Abstract
Genomic discovery efforts for hematological traits have been successfully conducted through genome-wide association study on samples of predominantly European ancestry. We sought to conduct unbiased genetic discovery for coding variants that influence hematological traits in a Han Chinese population. A total of 5257 Han Chinese subjects from Beijing, China were included in the discovery cohort and analyzed by an Illumina ExomeChip array. Replication analyses were conducted in 3827 independent Chinese subjects. We analyzed 12 hematological traits and identified 22 exome-wide significant single-nucleotide polymorphisms (SNP)–trait associations with 15 independent SNPs. Our study provides replication for two associations previously reported but not replicated. Further, one association was identified and replicated in the current study, of a coding variant in the myeloproliferative leukemia (MPL) gene, c.793C > T, p.Leu265Phe (L265F) with increased platelet count (β = 20.6 109 cells/l, Pmeta-analysis = 2.6 × 10−13). This variant is observed at ~2% population frequency in East Asians, whereas it has not been reported in gnomAD European or African populations. Functional analysis demonstrated that expression of MPL L265F in Ba/F3 cells resulted in enhanced phosphorylation of Stat3 and ERK1/2 as compared with the reference MPL allele, supporting altered activation of the JAK–STAT signal transduction pathway as the mechanism underlying the novel association between MPL L265F and platelet count.
Introduction
Blood cells, including white blood cells (WBC), red blood cells (RBC) and platelets, participate in vital physiological processes (1,2). Hematological traits are used clinically to detect abnormalities such as reduced oxygen carrying capacity, clotting potential and inflammation that can occur as a result of infection or other causes. Abnormal hematological values occur in several Mendelian diseases and are associated epidemiologically with more common disorders, such as cardiovascular disease (3–6).
Several genome-wide association studies (GWAS) have been conducted in populations with primarily European ancestry, and only a few studies have involved African and Asian ancestry samples. These prior studies have demonstrated that the count and volume of cellular elements in circulating blood are highly heritable and vary widely between individuals (7–11). Taken together, >1000 single-nucleotide polymorphisms (SNPs) have been previously associated with quantitative hematological traits, including blood counts and indices (7,8,12–16).
The degree of overlap between the variants and genes that contribute to variation in hematological traits in different populations is largely unexplored. Genetic drift and selection have contributed to the genetic architecture of these traits in different ancestry groups (16,17). To date, few GWAS or exome-wide association studies have explored complex genetic associations with hematological traits in Han Chinese (18–20). We report an exome-wide association study of hematological traits, focused on exonic variants, in 5257 Han Chinese subjects enrolled from a population cohort in Beijing. We replicated several known but not replicated associations, we conducted replication analyses for novel and previous reported but not replicated associations in independent samples. We identified and replicated an association with a low-frequency, coding variant in the myeloproliferative leukemia (MPL) gene, which encodes for the thrombopoietin (TPO) receptor, and studied its function in vitro to assess its ability to activate the JAK–STAT pathway.
Results
PUUMA Cohort Discovery Study samples
The Peking University–University and Michigan of Atherosclerosis (PUUMA) Study included 5257 subjects with hematological trait data. Hematological traits analyzed were: erythrocyte count (RBC), hemoglobin concentration (Hb), hematocrit (Hct), mean corpuscular hemoglobin (MCH), mean corpuscular hemoglobin concentration (MCHC), mean corpuscular volume (MCV), red cell distribution width coefficient of variation (RDWCV), platelet count, mean platelet volume (MPV), WBC count, count of neutrophil granulocyte (GRAN) and lymphocyte (LYMPH). Trait definitions are provided in Supplementary Material, Table S1. In the discovery PUUMA cohort, 62.0% of participants were female, and mean age was 57.2 years. Further clinical characteristics and trait summaries are presented in Table 1.
Table 1.
