Abstract
Agrobacterium tumefaciens is a pathogen of various plants which transfers its own DNA (T-DNA) to the host plants. It is used for producing genetically modified plants with this ability. To control T-DNA transfer to the right place, toxin-antitoxin (TA) systems of A. tumefaciens were used to control the target site of transfer without any unintentional targeting. Here, we describe a toxin-antitoxin system, Atu0939 (mazE-at) and Atu0940 (mazF-at), in the chromosome of Agrobacterium tumefaciens. The toxin in the TA system has 33.3% identity and 45.5% similarity with MazF in Escherichia coli. The expression of MazF-at caused cell growth inhibition, while cells with MazF-at co-expressed with MazE-at grew normally. In vivo and in vitro assays revealed that MazF-at inhibited protein synthesis by decreasing the cellular mRNA stability. Moreover, the catalytic residue of MazF-at was determined to be the 24th glutamic acid using site-directed mutagenesis. From the results, we concluded that MazF-at is a type II toxin-antitoxin system and a ribosome-independent endoribonuclease. Here, we characterized a TA system in A. tumefaciens whose understanding might help to find its physiological function and to develop further applications.
Keywords: Agrobacterium tumefaciens, TA system, mazF, MazE, mRNA endonuclease
1. Introduction
Many toxin-antitoxin (TA) systems are present on plasmids or chromosomes and consist of two small genetic elements, namely toxin and antitoxin [1]. The TA system is known to regulate cell growth or the arrest of bacteria for maintaining life under stressful environmental conditions, but its various proposed functions are still under debate, and its role is still unclear [1]. The TA systems were first identified in the plasmid F as ccdAB and in the plasmid R1 as hok-sok and kis-kid [2,3,4]. Among the type II TA systems on plasmid, the CcdAB TA system consisting of antitoxin CcdA and toxin CcdB was identified in the Escherichia coli F plasmid. CcdB inhibits DNA replication through the interaction with the DNA gyrase and leads to cell growth arrest, while the antitoxin CcdA neutralizes the CcdB toxin [5].
Recently, various TA systems were identified in bacterial chromosomes and classified into six different groups, from type I to VI, on the basis of the construction of antitoxins and the mode of action of the antitoxin to prevent the action of the toxin [6]. Among the TA system types, the type II TA system has been well studied for its mechanisms or physiological properties. Generally, type II TA systems consist of two proteins, a toxin and its cognate unstable antitoxin. The toxins in TA systems inhibit cell growth through cellular toxicity; in contrast, antitoxins neutralize the toxin activity [6]. Currently, 12 and 76 type II TA systems have been identified in E. coli K-12 and Mycobacterium tuberculosis H37Rv genomes, respectively. The mazEF TA system is classified as a type II TA system and has been identified on chromosomes from various bacterial species. The first mazEF TA system has been identified in the chromosome in E. coli, and its function has been reported to be programmed cell death in E. coli [7].
Moreover, toxins in type II TA systems not only inhibit DNA, RNA, or protein synthesis but are also involved in antibiotic resistance, biofilm formation, and persistent cell formation in the plasmid-based systems [5,8]. Some toxins in the TA systems target and cleave mRNA and are called mRNA interferases [9]. mRNA interferases are classified into ribosome-dependent or -independent toxins based on the ribosome dependency for their endoribonuclease activity [10]. MazF homologs are well known as ribosome-independent toxins and have sequence-specific endoribonucleases that can cleave mRNA sequences in a ribosome-independent manner [11]. RelE homologs have been reported to be ribosome-dependent and have an endoribonuclease activity under the interaction with bacterial ribosomes [12,13,14].
Agrobacterium tumefaciens is a gram-negative soil bacterium, and it is a serious plant pathogen. Recently, six candidates of TA modules or toxins (Atu0939-Atu0940, Atu1004, Atu2140, Atu3013, Atu6122, Atu6083) were identified using in an silico analysis of A. tumefaciens C58 [15]. Five of those six candidates showed cell growth inhibition, when overexpressed in A. tumefaciens gv3101 strain. A. tumefaciens is used for making genetically modified plants, because of the ability to transfer T-DNA of its Ti plasmid to host plants [15]. For this application, it is of important to control the target site of transfer without any unintentional targeting. A previous study showed the possibility of pemIK(Atu0939-Atu0940) to control the target site of T-DNA transfer. Moreover, ietA/iteS and yoeB/yefM TA systems have been identified on plasmids or chromosomes in A. tumefaciens [16,17]. Notably, A. tumefaciens YoeB toxin (AtYoeB) showed ribosome-independent non-specific nuclease activity cleaving both RNA and DNA, even though E. coli YoeB is a well-known RelE superfamily ribosome-dependent toxin [18,19]. In this study, we identified a novel TA system, MazF-at and MazE-at, in A. tumefaciens. MazF-at has endoribonuclease activity, and its cognate antitoxin MazE-at can neutralize the toxicity of MazF-at.
2. Materials and Methods
2.1. Bacterial Strains, Plasmids, Primers, and Chemicals
All the bacterial strains and plasmids used in this study are listed in Table 1. The A. tumefaciens EHA105 strain was grown in YEP (10 g yeast extract (BD, Sparks, MD, USA), 10 g Bacto peptone (BD, Sparks, MD, USA), and 5 g NaCl/L, pH 7.0 with spectinomycin 100 µg/mL (Duchefa Biochemie, Shanghai, China)) at 28 °C in a shaker incubator (180 rpm). DH5α, BW25113, and BL21(DE3) of E. coli strains were used for cloning, cell toxicity, and protein expression assays. pBAD24, pET28a (BBI Life Sciences, Novagen, Darmstadt, Germany), and pGEX 6p-1 (Merck, St. Louis, MO, USA) plasmids were used in this study. The antibiotics for E. coli ampicillin (Duchefa Biochemie, Haarlem, Netherlands) and kanamycin (Merck, St. Louis, MO, USA) were used at concentrations of 100 µg/mL and 50 µg/mL, respectively.
Table 1.
