Abstract
Tuberculosis (TB) is a top-ten cause of death worldwide. Successful treatment is often limited by insufficient diagnostic capabilities, especially at the point of care in low-resource settings. The ideal diagnostic must be fast, be cheap, and require minimal clinical resources while providing high sensitivity, selectivity, and the ability to differentiate live from dead bacteria. We describe here the development of a fast, luminescent, and affordable sensor of Hip1 (FLASH) for detecting and monitoring drug susceptibility of Mycobacterium tuberculosis (Mtb). FLASH is a selective chemiluminescent substrate for the Mtb protease Hip1 that, when processed, produces visible light that can be measured with a high signal-to-noise ratio using inexpensive sensors. FLASH is sensitive to fmol of recombinant Hip1 enzyme in vitro and can detect as few as thousands of Mtb cells in culture or in human sputum samples within minutes. The probe is highly selective for Mtb compared to other nontuberculous mycobacteria and can distinguish live from dead cells. Importantly, FLASH can be used to measure antibiotic killing of Mtb in culture with greatly accelerated timelines compared to traditional protocols. Overall, FLASH has the potential to enhance both TB diagnostics and drug resistance monitoring in resource-limited settings.
Short abstract
FLASH is a sensitive and selective chemiluminescent protease probe for quantification and drug susceptibility testing of Mycobacterium tuberculosis, the causative agent of tuberculosis.
Introduction
Tuberculosis (TB) is a top-ten cause of death worldwide, with an estimated 10 million new cases leading to nearly 1.5 million deaths yearly. Many of these deaths could be prevented by improving access to diagnostics and therapeutics, especially in low-resource countries that are disproportionally affected. This fact is exemplified by the disparity in disease burdens which range from five or fewer cases per 100 000 in the United States and Europe to more than 500 cases per 100 000 in some countries in Southern Africa and Southeastern Asia. Exacerbating this issue is the extraordinary number of cases that go undiagnosed. The World Health Organization estimates that 2.9 million cases (approximately 30% of all new cases) went unreported in 2019.1 Undiagnosed cases cannot be treated, nor can their spread be mitigated, leading to poor patient outcomes and increased infections. Additionally, diagnoses that take longer than a single clinical visit require the patient to return to receive the test result and begin antibiotic therapy. Diagnostic delay leads to further delays in treatment.2 Thus, one of the key pillars of the strategy for eradicating TB is increasing diagnostic capabilities. Rapid, simple, and accurate diagnosis of TB remains a challenge in resource-poor settings.
Current diagnostics are limited by their speed, sensitivity, cost, and ability to differentiate live from dead bacteria. Culture of Mycobacterium tuberculosis (Mtb), the causative agent of TB, in liquid or solid media is the gold standard for TB diagnosis. Culture methods offer the highest sensitivity and specificity but are expensive and slow, with conventional culture methods taking up to 8 weeks. In decentralized, resource-poor settings, the recommended diagnostic methods are sputum smear microscopy or the GeneXpert MTB/RIF assay.3 Sputum smear microscopy is relatively simple and inexpensive. However, the sensitivity of sputum smear microscopy is dependent on the sputum processing method and the experience of the user and thus varies widely between 0.32 and 0.97,4 leading to false negatives. The Xpert MTB/RIF assay, developed for the GeneXpert platform, is a nucleic acid amplification test (NAAT) that uses PCR to detect Mtb and mutations that confer resistance to rifampicin in under 2 h and has sensitivity greater than 0.86.5−7 However, infrastructure requirements, such as continuous electrical supply and trained personnel, prohibit the implementation of GeneXpert in peripheral health clinics, and the cost of each Xpert MTB/RIF test in addition to the capital cost for the GeneXpert System is too high for widespread use of this diagnostic method. Furthermore, NAATs like GeneXpert are susceptible to false positives when evaluating disease progression or treatment outcomes because they can amplify bacterial DNA from dead bacteria following antibiotic treatment. Therefore, there is a need for rapid, affordable, point-of-care diagnostics for TB that offer high sensitivity and selectivity. Ideally, new diagnostic methods should be able to detect small numbers of bacilli and be specific for Mtb as opposed to other bacterial species present in the sputum, airway, and oral sites.
The increasing prevalence of multi-drug-resistant TB also leads to poor health outcomes. In 2018, there were half a million new cases of rifampicin (RIF)-resistant TB.1 Early identification of resistance is important for developing proper antibiotic regimens for patients, yet drug susceptibility testing (DST) to determine resistance in clinical isolates can take from 10 days in liquid medium to 28–42 days on solid medium. Molecular tests such as Xpert MTB/RIF provide a rapid method of identifying the presence of mutations that confer resistance, but the design of these tests requires genetic information for each mutation. The development of new tests to detect new resistance mutations or resistance to novel drugs thus requires substantial investment in time and resources. Therefore, new rapid, inexpensive, and comprehensive methods for phenotypic DST have the potential to be transformative to clinical testing of Mtb infections.
The development of new diagnostics and methods for DST has attracted substantial attention.8,9 Promising diagnostics under investigation include those that use patient samples other than sputum such as RNA measurements from blood samples,10 detection of the Mtb cell wall component lipoarabinomannan in patient urine,11 and identification of volatile compounds from patient breath.12,13 New fluorescent probes have been developed to report on the activity of mycobacterial enzymes. For example, probes have been developed that target mycobacterial esterases,14 sulfatases,15 and trehalose mycolyltransesterases,16,17 but these probes are nonspecific since they also label nontuberculous mycobacteria (NTMs) that may also be present in sputum. Probes activated by the Mtb β-lactamase BlaC have been shown to specifically label Mtb in patient sputum samples.18−20 These enzyme-based tests offer an improvement in specificity compared to sputum smear microscopy; however, their reliance on fluorescence measurements necessitates imaging instruments that may not be practical for point-of-care use.
Luminescent probes serve as a promising alternative to fluorescence counterparts. Unlike fluorescence, detection of the luminescent signal does not require an excitation source nor advanced optics and can be achieved by a simple, inexpensive luminometer. The background signal is substantially lower because the components of biological samples that often produce undesired fluorescent signals do not spontaneously generate light. Recent advancements in the development of chemiluminescent reporters have made it possible to design aqueous soluble and stable probes that generate light only upon enzymatic cleavage.21−23 This approach has been used to create chemiluminescent sensors for β-galactosidase,24 cathepsin B,25Salmonella sp. esterases and Listeria monocytogenes phosphatidylinositol-specific phospholipase C,26 and carbapenemase activity in bacteria.27 The latter three probes exhibited rapid and sensitive detection of bacteria in culture.
To address diagnostic needs for TB, we have developed a luminescence-based probe that overcomes many of the limitations faced by existing diagnostics and clinical methods for DST. We sought to combine the sensitivity and selectivity of an enzyme-based probe with the ease of detection and low background of a chemiluminescent output. As an enzymatic target, we chose the Mtb hydrolase important for pathogenesis 1 (Hip1 or carboxylesterase A CaeA, Rv2224c). Hip1 is a cell-envelope-associated serine protease that is essential for Mtb virulence28 and its survival in macrophages.29 Hip1 cleaves the Mtb protein GroEL2, contributing to the suppression of early macrophage proinflammatory responses.30−32 Together, the following characteristics make Hip1 an attractive target: (i) Its presence on the cell surface makes it highly accessible to small-molecule probes. (ii) Its importance for pathogenesis suggests that it will be expressed during infection. (iii) The human genome does not encode a homologue of Hip1. (iv) Prior work from our group has identified an amino acid recognition sequence that is specifically cleaved by Hip1.33 Here, we describe a new diagnostic probe for detecting Mtb: fast, luminescent, and affordable sensor of Hip1 (FLASH). FLASH quantitatively reports on the presence of active Hip1 and can quantify and detect as few as 4000 Mtb cells in a 1 h measurement. In human sputum samples, FLASH can detect Mtb spiked in to final concentrations typically found in clinical specimens. Importantly, FLASH also differentiates live from dead bacteria and thus can be used to determine drug susceptibility of clinical isolates using a greatly accelerated and simplified workflow compared to current culture-based methods. Together, these data show that FLASH is a promising candidate for rapid TB diagnostics in point-of-care clinics and clinical DST.