Characteristics of participants in the discovery stage
| Characteristics | N (%) or mean ± SD |
|---|---|
| Women (N, %) | 3260 (62.01) |
| Mean age (years) | 57.18 ± 8.97 |
| Current smoker (N, %) | 1041 (19.80) |
| RBC count (1012 cells/l) | 4.26 ± 0.47 |
| Hb (g/l) | 132.48 ± 17.12 |
| Hct (%) | 39.81 ± 4.47 |
| MCH (pg) | 31.02 ± 2.22 |
| MCHC (%) | 331.85 ± 13.16 |
| MCV (fl) | 93.58 ± 5.13 |
| RDWCV | 13.99 ± 0.65 |
| WBC (109 cells/l) | 6.19 ± 1.59 |
| GRAN (109 cells/l) | 3.78 ± 1.24 |
| LYMPH (109 cells/l) | 1.95 ± 0.63 |
| PLT (109 cells/l) | 210.15 ± 49.30 |
| MPV (fl) | 8.10 ± 0.71 |
Values for age and each hematological trait are presented as mean ± SD. PLT, platelet count; SD, standard deviation.
PUUMA Cohort Discovery Stage exome-wide association study
After performing quality control (QC) of the genotypes as described previously (21), we analyzed 85 344 autosomal SNPs meeting QC and with a minor allele count filter of at least 3 in 5257 participants. We conducted an exome-wide association study for each hematological trait, using standard linear regression, adjusted for age, sex and 1–10 principal components (PCs), with lambda genomic control (GC) values 0.94–1.02. Manhattan plots of the exome-wide association studies and quantile–quantile plots in the discovery stage are shown in Supplementary Material, Figure S1. In total, we identified 22 locus–trait associations with minor allele count >40 that reached the Bonferroni-corrected genome-wide significance threshold of P < 5 × 10−7 in the discovery analysis, including two genotype–trait associations identified but not replicated in previous studies and one novel genotype–trait association. The other 19 associations had been previously discovered and replicated.
Previously reported SNP–trait associations within a ± 500 kb window were observed for 21 of the 22 genotype–trait associations identified. The SNP–trait associations in our study were the same as previously reported for 18 of the 21 previously reported associations (Supplementary Material, Table S2). For 3 of the 21 previously reported associations, known SNPs were identified within a ± 500 kb window and had an r2 > 0.3 (Supplementary Material, Table S3). Two of these 21 SNP–trait combinations, rs117656396 and rs78894077 associated with platelet count, were reported very recently but not replicated (16). We additionally identified a novel combination, rs138745983 associated with platelet count, which has not been previously reported (Supplementary Material, Table S4).
The association results of all genotype–trait associations with minor allele count >40 are shown in Supplementary Material, Table S5. The genotype–trait associations with minor allele count >3 and ≤40 are shown in Supplementary Material, Table S6.
Replication of novel SNP–trait associations
Two genotype–trait associations with platelet count (rs117656396 in MPL and rs78894077 in SH2B3) were identified but not replicated in previous studies and one novel genotype–trait association with platelet count (rs138745983 in TVP23A) with minor allele count >40 identified in discovery stage were carried forward to the replication stage. The replication results were shown in Table 2 and Supplementary Material, Table S7. We replicated a nonsynonymous SNP rs117656396 (chromosome 1:43805737-C-T, GRCh37) in MPL, which causes a Leucine at position 265 to be substituted with a Phenylalanine (L265F). This SNP association with platelet count was replicated in 1497 Chinese individuals from two hospital-based studies and 2330 individuals from the Hong Kong Theme-based Research Scheme cohort (21), with similar direction and magnitude of effect. Replication results are shown in Table 2, including the result of meta-analysis of the discovery and replication stage results at the chromosome 1p34.2 locus (rs117656396) in the MPL gene (P = 2.63 × 10−13). The effect size of per one T allele was 20.35 × 109 cells/l. The platelet count in participants with a gene type of CC, CT and TT was 209.5 ± 48.9, 230.5 ± 58.4 and 223.0 ± 58.4 109 cells/l, respectively (Supplementary Material, Table S8). The novel association of rs138745983 in TVP23A with platelet count was not replicated.
Table 2.