Strains and plasmids used in this study.
| Strain and Plasmid | Description | Source |
|---|---|---|
| Strains | ||
| A. tumefaciens EHA105 | Genomic DNA for cloning | Lab stock |
| E. coli DH5α | General cloning host | Lab stock |
| E. coli BL21(DE3) | General protein expression host | Lab stock |
| E. coli BW25113 | Wild type | Lab stock |
| Plasmids | ||
| pBAD24 | Gene expression vector, PBAD, amp | Lab stock |
| pBAD24-mazE-at | mazE in pBAD24 | This study |
| pBAD24- mazF-at | mazF in pBAD24 | This study |
| pBAD24- mazEF-at | mazEF in pBAD24 | This study |
| pBAD24- mazF-at (E24N) | Mutant MazF of E24N in pBAD24 | This study |
| pBAD24- mazF-at (E24Q) | Mutant MazF of E24Q in pBAD24 | This study |
| pBAD24- mazF-at (E24K) | Mutant MazF of E24K in pBAD24 | This study |
| pBAD24- mazF-at (E24D) | Mutant MazF of E24D in pBAD24 | This study |
| pBAD24- mazF-at (E24A) | Mutant MazF of E24A in pBAD24 | This study |
| pET28a | Protein expression vector, PIPTG, kan | Lab stock |
| pET28- mazE-at | mazE in pET28a | This study |
| pET28- mazF-at | mazF in pET28a | This study |
| pET28- mazEF-at | mazEF in pET28a | This study |
| pGEX6p-1 | Protein expression vector, PIPTG, amp | Lab stock |
| pGEX6p-1-mazE-at | mazE in pGEX6p-1 | This study |
2.2. Cloning of mazE, mazF, mazEF, and mazF Mutants
The mazE-at, mazF-at, and mazEF-at genes in the mazEF-at operon were separately amplified through PCR using A. tumefaciens gDNA as a template, and cloned into pBAD24, pET28a, and pGEX 6p-1 with specific primer sets (Table 2). The mazF-at mutants (MazF-at-E24N, -E24Q, -E24K, -E24D, and -E24A) were constructed by site-directed mutagenesis with the use of the mutagenic oligonucleotide primers (Table 2) using pBAD24- mazF-at as template. The PCR products were digested with DpnI to remove the template DNA and transformed into the E. coli DH5α strain. All point mutations were confirmed using DNA sequencing.
Table 2.
Primers used in this study.
| Primer | Sequence | Source |
|---|---|---|
| A-mazE-F | 5′-TATAGGATCCATGACCGTGACCACGA-3′ | This study |
| A-mazE-R | 5′-TATAAAGCTTTCACAACGCTTCTTTG-3′ | This study |
| T-mazF-F | 5′-TATACATATGGTCCGCAACCAGAT-3′ | This study |
| T-mazF-R | 5′-TATACTCGAGTCAAGCTGGATCGATCAT-3′ | This study |
| TA-mazEF-F | 5′-TATAGGATCCATGACCGTGACCACGA-3′ | This study |
| TA-mazEF-R | 5′-TATACTCGAGTCAAGCTGGATCGATCAT-3′ | This study |
| Pull-mazE-F | 5′-TATAGGATCCATGACCGTGACCACGA-3′ | This study |
| Pull-mazE-R | 5′-TATAGCGGCCGCTCACAACGCTTCTTTGC-3′ | This study |
| Mu-MazF-E24N-F | 5′-GTCGTAGGCAGCAACATCAAGGACGAA-3′ | This study |
| Mu-MazF-E24N-R | 5′-TTCGTCCTTGATGTTGCTGCCTACGAC-3′ | This study |
| Mu-MazF-E24Q-F | 5′-GTCGTAGGCAGCCAAATCAAGGACGAA-3′ | This study |
| Mu-MazF-E24Q-R | 5′-TTCGTCCTTGATTTGGCTGCCTACGAC-3′ | This study |
| Mu-MazF-E24K-F | 5′-GTCGTAGGCAGCAAAATCAAGGACGAA-3′ | This study |
| Mu-MazF-E24K-R | 5′-TTCGTCCTTGATTTCGCTGCCTACGAC-3′ | This study |
| Mu-MazF-E24D-F | 5′-GTCGTAGGCAGCGACATCAAGGACGAA-3′ | This study |
| Mu-MazF-E24D-R | 5′-TTCGTCCTTGATGTCGCTGCCTACGAC-3′ | This study |
| Mu-MazF-E24A-F | 5′-GTCGTAGGCAGCGCAATCAAGGACGAA-3′ | This study |
| Mu-MazF-E24A-R | 5′-TTCGTCCTTGATTGCGCTGCCTACGAC-3′ | This study |
| ompA1-F | 5′-GGTGCATACAAAGCTCAGGG-3′ | [6] |
| ompA1-R | 5′-GTGACTGCGTACTCAACACC-3′ | [6] |
| ompF1-F | 5′-CTTTGGTCTGGTTGATGGCC-3′ | [6] |
| ompF1-R | 5′-ACTTCAGACCAGTAGCCCAC-3′ | [6] |
2.3. Cell Toxicity on Plates and in Liquid Medium
The pBAD24-mazE-at, mazF-at, –mazEF-at, and -mazF-at mutants (MazF-E24N, -E24Q, -E24K, -E24D, and -E24A) were transformed into E. coli BW25113. To conduct the cell toxicity assay, transformed cells were grown on M9 agar plates with or without 0.2% L-arabinose (Merck, St. Louis, MO, USA) at 37 °C for 20 h. To construct the growth curve, transformed cells were grown in an M9 liquid medium with 0.5% glycerol at 37 °C in the presence or absence of 0.2% L-arabinose. The cell cultures were sampled at 0, 30, 60, 90, 120, 150, and 240 min after the induction.
2.4. Cellular Target Assay of DNA, RNA, and Protein Synthesis In Vivo
The pBAD24-mazF-at was transformed into E. coli BW25113. The transformed cells were inoculated in M9 medium with 0.5% glycerol and cultured at 37 °C. The culture was grown until the OD600nm of the culture reached 0.4, and then MazF-at was overexpressed by induction with arabinose to a final concentration of 0.2%. Each 0.4 mL cell culture was sampled at 0, 5, 10, 30, and 60 min after the induction. Each sample was mixed with 10 µCi of [3H]-thymidine, 10 µCi of [3H]-uridine, or 30 µCi of [35S]-methionine, and 30 µg of nonradioactive thymidine, 30 µg of nonradioactive uridine, or 80 µg of nonradioactive methionine, respectively, and incubated at 37 °C for 30 sec. The rates of DNA, RNA, and protein synthesis were analyzed as previously described [19].
