Abstract
Lysogenic phages can integrate into their bacterial host’s genome, potentially transferring any genetic information they possess including virulence or resistance genes, and are therefore routinely excluded from therapeutic applications. Lysogenic behavior is typically seen in phages that create turbid plaques or possess subpar bactericidal activity; yet, these are not definitive indicators. As a result, the presence of integrase genes is often used as a hallmark for lysogenic behavior; however, the accuracy of genetic screening for lysogeny depends on the quality of the extraction, sequencing and assembly of the phage genome, and database comparison. The present protocol describes a simple phenotypic test that can be used to screen therapeutically relevant phages for lysogenic behavior. This test relies on the identification of spontaneous phage release from their lysogenized host and can be reliably used in cases where no sequencing data are available. The protocol does not require specialized equipment, is not work-intensive, and is broadly applicable to any phage with an easily culturable bacterial host, making it particularly amenable to settings with limited resources.
Graphical abstract:
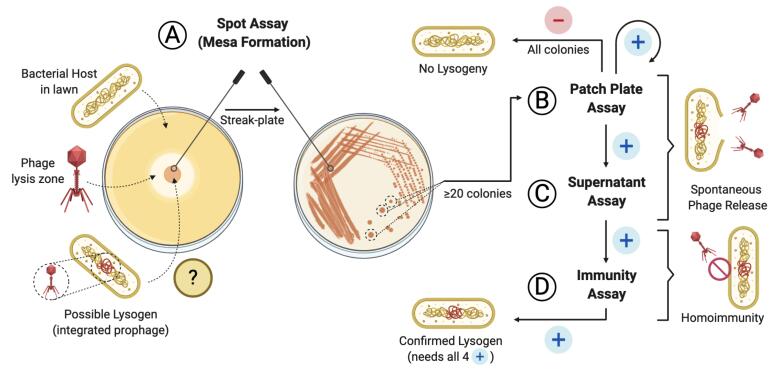
Screening pipeline for lysogen activity of a given phage
Keywords: Bacteriophage, Phage therapy, Lysogeny, Prophage integration, Prophages, Acinetobacter baumannii, Enterococcus faecium
Background
The field of phage therapy, which is the clinical use of viruses to kill bacteria, has grown exponentially over recent decades (Gordillo Altamirano and Barr, 2019). Prior to their selection for therapeutic use, phages are typically subjected to a thorough characterization pipeline ( Terwilliger et al., 2020 ). Lysogenic or temperate phages are those with the capacity to integrate themselves into their host’s genome, and are routinely excluded from therapeutic use due to their subpar bactericidal ability, the emergence of homoimmunity, and the potentially harmful consequences of lysogenic conversion (Fortier and Sekulovic, 2013; Hyman, 2019).
Temperate behavior is first suspected in phages that produce turbid plaques on bacterial lawns. However, plaque morphology can vary greatly depending on a variety of factors, including the growth medium used and the physiological state of the bacterial host ( Ramesh et al., 2019 ). Delayed or substandard bactericidal activity in bacterial growth assays can also serve as an indicator, but their interpretation is not always clear-cut. Alternatively, genotypic screening for genes that are essential for lysogeny, such as integrase and excisionase enzymes, are further indicators of temperate behavior (Hyman, 2019). However, extraction and sequencing of phage genomes can be challenging, and the accuracy of homology-based analyses is highly dependent on the quality of the sequencing data, assemblies, and available databases (Russell, 2018). Importantly, the presence of a predicted integrase in the genome of a phage may not be definite confirmation of the ability to lysogenize a host in vitro or in vivo (Howard- Varona et al., 2017 ).
Here, we outline a simple protocol, modified from Pope et al. (2013) , that evaluates the ability of a given phage to lysogenize its host in vitro. The assay can be reliably used in cases where no sequencing data are available. Furthermore, this protocol has the advantage of providing confirmation of lysogenic behavior for a given phage, instead of prediction, given by genome sequencing-based tests. It does not require specialized equipment and can complement analysis of plaque morphology and growth. Although we describe the protocol using an Acinetobacter baumannii-specific phage (Gordillo Altamirano et al., 2021 ), we also provide data from an Enterococcus faecium-specific phage (Figure 1), demonstrating that the methodology is broadly applicable to any phage with a bacterial host culturable on solid media. As such, incubation and media conditions may need be adapted to suit the host. Finally, in addition to excluding lysogenic phages from therapeutic use, the protocol can be used to produce lysogens (bacteria harboring a prophage) for downstream applications in phage biology and biotechnology research.