Association of the MPL c.793C > T, p.Leu265Phe (L265F) variant with platelet count.
| Chr:positiona | Trait | Gene | Discovery stage | Replication stage | Meta-analysis | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Hospital-based study | Hong Kong cohort | ||||||||||||
| MAF | β | P-value | MAF | β | P-value | MAF | β | P-value | β | P-value | |||
| 1:43805737 | PLT | MPL | 1.55% | 20.35 | 5.64 × 10−8 | 1.57% | 25.28 | 2.87 × 10−3 | 2.55% | 19.52 | 9.21 × 10−5 | 20.63 | 2.63 × 10−13 |
aAssembly version: hg19
In our PUUMA discovery cohort comprised entirely of individuals of Han Chinese ancestry, the minor allele frequency (MAF) for the MPL L265F variant was 1.55%. Among the 1000 Genomes super-populations, the frequency of this variant is highest in the East Asian population (MAF = 2.18%) and monomorphic in South Asian, African, European and American admixed populations. In GnomAD exome sequencing data, the frequency of this variant was 0.72% in Asian and not observed in others, and in GnomAD whole-genome sequencing data, the frequency of this variant in Asian and others were 2.56% and 0.28%, respectively. In silico algorithmic estimation of the effect on the MPL protein showed that variant is predicted to be tolerated by Sorting Intolerant From Tolerant (SIFT) (score = 0.26) (22) and benign by Polyphen2 (23), with Combined Annotation-Dependent Depletion (CADD) score 16.7 (24), indicating that this variant is not strongly deleterious for the predicted gene function. We evaluated linkage disequilibrium (LD) structure in the region around this variant using LDlink to interactively explore proxy and putatively functional variants. This demonstrated an intronic variant rs193129088 in gene PTPRF was in linkage disequilibrium (r2 = 0.75) with rs117656396, the MPL L265F variant. This SNP is not present in our analysis owing to its intronic location, and no association of rs193129088 with platelet count has been reported previously. Previously reported SNPs within 500 kb windows for the significant SNPs are listed in Supplementary Material, Table S9.
Functional analysis of the MPL L265F variant
A somatic activating pathogenic variant of MPL (W515L) has been described in the hematological disease, myelofibrosis with myeloid metaplasia previously (25). We therefore evaluated whether the MPL L265F mutant identified in the current study similarly affects its downstream signaling. We generated and studied MPL L265F-expressing Ba/F3 cells, which exhibited hypersensitivity to TPO as compared with wild-type (WT) MPL-expressing cells as evidenced by elevated ERK1/2 and STAT3 phosphorylation following TPO stimulation. Overexpression of the MPL L265F mutant protein in Ba/F3 cells also enhanced ERK1/2 and STAT3 phosphorylation prior to TPO stimulation (Fig. 1). Herein, we defined L265F as a novel activating pathogenic variant of MPL, consistent with an effect of upregulation of platelet production.
Figure 1.
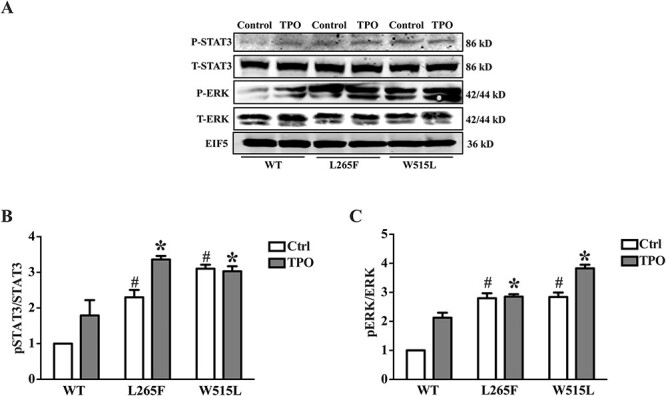
Functional analysis of MPL L265F on pSTAT3/STAT3, pAKT/AKT and pERK/ERK. Ba/F3 cells were infected with lentivirus overexpressing WT MPL, L265F or W515L pathogenic variant. Four days later, infected cells were treated with TPO (5 ng/ml) for 15 min. Representative (A) and quantification (B–C) of western blot is acquired from three independent experiments. Two-way analysis of variance was performed; *PBonferroni < 0.05.
Discussion
In this exome-wide association study of hematological traits in a Han Chinese cohort, we identified 22 locus–trait associations for 12 hematological traits in 5257 Chinese individuals, including an association with a low-frequency coding variant. Replication analysis of rs117656396 in the MPL gene, corresponding to a nonsynonymous variant causing an amino acid residue substitution, L265F, was successfully replicated. Functional analysis of the MPL L265F variant demonstrated a gain of function effect, with downstream activation of the JAK–STAT signaling pathway proposed as the mechanism for the association between MPL L265F genetic variant and platelet count. The novel association of rs138745983 in TVP23A with platelet count was not replicated.
