Abstract
MicroRNAs are short non-coding RNAs that regulate gene expression. Several microRNAs, useful for coronary artery disease assessment, have previously been identified. MicroRNA-33 is located within SREBP introns and controls cholesterol homeostasis. In order to find the possibility of microRNA-33 as a potential biomarker in high cholesterol disease, we developed a mouse model for coronary heart disease by feeding mice with a high-fat diet. The expression differences of microRNA-33, SREBP and ABCA1 genes in the liver, muscle, and lipid tissues were compared between a high-cholesterol group and control group in mice. The results showed that ABCA1 was up-regulated by high cholesterol conditions in liver, muscle and lipid tissues. SREBP1C was up-regulated by high cholesterol conditions in the liver and lipid tissues and down-regulated by high cholesterol conditions in the muscle tissue. MicroRNA-33 and SREBP2 were down-regulated by high cholesterol conditions in the liver and muscle tissues and up-regulated by high cholesterol conditions in the lipid tissue. Our study suggests that antisense therapeutic targeting of microRNA-33 may be a potential biomarker for cardiovascular disease.
1. Introduction
Cholesterol plays key role in many physiological process, and aberrant cholesterol content has been linked to disease, including coronary atherosclerosis and other diseases (Maxfield and Tabas, 2005; Moller and Kaufman, 2005). The classical transcription regulators, microRNAs, are important modulators of numerous cellular processes that impact disease (Williams et al., 2009). MicroRNAs, such as microRNA-126 (Fujii et al., 2016), microRNA-144 (Chen et al., 2018), and microRNA-33 (Kim et al., 2017), are a class of endogenous small, non-coding RNAs that bind to specific mRNAs to either inhibit their translation or promote their degradation (Lee et al., 1993; Reinhart et al., 2000; van Rooij and Olson, 2007). Circulating microRNAs are present within body fluids in a stable, cell-free form, and differing microRNA expression levels can be identified in various stages of coronary artery disease (CAD). Studies have shown a significant association between increased levels of plasma microRNA-33 and coronary artery disease, and therefore, microRNA-33 could be a useful biomarker for assessing CAD (Alrob et al., 2017; Gharipour and Sadeghi, 2013; Ono et al., 2015; Reddy et al., 2019; Rottiers and Naar, 2012; Rottiers et al., 2011).
MicroRNA-33a and microRNA-33b (the latter being present in primates but absent in rodents and lower species) are located within the introns of the sterol regulatory element binding protein (SREBP)-encoding genes. The SREBP cooperates with the proteins to control cholesterol homeostasis, fatty acid levels and expression of genes related to fat metabolism (Brown and Goldstein, 1997; Horie et al., 2010). SREBP-1c and SREBP-2 are two members of the SREBP family; there are differences in lengths. SREBP-1 regulated the gene expression which related to fatty acid synthesis. SREBP-2 regulated the gene expression which related to cholesterol uptake and synthesis (Moon et al., 2001).
Cholesterol plays a key role in many physiological processes. In lipid homeostasis, it has been linked to a number of diseases, including atherosclerosis (Najafi-Shoushtari et al., 2010). Cholesterol homeostasis in the liver is tightly controlled by pathways that regulate sterol synthesis and uptake from the plasma, which is mediated by SREBP and cholesterol catabolism. MicroRNA-33, encoded in an SREBP intron, targets the ATP-binding cassette transporter A1 (ABCA1) (an integration member protein) (Horie et al., 2010). ABCA1 can regulate high-density lipoprotein (HDL) synthesis and reverse cholesterol transport (Mao et al., 2014; Singaraja et al., 2002), implying that enhanced ABCA1 activity may protect against high cholesterol.
The microRNA-33, SREBP and ABCA1 genes have important role in cholesterol synthesis. Herein, we developed a mouse model of high cholesterol to investigate the functions of microRNA-33 and its related genes in mice.