Results
Design of a Chemiluminescent Substrate Probe for Mtb Hip1
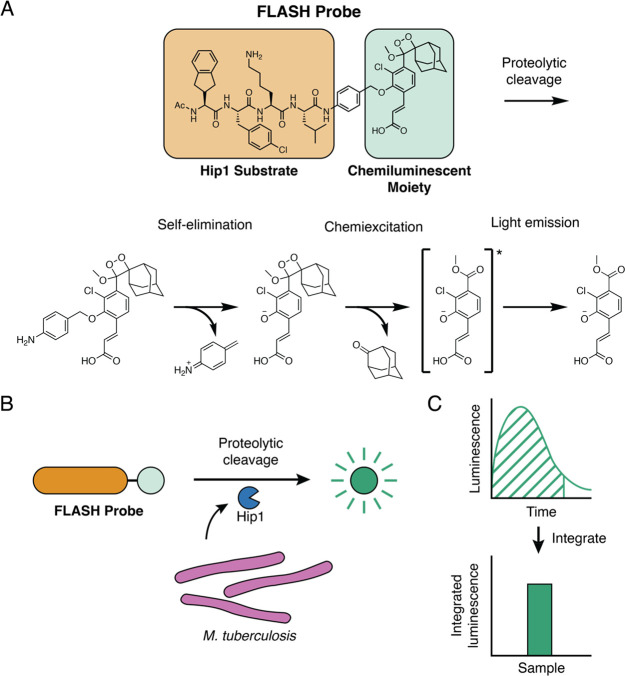
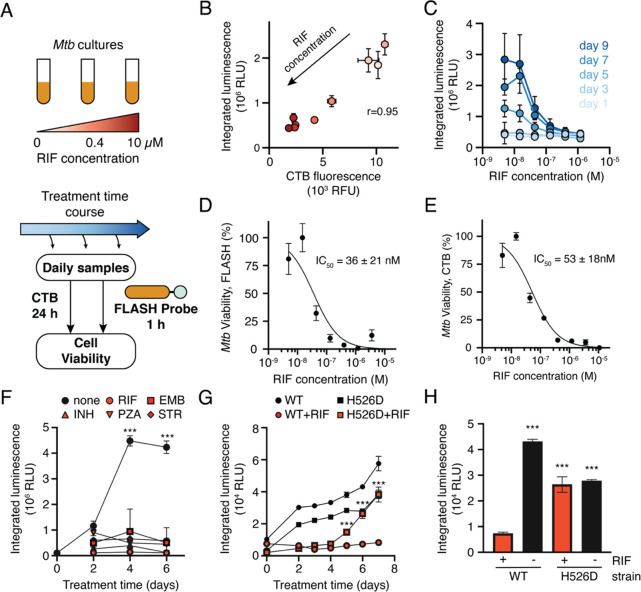
To generate a probe that produces light upon cleavage by Hip1, we combined a selective tetrapeptide Hip1 substrate with a p-amino-benzyl-alcohol self-eliminating linker and a phenoxy-dioxetane luminophore (Figure 1A). The tetrapeptide sequence was previously optimized for Hip1 cleavage and offers high selectivity for Hip1 compared to other enzymes from Mtb or humans.33 Upon enzymatic cleavage, the aniline linker undergoes spontaneous elimination, releasing the activated phenoxy-dioxetane luminophore. Subsequent chemiexcitation and decay processes result in the spontaneous generation of light. Live Mtb express active Hip1 which processes the probe to produce light (Figure 1B). Emitted light is measured with a sensitive luminometer and integrated over time to yield a total luminescent signal (Figure 1C).
Figure 1.
Fast luminescent affordable sensor of Hip1 (FLASH). (A) Following proteolytic cleavage of the FLASH probe by Mtb Hip1, self-elimination and chemiexcitation steps ultimately lead to light emission. (B) Mtb produces Hip1 protease which cleaves the FLASH probe, producing light. (C) Light produced by probe cleavage is measured over time. Total light output in a given time period (dashed area) is summed to yield integrated luminescence.
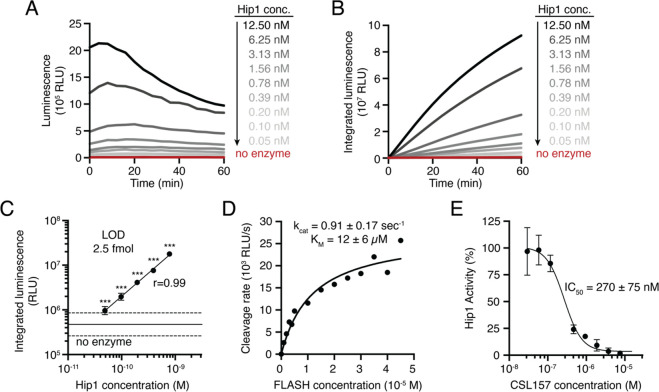
We first sought to test that our previously reported fluorogenic substrate probe for Hip1 could be converted to a luminescent reporter. To evaluate Hip1 activity toward the FLASH probe, we titrated recombinantly expressed enzyme and measured luminescent signal with a microplate reader. Light was detected immediately upon enzyme addition, and the magnitude of the signal depended on the amount of enzyme added (Figure 2A). Integration of luminescence yielded typical substrate processing curves (Figure 2B). Incubation of Hip1 with a negative control probe containing d-amino acids (D-FLASH, Figure S1) yielded no luminescent signal and provided a measure of background signal resulting from spontaneous release of the reporter due to probe instability (Figure S2). To evaluate the sensitivity of the probe, we compared the integrated FLASH luminescence after 60 min of measurement for each concentration of enzyme to the control samples that lacked enzyme. Samples containing as little as 2.5 fmol of enzyme (50 pM) produced a signal that was significantly above the background signal (Figure 2C). Integrated luminescence correlated linearly with total enzyme concentration (r = 0.99), indicating that FLASH is both highly sensitive and quantitative for detection of Hip1. Kinetic analysis of the titrated probe yielded a kcat/KM of 7.5 × 104 M–1 s–1 (Figure 2D). To verify that the FLASH probe responds only to active enzyme, we incubated Hip1 with the Hip1 inhibitor CSL157 (Figure S1)33 prior to the addition of the FLASH probe. We observed a dose-dependent reduction in luminescence with a calculated IC50 of 270 nM (Figure 2E), within error of the value measured using a fluorogenic Hip1 substrate (also 270 nM).33 Results obtained from the recombinant enzyme show that the FLASH probe is a sensitive, quantitative measure of active enzyme. For all FLASH experiments, raw luminescence data (as shown in Figure 2A) were integrated to yield total luminescence (as shown in Figure 2C).
Figure 2.
FLASH is a sensitive probe for Mtb Hip1 activity. (A) Light emitted by the FLASH probe upon incubation with various concentrations of Mtb Hip1. (B) Time course of integrated luminescence from part A. (C) Total integrated luminescence after 1 h of incubation with Mtb Hip1 (mean ± SD, n = 3). Horizontal lines show the mean (solid) ± 3 SD (dashed) of control samples lacking enzyme. Each enzyme concentration was compared to the control samples via one-way ANOVA with Dunnett’s test (***, p < 0.001). The best fit line shows linear regression to the log-transformed data. (D) Kinetic analysis of the FLASH probe. Data were fitted to the Michaelis–Menten equation to yield kinetic parameters. (E) Inhibition of Mtb Hip1 by CSL157 (mean ± SD, n = 3). Mtb Hip1 was preincubated with inhibitor for 30 min at 37 °C before the addition of the FLASH probe. Data were fitted to a four-parameter logistic equation to yield IC50. All parameters are reported as the 95% confidence interval.
FLASH is a Quantitative Probe for Mtb
To determine whether the FLASH probe can detect live Mtb, we added probe to bacterial cultures and measured luminescence. To optimize assay conditions for bacterial detection, the signal-to-noise ratio (SNR) was calculated for different integration times by dividing the integrated signal for cultures with and without Mtb cells (Figure S3A). 60 min of integration time yielded an SNR of greater than 15, and we used this measurement time for all subsequent experiments. The luminescent signal was reduced when cultures were preincubated with Hip1 inhibitor (Figure S3B), indicating that, as observed for experiments with the recombinant enzyme, probe cleavage in the presence of cells is dependent on active Hip1.