In addition to the JAK2 V617F and CALR pathogenic variants associated with myeloproliferative disorders (MPDs), pathogenic variants of MPL have also been described in these patients (26). Previous GWAS studies have found an association between MPL and platelet count (27–28), an MPL c.117G>T (rs17292650 with MAF 4.34%) variant encoding a p.Lys39Asn amino acid has been reported to be associated with platelet count (27–28). The MPL gene encodes for the TPO receptor, which is known to be involved in myelofibrosis and congenital thrombocytopenia syndromes, both of which are associated with abnormal platelet levels. Pathogenic variants in MPL were also found to be associated with other hematological disease (29–30). MPL has been shown to be reduced or absent in platelets of patients with polycythemia vera and in most patients with idiopathic myelofibrosis (31), consistent with the previous definition of myeloid progenitor cell expression of MPL. Pathogenic variants in the MPL gene have also been reported in congenital amegakaryocytic thrombocytopenia (32). The probability of this gene being intolerant to loss-of-function genetic variation is low, with the loss-of-function intolerance score for this variant was 0 (33), and algorithmic prediction of protein effects in silico supported that the MPL L265F variant is not strongly deleterious for TPO receptor function. Nevertheless, the MPL L265F that we identified impacted cellular signals triggered by TPO in functional studies that have an important role during the proliferation and differentiation of platelets.
To date, the pathogenic variant of MPL that has garnered the most interest is located at protein position 515, with at least four types of W515 pathogenic variants identified in MPD patients: W515L, W515K, W515R, W515A (29–30). Previous investigations of MPL variants in MPD patients have identified associations with other point pathogenic variants (Y591D, S204F) (34). Another pathogenic variant, MPL S505N, has been shown to be the cause of familial thrombocythemia and is associated with a high thrombotic risk (35). Here, we identified a pathogenic variant, MPL L265F associated with a high risk of increased platelet count in a community-based Chinese population exome-wide association study. In the ClinVar database (https://www.ncbi.nlm.nih.gov/clinvar/variation/134832), MPL L265F was determined to likely be benign, but the evidence was not specified.
Activation of the JAK–STAT signal transduction pathway leads to cytokine-independent proliferation in hematopoietic cells and is an important pathogenic event for many MPDs (25,29); MPL W515 pathogenic variants induce thrombocythemia in MPD patients also through the activation of downstream JAK–STAT signaling pathways (25). Thus, we hypothesized that activation of the downstream JAK–STAT signaling pathway was the mechanism behind the association between MPL L265F and platelet count. A functional study utilizing Ba/F3 cells demonstrated that MPL L265F-expressing cells displayed higher phosphorylation levels of ERK1/2 and Stat3 than cells expressing WT MPL. Taken together, these results suggest that activation of the JAK–STAT signal transduction pathway could underlie the association between MPL L265F and platelet count.
A review of reference allele frequency data shows relevant population-specific differences in the MAF for rs117656396 (MPL L265F) (https://gnomad.broadinstitute.org/variant/1-43805737-C-T?). There were no observations of the MPL L265F allele in European or African ancestry populations in GnomAD, and thus, this variant would not have been identified in previous GWAS efforts focused on these populations. In our study of Chinese individuals, the MAF of this variant was 1.55%. In a recently reported large-scale analysis of hematological traits, this variant was identified in an analysis of variants in non-European samples with MAF < 1% (16), consistent with our study’s findings.
In addition to the identification of the association with the MPL variant, we replicated several trait-loci associations described in previous studies (27,36–38) and observed a high degree of concordance of associations across hematological traits, and these traits are known to be highly correlated.
The limitations of this study include that the power to detect rare variant associations was limited because of sample size. Additionally, the coverage of some rare variants, identified through the sequencing of non-Asian samples, which are included on the exome array, was suboptimal for the Chinese population. We attempted to ameliorate this deficiency by incorporating custom content into the Illumina ExomeChip adding Asian-specific non-synonymous SNPs and previously identified SNPs from lipid and blood pressure GWAS. Finally, although we have demonstrated an effect of MPL L265F on JAK–STAT signaling, further experimental studies are needed to define its role in thrombopoiesis or maintenance of platelet counts. We identified a variant within the intron of a neighboring gene to be in LD with the MPL L265F variant, but this variant is not annotated to impact protein structure was not present in GTEx to detect an eQTL effect in that resource and otherwise has not been implicated in regulation of platelet count.