2.5. Protein Expression and Purification
To purify the N-terminal His6-tagged MazE-at, MazF-at, and MazEF-at complex, E. coli BL21(DE3) harboring pET28a-mazE-at, -mazF-at, or –mazEF-at was grown in LB medium at 37°C. The culture was grown until the OD600nm of the culture reached 0.5, and then MazE-at, MazF-at, or MazEF-at was overexpressed by the induction with isopropyl-β-d-1-thiogalactoside (IPTG) (Merck, St. Louis, MO, USA) to a final concentration of 0.5 mM. After the induction, the cells were grown for 6 h at 18°C. The cultured cells were harvested by centrifugation (4392× g for 30 min at 4°C) and disrupted using a French pressure cell press. After the centrifugation (12,000× g for 30 min at 4 °C), soluble fractions were applied to Ni-NTA agarose (Qiagen, Hilden, Germany) following the manufacturer’s protocol. The purified proteins were applied to 15% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to a polyvinylidene difluoride membrane (PVDF), and a Western Blot analysis was performed using the polyclonal anti-histidine tag antibody (#2365) (cell signaling Technology, Danvers, MA, USA).
2.6. mRNA Stability Analysis Using Northern Blot
The pBAD24-mazF-at was transfected into E. coli BW25113 cells. The transformed cells were cultured in M9 medium with 0.5% glycerol at 37 °C. The culture was grown until the OD600nm of the culture reached 0.4, then MazF-at was overexpressed by induction with arabinose to a final concentration of 0.2%. Non-induced samples were prepared without the induction by arabinose in the same growth conditions as the induced samples. Aliquots of the cell cultures were collected at 0, 5, 10, 30, and 60 min after the induction. Total RNA was extracted by TRIzol (Thermo Fisher Scientific, Waltham, MA, USA) using the manufacturer’s protocol. The 30 µg of total RNA was applied to each lane onto denatured agarose gel. Northern blot was performed as previously described, with the modification that the probes were labeled with biotin-14-dCTP [20].
2.7. Endoribonuclease Activity Assay of MazE-at, MazF-at, and MazEF-at In Vitro
The 0.5 μM MazF-at, MazEF-at, or MazF-ec(E24A) protein was incubated with 0.8 μg/µL MS2 phage RNA (Roche Applied Science, Penzberg, Germany) in 20 mM Tris-HCl (pH 8.0) including 0.5 μL of RNA inhibitor (Roche Applied Science, Penzberg, Germany) at 37°C. The reactions were sampled at the indicated time intervals and were stopped by adding RNA loading dye (95% formamide, 0.025% SDS, 0.025% bromophenol blue, 0.025% xylene cyanol FF, 0.5 mM EDTA), and then the reactions were incubated for 3 min at 70 °C and electrophoresed on a 1.5% agarose gel.
2.8. Secondary Structure Analysis by Circular Dichroism
Far-UV CD measurements were obtained using an automated Chirascan CD spectrometer (Applied Photophysics Ltd., Leatherhead, UK). The far-UV spectra were determined over a wavelength range of 190–260 nm using 0.5-mm path length cell at 25 °C. All experiments were performed in 20 mM Tris HCl, pH 7.5, and then adjusted to a 0.25 mg/mL protein concentration of purified MazE-at and MazF-at for CD analysis. The background CD spectrum of the buffer was subtracted from each spectrum. The percentage of secondary structure content was calculated using the CDNN software [21].
2.9. Protein Interaction Analysis by GST Pull-Down Assay with Beads
The pGEX6p-1 (Glutathione S-transferase (GST) only), pGEX-6-1-mazE-at (GST-MazE-at), and pET28-mazF-at (6H-MazF-at) were transformed into E. coli BL21(DE3) and cultured in LB medium with 100 µg/mL ampicillin for pGEX6p-1 and 50 µg/mL kanamycin for pET28. When the OD600nm of the cultures reached 0.5, 0.5 mM IPTG was added to induce the expression of GST, GST-MazE-at, and N-terminal His6-tagged MazF-at for 6 h at 18 °C. The cultured cells were harvested by centrifugation (4392× g for 30 min at 4 °C) and disrupted using a French pressure cell press. The cell debris were removed by centrifugation (12,000× g for 30 min at 4 °C), and soluble fractions were collected. GST only and GST-MazE-at were immobilized onto glutathione-Sepharose 4B beads by a gentle shaking motion on a rotating platform for 3 h at 4 °C, collected by centrifugation (1250× g for 1 min), and washed three times with PBS solution. The soluble fraction of MazF-at was incubated with GST beads by a gentle shaking motion on a rotating platform for 3 h at 4 °C and was collected by centrifugation (1250× g for 1 min). Beads were washed three times with PBS solution, separated by SDS-PAGE, and detected by Western Blotting with an anti-histidine antibody (#2365) (cell signaling Technology, Danvers, MA, USA).
3. Results
3.1. MazF-at Is Toxic in E. coli and Is Neutralized by MazE-at
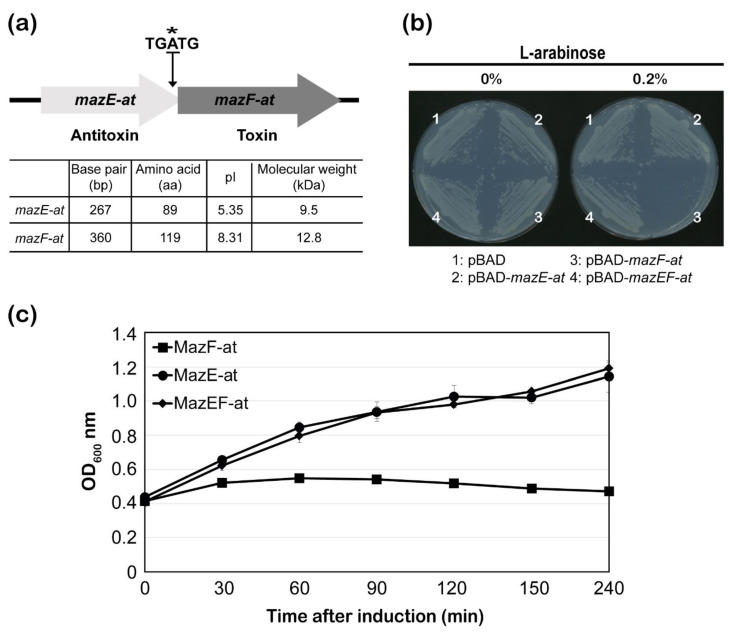
Using the toxin-antitoxin database (TADB 2.0), we identified 13 candidates (Table S1) for TA systems in A. tumefaciens C58. We found that six clones (Atu0940, Atu2017, Atu0674, Atu0934, Atu1004, and Atu8169) out of 13 showed toxicity (Figure S1, Table S1) [22]. With this screening of TA systems, we also found that two genes, mazE-at (Atu0939) and mazF-at (Atu0940), later classified as one of the mazEF homologs, had been predicted as a potential TA system in the chromosome of A. tumefaciens. The two genes, mazE-at and mazF-at, are located in the same operon, and the stop codon of mazE-at overlaps with the start codon of mazF-at by one nucleotide (Figure 1a, asterisk). The two proteins, MazE-at and MazF-at, consist of 89 and 119 amino acid residues, respectively, and the pI values are 5.35 and 8.31, respectively (Figure 1a). MazF-at (Atu0940) was identified as pemIK TA system in the previous study [15]. They found six candidates of TA modules in A. tumefaciens C58, and five of those six candidates showed growth inhibition in A. tumefaciens gv3101 strains [15]. In this study, we further evaluated whether the two genes, mazE-at and mazF-at, function as a TA system. mazE-at, mazF-at, and mazEF-at were amplified using genomic DNA extracted from A. tumefaciens and cloned into the pBAD24 vector. The plasmids pBAD24-mazE-at, mazF-at, and mazEF-at were transformed into E. coli BW25113 cells, and cell toxicity was tested on plates with or without 0.2 % L-arabinose. When MazE-at or MazEF-at were induced, transformant colonies were formed on the plate. However, when MazF-at was induced in the presence of 0.2 % arabinose, no colonies were formed. We also examined the effect of MazF-at inducing cell growth in a liquid culture (Figure 1b). As shown in Figure 1c, growth inhibition was immediately observed after the induction of MazF-at but not MazE-at or MazEF-at in the presence of 0.2% arabinose. The results indicated that MazF-at inhibits cell growth as a toxin of the TA system, and MazE-at neutralizes the MazF-at toxicity as an antitoxin of the TA system in the same operon.