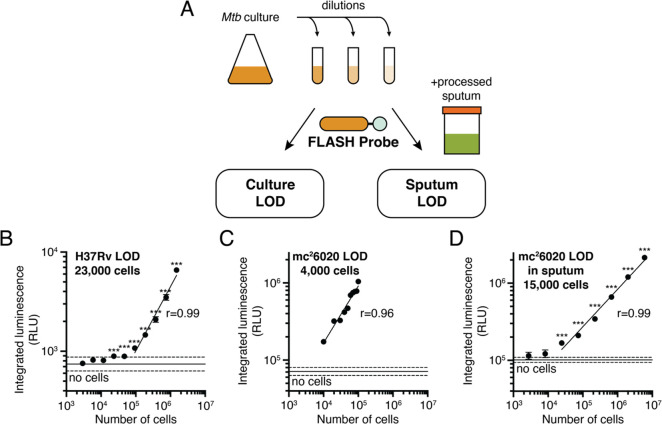
A key metric for evaluating FLASH as a potential diagnostic is the ability for the probe to detect the small numbers of bacteria usually found in sputum samples. To quantify the limit of detection of FLASH for Mtb, bacterial cultures were serially diluted into either culture medium or processed human sputum prior to adding the probe (Figure 3A). In all cases, absorbance (OD600) was used as a proxy for cell number. The conversion factor between absorbance and cell count was obtained by colony forming unit (CFU) plating (OD600 1 = 3 × 108 CFU/mL). The experiment was performed with two strains of Mtb: the laboratory strain, H37Rv, measured in a biosafety-level-3 (BSL3) facility; and the attenuated strain, mc26020,34 measured in a BSL2 facility. For both strains, the luminescent signal was linearly correlated with cell number (Figure 3B,C). Limits of detection (LODs) were calculated by extrapolating best fit lines to estimate the cell number for which the luminescent signal would equal 3 times the standard deviation of the negative controls lacking cells. LODs for H37Rv and for mc26020 were 23 000 and 4000 cells, respectively. We attribute the higher LOD in H37Rv to the less sensitive microplate reader available for use in the BSL3. SNRs were higher at all integration times for the more sensitive BSL2 microplate reader (Figure S3A), and there was not a significant difference between integrated luminescence for H37Rv and for mc26020 when both were measured on the BSL3 microplate reader (Figure S3C).
Figure 3.
FLASH is a quantitative probe for Mtb cells. (A) To determine the limit of detection, cultures of Mtb were serially diluted into medium or into processed human sputum and then incubated with the FLASH probe. Total integrated luminescence after 1 h of incubation of the FLASH probe with (B) Mtb H37Rv in 7H9 medium, (C) mc26020 in 7H9 medium, and (D) mc26020 in processed human sputum (mean ± SD, n = 3). Horizontal lines show the mean (solid) ± 3 SD (dashed) of control samples lacking cells. For all experiments, each sample was compared to the no-bacteria control via one-way ANOVA with Dunnett’s test (***, p < 0.001; for part C, all comparisons yielded p < 0.001). Best fit lines show linear regressions to the log-transformed data (excluding cell concentrations that are not significantly different from the control). Limits of detection were calculated by determining the cell number for which the best fit line intercepts the mean + 3 SD of the control samples.
To evaluate whether the high sensitivity of FLASH observed for bacteria in culture is likely to translate to diagnostic detection in clinical samples, we spiked mc26020 grown in culture into pooled human sputum collected from patients who had tested negative for TB. The sputum was processed using the standard NALC/NaOH protocol35 at the time of collection. Sputum processing was optimized by centrifuging sputum samples and resuspending the pellet in PBS to achieve neutral pH. This step resulted in an 8-fold increase in signal (Figure S3D). Using the centrifugation and resuspension protocol, the LOD for mc26020 in processed sputum was 15 000 cells, again with a linear correlation between cell number and luminescent signal. The higher LOD measured in sputum compared to culture is consistent with a slight loss of signal observed after the centrifugation step. Signal generated from bacteria in sputum was lower than the same amount of bacteria added directly to buffer, but this loss of signal was recapitulated by centrifugation of bacteria in culture (Figure S3D), suggesting that it was the centrifugation step and not the sputum itself that reduced the signal. Although Hip1 is reported to be surface-associated, one explanation for the decreased signal could be that some enzyme is released into the culture medium. To test this, we centrifuged bacterial cells and compared the luminescent signal among the original culture, the supernatant, and the resuspended pellet. Indeed, a substantial fraction of the culture signal was present in the supernatant (40%), with the remainder found in the pellet (55%; Figure S3E), suggesting that either free Hip1 enzyme or some fraction of cells with Hip1 activity are found in the supernatant following the centrifugation step.
The limit of detection for cells in processed sputum is encouraging for the ability of FLASH to diagnose TB. The concentration of Mtb bacilli in sputum ranges broadly across patients, according to disease severity, and throughout the course of antibiotic treatment. Measurements based on colony forming units (CFU) on a solid culture range from 101 to 107 CFU/mL sputum, but data from a variety of studies agree on average concentrations between 105 and 106 CFU/mL.36−38 With the concentration step routinely used in sputum processing, a typical 1 mL sample of sputum is expected to have at least an order of magnitude more cells than our limit of detection.
FLASH is a Selective Probe for Mtb
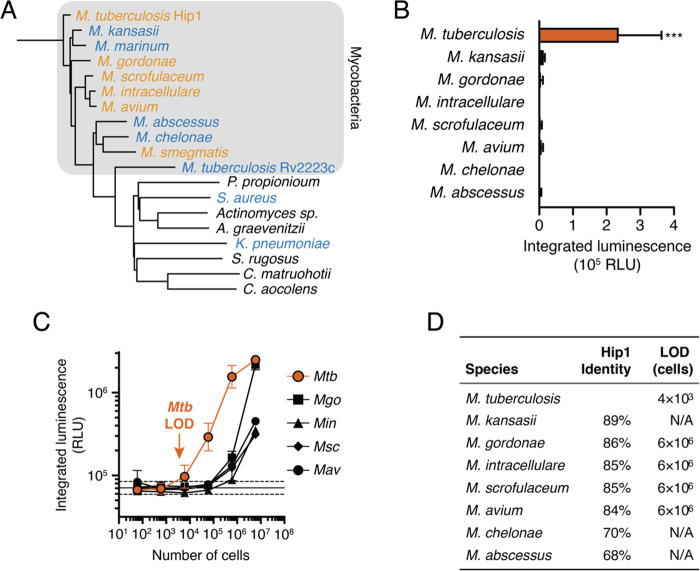
The ideal diagnostic for Mtb must not provide false negatives in the presence of Mtb nor false positives due to other bacteria or enzymes that may be present in clinical sputum samples. False negatives might occur if Mtb present in the lung does not express active Hip1. To account for this possibility, we searched the NCBI database of Mtb genome sequences for the presence of the hip1 gene. Of the 6514 genomes obtained from clinical isolates around the world, 98.6% (6420) encode for Hip1 and conserve the three key catalytic residues required for proteolytic activity. Although the presence of the gene does not guarantee expression of active Hip1 in these isolates, this result along with the fact that Hip1 activity is important for infection suggests that false negatives for this reason will be exceedingly rare.
Potential sources of false positives include host enzymes present in the saliva or sputum, constituent organisms of the healthy microbiota, or NTMs. The human genome does not encode a Hip1 homologue, and the low background signal observed in processed human sputum (Figure 3D) suggests that enzymes from the host or the microbiome do not activate the probe or do not survive the processing protocol. We used phylogenetic analysis of proteins with sequence similarity to Mtb Hip1 to identify organisms that may lead to false positives. We searched for proteins with sequence similarity to Hip1 in the Human Microbiome Project databases of oral and airway microbiota,39,40 a set of NTMs, and other pathogens known to infect the human lung (Staphylococcus aureus and Klebsiella pneumoniae). Putative Hip1 homologues from these organisms clustered into two groups: a set of closely related enzymes with at least 65% similarity found in NTMs and a more divergent set found in other bacteria (Figure 4A, Figure S4A). The latter group of enzymes shared higher sequence similarity with the Mtb α/β hydrolase Rv2223c than with Hip1. This is notable because our prior work showed that the fluorogenic precursor to the FLASH probe is not processed by a hip1 knockout strain of Mtb,29,33 suggesting that the peptide sequence is not a substrate for Rv2223c.
Figure 4.
FLASH is selective for Mtb. (A) Phylogenetic tree of potential Mtb Hip1 homologues found in NTMs, other lung pathogens, and commensal members of the human airway and oral microbiomes. Also included is Rv2223c, an uncharacterized peptidase encoded by Mtb with sequence similarity to Hip1. Bacteria are colored by their ability to process FLASH at high cell densities (orange, active; blue, inactive; black, not tested). (B) FLASH signal for 6 × 104 cells of each NTM in 7H9 medium (mean ± SD, n = 3). Each sample was compared to the no-bacteria control via one-way ANOVA with Dunnett’s test (***, p < 0.001). (C) FLASH signal for M. tuberculosis (Mtb), M. gordonae (Mgo), M. intracellulare (Min), M. scrofulaceum (Msc), and M. avium (Mav) at the indicated cell number (mean ± SD, n = 3). (D) Sequence similarity of potential homologues to Mtb Hip1 and limits of detection calculated from part C for each NTM.