In summary, a MPL variant, L265F was associated with platelet count in Han Chinese. Activation of the JAK–STAT signal transduction pathway may be the mechanism behind this association, and further studies to define the molecular basis of population-level association are warranted.
Materials and Methods
Study population
Frozen genomic DNA samples from the Peking University First Hospital biorepository that qualified for genotyping were used in the discovery stage (N = 5959) in the PUUMA Study. In silico replication resources from Hong Kong, Peking University First Hospital and Peking University Third hospital were used in the replication stage. The study protocol was approved by both Institutional Review Boards and Ethics Committees of Peking University Health Science Center and University of Michigan.
Discovery stage
Subjects from the PUUMA cohort were genotyped for data discovery (N = 5959). This cross-sectional survey on atherosclerosis risk factors was performed at a single research center from December 2011 to April 2012 by trained staff according to a standard protocol (39). In brief, 9540 Han Chinese residents ≥40 years old who lived in the Gucheng and Pingguoyuan communities of the Shijingshan district in Beijing were recruited through recruitment posters or invitation by phone calls. Phone calls were made to individuals who had medical records in community health centers. Samples of insufficient quality for genotyping and those that lacked phenotypic hematological data were excluded from further analysis, leaving 5257 independent subjects for inclusion in the discovery stage analyses.
Replication stage
Several sources were used for in silico replication: (1) a total of 1497 hospital-based samples stored in Peking University First Hospital and Peking University Third Hospital; (2) a total of 2330 samples collected from a Hong Kong population cohort.
Phenotype analysis
For laboratory-based testing and DNA analysis, blood samples were collected by venipuncture of the forearm from each participant after an overnight fast of at least 12 h. Physical examination details, history of disease and medication information were recorded for all participants. For the discovery stage, hematological traits including RBC count, WBC count, platelet count, Hb, Hct, MCH, MCHC, MCV, RDWCV, MPV and WBC subsets (GRAN; LYMPH) were determined using a BC-3000 auto hematology analyzer (Mindray Medical International, Inc. Shenzhou, Guangzhou, China) in the Department of Clinical Laboratory of Gucheng Community Health Service Center. For the replication stage, hematological traits were determined in Department of Clinical Laboratory of the hospitals from which the data were obtained. Phenotypes including age, sex and hematological traits were extracted from existing surveys and medical records databases.
Phenotype transformation
For normalization of traits, a rank-based inverse normal transformation was performed for MPV; natural log transformations were performed for MCH, MCHC, MCV, RDWCV, WBC count, GRAN; square root transformations were performed for RBC count and LYMPH.
Genotyping and QC
All subjects in both discovery and replication stages were genotyped using the Asian Exomechip, a specially designed exome array with a custom content of 58 317 variants on top of the standard Infinium Human Exome BeadChip (Illumina, CA); the details of this custom ExomeChip have been previously described (21). To obtain high-quality genotypes, strict criteria were applied to filter out low quality genotypes. We undertook plate-, individual- and variant-level checks to exclude poor-quality genotype calls from the dataset (21). Briefly, the individual-based QC criteria for filtering included a call rate of <99%, sex mismatch and excess heterozygosity. Passing above QC were 5959 samples. To ensure the independence of the samples, we conducted the Identity by Descent (IBD) analysis to explore the relatedness structures in our samples. Based on the pi-hat distribution, duplicated samples and close relatives were removed. The pi-hat≥0.35 was used as a cut off to remove higher than the first degree (based on the pi-hat distribution, the unbiased estimate is 0.5) of close relatives and duplicates. The sample with the highest call rate was kept for each family group. In total, 5257 participants having phenotypic hematological data were finally included in the analyses after excluding related samples using a greedy algorithm. Variant-level QC was performed to exclude variants with low cluster scores or low call rate (<99.9%) and those that deviated from the Hardy–Weinberg equilibrium (P < 1 × 10−4). Finally, 282 456 markers were retained after QC (21). We analyzed 85 344 SNPs with minor allele count of 3 or greater and meeting QC, using single-variant association tests.
Functional studies of the MPL L265F genetic variant
Expression vectors and cell culture
The retroviral vectors were generously provided by Beijing PREGENE Science and Technology Co., Ltd (Shenzhen, China). The MPL W515L and L265F pathogenic variants were generated using site-directed mutagenesis and confirmed by full-length DNA sequencing.