Figure 1.
The characterization of the MazEF-at TA system in Agrobacterium tumefaciens. (a) Gene map of the mazEF-a. The asterisk indicates the nucleotide where the orf of the two genes overlap. The table includes the gene length, number of amino acids, theoretical pI, and molecular weight of MazE-at and MazF-at; (b) Toxicity of MazF-at on plates. The cells of Escherichia coli BW25113 harboring pBAD24-mazE-at (2), -mazF-at toxin (3), -mazEF-at (4) or pBAD24 only (1) were incubated on the M9 plate with or without 0.2% L-arabinose; (c) Effect of MazF-at on cell growth induction. The culture of E. coli BW25113 harboring pBAD24-mazE-at (black circle), -mazF-at (black square), or –mazEF-at (black rhombus) was grown until the OD 600nm (optical density) of the culture reached ~0.4, followed by adding 0.2% L-arabinose at 0 min, and bacterial growth was measured for 6 h at OD600 nm. Error bars represent the standard error of the mean (SEM).
3.2. The MazF-at Belongs to the MazF Family and E24 of MazF-at Is an Active Residue for Its Toxicity
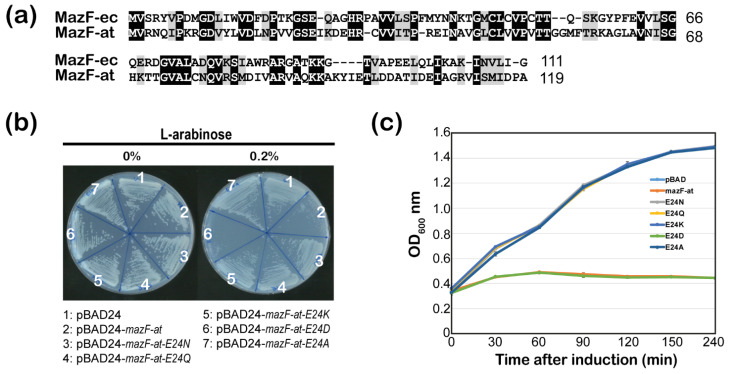
The MazF-at is one of the MazF homologs in A. tumefaciens. MazF is reported in E. coli as an mRNA endoribonuclease that has the ability to cleave mRNA. To explore this possibility, we analyzed the global sequence alignment using the amino acid sequences of MazF-at and MazF-ec. The results showed that these two proteins have 33.3% identity and 45.5% similarity (Figure 2a). From a previous study on the tertiary protein structure of MazF-ec, a purified MazF-ec E24A mutant was used because it is impossible to purify wild-type MazF-ec from E. coli due to its toxicity. The E24A mutant is shown to have less toxicity than the non-mutated form, and the glutamic acid (E24) of MazF-ec has been determined as an active residue for the mRNA endoribonuclease activity [23]. In a recent study on the crystal structure of the E24 mutant, it is shown that the E24A mutant of MazF-ec affected substrate recognition [24,25]. Therefore, we created MazF-at mutants such as E24N, E24Q, E24K, E24D, and E24A in MazF-at and tested the cell growth on plates and in liquid medium with or without 0.2% L-arabinose. Glutamic acid was replaced by a different type of amino acids, such as polar uncharged side chain (N and Q), positive charged side chain (K), negative charged chain (D), or hydrophobic side chain (A). The results showed that the MazF-at mutants (E24D) have only preserved their toxicity, like wild-type MazF-at, and that the other mutants, MazF-at E24N, E24Q, E24K, and E24A, did not inhibit cell growth under 0.2 % arabinose (Figure 2b). In a liquid culture, the growth curve also indicated that MazF-at (E24D) had a cell growth arrest similar to that of wild-type MazF-at (Figure 2c). This might be because aspartic acid is a negatively charged amino acid, as glutamic acid. These results suggest that the glutamic acid (E24) amino acid of MazF-at is one of the active residues responsible for the toxicity of MazF-at and the MazF homolog, which is an endoribonuclease.
Figure 2.
The identification of active residues in MazF-at for ribonuclease activity. (a) Sequence alignments between MazF-at and MazF-ec. Residues were shaded according to BLOSUM62 score. Dark- and grey- shaded residues represent identical and similar amino acid residues, respectively; (b) Toxicity of MazF-at mutants, MazF-at-E24N, -E24Q, -E24K, -E24D, and -E24A, on plates. The cells of E. coli BW25113 harboring each mazE-at mutant in pBAD24 were incubated on the M9 plate with or without 0.2% L-arabinose; (c) Effect of MazF-at mutants, MazF-at-E24N, -E24Q, -E24K, -E24D, and -E24A induction on cell growth. The culture of E. coli BW25113 harboring each mazE-at mutant in pBAD24 was grown until the OD600nm of the culture reached ~0.4, followed by adding 0.2% L-arabinose at 0 min, and bacterial growth was measured for 6 h at OD600 nm. Error bars represent standard error of the mean (SEM).