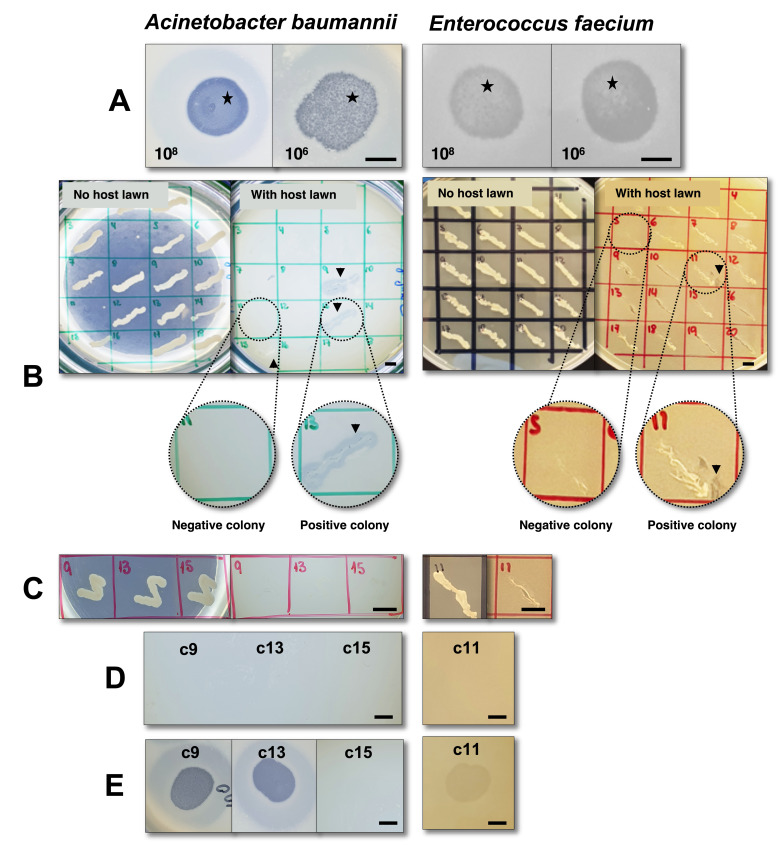
Figure 1. Screening for lysogenic behavior of phage øCO01 (Gordillo Altamirano et al., 2021 ) on host strain Acinetobacter baumannii A9844 ( Peleg et al., 2008 ) and phage øFG-VRE (unpublished) on host strain Enterococcus faecium ST796 ( Buultjens et al., 2017 ).
A. Spot assay with droplets of 108 and 106 PFU/ml phage lysate after a 48-h incubation. Black stars denote mesas. B. First patch assay using plates with and without lawns of the bacterial hosts. Black arrows (colonies 9, 13, and 15 on A. baumannii and colony 11 on E. faecium) show bacterial growth with surrounding lysis, indicating the presence of phage, possibly released from a lysogen. Zoomed sections are provided as examples of positive and negative colonies. For A. baumannii, the panel only shows half the total screened colonies (18/36). C. Second patch plate, after two rounds of single-colony purification of colonies 9, 13, and 15 (A. baumannii) and colony 11 (E. faecium), without indication of phage presence when patched onto plates containing bacterial host lawns. D. Spot assay performed with the filtered supernatant from cultures of the putative lysogenic colonies, with no indication of spontaneous phage release. E. Immunity assay of colonies 9 and 13 (A. baumannii) and colony 11 (E. faecium) indicates sensitivity to their respective phages, thereby excluding homoimmunity; for colony 15 (A. baumannii), the test demonstrates resistance to the phage, most likely due to a mechanism other than homoimmunity (see Table 1 for interpretation). Taken together, the tests demonstrate the inability of phages øCO01 and øFG-VRE to lysogenize host strains A. baumannii A9844 and E. faecium ST796, respectively. For assays with E. faecium, LB was replaced with BHI (brain heart infusion) media. Scale bars = 0.5 cm.
Materials and Reagents
15-ml polystyrene tubes (In Vitro Technologies, Falcon, catalog number: FAL352095)
Pipettes, serological pipettes, and pipette tips
10-ml sterile rimless glass test tubes (Thermo-Fisher, Youlyy, catalog number: LBSDCT13100)
Wire loop
10-ml syringes with detachable needle (McFarlane, Terumo, catalog number: 19046TE)
0.2-μm filters, attachable to syringes (Pall, catalog number: 4612)
-
Phage to be tested, high titer lysate (≥ 106 plaque-forming units [PFU]/ml)
Note: For a protocol on phage isolation and purification, see Bonilla et al., 2016.