To evaluate probe specificity, we measured the FLASH signal in bacterial cultures of NTMs and other common pathogens. Three commonly used laboratory strains of Mtb (H37Rv, CDC1551, and Erdman) as well as the disease-causing Mycobacterium bovis, which is a member of the Mtb complex, all yielded a signal substantially above background (Figure S4B). At high cell densities (3 × 108 CFU/mL; 1.2 × 107 cells), clinical isolates of Mycobacterium gordonae, Mycobacterium intracellulare (Min), Mycobacterium scrofulaceum (Msc), Mycobacterium avium (Mav), and the laboratory strain of Mycobacterium smegmatis (Msm) yielded a luminescent signal above background but substantially lower than for Mtb (Figure S4C,D). Furthermore, at a 200-fold lower cell density that is more representative of bacterial burdens in TB sputum (1.5 × 106 CFU/mL; 6 × 104 cells), only cultures of Mtb yielded a luminescent signal significantly greater than background (Figure 4B). To compare activation of the FLASH probe among NTMs, we calculated LODs for each species that showed some activation (Figure 4C). All LODs were 1000-fold higher than for Mtb (Figure 4D), indicating that FLASH is highly selective and is unlikely to give false positive signals in response to NTMs.
FLASH Enables Rapid Drug Susceptibility Testing
The ability to distinguish live from dead M. tuberculosis is another key feature of a TB diagnostic. Similarly, the ability to detect antibiotic killing of isolates is critical in clinical microbiology, especially for TB for which drug resistance is a growing global threat. We hypothesized that FLASH could be used to distinguish live from dead bacteria. Initial experiments showed that the luminescent signal was greatly reduced in heat-killed cultures or following treatment of cultures with the antibiotic rifampicin (RIF, Figure S5A). This observation suggests that cell death results in decreased levels of active Hip1, presumably due to protein degradation or instability combined with the cessation of new protein synthesis. To further evaluate the potential for FLASH to report on cell viability, we treated Mtb cultures with RIF over the course of 9 days. Cultures were sampled throughout the treatment course, and cell viability was measured using a 24 h treatment with resazurin (CellTiter-Blue, CTB). The same cultures were incubated with the FLASH probe for 1 h (Figure 5A). RIF itself absorbs light in the visible range (absorption maximum at 475 nm41), but at the concentrations tested, it had no effect on the luminescent signal from the dioxetane luminophore (Figure S5B). We observed a high correlation (r = 0.95) between FLASH luminescence and CTB fluorescence across the range of RIF concentrations (Figure 5B) indicating that FLASH is a quantitative indicator of cell viability. We generated dose–response curves by comparing FLASH signal to RIF concentration (Figure 5C). At early time points (days one and three), there was no difference between high and low concentrations of RIF, but at later time points we observed the expected dose response curves, with a low FLASH signal in the cultures treated with RIF concentrations that prevent growth. At day seven, the dose response curve generated using FLASH yielded an IC50 of 36 ± 21 nM (Figure 5D), matching the EC50 value calculated from CTB measurements of the same cultures (Figure 5E).
Figure 5.
FLASH provides a quantitative measure of Mtb viability. (A) Mtb cultures were treated with RIF for up to 9 days. Samples were removed throughout the treatment period and incubated with FLASH for 1 h, or with CellTiter-Blue (CTB) for 24 h. (B) FLASH and CTB measurements for cultures treated for 7 days with RIF (mean ± SD, n = 3). Marker colors correspond to RIF concentrations shown in part A. (C) FLASH signal dependence on RIF concentration for each day (mean ± SD, n = 3). Dose response for killing by RIF as measured by the FLASH probe (D) or CTB (E) (mean ± SD, n = 3). Data were normalized to DMSO (100% viability) and 10 μM RIF (0% viability) and fitted to a two-parameter logistic function. IC50 values are reported as 95% confidence intervals. (F) Time course of mc26020 treated with the critical concentrations of rifampicin (RIF, 1 μg/mL), ethambutol (EMB, 5 μg/mL), isoniazid (INH, 0.1 μg/mL), pyrazinamide (PZA, 100 μg/mL), or streptomycin (STR, 1 μg/mL). For all days, the signal from untreated cultures was compared to each of the treated cultures via a two-way ANOVA with Dunnett’s test (n = 3; ***, p < 0.001 for the comparison between untreated cultures and each of the antibiotic conditions). (G) Time course of H37Rv (WT) Mtb and RpoB H526D mutant Mtb (H526D) treated with DMSO (black) or the critical concentration of RIF (red). For each day, the RIF- and DMSO-treated conditions were compared via an independent t test (n = 3; ***, p < 0.001). (H) Luminescent signal from H37Rv (WT) or H526D after 6 days of culture in the presence or absence of RIF. Samples are compared to the WT Mtb strain treated with RIF via one-way ANOVA with Dunnett’s test (n = 3; ***, p < 0.001).
To evaluate the potential for FLASH as a tool for drug-susceptibility testing (DST), we treated cultures of mc26020 with five antibiotics with differing modes of action, each at the “critical concentration” used clinically to determine drug susceptibility. For each antibiotic, the FLASH signal was compared between treated and untreated cultures throughout the treatment period (Figure 5F). We observed a significantly higher signal in the untreated samples as soon as 4 days after treatment, for all antibiotics tested. For clinical use, a DST protocol must be able to distinguish susceptible and resistant strains. To test for its ability to identify drug resistance, we repeated the DST experiment and compared an RIF-susceptible (H37Rv) to an RIF-resistant strain (RpoB H526D mutant) (Figure 5G). Luminescence increased over time for the untreated cultures and for the resistant strain treated with RIF. As observed before, there was no change in signal for the susceptible strain treated with RIF. After 6 days of treatment, the signal for the resistant strain treated with RIF had increased substantially, and all samples yielded a significantly higher signal than the susceptible strain treated with RIF (Figure 5H). Similar results were obtained when comparing the RIF-susceptible strain CDC1551 to the RIF-resistant strain (Figure S5C). These results show that FLASH can be used to monitor growth inhibition by clinical antibiotics and can differentiate susceptible from resistant strains of Mtb after 5–6 days of culture with antibiotic—substantially faster than the weeks to months generally required for culture-based DST, and faster than newer methods for DST such as microscopic-observation drug-susceptibility (8 days)42 and Sensititer MycoTB plates (10–14 days).43
Discussion
Global health initiatives including The World Health Organization1 and the Stop TB Partnership44 have highlighted rapid point-of-care diagnostics and rapid DST as critical developments needed to reach the goal of reduced TB cases and better patient outcomes. FLASH has the potential to address these needs by providing a fast, sensitive, and selective tool for the detection of live Mtb. The labeling protocol is simple, requires minimal training, and does not require microscopy or other sophisticated laboratory instrumentation. Our experiments with processed sputum show that it is compatible with the standard NALC/NaOH protocol used for decontaminating samples. Because FLASH is selective for Mtb compared to other organisms that may be present in sputum samples, the readout is not sensitive to potential contaminants, and this decontamination step may be unnecessary. We expect that further optimization of sputum processing and concentration of bacteria will reduce the loss of signal due to centrifugation, decreasing the limit of detection in sputum samples. Though it remains to be tested, FLASH also has the potential to detect dormant bacteria, a physiological state in which bacteria persist without apparent replication. In hypoxic culture models of Mtb dormancy, an absolute proteome analysis showed that Hip1 is present in cultures,48 and an activity-based proteomic analysis of active hydrolases and proteases confirmed that Hip1 is both present and enzymatically active in nongrowing cells.14 These observations suggest that FLASH has the potential to be used to detect dormant infections.
The ability of FLASH to differentiate live from dead bacteria also gives the probe potential as a rapid readout for monitoring TB treatment. Despite their high sensitivity, NAATs like Xpert are problematic when used for diagnosis following treatment because they can amplify DNA left over from dead bacteria. These false positives can persist for years after treatment45,46 making it difficult for NAATs to accurately track decreasing disease burden during antibiotic therapy.47 In contrast, FLASH does not respond to dead bacteria and should be able to track the decrease in bacterial burden throughout a course of antibiotic treatment. The short time to result for FLASH compared to the culture-based method should enable rapid assessment of treatment outcomes, which is important for identifying and adapting to treatment failures due to antibiotic resistance.
