Abstract
N6-methyladenosine (m6A), the most abundant internal RNA modification, serves a critical role in cancer development. However, the clinical implications of m6A in hepatocellular carcinoma (HCC) remain unclear. The present study sought to reveal the potential roles of m6A readers, which recognize m6A, in HCC. A total of 177 HCC and paired non-cancerous liver tissues from patients who underwent hepatectomy were analysed using quantitative PCR for the expression of m6A readers: YT521-B homology domain family 1 (YTHDF1) and YT521-B homology domain family 2 (YTHDF2). The expression levels of both YTHDF1 and YTHDF2 were not significantly different between tumour and non-cancerous tissues (P=0.93 and P=0.7, respectively). Analysis of the association between clinical features and m6A reader expression revealed that YTHDF1 expression was associated with formation of capsule (P=0.02), whereas low YTHDF2 expression was associated with septal formation (P=0.02). Furthermore, high YTHDF1 expression and high YTHDF2 expression were significantly associated with shorter recurrence-free survival (RFS) [YTHDF1: Mean survival time (MST), 34.0 vs. 19.0 months, P=0.014; YTHDF2: MST, 30.1 vs. 12.9 months, P=0.0032], whereas YTHDF1 and YTHDF2 expression was not significantly associated with overall survival (OS) (YTHDF1: MST, 99.4 vs. 70.2 months, P=0.74; YTHDF2: MST, 98.4 vs. 64.1 months, P=0.28). According to multivariate analysis, serosal invasion [hazard ratio (HR), 2.39; 95% CI 1.30–4.42; P=0.005), portal vein or hepatic vein invasion (HR, 2.82; 95% CI 1.26–6.28; P=0.01) and YTHDF2 expression in HCC tissues (HR, 1.85; 95% CI 1.09–3.15; P=0.02) were identified as significant independent prognostic factors for RFS. α-fetoprotein (HR, 1.79; 95% CI 1.10–2.92; P=0.02), serosal invasion (HR, 1.99; 95% CI 1.17–3.34; P=0.01) and portal vein or hepatic vein invasion (HR, 3.02; 95% CI 1.38–6.61; P=0.006) were identified as significant independent prognostic factors for OS. In conclusion, the present study revealed that high YTHDF2 expression, an m6A reader, in HCC tissues was associated with cancer recurrence.
Keywords: hepatocellular carcinoma, methylation of N6 adenosine, YTH domain family, prognosis
Introduction
Hepatocellular carcinoma (HCC) is the main type of primary liver cancer and is one of the most common malignancies with poor survival (1). Hepatectomy is a potentially curative treatment, but the recurrence rate of HCC after surgery is remarkably high at approximately 70% (2). Therefore, further understanding of the molecular mechanisms of HCC development and recurrence is required.
More than 100 types of chemical modifications have been identified in RNA (3). Recently, internal modifications of mRNA have received attention for their roles in mRNA metabolism. N6-methyladenosine (m6A) is the most abundant modification of mRNA in eukaryotes and was first reported in the 1970s (4). Recent evidence suggests that m6A has various functions in RNA metabolism, such as pre-mRNA splicing, 3′-end processing, nuclear export, translation regulation, regulation of mRNA decay and noncoding RNA processing (5–7). Furthermore, m6A methylation has been revealed to have crucial roles in the initiation and progression of cancer (8).
m6A readers are the proteins that recognize and bind to m6A sites and thereby elicit multiple effects (9). YT521-B homology domain family 2 (YTHDF2) was the first identified m6A binding protein (10). YTHDF2 weakens mRNA stability by recognizing m6A, while YT521-B homology domain family 1 (YTHDF1) promotes mRNA translation efficiency (11). In solid cancer, YTHDF2 and YTHDF1 have been reported to have roles as both tumour promoters and suppressors (12–14). However, the significance of YTHDF1 and YTHDF2 in oncogenesis remains unclear.
In the current study, we assessed the expression of YTHDF1 and YTHDF2 in both resected HCC tissues and paired normal liver tissues collected from patients who underwent surgery with curative intent. We also sought to discover novel prognostic implications of m6A readers that could be used to predict prognosis in patients with resected HCC.
Patients and methods
Patients and samples
A total of 177 frozen tumour specimens and paired paratumor noncancerous tissues were collected from patients with HCC who underwent surgery at Nagoya University Hospital (Nagoya City, Japan) between January 1998 and April 2014. All fresh tissues were immediately frozen in liquid nitrogen and stored at −80°C until use. Patient characteristics are summarized in Table I. After surgery, all patients were monitored via blood examinations, ultrasonography, and computed tomography once every six months. Angiography was performed for further information whenever recurrence was suspected. The median follow-up duration of all patients was 48.8 months (range, 0.3 to 191 months). This study and all procedures were approved by the Institutional Review Board at Nagoya University (Nagoya City, Japan), and all patients provided written informed consent. All clinical investigations were conducted in accordance with the principles of the Declaration of Helsinki.
Table I.
Clinicopathological characteristics of patients with hepatocellular carcinoma (n=177).
| Characteristics | Value |
|---|---|
| Median age (range), years | 65 (37–84) |
| Sex, male:female, n (%) | 148 (84) : 29 (16) |
| Viral infection, HBV:HCV:non-HBV/HCV, n (%) | 41 (23) : 106 (60) : 30 (17) |
| Median albumin (range), g/dl | 3.9 (2.3–4.9) |
| Median total bilirubin (range), mg/dl | 0.7 (0.2–7.3) |
| Median PT (range), % | 88.7 (46.9–138) |
| Median ICG-R15 (range), % | 11.4 (1.6–70.5) |
| Child-Pugh classification, A:B, n (%) | 166 (94):10 (6) |
| Liver damage classification, A:B:C, n (%) | 142 (83):28 (16):1 (1) |
| Tumour multiplicity, solitary:multiple, n (%) | 138 (78):39 (22) |
| Median tumour size (range), cm | 3.5 (0.15–15) |
| Median AFP (range), ng/ml | 17 (0.8–119923) |
| Stage, I:II:III:IV, n (%) | 19 (11):91 (52):44 (25):21 (12) |
PT, prothronbin time; ICG-R15, indocyanine green 15-min clearance rate; AFP, α-fetoprotein; HBV, hepatitis B virus; HCV, hepatitis C virus.
