Abudu et al. report that mitochondrial β-barrel biogenesis protein SAMM50 recruits ATG8 proteins through an LIR motif and interacts with p62/SQSTM1 to mediate basal piecemeal mitophagy of SAM and MICOS components. Upon metabolic switch to oxidative phosphorylation, SAMM50 and p62 cooperate to mediate efficient mitophagy.
Abstract
Mitophagy is the degradation of surplus or damaged mitochondria by autophagy. In addition to programmed and stress-induced mitophagy, basal mitophagy processes exert organelle quality control. Here, we show that the sorting and assembly machinery (SAM) complex protein SAMM50 interacts directly with ATG8 family proteins and p62/SQSTM1 to act as a receptor for a basal mitophagy of components of the SAM and mitochondrial contact site and cristae organizing system (MICOS) complexes. SAMM50 regulates mitochondrial architecture by controlling formation and assembly of the MICOS complex decisive for normal cristae morphology and exerts quality control of MICOS components. To this end, SAMM50 recruits ATG8 family proteins through a canonical LIR motif and interacts with p62/SQSTM1 to mediate basal mitophagy of SAM and MICOS components. Upon metabolic switch to oxidative phosphorylation, SAMM50 and p62 cooperate to mediate efficient mitophagy.
Introduction
Mitochondria are involved in crucial cellular activities, including metabolism, signaling, pathogen defense, and energetics (Bratic and Trifunovic, 2010; Chandel, 2015; Tzameli, 2012; Weinberg et al., 2015). All mitochondrial functions require nuclear DNA-encoded proteins. Their precursors are transported through one or several channels in the outer mitochondrial membrane (OMM) and inner mitochondrial membrane (IMM). The translocase of the OMM (TOM) complex, including the channel-forming protein TOMM40 and several peripheral receptor proteins, is responsible for translocating precursors into the mitochondrial intermembrane space (IMS; Mokranjac and Neupert, 2015; Neupert and Herrmann, 2007; Rapaport, 2002; Shiota et al., 2015; Wiedemann et al., 2003; Wiedemann and Pfanner, 2017). Once there, proteins destined for the matrix are transported by the translocase of the IMM (TIM) complex in collaboration with the presequence translocase-associated motor (Banerjee et al., 2015; Horst et al., 1997; Kang et al., 1990; Lytovchenko et al., 2013; Neupert and Herrmann, 2007; Truscott et al., 2001; Wiedemann and Pfanner, 2017). Precursors of β-barrel proteins in the OMM, including TOMM40 and voltage-dependent anion channel proteins (VDACs), are transferred from the TOM complex to the sorting and assembly machinery (SAM) complex for incorporation into the OMM (Höhr et al., 2018; Humphries et al., 2005; Kozjak-Pavlovic et al., 2007; Kutik et al., 2008; Stojanovski et al., 2012; Ulrich and Rapaport, 2015; Wiedemann et al., 2004; Wiedemann et al., 2003). The SAM complex consists of the integral pore-forming protein SAMM50 and peripheral proteins metaxin 1 (MTX1) and MTX2 (Armstrong et al., 1997; Kozjak-Pavlovic et al., 2007). SAMM50 also regulates cristae stability by interacting with core proteins of the mitochondrial contact site and cristae organizing system (MICOS) complex (Darshi et al., 2011; Ding et al., 2015; Xie et al., 2007).
Mitochondrial integrity is also maintained by shuttling of OMM proteins and lipids to other organelles via mitochondria-derived vesicles (MDVs; McLelland and Fon, 2018; McLelland et al., 2014; Neuspiel et al., 2008; Soubannier et al., 2012a; Soubannier et al., 2012b; Sugiura et al., 2014). Defective mitochondria are degraded in the lysosome by selective autophagy processes called “mitophagy” (Kim et al., 2007a; Lemasters, 2005; Palikaras et al., 2018; Youle and Narendra, 2011). Dysfunctional mitochondria can trigger apoptosis or contribute to disease. Recent studies have revealed various forms of mitophagy, including basal mitophagy, programmed mitophagy, and stress- or damage-induced mitophagy (Palikaras et al., 2018; Pickles et al., 2018; Rodger et al., 2018).
Basal mitophagy is a steady-state housekeeping process that continuously recycles whole mitochondria or parts (piecemeal mitophagy) of mitochondria to maintain function. Using a transgenic mouse model expressing a pH-dependent mitochondrial tandem-tag fluorescent reporter, researchers demonstrated basal mitophagy in a number of tissues, particularly in tissues of high metabolic demand, like the heart, liver, and skeletal muscles (McWilliams et al., 2018). Subsequent studies using mitochondria-targeted Keima, a pH-sensitive fluorescent probe, showed basal mitophagy in Drosophila (Cornelissen et al., 2018; Kim et al., 2019). Using a combination of proximity labeling and quantitative proteomics, Behrends and co-workers reported a piecemeal form of basal mitophagy that specifically targets certain mitochondrial proteins to the lysosome in a process dependent on LC3C (Le Guerroué et al., 2017).
Mitophagy is induced in response to a variety of stress and damage stimuli. Most of these signals converge on the PTEN-induced kinase 1 (PINK1)–Parkin-mediated pathway. PINK1 and E3-ubiquitin ligase Parkin, whose mutations are linked to autosomal recessive Parkinson’s disease, function in this pathway to eliminate damaged mitochondria to protect neuronal cells (Ashrafi et al., 2014; Youle and Narendra, 2011). Basally, PINK1 is translocated into mitochondria and rapidly degraded by proteases (Palikaras et al., 2018; Sekine and Youle, 2018). Following mitochondrial damage or depolarization, PINK1 accumulates on the OMM and phosphorylates both Parkin and ubiquitin on Ser65. This activates Parkin’s ligase activity to ubiquitinate several OMM proteins, triggering a cascade of events that leads to recruitment of autophagy receptors. Autophagosome formation results, and the damaged mitochondria are delivered to the lysosome (Aguirre et al., 2017; Harper et al., 2018; Koyano et al., 2014; Lazarou et al., 2012; Lazarou et al., 2015; Pickles et al., 2018; Sekine and Youle, 2018). However, several cell types do not express, or express very low amounts of Parkin. Basal mitophagy independent of Pink1 or Parkin is widespread in humans and Drosophila (McWilliams et al., 2018, Lee et al., 2018).
Programmed mitophagy occurs during development and cellular differentiation, i.e., removal of mitochondria from RBCs during erythrocyte differentiation and elimination of paternal mitochondria from fertilized oocytes in early mouse embryo (Al Rawi et al., 2011; Rojansky et al., 2016; Sandoval et al., 2008; Schweers et al., 2007).
Relatively little is known about basal mitophagy. Here, we employed a combination of biochemical studies, bioimaging, CRISPR-mediated knockdown (KD), and lysosomal inhibition to show that SAMM50 acts as a receptor for basal piecemeal degradation of SAM and MICOS complex proteins through LC3-interacting region (LIR)–dependent recruitment of ATG8 proteins and the autophagy receptor p62/sequestosome 1 (SQSTM1). SAMM50 and p62 cooperate to mediate an efficient piecemeal mitophagy upon metabolic switch to oxidative phosphorylation (OXPHOS).
Results
SAMM50 interacts directly with p62/SQSTM1
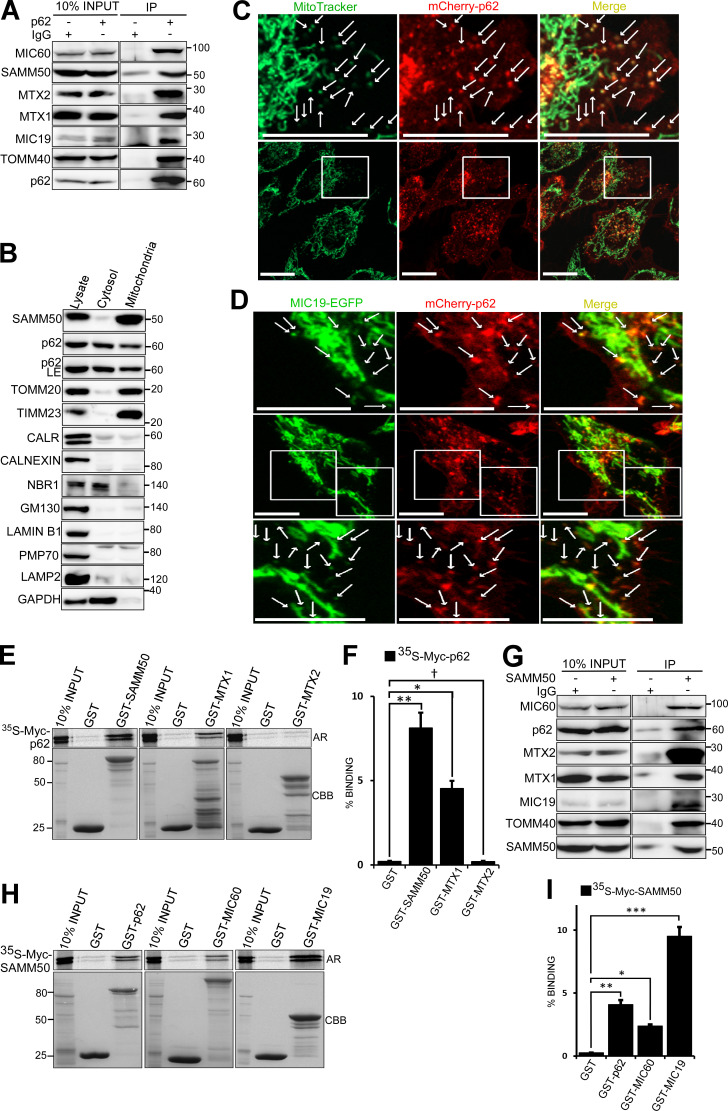
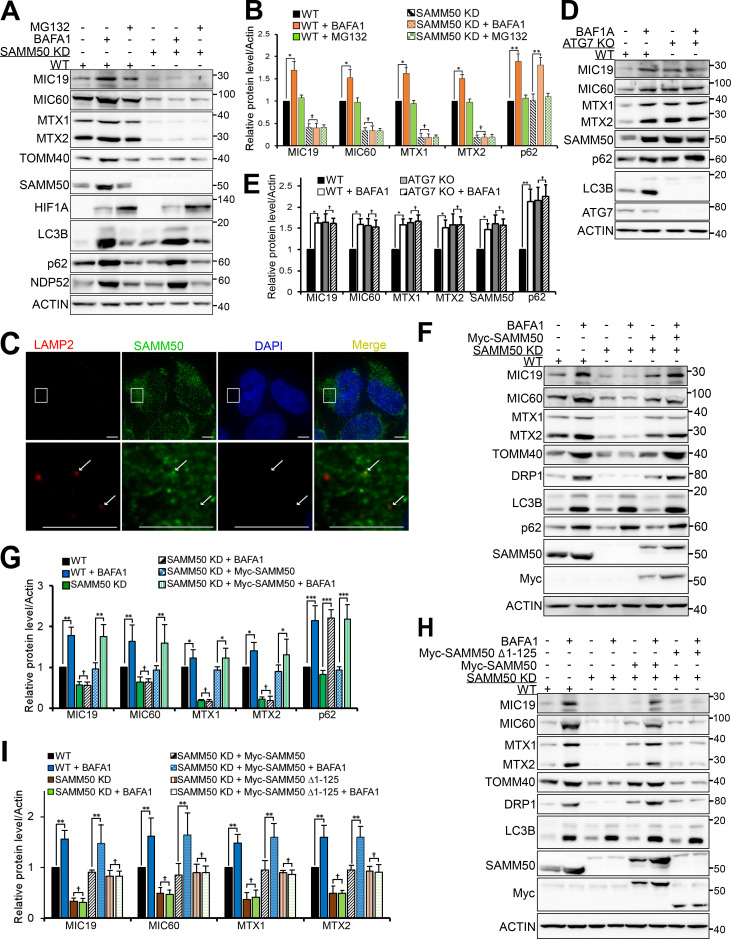
p62/SQSTM1 is both substrate and cargo receptor for selective macroautophagy (hereafter “autophagy”; Johansen and Lamark, 2011). To identify novel interaction partners and putative autophagic cargoes, we immunoprecipitated endogenous p62 and identified proteins interacting with p62 under basal conditions by mass spectrometry. Among the top candidates were several mitochondrial proteins, including matrix proteins 4-nitrophenylphosphatase domain and nonneuronal synaptosomal-associated protein 25-like protein 1 (NIPSNAP1) and NIPSNAP2 (Abudu et al., 2019); pore-forming OMM proteins VDAC1 and TOMM40; SAM complex proteins SAMM50, MTX1, and MTX2; and MICOS complex proteins MIC19 (CHCHD3) and MIC60 (mitofilin/IMMT; Fig. S1 A). We confirmed the presence of these proteins in immunoprecipitates of endogenous p62 (Fig. 1 A). Subcellular fractionation revealed p62 in the mitochondrial fraction (Fig. 1 B and Fig. S1 B). Live-cell imaging of HeLa cells stably expressing mCherry-p62 and stained with MitoTracker (Fig. 1 C) and diffraction-limited deconvolution (DV) analysis of HeLa cells stained with antibodies to p62 and SAMM50 (Fig. S1 C) revealed foci of p62 colocalized with MitoTracker and endogenous SAMM50, respectively. High-resolution live-cell confocal imaging of HeLa cells stably coexpressing mCherry-p62 and MIC19-EGFP showed p62 association with both tubular mitochondria and mitochondrial fragments (Fig. 1 D). SAMM50 and MTX1 interacted directly with p62, while MTX2, MIC19, MIC60, and VDAC1 did not (Fig. 1, E and F; and Fig. S1 D). SAMM50 regulates the biogenesis of β-barrel proteins, including TOMM40 and VDAC1 (Humphries et al., 2005; Kozjak-Pavlovic et al., 2007; Kutik et al., 2008). SAMM50 also regulates cristae stability by binding to MICOS complex proteins (Darshi et al., 2011; Ding et al., 2015; Xie et al., 2007). Mass spectrometry of proteins immunoprecipitated with endogenous SAMM50 revealed many of the same mitochondrial proteins found in p62 immunoprecipitates (Fig. S1 E). We verified this by immunoblotting (Fig. 1 G). GST-pulldown assays confirmed direct interaction between SAMM50 and p62, between SAMM50 and MICOS complex proteins MIC19 and MIC60 (Fig. 1, H and I), and between SAMM50 and MTX1 and -2 (Fig. S1 F).
Figure S1.
p62 associates with several mitochondrial proteins. (A) Endogenous p62 was immunoprecipitated from HeLa cells followed by mass spectrometry analysis of associated proteins. Only some mitochondrial related proteins and proteins showing specific interaction with p62 are presented here. (B) HEK293 cells were subjected to subcellular fractionation and immunoblotted with indicated antibodies. (C) Diffraction-limited DV microscopy images of HeLa cells co-stained with antibodies to endogenous p62 and SAMM50. Nuclear DNA was stained with DAPI. Boxes indicate enlarged images shown below. White arrows indicate colocalization events. Scale bars, 10 µm. (D) GST pulldown using in vitro translated Myc-tagged p62 in the presence of radioactive methionine with GST-tagged mitochondrial proteins. Myc-tagged protein binding was measured by AR while GST-tagged proteins were stained with CBB. (E) Endogenous SAMM50 was immunoprecipitated from HeLa cells followed by mass spectrometry analysis of associated proteins. (F) Myc-tagged SAMM50 was in vitro-translated and its interaction with GST-tagged mitochondrial proteins was tested in GST pulldown assays and analyzed by AR.
Figure 1.
p62 interacts directly with SAMM50. (A) Endogenous p62 was immunoprecipitated (IP) from HeLa cells followed by immunoblotting with antibodies for mitochondrial proteins identified in mass spectrometric analysis of endogenous p62 immunoprecipitates presented in Fig. S1 A. (B) Subcellular fractions of HeLa cells immunoblotted with the indicated antibodies. CALR, calreticulin. (C) High-resolution live-cell imaging of HeLa cells expressing mCherry-p62 stained with MitoTracker. Arrows indicate p62-containing mitochondrial fragments. Scale bars, 20 µm. (D) High-resolution imaging of HeLa cells stably coexpressing MIC19-EGFP and mCherry-p62 with enlarged images shown above and below. Arrows indicate association of p62 with mitochondria. Scale bars, 10 µm. (E) In vitro translated and [35S]-methionine–labeled Myc-p62 tested in GST-pulldown experiments for interaction with selected mitochondrial proteins. Bound p62 was visualized by autoradiography (AR), and immobilized GST or GST-tagged proteins were stained with CBB. (F) Quantitative analysis of GST pulldowns in E, based on three independent experiments using Science Lab Image Gauge software (Fujifilm). Values are mean ± SD. **, P < 0.005; *, P < 0.01; †, NS; one-way ANOVA. (G) HeLa cell extracts immunoprecipitated with antibody to endogenous SAMM50 and immunoblotted with antibodies to indicated proteins. (H) GST pulldowns of in vitro translated Myc-SAMM50 and indicated GST-tagged proteins as in E. (I) Quantitative analysis of three independent GST pulldowns in H. Statistical values are mean ± SD. ***, P < 0.001; **, P < 0.005; *, P < 0.01; one-way ANOVA.
Depletion of SAMM50 reduces SAM and MICOS complex levels and destabilizes cristae
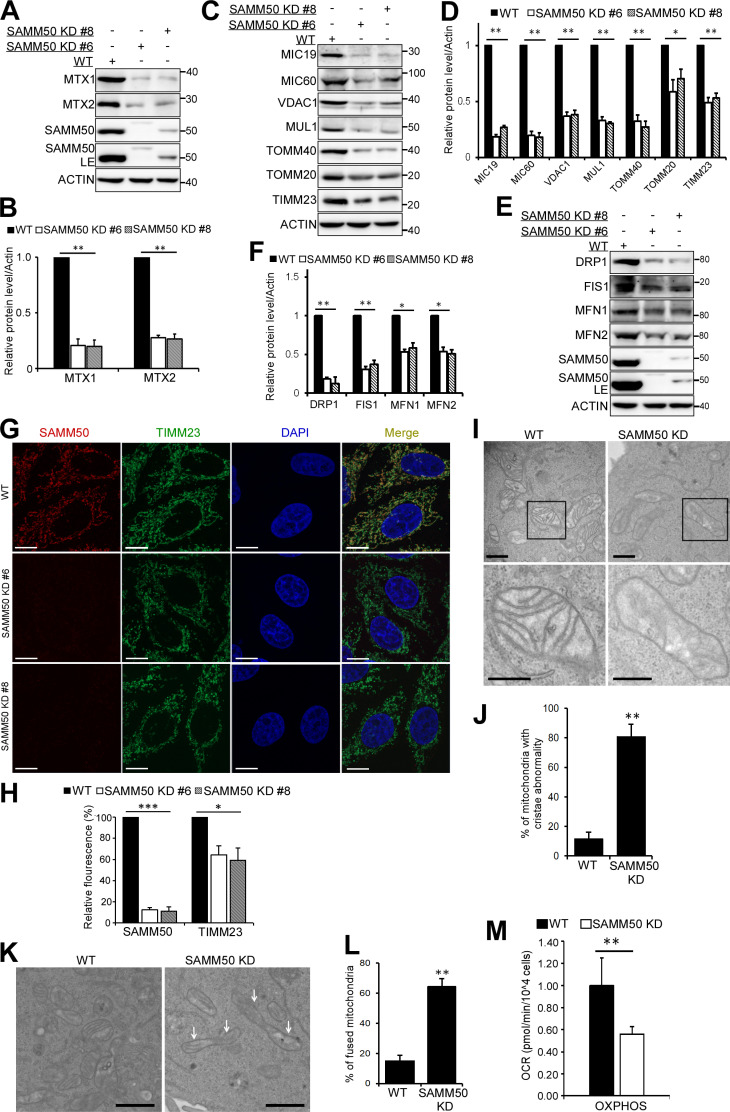
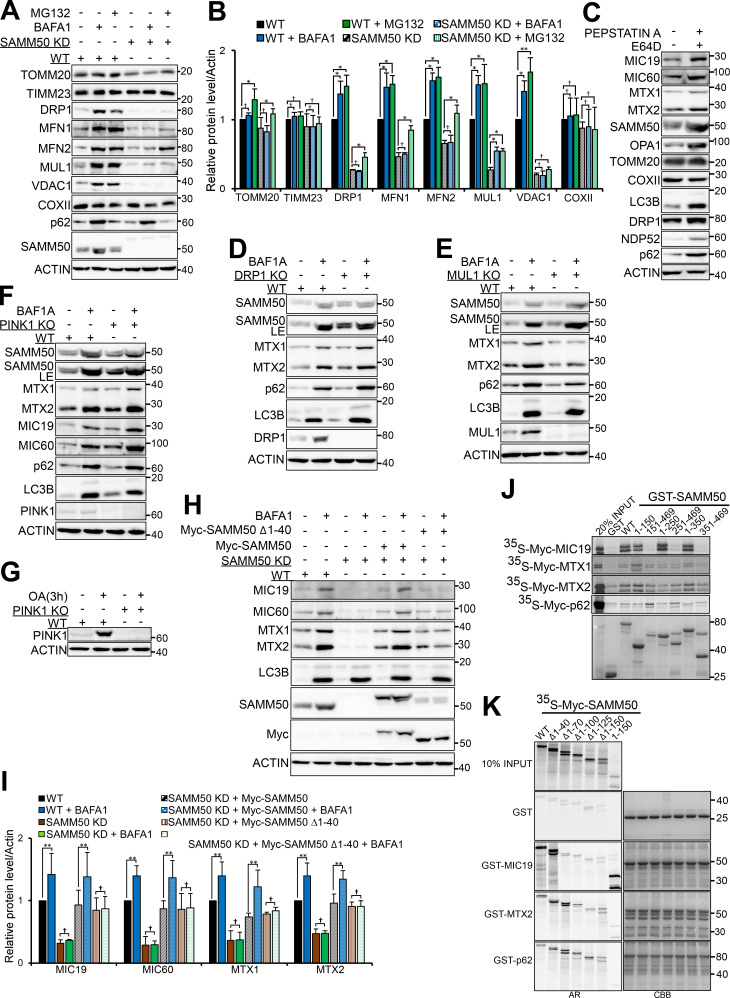
We generated SAMM50 KD HeLa cell clones using CRISPR/CRISPR-associated protein 9 (Cas9). In agreement with earlier studies (Kozjak-Pavlovic et al., 2007), we observed a substantial reduction of SAM complex proteins MTX1 and MTX2 (Fig. 2, A and B). MICOS complex proteins MIC60 and MIC19 were also reduced (Fig. 2, C and D). TOMM40 and TOMM20 of the TOM complex were also significantly reduced (Fig. 2, C and D). Other proteins with significantly reduced levels were OMM porin VDAC1; E3 ligase mitochondrial ubiquitin ligase activator of nuclear factor-κB 1 (MUL1); and the channel-forming component of the TIM complex, TIMM23 (Fig. 2, C and D). There was also a significant reduction in the GTPase DRP1 and mitofusins, MFN1 and MFN2. Cytochrome C oxidase subunit 2 (COXII/MT-CO2), a component of the respiratory chain, was also significantly reduced (Fig. S2, A and B). However, superoxide dismutase 2, involved in catabolism of superoxide anion radicals and reactive oxygen species (Flynn and Melov, 2013), and FKBP8, a receptor for Parkin-independent mitophagy (Bhujabal et al., 2017), were only mildly affected (Fig. S2, A and B). Other proteins with significantly reduced levels included choline dehydrogenase, NIPSNAP1, and NIPSNAP2, implicated in Parkin-dependent mitophagy (Abudu et al., 2019; Park et al., 2014); phosphatidylserine decarboxylase, an enzyme that converts phosphatidylserine to phosphatidylethanolamine (Vance and Tasseva, 2013); and FUNDC1, a receptor for hypoxia-induced mitophagy (Fig. S2, A and B; Liu et al., 2012). In agreement with immunoblot analyses, confocal microscopy showed a reduced staining of TIMM23 (Fig. 2, G and H). Antibody staining of mitochondrial DNA revealed no decrease in the number of mitochondria per cell in SAMM50-depleted cells relative to WT (Fig. S2, C and D). SAMM50 depletion affected protein content, but mitochondrial DNA nucleoids were preserved.