In this study, we tested three different plate readers to evaluate the sensitivity and selectivity of FLASH, and we note that differences in instrumentation lead to differences in the limits of detection and signal-to-noise ratios (Figure 3B,C, Figure S3A). For point-of-care diagnostics using FLASH, it will be critical to use a sensitive, inexpensive, and low-power luminometer. A number of devices that meet these criteria have been evaluated for other purposes including handheld, battery-operated luminometers49 and adapters for use with smartphone cameras.50,51 Additional studies will be required to evaluate the ability of FLASH to (i) sensitively and specifically detect TB at the point of care, (ii) track disease progression throughout the course of antibiotic treatment, and (iii) reliably determine the antibiotic susceptibility of clinical isolates more quickly than existing DST approaches.
Finally, we note that FLASH may be a useful tool for tracking bacterial viability, bacterial growth, and the activity of Hip1 in the context of infection. Because of its low LOD, FLASH could serve as a straightforward and quantitative measurement of live Mtb cells in bacterial culture, and cell culture or animal models of infection. In addition, FLASH may serve as a tool for addressing outstanding questions about how Hip1 activity changes under different growth conditions, throughout the course of infection, and in response to the host immune system. In our hands, a simple 1 h measurement in a microplate reader can report on Hip1 activity in many parallel experiments (e.g., in a 384-well plate). We note that FLASH could also be used to measure Hip1 activity with spatial resolution in the context of infection using chemiluminescent imaging.
This study along with other examples of luminescent enzyme probes for pathogen detection26 highlight the versatility and adaptability of turn-on dioxetane luminophores. Key advantages of luminescent measurements are the low background signal in biological samples and the simplicity of detection. The design of these probes is straightforward—any enzymatic or chemical unmasking event can lead to light emission. Proteases offer attractive targets for such probes because of the flexibility to design peptidic substrates with great selectivity. By carefully choosing the appropriate enzyme target and cognate substrate, we expect luminescent turn-on probes will serve as effective point-of-care diagnostics for other infectious diseases in addition to TB.
Materials and Methods
Chemical Synthesis
Methods for the synthesis and characterization of the FLASH and D-FLASH probes are presented in the Supporting Information.54,55
Bacterial Culture
M. tuberculosis H37Rv, Mycobacterium marinum M, and Mycobacterium smegmatis mc2155 were a gift from Carolyn Bertozzi (Stanford University). M. tuberculosis mc26020 was a gift from Niaz Banaei (Stanford University). M. tuberculosis Erdman CDC1551 and the CDC1551-derived RpoB H526D rifampicin-resistant mutant52 were obtained from the Center for Tuberculosis Research (Johns Hopkins University). The following clinical isolates of NTMs were a gift from Dr. Nicole Parrish and Derek Armstrong (Department of Pathology, the Johns Hopkins University School of Medicine): Mycobacterium kansasii, Mycobacterium gordonae, Mycobacterium intracellulare, Mycobacterium scrofulaceum, Mycobacterium avium, Mycobacterium chelonae, and Mycobacterium abscessus.
M. tuberculosis strains (except mc26020) and all NTMs were cultured in liquid 7H9/OADC medium (4.7 g/L 7H9 powder, 0.2% w/v glycerol, 0.05% w/v Tween-80, and 10% v/v OADC supplement) or on solid 7H10 agar plates (19 g/L 7H10 powder, 1% w/v glycerol, 10% OADC supplement). M. tuberculosis mc26020 was cultured in liquid 7H9/OADC medium supplemented with 24 mg/L pantothenate, 80 mg/L l-lysine, and 0.2% w/v casamino acids or solid 7H9 plates (15 g of agar, 4.7 g of 7H9 powder, 0.1% w/v glycerol, 0.2% w/v casamino acids, 24 mg/L pantothenate, 80 mg/L l-lysine, 10% OADC supplement). OADC supplement contained 0.5 g/L oleic acid, 50 g/L albumin fraction V, 20 g/L dextrose, 40 mg/L catalase, and 8.5 g/L NaCl. Cultures were inoculated from frozen glycerol stocks or from agar plates and cultured at 37 °C with shaking for at least 1 week. To estimate the number of cells used for each experiment, a conversion factor was calculated by plating serial dilutions of cultures with known OD600 values onto agar plates. After 3–5 weeks of growth at 37 °C, individual colonies were counted to determine CFU/mL. Two separate experiments yielded the same factor of OD600 1 = 3 × 108 CFU/mL. For each experiment, OD600 was measured in a spectrophotometer, and cultures were diluted to the desired cell density based on the conversion factor.
FLASH Measurements and Data Analysis
All experiments were performed with biological triplicates, unless otherwise indicated. For experiments with live Mtb, measurements were made of independent bacterial cultures. For experiments with recombinant Hip1, measurements were made of independent mixtures using the same enzyme preparation. All chemiluminescence assays were performed in white, opaque flat-bottom 384- or 96-well plates. Luminescence was measured in different microplate readers, depending on the laboratory location. Measurements of H37Rv, mc26020, and M. marinum were obtained on a SpectraMax M3 instrument (Molecular Devices) at 25 °C. Measurements of recombinant Hip1, mc26020, NTMs, and other bacteria were obtained on a Cytation 3 instrument (Biotek) at 37 °C. Measurements of rifampicin susceptibility of the H37Rv and RpoB H526D mutant were obtained on a FLUOstar Omega instrument (BMG Labtech) at 37 °C. For all experiments, luminescence measurements began within 5 min after the addition of the FLASH probe and were continued for at least 1 h. Measurements were made without an emission filter, using a 1 s integration time. For each sample, luminescence measurements from the first hour were summed to yield integrated luminescence.
Detection of Hip1 Enzyme Activity
Recombinant Hip1 was purified as previously described.33 For each experiment, 40 μL of Hip1 in Hip1 buffer (0.01% Triton X-100 in PBS) was combined with 5 μL of 9× FLASH probe in 1:1 DMSO/Hip1 buffer. To determine the limit of detection, 40 μL of 2-fold series dilutions of recombinant Hip1 (final concentrations: 0.05–12.5 nM) was combined with 5 μL of 90 μM FLASH probe (final concentration 10 μM). To determine kinetic parameters of the probe, 40 μL of 3 nM Hip1 in Hip1 buffer was combined with a dilution series of FLASH probe concentrations (final concentrations: 0–50 μM). To measure enzyme inhibition, 37.5 μL of 3 nM Hip1 in Hip1 buffer were combined with 2.5 μL of CSL157 in DMSO (final concentrations: 20 nM to 7 μM) and incubated for 30 min at 37 °C before addition of 5 μL of FLASH probe (final concentration: 10 μM). To test D-FLASH, 40 μL of 3 nM Hip1 in Hip1 buffer was combined with 10 μM D-FLASH probe. All conditions were tested in triplicate.
Analysis of Hip1 Homologues
The protein sequence of Hip1 (CaeA, Rv2224c) from M. tuberculosis H37Rv was used as the query sequence for BLASTP analysis. The search set comprised whole genome sequences from Mtb isolates from the National Center for Biotechnology Information (https://www.ncbi.nlm.nih.gov, retrieved January 14, 2021), NTMs, airway and oral subsets from the Human Microbiome Project Reference Genome (https://www.hmpdacc.org/hmp/HMRGD/, retrieved on April 28, 2020), Staphylococcus aureus, and Klebsiella pneumoniae. Hip1 was queried against the protein sequences by BLASTP (2.9.0+), requiring a minimum e-value of 1 × 10–4. The sequences of Hip1, M. tuberculosis Rv2223c, and the closest homologue of Hip1 in NTMs, S. aureus, K. pneumoniae, and select reference bacteria from the oral and airway microbiomes were aligned using Clustal Omega (https://www.ebi.ac.uk/Tools/msa/clustalo/),53 and the phylogenetic tree was generated using FigTree (1.4.4).
Detection of Bacteria in Culture
Cultures were grown until reaching an OD600 of 0.4–1.0 and then were diluted in growth medium to reach the desired cell concentration for each condition. To determine the limit of detection, 40 μL of serially diluted cultures was added to a 384-well plate. Heat-killed control samples were prepared by heating 0.5 mL of culture in an O-ring tube at 95 °C for 15 min. Hip1 inhibition in live cells was tested by treating 3 × 109 CFU/mL H37Rv with 10 μM CSL157 for 1 h at 37 °C before the addition of FLASH. Where indicated, cultures were centrifuged for 10 min at 8000 rcf; the supernatant was removed by pipetting, and the cell pellet was resuspended in PBS. For all experiments, 5 μL of 225 μM FLASH probe in 1:1 DMSO/Hip1 buffer (final concentration 25 μM) was added to 40 μL of bacterial sample. All conditions were tested in triplicate.