RNA isolation and RT-qPCR
Total RNA was extracted from tissue samples using a Qiagen miRNeasy mini-kit (Qiagen, Hilden, Germany). We used DNAse to avoid contamination, and RNA quality was analysed by a NanoDrop (Thermo Scientific Fisher, Waltham, MA, USA). Total RNA was converted to complementary DNA by reverse transcription with M-MLV Reverse Transcriptase (Invitrogen, Carlsbad, CA, USA). This total cDNA was used as a template for the next step of quantitative PCR (qPCR). qPCR was performed using SYBR Premix Ex Taq II (Takara Clontech, Kyoto, Japan) under the following conditions: Denaturation at 95°C for 10 sec and 40 cycles of denaturation at 95°C for 5 sec and annealing/extension at 60°C for 30 sec. The SYBR Green signal was detected in real-time using a StepOne Plus Real-Time PCR System (Life Technologies, Carlsbad, CA, USA). The relative quantification method was used, and the expression level of each gene was normalized to the expression level of the control gene glyceraldehyde-3-phosphate dehydrogenase (GAPDH) for each sample. The relative gene expression levels were determined using the comparative threshold cycle (2−ΔCT) method.
The PCR primers used in the current study were specific for the 78-base-pair fragment of YTHDF1 (sense, 5′-TCCATCTTCGACGACTTTGCT-3′; antisense, 5′-TCGACTCTGCCGTTCCTTG-3′) and for the 50-base-pair fragment of YTHDF2 (sense, 5′-GAGGATCTGAGAGCCATGTCG-3′; antisense, 5′-ATTTTGTACTGCTCCAAGAGGC-3′). GAPDH primers (sense, 5′-GAGTCCACTGGCGTCTTCAC-3′; antisense, 5′-GTTCACACCCATGACGAACA-3′) were used to quantify the expression in each sample as an internal control. The primers were designed as intron spanning. All qPCR experiments were performed in duplicate, including the template-omitted negative controls.
Acquisition of publicly available data
Normalized TCGA RNA-sequencing data of HCC were downloaded from the Broad GDAC Firehose (http://gdac.broadinstitute.org/, accessed on January 1st, 2020). This dataset consists of 50 noncancerous cases and 360 HCC cases, including seven HCC cases mixed with hepatocholangiocarcinoma and two cases with fibrolamellar carcinoma. Of the 360 cases, there were 266 cases with recurrence-free survival (RFS) information and 336 cases with overall survival (OS) information.
Statistical analysis
Continuous variables are expressed as the median (range), and the expression of each target gene was compared by a Wilcoxon signed-rank test and paired t-test. Categorical variables were compared using χ2 or Fisher's exact tests, as appropriate. The OS and RFS rates at each point of the follow-up time were estimated using the Kaplan-Meier method and compared using a log-rank test. A Cox proportional hazard regression model was used to perform univariate analysis and multivariate analysis for OS and RFS. In the multivariate analysis, variables that showed statistical significance in the univariate Cox proportional hazard regression were included. All statistical analyses were performed using R version 3.5.3 (http//www.r-project.org/), and P<0.05 obtained using two-tailed tests was considered to indicate statistical significance.
Results
YTHDF1 and YTHDF2 in resected specimens from HCC patients
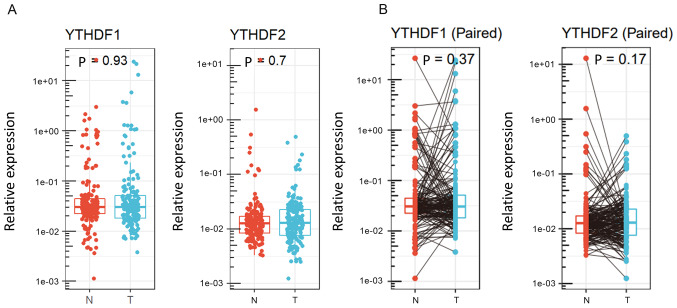
First, expression analyses of m6A readers were conducted with our surgically resected specimens. The expression levels of YTHDF1 and YTHDF2 were measured by qPCR. The expression of YTHDF1 and YTHDF2 was not significantly different between tumour tissues and noncancerous tissues (P=0.93 and P=0.7, respectively, Fig. 1A). Fig. 1B shows individual changes in YTHDF1 and YTHDF2 expression in paired analysis. Based on the results obtained by qPCR, 177 HCC cases were divided into two groups according to YTHDF1 and YTHDF2 expression in tumour tissues. We selected the cut-off values that showed the best statistical difference. Clinical features of the groups stratified by YTHDF1 and YTHDF2 expression are shown in Table II. In HCC tissues, low YTHDF1 expression was associated with a lack of capsule (65% vs. 81%, P=0.02), whereas low YTHDF2 expression was associated with septal formation (73% vs. 53%, P=0.02).
Figure 1.
Relative expression levels of YTHDF1 and YTHDF2 in (A) individual and (B) paired hepatocellular carcinoma and adjacent tissues. YTHDF1/2, YT521-B homology domain family 1/2; T, tumour; N, non-cancerous.
Table II.