Figure 2.
SAMM50 KD alters mitochondrial morphology and depletes a subset of mitochondrial proteins. (A) Expression of SAM complex proteins in lysates from WT and two clones of SAMM50 CRISPR KD HeLa cells. LE, long exposure. (B) Densitometric analysis of MTX1 and MTX2 levels from A. Values are mean ± SD from three independent experiments. **, P < 0.005; one-way ANOVA. (C–F) Whole-cell lysates from WT and SAMM50 KD cells analyzed for expression of indicated mitochondrial proteins (C and E). LE, long exposure. Relative expression levels quantified (D and F) with mean ± SD from three different experiments. **, P < 0.005; *, P < 0.01; †, NS; one-way ANOVA. (G) High-resolution confocal images of WT and SAMM50 KD cells stained with antibodies to endogenous SAMM50 and TIMM23. DNA was stained with DAPI. Scale bars, 10 µm. (H) Quantification of relative fluorescence from G. Fluorescence intensity from 60–80 cells was quantified per sample using ImageJ software. Values are mean ± SD. ***, P < 0.001; *, P < 0.01; one-way ANOVA. (I) Mitochondrial cristae structure in WT and SAMM50 KD cells visualized by TEM. Scale bars, 0.2 µm. (J) Percentage of mitochondria with abnormal cristae were scored in I, based on 200 mitochondria from 8–10 micrographs per sample (see Materials and methods). Values are mean ± SD. **, P < 0.005; one-way ANOVA. (K and L) WT and SAMM50 KD HeLa cells analyzed for mitochondrial shape by TEM. Arrows indicate fused mitochondria (K). Percentage of fused mitochondria was scored in L (see Materials and methods). Values are mean ± SD. **, P < 0.005; one-way ANOVA. Scale bars, 0.5 µm. (M) OXPHOS measured in WT and SAMM50 KD cells using a Seahorse XFp flux analyzer. Graphs show one representative example from three independent experiments. Values are mean ± SD from three replicates. **, P < 0.005; one-way ANOVA.
Figure S2.
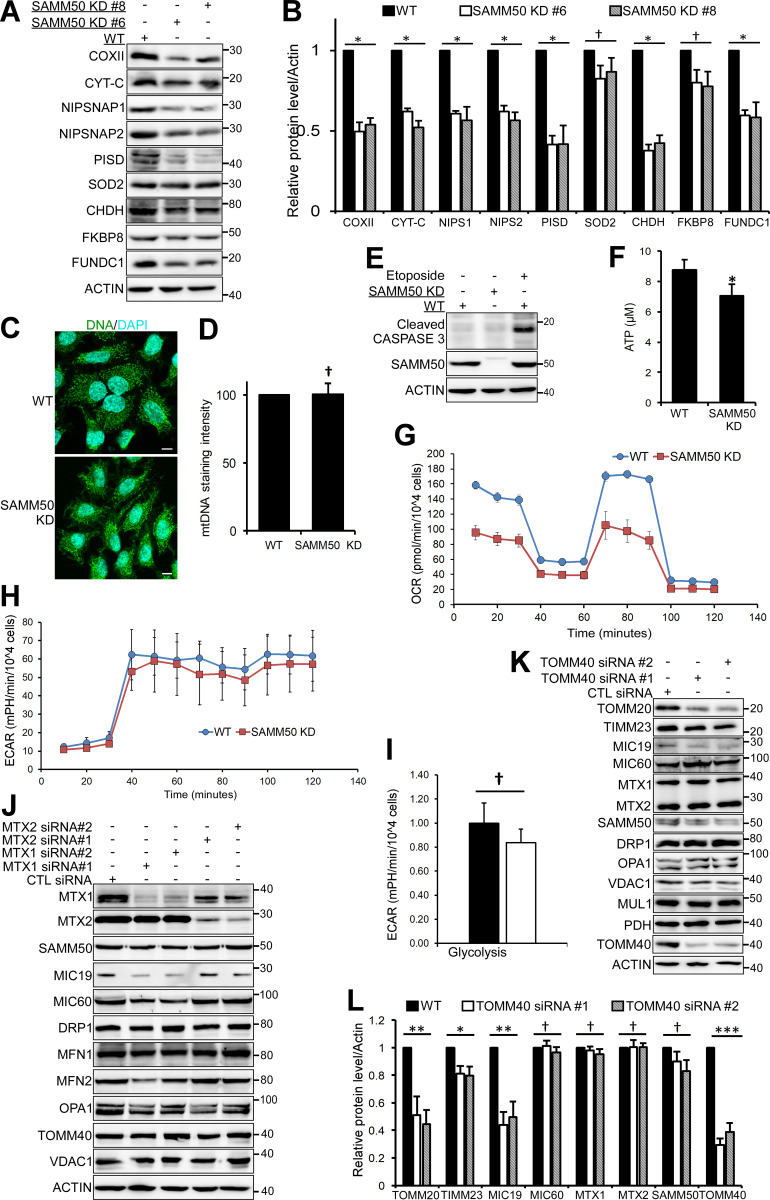
SAMM50 knockdown affects oxidative phosphorylation. (A and B) Lysates from WT and SAMM50 CRISPR KD HeLa cells were analyzed for mitochondrial protein expression by immunoblotting (A) and quantified (B). Values are mean ± SD from three different experiments. *, P < 0.01; †, NS; one-way ANOVA. (NIPS1:NIPSNAP1, NIPS2:NIPSNAP2). (C and D) WT and SAMM50 KD HeLa cells were immunostained with antibody to mtDNA and nuclei stained with DAPI. Mitochondrial DNA (mtDNA) nucleoids were visualized by fluorescence confocal microscopy (C) and staining intensity quantified (D) from 100 cells per sample using the ImageJ software. Values are mean ± SD. †, NS; one-way ANOVA. Scale bars, 10 µm. (E) WT and SAMM50 KD HeLa cells lysates were analyzed for presence of cleaved caspase 3 by immunoblotting. Lysate from WT HeLa cells treated with 100 μM etoposide for 24 h is used as a control. (F) Relative ATP levels from WT and SAMM50 KD cells were measured using the ATP determination kit from three independent experiments. Values are mean ± SD. *, P < 0.01; one-way ANOVA. (G) Direct measurement of OCR was done using a Seahorse XFp flux analyzer. Basal respiration was measured for 30 min. Oligomycin was injected at 30 min, blocking ATP production due to oxidative phosphorylation. FCCP was injected at 60 min, followed by complex I and III inhibitors at 90 min, showing differences in maximal mitochondrial capacity. (H) Direct measurements of ECAR was done using a Seahorse XFp flux analyzer. (I) Glycolytic rates in WT and SAMM50 KD HeLa cells were measured using a Seahorse XFp flux analyzer. Graphs show one representative from three independent experiments. Values are ± SD from three replicates. †, NS. (J) Lysates from HeLa cells treated with either control (CTL) siRNA or two different siRNA to MTX1 and MTX2 for 6 d (three pulses of 48 h each) were immunoblotted with the indicated antibodies. (K and L) Lysates from HeLa cells treated with control siRNA or two different siRNAs to TOMM40 for 4 d (two pulses of 48 h each) were immunoblotted with indicated antibodies (K) and quantified (L). Values are mean ± SD from three different experiments. ***, P < 0.001; **, P < 0.005; *, P < 0.01; †, NS; one-way ANOVA.
The MICOS complex maintains cristae stability and respiratory complexes for OXPHOS (Ding et al., 2015; Ott et al., 2012). Transmission EM (TEM) showed that almost 80% of mitochondria in SAMM50 KD cells had abnormal, distorted cristae (Fig. 2, I and J). This is consistent with a substantial reduction in key MICOS proteins MIC60 and MIC19. The fraction of fused mitochondria was increased in SAMM50 KD cells (Fig. 2, K and L). The levels of DRP1 were profoundly more reduced than those of the mitofusins, likely tipping the balance toward mitochondrial fusion. SAMM50 depletion did not induce apoptosis, but it significantly reduced cellular ATP production (Fig. S2, E and F). SAMM50 KD cells displayed a significant reduction in OXPHOS relative to WT cells (Fig. 2 M and Fig. S2 G). However, SAMM50 KD did not significantly affect glycolysis (Fig. S2, H and I). siRNA-mediated KD of MTX1 and -2 in the SAM complex did not have a profound effect on the overall protein content of mitochondria (Fig. S2 J). HeLa cells treated for 6 d with two separate siRNAs targeting MTX1 and MTX2, respectively, showed some reduction in MIC19 and MIC60 with MTX1 siRNA but not with MTX2 siRNA. MTX1 and MTX2 KD did not significantly affect levels of SAMM50, TOMM40, or VDAC1. Levels of MTX1 were reduced upon depletion of MTX2, but depletion of MTX1 did not affect MTX2 (Fig. S2 J). We conclude that MTX1 and MTX2 play a more subtle role in the function of the SAM complex.
The TOM complex imports nuclear-encoded proteins (Neupert and Herrmann, 2007; Wiedemann et al., 2003). HeLa cells treated for 4 d (longer treatment killed the cells) with two different TOMM40 siRNAs did not show substantial reduction in most mitochondrial proteins analyzed, except for TOMM20 and MIC19 (Fig. S2, K and L). Thus, the profound reduction in several mitochondrial proteins in SAMM50 KD cells is only partly dependent on the defect in the TOM complex.
As reported (Jian et al., 2018), SAMM50 KD stabilized PINK1 and increased processing of LC3B-I to LC3B-II (Fig. S3, A and B). A similar accumulation of PINK1 and increased LC3B-II processing occurred in cells treated with siRNAs to TOMM40 (Fig. S3, C and D). TOMM40 is required for PINK1 localization to the OMM (Okatsu et al., 2015). Thus, SAMM50 KD–dependent reduction of TOMM40 may partially account for the PINK1 stabilization. Since HeLa cells do not express detectable Parkin, reduction of mitochondrial proteins upon SAMM50 KD is not due to PINK1/Parkin-dependent mitophagy. Treatment of WT and SAMM50 KD cells with oligomycin and antimycin A (OA) for 3 h stabilized PINK1 in the absence of Parkin (Fig. S3, E and F).
Figure S3.
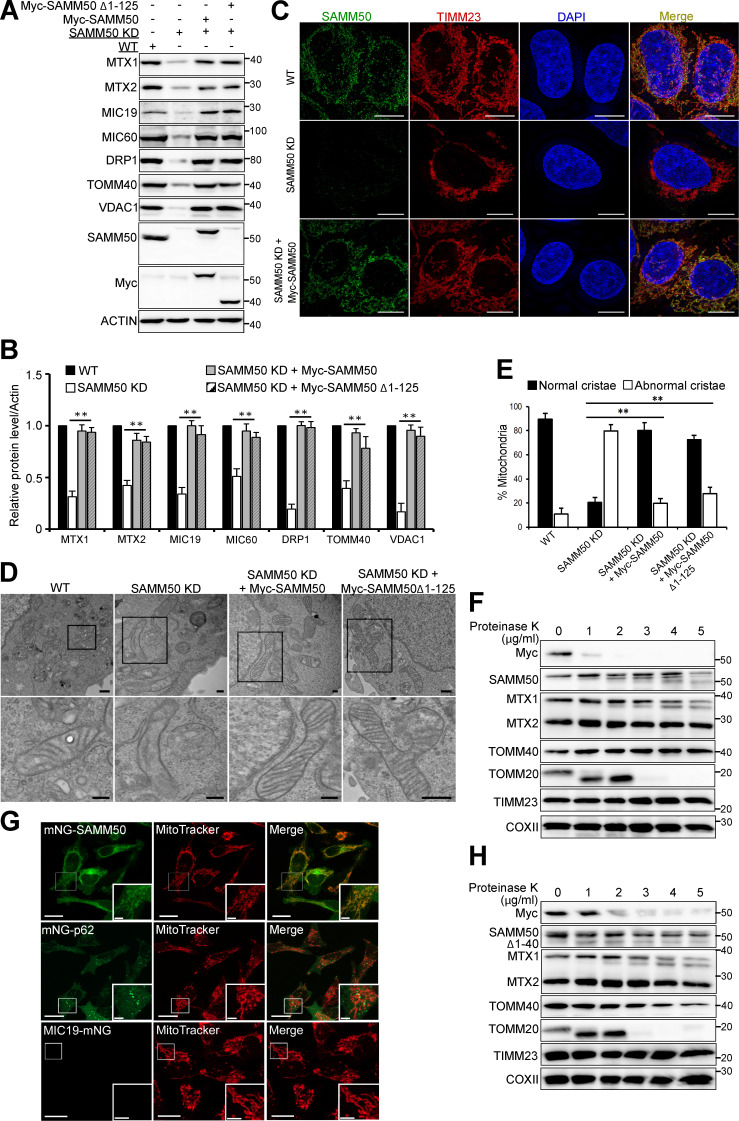
The SAMM50 N-terminal region with the NTS and POTRA domain is not required for mitochondrial protein biogenesis. (A and B) Whole cell lysates from WT cells and two clones of SAMM50 KD cells were immunoblotted to reveal PINK1, p62 and LC3B protein levels (A) and quantified (B). Values are mean ± SD from three different experiments. **, P < 0.005; *, P < 0.01; †, NS; one-way ANOVA. (C and D) HeLa cells were either treated with CTL siRNA or two different siRNA to TOMM40 and analyzed for the levels of indicated proteins by immunoblotting (C) and quantified (D). Values are mean ± SD from three different experiments. **, P < 0.005; *, P < 0.01; one-way ANOVA. (E and F) WT and SAMM50 KD cells were treated with a combination of OA for 3h and protein levels of PINK1 were analyzed by immunoblotting (E) and quantified (F). Values are mean ± SD from three different experiments. ***, P < 0.001; **, P < 0.005; one-way ANOVA. (G) Domain architecture of SAMM50 showing WT and various deletion constructs used in reconstituting SAMM50 KD cells. (H) Expression of the indicated mitochondrial proteins in WT, SAMM50 KD, AND SAMM50 KD cells reconstituted with WT SAMM50 or SAMM50 Δ1-125 mutant was analyzed by immunoblotting. (I and J) WT, SAMM50 KD, and SAMM50 KD cells reconstituted with Myc-SAMM50 and indicated deletion mutants were analyzed for expression of the indicated mitochondrial proteins by immunoblotting (I) and quantified (J). Values are mean ± SD from three different experiments. ***, P < 0.001; **, P < 0.005; one-way ANOVA. (K) Isolated mitochondria from SAMM50 KD cells reconstituted with Myc-SAMM50 were subjected to digestion with different concentration of proteinase K. Mitochondria protein levels were analyzed by immunoblotting with indicated antibodies.
The polypeptide transport-associated (POTRA) domain of SAMM50 is dispensable for β-barrel assembly and biogenesis
Most studies on β-barrel protein assembly have been performed in bacteria, yeast, and chloroplasts (Dolezal et al., 2006; Gentle et al., 2004; Höhr et al., 2018; Kim et al., 2007b; Paschen et al., 2003; Sklar et al., 2007; Wiedemann and Pfanner, 2017). Assembly and integration of β-barrel proteins is mediated by a highly conserved protein of the OMM protein of 85 kD (omp85) protein family. The bacterial omp85 protein (aka YaeT or BamA), yeast Sam50/Tob55, and chloroplast Toc75 have a large C-terminal channel forming a β-barrel domain (Paschen et al., 2003; Robert et al., 2006) and an N-terminal α-helical domain containing one to five POTRA domains, present in five copies in bacterial omp85 and one copy in Sam50 and Toc75, respectively (Dolezal et al., 2006; Habib et al., 2007; Kim et al., 2007b). The POTRA domain is thought to play a central role as a receptor in recognition and assembly of β-barrel proteins (Habib et al., 2007; Knowles et al., 2009; Koenig et al., 2010; O’Neil et al., 2017; Paila et al., 2016). The human homologue SAMM50 has a POTRA domain with two α-helical and three β-sheet sequences from aa 45 to 125 (Sánchez-Pulido et al., 2003).
To study the role of the POTRA domain in assembly and OMM integration of β-barrel proteins, we reconstituted SAMM50 KD cells with several N-terminal deletions (Fig. S3 G). Both WT and a mutant lacking the N-terminal region with the POTRA domain rescued mitochondrial protein levels affected by SAMM50 KD (Fig. 3, A and B; and Fig. S3 H). Hence, the POTRA domain of SAMM50 is dispensable for mitochondrial membrane protein biogenesis and assembly. All N-terminal deletions tested (Δ1–40, Δ1–70, and Δ1–100) rescued levels of TOMM40, VDAC, MIC60, and MIC19 (Fig. S3, I and J). Structured illumination microscopy (SIM) imaging showed restored TIMM23 intensity and volume in SAMM50 KD cells reconstituted with WT SAMM50 (Fig. 3 C). TEM showed cristae organization and structure fully restored in cells reconstituted with SAMM50 WT or Δ1–125 N-terminal deletion mutant (Fig. 3, D and E). Thus, we provide the first evidence that the POTRA domain is dispensable for SAMM50’s role in assembly and membrane integration of β-barrel proteins and maintenance of normal cristae morphology in human mitochondria.
Figure 3.
The N-terminal domain of SAMM50 is dispensable for its activity and is oriented to the cytoplasm. (AandB) Whole-cell lysates of WT, SAMM50 KD, and SAMM50 KD HeLa cells reconstituted with Myc-tagged SAMM50 WT and SAMM50 Δ1–125 mutant analyzed for mitochondrial protein expression by immunoblotting (A), and relative expression levels were quantified (B). Values are mean ± SD from three independent experiments. **, P < 0.005; one-way ANOVA. (C) SIM of WT, SAMM50 KD, and SAMM50 KD cells reconstituted with Myc-SAMM50 and stained with antibodies to SAMM50 and TIMM23. Scale bars, 10 µm. (D) TEM of mitochondrial ultrastructure of WT, SAMM50 KD, and SAMM50 KD cells reconstituted with either WT Myc-SAMM50 or Myc-SAMM50 Δ1–125. Scale bars, 0.2 µm. (E) Cristae morphology shown in D was scored based on 150 mitochondria from 9–10 micrographs per sample (see Materials and methods). Values are mean ± SD. **, P < 0.005; one-way ANOVA. (F) Mitochondria from SAMM50 KD cells reconstituted with Myc-SAMM50 subjected to increasing concentrations of proteinase K analyzed by immunoblotting. (G) SAMM50 and p62 were N-terminally tagged, while MIC19 was C-terminally tagged, with the 11th β-sheet of split fluorescent modified mNG11 and stably coexpressed, respectively, in HeLa cells stably expressing the first 10 β-sheets of modified mNG1-10 in a Tet-off/on system. Fluorescence complementation was induced with 1 µg/ml Tet overnight and analyzed by live-cell imaging. Mitochondria were imaged with MitoTracker Deep Red. Scale bars, 20 µm (main), 5 µm (inset). (H) Mitochondria from SAMM50 KD cells reconstituted with Myc-SAMM50 Δ1–40 treated with increasing concentrations of proteinase K and immunoblotted with the indicated antibodies.
The N-terminal segment (NTS) preceding the POTRA domain is oriented toward the cytosol
Recently reported cryo-EM structures of Sam complexes from Saccharomyces cerevisiae (Takeda et al., 2021) and the thermophilic fungus Thermothelomyces thermophilus (Diederichs et al., 2020) show the POTRA domain located in the IMS. To examine the orientation of the human SAMM50 N-terminal region, we used a combination of protease protection and split fluorescent self-complementation assays (Feng et al., 2017). Isolated mitochondria from SAMM50 KD cells reconstituted with N-terminal Myc-tagged SAMM50 subjected to increasing concentrations of proteinase K showed loss of Myc immunostaining at 2 µg/ml of proteinase K, whereas staining with SAMM50 antibody still showed the presence of SAMM50 in mitochondria (Fig. 3 F). The loss of Myc signal was similar to TOMM20, a peripheral OMM receptor on the cytoplasmic side of the TOM complex (Wiedemann and Pfanner, 2017). MTX1 was partially degraded as the concentration of proteinase K increased, while MTX2 remained largely untouched. Increasing proteinase K to 10 and 20 µg/ml degraded both SAMM50 and TOMM40 (Fig. S3 K).
We next performed bimolecular fluorescence complementation assays using the self-complementing split fluorescent monomeric NeonGreen1-10/11 (mNG1-10/11) system (Cabantous et al., 2005; Feng et al., 2017). We generated HeLa cells stably expressing either a tetracycline-controlled (Tet-off/on) mNG1-10 segment and N-terminally tagged SAMM50 or a C-terminally tagged MIC19 or p62 with the smaller mNG11. When expressed in cells, the larger mNG1-10 segment is only in the cytosol. Upon induction with Tet, we observed green fluorescence in cells with both mNG11-tagged SAMM50 and the p62 control. Cells only expressing mNG1-10 showed no fluorescence. Neither did cells coexpressing mNG1-10 and mNG11-MIC19 (Fig. 3 G). The green structures colocalized with MitoTracker in cells with mNG11-SAMM50, confirming that the N-terminus of mitochondrial SAMM50 is exposed to the cytosol. We denote aa 1–40 of human SAMM50 the NTS (Fig. S3 G). Proteinase K treatment of isolated mitochondria from SAMM50 KD cells reconstituted with SAMM50 lacking the NTS showed the Myc tag to be more protected in SAMM50 Δ1–40 (Fig. 3 H) than WT (Fig. 3 F). Hence, the POTRA domain is likely oriented more toward the IMS. These results are consistent with a topology where the SAMM50 C-terminal β-barrel domain is incorporated within the OMM lipid bilayer with most of the POTRA domain proximal to the β-barrel domain located in the IMS, while the NTS (aa 1–40) is exposed to the cytosol.