2. Materials and methods
2.1. Animal experiment protocols
The animals used in this study were reared and sacrificed in compliance with national regulations for the humane care and use of animals in research (China Administration Rule of Laboratory Animals, Operating Procedure of Cattle Slaughtering GB/T 19477-2004). A total of 36 male C57BL mice weighing 18–20 g and aged 6 weeks were separated into two groups: one group was given water ad libitum and fed a high-fat diet containing 80 % ordinary feed, 10 % lard, 6 % egg yolk powder, 4 % cholesterol and 0.5 % sodium deoxycholate; another group was given water ad libitum and fed a standard laboratory chow diet for 30 d. On day 30, these mice were fasted for 12 h, weighed and anesthetized. Blood was collected from the mice to determine the serum levels of total cholesterol (TC), total triglyceride (TG), high-density lipoprotein (HDL) and low-density lipoprotein (LDL). Liver and muscle tissue were cut into small pieces and frozen in liquid nitrogen until the next step to detect gene expression.
2.2. RNA extraction
Total RNA was extracted using TRIzol Reagent (Life Technologies, Inc., Gaithersburg, MD, USA) in accordance with the manufacturer's instructions and stored at C. The purity and integrity of RNA was evaluated via electrophoresis, ethidium bromide staining, and optical density (OD) absorption ratios OD260 OD280 and rRNA (28S 18S).
2.3. Quantitative real-time PCR
Expression of the selected genes was analyzed using the Fast SYBR® Green Master Mix Bulk Pack (4385614; Invitrogen, USA). The following primers were used for quantitative reverse-transcription PCR (qRT-PCR): ABCA1, sense, 5-CATTTCGAAGGAGACAAACATGTCA-3 and antisense, 5-CATGGCTTTATTCGGAAAGTGGCAC-3; SREBP1C sense, 5-GGCTGTTGTCATCCATAAGC-3 and antisense 5-AGGAAGAAACATGTCAAGAA-3; SREBP2, sense 5-TTCCCTTGTGTTGACCACGC-3 and antisense 5-TGTGGTCAGAATGGTCCCGT-3; and -actin, sense 5-TCTCCACCTTCCAGCAGATGT-3 and antisense 5-AGCTCAGTAACAGTCCGCCTAGA-3 (Beijing Dingguo Changsheng Biotechnology Co. Ltd.). Three replicates were used for each experiment. Relative mRNA expression levels were measured using the comparative Ct method (Ct) with -actin as the internal control. The microRNA-33-3p sense sequence was CGCGCAATGTTTCCACAGTG, and the antisense sequence was AGTGCAGGGTCCGAGGTATT. The microRNA-33-3p-RT sequence was GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACGTGATG.
Figure 1.
Mice in the experimental group.
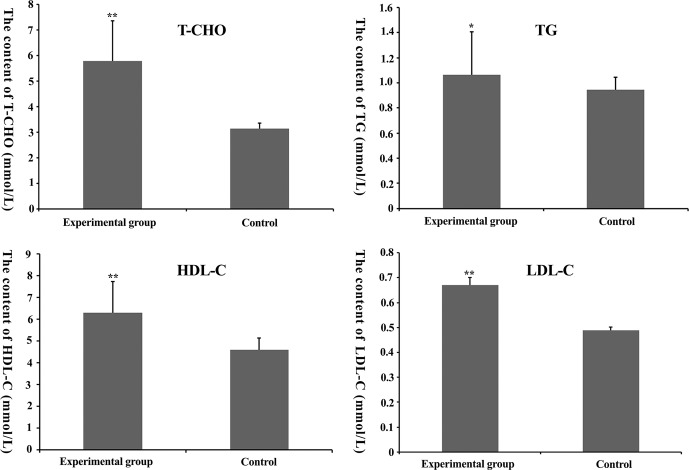
Figure 2.
Total cholesterol (TC), total triglyceride (TG), high-density lipoprotein-cholesterol (HDL-C) and low-density lipoprotein-cholesterol (LDL-C) content in mouse serum. denotes . denotes .