Clinical features of 177 patients with hepatocellular carcinoma according to YTHDF1 and YTHDF2 expression.
| YTHDF1 expression (n=172)a | YTHDF2 expression (n=174)b | |||||
|---|---|---|---|---|---|---|
| Variables | Low (n=108) | High (n=64) | P-value | Low (n=137) | High (n=37) | P-value |
| Age, years | ||||||
| <65 | 50 | 34 | 0.43 | 67 | 18 | 0.98 |
| ≥65 | 58 | 30 | 70 | 19 | ||
| Sex | ||||||
| Female | 14 | 13 | 0.28 | 22 | 6 | 0.98 |
| Male | 94 | 51 | 115 | 31 | ||
| Virus infection | ||||||
| Others | 44 | 25 | 0.87 | 58 | 13 | 0.46 |
| HCV | 64 | 39 | 79 | 24 | ||
| Albumin, g/dl | ||||||
| ≥3.5 | 85 | 54 | 0.54 | 110 | 30 | 0.98 |
| <3.5 | 22 | 10 | 26 | 7 | ||
| NA | 1 | 0 | 1 | 0 | ||
| PT, % | ||||||
| ≥70 | 91 | 57 | 0.50 | 114 | 34 | 0.29 |
| <70 | 16 | 7 | 22 | 3 | ||
| NA | 1 | 0 | 1 | 0 | ||
| ICG-R15, % | ||||||
| <15 | 58 | 35 | 0.91 | 75 | 19 | 0.15 |
| ≥15 | 19 | 12 | 21 | 11 | ||
| NA | 31 | 17 | 41 | 7 | ||
| Liver cirrhosis | ||||||
| Negative | 72 | 39 | 0.51 | 90 | 20 | 0.25 |
| Positive | 36 | 25 | 47 | 17 | ||
| Child-Pugh classification | ||||||
| A | 100 | 61 | 0.74 | 128 | 35 | 0.91 |
| B | 7 | 3 | 8 | 2 | ||
| NA | 1 | 0 | 1 | 0 | ||
| Liver damage | ||||||
| A | 84 | 54 | 0.53 | 110 | 29 | 0.80 |
| B or C | 19 | 9 | 22 | 7 | ||
| NA | 5 | 1 | 5 | 1 | ||
| Tumour number | ||||||
| Solitary | 86 | 48 | 0.57 | 109 | 27 | 0.38 |
| Multiple | 22 | 16 | 28 | 10 | ||
| Tumour size, cm | ||||||
| <2 | 14 | 10 | 0.82 | 17 | 6 | 0.79 |
| ≥2 | 78 | 50 | 100 | 29 | ||
| NA | 16 | 4 | 20 | 2 | ||
| AFP, ng/ml | ||||||
| <20 | 58 | 35 | 0.91 | 68 | 25 | 0.06 |
| ≥20 | 48 | 28 | 67 | 11 | ||
| NA | 2 | 1 | 2 | 1 | ||
| Differentiation | ||||||
| Good or moderate | 98 | 56 | 0.77 | 121 | 35 | 0.08 |
| Poor | 8 | 6 | 14 | 0 | ||
| NA | 2 | 2 | 2 | 2 | ||
| Growth form | ||||||
| Expansive | 88 | 54 | 0.52 | 114 | 31 | 0.42 |
| Infiltrative | 18 | 8 | 22 | 3 | ||
| NA | 2 | 2 | 1 | 3 | ||
| Formation of capsule | ||||||
| Positive | 70 | 52 | 0.02 | 97 | 27 | 0.84 |
| Negative | 38 | 12 | 40 | 10 | ||
| Infiltration to capsule | ||||||
| Negative | 52 | 22 | 0.08 | 57 | 17 | 0.71 |
| Positive | 55 | 42 | 79 | 20 | ||
| NA | 1 | 0 | 1 | 0 | ||
| Septal formation | ||||||
| Positive | 74 | 42 | 0.61 | 98 | 19 | 0.02 |
| Negative | 31 | 21 | 36 | 17 | ||
| NA | 3 | 1 | 3 | 1 | ||
| Serosal invasion | ||||||
| Negative | 85 | 50 | 0.69 | 107 | 31 | 0.81 |
| Positive | 19 | 14 | 26 | 6 | ||
| NA | 4 | 0 | 4 | 0 | ||
| Portal vein or hepatic vein invasion | ||||||
| Negative | 80 | 45 | 0.60 | 101 | 26 | 0.68 |
| Positive | 28 | 19 | 36 | 11 | ||
| Surgical margin | ||||||
| Negative | 91 | 53 | 0.66 | 114 | 31 | 0.92 |
| Positive | 15 | 11 | 21 | 6 | ||
| NA | 2 | 0 | 2 | 0 | ||
| Stage | ||||||
| <III | 65 | 42 | 0.62 | 85 | 24 | 0.83 |
| ≥III | 41 | 22 | 50 | 13 | ||
| NA | 2 | 0 | 2 | 0 | ||
The data for YTHDF1 expression were not available for 5 patients.
The data for YTHDF2 expression were not available for 3 patients. PT, prothrombin time; ICG-R15, indocyanine green 15-min clearance rate; AFP, α-fetoprotein; HCV, hepatitis C virus; YTHDF1/2, YT521-B homology domain family 1/2; NA, not available.