1× phosphate-buffered saline (PBS) solution (Merck Australia, OmniPur, catalog number: US16506)
BactoTM Tryptone (Gibco, catalog number: 211705)
Granulated yeast extract (Merck Australia, Millipore, catalog number: 1037530500)
NaCl (Merck Australia, Supelco, catalog number: 1064040500)
Agar powder (Merck Australia, Millipore, catalog number: 05040)
Overnight culture in LB of the phage’s bacterial host (see Recipes for LB medium)
LB medium top agar (see Recipes)
-
LB medium agar plates (see Recipes)
Note: The growth media in 13-15 can be replaced to cater for the bacterial host’s growth requirements.
Equipment
Microwave (Panasonic, model: 32 L stainless steel, catalog number: NN-ST67JSQPQ)
37°C incubator
Shaking platform (Thermo Scientific, model: CO2-resistant shaker, catalog number: 88881102)
Bunsen burner
Benchtop centrifuge (Eppendorf South Pacific, model: 5810 R, catalog number: 5811000487)
Autoclave (Tuttnauer, model: vertical 110L, catalog number: 5050ELV-D)
Procedure
-
Coincubation of phages and bacterial hosts (spot assay)
Note: The estimated time to complete this section is 45 min of benchwork followed by overnight incubation.
-
In a sterile glass test tube, mix 1 ml bacterial culture with 3 ml molten top agar.
Note: The top agar can be liquified using a microwave. Be careful: too hot a temperature of the top agar will kill the bacterial population, resulting in a poor lawn. The top agar should be liquid but pleasantly warm to the touch (approximately 50°C).
Pour the mixture onto a room-temperature LB agar plate and allow to solidify.
-
Prepare two or three serial dilutions (~ 1 ml) of the high-titer phage lysate in PBS.
Note: We recommend using at least two concentrations of phage, for example 108 and 106 PFU/ml.
-
Spot 10-μl drops of each phage lysate dilution onto the surface of the prepared plate.
Note: Spots of further dilutions or replicates of the same dilutions can be added to the same plate.
Leave the plate, lid partially on, close to a Bunsen burner until the spots are completely dry; this can take up to 30 min. Alternatively, the plates can be left to dry in a biosafety hood.
-
Incubate at 37°C for up to 4 days, with daily checks to assess the formation of “mesas” (Figure 1A) ( Pope et al., 2013 ).
Note: Mesas are zones of confluent bacterial growth in the center of the lysis spots. Bacteria growing on mesas have become resistant to the phage, possibly (but not exclusively) due to homoimmunity. While checking for mesas, also look for agar dehydration due to prolonged incubation, which could hamper bacterial viability. Desiccation can be further avoided by incubating the plates in a sealed container with a wet paper towel, creating a humid chamber.
Using a sterile wire loop, scrape one loopful of bacterial growth from a mesa and use it as the primary inoculum for a streak plate on fresh LB agar (see Graphical abstract). Repeat for each mesa.
-
Incubate at 37°C overnight. Assess the growth of clearly isolated colonies.
Note: Any of these colonies could contain the integrated prophage. We routinely test 10 colonies obtained from each of at least two mesas, for a total of at least 20 colonies.
-
-
Patch plate screen
Note: The estimated time to complete this section is 30 min of benchwork and multiple overnight incubations.
-
Label and divide two LB agar plates into a grid with as many sections as colonies to be screened. Number the sections on both plates identically from 1 to 20.
Note: A single plate can comfortably fit 20 colonies.
Prepare one of the plates with a lawn of bacterial host following Steps A1 and A2. The other plate is used without further preparation.
-
Using a sterile wire loop, a disposable pipette tip, or a sterile toothpick, take a random colony from the streak plate (prepared in Step A7), gently patch it onto section 1 of the LB plate, and then onto its corresponding section of the LB plate with the bacterial host lawn (Step B2).
Note: Avoid tearing the delicate agar while performing the patches.
Repeat Step B3 with as many colonies as needed.
Incubate the plates at 37°C for up to 2 days, with daily checks for interpretation.
-
To interpret the test, look closely at the plate with the host lawn. Positive colonies, those potentially harboring the prophage, will present signs of lysis (plaques, halos, loss of turbidity) on the lawn around them, which is caused by the spontaneous release of the phage (Figure 1B).