3.3. MazF-at Inhibits Protein Synthesis
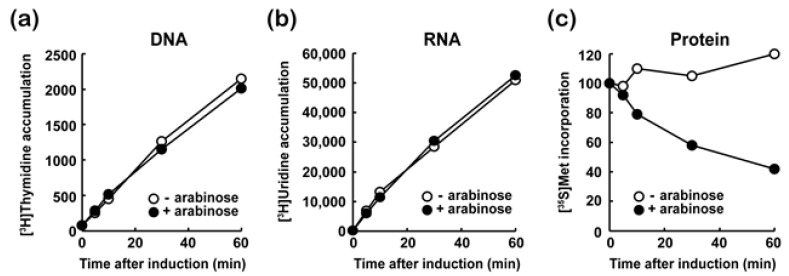
Toxins in TA systems have various targets, such as the essential cellular process of DNA, RNA, or protein synthesis to regulate cell growth according to environmental conditions. Therefore, to evaluate the effect of MazF-at on cell growth, we examined the effect of MazF-at induction with or without 0.2 % arabinose on DNA, RNA, and protein synthesis using [3H]-thymidine, [3H]-uridine, or [35S]-methionine in E. coli BW25113 cells, respectively. The incorporation of [3H]-thymidine (Figure 3a) or [3H]-uridine (Figure 3b) was not significantly affected upon MazF-at induction. However, MazF-at blocked the incorporation of [35S]-methionine into cellular proteins within 10 min of MazF-at induction (Figure 3c). This indicates that MazF-at inhibits protein synthesis, but not RNA and DNA synthesis.
Figure 3.
Effect of MazF-at induction on (a) DNA, (b) RNA, and (c) protein synthesis in vivo. The culture harboring pBAD24-mazF-at was grown until the OD600nm of the culture reached 0.4, then MazF-at was overexpressed by induction with arabinose to a final concentration of 0.2%. Each 0.4 mL cell culture was sampled at 0, 5, 10, 30, and 60 min after the induction and was mixed with [3H]-thymidine (a), [3H]-uridine (b), or [35S]-methionine (c).
3.4. MazF-at Is an Endoribonuclease
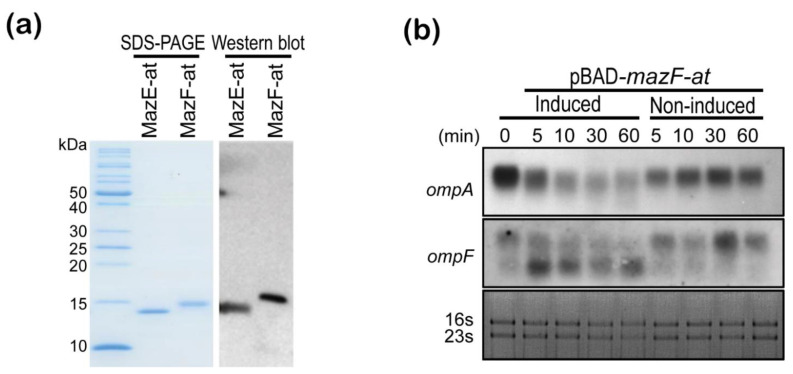
Next, we purified MazE-at, MazF-at, and MazEF-at complex proteins using pET28-mazE-at, -mazF-at, and –mazEF-at with a His6-tag at the N-terminal end to confirm the activity of MazF-at in vitro. The molecular weight and purity of purified MazE-at, MazF-at, and MazEF-at were confirmed by SDS-PAGE and Western blot (Figure 4a). In addition, we evaluated whether MazF-at inhibited protein synthesis by decreasing cellular mRNA stability as MazF or YhaV does in E. coli using Northern blot. When MazF-at was overexpressed in the presence of 0.2% arabinose, the degradation of cellular mRNAs (ompA, ompF) was observed 5 min after induction (Figure 4b). These results suggest that MazF-at in the MazEF-at TA system has potential as an endoribonuclease in A. tumefaciens.
Figure 4.
(a) Protein purification and Western blot assay using anti-his6 antibody of MazE-at and MazF-at. 1.5 ng of purified MazE-at and MazF-at proteins with N-terminal His6-tag were separated by 15% SDS-PAGE gel, and the gel was stained with Coomassie blue (left panel). Western blot analysis was performed on PVDF, and the blots were detected by anti-his6 antibody (right panel); (b) Effect of MazF-at on cellular mRNA stability using Northern blot assay. The total RNAs were extracted from E. coli BW25113 cells harboring pBAD24-mazF-at with (induced) or without 0.2% L-arabinose (non-induced) at 0, 5, 10, 30, and 60 min after the induction, and Northern blotting was performed using the ompF or ompA mRNA specific probes labeled with biotin-14-dCT.
Next, to evaluate the in vitro endoribonuclease activity of purified MazF-at, the MazF-at or MazEF-at complex was incubated with MS2 phage RNA, and its cleavage patterns were compared with those of MazF-ec(E24A) (Figure 5c). MazF-at cleaved MS2 phage RNA from 1 min and completely digested the full-size MS2 phage RNA within 10 min (Figure 5a). Notably, MazEF-at did not cleave MS2 phage RNA even after 30 min of incubation (Figure 5b). The results indicated that MazF-at has in vitro endoribonuclease activity and its cognate antitoxin, MazE-at, can inhibit the activity of MazF-at (Figure 5).
Figure 5.
Endoribonuclease activity assay of (a) MazF-at, (b) MazEF-at complex, and (c) MazF-ec on MS2 phage RNA. 0.5 μM of MazF-at, MazEF-at, or MazF-ec protein was incubated with 0.8 μg/µL MS2 phage RNA, and the reaction was loaded on 1.5% agarose gel. The arrowheads indicate the full-length of MS2 phage RNA.
3.5. Characterization of Purified MazE-at and MazF-at Proteins
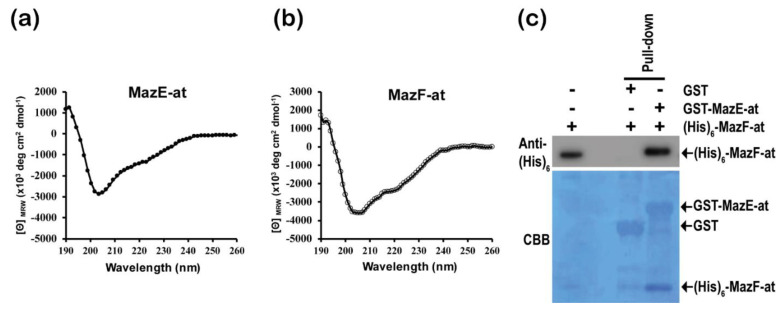
To investigate the secondary structure of MazE-at and MazF-at, we used far-UV circular dichroism (CD) with 0.25 mg/mL of purified proteins. The CD spectrum of MazE-at showed two negative peaks at approximately 208 and 222 nm that consist of α-helices (30.9%), β-sheets (31.5%), turns (19.7%), and random coils (18.0%) (Figure 6a). Moreover, MazF-at also has two negative peaks at approximately 208 and 222 nm, which consist of α-helices (28.9%), β-sheets (32.1%), turns (19.6%), and random coils (21.9%) (Figure 6b). The components of the secondary structures were calculated through CDNN [21]. These results suggest that the two proteins have similar secondary structure compositions. Normally, the antitoxin of type II TA systems has the ability to block the activity of its cognate toxins by forming a TA complex through protein-protein interactions. To confirm the in vitro interaction of the antitoxin MazE-at with MazF-at, we performed a pull-down assay using GST-MazE-at fusion protein. When MazF-at was incubated with GST or GST-MazE-at immobilized beads, GST-MazE-at only showed the band of MazF-at with His6-tag at the N-terminal end from the Western blot assay using anti-his6 antibody. This result showed that MazE-at directly interacts with MazF-at in vitro (Figure 6c).