Western blot analysis
Ba/F3 cells were cultured in RPMI1640 medium including 10% fetal bovine serum and IL-3 (10 ng/ml). Lentivirus infections were performed by adding 100 MOI virus and polybrene (6 μg/ml) into the 70% confluent Ba/F3 cells. The medium was changed 6 h later to remove the virus. Infected cells were used for further experiments 4 days following infection.
Infected cells were activated by TPO (5 ng/ml) for 15 min. Cells were harvested in RIPA lysis buffer. Protein samples were separated by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred to a nitrocellulose membrane. The membranes were blocked using 5% fat-free milk and then incubated with various primary antibodies (1:1000), including rabbit anti-pERK, ERK, pStat3 and Stat3 (Cell Signaling Technology, Danvers, MA), followed with IRDye-conjugated anti-rabbit IgG secondary antibodies (1:10 000, Rockland Inc., Gilbertsville, PA). The protein bands were visualized by an Odyssey infrared imaging system (LICOR Biosciences, Lincoln, NB) and quantified using NIH FIJI software.
Statistical analysis
For both the discovery and replication stages, single-variant association tests were performed using linear regression with PLINK. Age, sex, current smoker (yes or no) and ancestry informative PC 1–10 derived from all common genetic variants were adjusted as covariates for the main hematological traits: RBC count, Hct, Hb, MCHC, MCH, MCV, RDWCV and WBC count as well as the WBC subsets: LYMPH and GRAN. For platelet count and MPV, age, sex and the first 10 PCs were used as covariates. In the discovery stage, for single-variant association testing, we used a threshold of P < 5 × 10−7 corresponding to the Bonferroni correction for 85 344 variants. In the replication stage, we used the significance threshold of P < 0.017 corresponding to the Bonferroni correction for the three variants for which replication was sought.
Data Sharing Statement
The summary statistics will be made available through the GWAS catalog (Accession #GCST90002413). Individual-level data cannot be shared publicly because of regulatory restrictions.
Conflict of Interest statement. None declared.
Supplementary Material
Funding
UM-PUHSC Joint Institute for Translational and Clinical Research (BMU20110177, BMU20160530); Key Laboratory of Molecular Cardiovascular Sciences (Peking University), Ministry of Education to Y.Z; National Health Commission Key Laboratory of Cardiovascular Molecular Biology and Regulatory Peptides to Y.Z; the Fundamental Research Funds for the Central Universities; National Institutes of Health (R01 HL122684 to S.K.G); Hong Kong Research Grant Council: Theme Based Research Scheme (T12-705/11) to C.Y.C.
References
- 1. Weiss, G. and Goodnough, L.T. (2005) Anemia of chronic disease. N. Engl. J. Med., 352, 1011–1023. [DOI] [PubMed] [Google Scholar]
- 2. Ross, R. (1999) Atherosclerosis--an inflammatory disease. N. Engl. J. Med., 340, 115–126. [DOI] [PubMed] [Google Scholar]
- 3. Mohandas, N. and Gallagher, P.G. (2008) Red cell membrane: past, present, and future. Blood, 112, 3939–3948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Patel, K.V., Ferrucci, L., Ershler, W.B., Longo, D.L. and Guralnik, J.M. (2009) Red blood cell distribution width and the risk of death in middle-aged and older adults. Arch. Intern. Med., 169, 515–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Patti, G., Di Martino, G., Ricci, F., Renda, G., Gallina, S., Hamrefors, V., Melander, O., Sutton, R., Engstrom, G., De Caterina, R. et al. (2019) Platelet indices and risk of death and cardiovascular events: results from a large population-based cohort study. Thromb. Haemost., 119, 1773–1784. [DOI] [PubMed] [Google Scholar]
- 6. Santos-Silva, A., Castro, E.M., Teixeira, N.A., Guerra, F.C. and Quintanilha, A. (1995) Altered erythrocyte membrane band 3 profile as a marker in patients at risk for cardiovascular disease. Atherosclerosis, 116, 199–209. [DOI] [PubMed] [Google Scholar]