SAMM50 mediates basal lysosomal degradation of SAM and MICOS complex proteins
Given the interaction of SAMM50 with the autophagy receptor p62, it was of interest to explore if SAMM50 had any role in mitophagy. To analyze this, we treated both WT and SAMM50 KD cells with bafilomycin A1 (BafA1), a potent lysosomal inhibitor, and with MG132 (carbobenzoxy-Leu-Leu-leucinal), a membrane-permeable proteasome inhibitor, and analyzed protein levels by immunoblotting (Fig. 4, A and B). MICOS complex proteins MIC19 and MIC60, SAM complex proteins SAMM50, MTX1 and MTX2, and TOM complex protein TOMM40 accumulated upon BafA1 treatment of WT cells. Strikingly, SAMM50 KD cells had no accumulation of these proteins. These proteins were unaffected by MG132 treatment in WT and SAMM50 KD cells. TOMM20, MFN1, MFN2, and DRP1 accumulated with both BafA1 and MG132 in WT cells, but they only accumulated upon proteasomal inhibition in SAMM50 KD cells (Fig. S4, A and B). MUL1 accumulated with BafA1 and MG132 in both WT and SAMM50 KD cells (Fig. S4, A and B). IMM proteins TIMM23 and COXII were not affected by BafA1 or MG132. Clearly, basal autophagic degradation of selected mitochondrial proteins depends on SAMM50.
Figure 4.
SAMM50 mediates basal lysosomal degradation of members of the SAM and MICOS complex. (A) Immunoblots of lysates of WT and SAMM50 KD HeLa cells untreated or treated with BafA1 or MG132 for 24 h. (B) Densitometric analysis of protein levels for A from four independent experiments. Values are mean ± SD. **, P < 0.005; *, P < 0.01; †, NS; one-way ANOVA. (C) Diffraction-limited DV microscopic images of WT HeLa cells stained with antibodies to LAMP2 and SAMM50. Arrows indicate colocalization. Scale bars, 10 µm. (D and E) Immunoblots of extracts from WT and ATG7 KO HeLa cells untreated or treated with BafA1 for 24 h (D) and quantified (E). Values are mean ± SD. **, P < 0.005; *, P < 0.01; †, NS; one-way ANOVA. (F) Immunoblots of extracts of WT, SAMM50 KD, and SAMM50 KD cells reconstituted with Myc-SAMM50 untreated or treated with BafA1 for 24 h. (G) Relative protein levels for F from three independent experiments. Values are mean ± SD. ***, P < 0.001; **, P < 0.005; *, P < 0.01; †, NS; one-way ANOVA. (H) Immunoblots of lysates from WT, SAMM50 KD, and SAMM50 KD reconstituted with either Myc-SAMM50 WT or SAMM50 Δ1–125, untreated or treated with BafA1 for 24 h. (I) Relative protein levels for H from three independent experiments. Values are mean ± SD. **, P < 0.005; †, NS; one-way ANOVA.
Figure S4.
SAMM50 is important for basal mitophagy. (A and B) Lysates from WT and SAMM50 KD HeLa cells left untreated or treated with either BafA1 or MG132 for 24 h, respectively, were immunoblotted with indicated antibodies (A) and quantified (B). Values are mean ± SD from three different experiments. **, P < 0.005; *, P < 0.01; †, NS; one-way ANOVA. (C) Lysates from HeLa cells treated with a combination of Pepstatin A and E64d for 24 h to block lysosomal protein degradation were immunoblotted using the indicated antibodies. (D and E) WT, DRP1 KO (D), and MUL1 KO (E) cells were treated with BafA1 for 24 h followed by immunoblotting with indicated antibodies. (F and G) WT and PINK1 KO cells were left untreated or treated with either BafA1 for 24h (F) or a combination of OA for 3h (G). Indicated protein levels were analyzed by immunoblotting. (H) WT, SAMM50 KD, and SAMM50 KD HeLa cells reconstituted with Myc-SAMM50 WT and Myc-SAMM50 Δ1-40 mutant were either untreated or treated with BafA1 for 24 h. Lysates were prepared and used for immunoblotting with the indicated antibodies. (I) Densitometric analysis of relative protein levels for (H) from three independent experiments. Values are mean ± SD. **, P < 0.005; †, NS; one-way ANOVA. (J and K) Mapping of binding site on SAMM50 for MIC19, MTX1, MTX2, and p62. (J) Myc-tagged proteins were in vitro translated in the presence of radioactive methionine and used in GST-pulldown assay with GST-tagged SAMM50 WT and indicated mutants. (K) In vitro translated SAMM50 WT and indicated mutants with GST and GST-tagged MIC19, MTX2, and p62. Bound Myc-tagged proteins were detected by AR while GST proteins were stained with CBB.
To rule out that accumulation of these mitochondrial proteins was due to impairment of iron degradation caused by inhibition of H+ ATPases (Miles et al., 2017), we treated cells with the lysosomal protease inhibitors pepstatin A and E64D. A similar accumulation of mitochondrial proteins was seen, confirming lysosomal degradation (Fig. S4 C). Diffraction-limited DV microscopic analysis showed colocalization between SAMM50 and LAMP2 under basal conditions (Fig. 4 C). The sites of colocalization appeared as dots, pointing to piecemeal mitophagy. Consistent with selective autophagic turnover of SAM and MICOS complex proteins, these proteins accumulated in ATG7 knockout (KO) cells with no further accumulation upon treatment with BafA1 (Fig. 4, D and E).
SAMM50-dependent basal mitophagy is independent of MDVs
Next, we asked if the SAMM50-dependent mitophagy targeting SAM and MICOS complex proteins could involve MDVs (Neuspiel et al., 2008; Soubannier et al., 2012a). A major characteristic of MDV is independence from DRP1-mediated fission (Neuspiel et al., 2008; Soubannier et al., 2012a). Hence, we made DRP1 KO cells and found basal lysosomal degradation of SAM complex proteins to occur in DRP1 KO cells, suggesting DRP1 is dispensable (Fig. S4 D). MUL1 expression stimulates MDV formation and the formation of TOMM20-containing vesicles (Neuspiel et al., 2008), but TOMM20 was not a substrate for SAMM50-dependent basal mitophagy (Fig. S4, A and B). We made MUL1 KO cells and observed lysosomal turnover in MUL1 KO cells comparable to WT, suggesting MUL1 is also dispensable (Fig. S4 E). Thus, although SAMM50-dependent basal piecemeal mitophagy shares some features with MUL1-induced MDVs, these two pathways are very likely distinct processes involved in mitochondrial quality control. SAMM50 KD cells showed stabilization of PINK1 (Fig. S3, A and B). So, we asked if PINK1 is involved in SAMM50-dependent basal piecemeal mitophagy. We made PINK1 KO HeLa cells and subjected them to lysosomal inhibition for 24 h (Fig. S4 F). As reported earlier (McWilliams et al., 2018), basal mitophagy was comparable between WT cells and PINK1 KO cells (Fig. S4, F and G). Hence, PINK1 is dispensable for this form of basal mitophagy.
The SAMM50 NTS is required for basal mitophagy
Recently, a novel piecemeal mitophagy that degrades selected proteins to maintain mitochondrial network morphology involving LC3C and p62 was reported (Le Guerroué et al., 2017). We hypothesized that this process may be regulated and mediated by SAMM50. The lack of basal lysosomal degradation of selected mitochondrial proteins in SAMM50 KD cells relative to WT was restored upon reconstitution with WT SAMM50 (Fig. 4, F and G). While reconstitution with WT SAMM50 rescued lysosomal degradation, reconstitution with Δ1–125 mutant did not (Fig. 4, H and I), nor did SAMM50 with NTS deleted (SAMM50 Δ1–40; Fig. S4, H and I). Thus, the NTS plays an essential role in basal piecemeal mitophagy of members of SAM and MICOS complexes. We mapped the binding of some of these proteins to SAMM50 (Fig. S4, J and K). MIC19 bound to the POTRA domain, and p62 bound mainly to the β-barrel domain, while MTX1 and -2 required both the N-terminal region and parts of the β-barrel domain. Thus, SAMM50 lacking the NTS bound to MIC19 and the other substrates but was unable to mediate their degradation (Fig. S4, H–K).
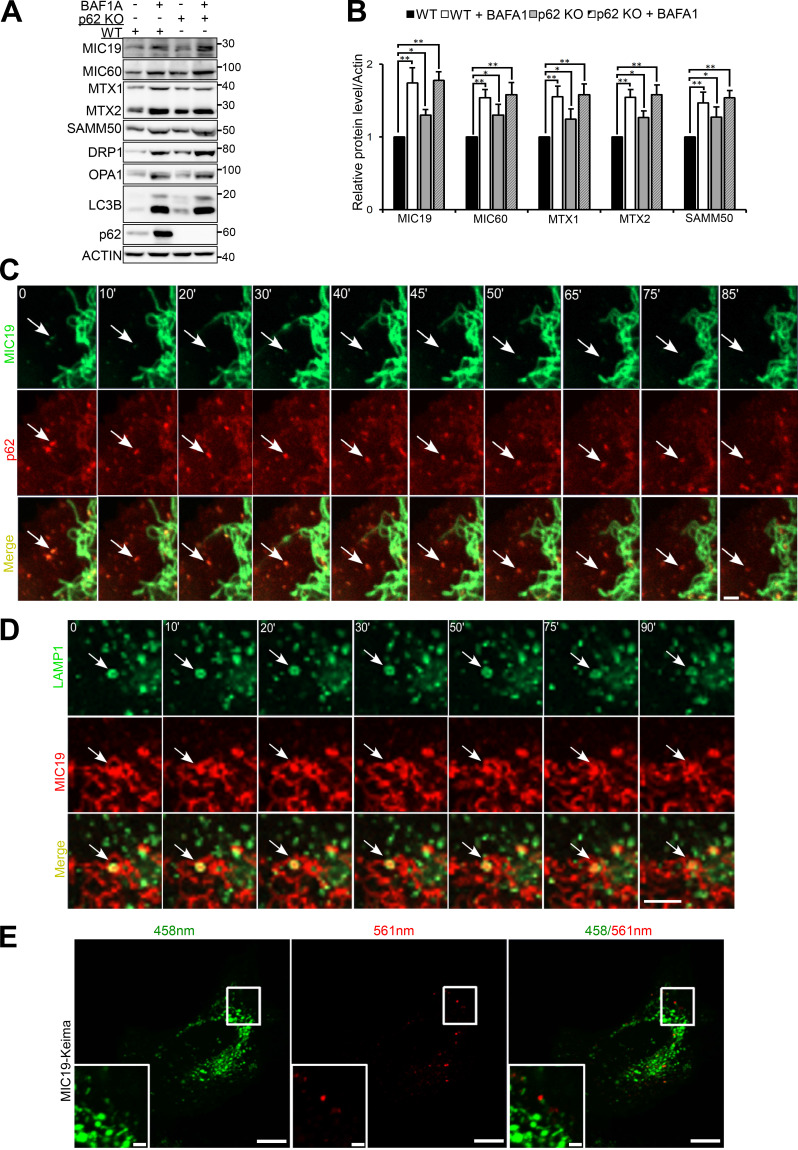
SAMM50 and p62 interact directly (Fig. 1 G and Fig. S4, J and K), and p62 is in the mitochondrial fraction (Fig. 1 B and Fig. S1 B). Thus, we asked if p62 could act as a mitophagy receptor under basal conditions. To test this, we made p62 KO HeLa cells and treated them with BafA1 for 24 h (Fig. 5, A and B). We observed a significant increase of these mitochondrial proteins in p62 KO cells, but there was further accumulation with BafA1. Hence, p62 contributes to this form of basal mitophagy but is dispensable.
Figure 5.
SAMM50-dependent basal piecemeal mitophagy requires p62 and hATG8 proteins. (A and B) Immunoblots of extracts of WT and p62 KO HeLa cells treated with BafA1 for 24 h (A) and quantified (B). Values are mean ± SD from three different experiments. **, P < 0.005; *, P < 0.01; one-way ANOVA. (C) Time-lapse (seconds) live-cell confocal imaging of HeLa cells stably expressing mCherry-p62 and MIC19-EGFP. Arrows show mitochondrial fragment colocalized with p62 and subsequently degraded (see Video 1). Scale bar, 5 µm. (D) Time-lapse (seconds) live-cell confocal imaging of HeLa cells stably coexpressing LAMP1-EGFP and MIC19-mCherry. Arrows indicate mitochondrial fragment engulfed by a lysosome (see Video 2). Scale bar, 2 µm. (E) Live-cell images of HeLa cells stably expressing MIC19-Keima excited at 458 nm in a neutral environment (mitochondria; green) and at 561 nm in an acidic environment (lysosome; red). Scale bars, 10 µm (main), 2 µm (inset).
Time-lapse imaging of HeLa cells stably expressing mCherry-p62 and MIC19-EGFP revealed recruitment of p62 to mitochondrial fragments that were subsequently degraded, as monitored by loss of green fluorescence from MIC19-EGFP and persistence of red fluorescence from mCherry-p62 due to the stability of mCherry in lysosomes (Fig. 5 C and Videos 1 and 2). Time-lapse confocal imaging of HeLa cells stably expressing MIC19-mCherry and EGFP-LAMP1 revealed a lysosome containing MIC19-mCherry, confirming basal piecemeal mitophagy (Fig. 5 D and Video 3). The mitochondrial fragments were seen as distinct entities moving away from the mitochondrial network enclosed inside a LAMP1-positive vesicle or lysosome. These fragments are similar in size (∼1 µm in diameter) to those colocalizing and degraded with p62 (Fig. 5 C). This supports the notion of involvement of p62 in basal piecemeal mitophagy (Fig. 5, A and B; Le Guerroué et al., 2017).
Video 1.
Mitochondrial fragments are recruited to p62-positive puncta and then degraded. Related to Fig. 5 C. This is a video image of a segment of HeLa cell stably co-expressing mCherry-p62 and MIC19-EGFP that were subjected to confocal live cell imaging. Scale bars, 5µm.
Video 2.
This video shows the insert presented in Fig. 5 C of confocal live cell imaging of HeLa cells stably co-expressing mCherry-p62 and MIC19-EGFP. The insert was made from Video 1. Scale bars, 5µm.
Video 3.
Mitochondria fragment engulfed by the lysosome. Related to Fig. 5 D. Confocal live cell imaging of HeLa cells stably co-expressing LAMP1-EGFP and MIC19-mCherry. Scale bars, 2 µm.
In addition, by tagging MIC19 with Keima, a pH-sensitive fluorescent probe, we were able to observe delivery of MIC19-positive mitochondrial fragments to the lysosome (Fig. 5 E).
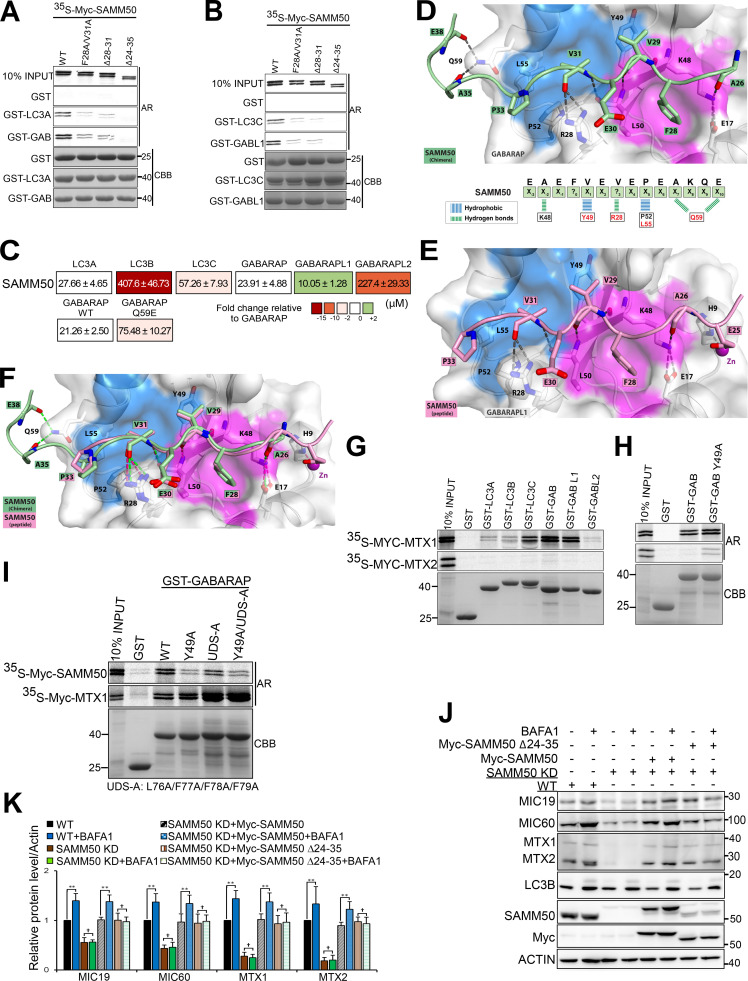
SAMM50 interacts with ATG8 proteins via an LIR motif in the NTS to mediate piecemeal basal mitophagy
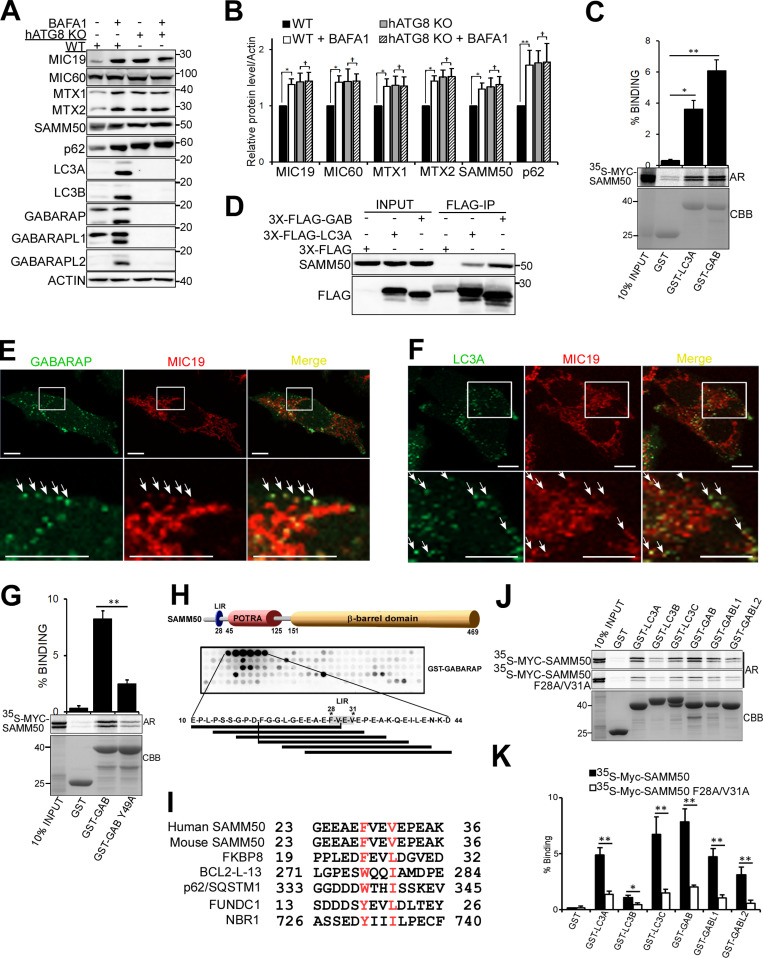
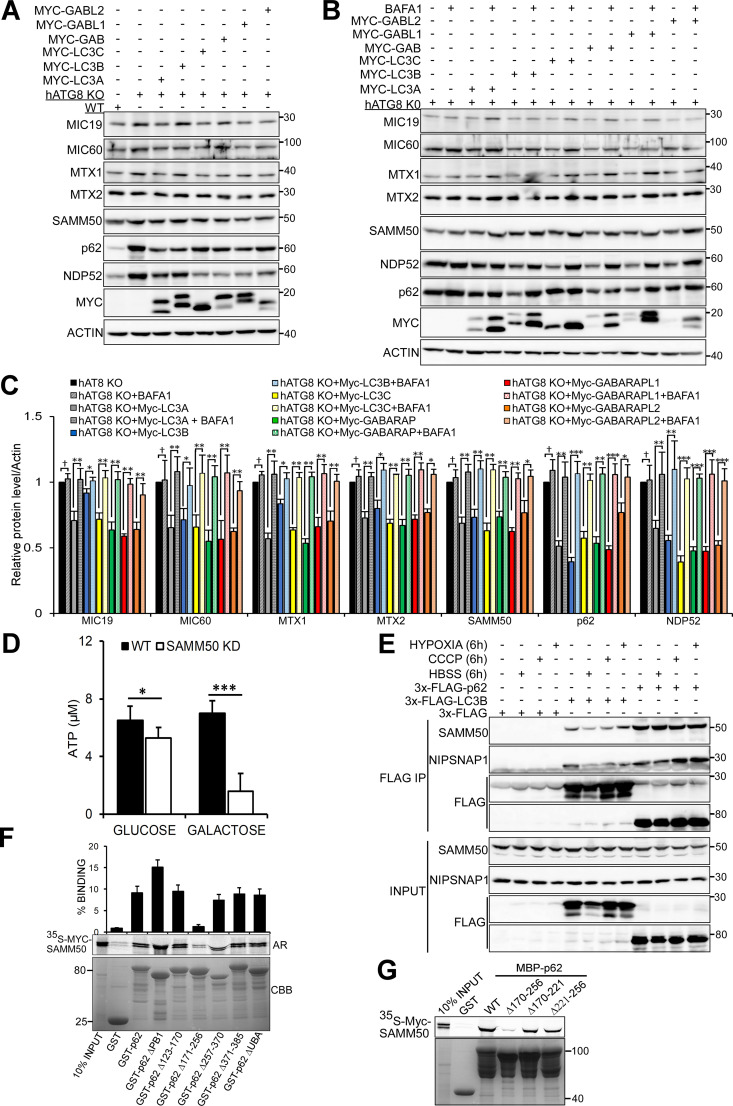
The six human ATG8 family proteins (hATG8s) LC3A, LC3B, LC3C, GABARAP, GABARAPL1, and GABARAPL2 play crucial roles in autophagy (Johansen and Lamark, 2020; Kriegenburg et al., 2018; Shpilka et al., 2011). Interaction between hATG8s and autophagy proteins (receptors, adaptors, and enzymes) is mediated by a short sequence motif first identified in p62 and known as the LIR (Birgisdottir et al., 2013; Kirkin et al., 2009; Pankiv et al., 2007; von Muhlinen et al., 2012). Several mitophagy receptors have LIRs that facilitate degradation of the mitochondria (Bhujabal et al., 2017; Liu et al., 2012; Novak et al., 2010). We used CRISPR/Cas9 to knock out all six ATG8 proteins in HeLa cells and treated the cells with BafA1 for 24 h (Fig. 6, A and B). Basal degradation of SAMM50, MTX1, MTX2, MIC60, and MIC19 was blocked in hATG8 KO cells. We speculated that SAMM50 interacts with ATG8s to selectively deliver mitochondrial fragments to the lysosome. In vitro GST-pulldown assays confirmed direct interaction between SAMM50 and hATG8 family proteins (Fig. 6 C). Endogenous SAMM50 was recovered from immunoprecipitates of cells overexpressing 3xFLAG-tagged LC3A or GABARAP (Fig. 6 D). High-resolution live-cell imaging showed colocalization between stably expressed EGFP-GABARAP and MIC19-mCherry (Fig. 6 E and Videos 4 and 5) and EGFP-LC3A and MIC19-mCherry (Fig. 6 F and Videos 6 and 7), mostly in puncta we suggest are mitochondrial fragments undergoing basal piecemeal mitophagy.