Note: If all the screened colonies are negative for the patch test, it can be concluded that the phage is unable to lysogenize the host (see Table 1). False positive results here are possible due to carry-over of phage particles from the mesa onto the streak plate (Step A7).
For each positive colony, return to the LB plate without the host lawn (prepared in Steps B3-B4), pick a section of the patch with a wire loop, and streak onto a fresh LB plate.
Incubate overnight at 37°C.
Pick a single colony with a wire loop and re-streak onto a fresh LB plate.
-
Incubate overnight at 37°C.
Note: You have now performed a double single-colony purification. Using colonies from this plate (B9), repeat Procedure B (Figure 1C) and perform tests C and D to confirm lysogenic behavior.
-
-
Spontaneous phage release in liquid culture test (Supernatant assay)
Note: The estimated time to complete this section is 45 min of benchwork followed by overnight incubation.
Following purification of the colony of interest with two consecutive streak plates, inoculate the colony into a 10-ml Falcon tube containing ~4 ml LB.
-
Incubate overnight at 37°C with aeration.
Note: Aeration, achieved by leaving enough head space in the tube and incubating on a shaking platform at 150 rpm, is needed for optimal growth of the host used in this example. Growth conditions can be modified to cater to different hosts.
Centrifuge the culture at 3,500 × g for 10 min.
Collect the supernatant using a 10-ml syringe.
-
Attach a 0.2-μm filter to the syringe and carefully depress the plunger to filter the supernatant into a fresh tube.
Note: This step is optional. If not performed, we recommend repeating Steps C3 and C4.
Perform a spot assay (Procedure A), replacing the phage lysate with the filtered supernatant from C5.
For test interpretation, look for signs of lysis on the spot assay caused by the spontaneous release of phages from the lysogens into the supernatant of the liquid culture (Figure 1D).
-
Immunity assay
Note: The estimated time to complete this section is 15 min of benchwork followed by overnight incubation.
Perform a spot assay (Procedure A) replacing the original bacterial host with the candidate lysogen purified in Step B9.
-
For test interpretation, look for the absence of lysis caused by homoimmunity (no clearing, plaques, or loss of turbidity on the bacterial lawn) (Figure 1E).
Note: An immunity assay showing resistance to the original phage, without positive findings of lysogeny on Procedures B and C, is suggestive of alternative mechanisms of phage resistance. Conversely, a confirmed lysogen will test positive in Procedures B, C, and D. For an expanded explanation regarding interpretation of the procedures, see Figure 1 and Table 1.
Table 1. Interpretation guide.
Most commonly encountered combinations of results and their interpretation.
|
Procedure Subject |
A | B | C | D | Interpretation / Troubleshooting | |
| Mesa formation | First patch test | Second patch test | Supernatantassay | Immunity assay | ||
| Phage/host 1 | - | N/A | N/A | N/A | N/A | Phage unable to lysogenize host (?)Repeat A with lower phage titers and/or a longer incubation time |
| Phage/host 2 | + | -* | N/A | N/A | N/A | Phage unable to lysogenize host |
| Colony 1 | N/A | + | - | - | - | Phage carried-over from the streak plate, no lysogen production |
| Colony 2 | N/A | + | + | + | + | Successful lysogen production |
| Colony 3 | N/A | - | - | - | + | Phage resistance due to other mechanisms |
| Colony 4 | N/A | + | - | - | + | False positive on the first patch test, phage resistance due to other mechanisms |
* At least 20 colonies screened, 10 from each of 2 different mesas. +: positive test. -: negative test.
Data analysis
A summary of the most frequent combinations of results in each of the four described procedures and a guide to their interpretation are included in Table 1. Briefly, lysogenic behavior of a phage can be safely excluded when at least 20 host colonies from at least two different mesas are negative in a patch test (Procedure B). Additionally, for a host colony to be confirmed as harboring the prophage, it must undergo double single-colony purification and subsequently test positive in Procedures B, C, and D.
Recipes
-
LB medium
10 g BactoTM Tryptone
10 g granulated yeast extract
5 g NaCl
500 ml deionized water
Note: Autoclave, cool down, and store at room temperature.
-
LB agar plates
7.5 g agar powder
500 ml sterile LB medium
Note: Autoclave, allow to cool, pour ~20 ml per petri dish, allow to solidify, and store at 4°C.