Prognostic significance of YTHDF1 and YTHDF2 in resected HCC cases
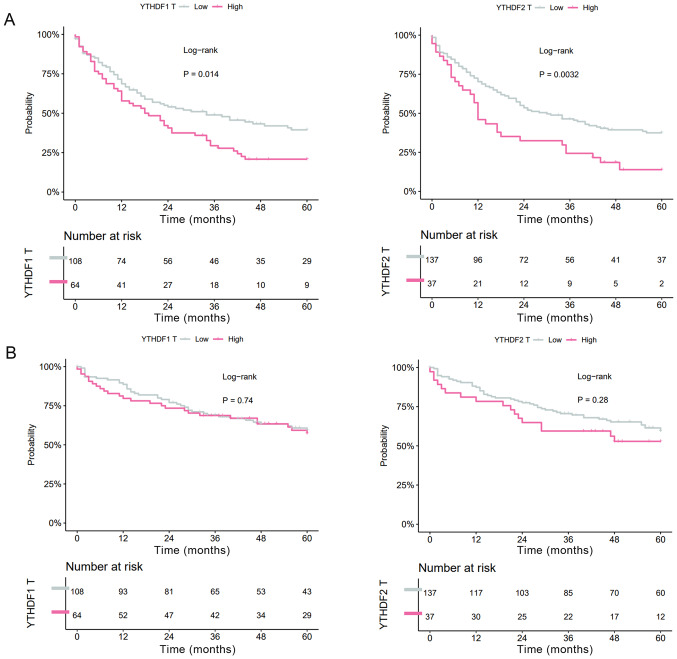
Next, the effects of the expression levels on RFS and OS were evaluated. In HCC tissues, both high YTHDF1 expression and high YTHDF2 expression were significantly correlated with shorter RFS (YTHDF1: MST=34.0 vs. 19.0 months, P=0.014; YTHDF2: MST=30.1 vs. 12.9 months, P=0.0032, Fig. 2A), whereas YTHDF1 expression and YTHDF2 expression were not correlated with OS (YTHDF1: MST=99.4 vs. 70.2 months, P=0.74; YTHDF2: MST=98.4 vs. 64.1 months, P=0.28, Fig. 2B).
Figure 2.
Kaplan-Meier analysis of (A) recurrence-free survival and (B) overall survival for 177 patients with hepatocellular carcinoma based on YTHDF1 and YTHDF2 expression. YTHDF1/2, YT521-B homology domain family 1/2.
YTHDF1 and YTHDF2 expression levels and their correlation with HCC prognosis in a publicly available dataset
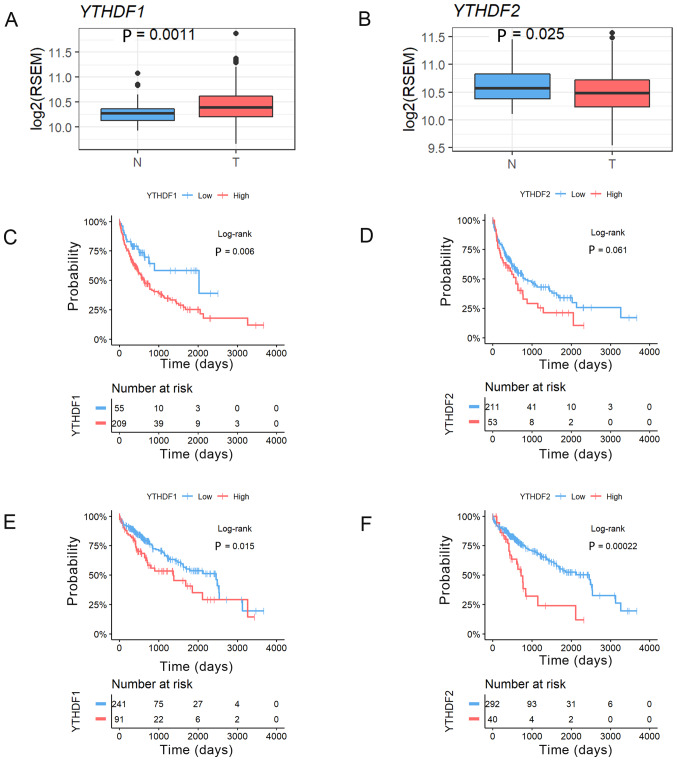
We analysed the expression levels of YTHDF1 and YTHDF2 in HCC and noncancerous tissues using a TCGA RNA-sequence dataset. This analysis revealed that the expression of YTHDF1 was significantly higher in HCC tumour tissues and that YTHDF2 expression was significantly lower in HCC tumour tissues than in noncancerous tissues (YTHDF1, P=0.0011; YTHDF2, P=0.025, Fig. 3A and B). We then confirmed the prognostic impact of YTHDF1 and YTHDF2 expression in resected HCC patients using the same TCGA dataset. HCC cases were divided into two groups according to the YTHDF1 and YTHDF2 expression in HCC tissues in the normalized RNA-sequencing data. We also selected the cut-off values that showed the best statistical difference. This analysis revealed that the patients with high YTHDF1 expression had significantly worse RFS (MST=754 vs. 489 days, P=0.006, Fig. 3C), and the patients with high YTHDF2 expression tended to have worse RFS (MST=636 vs. 315 days, P=0.06, Fig. 3D). In addition, the patients with high YTHDF1 expression had significantly worse OS (MST=2456 vs. 1,372 days, P=0.015, Fig. 3E), and the patients with high YTHDF2 expression had significantly worse OS (MST=2,456 vs. 724 days, P=0.0002, Fig. 3F).
Figure 3.
Relative (A) YTHDF1 and (B) YTHDF2 expression in 360 paired hepatocellular carcinoma tissues and 50 adjacent tissues from TCGA dataset. Survival analysis using Kaplan-Meier curves for recurrence-free survival in TCGA dataset based on (C) YTHDF1 and (D) YTHDF2 expression. Survival analysis using Kaplan-Meier curves for overall survival in TCGA dataset based on (E) YTHDF1 and (F) YTHDF2 expression. TCGA, The Cancer Genome Atlas; YTHDF1/2, YT521-B homology domain family 1/2; T, tumour; N, non-cancerous; RSEM, RNA-Sequencing by Expectation Maximization.