Figure 6.
SAMM50 interacts with hATG8 proteins to regulate basal mitophagy. (A) Immunoblots of extracts of WT and HeLa cells with KO of all six hATG8 proteins untreated or treated with BafA1 for 24 h. (B) Relative protein levels for A from three independent experiments. Values are mean ± SD. **, P < 0.005; *, P < 0.01; †,NS; one-way ANOVA. (C) GST-pulldown assays with in vitro translated Myc-SAMM50 and recombinant GST, GST-LC3A, and GST-GABARAP. Values are mean ± SD from three independent experiments. **, P < 0.005; *, P < 0.01; one-way ANOVA. (D) Coimmunoprecipitation of endogenous SAMM50 from HeLa cells transiently transfected with 3x-Flag-LC3A or 3x-Flag-GABARAP. (E) Live-cell images of HeLa cells stably expressing EGFP-GABARAP and MIC19-mCherry. Arrows indicate colocalization between mitochondrial fragments and GABARAP. Scale bars, 10 µm. See Video 3. (F) Live-cell images of HeLa cells stably expressing EGFP-LC3A and MIC19-mCherry. Arrows indicate colocalization between mitochondrial fragments and LC3A. Scale bars, 10 µm. See Video 4. (G) GST-pulldown assay with in vitro translated Myc-SAMM50 and recombinant GST, GST-GABARAP, and GST-GABARAP F49A LDS mutant. Values are mean ± SD from three independent experiments. **, P < 0.005; one-way ANOVA. (H) Domain architecture of SAMM50 with positions of LIR, POTRA, and β-barrel domains. Peptide array of 20-mer peptides spanning full-length SAMM50 probed with GST-GABARAP and developed with GST antibody. Each peptide was moved three amino acids relative to the previous one. (I) Sequence alignment of canonical LIR motifs in human and mouse SAMM50 and other mitophagy receptors FKBP8, BCL2-L-13, FUNDC1, and autophagy receptors p62 and NBR1. (J and K) GST pulldowns with GST-tagged hATG8s and in vitro translated Myc-SAMM50 WT and the F28A/V31A mutant (J) quantified in K from three independent experiments. Values are mean ± SD. **, P < 0.005; *, P < 0.01; one-way ANOVA.
Video 4.
Mitochondrial fragments colocalize with GABARAP. Related to Fig. 6 E. Confocal live cell imaging of HeLa cells stably co-expressing EGFP-GABARAP and MIC19-mCherry. Scale bars, 5 µm.
Video 5.
This video shows the insert presented in Fig. 6 E of confocal live cell imaging of HeLa cells stably co-expressing EGFP-GABARAP and MIC19-mCherry. The insert was made from Video 4. Scale bars, 5 µm.
Video 6.
Mitochondrial fragments colocalize with LC3A. Related to Fig. 6 F. Confocal live cell imaging of HeLa cells stably co-expressing EGFP-LC3A and MIC19-mCherry. Scale bars, 5 µm.
Video 7.
This video shows the insert presented in Fig. 6 F of confocal live cell imaging of HeLa cells stably co-expressing EGFP-LC3A and MIC19-mCherry. The insert is a portion of Video 6. Scale bars, 5 µm.
ATG8s interact with LIR-containing proteins via the LIR docking site (LDS; Birgisdottir et al., 2013; Ichimura et al., 2008; Noda et al., 2008; Skytte Rasmussen et al., 2017). GST-pulldown assays between in vitro translated SAMM50 and GST-tagged GABARAP WT and the LDS mutant GABARAP Y49A revealed >60% reduction in binding of SAMM50 to the LDS mutant relative to WT, suggesting the presence of an LIR in SAMM50 (Fig. 6 G). An unbiased peptide array scan of SAMM50, using overlapping 20-mer peptides, moved a window of 3 aa to cover the full-length sequence, probed with GST-GABARAP, which revealed a strong single LIR motif. The core consensus sequence FVEV is located between aa 28 and 31 in the NTS (Fig. 6 H). The LIR and surrounding sequences are conserved in both human and mouse (Fig. 6 I). Significant reduction in binding of SAMM50 to all hATG8s was seen upon mutation of core LIR residues F28 and V31 to alanines (Fig. 6, J and K). SAMM50 bound strongly to LC3A, LC3C, and GABARAP proteins (Fig. 6 K). Mutations of core LIR residues did not completely abolish binding, suggesting that several acidic amino acids N-terminal to the core LIR may engage in electrostatic interactions with the basic residues surrounding the LDS. Such electrostatic interactions contribute to the stability and strength of LIR–LDS interactions (Skytte Rasmussen et al., 2017). Hence, binding of SAMM50 to hATG8 proteins was completely abolished when both the LIR and surrounding sequences (Δ24–35) were deleted (Fig. 7, A and B). Deleting only the core LIR (Δ28–31) had the same effect as the point mutations. This confirms the contribution of flanking acidic sequences to the overall stability of the LIR–LDS interaction.
Figure 7.
SAMM50 binds to ATG8 proteins via an LIR motif in the NTS. (A and B) GST pulldowns with GST-hATG8 proteins and in vitro translated WT or LIR mutant Myc-SAMM50. (C) Affinities (Kd values) of SAMM50 LIR peptides to hATG8 proteins determined by BLI. Color code indicates fold changes relative to WT GABARAP. (D and E) Structure of the SAMM50 LIR bound to GABARAP and GABARAPL1. (D) Close-up of chimera structure of SAMM50 LIR bound to GABARAP. SAMM50 LIR (aa 24–38) is in green ribbon with interacting residues as sticks and GABARAP in white cartoon and transparent surface with HP1 and HP2 colored in pink and blue surfaces, respectively. (E) Close-up of SAMM50 LIR bound to GABARAPL1. The LIR (aa 24–38) is in pink ribbon with interacting residues as sticks. GABARAPL1 is in white cartoon, and transparent surface with HP1 and HP2 colored in pink and blue, respectively. (F) Superposition of chimera structure of SAMM50 LIR chimera (green) bound to GABARAP and SAMM50 LIR peptide (pink) bound to GABARAPL1. Both LIRs are in cartoon with interacting residues as sticks. GABARAP and GABARAPL1 are in white cartoon, and transparent surface with HP1 and HP2 colored in pink and blue, respectively. (G and H) GST pulldowns of in vitro translated Myc-MTX1 and Myc-MTX2 with recombinant GST-hATG8s (G) or with GST-GABARAP WT and the Y49A mutant (H). (I) GST pulldowns of in vitro translated Myc-SAMM50 and Myc-MTX1 with recombinant GST-GABARAP and indicated mutants. (J) Immunoblots of lysates of WT, SAMM50 KD, and SAMM50 KD HeLa cells reconstituted with Myc-SAMM50 WT and Myc-SAMM50 Δ24–35 mutant untreated or treated with BafA1 for 24 h. (K) Relative protein levels in J from three independent experiments. Values are mean ± SD. **, P < 0.005; †, NS; one-way ANOVA. UDS, ubiquitin-interacting motif docking site.
Using biolayer interferometry (BLI), we found that the SAMM50 LIR bound the strongest to GABARAPL1, GABARAP, and LC3A with dissociation constant (Kd) values of 10.0, 23.9, and 27.6 µM, respectively, followed by LC3C (Kd of 57.2 µM; Fig. 7 C). To elucidate the molecular basis of the SAMM50-LIR binding to ATG8, we solved the structure of the chimeric protein consisting of GABARAP N-terminally fused to the SAMM5024-38 sequence with a G/S linker. The structure was solved at a resolution of 1.10 Å (Fig. 7 D and Table S1) and displayed canonical LIR interactions comprising SAMM50 hydrophobic residues F28SAMM50 (Θ0) and V31SAMM50 (Γ3) deeply bound to HP1 and HP2 and three hydrogen bonds between the main chains of SAMM50 LIR residues V29SAMM50 (X1) and V31SAMM50 (Γ3) with main chains of GABARAP K48 and L50. Additional specific contacts were observed. Within core LIR, V29SAMM50 in position X1 formed a hydrophobic interaction with Y49GAB of HP2, and V31 (X3) carbonyl is engaged in two hydrogen bonds with R28GAB guanidinium. The adjacent C-terminal region (X4-10) also contributes to binding with P23SAMM50 (X5), forming hydrophobic interactions with P52/L55GAB from HP2, and the carbonyls of A35/E38SAMM50 (X7/10), forming two hydrogen bonds with the side chain of Q59GAB (Fig. 7 D). Similar interactions between the C-terminal region of the LIR motif and hATG8 were observed and discussed recently for other LIR motifs (Wirth et al., 2019). Importantly, there is hydrophobic contact between C-terminal regions of LIR motifs of several LIR-containing proteins with the edge of HP2 of the LDS. Relevant in SAMM50 is the formation of two hydrogen bonds between A35 and E38 of SAMM50 with GABARAP Q59, which is conserved only in GABARAPs and LC3C. Mutation of GABARAP Q59 to glutamate (Q59E) significantly reduced the binding of SAMM50 to GABARAP (>3.5-fold; Fig. 7 C). We also solved the cocrystal structures of SAMM5024-35 peptide (EEAEFVEVEPEA) bound to GABARAPL1 at a resolution of 1.06 Å (Fig. 7 E and Table S1). All interactions in SAMM50:GABARAP chimera structure are conserved in this structure (Fig. 7 F). The LIR sequence used is shorter (lacking X8-10). Thus, the interaction between the C-terminal region of SAMM50 LIR (X7/10) and Q59GABL1 was not seen. We observed an additional interaction bridged by a Zn cation between E25SAMM50 (X-3) and H9GABL1 (Fig. 7 E). The Zn cation is coming from the crystallization solution. We do not think this interaction is physiologically relevant.
GST pulldowns showed a direct interaction between MTX1 and hATG8s with preference for LC3C, GABARAP, and GABARAPL1 (Fig. 7 G). MTX2 did not interact with hATG8s (Fig. 7 G). MTX1 bound equally well to the GABARAP LDS mutant (Y49A), suggesting LIR-independent binding (Fig. 7 H). Interactions with ATG8 can also occur via a unique hydrophobic surface C-terminal to the LDS. This region binds to ubiquitin-interacting motif (UIM)–like sequences rather than to canonical LIR sequences (Marshall et al., 2019). We mutated this UIM-docking site alone or in concert with LDS and tested interaction with MTX1 and SAMM50 (Fig. 7 I). MTX did not interact via either the UIM-docking site or LDS, while SAMM50 required the LDS. In fact, MTX1 bound more strongly to the GABARAP LDS mutant (Fig. 7, H and I). This suggests a scenario where first SAMM50 binds to hATG8 in an LIR-dependent manner. This LIR–LDS interaction allows a stronger binding of MTX1 to another site on the same hATG8, providing the stability and strength required to recruit the mitochondrial piecemeal substrate(s) to forming autophagosomes. To test the role of SAMM50 LIR in basal piecemeal mitophagy, we reconstituted SAMM50 KD cells with the deletion mutant of both the core LIR and surrounding regions (Δ24–35). Treatment with BafA1 for 24 h revealed restoration of basal piecemeal mitophagy in cells reconstituted with WT but not with the LIR deletion mutant (Fig. 7, J and K).
Upon reconstitution of hATG8 KO cells with individual Myc-tagged ATG8 proteins, we observed reduction in MICOS and SAM complex proteins with all ATG8 proteins (Fig. S5 A). BafA1 treatment accumulated SAMM50, MTX1, MTX2, MIC19, and MIC60 in cells rescued with individual ATG8 proteins (Fig. S5, B and C). The accumulation was most efficient in cells rescued with LC3A, LC3C, and GABARAP proteins. This is in line with the binding preference of SAMM50 for LC3A, LC3C, and GABARAP proteins (Fig. 6, J and K). Together, these results suggest that SAMM50-dependent basal piecemeal mitophagy depends on SAMM50 LIR–hATG8 interactions with support from MTX1.
Figure S5.
hATG8 proteins are required for basal mitophagy. (A) Lysates from WT, hATG8 KO, and hATG8 KO HeLa cells reconstituted with individual Myc-tagged hATG8 proteins were immunoblotted with indicated antibodies. (B and C) hATG8 KO cells were reconstituted with individual Myc-tagged human ATG8 proteins, and cells were treated or not with BafA1 for 24h. Lysates were immunoblotted with indicated antibodies (B), and the ability of individual ATG8 proteins to restore basal mitophagy monitored as an increase in mitochondrial protein level upon treatment with BafA1 were quantified (C). Values are mean ± SD. ***, P < 0.001; **, P < 0.005; *, P < 0.01; †, NS; one-way ANOVA. (D) Relative ATP levels from WT and SAMM50 KD cells grown in either glucose or galactose media were measured with an ATP determination kit. Values are mean ± SD based on three independent experiments. ***, P < 0.001; *, P < 0.01; one-way ANOVA. (E) Extracts from HeLa cells transiently transfected with 3x-FLAG, 3x-FLAG-LC3B, and 3x-FLAG-p62 expression vectors and treated with CCCP, hypoxia (1% oxygen), and HBSS for 6h were immunoprecipitated with FLAG resin. Co-immunoprecipitation of endogenous NIPSNAP1 and SAMM50 was analyzed by immunoblotting. (F and G) Mapping of the SAMM50 binding site on p62. Myc-SAMM50 in vitro translated in the presence of radioactive methionine was incubated with recombinant GST, GST-p62 WT, and indicated deletion constructs (F) or with recombinant MBP, MBP-p62, and indicated mutants (G). Bound SAMM50 was analyzed by AR, and GST-tagged proteins were stained with CBB. The graphs in F represent percentage binding of in vitro translated Myc-SAMM50 to recombinant GST proteins. Values are mean ± SD based on three independent experiments.
p62 cooperates with SAMM50 to mediate OXPHOS-induced mitophagy
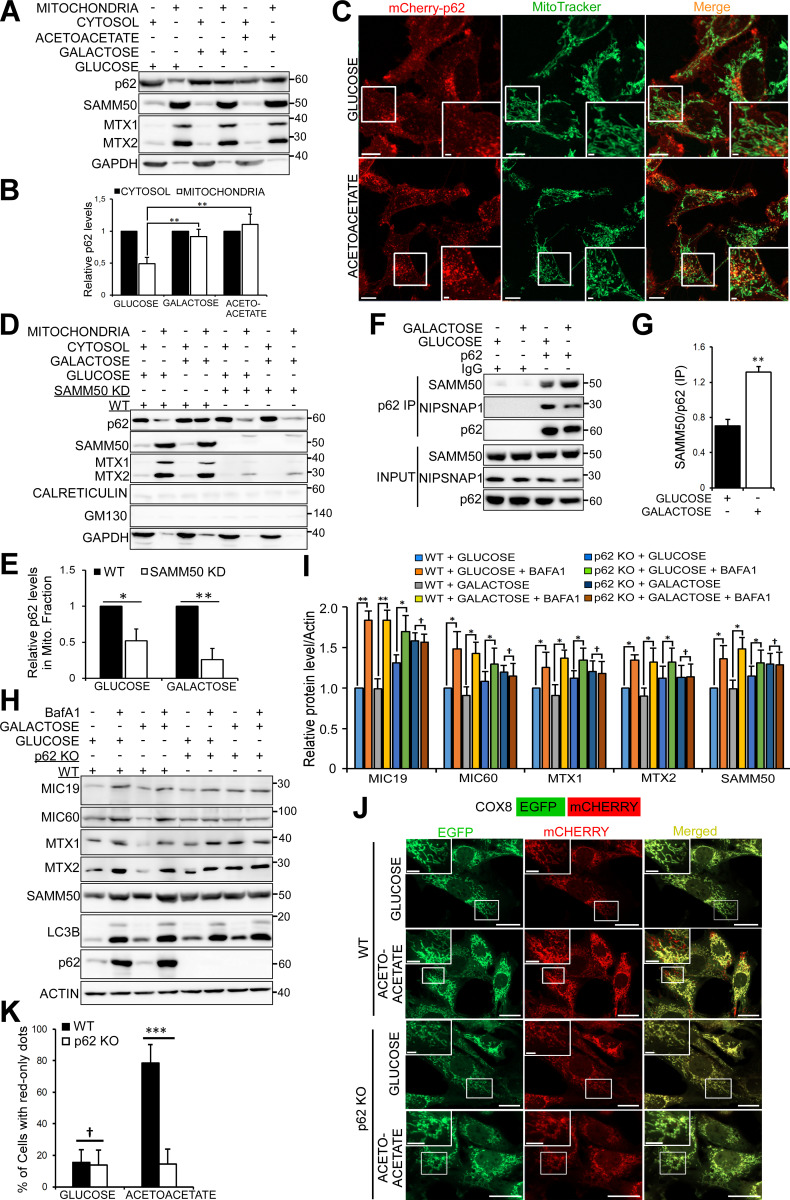
Most normal cells generate ATP through mitochondrial OXPHOS (Stefano and Kream, 2015). Glucose-free media containing galactose or acetoacetate induce cells to up-regulate OXPHOS (MacVicar and Lane, 2014; Melser et al., 2013; Mishra et al., 2014). OXPHOS induces basal mitophagy (Melser et al., 2013), allowing cells to accommodate oxidative by-products while maintaining cellular homeostasis. When both normal and tumor cells are grown in glucose-free media, the dependence of these cells on OXPHOS or the metabolic shift from aerobic glycolysis to OXPHOS leads to increased basal mitophagy. p62 has been implicated in OXPHOS-dependent basal mitophagy (Rojansky et al., 2016). We hypothesized that p62 and SAMM50 cooperate in mediating basal mitophagy during metabolic switch to OXPHOS. Consistent with this notion, we observed an increase of p62 in the mitochondrial fraction in cells grown in glucose-free media containing either galactose or acetoacetate (Fig. 8, A and B). Live-cell imaging revealed an increase in p62 puncta colocalized with fragmented mitochondria (Fig. 8 C), suggesting that OXPHOS-induced basal mitophagy is executed in a piecemeal fashion. However, there was a substantial reduction in the relative amount of p62 in the mitochondrial fraction in SAMM50 KD cells compared with WT in OXPHOS-inducing conditions (Fig. 8, D and E). Immunoprecipitation of endogenous p62 from cells grown in galactose-containing media showed an increased interaction between p62 and SAMM50 compared with cells grown in glucose-containing media (Fig. 8, F and G).
Figure 8.
p62/SQSTM1 is indispensable for OXPHOS-induced mitophagy. (A) Immunoblots of cytosolic and mitochondrial fractions of HeLa cells grown in media with glucose, galactose, or acetoacetate as the sole sugar source. (B) p62 protein levels in A from three independent experiments with mean ± SD. **, P < 0.005; one-way ANOVA. (C) Live-cell imaging of HeLa cells stably expressing mCherry-p62 and stained with MitoTracker. Cells were grown in media with either glucose or acetoacetate. Scale bars, 10 µm (main), 2 µm (inset). (D) Immunoblots of cytosolic and mitochondrial fractions from WT and SAMM50 KD cells grown in glucose or galactose media. (E) Quantification of p62 recruitment to mitochondria (Mito.) in D. Values are mean ± SD from three different experiments. **, P < 0.005; *, P < 0.01; one-way ANOVA. (F) Immunoprecipitates (IP) of endogenous p62 from HeLa cells grown in glucose or galactose media analyzed for coprecipitation of endogenous SAMM50 and NIPSNAP1 by immunoblotting. (G) Quantification of coimmunoprecipitated (IP) SAMM50 in F from three independent experiments. Values are mean ± SD. **, P < 0.005; one-way ANOVA. (H and I) Immunoblots of WT and p62 KO HeLa cells grown in glucose or galactose media untreated or treated with BafA1 for 24 h (H) with quantifications from three independent experiments (I). Values are mean ± SD. **, P < 0.005; *, P < 0.01; †, NS; one-way ANOVA. (J) Live-cell images of WT and p62 KO MEFs stably expressing COX8-EGFP-mCherry grown in glucose media or glucose-free acetoacetate-containing media for 96 h. Scale bars, 20 µm. (K) Percentage of cells with red-only dots signifying mitophagy quantified in J from three independent experiments. Values are mean ± SD. ***, P < 0.001; †, NS; one-way ANOVA.
In glucose-containing media, there was only a modest reduction in ATP levels between WT and SAMM50 KD cells (Figs. S2 F and S5 D). However, SAMM50 KD cells grown in glucose-free, galactose-containing media displayed a substantial reduction in ATP levels (Fig. S5 D). The shift in ATP production from glycolysis to OXPHOS drives the need for OXPHOS-related mitochondrial proteins as well as degradation and replenishment of selected proteins to maintain mitochondrial morphology and function (Melser et al., 2013). Depletion of SAMM50 may block both biogenesis and integration of core OXPHOS-related proteins as well as piecemeal mitophagy required to replenish worn-out proteins. Subsequently, the efficiency of mitochondrial OXPHOS will be reduced. We have reported that p62 and other sequestosome-like receptors interact with the mitochondrial proteins NIPSNAP1 and NIPSNAP2 to regulate Parkin-dependent mitophagy (Abudu et al., 2019). Consistently, immunoprecipitation of FLAG-p62 showed an increased interaction between p62 and NIPSNAP1 when cells were treated with carbonyl cyanide 3-chlorophenylhydrazone (CCCP) or exposed to hypoxia. However, there was no increased interaction between p62 and SAMM50, ruling out a role for SAMM50-dependent mitophagy during these conditions (Fig. S5 E).