-
LB top agar
1.5 g agar powder
200 ml sterile LB medium
Note: Autoclave, cool down, and store at room temperature. When needed, warm up in a microwave for ~3 min.
Acknowledgments
Fernando L. Gordillo Altamirano acknowledges the support received from Monash University through the Monash Postgraduate Research Scholarship funding his doctoral studies. This work, including the efforts of Jeremy J. Barr, was funded by the Australian Research Council (ARC), Discovery Early Career Researcher Award (DECRA) (DE170100525), National Health and Medical Research Council (NHMRC: 1156588), and the Perpetual Trustees Australia award (2018HIG00007).
We thank Prof Anton Y. Peleg and Prof Tim P. Stinear for providing the bacterial strains used in the present study. This protocol is derived from previous work by Broussard et al., 2013 and Pope et al., 2013 and is previously described in a summarized form by Gordillo Altamirano et al., 2021 .
Competing interests
The authors declare not to have any competing interests.
Citation
Readers should cite both the Bio-protocol article and the original research article where this protocol was used.
References
- 1. Bonilla N., Rojas M. I., G. Netto Flores Cruz, Hung S. H., Rohwer F. and Barr J. J.(2016). Phage on tap-a quick and efficient protocol for the preparation of bacteriophage laboratory stocks. PeerJ 4: e2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Broussard G. W., Oldfield L. M., Villanueva V. M., Lunt B. L., Shine E. E. and Hatfull G. F.(2013). Integration-dependent bacteriophage immunity provides insights into the evolution of genetic switches. Mol Cell 49(2): 237-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Buultjens A. H., Lam M. M., Ballard S., Monk I. R., Mahony A. A., Grabsch E. A., Grayson M. L., Pang S., Coombs G. W., Robinson J. O., Seemann T., Johnson P. D., Howden B. P. and Stinear T. P.(2017). Evolutionary origins of the emergent ST796 clone of vancomycin resistant Enterococcus faecium . PeerJ 5: e2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Fortier L. C. and Sekulovic O.(2013). Importance of prophages to evolution and virulence of bacterial pathogens. Virulence 4(5): 354-365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gordillo Altamirano F. L., Forsyth J. H., Patwa R., Kostoulias X., Trim M., Subedi D., Archer S., Morris F. C., Oliveira C., Kieltyet al. (2021). Bacteriophage-resistant Acinetobacter baumannii are resensitized to antimicrobials . Nat Microbiol doi: 10.1038/s41564-020-00830-7 [DOI] [PubMed] [Google Scholar]
- 6. Gordillo Altamirano F. L. and Barr J. J.(2019). Phage Therapy in the Postantibiotic Era. Clin Microbiol Rev 32(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Howard-Varona C., Hargreaves K. R., Abedon S. T. and Sullivan M. B.(2017). Lysogeny in nature: mechanisms, impact and ecology of temperate phages. ISME J 11(7): 1511-1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hyman P.(2019). Phages for Phage Therapy: Isolation, Characterization, and Host Range Breadth. Pharmaceuticals(Basel) 12(1): 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Peleg A. Y., Tampakakis E., Fuchs B. B., Eliopoulos G. M., Moellering R. C. Jr.and Mylonakis E.(2008). Prokaryote-eukaryote interactions identified by using Caenorhabditis elegans . Proc Natl Acad Sci U S A 105(38): 14585-14590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Pope W., Sarkis G., Hatfull G. and Broussard G.(2013). Lysogeny experiments. Retrieved from https://digitalcommons.library.umaine.edu/cgi/viewcontent.cgi?article=1261&context=honors.
- 11. Ramesh N., Archana L., Madurantakam Royam M., Manohar P. and Eniyan K.(2019). Effect of various bacteriological media on the plaque morphology of Staphylococcus and Vibrio phages . Access Microbiol 1(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Russell D. A.(2018). Sequencing, Assembling, and Finishing Complete Bacteriophage Genomes. Methods Mol Biol 1681: 109-125. [DOI] [PubMed] [Google Scholar]
- 13. Terwilliger A. L., Gu Liu C., Green S. I., Clark J. R., Salazar K. C., Hernandez Santos H., Heckmann E. R., Trautner B. W., Ramig R. F. and Maresso A. W.(2020). Tailored Antibacterials and Innovative Laboratories for Phage(Phi) Research: Personalized Infectious Disease Medicine for the Most Vulnerable At-Risk Patients. Phage 1(2): 66-74. [DOI] [PMC free article] [PubMed] [Google Scholar]