Cox regression analysis of HCC survival
Since the survival curves showed that YTHDF1 and YTHDF2 expression levels in HCC tissues were correlated with RFS, we performed Cox proportional hazards analyses to further investigate the prognostic value of YTHDF1 and YTHDF2 expression. The multivariate analysis identified serosal invasion (hazard ratio (HR): 2.39, 95% confidence interval (95% CI): 1.30–4.42, P=0.005), portal vein or hepatic vein invasion (HR, 2.82, 95% CI: 1.26–6.28, P=0.01) and YTHDF2 expression in HCC tissues (HR, 1.85, 95% CI: 1.09–3.15, P=0.02) as significant independent factors for RFS (Table III) and AFP (HR, 1.79, 95% CI: 1.10–2.92, P=0.02), serosal invasion (HR, 1.99, 95% CI: 1.17–3.34, P=0.01), and portal vein or hepatic vein invasion (HR, 3.02, 95% CI: 1.38–6.61, P=0.006) as significant independent factors for OS (Table IV). Consequently, high expression of YTHDF2 in HCC tissues was significantly associated with recurrence after HCC surgery.
Table III.
Univariate and multivariate Cox proportional-hazard regression analysis of recurrence free survival in patients with hepatocellular carcinoma.
| Univariate analysis | Multivariate analysis | |||||
|---|---|---|---|---|---|---|
| Characteristic | HR | 95% CI | P-value | HR | 95% CI | P-value |
| Age, ≥65 vs. <65 years | 1.01 | 0.71–1.43 | 0.98 | |||
| Sex, male vs. female | 1.27 | 0.77–2.10 | 0.35 | |||
| Virus infection, HCV vs. others | 1.28 | 0.88–1.85 | 0.19 | |||
| Albumin, <3.5 vs. ≥3.5 g/dl | 1.74 | 1.13–2.68 | 0.01 | 1.17 | 0.59–2.35 | 0.65 |
| PT, <70 vs. ≥70% | 1.12 | 0.67–1.84 | 0.67 | |||
| ICG-R15, ≥15 vs. <15% | 2.06 | 1.31–3.26 | 0.002 | 1.17 | 0.60–2.29 | 0.64 |
| Liver cirrhosis, (+) vs. (−) | 1.31 | 0.91–1.88 | 0.14 | |||
| Child-Pugh classification, B vs. A | 1.24 | 0.60–2.53 | 0.56 | |||
| Liver damage, B or C vs. A | 1.97 | 1.26–3.08 | 0.003 | 1.91 | 0.88–4.14 | 0.10 |
| Tumour number, multiple vs. solitary | 1.61 | 1.07–2.42 | 0.02 | 1.44 | 0.66–3.17 | 0.35 |
| Tumour size, ≥2 vs. <2 cm | 1.73 | 0.97–3.09 | 0.06 | |||
| AFP, ≥20 vs. <20 ng/ml | 1.46 | 1.02–2.08 | 0.04 | 1.43 | 0.88–2.34 | 0.15 |
| Differentiation, poor vs. good/moderate | 1.58 | 0.85–2.94 | 0.15 | |||
| Growth form, infiltrative vs. expansive | 1.49 | 0.92–2.42 | 0.10 | |||
| Formation of capsule, (−) vs. (+) | 1.27 | 0.84–1.91 | 0.25 | |||
| Infiltration to capsule, (+) vs. (−) | 1.06 | 0.74–1.51 | 0.77 | |||
| Septal formation, (−) vs. (+) | 1.00 | 0.68–1.47 | 0.99 | |||
| Serosal invasion, (+) vs. (−) | 2.00 | 1.33–3.02 | 0.0009 | 2.39 | 1.30–4.42 | 0.005 |
| Portal vein or hepatic vein invasion, (+) vs. (−) | 2.36 | 1.57–3.54 | <.0001 | 2.82 | 1.26–6.28 | 0.01 |
| Surgical margin, (+) vs. (−) | 1.32 | 0.81–2.13 | 0.26 | |||
| Stage, III/IV vs. I/II | 1.46 | 1.01–2.10 | 0.04 | 0.65 | 0.28–1.51 | 0.32 |
| YTHDF1 expression, high vs. low | 1.60 | 1.11–2.31 | 0.01 | 1.37 | 0.83–2.27 | 0.21 |
| YTHDF2 expression, high vs. low | 1.82 | 1.20–2.76 | 0.004 | 1.85 | 1.09–3.15 | 0.02 |
PT, prothrombin time; ICG-R15, indocyanine green 15-min clearance rate; AFP, α-fetoprotein; HCV, hepatitis C virus; YTHDF1/2, YT521-B homology domain family 1/2; HR, hazard ratio.
Table IV.
Univariate and multivariate cox proportional-hazard regression analysis of overall survival in patients with hepatocellular carcinoma.