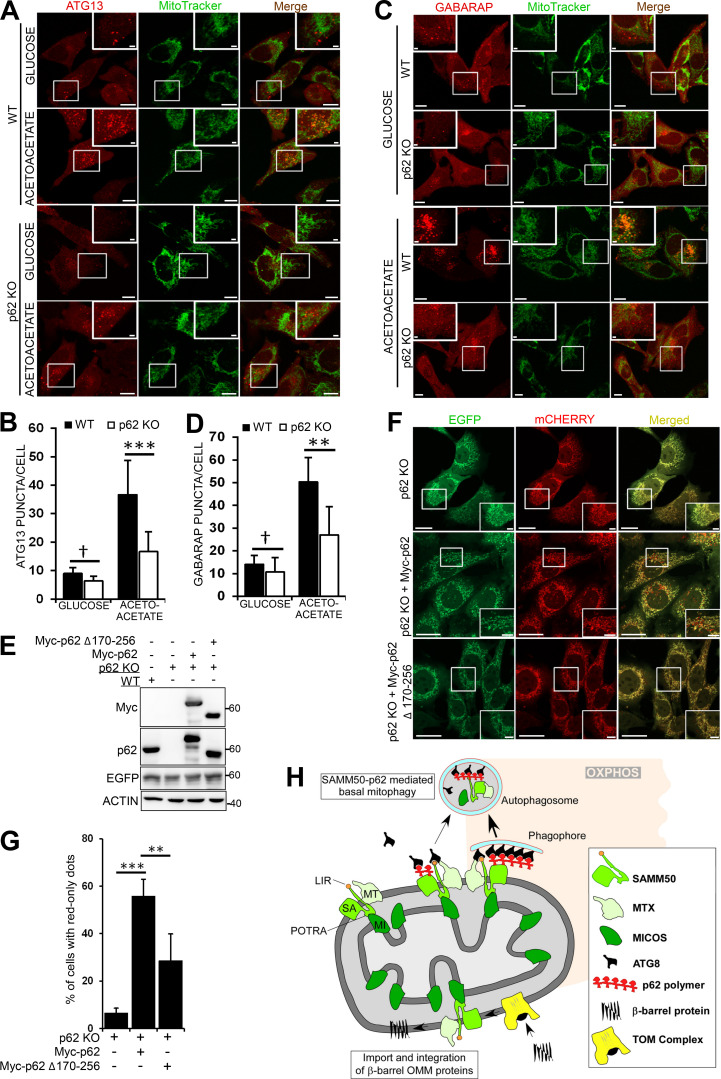
Consistent with a role of p62 in OXPHOS-dependent basal mitophagy (Rojansky et al., 2016), there was no mitophagy in p62 KO cells grown in galactose media (Fig. 8, H and I). We stably expressed a tandem tag COX8-EGFP-mCherry mitophagy reporter in WT and p62 KO mouse embryonic fibroblast (MEF) cells grown in glucose-free, acetoacetate-containing media for 96 h. Live-cell imaging revealed red-only dots in almost 80% of WT MEF cells reporting up-regulated basal mitophagy following metabolic switch (Fig. 8, J and K). Very few red-only dots appeared in p62 KO cells. ATG13 puncta are representative of autophagosome formation sites (Alers et al., 2014; Karanasios et al., 2013; Karanasios et al., 2016; Mizushima et al., 2011; Nishimura and Mizushima, 2017). In glucose-containing media, very few ATG13 dots were seen in both WT and p62 KO cells stably expressing mCherry-tagged ATG13 (Fig. 9, A and B). Upon switch to acetoacetate-containing media, there was a substantial increase in ATG13 puncta in WT cells, indicating induction of autophagy, but only few ATG13 puncta in p62 KO MEF cells (Fig. 9, A and B). Several ATG13 puncta colocalized or were in close proximity to mitochondria (Fig. 9 A, right panel). There was also an increase in GABARAP puncta in WT cells grown in acetoacetate-containing media compared with p62 KO cells (Fig. 9, C and D). p62 interacts with FIP200 of the ULK complex (Turco et al., 2019). Thus, p62 puncta may act as sites of autophagosomal biogenesis on mitochondria during switch to OXPHOS. Consistently, mitochondrial fragments colocalize with p62 in distinct puncta before being degraded (Fig. 5 C). We mapped the binding site of SAMM50 to aa 170–256 of p62 (Fig. S5, F and G). This confirms direct binding and that ubiquitination, or ubiquitin binding, may not be required. We reconstituted p62 KO MEFs with myc-tagged p62 WT or SAMM50-binding deficient mutant (Δ170–256; Fig. 9 E) and grew the cells in acetoacetate-containing media for 96 h. Live-cell imaging revealed red-only dots in cells reconstituted with p62 WT, showing restoration of mitophagy (Fig. 9, F and G). A significant reduction in red-only dots in cells reconstituted with the SAMM50 binding–deficient p62 mutant suggests a role for the p62 interaction with SAMM50 in OXPHOS-induced mitophagy (Fig. 9, F and G). Together, these results suggest a cooperation between p62 and SAMM50 to selectively deliver mitochondrial proteins (or fragments) to the lysosome during metabolic switch to OXPHOS.
Figure 9.
The interaction between SAMM50 and p62/SQSTM1 is important for OXPHOS-induced mitophagy. (A) Live cell images of WT and p62 KO MEF cells stably expressing mCherry-ATG13 grown in glucose media or glucose-free acetoacetate-containing media for 96 h. Cells were stained with MitoTracker. Enlarged insets are indicated. Scale bars, 10 µm (main), 2 µm (inset). (B) ATG13 puncta quantified per cell in A in ∼60 cells for each treatment. Values are mean ± SD. ***, P < 0.001; †, NS; one-way ANOVA. (C) Live-cell images of WT and p62 KO MEFs stably expressing mCherry-GABARAP grown in glucose or acetoacetate media for 96 h and stained with MitoTracker. Scale bars, 10 µm (main), 2 µm (inset). (D) GABARAP puncta per cell quantified in C in ∼60 cells for each treatment. Values are mean ± SD. **, P < 0.005; †, NS; one-way ANOVA. (E) Immunoblots of lysates of WT, p62 KO, and p62 KO MEFs reconstituted with Myc-p62 or Myc-p62 Δ170–256 stably expressing COX8-EGFP-mCherry. (F) Live-cell images of p62 KO MEFs and p62 KO MEFs reconstituted with Myc-p62 or Myc-p62 Δ170–256 stably expressing COX8-EGFP-mCherry grown in media containing acetoacetate for 96 h. Scale bars, 20 µm (main), 5 µm (inset). (G) Percentage of cells with red-only dots signifying mitophagy quantified in F from three different experiments. Values are mean ± SD. ***, P < 0.001; **, P < 0.005; one-way ANOVA. (H) Model for basal piecemeal and OXPHOS-induced mitophagy. SAMM50 interacts with SAM and MICOS complex proteins and recruits hATG8 and p62 to these fragments. The fragments are recruited to p62 puncta, which mark sites of forming autophagosomes. The fragmented mitochondria are enclosed in the autophagosome and subsequently degraded in the lysosome.
Discussion
We describe a SAMM50-dependent basal piecemeal mitophagy of SAM and MICOS complex components. SAMM50 has a dual role in mitochondrial quality control by exerting basal piecemeal mitophagy to replace “worn out” MICOS and SAM complexes and facilitate membrane insertion of newly synthesized β-barrel proteins into the OMM. We present a model (Fig. 9 H) where SAMM50 induces mitophagosome formation at the OMM in regions containing SAM and MICOS complex proteins that need turnover. By direct interactions between SAMM50 and components of the SAM and MICOS complexes and subsequent binding to hATG8s supported by MTX1, these basal piecemeal cargoes are recruited to p62-generating sites of autophagosome formation. The autophagosomes fuse with lysosomes while still proximal to the mitochondrial network.
The LIR motifs of selective autophagy receptors mediate binding to hATG8 proteins acting as bridges between receptor-bound cargo and the forming autophagosome (Birgisdottir et al., 2013; Kirkin et al., 2009; Pankiv et al., 2007; von Muhlinen et al., 2012). Consistent with this model, we observed a block in basal piecemeal mitophagy in ATG8 KO cells and identified an LIR motif in the NTS of SAMM50 required for interaction with hATG8 proteins. The SAMM50 LIR binds best to GABARAPL1, GABARAP, LC3C, and LC3A (Kd values from 10.0 to 57 µM) and more poorly to LC3B. We solved two crystal structures at high resolution, showing that, in addition to the canonical interaction formed between SAMM50 core LIR motif and GABARAP, the C-terminal region of the LIR motif (residues X5, X7, and X10) is important for GABARAP binding. X5 is engaged in hydrophobic interactions with the edge of HP2, while X7/X10 form hydrogen bond with Q59GAB. A recent study (Wirth et al., 2019) highlighted a critical role of the C-terminal region of various LIR motifs in hATG8 binding and selectivity. Similarly to SAMM50 LIR, they observed hydrophobic contact between the C-terminal region of LIR motifs with the edge of HP2 and some hydrogen bonds with Q59, which is only conserved among GABARAPs and LC3C (E62 in LC3A and LC3B). Interestingly, mutation of Q59 by a glutamate (Q59EGAB) reduced SAMM50 binding by >3.5-fold.
MTX1 strongly interacted with hATG8 proteins through an unknown interaction surface. Binding of MTX1 to GABARAP increased upon disruption of the LDS. Hence, LIR binding of SAMM50 to an hATG8 protein may induce a structural change in the hATG8 protein, allowing a strong binding of MTX1 to the same hATG8 protein. Recently, a novel LC3C-dependent piecemeal mitophagy that selectively removes MTX1 and MIC60 to maintain mitochondrial morphology was reported (Le Guerroué et al., 2017). Consistent with this work, we discovered that basal lysosomal degradation of MTX1, MTX2, MIC19, and MIC60 was dependent on SAMM50.
Upon switch from glycolysis to OXPHOS, activity and tension on the cristae and MICOS proteins will increase, since both MICOS and SAM complex proteins are involved in assembly of respiratory complexes (Ding et al., 2015; Ott et al., 2012). Hence, piecemeal mitophagy is required to maintain a healthy mitochondrial network because cells rely on OXPHOS. We showed that SAMM50 cooperates with p62/SQSTM1 to selectively deliver MICOS and SAM complex proteins to the lysosome to maintain mitochondrial integrity. Upon switch to OXPHOS, interaction between p62 and SAMM50 increases, and depletion of SAMM50 reduces, recruitment of p62 to mitochondria. Using the COX8-EGFP-mCherry mitophagy flux reporter, we discovered that basal mitophagy increased in WT MEFs upon switch to OXPHOS with no substantial increase in MEFs lacking p62. We saw a substantial reduction in ATG13 and GABARAP puncta in p62 KO MEFs upon metabolic switch. Many of these puncta were close to the mitochondrial network, indicating a role for p62 in autophagosome formation on mitochondria. Consistently, p62 is reported to interact with FIP200 to promote autophagosome formation (Turco et al., 2019). p62 is involved in metabolic reprogramming during neurodifferentiation by controlling expression of key enzymes of glycolysis and OXPHOS (Calvo-Garrido et al., 2019). We suggest that p62 also assembles early autophagic complexes to induce autophagosome formation during OXPHOS-induced mitophagy. Consistently, mitochondrial fragments were recruited to p62-positive puncta for degradation during basal piecemeal mitophagy. The ability of p62 to polymerize and create multivalent binding surfaces for ATG8s and SAMM50 may give the process the effectiveness required during OXPHOS.
We found the POTRA domain to be dispensable for mitochondrial β-barrel protein biogenesis and integration. The recent cryo-EM structures of fungal Sam50 showed the POTRA domain located in the IMS. Using a combination of protease protection and split fluorescent complementation assays, we suggest a topology of human SAMM50 with the POTRA domain in the IMS, but with the NTS traversing the membrane, exposing the LIR to the cytosol.
SAMM50-dependent basal piecemeal mitophagy shares similarities with MDVs in cargo selectivity and DRP1 independence (Neuspiel et al., 2008; Soubannier et al., 2012a) but is MUL1 independent, and it differs in terms of cargo such as TOMM20. PINK1 is also dispensable for basal piecemeal mitophagy.
We reveal a role of SAMM50 in basal piecemeal mitophagy of both the SAM and MICOS complexes that is very likely vital for an efficient quality control of these complexes. This is particularly the case upon switch to OXPHOS, where the dependency on p62/SQSTM1 is clearly manifested.
Materials and methods
Antibodies and reagents
The following primary antibodies were used: mouse anti-DLP1 mAb (BD Biosciences; 611112), mouse anti-CHCHD3 polyclonal antibody (Abcam; ab69328), rabbit anti-mitofilin polyclonal antibody (Novus Biologicals; NB100-1919), mouse anti-MTX1 mAb (Santa Cruz Biotechnology; sc-514469), mouse anti-MTX2 mAb (Santa Cruz Biotechnology; sc-514231), rabbit anti-SAMM50 polyclonal antibody (Novus Biologicals; NBP1-84509), rabbit anti-SAMM50 polyclonal antibody (Novus Biologicals; NBP2-20257), mouse anti-TOMM40 mAb (Santa Cruz Biotechnology; sc-365467), mouse anti-TOMM20 mAb (Santa Cruz Biotechnology; sc-17764), rabbit anti-TOMM20 polyclonal antibody (Santa Cruz Biotechnology; sc-11415), mouse anti–p62-Lck ligand mAb (BD Biosciences; #610833), mouse anti-TIMM23 mAb (BD Biosciences; 611223), rabbit anti-NIPSNAP1 mAb (Cell Signaling Technology; 13226), rabbit anti-GAPDH polyclonal antibody (Sigma-Aldrich; G9545), mouse anti–Myc-tag mAb (Cell Signaling Technology; 2276), rabbit anti-FIS1 polyclonal antibody (Abcam; ab71498), rabbit anti–mitochondrial fission factor polyclonal antibody (Abcam; ab81127), mouse anti-MFN1 mAb (Santa Cruz Biotechnology; sc-166644), mouse anti-MFN2 mAb (Santa Cruz Biotechnology; sc-100560), mouse anti-OPA1 mAb (BD Biosciences; 612607), mouse anti-VDAC1 mAb (Abcam; ab14734), rabbit anti-actin polyclonal antibody (Sigma-Aldrich; A2066), rabbit anti-LC3B polyclonal antibody (Novus Biologicals; NB100-2220), rabbit anti-GFP polyclonal antibody (Abcam; ab290), rabbit anti-MUL1 polyclonal antibody (Sigma-Aldrich; HPA026827), rabbit anti-MUL1 polyclonal antibody (Sigma-Aldrich; SAB2702071), mouse anti-DNA mAb (Progen; AC-30-10), mouse anti-MTCO2 mAb (Abcam; ab110258), mouse anti–cytochrome C mAb (Abcam; ab110325), mouse anti-GBAS mAb (LSBio; LS-B13280), rabbit anti–phosphatidylserine decarboxylase polyclonal antibody (Santa Cruz Biotechnology; sc-86197), rabbit anti–superoxide dismutase 2 polyclonal antibody (Santa Cruz Biotechnology; sc-30080), rabbit anti–choline dehydrogenase polyclonal antibody (Santa Cruz Biotechnology; sc-102442), rabbit anti-FKBP8 polyclonal antibody (Abcam; ab96322), mouse anti-FLAG M2 mAb (Sigma-Aldrich; F1804), rabbit anti-FUNDC1 polyclonal antibody (AvivaSystems; ARP53280-P050), rabbit anti-cleaved caspase-3 mAb (Cell Signaling Technology; 9661), rabbit anti–pyruvate dehydrogenase mAb (Cell Signaling Technology; 2784), rabbit anti-PINK1 mAb (Cell Signaling Technology; 6946), mouse anti–hypoxia-inducible factor 1A mAb (BD Biosciences; 610959), rabbit anti-NDP52 mAb (Cell Signaling Technology; 60732), rabbit anti-ATG7 mAb (Cell Signaling Technology; 8558), rabbit anti-LC3A mAb (Cell Signaling Technology; 4599), mouse anti-GABARAP mAb (MBL; M135-3), rabbit anti-GABARAPL1 mAb (Cell Signaling Technology; 26632), mouse anti-GABARAPL2 mAb (MBL; PM038), rabbit anti-ATG7 mAb (Cell Signaling Technology; 8558), mouse anti-LAMP2 mAb (Santa Cruz Biotechnology; sc-18822), rabbit anti-GST polyclonal antibody (Santa Cruz Biotechnology; sc-459), rabbit mAb IgG isotype control (Cell Signaling Technology; 3900), and mouse IgG isotype control antibody (LSBio; LS-C355904).
Secondary antibodies used included HRP-conjugated anti-GST antibody (Sigma-Aldrich; GERPN1236), HRP-conjugated goat anti-rabbit IgG (BD Biosciences; 554021), HRP-conjugated goat anti-mouse IgG (BD Biosciences; 554002), Alexa Fluor 555–conjugated goat anti-rabbit IgG (Thermo Fisher Scientific; A-21428), Alexa Fluor 488–conjugated goat anti-rabbit IgG (Thermo Fisher Scientific; A-11008), Alexa Fluor 555–conjugated goat anti-mouse IgG (Thermo Fisher Scientific; A-21424), and Alexa Fluor 488–conjugated goat anti-mouse IgG (Thermo Fisher Scientific; A-11029).
Reagents used were as follows: DAPI (Thermo Fisher Scientific; 62248), tetramethylrhodamine ethyl ester (TMRE; Thermo Fisher Scientific; T669), MitoTracker Deep Red FM (Thermo Fisher Scientific; M22426), CCCP (Sigma-Aldrich; C2759), oligomycin (Sigma-Aldrich; O4876), antimycin A (Sigma-Aldrich; A8674), Mito Stress Test (Agilent Technologies; 103010-100), pepstatin A (Sigma-Aldrich; P5318), E-64d (Sigma-Aldrich; E8640), BafA1 (Sigma-Aldrich; B1793), MG132 (Z-Leu-Leu-Leu-al; Sigma-Aldrich; C2211), etoposide (Sigma-Aldrich; E1383), proteinase K (Sigma-Aldrich, P2308; Takara, 9033), HBSS (Sigma-Aldrich; H8264), hygromycin (Thermo Fisher Scientific; 10687-010), Tet (Sigma-Aldrich; 87128), and doxycycline (Sigma-Aldrich; D9891).
Plasmids
Plasmids used in this study are presented in Table 1. Details are available on request. Gateway (GW) BP and LR recombination reactions were done according to the instructions in the GW-cloning manual (Thermo Fisher Scientific). Point mutations and deletions were performed by site-directed mutagenesis. PCR and sequencing oligonucleotides were designed and ordered from Sigma-Aldrich. Restriction digestion and DNA sequencing (BigDye, 4337455; Applied Biosystems) were used to verify all plasmids.
Table 1. Plasmids used in this study.