Figure 6.
Circular dichroism (CD) analysis and the GST pull-down assay of MazE-at and MazF-at. The CD spectrum of MazE-at (a) and MazF-at (b) were measured from 190 to 260 nm wavelength (far-UV), and 0.25 mg/mL of purified protein was used; (c) Western blot (upper panel) and Coomassie blue staining (down panel) for GST pull-down assay of GST only, GST-MazE-at, and N-terminal His6 tag-MazF-at.
4. Discussion
Here, we identified a MazEF TA system candidate in the A. tumefaciens genome and showed that MazF-at is a toxin and that the gene overlaps by one base with MazE-at, which functions as an antitoxin for MazF-at. The PemK family in the conserved domain database includes different toxins, such as MazF, Kid, PemK, ChpA, ChpB, and ChpAK in TA systems [26]. The Kis-Kid TA system is also known as the PemIK TA system. The Kis-Kid in plasmid R1 is known to encode the addiction modules to prevent the loss of plasmids through a mechanism known as post-segregational killing [7,25,26]. Recently, a MazEF homolog (MazF-dr) in Deinococcus radiodurans responds by an increase in ROS accumulation upon DNA damage stress [27]. Moreover, MazF in Staphylococcus aureus may contribute to reversible bacteria dormancy through activation of translation rescue, ribosome hibernation, the increase of cell wall thickness, and the decrease of cell division [28].
The amino acid alignment showed a high homology between MazF-ec from E. coli and MazF-at with 33.3% identity and 45.5% similarity. In addition, MazF-at has a high structural similarity with MazF-ec using a J-pred server (Figure S2 and Table S2) [29]. The active residue of MazF-ec was predicted to be the 24th glutamic acid because the toxicity of MazF-ec was reduced tenfold when the 24th glutamic acid was replaced by alanine (E24A) [23]. Based on this result, we changed the 24th glutamic acid of MazF-at (MazF-at-E24A) to alanine and confirmed that the E24 of MazF-at is an active residue for its toxicity using site-directed mutagenesis in a manner similar to MazF-ec. However, the MazF-homolog from Bacillus subtilis has its catalytic key residues at R25 and T48 from the crystal structure and site-directed mutagenesis assay. Therefore, it is necessary to study whether there are more active residues of MazF-at besides E24. In addition, it is not known whether they are involved in binding to target RNA, endoribonuclease activity, or both. In order to prove this, further studies are required to determine the tertiary structure of MazF-at.
Previous studies have shown that MazF homologs in other bacteria have mRNA interferase activity to cleave mRNA. Here, we evaluated whether MazF-at also has endoribonuclease activity, as MazF does in E. coli. First, we examined whether MazF-at affects cell growth by inhibiting important cellular processes and found that MazF-at is an inhibitor for the synthesis of all cellular proteins, but not DNA and RNA synthesis. In addition, to evaluate whether MazF-at inhibits protein synthesis by decreasing cellular mRNA stability, MazF-at was overexpressed in vivo, and we found that the cellular mRNAs (ompA, ompF) were degraded following specific time intervals. To confirm that this RNA degradation was due to MazF-at, we purified MazE-at, MazF-at, MazEF-at complex, and MazF-E24A, and the decrease in RNA stability after the incubation of purified MazF-at with MS2 phage RNA was evaluated in vitro. Ideally, all of these experiments should be done in its original host, A. tumefaciens. However, it has been proven to have toxicity with overexpressed MazF-at in A. tumefaciens, and it is difficult to obtain a sufficient amount of proteins from A. tumefaciens for in vitro studies, because its doubling time is 2.5–4 hrs. For this reason, this study was conducted using E. coli, although MazF-at may have different physiological function in A. tumefaciens. From these results, we showed that MazF-at has a ribosome-independent endoribonuclease activity because MazF-at showed endoribonuclease activity without ribosomes in vitro. Usually, ribosome-independent mRNA interferases recognize and cleave specific RNA sequences and specific lengths. The MqsR homolog from E. coli specifically cleaves intracellular mRNAs at a GCU sequence [30]. Recently, it was reported that an MazF homolog from E. coli recognized about seven nucleotide regions with extended recognition specificity for ACA and its flaking sequences [31]. The protein purification of MazF-ec or MqsR has not been possible without the modification of the protein, but an MazF homolog (MazF-cd) from Clostridium difficile, which cleaves 5bp UACAU sequences of mRNA, was successfully purified [32]. This might be because of the different level of toxicity of those toxins, but what makes this difference in toxicity is still unknown. Furthermore, the global analysis of the cleavage site of MazF showed that E. coli MazF cleaved rRNA precursors and ribosomal protein transcripts, which may inhibit ribosome biogenesis [31,33]. However, AtYoeB showed ribosome-independent non-specific nuclease activity cleaving both RNA and DNA [19]. Therefore, further study is needed to identify whether MazF-at has specific recognition sites and the length of the cleavage site.
The antitoxin of the type II TA system inhibits toxin activity by forming a TA complex through protein-protein binding. To confirm whether MazE-at also binds to MazF-at through direct protein-protein interaction, we examined the pull-down assay, which showed the direct binding between MazE-at and MazF-at. Moreover, we evaluated the secondary structure of MazE-at and MazF-at. The two proteins have typical helix structures. Usually, although many antitoxins in type II TA systems have an unstructured region of free antitoxin, the disordered domain of antitoxin is highly susceptible to stress-induced proteases [34]. Interestingly, MazE-at has a stable secondary structure, unlike other antitoxins. Almost all antitoxins block the toxicity of their cognate toxins by direct binding to their active sites. MazE-at antitoxin binding to MazF-at may induce a conformational change to inhibit the activity of MazF-at.