| Univariate analysis | Multivariate analysis | |||||
|---|---|---|---|---|---|---|
| Characteristic | HR | 95% CI | P-value | HR | 95% CI | P-value |
| Age, ≥65 vs. <65 years | 1.34 | 0.87–2.06 | 0.18 | |||
| Sex, female vs. male | 1.01 | 0.57–1.80 | 0.96 | |||
| Virus infection, HCV vs. others | 1.32 | 0.84–2.07 | 0.23 | |||
| Albumin, <3.5 vs. ≥3.5 g/dl | 1.75 | 1.05–2.92 | 0.03 | 1.34 | 0.63–2.85 | 0.45 |
| PT, <70 vs. ≥70% | 1.51 | 0.87–2.61 | 0.14 | |||
| ICG-R15, ≥15 vs. <15% | 1.69 | 0.95–2.98 | 0.07 | |||
| Liver cirrhosis, (+) vs. (−) | 1.38 | 0.90–2.13 | 0.14 | |||
| Child-Pugh classification, B vs. A | 1.33 | 0.58–3.06 | 0.50 | |||
| Liver damage, B or C vs. A | 2.08 | 1.24–3.49 | 0.005 | 1.73 | 0.80–3.75 | 0.16 |
| Tumour number, multiple vs. solitary | 1.86 | 1.16–2.97 | 0.009 | 1.63 | 0.81–3.29 | 0.17 |
| Tumour size, ≥2 vs. <2 cm | 1.78 | 0.82–3.89 | 0.14 | |||
| AFP, ≥20 vs. <20 ng/ml | 2.30 | 1.48–3,58 | 0.0002 | 1.79 | 1.10–2.92 | 0.02 |
| Differentiation, poor vs. good/moderate | 2.02 | 1.04–3.93 | 0.04 | 1.18 | 0.50–2.76 | 0.71 |
| Growth form, infiltrative vs. expansive | 1.69 | 0.99–2.90 | 0.05 | |||
| Formation of capsule, (−) vs. (+) | 1.03 | 0.64–1.66 | 0.89 | |||
| Infiltration to capsule, (−) vs. (+) | 1.10 | 0.71–1.70 | 0.66 | |||
| Septal formation, (−) vs. (+) | 1.04 | 0.65–1.65 | 0.87 | |||
| Serosal invasion, (+) vs. (−) | 1.90 | 1.17–3.09 | 0.008 | 1.99 | 1.17–3.34 | 0.01 |
| Portal vein or hepatic vein invasion, (+) vs. (−) | 2.55 | 1.61–4.05 | <.0001 | 3.02 | 1.38–6.61 | 0.006 |
| Surgical margin, (+) vs. (−) | 1.77 | 1.04–3.02 | 0.04 | 1.68 | 0.90–3.13 | 0.10 |
| Stage, III/IV vs. I/II | 1.68 | 1.09–2.59 | 0.02 | 2.22 | 0.91–5.42 | 0.08 |
| YTHDF1 expression, high vs. low | 1.22 | 0.78–1.90 | 0.38 | |||
| YTHDF2 expression, high vs. low | 1.48 | 0.90–2.43 | 0.12 | |||
PT, prothrombin time; ICG-R15, indocyanine green 15-min clearance rate; AFP, α-fetoprotein; HCV, hepatitis C virus; YTHDF1/2, YT521-B homology domain family 1/2; HR, hazard ratio.
Discussion
In this study, we primarily evaluated the clinical effects of m6A readers in resected HCC patients. The expression of YTHDF1 and YTHDF2 in HCC tissues was correlated with tumour recurrence. Furthermore, YTHDF2 was an independent prognostic factor in resected HCC patients. Members of the YT521-B homology (YTH) domain family, including YTHDF1, YTHDF2, YTHDF3, YTHDC1 and YTHDC2, all have a conserved m6A-binding domain and preferentially bind to m6A-modified RNA at the RRm6ACH consensus sequence (15). YTHDF2, the first characterized m6A reader, accelerates the decay of m6A-modified transcripts by facilitating the recruitment of the CCR4-NOT complex directly (10). In contrast, YTHDF1 was initially demonstrated to bind to m6A sites around the stop codon and then cooperate with the translation initiation machinery to improve the translation efficacy of target RNAs in mammals (11).
We first compared the expression levels of YTHDF1 and YTHDF2 in HCC and noncancerous liver tissues from patients who underwent hepatectomy with curative intent at our institution. Neither YTHDF1 expression nor YTHDF2 expression was significantly different between HCC tissues and noncancerous tissues. In the paired analysis, patients with high expression in tumour tissues tended to have low expression in noncancerous tissues. In another cohort in the TCGA RNA-sequence dataset, however, the expression of YTHDF1 was found to be significantly higher in HCC tissues, and YTHDF2 was significantly lower in HCC tissues. Li et al reported that upregulation of YTHDF2 was observed in TCGA prostate cancer tissues compared with normal controls (16). Bai et al also reported that YTHDF1 is significantly upregulated in tumour compared with adjacent normal tissues in colorectal cancer (17). In the TCGA dataset, there were relatively small numbers of noncancerous tissues available, but we studied the expression of YTHDF1 and YTHDF2 in both tumour and noncancerous tissues from 177 HCC patients. This might have caused the discrepancy in results between the TCGA cases and our cases. In addition, our data showed that the reader expression in tumour tissues was different from that in noncancerous tissues. This change could be more important than the absolute value. In addition, the prognostic analysis stratified by YTHDF1 and YTHDF2 expression in our study revealed that the expression of these two m6A readers in HCC tissues is not associated with OS. On the other hand, high expression of both YTHDF1 and YTHDF2 in HCC tissues was associated with significantly worse RFS. In the public dataset, high expression of YTHDF1 or YTHDF2 was associated with worse prognosis than low expression. In particular, our Cox regression analysis showed that YTHDF2 was an independent risk factor for recurrence in resected HCC. Thus, YTHDF1 and YTHDF2 might have inherent effects in HCC carcinogenesis and influence the long-term outcome after HCC resection, for example, by causing sporadic recurrence.