| Vector | Description | Source |
|---|---|---|
| pDONR221 | Gateway donor vector | Thermo Fisher Scientific |
| pENTR1A | Gateway entry vector | Thermo Fisher Scientific |
| pDestMyc | Mammalian N-terminal Myc-tag fusion expression vector with CMV and T7 promoters | (Lamark et al., 2003) |
| pDest-3xFLAG | Mammalian N-terminal 3xFlag-tag fusion expression vector with CMV promoter | (Jain et al., 2010) |
| pDest15 | Bacterial GST fusion expression vector with a T7 promoter | Thermo Fisher Scientific |
| pENTR223-SAMM50 | Human SAMM50 in Gateway entry vector | This study |
| pDestMyc-SAMM50 | Human SAMM50 with N-terminal Myc tag | This study |
| pDest15 SAMM50 | Human SAMM50 with N-terminal GST fusion tag | This study |
| pDest15 p62 | Human p62 with N-terminal GST fusion tag | (Jain et al., 2010) |
| pDest 3xFLAG | Mammalian triple FLAG tag fusion expression vector with CMV promoter | (Jain et al., 2010) |
| pDest 3xFLAG p62 | Human p62 with N-terminal 3xFLAG tag | (Jain et al., 2010) |
| pDest 3xFLAG LC3B | Human LC3B with N-terminal 3xFLAG tag | This study |
| pDest 3xFLAG GABARAP | Human GABARAP with N-terminal 3xFLAG tag | This study |
| pDest15 p62 ΔPB1 | Human p62 with PB1 domain deletion and N-terminal GST fusion tag | This study |
| pDest15 p62 Δ123–170 | Human p62 with 123–170 deletion and N-terminal GST fusion tag | This study |
| pDest15 p62 Δ170–256 | Human p62 with 170–256 deletion and N-terminal GST fusion tag | This study |
| pDest15 p62 Δ256–370 | Human p62 with 256–370 deletion and N-terminal GST fusion tag | This study |
| pDest15 p62 Δ371–385 | Human p62 with 371–385 deletion and N-terminal GST fusion tag | This study |
| pDest15 p62 ΔUBA | Human p62 with UBA domain deletion and N-terminal GST fusion tag | This study |
| pDONR221 MIC60 | Human MIC60 in Gateway donor vector | Harvard |
| pDest15 MIC60 | Human MIC60 with N-terminal GST fusion tag | This study |
| pENTR223 MIC19 | Human MIC19 in Gateway entry vector | Harvard |
| pDest15 MIC19 | Human MIC19 with N-terminal GST fusion tag | This study |
| pENTR VDAC1 | Human VDAC1 in Gateway entry vector | This study |
| pDest15 VDAC1 | Human VDAC1 with N-terminal GST fusion tag | This study |
| pENTR MTX1 | Human MTX1 in Gateway entry vector | This study |
| pDest15 MTX1 | Human MTX1 with N-terminal GST fusion tag | This study |
| pDestMyc-MTX1 | Human MTX1 with N-terminal Myc tag | This study |
| pENTR MTX2 | Human MTX2 in Gateway entry vector | This study |
| pDestMyc-MTX2 | Human MTX2 with N-terminal Myc tag | This study |
| pDest15 MTX2 | Human MIC60 with N-terminal GST fusion tag | This study |
| pDONR221 SAMM50 Δ1–40 | Human SAMM50 with 1–40 deletion in Gateway donor vector | This study |
| pDONR221 SAMM50 Δ1–70 | Human SAMM50 with 1–70 deletion in Gateway donor vector | This study |
| pDONR221 SAMM50 Δ1–100 | Human SAMM50 with 1–100 deletion in Gateway donor vector | This study |
| pDONR221 SAMM50 Δ1–125 | Human SAMM50 with 1–125 deletion in Gateway donor vector | This study |
| pDestMyc SAMM50 Δ1–40 | Human SAMM50 with 1–40 deletion and N-terminal Myc tag | This study |
| pDestMyc SAMM50 Δ1–70 | Human SAMM50 with 1–70 deletion and N-terminal Myc tag | This study |
| pDestMyc SAMM50 Δ1–100 | Human SAMM50 with 1–100 deletion and N-terminal Myc tag | This study |
| pDestMyc SAMM50 Δ1–125 | Human SAMM50 with 1–125 deletion and N-terminal Myc tag | This study |
| pMXS-puro | Retroviral vector with Moloney murine leukemia virus promoter | CellBiolabs RTV-012 |
| pMRXIP | Retroviral vector and MMLV promoter (backbone is pMRXIP-GFP-STX17; Addgene, 45909) | This study |
| pMXs puro-mCherry PARKIN | Retroviral vector with Human PARKIN and N-terminal mCherry fusion tag | This study |
| pMXs puro-Myc-SAMM50 | Retroviral vector with Human SAMM50 and N-terminal Myc tag | This study |
| pMXs puro-Myc SAMM50 Δ1–40 | Retroviral vector with Human SAMM50 Δ1–40 and N-terminal Myc tag | This study |
| pMXs puro-Myc SAMM50 Δ1–70 | Retroviral vector with Human SAMM50 Δ1–70 and N-terminal Myc tag | This study |
| pMXs puro-Myc SAMM50 Δ1–100 | Retroviral vector with Human SAMM50 Δ1–100 and N-terminal Myc tag | This study |
| pMXs puro-Myc SAMM50 Δ1–125 | Retroviral vector with Human SAMM50 Δ1–125 and N-terminal Myc tag | This study |
| pMRXIP Myc-SAMM50 | Retroviral vector with Human SAMM50 and N-terminal Myc tag | This study |
| pMRXIP Myc-SAMM50 Δ1–40 | Retroviral vector with Human SAMM50 Δ1–40 and N-terminal Myc tag | This study |
| pMRXIP Myc-SAMM50 Δ1–70 | Retroviral vector with Human SAMM50 Δ1–70 and N-terminal Myc tag | This study |
| pMRXIP Myc-SAMM50 Δ1–100 | Retroviral vector with Human SAMM50 Δ1–100 and N-terminal Myc tag | This study |
| pMRXIP Myc-SAMM50 Δ1–125 | Retroviral vector with Human SAMM50 Δ1–125 and N-terminal Myc tag | This study |
| pMRXIP GFP-SAMM50 | Retroviral vector with Human SAMM50 and N-terminal GFP fusion tag | This study |
| pMRXIP GFP-SAMM50 Δ1–125 | Retroviral vector with Human SAMM50 Δ1–125 and N-terminal GFP fusion tag | This study |
| pDest15 LC3A | Human LC3A with N-terminal GST fusion tag | (Pankiv et al., 2007) |
| pDest15 LC3B | Human LC3B with N-terminal GST fusion tag | Pankiv et al., 2007 |
| pDest15 LC3C | Human LC3C with N-terminal GST fusion tag | (Pankiv et al., 2007) |
| pDest15 GABARAP | Human GABARAP with N-terminal GST fusion tag | (Pankiv et al., 2007) |
| pDest15 GABARAPL1 | Human GABARAPL1 with N-terminal GST fusion tag | (Pankiv et al., 2007) |
| pDest15 GABARAPL2 | HumanGABARAPL2 with N-terminal GST fusion tag | (Pankiv et al., 2007) |
| pDest15 GABARAP Y49A | Human GABARAP Y49A with N-terminal GST fusion tag | This study |
| pDONR221 SAMM50 F28A/V31A | Human SAMM50 with F28A/V31A point mutations in Gateway donor vector | This study |
| pDONR221 SAMM50 Δ28–31 | Human SAMM50 with 28–31 deletion in Gateway donor vector | This study |
| pDONR221 SAMM50 Δ24–35 | Human SAMM50 with 24–35 deletion in Gateway donor vector | This study |
| pDestMyc SAMM50 F28A/V31A | Human SAMM50 with F28A/V31A point mutations and N-terminal Myc tag | This study |
| pDestMyc SAMM50 Δ28–31 | Human SAMM50 with 28–31 deletion and N-terminal Myc tag | This study |
| pDestMyc SAMM50 Δ24–35 | Human SAMM50 with 24–35 deletion and N-terminal Myc tag | This study |
| pMXs-puro mCherry-EGFP OMP25TM | Retroviral vector with tandem tag mCherry-EGFP OMP25TM mitophagy reporter | This study |
| pMRXIP Myc-LC3A | Retroviral vector with Human LC3A and N-terminal Myc tag | This study |
| pMRXIP Myc-LC3B | Retroviral vector with Human LC3B and N-terminal Myc tag | This study |
| pMRXIP Myc-LC3C | Retroviral vector with Human LC3C and N-terminal Myc tag | This study |
| pMRXIP Myc-GABARAP | Retroviral vector with Human GABARAP and N-terminal Myc tag | This study |
| pMRXIP Myc-GABARAPL1 | Retroviral vector with Human GABARAPL1 and N-terminal Myc tag | This study |
| pMRXIP Myc-GABARAPL2 | Retroviral vector with Human GABARAPL2and N-terminal Myc tag | This study |
| pMRXIP Myc-SAMM50 | Retroviral vector with Human SAMM50 and N-terminal Myc tag | This study |
| pMRXIP Myc-SAMM50 Δ1–40 | Retroviral vector with Human SAMM50 Δ1–40 and N-terminal Myc tag | This study |
| pMRXIP Myc-SAMM50 Δ1–70 | Retroviral vector with Human SAMM50 Δ1–70 and N-terminal Myc tag | This study |
| pMRXIP Myc-SAMM50 Δ1–100 | Retroviral vector with Human SAMM50 Δ1–100 and N-terminal Myc tag | This study |
| pMRXIP Myc-SAMM50 Δ1–125 | Retroviral vector with Human SAMM50 Δ1–125 and N-terminal Myc tag | This study |
| pMRXIP COX8-EGFP-mCherry | Retroviral vector with tandem tag COX8-EGFP-mCherry mitophagy reporter | This study |
| pCDNA5/FRT-mNG1-10 | Flip-In vector with modified mNeonGreen β-sheet 1–10 (mNG1-10) | This study |
| pMRXIP-mNG11-SAMM50 | Retroviral vector with the Human SAMM50 and N-terminal mNG11 tag | This study |
| pMRXIP-mNG11-SAMM50 | Retroviral vector with the Human p62 and N-terminal mNG11 tag | This study |
| pCDNA5/FRT-MIC19-EGFP | Flip-In vector with Human MIC19 and C-terminal EGFP tag | This study |
| pMRXIP-MIC19-mCherry | Retroviral vector with Human MIC19 and C-terminal mCherry tag | This study |
| pCDNA5/FRT-LAMP1-mCherry | Flip-In vector with Human LAMP1 and C-terminal mCherry tag | This study |
| pMRXIP-LAMP1-EGFP | Retroviral vector with Human LAMP1 and C-terminal EGFP tag | This study |
| pMRXIP-EGFP-p62 | Retroviral vector with Human p62 and N-terminal EGFP tag | This study |
| pMRXIP-EGFP-LC3A | Retroviral vector with Human LC3A and N-terminal EGFP tag | This study |
| pMRXIP-EGFP-GABARAP | Retroviral vector with Human GABARAP and N-terminal EGFP tag | This study |
| pCDH-EF1α-GW-IRES-puromycin | Lentiviral vector with eukaryotic elongation factor 1 alpha (EF1α) promoter | System Biosciences |
| pCDH-EF1α-GW-IRES-puromycin-mCherry-p62 | Lentiviral vector with p62 and N-terminal mCherry tag | This study |
| pCDH-EF1α-GW-IRES-puromycin-Myc-p62 | Lentiviral vector with p62 and N-terminal Myc tag | This study |
| pCDH-EF1α-GW-IRES-puromycin-Myc-p62 Δ170–256 | Lentiviral vector with p62 Δ170-256 and N-terminal mCherry tag | This study |
| pCDH-EF1α-GW-IRES-puromycin-mCherry-ATG13 | Lentiviral vector with ATG13 and N-terminal mCherry tag | This study |
| pCDH-EF1α-GW-IRES-puromycin-mCherry-GABARAP | Lentiviral vector with GABARAP and N-terminal mCherry tag | This study |
Cell culture and treatments
HeLa (American Type Culture Collection; CCL2) and HeLa FlpIn T-REx cells (Thermo Fisher Scientific; R714-07) were grown in DMEM (Sigma-Aldrich; D5796) supplemented with 10% FBS (Biochrom AG; S0615) and 1% streptomycin-penicillin (Sigma-Aldrich; P4333). HEK293 (American Type Culture Collection; CRL-1573) and MEF cells were grown in DMEM (Sigma-Aldrich; D6046) supplemented with 10% FBS and 1% streptomycin-penicillin. For OXPHOS experiments, cells were grown in customized media as described by Mishra et al. (2014). The base medium DMEM (Thermo Fisher Scientific; A14430) lacking glucose, pyruvate, and glutamine was used. For customized glucose-containing media, DMEM was supplemented with 10% FBS (Biowest; S181B), 10 mM glucose (Sigma-Aldrich; 49159), 4 mM glutamine (Sigma-Aldrich; G7513), and 1% streptomycin-penicillin. For glucose-free, galactose-containing media, DMEM was supplemented with 10% FBS, 10 mM galactose (Sigma-Aldrich; G0750), 4 mM glutamine, and 1% streptomycin-penicillin. For acetoacetate-containing media, DMEM was supplemented with 10% FBS, 10 mM acetoacetate (Sigma-Aldrich; A8509), 4 mM glutamine, and 1% streptomycin-penicillin. Cells were tested regularly for mycoplasma. Unless otherwise indicated, all treatments were done for 24 h. Cells were treated with 0.2 µM BafA1, 10 µM MG132, combination of 10 µg/ml pepstatin A and 10 µg/ml E-64d, 10 µM CCCP, combination of 10 µM oligomycin and 4 µM antimycin A, 100 µM etoposide, or exposed to hypoxia (1% oxygen). Treatment with MG132 was supplemented with 20 µM Q-VD-OPh hydrate (APExBIO; A1901) to prevent apoptosis.
siRNA and transient transfections
siRNAs were either ordered from Sigma-Aldrich as custom-made oligonucleotides (CMOs) or purchased from Santa Cruz Biotechnology. The following siRNAs were used: control siRNA, 5′-GAUCCGCAGCGACAUCAACCUGA-3′ (CMO; Santa Cruz Biotechnology; sc37007); MTX1, 5′-AAGUGGUAUGCAGAGGCUAUG-3′ (CMO; Santa Cruz Biotechnology; sc-88250); MTX2, 5′-GGGAAGUCAAACGUAAGAUGA-3′ (CMO; Santa Cruz Biotechnology; sc-9503); TOMM40, 5′-GGAGCUGUUUCCCAUUCAG-3′ (CMO; Santa Cruz Biotechnology; sc-61697). For siRNA experiments, cells seeded in 6-cm plates and grown to 50–60% confluency were transfected with the indicated siRNA using Lipofectamine RNAiMAX (Thermo Fisher Scientific; 13778) according to the manufacturer’s instructions. Media were changed 5 h after transfection. Transfection was repeated every 48 h (pulse), and finally cells were harvested 48 h after final transfection or pulse. For transient overexpression, cells were transfected with the indicated expression vector using Metafectene Pro (Biontex; T040).
CRISPR/Cas9 KO, KD, and generation of stable cell lines
CRISPR-mediated KO and KD were generated as described (Ran et al., 2013). gRNAs targeting different exons of the indicated proteins were annealed and ligated into Bbs1 linearized vectors carrying a WT Cas9 and puromycin resistance gene (Addgene; 62988). HeLa cells were seeded into 6-cm plates and transfected with the gRNA-containing Cas9 vector using Metafectene Pro (Biontex; T040). 24 h after transfection, cells were selected by treatment with 1 µg/ml puromycin for 36 h. Single clones were sorted into 96-well plates and expanded. KD and/or KO were screened by immunoblotting and/or genomic analysis. For genomic analysis, DNAs were extracted using the GenElute Mammalian Genomic DNA Miniprep Kit (Sigma-Aldrich; G1N350), and the area of interest was amplified by PCR. The PCR-amplified region of interest was ligated into the pGEM-T vector (Promega; A3600) and sequenced to identify insertions or deletions.
CRISPR gRNA primers used included the following: SAMM50, 5′-GTAAGAGGCCCAGCGACCCG-3′, 5′-CATGGGGACTGTGCACGCCC-3′; ATG7, 5′-AGAAGAAGCTGAACGAGTAT-3′; DRP1, 5′-GAGGTACTGGAATTGTCACC-3′; MUL1, 5′-CTTGAGCTCTTGGGAGACCC-3′; p62, 5′-GTGAGCGACGCCATAGCGAG-3′; LC3A, 5′-CAGACCGGCCTTTCAAGCAG-3′; LC3B, 5′-GAGACTACCTACCGGGATTT-3′; LC3C, 5′-ACGCTTGGGATTTTCTGTGG-3′; GABARAP, 5′-GGATCTTCTCGCCCTCAGAG-3′; GABARAPL1, 5′-GCATTACCAGTAAGGTCAGA-3′; GABARAPL2, 5′-GAACATCCACTTCATGGCGG-3′; PINK1, 5′-CGCCACCATGGCGGTGCGAC-3′. Stable and reconstituted cell lines were generated using either the FlpIn TREx system (Thermo Fisher Scientific; R71407) or viral transduction. For FlpIn TREx cell lines, cDNAs were PCR amplified and ligated into the inducible FlpIn expression vector pCDNA5/FRT/TO. FlpIn TREx cells were cotransfected with the cDNA-containing FlpIn expression vector and the Flp recombinase vector pOG44 in a ratio of 1:3. Cells were selected by treatment with 200 µg/ml hygromycin, and protein expression was verified by induction with Tet or doxycycline. For retroviral transduction, pMXs vectors (CellBiolabs; RTV-011 and RTV-012) and pMRXIP (Backbone; Addgene, 45909) were used. For lentiviral transduction, pCDH-EF1α-GW-IRES-PURO vector (Sneeggen et al., 2019) was used. cDNAs were either amplified by PCR and ligated into the viral vectors or inserted by GW recombination. These vectors were transfected into either the HEK293 Phoenix cells for retroviral packaging or HEK293T, together with the packaging vectors pMD2.G and psPAX2 for lentiviral packaging. The resulting viral particles were mixed with 8 µg/ml hexadimethrine bromide (Sigma-Aldrich; H9268) and used to transduce cells. Cells were selected and maintained in appropriate antibiotics to optimize protein expression.
Immunoblotting, immunoprecipitation, and mass spectrometry
HeLa cells were seeded into 6-cm plates and treated as indicated. Cells were lysed in 2× SDS buffer (50 mM Tris, pH 7.4, 2% SDS, 10% glycerol) supplemented with 200 mM DTT (Sigma-Aldrich; D0632) and heated at 100°C for 10 min. Protein concentration was measured using the Pierce BCA Protein Assay Kit (Thermo Fisher Scientific; 23227). 10–40 µg of protein was resolved by SDS-PAGE and transferred to nitrocellulose membrane (Sigma-Aldrich; GE10600003). The membrane was first blocked with 5% nonfat dry milk or BSA (Sigma-Aldrich; A7906) in 1% TBS-T (0.2 M Tris, pH 8.0, 1.5 M NaCl, and 0.05% Tween-20; Sigma-Aldrich; P9416) and then incubated with the indicated primary antibodies for 24 h. The membrane was washed three times for 10 min each with TBS-T followed by incubation with secondary antibody for 1 h. The membrane was washed three times for 10 min and analyzed by ECL using the ImageQuant LAS 4000 system (GE Healthcare). For immunoprecipitation, HeLa cells were lysed in modified radioimmunoprecipitation assay buffer (50 mM Tris-HCl, pH 7.4, 120 mM NaCl, 1 mM EDTA, pH 8.0, 1% NP-40, 0.25% Triton X-100) supplemented with cOmplete Mini EDTA-free protease inhibitor cocktail tablets (Roche Applied Science; 11836170001) and phosphatase inhibitor cocktail (Merck Millipore; 524625) by shaking at 4°C for 30 min. The cell lysate was then centrifuged at 10,000 × g for 10 min. The resulting supernatant was incubated with antibody to endogenous proteins for endogenous immunoprecipitation or with Myc-Trap (Chromotek; yta-20) for cells stably expressing Myc-tagged proteins or with anti-FLAG M2 affinity gel (Sigma-Aldrich; A2220) for cells transiently transfected with 3xFLAG-tagged proteins. This was followed by either immunoblotting or protein identification by mass spectrometry. Protein identification by liquid chromatography–tandem mass spectrometry was performed as described earlier (Abudu et al., 2019).
Recombinant protein expression, peptide array, and pulldown assays
GST and maltose-binding protein (MBP) fusion proteins were expressed in competent Escherichia coli SoluBL21 (Genlantis; C700200) and BL21(DE3)pLysS (Promega; L1195). Protein expression was induced by treating overnight bacterial culture with 50 µg/ml IPTG, and expressed protein was purified by immobilization on either Glutathione Sepharose 4 Fast Flow beads (GE Healthcare; 17-5132-01) for GST fusion protein or Amylose Resin (New England Biolabs; E8021L) for MBP fusion protein. Peptide arrays were synthesized using the MultiPrep peptide synthesizer (Intavis Bioanalytical Instruments AG). Membranes containing the arrays were first blocked with 5% nonfat dry milk and incubated with 1–2 µg/ml GST-GABARAP in TBS-T for 24 h. The membrane was then visualized by immunoblotting with anti-GST antibody. For pulldown assay, Myc-tagged proteins were translated in vitro using the TNT T7 Reticulocyte Lysate System (Promega; L4610) in the presence of radioactive 35S-methionine. 10 µl of the translated protein were first precleared with either empty glutathione sepharose or amylose beads in NETN buffer (50 mM Tris, pH 8.0, 150 mM NaCl, 1 mM EDTA, 0.5% NP-40) supplemented with cOmplete Mini EDTA-free protease inhibitor cocktail tablets at 4°C for 30 min to remove unspecific binding. This was followed by incubation with the GST or MBP fusion proteins at 4°C for 1–2 h. The beads were washed five times with 500 µl NETN buffer by centrifugation at 2,500 × g for 2 min. 2× SDS gel loading buffer (100 mM Tris, pH 7.4, 4% SDS, 20% glycerol, 0.2% bromophenol blue, and 200 mM DTT; Sigma-Aldrich; D0632) was added, and the mixture was heated for 10 min at 100°C. The assay was resolved by SDS-PAGE, and the gel was stained with Coomassie Brilliant Blue (CBB) R-250 Dye (Thermo Fisher Scientific; 20278) to visualize the fusion proteins. The stained gel was vacuum dried, and the radioactive signal was detected by a Fujifilm bioimaging analyzer BAS-5000 (Fujifilm).
Confocal and super-resolution microscopy
For live-cell imaging, cells were seeded on an eight-well Lab-Tek chamber coverglass (Thermo Fisher Scientific; 155411) and grown as indicated. Cells were maintained in an incubation chamber on either the Zeiss LSM780 or LSM800 system during imaging. Images were sequentially acquired as either Z-stack or time-lapse scans using the 40×/1.20 water immersion objective. For Keima, live cells were visualized with two sequential excitation lasers at 458 nm and 561 nm and an emission spectrum of 590–680 nm. Keima is predominantly excited by at 458 nm in a neutral environment and at 561 nm in an acidic environment.
For fixed cells, HeLa cells seeded on coverslips (VWR; 631-0150) in 24-well plates were treated as indicated. Cells were fixed in 4% PFA for 10 min at 37°C and permeabilized in 0.1% Triton X-100 for 5 min at RT. Cells were blocked in 3% goat serum for 30 min before incubation in primary antibodies for 1 h at RT, followed by washing five times with PBS for 5 min each. The cells were then incubated with the corresponding Alexa Fluor–conjugated secondary antibodies for 30 min at RT followed by washing with PBS five times. Finally, the cells were incubated with 10 µg/ml DAPI for 10 min. Slides were mounted with VECTASHIELD antifade mounting media (Vector Laboratories; H-1000). For live-cell imaging, cells were grown in 8-well Lab-Tek II chambered cover glasses (Thermo Fisher Scientific; 155360) and subjected to the indicated treatments. Images were obtained using a 40×/1.2 water immersion objective or a 63×/1.4 oil immersion objective on either the LSM780 or LSM800 system (Carl Zeiss Microscopy).
For quantitative analysis, 60–100 cells were randomly selected per sample and quantitatively processed. For mitochondria, cells were visually scored into three classifications (elongated, tubular, and fragmented). Super-resolution and DV images were obtained with a DeltaVision OMX V4 Blaze imaging system (GE Healthcare) equipped with a 60×/1.42 NA oil immersion objective (Olympus) and three scientific complementary metal–oxide–semiconductor cameras. DV and SIM image reconstruction were completed using the manufacturer-supplied SoftWoRx program (GE Healthcare). Fluorescence intensity was quantified using ImageJ software.
TEM
Cells were seeded on 35-mm plates and grown to 50–60% confluency. Cells were fixed directly in prewarmed PHEM buffer (60 mM Pipes, 25 mM Hepes, 10 mM EGTA, 4 mM MgSO4·7H2O) containing 4% PFA, 0.5% glutaraldehyde, and 0.05% malachite green using a microwave protocol (microwave processor; Ted Pella). The cells were further processed in 1% osmium tetroxide/0.8% K3Fe(CN)6 in PHEM buffer, followed by 1% tannic acid and 1% uranyl acetate in double-distilled water. Finally, cells were processed through stepwise ethanol dehydration and embedded in EPON resin. The resin was polymerized at 60°C for 48 h, and 70-nm sections were cut using a diamond knife (Diatome) on a Reichert Ultracut S ultramicrotome (Leica Microsystems). Sections were imaged using a JEM-1010 TEM microscope (JEOL) equipped with a Morada charge-coupled device camera and iTEM software (Olympus). For quantification, 150–200 mitochondria from at least 10 micrographs were used per sample. Mitochondrial fusion and cristae morphology were visually scored.