Detection of Bacteria in Sputum
Sputum samples were obtained from the Johns Hopkins Medical Microbiology Laboratory as per standard of care. Sputum was processed using a standard decontamination and concentration protocol. Sputum was transferred to a 50 mL centrifuge tube, combined with an equal volume of Snap n’ Digest (Scientific Device), vortexed, and incubated at 25 °C for 15 min. Samples were neutralized by addition of PBS to a final total volume of 45 mL. Samples were centrifuged at 3200 rcf, and the supernatant was removed. Pellets were resuspended in up to 5 mL of PBS. Decontaminated samples from multiple patients were pooled, aliquoted, and frozen at −80 °C until use. Cultures of mc26020 were grown until reaching an OD600 of 0.2. Cultures were diluted in growth medium to reach the desired cell concentration for each condition and added to an equal volume of processed human sputum. Samples were neutralized by addition of PBS and then centrifuged for 17 min at 3200 rcf. Supernatant was removed, and the pellet was resuspended in 130 μL of PBS. For all experiments, 5 μL of 225 μM FLASH probe in 1:1 DMSO/Hip1 buffer (final concentration 25 μM) was added to 40 μL of bacterial sample. All conditions were tested in triplicate.
Drug Susceptibility Testing
Cultures of M. tuberculosis were grown until reaching an OD600 of 0.4–1.0 and diluted to an OD600 of 0.2 in growth medium. The diluted culture was aliquoted for each treatment condition into 5–10 mL cultures. Antibiotics were added from 100× stock made up in DMSO to the desired concentration. Cultures were shaken at 37 °C for the duration of the experiment. At each time point, an aliquot of culture was transferred to a 384-well plate. For all experiments, 5 μL of 225 μM FLASH probe in 1:1 DMSO/Hip1 buffer (final concentration 25 μM) was added to 40 μL of bacterial sample. To test for cell viability, 100 μL of each culture was transferred to a 96-well plate, treated with 20 μL of CellTiter-Blue (Promega), and incubated for 24 h at 37 °C. CellTiter-Blue fluorescence was measured with 560 nm excitation and 590 nm emission. All conditions were tested in triplicate.
Safety Statement
All experiments with infectious strains of Mtb (H37Rv, Edrman, and CDC1551) carry some risk of infection and were performed in BSL3 laboratories. The protocols used do not pose a high risk for aerosolization of bacteria. No other unexpected or unusually high safety hazards were encountered.
Acknowledgments
We thank Dr. Carolyn Bertozzi and Dr. Douglas Fox (Stanford University) for sharing bacterial strains and for training and use of the Stanford BSL3 facility. We thank Dr. Niaz Banaei (Stanford University) for sharing bacterial strains. We thank Dr. Nicole Parrish and Derek Armstrong (Department of Pathology, the Johns Hopkins University School of Medicine) for sharing bacterial strains and deidentified human sputum samples.
Glossary
Abbreviations
- TB
tuberculosis
- Mtb
Mycobacterium tuberculosis
- NAAT
nucleic acid amplification test
- DST
drug susceptibility testing
- NTM
nontuberculous mycobacteria
- FLASH
fast, luminescent, affordable, sensor of Hip1
- SNR
signal to noise ratio
- LOD
limit of detection
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acscentsci.0c01345.
Supporting methods and additional figures including chemical structures, activities of Hip1 toward FLASH and D-FLASH, comparison of M. tuberculosis strains and sputum treatment conditions, comparison of the FLASH signal from NTMs and other bacteria, and FLASH and CTB measurements of RIF-treated cultures (PDF)
Author Contributions
B.M.B. contributed to the conceptualization, formal analysis, investigation, visualization, and writing of the original draft. G.F-C. contributed to the conceptualization, formal analysis, funding acquisition, investigation, resources, and writing (review and editing). J.S. contributed to the investigation, formal analysis, and writing (review and editing). O.G. contributed to the investigation and resources. A.A.O. contributed to the conceptualization, investigation, funding acquisition, and writing (review and editing). M.L.T. contributed to the investigation. L.J.K. contributed to the investigation and writing (review and editing). S.K.J. contributed to the resources, funding acquisition, and writing (review and editing). D.S. contributed to the conceptualization, funding acquisition, supervision, and writing (review and editing). M.B. contributed to the conceptualization, funding acquisition, supervision, and writing (review and editing).
B.M.B. was supported by the A. P. Giannini Foundation. G.F.-C. was supported by a Research Supplement to Promote Diversity in Health-Related Research for NIH grant R01CA179253. L.J.K. was supported by the Stanford ChEM-H Chemistry/Biology Interface Predoctoral Training Program, Stanford Molecular Pharmacology Training Grant, and Stanford Graduate Fellowship. D.S. thanks the Israel Science Foundation (ISF) for financial support. Research reported in this publication was supported by the National Center for Advancing Translational Sciences of the National Institutes of Health under Award UL1TR003142. This work was also funded in part by grants from the National Institutes of Health (T32AI07328 to B.M.B., R21AI149760 to S.K.J. and A.A.O., and R01EB026332 to M.B.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
The authors declare no competing financial interest.
Supplementary Material
References
- World Health Organization . Global Tuberculosis Report 2019; World Health Organization: Geneva, Switzerland, 2019.
- Storla D. G.; Yimer S.; Bjune G. A. A systematic review of delay in the diagnosis and treatment of tuberculosis. BMC Public Health 2008, 8, 15. 10.1186/1471-2458-8-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization . Implementing Tuberculosis Diagnostics: Policy Framework; World Health Organization: Geneva, Switzerland, 2015.
- Steingart K. R.; Ng V.; Henry M.; Hopewell P. C.; Ramsay A.; Cunningham J.; Urbanczik R.; Perkins M. D.; Aziz M. A.; Pai M. Sputum processing methods to improve the sensitivity of smear microscopy for tuberculosis: a systematic review. Lancet Infect. Dis. 2006, 6, 664–674. 10.1016/S1473-3099(06)70602-8. [DOI] [PubMed] [Google Scholar]
- Agrawal M.; Bajaj A.; Bhatia V.; Dutt S. Comparative Study of GeneXpert with ZN Stain and Culture in Samples of Suspected Pulmonary Tuberculosis. J. Clin. Diagn. Res. 2016, 10, DC09–DC12. 10.7860/JCDR/2016/18837.7755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorman S. E.; Schumacher S. G.; Alland D.; Nabeta P.; Armstrong D. T.; King B.; Hall S. L.; Chakravorty S.; Cirillo D. M.; Tukvadze N.; Bablishvili N.; Stevens W.; Scott L.; Rodrigues C.; Kazi M. I.; Joloba M.; Nakiyingi L.; Nicol M. P.; Ghebrekristos Y.; Anyango I.; Murithi W.; Dietze R.; Peres R. L.; Skrahina A.; Auchynka V.; Chopra K. K.; Hanif M.; Liu X.; Yuan X.; Boehme C. C.; Ellner J. J.; Denkinger C. M.; team s.; Dorman S. E.; Schumacher S. G.; Alland D.; Nabeta P.; Armstrong D. T.; King B.; Hall S. L.; Chakravorty S.; Cirillo D. M.; Tukvadze N.; Bablishvili N.; Stevens W.; Scott L.; Rodrigues C.; Kazi M. I.; Joloba M.; Nakiyingi L.; Nicol M. P.; Ghebrekristos Y.; Anyango I.; Murithi W.; Dietze R.; Peres R. L.; Skrahina A.; Auchynka V.; Chopra K. K.; Hanif M.; Liu X.; Yuan X.; Boehme C. C.; Ellner J. J.; Denkinger C. M.; Manabe Y. C.; Hom D.; Aspindzelashvili R.; David A.; Surve U.; Kamulegeya L. H.; Nabweyambo S.; Surtie S.; Hapeela N.; Cain K. P.; Agaya J.; McCarthy K. D.; Marques-Rodrigues P.; Castellani L. G. S.; Almeida P. S.; Aguiar P. P. L. d.; Solodovnikova V.; Ruan X.; Liang L.; Zhang G.; Zhu H.; Xie Y. Xpert MTB/RIF Ultra for detection of Mycobacterium tuberculosis and rifampicin resistance: a prospective multicentre diagnostic accuracy study. Lancet Infect. Dis. 2018, 18, 76–84. 10.1016/S1473-3099(17)30691-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization . Xpert MTB/RIF Implementation Manual; World Health Organization: Geneva, Switzerland, 2014.