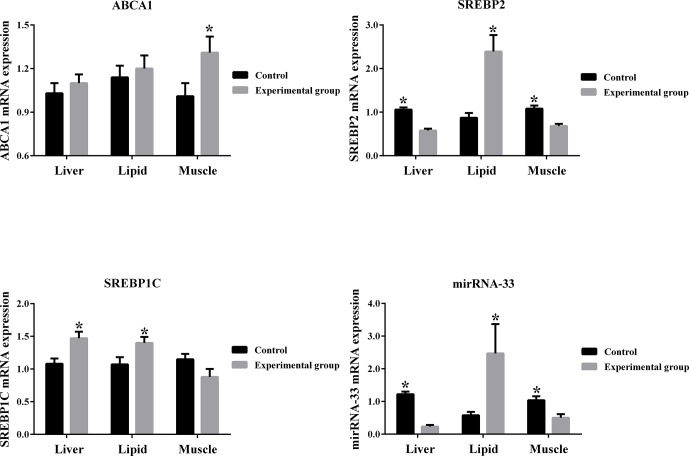
Figure 3.
Gene expression in different mouse tissues between the control and experimental groups.
Table 1.
The correlation between miRNA-33 and TG, TC, HDL-C and LDL-C.
| TC | TG | LDL-C | HDL-C | ||
|---|---|---|---|---|---|
| Liver miRNA-33 | Pearson correlation | 0.690 | 0.828 | 0.706 | 0.844 |
| |
Significance |
0.040 |
0.006 |
0.034 |
0.004 |
| Lipid miRNA-33 | Pearson correlation | 0.573 | 0.797 | 0.708 | 0.540 |
| |
Significance |
0.107 |
0.010 |
0.033 |
0.134 |
| Muscle miRNA-33 | Pearson correlation | 0.498 | 0.542 | 0.528 | 0.650 |
| Significance | 0.070 | 0.045 | 0.052 | 0.012 |
2.4. Blood lipid analysis
TC, TG, HDL-C and LDL-C levels were measured in the plasma using detection kits specific to each cholesterol type (TC kit, TG kit, HDL-C kit and LDL-C kit; Nanjing Jiancheng Biology Institute, Nanjing, China).
2.5. Statistical analysis
SPSS software version 20.0 was used to perform statistical analysis. Data differences in gene expression among multiple groups were calculated using an analysis of variance, followed by a Bonferroni test. Comparisons between two groups were analyzed using the independent-sample test. The Pearson correlation was used to analyze the correlation between genes and blood indexes.
3. Results
3.1. Animal experiment
After the experiment, mouse appearance was observed. From Fig. 1, we find that most of the mice in the experimental group lost their hair more or less by being fed a high cholesterol diet. The percent value of hair loss can be as high as 60 %.
3.2. TC, TG, HDL-C and LDL-C content in serum of mice
From Fig. 2, we find that T-C, TG, HDL-C and LDL-C content in the experimental group was higher than that in the control group (), suggesting that the feasibility of the model was founded on nutrition.
3.3. Difference in gene expression in different tissues of mice between experimental group and control group
From Fig. 3 it can be seen that ABCA1 is up-regulated by high cholesterol conditions in the liver, muscle and lipid tissues of mice. SREBP1C is up-regulated by high cholesterol conditions in the liver and lipid tissues and down-regulated by high cholesterol conditions in the muscle tissue. Additionally, microRNA-33 and SREBP2 are down-regulated by high cholesterol conditions in the liver and muscle tissues and up-regulated by high cholesterol conditions in the lipid tissue.
3.4. Correlation between miRNA-33 and TG, TC, HDL-C and LDL-C
From Table 1 we observe a significant negative correlation between microRNA-33 and TC, TG, HDL-C and LDL-C in the liver and between microRNA-33 and TG, HDL-C in muscle tissues. However, there are significant positive correlations between microRNA-33 and TG as well as between microRNA-33 and LDL-C in the lipid tissue.
4. Discussion
Altered cholesterol homeostasis is involved in the pathogenesis of cicatricial alopecia; the mutation of cholesterol transporter is associated with congenital hypertrichosis (Palmer et al., 2020). The control of cellular cholesterol in the hair follicle and the potential impact on the hair cycle may identify novel target for regulating hair growth and the treatment of hair disorders linked to disordered sterol homeostasis or sterol-sensitive signaling pathways (DeStefano et al., 2014; Evers et al., 2010). To the best of our knowledge, this is one of the few studies wherein diet appeared to affect the hair loss of mice. In the current study, the ApoE mice (fed diets rich in cholesterol and fat) showed skin inflammation, hair discoloration and hair loss (Bedja et al., 2018). It is not clear whether such hair loss will appear in humans in the future, but this must be tested clinically. Although this result is rarely discussed in the literature, we believe that it is an important theory to be tested in future research.