Subcellular fractionation and protease protection assay
Subcellular fractionation was performed using the QProteome mitochondria isolation kit (Qiagen; 37612) in accordance with the manufacturer’s instructions. For protease protection assay, isolated mitochondria were resuspended in assay buffer (210 mM mannitol, 70 mM sucrose, 10 mM Hepes, 1 mM EDTA, pH 7.5) containing the indicated concentration of proteinase K and incubated for 30 min at RT. The reaction was stopped by addition of 5 µM PMSF. 2× SDS was added, and the reaction was heated for 10 min at 100°C, followed by immunoblotting with the indicated antibodies.
Bimolecular fluorescence complementation assay
The self-complementing split fluorescent mNG1-10/11 system is composed of two segments engineered by a split between the 10th and 11th β-strands of super-folder GFP into a large segment with 10 β-strands (mNG1-10) and a small 16-aa 11th β-strand (mNG11) segment. Expressed alone, individual segments show no fluorescence, while bright green fluorescence results when they come together. The segments can be linked to proteins to determine their interaction or subcellular localization. The mNG first 10 β-sheets (mNG1-10) were cloned into the Tet-off/on FlpIn vector pCDNA5/FRT/TO and stably expressed in the FlpIn HeLa TREx cells (Thermo Fisher Scientific; R71407) following the manufacturer’s instructions. mNG11-tagged SAMM50 and p62 were cloned into the pMRXIP vector (Addgene; 45909) using Pac1 and Not1 sites. (Pac1 sites were introduced into the vector.) These vectors were stably expressed in the HeLa TREx cells with the mNG1-10 using retroviral transduction. To induce bimolecular fluorescence complementation, cells were treated with 1 µg/ml Tet or doxycycline for 24 h followed by live-cell confocal imaging.
ATP levels and membrane potential
Cellular ATP levels were measured using the ATP determination kit (Thermo Fisher Scientific; A22066) according to the manufacturer’s instructions. For measurement of mitochondrial membrane potential, cells were grown in eight-well Lab-Tek chamber cover glasses (Thermo Fisher Scientific; 155409, 155411) and incubated with 100 nM TMRE in DMEM for 30 min at 37°C. The TMRE media were removed and replaced with DMEM, and live cells were analyzed by confocal microscopy.
Measurements of OXPHOS and glycolysis
Oxygen consumption rate (OCR) and extracellular acidification rate (ECAR) were measured using the mitochondrial stress test assay, conducted per the manufacturer’s instructions (Agilent Technologies). Briefly, 103 cells were plated on fibronectin-coated XFp Flux Analyzer Miniplates (103025) and incubated in unbuffered DMEM (103575) supplemented with 2 mM glutamine (103579), 1 mM sodium pyruvate (103578), and 10 mM glucose (pH 7.4; 103577) for 1 h at 37°C in a non-CO2 incubator. The basal OCR was measured for 30 min followed by injection of 1.5 µM oligomycin, a mitochondrial complex V inhibitor. The decrease in OCR due to oligomycin treatment was defined as the OXPHOS rate. Glycolytic rate was defined as the basal ECAR. 1 µM carbonyl cyanide 4-(trifluoromethoxy)phenylhydrazone, an uncoupling agent, was subsequently added at 60 min to measure maximal mitochondrial activity. A mixture of 0.5 µM antimycin A and rotenone, mitochondrial complex III and mitochondrial complex I inhibitors, was injected at 90 min for complete inhibition of OXPHOS. OCR and extracellular acidification rates were normalized to cell number. Total cell number in each well was determined by nuclear staining with DAPI (Sigma-Aldrich) of cells fixed in 4% formaldehyde. Imaging was performed by tile imaging/stitching of the entire well surface using an Axio Observer 7 widefield fluorescence microscope (Carl Zeiss Microscopy), and nuclear quantification was done using an ImageJ software particle counter.
Protein expression and purification for crystallization
All six human ATG8 proteins and GABARAP-SAMM5024-38 chimera were expressed in E. coli Rosetta (DE3) pLysS (Merck; 70956) at 25°C overnight. The bacteria pellets were harvested by centrifugation and resuspended in lysis buffer containing 50 mM Tris-HCl, pH 8.0, 500 mM NaCl, 0.5 mM Tris(2-carboxyethyl)phosphine (TCEP), 15 µg/ml benzamidine, and 0.4 mM 4-(2-aminoethyl)benzenesulfonyl fluoride hydrochloride. The bacteria were then lysed using a French press. After the removal of cell debris, the overexpressed GST-tagged proteins in cell lysate were batch absorbed onto Glutathione Sepharose 4B affinity matrix (GE Healthcare) and recovered with 3C protease at 4°C overnight. The recovered proteins were further purified by size exclusion chromatography using a Superdex 26/600 column (GE Healthcare) running in buffer containing 25 mM Tris-HCl, pH 8.0, 150 mM NaCl, and 0.5 mM TCEP.
Crystallization and data analysis
GABARAPL1-SAMM5024-35 LIR complex was prepared by mixing full-length GABARAPL1 and SAMM50 synthetic peptide (residues 24–35, defined hereafter as SAMM50 LIR; acetyl-EEAEFVEVEPEA-amide) at a 1:2 M ratio. The complex was then loaded onto a HiTrap desalting column (5 ml; GE Healthcare) equilibrated in 25 mM Tris-HCl, pH 8.0, 150 mM NaCl, and 0.5 mM TCEP buffer. Both the GABARAP-SAMM5024-38 chimera and the GABARAPL1-SAMM5024-35 LIR complex were then concentrated to 35 mg/ml using Vivacon2 (2,000 molecular weight cutoff; Sartorius) and crystallized at 20°C using the sitting-drop vapor diffusion method. The initial crystallization trial was performed using JCSG core 1–4, AMSO4 (Qiagen), PACT, Wizard 1–4 (Molecular Dimensions), and Pi-PEG (Jena Bioscience). In all cases, the drop included 0.5 µl of protein and 0.5 µl of mother liquor. For GABARAP-SAMM5024-38 chimera, crystals grew in 20% vol/vol isopropanol and 0.1 M Na-acetate, pH 5.5. For GABARAPL1-SAMM5024-35, LIR complex crystals grew in 24% wt/vol PEG 6000, 10 mM ZnCl2, and 0.1 M Tris, pH 7.5.
Crystals were flash frozen in liquid nitrogen, and x-ray datasets were collected at 100 K at the I04 and I04-1 beamlines (mx18566) of the Diamond Light Source synchrotron (Oxford, UK). Data collection and refinement statistics are summarized in Table S1. The datasets were indexed and scaled with xia (Winter et al., 2013). Molecular replacement was achieved by using the atomic coordinates of the peptide-free GABARAP (Protein Data Bank accession no. 1GNU) and GABARAPL1 (Protein Data Bank accession no. 2R2Q) in PHASER (McCoy et al., 2007). Refinement was performed using Phenix (Adams et al., 2010). Model building was performed in COOT (Emsley et al., 2010). Model validation used PROCHECK (Vaguine et al., 1999), and figures were prepared using the graphics program PYMOL 2.3.4 (https://pymol.org/2/).
BLI binding affinity determinations
BLI is an optical dip-and-read technique to monitor real-time interaction between ligand, which is immobilized on biosensor tips, and an analyte, which is in solution. The binding of analyte to immobilized ligand thickens the layer of biosensor tips and causes a wavelength shift of the reflected light, which is used to determine the binding affinity. Binding affinities of SAMM50 LIR (residues 18–41) with six human ATG8 proteins were measured using the Octet RED96 (ForteBio). 50 µg/ml biotinylated SAMM50 LIR peptide was immobilized on streptavidin-coated biosensors (ForteBio) and then incubated with different concentrations of purified ATG8 proteins. ATG8 proteins were diluted in assay buffer containing 25 mM Tris-HCl, pH 8.0, 150 mM NaCl, 0.5 mM TCEP, 0.1% Tween 20 (Sigma-Aldrich), and 1 mg/ml BSA. All binding assays were performed in 96-well microtiter plates agitating at 1,000 rpm and 25°C. Kd values for SAMM50-ATG8s were calculated by plotting the increase of response in BLI, which represents the spectral shift in nm during an association step, as a function of protein concentration. The datasets were fitting to a nonlinear regression using ForteBio 7.1 data analysis and further analyzed in GraphPad Prism 8 software.
Statistical analyses
Densitometric analyses were performed by Science Lab Image Gauge software (Fujifilm) and ImageJ (National Institutes of Health). All data are presented as mean ± SD. Statistical comparison was analyzed by one-way ANOVA, and comparison analysis was performed with post hoc Tukey’s test. Statistical significance displayed as ***, P < 0.001; **, P < 0.005; *, P < 0.01; †, NS.
Online supplemental material
Fig. S1 shows interaction of p62 with several mitochondrial proteins. Fig. S2 shows analysis of the effect of SAMM50 KD on apoptosis and OXPHOS. Fig. S3 shows the effect of the POTRA domain of SAMM50 on mitochondrial protein biogenesis. Fig. S4 shows the analysis of the effect of SAMM50 NTS deletion, DRP1, MUL1, and PINK1 on basal mitophagy. Fig. S5 shows the role of individual hATG8 protein in basal mitophagy. Video 1 shows recruitment of mitochondrial fragments to p62-positive puncta. Video 2 shows the insert presented in Fig. 5 C of confocal live cell imaging of HeLa cells stably co-expressing mCherry-p62 and MIC19-EGFP. Video 3 shows engulfment of mitochondrial fragment by the lysosome. Video 4 shows colocalization of several mitochondrial fragments with GABARAP. Video 5 shows the insert presented in Fig. 6 E of confocal live cell imaging of HeLa cells stably co-expressing EGFP-GABARAP and MIC19-mCherry. Video 6 shows mitochondrial fragments that colocalize with LC3A. Video 7 shows the insert presented in Fig. 6 F of confocal live cell imaging of HeLa cells stably co-expressing EGFP-LC3A and MIC19-mCherry. Table S1 shows parameters for the crystal structures of SAMM50 LIR bound to GABARAP and GABARAPL1.
Supplementary Material
shows parameters for the crystal structures of SAMM50 LIR bound to GABARAP and GABARAPL1.
Acknowledgments
We thank Jack-Ansgar Bruun, Ilona Urbarova, and Toril Anne Grønset at Tromsø University Proteomics Platform for help with mass spectrometry analyses; the Advanced Microscopy Core Facility (University of Tromsø–The Arctic University of Norway) for use of instruments and assistance; Francis Crick Fermentation Science Technology Platform (STP) for protein expression; Francis Crick Structural Biology STP for technical support; Francis Crick Peptide Chemistry STP for peptide synthesis; and Diamond Light Source synchrotron for access to beamlines I04 and I04-1 (MX18566). We thank Ola Rumohr Blingsmo, Biotechnology Centre of Oslo, for synthesizing peptide arrays and Gry Evjen for probing the arrays.
The work was supported by the Norwegian Cancer Society (190214) and the Research Council of Norway (249884) to T. Johansen. S.A. Tooze, W. Zhang, and S. Mouilleron were supported by the Francis Crick Institute, receiving its core funding from Cancer Research UK (FC001187 and FC001999); by the Medical Research Council (FC001187 and FC001999); and by Wellcome Trust (FC001187 and FC001999).
The authors declare no competing financial interests.
Author contributions: Y.P. Abudu, T. Lamark, and T. Johansen conceptualized the experiments. Y.P. Abudu performed most experiments. A. Palara and H.B. Brenne performed some of the experiments. B.K. Shrestha generated human ATG8 KO cells. K.B. Larsen performed TEM. W. Zhang and S. Mouilleron performed crystallization, structural data analysis, and affinity measurements. G. Dumitriu, C.I. Øie, and G. Levy performed Seahorse experiments. D.L. Wolfson did super-resolution and deconvolution imaging analysis (SIM and DV). C. Behrends contributed cell lines and reagents. T. Lamark, S.A. Tooze, B.S. Ahluwalia, and C. Behrends provided critical discussions and edited the manuscript. Y.P. Abudu and T. Johansen wrote the final version of the manuscript with help from T. Lamark.
Data availability
Atomic coordinates and crystallographic structure factors have been deposited in the Protein Data Bank under accession nos. 6YOO (SAMM5024-35-GABARAPL1) and 6YOP (SAMM5024-38-GABARAP).
References
- Abudu, Y.P., Pankiv S., Mathai B.J., Håkon Lystad A., Bindesbøll C., Brenne H.B., Yoke Wui Ng M., Thiede B., Yamamoto A., Mutugi Nthiga T., et al. 2019. NIPSNAP1 and NIPSNAP2 act as “eat me” signals for mitophagy. Dev. Cell. 49:509–525.e12. 10.1016/j.devcel.2019.03.013 [DOI] [PubMed] [Google Scholar]
- Adams, P.D., Afonine P.V., Bunkóczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W., et al. 2010. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66:213–221. 10.1107/S0907444909052925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguirre, J.D., Dunkerley K.M., Mercier P., and Shaw G.S.. 2017. Structure of phosphorylated UBL domain and insights into PINK1-orchestrated parkin activation. Proc. Natl. Acad. Sci. USA. 114:298–303. 10.1073/pnas.1613040114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al Rawi, S., Louvet-Vallée S., Djeddi A., Sachse M., Culetto E., Hajjar C., Boyd L., Legouis R., and Galy V.. 2011. Postfertilization autophagy of sperm organelles prevents paternal mitochondrial DNA transmission. Science. 334:1144–1147. 10.1126/science.1211878 [DOI] [PubMed] [Google Scholar]
- Alers, S., Wesselborg S., and Stork B.. 2014. ATG13: just a companion, or an executor of the autophagic program? Autophagy. 10:944–956. 10.4161/auto.28987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong, L.C., Komiya T., Bergman B.E., Mihara K., and Bornstein P.. 1997. Metaxin is a component of a preprotein import complex in the outer membrane of the mammalian mitochondrion. J. Biol. Chem. 272:6510–6518. 10.1074/jbc.272.10.6510 [DOI] [PubMed] [Google Scholar]
- Ashrafi, G., Schlehe J.S., LaVoie M.J., and Schwarz T.L.. 2014. Mitophagy of damaged mitochondria occurs locally in distal neuronal axons and requires PINK1 and Parkin. J. Cell Biol. 206:655–670. 10.1083/jcb.201401070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee, R., Gladkova C., Mapa K., Witte G., and Mokranjac D.. 2015. Protein translocation channel of mitochondrial inner membrane and matrix-exposed import motor communicate via two-domain coupling protein. eLife. 4:e11897. 10.7554/eLife.11897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhujabal, Z., Birgisdottir A.B., Sjøttem E., Brenne H.B., Øvervatn A., Habisov S., Kirkin V., Lamark T., and Johansen T.. 2017. FKBP8 recruits LC3A to mediate Parkin-independent mitophagy. EMBO Rep. 18:947–961. 10.15252/embr.201643147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birgisdottir, A.B., Lamark T., and Johansen T.. 2013. The LIR motif – crucial for selective autophagy. J. Cell Sci. 126:3237–3247. 10.1242/jcs.126128 [DOI] [PubMed] [Google Scholar]
- Bratic, I., and Trifunovic A.. 2010. Mitochondrial energy metabolism and ageing. Biochim. Biophys. Acta. 1797:961–967. 10.1016/j.bbabio.2010.01.004 [DOI] [PubMed] [Google Scholar]
- Cabantous, S., Terwilliger T.C., and Waldo G.S.. 2005. Protein tagging and detection with engineered self-assembling fragments of green fluorescent protein. Nat. Biotechnol. 23:102–107. 10.1038/nbt1044 [DOI] [PubMed] [Google Scholar]
- Calvo-Garrido, J., Maffezzini C., Schober F.A., Clemente P., Uhlin E., Kele M., Stranneheim H., Lesko N., Bruhn H., Svenningsson P., et al. 2019. SQSTM1/p62-directed metabolic reprogramming is essential for normal neurodifferentiation. Stem Cell Reports. 12:696–711. 10.1016/j.stemcr.2019.01.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandel, N.S.2015. Evolution of mitochondria as signaling organelles. Cell Metab. 22:204–206. 10.1016/j.cmet.2015.05.013 [DOI] [PubMed] [Google Scholar]
- Cornelissen, T., Vilain S., Vints K., Gounko N., Verstreken P., and Vandenberghe W.. 2018. Deficiency of parkin and PINK1 impairs age-dependent mitophagy in Drosophila. eLife. 7:e35878. 10.7554/eLife.35878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darshi, M., Mendiola V.L., Mackey M.R., Murphy A.N., Koller A., Perkins G.A., Ellisman M.H., and Taylor S.S.. 2011. ChChd3, an inner mitochondrial membrane protein, is essential for maintaining crista integrity and mitochondrial function. J. Biol. Chem. 286:2918–2932. 10.1074/jbc.M110.171975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diederichs, K.A., Ni X., Rollauer S.E., Botos I., Tan X., King M.S., Kunji E.R.S., Jiang J., and Buchanan S.K.. 2020. Structural insight into mitochondrial β-barrel outer membrane protein biogenesis. Nat. Commun. 11:3290. 10.1038/s41467-020-17144-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding, C., Wu Z., Huang L., Wang Y., Xue J., Chen S., Deng Z., Wang L., Song Z., and Chen S.. 2015. Mitofilin and CHCHD6 physically interact with Sam50 to sustain cristae structure. Sci. Rep. 5:16064. 10.1038/srep16064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolezal, P., Likic V., Tachezy J., and Lithgow T.. 2006. Evolution of the molecular machines for protein import into mitochondria. Science. 313:314–318. 10.1126/science.1127895 [DOI] [PubMed] [Google Scholar]
- Emsley, P., Lohkamp B., Scott W.G., and Cowtan K.. 2010. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66:486–501. 10.1107/S0907444910007493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng, S., Sekine S., Pessino V., Li H., Leonetti M.D., and Huang B.. 2017. Improved split fluorescent proteins for endogenous protein labeling. Nat. Commun. 8:370. 10.1038/s41467-017-00494-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn, J.M., and Melov S.. 2013. SOD2 in mitochondrial dysfunction and neurodegeneration. Free Radic. Biol. Med. 62:4–12. 10.1016/j.freeradbiomed.2013.05.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentle, I., Gabriel K., Beech P., Waller R., and Lithgow T.. 2004. The Omp85 family of proteins is essential for outer membrane biogenesis in mitochondria and bacteria. J. Cell Biol. 164:19–24. 10.1083/jcb.200310092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habib, S.J., Waizenegger T., Niewienda A., Paschen S.A., Neupert W., and Rapaport D.. 2007. The N-terminal domain of Tob55 has a receptor-like function in the biogenesis of mitochondrial beta-barrel proteins. J. Cell Biol. 176:77–88. 10.1083/jcb.200602050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper, J.W., Ordureau A., and Heo J.M.. 2018. Building and decoding ubiquitin chains for mitophagy. Nat. Rev. Mol. Cell Biol. 19:93–108. 10.1038/nrm.2017.129 [DOI] [PubMed] [Google Scholar]
- Höhr, A.I.C., Lindau C., Wirth C., Qiu J., Stroud D.A., Kutik S., Guiard B., Hunte C., Becker T., Pfanner N., et al. 2018. Membrane protein insertion through a mitochondrial β-barrel gate. Science. 359:eaah6834. 10.1126/science.aah6834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horst, M., Oppliger W., Rospert S., Schönfeld H.J., Schatz G., and Azem A.. 1997. Sequential action of two hsp70 complexes during protein import into mitochondria. EMBO J. 16:1842–1849. 10.1093/emboj/16.8.1842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphries, A.D., Streimann I.C., Stojanovski D., Johnston A.J., Yano M., Hoogenraad N.J., and Ryan M.T.. 2005. Dissection of the mitochondrial import and assembly pathway for human Tom40. J. Biol. Chem. 280:11535–11543. 10.1074/jbc.M413816200 [DOI] [PubMed] [Google Scholar]
- Ichimura, Y., Kumanomidou T., Sou Y.S., Mizushima T., Ezaki J., Ueno T., Kominami E., Yamane T., Tanaka K., and Komatsu M.. 2008. Structural basis for sorting mechanism of p62 in selective autophagy. J. Biol. Chem. 283:22847–22857. 10.1074/jbc.M802182200 [DOI] [PubMed] [Google Scholar]
- Jain, A., Lamark T., Sjøttem E., Larsen K.B., Awuh J.A., Øvervatn A., McMahon M., Hayes J.D., and Johansen T.. 2010. p62/SQSTM1 is a target gene for transcription factor NRF2 and creates a positive feedback loop by inducing antioxidant response element-driven gene transcription. J. Biol. Chem. 285:22576–22591. 10.1074/jbc.M110.118976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jian, F., Chen D., Chen L., Yan C., Lu B., Zhu Y., Chen S., Shi A., Chan D.C., and Song Z.. 2018. Sam50 regulates PINK1-Parkin-mediated mitophagy by controlling PINK1 stability and mitochondrial morphology. Cell Rep. 23:2989–3005. 10.1016/j.celrep.2018.05.015 [DOI] [PubMed] [Google Scholar]
- Johansen, T., and Lamark T.. 2011. Selective autophagy mediated by autophagic adapter proteins. Autophagy. 7:279–296. 10.4161/auto.7.3.14487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansen, T., and Lamark T.. 2020. Selective autophagy: ATG8 family proteins, LIR motifs and cargo receptors. J. Mol. Biol. 432:80–103. 10.1016/j.jmb.2019.07.016 [DOI] [PubMed] [Google Scholar]
- Kang, P.J., Ostermann J., Shilling J., Neupert W., Craig E.A., and Pfanner N.. 1990. Requirement for hsp70 in the mitochondrial matrix for translocation and folding of precursor proteins. Nature. 348:137–143. 10.1038/348137a0 [DOI] [PubMed] [Google Scholar]
- Karanasios, E., Stapleton E., Manifava M., Kaizuka T., Mizushima N., Walker S.A., and Ktistakis N.T.. 2013. Dynamic association of the ULK1 complex with omegasomes during autophagy induction. J. Cell Sci. 126:5224–5238. 10.1242/jcs.132415 [DOI] [PubMed] [Google Scholar]