Evidence of RNA modifications in cancer development and progression has been increasing. The RNA methyltransferase METTL3 is the first characterized component of the m6A methyltransferase complex. METTL3 promotes tumour proliferation and invasion in several cancers (18–22). The m6A demethylases FTO and ALKBH5 were identified in the 2010s. FTO and ALKBH5 also play an important role in human cancer (23–25). However, the functions of YTHDF1 and YTHDF2 in HCC have not been uncovered. Zhao et al reported that YTHDF1 played a vital role in the regulation of HCC metabolism (26). Qu et al reported that m6A RNA methylation modulators, including YTHDF1, affected OS in HCC patients (27). YTHDF2 was able to degrade both tumour promoter and suppressor gene mRNAs. Zhang et al reported that YTHDF2 promotes the cancer stem cell liver phenotype and cancer metastasis by modulating the m6A methylation of OCT4 mRNA (28). In contrast, YTHDF2 may act as a tumour suppressor to repress cell proliferation and growth by destabilizing EGFR mRNA in HCC (29). Further investigations are required to reveal the role of YTHDF1 and YTHDF2 in HCC.
Although we showed important aspects of YTHDF1 and YTHDF2, there are some inherent limitations to the present study. First, more data are necessary because we used specimens from a single institute in this study. Second, more detailed molecular mechanisms through which specific m6A methylation enhances HCC development need to be discovered. Further investigation is necessary before the clinical utility of our findings can be determined.
In conclusion, our study revealed that high YTHDF2 expression in HCC tissues is related to cancer recurrence. Our results may pave the way for discovering the clinical utility of m6A methylation and associated genes in HCC therapy in the future.
Acknowledgements
The authors would like to thank Dr Raju Kandimalla (Bristol Myers Squibb, San Diego, CA, USA) for useful discussions and Ms. Yoko Nishikawa (Nagoya University Graduate School of Medicine, Nagoya, Japan) for technical assistance with the experiments.
Funding Statement
The present study was partly supported by the Japan Society for the Promotion of Science KAKENHI Grant-in-Aid for Scientific Research (grant nos. 18H06176 and 19K18110).
Funding
The present study was partly supported by the Japan Society for the Promotion of Science KAKENHI Grant-in-Aid for Scientific Research (grant nos. 18H06176 and 19K18110).
Availability of data and materials
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
NN, FS and YK conceived and designed the study. FS and YK provided financial support. SY and YK provided administrative support. SY, FS, YS, YI, HT, MH, MKa, CT, GN and MKo provided study materials and patients. SY, FS, YS, YI, HT, MH, MKa, CT, GN and MKo assisted with analyses and manuscript preparation. NN, FS, KT and YS collected and assembled the data. NN and KT performed quantitative PCR data analysis and interpretation. NN, FS and YK wrote the manuscript and confirmed the authenticity of all the raw data. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The study and all procedures were approved by the Institutional Review Board at Nagoya University (approval no. 2013-0295; Nagoya, Japan), and all patients provided written informed consent. All clinical investigations were conducted in accordance with the principles of the Declaration of Helsinki.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70:7–30. doi: 10.3322/caac.21601. [DOI] [PubMed] [Google Scholar]
- 2.Tabrizian P, Jibara G, Shrager B, Schwartz M, Roayaie S. Recurrence of hepatocellular cancer after resection: Patterns, treatments, and prognosis. Ann Surg. 2015;261:947–955. doi: 10.1097/SLA.0000000000000710. [DOI] [PubMed] [Google Scholar]
- 3.Roundtree IA, Evans ME, Pan T, He C. Dynamic RNA modifications in gene expression regulation. Cell. 2017;169:1187–1200. doi: 10.1016/j.cell.2017.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Adams JM, Cory S. Modified nucleosides and bizarre 5′-termini in mouse myeloma mRNA. Nature. 1975;255:28–33. doi: 10.1038/255028a0. [DOI] [PubMed] [Google Scholar]
- 5.Camper SA, Albers RJ, Coward JK, Rottman FM. Effect of undermethylation on mRNA cytoplasmic appearance and half-life. Mol Cell Biol. 1984;4:538–543. doi: 10.1128/MCB.4.3.538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Finkel D, Groner Y. Methylations of adenosine residues (m6A) in pre-mRNA are important for formation of late simian virus 40 mRNAs. Virology. 1983;131:409–425. doi: 10.1016/0042-6822(83)90508-1. [DOI] [PubMed] [Google Scholar]
- 7.Gilbert WV, Bell TA, Schaening C. Messenger RNA modifications: Form, distribution, and function. Science. 2016;352:1408–1412. doi: 10.1126/science.aad8711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ma S, Chen C, Ji X, Liu J, Zhou Q, Wang G, Yuan W, Kan Q, Sun Z. The interplay between m6A RNA methylation and noncoding RNA in cancer. J Hematol Oncol. 2019;12:121. doi: 10.1186/s13045-019-0805-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Allis CD, Jenuwein T. The molecular hallmarks of epigenetic control. Nat Rev Genet. 2016;17:487–500. doi: 10.1038/nrg.2016.59. [DOI] [PubMed] [Google Scholar]
