubMed] [Google Scholar]
- 15. Graham F. L., Smiley J., Russell W. C. and Nairn R.(1977). Characteristics of a human cell line transformed by DNA from human adenovirus type 5. J Gen Virol 36(1): 59-74. [DOI] [PubMed] [Google Scholar]
- 16. D. L’Abbé, Bisson L., Gervais C., Grazzini E. and Durocher Y.(2018). Transient Gene Expression in Suspension HEK293-EBNA1 Cells. In: Recombinant Protein Expression in Mammalian Cells: Methods and Protocols. Hacker,D. L.(Ed.). New York, NY, Springer New York: 1-16. [DOI] [PubMed] [Google Scholar]
- 17. Longo P. A., Kavran J. M., Kim M. S. and Leahy D. J.(2013). Transient mammalian cell transfection with polyethylenimine(PEI). Methods Enzymol 529: 227-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Nettleship J. E., Watson P. J., Rahman-Huq N., Fairall L., Posner M. G., Upadhyay A., Reddivari Y., Chamberlain J. M., Kolstoe S. E., Bagby S., Schwabe J. W. and Owens R. J.(2015). Transient expression in HEK 293 cells: an alternative to E. coli for the production of secreted and intracellular mammalian proteins . Methods Mol Biol 1258: 209-222. [DOI] [PubMed] [Google Scholar]
- 19. Olsen D., Yang C., Bodo M., Chang R., Leigh S., Baez J., Carmichael D., Perala M., Hamalainen E. R., Jarvinen M. and Polarek J.(2003). Recombinant collagen and gelatin for drug delivery. Adv Drug Deliv Rev 55(12): 1547-1567. [DOI] [PubMed] [Google Scholar]
- 20. Pham P. L., Kamen A. and Durocher Y.(2006). Large-scale transfection of mammalian cells for the fast production of recombinant protein. Mol Biotechnol 34(2): 225-237. [DOI] [PubMed] [Google Scholar]
- 21. Pham P. L., Perret S., Cass B., Carpentier E., St-Laurent G., Bisson L., Kamen A. and Durocher Y.(2005). Transient gene expression in HEK293 cells: peptone addition posttransfection improves recombinant protein synthesis. Biotechnol Bioeng 90(3): 332-344. [DOI] [PubMed] [Google Scholar]
- 22. Pham P. L., Perret S., Doan H. C., Cass B., St-Laurent G., Kamen A. and Durocher Y.(2003). Large-scale transient transfection of serum-free suspension-growing HEK293 EBNA1 cells: peptone additives improve cell growth and transfection efficiency. Biotechnol Bioeng 84(3): 332-342. [DOI] [PubMed] [Google Scholar]
- 23. Pollard H., Remy J. S., Loussouarn G., Demolombe S., Behr J. P. and Escande D.(1998). Polyethylenimine but not cationic lipids promotes transgene delivery to the nucleus in mammalian cells. J Biol Chem 273(13): 7507-7511. [DOI] [PubMed] [Google Scholar]
- 24. Portolano N., Watson P. J., Fairall L., Millard C. J., Milano C. P., Song Y., Cowley S. M. and Schwabe J. W.(2014). Recombinant protein expression for structural biology in HEK 293F suspension cells: a novel and accessible approach. J Vis Exp(92): e51897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Reeves P. J., Callewaert N., Contreras R. and Khorana H. G.(2002). Structure and function in rhodopsin: high-level expression of rhodopsin with restricted and homogeneous N-glycosylation by a tetracycline-inducible N-acetylglucosaminyltransferase I-negative HEK293S stable mammalian cell line. Proc Natl Acad Sci U S A 99(21): 13419-13424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ringenbach L., Bohbot A., Tiberghien P., Oberling F. and Feugeas O.(1998). Polyethylenimine-mediated transfection of human monocytes with the IFN-gamma gene: an approach for cancer adoptive immunotherapy. Gene Ther 5(11): 1508-1516. [DOI] [PubMed] [Google Scholar]
- 27. Scietti L., Chiapparino A., De Giorgi F., Fumagalli M., Khoriauli L., Nergadze S., Basu S., Olieric V., Cucca L., Banushi B., Profumo A., Giulotto E., Gissen P. and Forneris F.(2018). Molecular architecture of the multifunctional collagen lysyl hydroxylase and glycosyltransferase LH3. Nat Commun 9(1): 3163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Swiech K., Picanco-Castro V. and Covas D. T.(2012). Human cells: new platform for recombinant therapeutic protein production. Protein Expr Purif 84(1): 147-153. [DOI] [PubMed] [Google Scholar]
- 29. Thomas P. and Smart T. G.(2005). HEK293 cell line: a vehicle for the expression of recombinant proteins. J Pharmacol Toxicol Methods 51(3): 187-200. [DOI] [PubMed] [Google Scholar]
- 30. Wurm F. M.(2004). Production of recombinant protein therapeutics in cultivated mammalian cells. Nat Biotechnol 22(11): 1393-1398. [DOI] [PubMed] [Google Scholar]