- Walzl G.; McNerney R.; Plessis N. d.; Bates M.; McHugh T. D.; Chegou N. N.; Zumla A. Tuberculosis: advances and challenges in development of new diagnostics and biomarkers. Lancet Infect. Dis. 2018, 18, e199–e210. 10.1016/S1473-3099(18)30111-7. [DOI] [PubMed] [Google Scholar]
- Pai M.; Nicol M. P.; Boehme C. C. Tuberculosis Diagnostics: State of the Art and Future Directions. Microbiol. Spectrum 2016, 4, 4. 10.1128/microbiolspec.TBTB2-0019-2016. [DOI] [PubMed] [Google Scholar]
- Warsinske H.; Vashisht R.; Khatri P. Host-response-based gene signatures for tuberculosis diagnosis: A systematic comparison of 16 signatures. PLoS Med. 2019, 16, e1002786 10.1371/journal.pmed.1002786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawn S. D. Point-of-care detection of lipoarabinomannan (LAM) in urine for diagnosis of HIV-associated tuberculosis: a state of the art review. BMC Infect. Dis. 2012, 12, 103. 10.1186/1471-2334-12-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruins M.; Rahim Z.; Bos A.; Sande W. W. J. v. d.; Endtz H. P.; Belkum A. v. Diagnosis of active tuberculosis by e-nose analysis of exhaled air. Tuberculosis 2013, 93, 232–238. 10.1016/j.tube.2012.10.002. [DOI] [PubMed] [Google Scholar]
- Teixeira R. C.; Rodríguez M.; Romero N. J. d.; Bruins M.; Gómez R.; Yntema J. B.; Abente G. C.; Gerritsen J. W.; Wiegerinck W.; Bejerano D. P.; Magis-Escurra C. The potential of a portable, point-of-care electronic nose to diagnose tuberculosis. J. Infect. 2017, 75, 441–447. 10.1016/j.jinf.2017.08.003. [DOI] [PubMed] [Google Scholar]
- Tallman K. R.; Levine S. R.; Beatty K. E. Small-Molecule Probes Reveal Esterases with Persistent Activity in Dormant and Reactivating Mycobacterium tuberculosis. ACS Infect. Dis. 2016, 2, 936–944. 10.1021/acsinfecdis.6b00135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beatty K. E.; Williams M.; Carlson B. L.; Swarts B. M.; Warren R. M.; Helden P. D. v.; Bertozzi C. R. Sulfatase-activated fluorophores for rapid discrimination of mycobacterial species and strains. Proc. Natl. Acad. Sci. U. S. A. 2013, 110, 12911–12916. 10.1073/pnas.1222041110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamariza M.; Shieh P.; Ealand C. S.; Peters J. S.; Chu B.; Rodriguez-Rivera F. P.; Sait M. R. B.; Treuren W. V.; Martinson N.; Kalscheuer R.; Kana B. D.; Bertozzi C. R. Rapid detection of Mycobacterium tuberculosis in sputum with a solvatochromic trehalose probe. Sci. Transl. Med. 2018, 10, eaam6310 10.1126/scitranslmed.aam6310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamariza M.; Keyser S. G. L.; Utz A.; Knapp B. D.; Ahn G.; Cambier C. J.; Chen T.; Huang K. C.; Bertozzi C. R.. Towards Mycobacterium tuberculosis detection at the point-of-care: a brighter solvatochromic probe permits the detection of mycobacteria within minutes. bioRxiv, 2020, 2020.2005.2029.124008. https://www.biorxiv.org/content/10.1101/2020.05.29.124008v2. [DOI] [PMC free article] [PubMed]
- Cheng Y.; Xie J.; Lee K.-H.; Gaur R. L.; Song A.; Dai T.; Ren H.; Wu J.; Sun Z.; Banaei N.; Akin D.; Rao J. Rapid and specific labeling of single live Mycobacterium tuberculosis with a dual-targeting fluorogenic probe. Sci. Transl. Med. 2018, 10, eaar4470 10.1126/scitranslmed.aar4470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sule P.; Tilvawala R.; Mustapha T.; Hassounah H.; Noormohamed A.; Kundu S.; Graviss E. A.; Walkup G. K.; Kong Y.; Cirillo J. D. Rapid Tuberculosis Diagnosis Using Reporter Enzyme Fluorescence. J. Clin. Microbiol. 2019, 57, 01462-19. 10.1128/JCM.01462-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie H.; Mire J.; Kong Y.; Chang M.; Hassounah H. A.; Thornton C. N.; Sacchettini J. C.; Cirillo J. D.; Rao J. Rapid point-of-care detection of the tuberculosis pathogen using a BlaC-specific fluorogenic probe. Nat. Chem. 2012, 4, 802–809. 10.1038/nchem.1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gnaim S.; Green O.; Shabat D. The emergence of aqueous chemiluminescence: new promising class of phenoxy 1,2-dioxetane luminophores. Chem. Commun. 2018, 54, 2073–2085. 10.1039/C8CC00428E. [DOI] [PubMed] [Google Scholar]
- Hananya N.; Shabat D. A Glowing Trajectory between Bio- and Chemiluminescence: From Luciferin-Based Probes to Triggerable Dioxetanes. Angew. Chem., Int. Ed. 2017, 56, 16454–16463. 10.1002/anie.201706969. [DOI] [PubMed] [Google Scholar]
- Hananya N.; Shabat D. Recent Advances and Challenges in Luminescent Imaging: Bright Outlook for Chemiluminescence of Dioxetanes in Water. ACS Cent. Sci. 2019, 5, 949–959. 10.1021/acscentsci.9b00372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green O.; Eilon T.; Hananya N.; Gutkin S.; Bauer C. R.; Shabat D. Opening a Gateway for Chemiluminescence Cell Imaging: Distinctive Methodology for Design of Bright Chemiluminescent Dioxetane Probes. ACS Cent. Sci. 2017, 3, 349–358. 10.1021/acscentsci.7b00058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth-Konforti M. E.; Bauer C. R.; Shabat D. Unprecedented Sensitivity in a Probe for Monitoring Cathepsin B: Chemiluminescence Microscopy Cell-Imaging of a Natively Expressed Enzyme. Angew. Chem. 2017, 129, 15839–15844. 10.1002/ange.201709347. [DOI] [PubMed] [Google Scholar]
- Roth-Konforti M.; Green O.; Hupfeld M.; Fieseler L.; Heinrich N.; Ihssen J.; Vorberg R.; Wick L.; Spitz U.; Shabat D. Ultrasensitive Detection of Salmonella and Listeria monocytogenes by Small-Molecule Chemiluminescence Probes. Angew. Chem., Int. Ed. 2019, 58, 10361. 10.1002/anie.201904719. [DOI] [PubMed] [Google Scholar]
- Das S.; Ihssen J.; Wick L.; Spitz U.; Shabat D. Chemiluminescent Carbapenem-Based Molecular Probe for Detection of Carbapenemase Activity in Live Bacteria. Chem. - Eur. J. 2020, 26, 3647–3652. 10.1002/chem.202000217. [DOI] [PubMed] [Google Scholar]
- Lun S.; Bishai W. R. Characterization of a Novel Cell Wall-anchored Protein with Carboxylesterase Activity Required for Virulence in Mycobacterium tuberculosis. J. Biol. Chem. 2007, 282, 18348–18356. 10.1074/jbc.M700035200. [DOI] [PubMed] [Google Scholar]
- Rengarajan J.; Murphy E.; Park A.; Krone C. L.; Hett E. C.; Bloom B. R.; Glimcher L. H.; Rubin E. J. Mycobacterium tuberculosis Rv2224c modulates innate immune responses. Proc. Natl. Acad. Sci. U. S. A. 2008, 105, 264–269. 10.1073/pnas.0710601105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madan-Lala R.; Peixoto K. V.; Re F.; Rengarajan J. Mycobacterium tuberculosis Hip1 Dampens Macrophage Proinflammatory Responses by Limiting Toll-Like Receptor 2 Activation. Infect. Immun. 2011, 79, 4828–4838. 10.1128/IAI.05574-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naffin-Olivos J. L.; Georgieva M.; Goldfarb N.; Madan-Lala R.; Dong L.; Bizzell E.; Valinetz E.; Brandt G. S.; Yu S.; Shabashvili D. E.; Ringe D.; Dunn B. M.; Petsko G. A.; Rengarajan J. Mycobacterium tuberculosis Hip1Modulates Macrophage Responses through Proteolysis of GroEL2. PLoS Pathog. 2014, 10, e1004132 10.1371/journal.ppat.1004132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madan-Lala R.; Sia J. K.; King R.; Adekambi T.; Monin L.; Khader S. A.; Pulendran B.; Rengarajan J. Mycobacterium tuberculosis Impairs Dendritic Cell Functions through the Serine Hydrolase Hip1. J. Immunol. 2014, 192, 4263–4272. 10.4049/jimmunol.1303185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lentz C. S.; Ordonez A. A.; Kasperkiewicz P.; La Greca F.; O’Donoghue A. J.; Schulze C. J.; Powers J. C.; Craik C. S.; Drag M.; Jain S. K.; Bogyo M. Design of Selective Substrates and Activity-Based Probes for Hydrolase Important for Pathogenesis 1 (HIP1) from Mycobacterium tuberculosis. ACS Infect. Dis. 2016, 2, 807–815. 10.1021/acsinfecdis.6b00092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambandamurthy V. K.; Derrick S. C.; Jalapathy K. V.; Chen B.; Russell R. G.; Morris S. L.; Jacobs W. R. Jr. Long-term protection against tuberculosis following vaccination with a severely attenuated double lysine and pantothenate auxotroph of Mycobacterium tuberculosis. Infect. Immun. 2005, 73, 1196–1203. 10.1128/IAI.73.2.1196-1203.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Global Laboratory Initiative . Mycobacterial Laboratory Manual; Global Laboratory Initiative: Geneva, Switzerland, 2014.