In this study, we showed that the MazEF-at toxin-antitoxin system is located in the A. tumefaciens chromosome and one of the MazF homologs. MazF-at in the MazEF TA system has endoribonuclease nuclease activity, and its cognate antitoxin MazE-at may neutralize the toxicity of MazF-at. This might help us to find the function of TA systems in A. tumefaciens and to develop its application.
Acknowledgments
We thank Munjin Kwon for the critical reading of this manuscript.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/microorganisms9051107/s1, Figure S1: Effect of 13 candidates’ induction for TA systems identified in A. tumefaciens C58 on E. coli BW25113 cell growth. The pBAD24, pBAD24-aut0940, -aut1078, -aut1311, -aut1628, -aut8169, -aut2033, -aut2326, -aut2017, -aut0246, -aut0674, -aut0849, -aut0934, and –aut1004 were transformed into E. coli BW25113. To conduct the cell toxicity assay, transformed cells were grown on M9 agar plates with or without 0.2% L-arabinose at 37 °C for 20 h; Figure S2: Secondary structure prediction of MazF-at using Jpred 4, a protein secondary structure prediction server. Lupas_21, Lupas_14, and Lupas_28 represent coiled-coil predictions for the sequence. JNetPRED and JNetCONF represent the consensus prediction and the confidence estimate for the prediction (high values mean high confidence), respectively. JNETSOL25, JNETSOL5, and JNETSOL0 are the binary predictions of 25%, 5%, and 0% solvent accessibility, respectively. JNetHMM and JNETPSSM show the HMM-profile-based prediction and PSSM-based prediction, respectively. JNETJURY indicates the locations to rationalize the significantly different prediction residues with an asterisk; Table S1: TA system candidates from A. tumefaciens C58 genome using the toxin-antitoxin database (TADB 2.0); Table S2: High structural similarity homologs of MazF-at using a J-pred server.
Author Contributions
Conceptualization, W.C., Y.Y., M.I., M.-H.Y. and J.-H.P.; methodology, W.C., Y.Y., J.-Y.P., S.-H.P., H.-W.L. and B.-K.L.; software, W.C.; validation, W.C., M.O., M.-H.Y. and J.-H.P.; writing—original draft preparation, W.C. and J.-H.P.; writing—review and editing, W.C., M.O., and J.-H.P. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by grants from the Korea Research Institute of Bioscience & 415 Biotechnology (KRIBB) Research Initiative Program (KGM5362113 and Korea Biomedical Scientist 416 Fellowship Program) and by the Intramural Research Program of the National Institute of Allergy 417 and Infectious Diseases (NIAID), U.S. National Institutes of Health (NIH) (Project number 1 ZIA 418 AI000904, to MO.), and was carried out with the support of "Cooperative Research Program for 419 Agriculture Science and Technology Development (Project No. PJ01595802)" Rural Development 420 Administration, Republic of Korea.
Data Availability Statement
Data are contained within the article or Supplementary Materials.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Fraikin N., Goormaghtigh F., Van Melderen L. Type II Toxin-Antitoxin Systems: Evolution and Revolutions. J. Bacteriol. 2020;202 doi: 10.1128/JB.00763-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ogura T., Hiraga S. Mini-F plasmid genes that couple host cell division to plasmid proliferation. Proc. Natl. Acad. Sci. USA. 1983;80:4784–4788. doi: 10.1073/pnas.80.15.4784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gerdes K., Rasmussen P.B., Molin S. Unique type of plasmid maintenance function: Postsegregational killing of plasmid-free cells. Proc. Natl. Acad. Sci. USA. 1986;83:3116–3120. doi: 10.1073/pnas.83.10.3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bravo A., de Torrontegui G., Diaz R. Identification of components of a new stability system of plasmid R1, ParD, that is close to the origin of replication of this plasmid. Mol. Gen. Genet. 1987;210:101–110. doi: 10.1007/BF00337764. [DOI] [PubMed] [Google Scholar]
- 5.Tripathi A., Dewan P.C., Barua B., Varadarajan R. Additional role for the ccd operon of F-plasmid as a transmissible persistence factor. Proc. Natl. Acad. Sci. USA. 2012;109:12497–12502. doi: 10.1073/pnas.1121217109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harms A., Brodersen D.E., Mitarai N., Gerdes K. Toxins, Targets, and Triggers: An Overview of Toxin-Antitoxin Biology. Mol. Cell. 2018;70:768–784. doi: 10.1016/j.molcel.2018.01.003. [DOI] [PubMed] [Google Scholar]
- 7.Aizenman E., Engelberg-Kulka H., Glaser G. An Escherichia coli chromosomal “addiction module” regulated by guanosine [corrected] 3′,5′-bispyrophosphate: A model for programmed bacterial cell death. Proc. Natl. Acad. Sci. USA. 1996;93:6059. doi: 10.1073/pnas.93.12.6059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kamruzzaman M., Iredell J. A ParDE-family toxin antitoxin system in major resistance plasmids of Enterobacteriaceae confers antibiotic and heat tolerance. Sci. Rep. 2019;9:9872. doi: 10.1038/s41598-019-46318-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kang S.M., Koo J.S., Kim C.M., Kim D.H., Lee B.J. mRNA Interferase Bacillus cereus BC0266 Shows MazF-Like Characteristics Through Structural and Functional Study. Toxins. 2020;12:380. doi: 10.3390/toxins12060380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cook G.M., Robson J.R., Frampton R.A., McKenzie J., Przybilski R., Fineran P.C., Arcus V.L. Ribonucleases in bacterial toxin-antitoxin systems. Biochim. Biophys. Acta. 2013;1829:523–531. doi: 10.1016/j.bbagrm.2013.02.007. [DOI] [PubMed] [Google Scholar]
- 11.Zhang Y., Zhang J., Hara H., Kato I., Inouye M. Insights into the mRNA cleavage mechanism by MazF, an mRNA interferase. J. Biol. Chem. 2005;280:3143–3150. doi: 10.1074/jbc.M411811200. [DOI] [PubMed] [Google Scholar]
- 12.Pedersen K., Zavialov A.V., Pavlov M.Y., Elf J., Gerdes K., Ehrenberg M. The bacterial toxin RelE displays codon-specific cleavage of mRNAs in the ribosomal A site. Cell. 2003;112:131–140. doi: 10.1016/S0092-8674(02)01248-5. [DOI] [PubMed] [Google Scholar]