MicroRNA-33 has been shown to be up-regulated by low sterol conditions, including lipid deprivation and statin treatment, and down-regulated by lipid loading (Marquart et al., 2010; Najafi-Shoushtari et al., 2010; Rayner et al., 2010). MicroRNA-33 levels were positively correlated with the TC and LDL-C levels but negatively correlated with the HDL-C levels in blood (Chen et al., 2016). Long-term anti-microRNA-33 therapy significantly reduces the progression of atherosclerosis and improves HDL functionality (Rotllan et al., 2013). Further, RT-PCR analysis of miRNA-33 in the blood plasma of patients with CAD showed a 2.9-fold increase compared to that in the blood plasma of the control group, supporting the use of microRNA-33 as a potential biomarker for CAD (Reddy et al., 2019). Additionally, hepatic microRNA-33 levels show a 1.5–2.5-fold increase under physiological conditions that alter SREBP-2 expression (Tarling et al., 2015).
Consistent with the results of studies demonstrating the coordinated regulation of SREBP2 and microRNA-33 (Elizabeth et al., 2015), our results indicate that these two genes have the same expression trends in the three tissues when the high cholesterol diets also increase/decrease the microRNA-33 expression in parallel with the alteration in SREBP-2 mRNA levels in the tissues. We found that microRNA-33 and SREBP2 were down-regulated by high cholesterol conditions in the liver and muscle tissues and up-regulated by high cholesterol conditions in the lipid tissue.
In summary, our results indicate that the key regulators of cholesterogenic and lipogenic genes combine with microRNA-33 to govern intracellular cholesterol levels and cholesterol homeostasis in mice. We propose antisense therapeutic targeting of microRNA-33 as a potential strategy for the management of cardiovascular disease.
5. Conclusion
Our study suggests that antisense therapeutic targeting of microRNA-33 may be a potential biomarker for cardiovascular disease.
Acknowledgements
This work was supported by Young Talents Training Program of Shandong Academy of Agricultural Science, Agricultural Science and National Science Foundation of China (grant no. 31772649), Technology Innovation Project of Shandong Academy of Agricultural Sciences (grant nos. CXGC2017B02, CXGC2018E10).
Contributor Information
Haichao Lin, Email: haichaolin@gmail.com.
Guifen Liu, Email: liuguifen126@126.com.
Code availability
SPSS software (version 20.0) is available at (IBM, 2021).
Data availability
The data are available from the corresponding author upon request.
Author contributions
All authors made substantial contributions to the study. HL and GL conceived and designed the research. XZ, HZ, QS, XL and WY performed the material preparation and experiments. XZ, HZ and XL performed the data collection and analysis. XZ, HL and GL drafted the manuscript. All authors read and approved the final article.
Competing interests
The authors declare that they have no conflict of interest.
Financial support
This research has been supported by the Shandong Academy of Agricultural Science, Agricultural Science and National Science Foundation of China (grant no. 31772649) and the Shandong Academy of Agricultural Sciences (grant nos. CXGC2017B02 and CXGC2018E10).
Review statement
This paper was edited by Steffen Maak and reviewed by Koray Kirikçi and one anonymous referee.