- 10.Du H, Zhao Y, He J, Zhang Y, Xi H, Liu M, Ma J, Wu L. YTHDF2 destabilizes m(6)A-containing RNA through direct recruitment of the CCR4-NOT deadenylase complex. Nat Commun. 2016;7:12626. doi: 10.1038/ncomms12626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang X, Zhao BS, Roundtree IA, Lu Z, Han D, Ma H, Weng X, Chen K, Shi H, He C. N(6)-methyladenosine modulates messenger RNA translation efficiency. Cell. 2015;161:1388–1399. doi: 10.1016/j.cell.2015.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen J, Sun Y, Xu X, Wang D, He J, Zhou H, Lu Y, Zeng J, Du F, Gong A, Xu M. YTH domain family 2 orchestrates epithelial-mesenchymal transition/proliferation dichotomy in pancreatic cancer cells. Cell Cycle. 2017;16:2259–2271. doi: 10.1080/15384101.2017.1380125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nishizawa Y, Konno M, Asai A, Koseki J, Kawamoto K, Miyoshi N, Takahashi H, Nishida N, Haraguchi N, Sakai D, et al. Oncogene c-Myc promotes epitranscriptome m6A reader YTHDF1 expression in colorectal cancer. Oncotarget. 2018;9:7476–7486. doi: 10.18632/oncotarget.23554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shi Y, Fan S, Wu M, Zuo Z, Li X, Jiang L, Shen Q, Xu P, Zeng L, Zhou Y, et al. YTHDF1 links hypoxia adaptation and non-small cell lung cancer progression. Nat Commun. 2019;10:4892. doi: 10.1038/s41467-019-12801-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang X, Lu Z, Gomez A, Hon GC, Yue Y, Han D, Fu Y, Parisien M, Dai Q, Jia G, et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature. 2014;505:117–120. doi: 10.1038/nature12730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li J, Xie H, Ying Y, Chen H, Yan H, He L, Xu M, Xu X, Liang Z, Liu B, et al. YTHDF2 mediates the mRNA degradation of the tumor suppressors to induce AKT phosphorylation in N6-methyladenosine-dependent way in prostate cancer. Mol Cancer. 2020;19:152. doi: 10.1186/s12943-020-01267-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bai Y, Yang C, Wu R, Huang L, Song S, Li W, Yan P, Lin C, Li D, Zhang Y. YTHDF1 regulates tumorigenicity and cancer stem cell-like activity in human colorectal carcinoma. Front Oncol. 2019;9:332. doi: 10.3389/fonc.2019.00332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Han J, Wang JZ, Yang X, Yu H, Zhou R, Lu HC, Yuan WB, Lu JC, Zhou ZJ, Lu Q, et al. METTL3 promote tumor proliferation of bladder cancer by accelerating pri-miR221/222 maturation in m6A-dependent manner. Mol Cancer. 2019;18:110. doi: 10.1186/s12943-019-1036-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lin S, Choe J, Du P, Triboulet R, Gregory RI. The m(6)A Methyltransferase METTL3 promotes translation in human cancer cells. Mol Cell. 2016;62:335–345. doi: 10.1016/j.molcel.2016.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miao W, Chen J, Jia L, Ma J, Song D. The m6A methyltransferase METTL3 promotes osteosarcoma progression by regulating the m6A level of LEF1. Biochem Biophys Res Commun. 2019;516:719–725. doi: 10.1016/j.bbrc.2019.06.128. [DOI] [PubMed] [Google Scholar]
- 21.Wang Q, Chen C, Ding Q, Zhao Y, Wang Z, Chen J, Jiang Z, Zhang Y, Xu G, Zhang J, et al. METTL3-mediated m6A modification of HDGF mRNA promotes gastric cancer progression and has prognostic significance. Gut. 2020;69:1193–1205. doi: 10.1136/gutjnl-2019-319639. [DOI] [PubMed] [Google Scholar]
- 22.Xia T, Wu X, Cao M, Zhang P, Shi G, Zhang J, Lu Z, Wu P, Cai B, Miao Y, Jiang K. The RNA m6A methyltransferase METTL3 promotes pancreatic cancer cell proliferation and invasion. Pathol Res Pract. 2019;215:152666. doi: 10.1016/j.prp.2019.152666. [DOI] [PubMed] [Google Scholar]
- 23.Li J, Han Y, Zhang H, Qian Z, Jia W, Gao Y, Zheng H, Li B. The m6A demethylase FTO promotes the growth of lung cancer cells by regulating the m6A level of USP7 mRNA. Biochem Biophys Res Commun. 2019;512:479–485. doi: 10.1016/j.bbrc.2019.03.093. [DOI] [PubMed] [Google Scholar]
- 24.Niu Y, Lin Z, Wan A, Chen H, Liang H, Sun L, Wang Y, Li X, Xiong XF, Wei B, et al. RNA N6-methyladenosine demethylase FTO promotes breast tumor progression through inhibiting BNIP3. Mol Cancer. 2019;18:46. doi: 10.1186/s12943-019-1004-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang S, Zhao BS, Zhou A, Lin K, Zheng S, Lu Z, Chen Y, Sulman EP, Xie K, Bögler O, et al. m6A Demethylase ALKBH5 maintains tumorigenicity of glioblastoma stem-like cells by sustaining FOXM1 expression and cell proliferation program. Cancer Cell. 2017;31:591–606.e6. doi: 10.1016/j.ccell.2017.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhao X, Chen Y, Mao Q, Jiang X, Jiang W, Chen J, Xu W, Zhong L, Sun X. Overexpression of YTHDF1 is associated with poor prognosis in patients with hepatocellular carcinoma. Cancer Biomark. 2018;21:859–868. doi: 10.3233/CBM-170791. [DOI] [PubMed] [Google Scholar]
- 27.Qu N, Qin S, Zhang X, Bo X, Liu Z, Tan C, Wen G, Jiang H. Multiple m6A RNA methylation modulators promote the malignant progression of hepatocellular carcinoma and affect its clinical prognosis. BMC Cancer. 2020;20:165. doi: 10.1186/s12885-020-6638-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang C, Huang S, Zhuang H, Ruan S, Zhou Z, Huang K, Ji F, Ma Z, Hou B, He X. YTHDF2 promotes the liver cancer stem cell phenotype and cancer metastasis by regulating OCT4 expression via m6A RNA methylation. Oncogene. 2020;39:4507–4518. doi: 10.1038/s41388-020-1303-7. [DOI] [PubMed] [Google Scholar]
- 29.Zhong L, Liao D, Zhang M, Zeng C, Li X, Zhang R, Ma H, Kang T. YTHDF2 suppresses cell proliferation and growth via destabilizing the EGFR mRNA in hepatocellular carcinoma. Cancer Lett. 2019;442:252–261. doi: 10.1016/j.canlet.2018.11.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.