- Karanasios, E., Walker S.A., Okkenhaug H., Manifava M., Hummel E., Zimmermann H., Ahmed Q., Domart M.C., Collinson L., and Ktistakis N.T.. 2016. Autophagy initiation by ULK complex assembly on ER tubulovesicular regions marked by ATG9 vesicles. Nat. Commun. 7:12420. 10.1038/ncomms12420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, I., Rodriguez-Enriquez S., and Lemasters J.J.. 2007a. Selective degradation of mitochondria by mitophagy. Arch. Biochem. Biophys. 462:245–253. 10.1016/j.abb.2007.03.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, S., Malinverni J.C., Sliz P., Silhavy T.J., Harrison S.C., and Kahne D.. 2007b. Structure and function of an essential component of the outer membrane protein assembly machine. Science. 317:961–964. 10.1126/science.1143993 [DOI] [PubMed] [Google Scholar]
- Kim, Y.Y., Um J.H., Yoon J.H., Kim H., Lee D.Y., Lee Y.J., Jee H.J., Kim Y.M., Jang J.S., Jang Y.G., et al. 2019. Assessment of mitophagy in mt-Keima Drosophila revealed an essential role of the PINK1-Parkin pathway in mitophagy induction in vivo. FASEB J. 33:9742–9751. 10.1096/fj.201900073R [DOI] [PubMed] [Google Scholar]
- Kirkin, V., Lamark T., Sou Y.S., Bjørkøy G., Nunn J.L., Bruun J.A., Shvets E., McEwan D.G., Clausen T.H., Wild P., et al. 2009. A role for NBR1 in autophagosomal degradation of ubiquitinated substrates. Mol. Cell. 33:505–516. 10.1016/j.molcel.2009.01.020 [DOI] [PubMed] [Google Scholar]
- Knowles, T.J., Scott-Tucker A., Overduin M., and Henderson I.R.. 2009. Membrane protein architects: the role of the BAM complex in outer membrane protein assembly. Nat. Rev. Microbiol. 7:206–214. 10.1038/nrmicro2069 [DOI] [PubMed] [Google Scholar]
- Koenig, P., Mirus O., Haarmann R., Sommer M.S., Sinning I., Schleiff E., and Tews I.. 2010. Conserved properties of polypeptide transport-associated (POTRA) domains derived from cyanobacterial Omp85. J. Biol. Chem. 285:18016–18024. 10.1074/jbc.M110.112649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyano, F., Okatsu K., Kosako H., Tamura Y., Go E., Kimura M., Kimura Y., Tsuchiya H., Yoshihara H., Hirokawa T., et al. 2014. Ubiquitin is phosphorylated by PINK1 to activate parkin. Nature. 510:162–166. 10.1038/nature13392 [DOI] [PubMed] [Google Scholar]
- Kozjak-Pavlovic, V., Ross K., Benlasfer N., Kimmig S., Karlas A., and Rudel T.. 2007. Conserved roles of Sam50 and metaxins in VDAC biogenesis. EMBO Rep. 8:576–582. 10.1038/sj.embor.7400982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriegenburg, F., Ungermann C., and Reggiori F.. 2018. Coordination of autophagosome-lysosome fusion by Atg8 family members. Curr. Biol. 28:R512–R518. 10.1016/j.cub.2018.02.034 [DOI] [PubMed] [Google Scholar]
- Kutik, S., Stojanovski D., Becker L., Becker T., Meinecke M., Krüger V., Prinz C., Meisinger C., Guiard B., Wagner R., et al. 2008. Dissecting membrane insertion of mitochondrial beta-barrel proteins. Cell. 132:1011–1024. 10.1016/j.cell.2008.01.028 [DOI] [PubMed] [Google Scholar]
- Lamark, T., Perander M., Outzen H., Kristiansen K., Øvervatn A., Michaelsen E., Bjørkøy G., and Johansen T.. 2003. Interaction codes within the family of mammalian Phox and Bem1p domain-containing proteins. J. Biol. Chem. 278:34568–34581. 10.1074/jbc.M303221200 [DOI] [PubMed] [Google Scholar]
- Lazarou, M., Jin S.M., Kane L.A., and Youle R.J.. 2012. Role of PINK1 binding to the TOM complex and alternate intracellular membranes in recruitment and activation of the E3 ligase Parkin. Dev. Cell. 22:320–333. 10.1016/j.devcel.2011.12.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarou, M., Sliter D.A., Kane L.A., Sarraf S.A., Wang C., Burman J.L., Sideris D.P., Fogel A.I., and Youle R.J.. 2015. The ubiquitin kinase PINK1 recruits autophagy receptors to induce mitophagy. Nature. 524:309–314. 10.1038/nature14893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Guerroué, F., Eck F., Jung J., Starzetz T., Mittelbronn M., Kaulich M., and Behrends C.. 2017. Autophagosomal content profiling reveals an LC3C-dependent piecemeal mitophagy pathway. Mol. Cell. 68:786–796.e6. 10.1016/j.molcel.2017.10.029 [DOI] [PubMed] [Google Scholar]
- Lee, J.J., Sanchez-Martinez A., Martinez Zarate A., Benincá C., Mayor U., Clague M.J., and Whitworth A.J.. 2018. Basal mitophagy is widespread in Drosophila but minimally affected by loss of Pink1 or parkin. J. Cell Biol. 217:1613–1622. 10.1083/jcb.201801044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemasters, J.J.2005. Selective mitochondrial autophagy, or mitophagy, as a targeted defense against oxidative stress, mitochondrial dysfunction, and aging. Rejuvenation Res. 8:3–5. 10.1089/rej.2005.8.3 [DOI] [PubMed] [Google Scholar]
- Liu, L., Feng D., Chen G., Chen M., Zheng Q., Song P., Ma Q., Zhu C., Wang R., Qi W., et al. 2012. Mitochondrial outer-membrane protein FUNDC1 mediates hypoxia-induced mitophagy in mammalian cells. Nat. Cell Biol. 14:177–185. 10.1038/ncb2422 [DOI] [PubMed] [Google Scholar]
- Lytovchenko, O., Melin J., Schulz C., Kilisch M., Hutu D.P., and Rehling P.. 2013. Signal recognition initiates reorganization of the presequence translocase during protein import. EMBO J. 32:886–898. 10.1038/emboj.2013.23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacVicar, T.D., and Lane J.D.. 2014. Impaired OMA1-dependent cleavage of OPA1 and reduced DRP1 fission activity combine to prevent mitophagy in cells that are dependent on oxidative phosphorylation. J. Cell Sci. 127:2313–2325. 10.1242/jcs.144337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall, R.S., Hua Z., Mali S., McLoughlin F., and Vierstra R.D.. 2019. ATG8-binding UIM proteins define a new class of autophagy adaptors and receptors. Cell. 177:766–781.e24. 10.1016/j.cell.2019.02.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy, A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C., and Read R.J.. 2007. Phaser crystallographic software. J. Appl. Cryst. 40:658–674. 10.1107/S0021889807021206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLelland, G.L., and Fon E.A.. 2018. Principles of mitochondrial vesicle transport. Curr. Opin. Physiol. 3:25–33. 10.1016/j.cophys.2018.02.005 [DOI] [Google Scholar]
- McLelland, G.L., Soubannier V., Chen C.X., McBride H.M., and Fon E.A.. 2014. Parkin and PINK1 function in a vesicular trafficking pathway regulating mitochondrial quality control. EMBO J. 33:282–295. 10.1002/embj.201385902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McWilliams, T.G., Prescott A.R., Montava-Garriga L., Ball G., Singh F., Barini E., Muqit M.M.K., Brooks S.P., and Ganley I.G.. 2018. Basal mitophagy occurs independently of PINK1 in mouse tissues of high metabolic demand. Cell Metab. 27:439–449.e5. 10.1016/j.cmet.2017.12.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melser, S., Chatelain E.H., Lavie J., Mahfouf W., Jose C., Obre E., Goorden S., Priault M., Elgersma Y., Rezvani H.R., et al. 2013. Rheb regulates mitophagy induced by mitochondrial energetic status. Cell Metab. 17:719–730. 10.1016/j.cmet.2013.03.014 [DOI] [PubMed] [Google Scholar]
- Miles, A.L., Burr S.P., Grice G.L., and Nathan J.A.. 2017. The vacuolar-ATPase complex and assembly factors, TMEM199 and CCDC115, control HIF1α prolyl hydroxylation by regulating cellular iron levels. eLife. 6:e22693. 10.7554/eLife.22693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra, P., Carelli V., Manfredi G., and Chan D.C.. 2014. Proteolytic cleavage of Opa1 stimulates mitochondrial inner membrane fusion and couples fusion to oxidative phosphorylation. Cell Metab. 19:630–641. 10.1016/j.cmet.2014.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima, N., Yoshimori T., and Ohsumi Y.. 2011. The role of Atg proteins in autophagosome formation. Annu. Rev. Cell Dev. Biol. 27:107–132. 10.1146/annurev-cellbio-092910-154005 [DOI] [PubMed] [Google Scholar]
- Mokranjac, D., and Neupert W.. 2015. Architecture of a protein entry gate. Nature. 528:201–202. 10.1038/nature16318 [DOI] [PubMed] [Google Scholar]
- Neupert, W., and Herrmann J.M.. 2007. Translocation of proteins into mitochondria. Annu. Rev. Biochem. 76:723–749. 10.1146/annurev.biochem.76.052705.163409 [DOI] [PubMed] [Google Scholar]
- Neuspiel, M., Schauss A.C., Braschi E., Zunino R., Rippstein P., Rachubinski R.A., Andrade-Navarro M.A., and McBride H.M.. 2008. Cargo-selected transport from the mitochondria to peroxisomes is mediated by vesicular carriers. Curr. Biol. 18:102–108. 10.1016/j.cub.2007.12.038 [DOI] [PubMed] [Google Scholar]
- Nishimura, T., and Mizushima N.. 2017. The ULK complex initiates autophagosome formation at phosphatidylinositol synthase-enriched ER subdomains. Autophagy. 13:1795–1796. 10.1080/15548627.2017.1358344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noda, N.N., Kumeta H., Nakatogawa H., Satoo K., Adachi W., Ishii J., Fujioka Y., Ohsumi Y., and Inagaki F.. 2008. Structural basis of target recognition by Atg8/LC3 during selective autophagy. Genes Cells. 13:1211–1218. 10.1111/j.1365-2443.2008.01238.x [DOI] [PubMed] [Google Scholar]
- Novak, I., Kirkin V., McEwan D.G., Zhang J., Wild P., Rozenknop A., Rogov V., Löhr F., Popovic D., Occhipinti A., et al. 2010. Nix is a selective autophagy receptor for mitochondrial clearance. EMBO Rep. 11:45–51. 10.1038/embor.2009.256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neil, P.K., Richardson L.G.L., Paila Y.D., Piszczek G., Chakravarthy S., Noinaj N., and Schnell D.. 2017. The POTRA domains of Toc75 exhibit chaperone-like function to facilitate import into chloroplasts. Proc. Natl. Acad. Sci. USA. 114:E4868–E4876. 10.1073/pnas.1621179114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okatsu, K., Kimura M., Oka T., Tanaka K., and Matsuda N.. 2015. Unconventional PINK1 localization to the outer membrane of depolarized mitochondria drives Parkin recruitment. J. Cell Sci. 128:964–978. 10.1242/jcs.161000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott, C., Ross K., Straub S., Thiede B., Götz M., Goosmann C., Krischke M., Mueller M.J., Krohne G., Rudel T., et al. 2012. Sam50 functions in mitochondrial intermembrane space bridging and biogenesis of respiratory complexes. Mol. Cell. Biol. 32:1173–1188. 10.1128/MCB.06388-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paila, Y.D., Richardson L.G., Inoue H., Parks E.S., McMahon J., Inoue K., and Schnell D.J.. 2016. Multi-functional roles for the polypeptide transport associated domains of Toc75 in chloroplast protein import. eLife. 5:e12631. 10.7554/eLife.12631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palikaras, K., Lionaki E., and Tavernarakis N.. 2018. Mechanisms of mitophagy in cellular homeostasis, physiology and pathology. Nat. Cell Biol. 20:1013–1022. 10.1038/s41556-018-0176-2 [DOI] [PubMed] [Google Scholar]
- Pankiv, S., Clausen T.H., Lamark T., Brech A., Bruun J.A., Outzen H., Øvervatn A., Bjørkøy G., and Johansen T.. 2007. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J. Biol. Chem. 282:24131–24145. 10.1074/jbc.M702824200 [DOI] [PubMed] [Google Scholar]
- Park, S., Choi S.G., Yoo S.M., Son J.H., and Jung Y.K.. 2014. Choline dehydrogenase interacts with SQSTM1/p62 to recruit LC3 and stimulate mitophagy. Autophagy. 10:1906–1920. 10.4161/auto.32177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paschen, S.A., Waizenegger T., Stan T., Preuss M., Cyrklaff M., Hell K., Rapaport D., and Neupert W.. 2003. Evolutionary conservation of biogenesis of beta-barrel membrane proteins. Nature. 426:862–866. 10.1038/nature02208 [DOI] [PubMed] [Google Scholar]
- Pickles, S., Vigié P., and Youle R.J.. 2018. Mitophagy and quality control mechanisms in mitochondrial maintenance. Curr. Biol. 28:R170–R185. 10.1016/j.cub.2018.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran, F.A., Hsu P.D., Wright J., Agarwala V., Scott D.A., and Zhang F.. 2013. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 8:2281–2308. 10.1038/nprot.2013.143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapaport, D.2002. Biogenesis of the mitochondrial TOM complex. Trends Biochem. Sci. 27:191–197. 10.1016/S0968-0004(02)02065-0 [DOI] [PubMed] [Google Scholar]
- Robert, V., Volokhina E.B., Senf F., Bos M.P., Van Gelder P., and Tommassen J.. 2006. Assembly factor Omp85 recognizes its outer membrane protein substrates by a species-specific C-terminal motif. PLoS Biol. 4:e377. 10.1371/journal.pbio.0040377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodger, C.E., McWilliams T.G., and Ganley I.G.. 2018. Mammalian mitophagy – from in vitro molecules to in vivo models. FEBS J. 285:1185–1202. 10.1111/febs.14336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojansky, R., Cha M.Y., and Chan D.C.. 2016. Elimination of paternal mitochondria in mouse embryos occurs through autophagic degradation dependent on PARKIN and MUL1. eLife. 5:e17896. 10.7554/eLife.17896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-Pulido, L., Devos D., Genevrois S., Vicente M., and Valencia A.. 2003. POTRA: a conserved domain in the FtsQ family and a class of beta-barrel outer membrane proteins. Trends Biochem. Sci. 28:523–526. 10.1016/j.tibs.2003.08.003 [DOI] [PubMed] [Google Scholar]
- Sandoval, H., Thiagarajan P., Dasgupta S.K., Schumacher A., Prchal J.T., Chen M., and Wang J.. 2008. Essential role for Nix in autophagic maturation of erythroid cells. Nature. 454:232–235. 10.1038/nature07006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweers, R.L., Zhang J., Randall M.S., Loyd M.R., Li W., Dorsey F.C., Kundu M., Opferman J.T., Cleveland J.L., Miller J.L., et al. 2007. NIX is required for programmed mitochondrial clearance during reticulocyte maturation. Proc. Natl. Acad. Sci. USA. 104:19500–19505. 10.1073/pnas.0708818104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekine, S., and Youle R.J.. 2018. PINK1 import regulation; a fine system to convey mitochondrial stress to the cytosol. BMC Biol. 16:2. 10.1186/s12915-017-0470-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiota, T., Imai K., Qiu J., Hewitt V.L., Tan K., Shen H.H., Sakiyama N., Fukasawa Y., Hayat S., Kamiya M., et al. 2015. Molecular architecture of the active mitochondrial protein gate. Science. 349:1544–1548. 10.1126/science.aac6428 [DOI] [PubMed] [Google Scholar]
- Shpilka, T., Weidberg H., Pietrokovski S., and Elazar Z.. 2011. Atg8: an autophagy-related ubiquitin-like protein family. Genome Biol. 12:226. 10.1186/gb-2011-12-7-226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sklar, J.G., Wu T., Gronenberg L.S., Malinverni J.C., Kahne D., and Silhavy T.J.. 2007. Lipoprotein SmpA is a component of the YaeT complex that assembles outer membrane proteins in Escherichia coli. Proc. Natl. Acad. Sci. USA. 104:6400–6405. 10.1073/pnas.0701579104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skytte Rasmussen, M., Mouilleron S., Kumar Shrestha B., Wirth M., Lee R., Bowitz Larsen K., Abudu Princely Y., O’Reilly N., Sjøttem E., Tooze S.A., et al. 2017. ATG4B contains a C-terminal LIR motif important for binding and efficient cleavage of mammalian orthologs of yeast Atg8. Autophagy. 13:834–853. 10.1080/15548627.2017.1287651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sneeggen, M., Pedersen N.M., Campsteijn C., Haugsten E.M., Stenmark H., and Schink K.O.. 2019. WDFY2 restrains matrix metalloproteinase secretion and cell invasion by controlling VAMP3-dependent recycling. Nat. Commun. 10:2850. 10.1038/s41467-019-10794-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soubannier, V., McLelland G.L., Zunino R., Braschi E., Rippstein P., Fon E.A., and McBride H.M.. 2012a. A vesicular transport pathway shuttles cargo from mitochondria to lysosomes. Curr. Biol. 22:135–141. 10.1016/j.cub.2011.11.057 [DOI] [PubMed] [Google Scholar]
- Soubannier, V., Rippstein P., Kaufman B.A., Shoubridge E.A., and McBride H.M.. 2012b. Reconstitution of mitochondria derived vesicle formation demonstrates selective enrichment of oxidized cargo. PLoS One. 7:e52830. 10.1371/journal.pone.0052830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefano, G.B., and Kream R.M.. 2015. Cancer: mitochondrial origins. Med. Sci. Monit. 21:3736–3739. 10.12659/MSM.895990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stojanovski, D., Bohnert M., Pfanner N., and van der Laan M.. 2012. Mechanisms of protein sorting in mitochondria. Cold Spring Harb. Perspect. Biol. 4:a011320. 10.1101/cshperspect.a011320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiura, A., McLelland G.L., Fon E.A., and McBride H.M.. 2014. A new pathway for mitochondrial quality control: mitochondrial-derived vesicles. EMBO J. 33:2142–2156. 10.15252/embj.201488104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda, H., Tsutsumi A., Nishizawa T., Lindau C., Busto J.V., Wenz L.-S., Ellenrieder L., Imai K., Straub S.P., Mossmann W., et al. 2021. Mitochondrial sorting and assembly machinery operates by β-barrel switching. Nature. 590:163–169. 10.1038/s41586-020-03113-7 [DOI] [PubMed] [Google Scholar]
- Truscott, K.N., Kovermann P., Geissler A., Merlin A., Meijer M., Driessen A.J., Rassow J., Pfanner N., and Wagner R.. 2001. A presequence- and voltage-sensitive channel of the mitochondrial preprotein translocase formed by Tim23. Nat. Struct. Biol. 8:1074–1082. 10.1038/nsb726 [DOI] [PubMed] [Google Scholar]
- Turco, E., Witt M., Abert C., Bock-Bierbaum T., Su M.Y., Trapannone R., Sztacho M., Danieli A., Shi X., Zaffagnini G., et al. 2019. FIP200 claw domain binding to p62 promotes autophagosome formation at ubiquitin condensates. Mol. Cell. 74:330–346.e11. 10.1016/j.molcel.2019.01.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzameli, I.2012. The evolving role of mitochondria in metabolism. Trends Endocrinol. Metab. 23:417–419. 10.1016/j.tem.2012.07.008 [DOI] [PubMed] [Google Scholar]
- Ulrich, T., and Rapaport D.. 2015. Biogenesis of beta-barrel proteins in evolutionary context. Int. J. Med. Microbiol. 305:259–264. 10.1016/j.ijmm.2014.12.009 [DOI] [PubMed] [Google Scholar]
- Vaguine, A.A., Richelle J., and Wodak S.J.. 1999. SFCHECK: a unified set of procedures for evaluating the quality of macromolecular structure-factor data and their agreement with the atomic model. Acta Crystallogr. D Biol. Crystallogr. 55:191–205. 10.1107/S0907444998006684 [DOI] [PubMed] [Google Scholar]
- Vance, J.E., and Tasseva G.. 2013. Formation and function of phosphatidylserine and phosphatidylethanolamine in mammalian cells. Biochim. Biophys. Acta. 1831:543–554. 10.1016/j.bbalip.2012.08.016 [DOI] [PubMed] [Google Scholar]
- von Muhlinen, N., Akutsu M., Ravenhill B.J., Foeglein Á., Bloor S., Rutherford T.J., Freund S.M., Komander D., and Randow F.. 2012. LC3C, bound selectively by a noncanonical LIR motif in NDP52, is required for antibacterial autophagy. Mol. Cell. 48:329–342. 10.1016/j.molcel.2012.08.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg, S.E., Sena L.A., and Chandel N.S.. 2015. Mitochondria in the regulation of innate and adaptive immunity. Immunity. 42:406–417. 10.1016/j.immuni.2015.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedemann, N., and Pfanner N.. 2017. Mitochondrial machineries for protein import and assembly. Annu. Rev. Biochem. 86:685–714. 10.1146/annurev-biochem-060815-014352 [DOI] [PubMed] [Google Scholar]
- Wiedemann, N., Kozjak V., Chacinska A., Schönfisch B., Rospert S., Ryan M.T., Pfanner N., and Meisinger C.. 2003. Machinery for protein sorting and assembly in the mitochondrial outer membrane. Nature. 424:565–571. 10.1038/nature01753 [DOI] [PubMed] [Google Scholar]
- Wiedemann, N., Frazier A.E., and Pfanner N.. 2004. The protein import machinery of mitochondria. J. Biol. Chem. 279:14473–14476. 10.1074/jbc.R400003200 [DOI] [PubMed] [Google Scholar]
- Winter, G., Lobley C.M., and Prince S.M.. 2013. Decision making in xia2. Acta Crystallogr. D Biol. Crystallogr. 69:1260–1273. 10.1107/S0907444913015308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirth, M., Zhang W., Razi M., Nyoni L., Joshi D., O’Reilly N., Johansen T., Tooze S.A., and Mouilleron S.. 2019. Molecular determinants regulating selective binding of autophagy adapters and receptors to ATG8 proteins. Nat. Commun. 10:2055. 10.1038/s41467-019-10059-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, J., Marusich M.F., Souda P., Whitelegge J., and Capaldi R.A.. 2007. The mitochondrial inner membrane protein mitofilin exists as a complex with SAM50, metaxins 1 and 2, coiled-coil-helix coiled-coil-helix domain-containing protein 3 and 6 and DnaJC11. FEBS Lett. 581:3545–3549. 10.1016/j.febslet.2007.06.052 [DOI] [PubMed] [Google Scholar]
- Youle, R.J., and Narendra D.P.. 2011. Mechanisms of mitophagy. Nat. Rev. Mol. Cell Biol. 12:9–14. 10.1038/nrm3028 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
shows parameters for the crystal structures of SAMM50 LIR bound to GABARAP and GABARAPL1.
Data Availability Statement
Atomic coordinates and crystallographic structure factors have been deposited in the Protein Data Bank under accession nos. 6YOO (SAMM5024-35-GABARAPL1) and 6YOP (SAMM5024-38-GABARAP).