References
- Alrob OA, Khatib S, Naser SA. MicroRNAs 33, 122, and 208: a potential novel targets in the treatment of obesity, diabetes, and heart-related diseases. J Physiol Biochem. 2017;73:307–314. doi: 10.1007/s13105-016-0543-z. [DOI] [PubMed] [Google Scholar]
- Bedja D, Yan W, Lad V, Iocco D, Sivakumar N, Bandaru VVR, Chatterjee S. Inhibition of glycosphingolipid synthesis reverses skin inflammation and hair loss in ApoE mice fed western diet. Sci Rep-UK. 2018;8:11463. doi: 10.1038/s41598-018-28663-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown MS, Goldstein JL. The SREBP pathway: regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell. 1997;89:331–340. doi: 10.1016/s0092-8674(00)80213-5. [DOI] [PubMed] [Google Scholar]
- Chen B, Luo L, Wei X, Gong D, Jin L. Altered Plasma miR-144 as a Novel Biomarker for Coronary Artery Disease. Ann Clin Lab Sci. 2018;48:440–445. [PubMed] [Google Scholar]
- Chen WM, Sheu WH, Tseng PC, Lee TS, Lee WJ, Chang PJ, Chiang AN. Modulation of microRNA Expression in Subjects with Metabolic Syndrome and Decrease of Cholesterol Efflux from Macrophages via microRNA-33-Mediated Attenuation of ATP-Binding Cassette Transporter A1 Expression by Statins. Plos One. 2016;11:e0154672. doi: 10.1371/journal.pone.0154672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeStefano GM, Kurban M, Anyane-Yeboa K, Dall'Armi C, Di Paolo G, Feenstra H, Silverberg N, Rohena L, López-Cepeda LD, Jobanputra V, Fantauzzo KA, Kiuru M, Tadin-Strapps M, Sobrino A, Vitebsky A, Warburton D, Levy B, Salas-Alanis JC, Christiano AM. Mutations in the cholesterol transporter gene ABCA5 are associated with excessive hair overgrowth. Plos Genet. 2014;15:e1004333. doi: 10.1371/journal.pgen.1004333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elizabeth JT, Hannah A, Thomas QAV. The Nuclear Receptor FXR Uncouples the Actions of miR-33 from SREBP-2. Arterioscl Throm Vas. 2015;35:787–795. doi: 10.1161/ATVBAHA.114.304179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evers BM, Farooqi MS, Shelton JM, Richardson JA, Goldstein JL, Brown MS, Liang G. Hair growth defects in Insig-deficient mice caused by cholesterol precursor accumulation and reversed by simvastatin. J Invest Dermatol. 2010;130:1237–1248. doi: 10.1038/jid.2009.442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii S, Sugiura T, Dohi Y, Ohte N. MicroRNA in atherothromobosis: is it useful as a disease marker? Thromb J. 2016;14:21. doi: 10.1186/s12959-016-0112-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gharipour M, Sadeghi M. Pivotal role of microRNA-33 in metabolic syndrome: A systematic review. ARYA Atheroscler. 2013;9:372–376. [PMC free article] [PubMed] [Google Scholar]
- Horie T, Ono K, Horiguchi M, Nishi H, Nakamura T, Nagao K, Kinoshita M, Kuwabara Y, Marusawa H, Iwanaga Y, Hasegawa K, Yokode M, Kimura T, Kita T. MicroRNA-33 encoded by an intron of sterol regulatory element-binding protein 2 (Srebp2) regulates HDL in vivo. P Natl Acad Sci USA. 2010;107:17321–17326. doi: 10.1073/pnas.1008499107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- IBM [last access: 17 March 2021];SPSS software (version 20.0) 2021 available at: https://www.ibm.com/analytics/spss-statistics-software .