- 13.Jurenaite M., Markuckas A., Suziedeliene E. Identification and characterization of type II toxin-antitoxin systems in the opportunistic pathogen Acinetobacter baumannii. J. Bacteriol. 2013;195:3165–3172. doi: 10.1128/JB.00237-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Korch S.B., Contreras H., Clark-Curtiss J.E. Three Mycobacterium tuberculosis Rel toxin-antitoxin modules inhibit mycobacterial growth and are expressed in infected human macrophages. J. Bacteriol. 2009;191:1618–1630. doi: 10.1128/JB.01318-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Denkovskienė E., Paškevičius Š., Stankevičiūtė J., Gleba Y., Ražanskienė A. Control of T-DNA Transfer from Agrobacterium tumefaciens to Plants Based on an Inducible Bacterial Toxin-Antitoxin System. Mol. Plant-Microbe Interact. 2020;33:1142–1149. doi: 10.1094/MPMI-03-20-0067-R. [DOI] [PubMed] [Google Scholar]
- 16.Yamamoto S., Kiyokawa K., Tanaka K., Moriguchi K., Suzuki K. Novel toxin-antitoxin system composed of serine protease and AAA-ATPase homologues determines the high level of stability and incompatibility of the tumor-inducing plasmid pTiC58. J. Bacteriol. 2009;191:4656–4666. doi: 10.1128/JB.00124-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ames J.R., McGillick J., Murphy T., Reddem E., Bourne C.R. Identifying a Molecular Mechanism That Imparts Species-Specific Toxicity to YoeB Toxins. Front. Microbiol. 2020;11:959. doi: 10.3389/fmicb.2020.00959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pavelich I.J., Maehigashi T., Hoffer E.D., Ruangprasert A., Miles S.J., Dunham C.M. Monomeric YoeB toxin retains RNase activity but adopts an obligate dimeric form for thermal stability. Nucleic Acids Res. 2019;47:10400–10413. doi: 10.1093/nar/gkz760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McGillick J., Ames J.R., Murphy T., Bourne C.R. A YoeB toxin cleaves both RNA and DNA. Sci. Rep. 2021;11:3592. doi: 10.1038/s41598-021-82950-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baker K.E., Mackie G.A. Ectopic RNase E sites promote bypass of 5’-end-dependent mRNA decay in Escherichia coli. Mol. Microbiol. 2003;47:75–88. doi: 10.1046/j.1365-2958.2003.03292.x. [DOI] [PubMed] [Google Scholar]
- 21.Bohm G., Muhr R., Jaenicke R. Quantitative analysis of protein far UV circular dichroism spectra by neural networks. Protein Eng. 1992;5:191–195. doi: 10.1093/protein/5.3.191. [DOI] [PubMed] [Google Scholar]
- 22.Shao Y., Harrison E.M., Bi D., Tai C., He X., Ou H.Y., Rajakumar K., Deng Z. TADB: A web-based resource for Type 2 toxin-antitoxin loci in bacteria and archaea. Nucleic Acids Res. 2011;39:D606–D611. doi: 10.1093/nar/gkq908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li G.Y., Zhang Y., Chan M.C., Mal T.K., Hoeflich K.P., Inouye M., Ikura M. Characterization of dual substrate binding sites in the homodimeric structure of Escherichia coli mRNA interferase MazF. J. Mol. Biol. 2006;357:139–150. doi: 10.1016/j.jmb.2005.12.035. [DOI] [PubMed] [Google Scholar]
- 24.Zorzini V., Mernik A., Lah J., Sterckx Y.G., De Jonge N., Garcia-Pino A., De Greve H., Versees W., Loris R. Substrate Recognition and Activity Regulation of the Escherichia coli mRNA Endonuclease MazF. J. Biol. Chem. 2016;291:10950–10960. doi: 10.1074/jbc.M116.715912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sterckx Y.G., De Gieter S., Zorzini V., Hadzi S., Haesaerts S., Loris R., Garcia-Pino A. An efficient method for the purification of proteins from four distinct toxin-antitoxin modules. Protein Expr. Purif. 2015;108:30–40. doi: 10.1016/j.pep.2015.01.001. [DOI] [PubMed] [Google Scholar]
- 26.Lu S., Wang J., Chitsaz F., Derbyshire M.K., Geer R.C., Gonzales N.R., Gwadz M., Hurwitz D.I., Marchler G.H., Song J.S., et al. CDD/SPARCLE: The conserved domain database in 2020. Nucleic Acids Res. 2020;48:D265–D268. doi: 10.1093/nar/gkz991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dai J., Chen Z., Hou J., Wang Y., Guo M., Cao J., Wang L., Xu H., Tian B., Zhao Y. MazEF Toxin-Antitoxin System-Mediated DNA Damage Stress Response in Deinococcus radiodurans. Front. Genet. 2021;12:632423. doi: 10.3389/fgene.2021.632423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bezrukov F., Prados J., Renzoni A., Panasenko O.O. MazF toxin causes alterations in Staphylococcus aureus transcriptome, translatome and proteome that underlie bacterial dormancy. Nucleic Acids Res. 2021;49:2085–2101. doi: 10.1093/nar/gkaa1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Drozdetskiy A., Cole C., Procter J., Barton G.J. JPred4: A protein secondary structure prediction server. Nucleic Acids Res. 2015;43:W389–W394. doi: 10.1093/nar/gkv332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chowdhury N., Kwan B.W., McGibbon L.C., Babitzke P., Wood T.K. Toxin MqsR cleaves single-stranded mRNA with various 5’ ends. Microbiologyopen. 2016;5:370–377. doi: 10.1002/mbo3.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Culviner P.H., Laub M.T. Global Analysis of the E. coli Toxin MazF Reveals Widespread Cleavage of mRNA and the Inhibition of rRNA Maturation and Ribosome Biogenesis. Mol. Cell. 2018;70:868–880.e10. doi: 10.1016/j.molcel.2018.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rothenbacher F.P., Suzuki M., Hurley J.M., Montville T.J., Kirn T.J., Ouyang M., Woychik N.A. Clostridium difficile MazF toxin exhibits selective, not global, mRNA cleavage. J. Bacteriol. 2012;194:3464–3474. doi: 10.1128/JB.00217-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mets T., Lippus M., Schryer D., Liiv A., Kasari V., Paier A., Maivali U., Remme J., Tenson T., Kaldalu N. Toxins MazF and MqsR cleave Escherichia coli rRNA precursors at multiple sites. RNA Biol. 2017;14:124–135. doi: 10.1080/15476286.2016.1259784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee K.Y., Lee B.J. Structure, Biology, and Therapeutic Application of Toxin-Antitoxin Systems in Pathogenic Bacteria. Toxins. 2016;8:305. doi: 10.3390/toxins8100305. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data are contained within the article or Supplementary Materials.