- Yajko D. M.; Wagner C.; Tevere V. J.; Kocagöz T.; Hadley W. K.; Chambers H. F. Quantitative culture of Mycobacterium tuberculosis from clinical sputum specimens and dilution endpoint of its detection by the Amplicor PCR assay. J. Clin. Microbiol. 1995, 33, 1944–1947. 10.1128/JCM.33.7.1944-1947.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palaci M.; Dietze R.; Hadad D. J.; Ribeiro F. K. C.; Peres R. L.; Vinhas S. A.; Maciel E. L. N.; Dettoni V. d. V.; Horter L.; Boom W. H.; Johnson J. L.; Eisenach K. D. Cavitary Disease and Quantitative Sputum Bacillary Load in Cases of Pulmonary Tuberculosis▽. J. Clin. Microbiol. 2007, 45, 4064–4066. 10.1128/JCM.01780-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayigire X. A.; Friedrich S. O.; Venter A.; Dawson R.; Gillespie S. H.; Boeree M. J.; Heinrich N.; Hoelscher M.; Diacon A. H. Direct Comparison of Xpert MTB/RIF Assay with Liquid and Solid Mycobacterial Culture for Quantification of Early Bactericidal Activity. J. Clin. Microbiol. 2013, 51, 1894–1898. 10.1128/JCM.03290-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Human Microbiome Project Consortium . Structure, function and diversity of the healthy human microbiome. Nature 2012, 486, 207–214, 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Human Microbiome Project Consortium . A framework for human microbiome research. Nature 2012, 486, 215–221, 10.1038/nature11209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asadpour-Zeynali K.; Saeb E. Simultaneous Spectrophotometric Determination of Rifampicin, Isoniazid and Pyrazinamide in a Single Step. Iran J. Pharm. Res. 2016, 15, 713–723. [PMC free article] [PubMed] [Google Scholar]
- Moore D. A.; Mendoza D.; Gilman R. H.; Evans C. A.; Hollm Delgado M. G.; Guerra J.; Caviedes L.; Vargas D.; Ticona E.; Ortiz J.; Soto G.; Serpa J.; Tuberculosis Working Group in P. Microscopic observation drug susceptibility assay, a rapid, reliable diagnostic test for multidrug-resistant tuberculosis suitable for use in resource-poor settings. J. Clin. Microbiol. 2004, 42, 4432–4437. 10.1128/JCM.42.10.4432-4437.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall L.; Jude K. P.; Clark S. L.; Dionne K.; Merson R.; Boyer A.; Parrish N. M.; Wengenack N. L. Evaluation of the Sensititre MycoTB plate for susceptibility testing of the Mycobacterium tuberculosis complex against first- and second-line agents. J. Clin. Microbiol. 2012, 50, 3732–3734. 10.1128/JCM.02048-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stop TB Partnership . Global Plan to Stop TB; Stop TB Partnership: Geneva, Switzerland, 2015.
- Theron G.; Venter R.; Calligaro G.; Smith L.; Limberis J.; Meldau R.; Chanda D.; Esmail A.; Peter J.; Dheda K. Xpert MTB/RIF Results in Patients With Previous Tuberculosis: Can We Distinguish True From False Positive Results?. Clin. Infect. Dis. 2016, 62, 995–1001. 10.1093/cid/civ1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theron G.; Venter R.; Smith L.; Esmail A.; Randall P.; Sood V.; Oelfese S.; Calligaro G.; Warren R.; Dheda K. False-Positive Xpert MTB/RIF Results in Retested Patients with Previous Tuberculosis: Frequency, Profile, and Prospective Clinical Outcomes. J. Clin. Microbiol. 2018, 56, 01696-17. 10.1128/JCM.01696-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrich S. O.; Rachow A.; Saathoff E.; Singh K.; Mangu C. D.; Dawson R.; Phillips P. P.; Venter A.; Bateson A.; Boehme C. C.; Heinrich N.; Hunt R. D.; Boeree M. J.; Zumla A.; McHugh T. D.; Gillespie S. H.; Diacon A. H.; Hoelscher M. Pan African Consortium for the Evaluation of Anti-Tuberculosis A. Assessment of the sensitivity and specificity of Xpert MTB/RIF assay as an early sputum biomarker of response to tuberculosis treatment. Lancet Respir. Med. 2013, 1, 462–470. 10.1016/S2213-2600(13)70119-X. [DOI] [PubMed] [Google Scholar]
- Schubert O. T.; Ludwig C.; Kogadeeva M.; Zimmermann M.; Rosenberger G.; Gengenbacher M.; Gillet L. C.; Collins B. C.; Rost H. L.; Kaufmann S. H.; Sauer U.; Aebersold R. Absolute Proteome Composition and Dynamics during Dormancy and Resuscitation of Mycobacterium tuberculosis. Cell Host Microbe 2015, 18, 96–108. 10.1016/j.chom.2015.06.001. [DOI] [PubMed] [Google Scholar]
- Pierce J.; Hiebert J. B.; Mahoney D.; Shen Q.; Peltzer J.; Rahman F.; Johnson S.; Pierce J. T. Development of a Point-of-Contact Technique to Measure Adenosine Triphosphate: A Quality Improvement Study. Ann. Med. Surg. 2019, 41, 29–32. 10.1016/j.amsu.2019.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roda A.; Michelini E.; Cevenini L.; Calabria D.; Calabretta M. M.; Simoni P. Integrating Biochemiluminescence Detection on Smartphones: Mobile Chemistry Platform for Point-of-Need Analysis. Anal. Chem. 2014, 86, 7299–7304. 10.1021/ac502137s. [DOI] [PubMed] [Google Scholar]
- Habimana J. d. D.; Ji J.; Sun X. Minireview: Trends in Optical-Based Biosensors for Point-Of-Care Bacterial Pathogen Detection for Food Safety and Clinical Diagnostics. Anal. Lett. 2018, 51, 2933–2966. 10.1080/00032719.2018.1458104. [DOI] [Google Scholar]
- Campodónico V. L.; Rifat D.; Chuang Y.-M.; Ioerger T. R.; Karakousis P. C. Altered Mycobacterium tuberculosis Cell Wall Metabolism and Physiology Associated With RpoB Mutation H526D. Front. Microbiol. 2018, 9, 494. 10.3389/fmicb.2018.00494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers F.; Wilm A.; Dineen D.; Gibson T. J.; Karplus K.; Li W.; Lopez R.; McWilliam H.; Remmert M.; Söding J.; Thompson J. D.; Higgins D. G. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akinboye E. S.; Rosen M. D.; Bakare O.; Denmeade S. R. Anticancer activities of emetine prodrugs that are proteolytically activated by the prostate specific antigen (PSA) and evaluation of in vivo toxicity of emetine derivatives. Bioorg. Med. Chem. 2017, 25, 6707–6717. 10.1016/j.bmc.2017.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hananya N.; Reid J. P.; Green O.; Sigman M. S.; Shabat D. Rapid chemiexcitation of phenoxy-dioxetane luminophores yields ultrasensitive chemiluminescence assays. Chem. Sci. 2019, 10, 1380–1385. 10.1039/C8SC04280B. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.