- Kim SH, Kim GJ, Umemura T, Lee SG, Cho KJ. Aberrant expression of plasma microRNA-33a in an atherosclerosis-risk group. Mol Biol Rep. 2017;44:79–88. doi: 10.1007/s11033-016-4082-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- Mao M, Lei H, Liu Q, Chen Y, Zhao L, Li Q, Luo S, Zuo Z, He Q, Huang W, Zhang N, Zhou C, Ruan XZ. Effects of miR-33a-5P on ABCA1/G1-mediated cholesterol efflux under inflammatory stress in THP-1 macrophages. Plos One. 2014;9:e109722. doi: 10.1371/journal.pone.0109722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquart TJ, Allen RM, Ory DS, Baldan A. miR-33 links SREBP-2 induction to repression of sterol transporters. P Natl Acad Sci USA. 2010;107:12228–12232. doi: 10.1073/pnas.1005191107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxfield FR, Tabas I. Role of cholesterol and lipid organization in disease. Nature. 2005;438:612–621. doi: 10.1038/nature04399. [DOI] [PubMed] [Google Scholar]
- Moon YA, Shah NA, Mohapatra S, Warrington JA, Horton JD. Identification of a mammalian long chain fatty acyl elongase regulated by sterol regulatory element-binding proteins. J Biol Chem. 2001;276:45358–45366. doi: 10.1074/jbc.M108413200. [DOI] [PubMed] [Google Scholar]
- Moller DE, Kaufman KD. Metabolic syndrome: a clinical and molecular perspective. Annu Rev Med. 2005;56:45–62. doi: 10.1146/annurev.med.56.082103.104751. [DOI] [PubMed] [Google Scholar]
- Najafi-Shoushtari SH, Kristo F, Li Y, Shioda T, Cohen DE, Gerszten RE, Naar AM. MicroRNA-33 and the SREBP host genes cooperate to control cholesterol homeostasis. Science. 2010;328:1566–1569. doi: 10.1126/science.1189123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono K, Horie T, Nishino T, Baba O, Kuwabara Y, Yokode M, Kita T, Kimura T. MicroRNA-33a/b in lipid metabolism – novel “thrifty” models. Circ J. 2015;79:278–284. doi: 10.1253/circj.CJ-14-1252. [DOI] [PubMed] [Google Scholar]
- Palmer MA, Blakeborough L, Harries M, Haslam IS. Cholesterol homeostasis: Links to hair follicle biology and hair disorders. Exp Dermatol. 2020;29:299–311. doi: 10.1111/exd.13993. [DOI] [PubMed] [Google Scholar]
- Rayner KJ, Suarez Y, Davalos A, Parathath S, Fitzgerald ML, Tamehiro N, Fisher EA, Moore KJ, Fernandez-Hernando C. MiR-33 contributes to the regulation of cholesterol homeostasis. Science. 2010;328:1570–1573. doi: 10.1126/science.1189862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy LL, Shah SAV, Ponde CK, Rajani RM, Ashavaid TF. Circulating miRNA-33: a potential biomarker in patients with coronary artery disease. Biomarkers. 2019;24:36–42. doi: 10.1080/1354750X.2018.1501760. [DOI] [PubMed] [Google Scholar]
- Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR, Ruvkun G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- Rotllan N, Ramirez CM, Aryal B, Esau CC, Fernandez-Hernando C. Therapeutic silencing of microRNA-33 inhibits the progression of atherosclerosis in Ldlr mice–brief report. Arterioscl Throm Vas. 2013;33:1973–1977. doi: 10.1161/ATVBAHA.113.301732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rottiers V, Naar AM. MicroRNAs in metabolism and metabolic disorders. Nat Rev Mol Cell Biol. 2012;13:239–250. doi: 10.1038/nrm3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rottiers V, Najafi-Shoushtari SH, Kristo F, Gurumurthy S, Zhong L, Li Y, Cohen DE, Gerszten RE, Bardeesy N, Mostoslavsky R, Naar AM. MicroRNAs in metabolism and metabolic diseases. Cold Spring Harb Symp Quant Biol. 2011;76:225–233. doi: 10.1101/sqb.2011.76.011049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singaraja RR, Fievet C, Castro G, James ER, Hennuyer N, Clee SM, Bissada N, Choy JC, Fruchart JC, McManus BM, Staels B, Hayden MR. Increased ABCA1 activity protects against atherosclerosis. J Clin Invest. 2002;110:35–42. doi: 10.1172/JCI15748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarling EJ, Ahn H, de Aguiar Vallim TQ. The nuclear receptor FXR uncouples the actions of miR-33 from SREBP-2. Arterioscl Throm Vas. 2015;35:787–795. doi: 10.1161/ATVBAHA.114.304179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Rooij E, Olson EN. MicroRNAs: powerful new regulators of heart disease and provocative therapeutic targets. J Clin Invest. 2007;117:2369–2376. doi: 10.1172/JCI33099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams AH, Liu N, van Rooij E, Olson EN. MicroRNA control of muscle development and disease. Curr Opin Cell Biol. 2009;21:461. doi: 10.1016/j.ceb.2009.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data are available from the corresponding author upon request.