- 7. van der Harst, P., Zhang, W., Mateo, L.I., Rendon, A., Verweij, N., Sehmi, J., Paul, D.S., Elling, U., Allayee, H., Li, X. et al. (2012) Seventy-five genetic loci influencing the human red blood cell. Nature, 492, 369–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hodonsky, C.J., Jain, D., Schick, U.M., Morrison, J.V., Brown, L., McHugh, C.P., Schurmann, C., Chen, D.D., Liu, Y.M., Auer, P.L. et al. (2017) Genome-wide association study of red blood cell traits in Hispanics/Latinos: the Hispanic community health study/study of Latinos. PLoS Genet., 13, e1006760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Schick, U.M., Jain, D., Hodonsky, C.J., Morrison, J.V., Davis, J.P., Brown, L., Sofer, T., Conomos, M.P., Schurmann, C., McHugh, C.P. et al. (2016) Genome-wide association study of platelet count identifies ancestry-specific loci in Hispanic/Latino Americans. Am. J. Hum. Genet., 98, 229–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Shameer, K., Denny, J.C., Ding, K., Jouni, H., Crosslin, D.R., de Andrade, M., Chute, C.G., Peissig, P., Pacheco, J.A., Li, R. et al. (2014) A genome- and phenome-wide association study to identify genetic variants influencing platelet count and volume and their pleiotropic effects. Hum. Genet., 133, 95–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Qayyum, R., Snively, B.M., Ziv, E., Nalls, M.A., Liu, Y., Tang, W., Yanek, L.R., Lange, L., Evans, M.K., Ganesh, S. et al. (2012) A meta-analysis and genome-wide association study of platelet count and mean platelet volume in African Americans. PLoS Genet., 8, e1002491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Astle, W.J., Elding, H., Jiang, T., Allen, D., Ruklisa, D., Mann, A.L., Mead, D., Bouman, H., Riveros-Mckay, F., Kostadima, M.A. et al. (2016) The allelic landscape of human blood cell trait variation and links to common complex disease. Cell, 167, 1415–1429.e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kamatani, Y., Matsuda, K., Okada, Y., Kubo, M., Hosono, N., Daigo, Y., Nakamura, Y. and Kamatani, N. (2010) Genome-wide association study of hematological and biochemical traits in a Japanese population. Nat. Genet., 42, 210–215. [DOI] [PubMed] [Google Scholar]
- 14. Ganesh, S.K., Zakai, N.A., van Rooij, F.J., Soranzo, N., Smith, A.V., Nalls, M.A., Chen, M.H., Kottgen, A., Glazer, N.L., Dehghan, A. et al. (2009) Multiple loci influence erythrocyte phenotypes in the CHARGE consortium. Nat. Genet., 41, 1191–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Soranzo, N., Spector, T.D., Mangino, M., Kuhnel, B., Rendon, A., Teumer, A., Willenborg, C., Wright, B., Chen, L., Li, M. et al. (2009) A genome-wide meta-analysis identifies 22 loci associated with eight hematological parameters in the HaemGen consortium. Nat. Genet., 41, 1182–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chen, M.H., Raffield, L.M., Mousas, A., Sakaue, S., Huffman, J.E., Moscati, A., Trivedi, B., Jiang, T., Akbari, P., Vuckovic, D. et al. (2020) Trans-ethnic and ancestry-specific blood-cell genetics in 746,667 individuals from 5 global populations. Cell, 182, 1198–1213.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Scheinfeldt, L.B. and Tishkoff, S.A. (2013) Recent human adaptation: genomic approaches, interpretation and insights. Nat. Rev. Genet, 14, 692–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. van Rooij, F., Qayyum, R., Smith, A.V., Zhou, Y., Trompet, S., Tanaka, T., Keller, M.F., Chang, L.C., Schmidt, H., Yang, M.L. et al. (2017) Genome-wide trans-ethnic meta-analysis identifies seven genetic loci influencing erythrocyte traits and a role for RBPMS in erythropoiesis. Am. J. Hum. Genet., 100, 51–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Andiappan, A.K., Melchiotti, R., Poh, T.Y., Nah, M., Puan, K.J., Vigano, E., Haase, D., Yusof, N., San, L.B., Lum, J. et al. (2015) Genome-wide analysis of the genetic regulation of gene expression in human neutrophils. Nat. Commun., 6, 7971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Keller, M.F., Reiner, A.P., Okada, Y., van Rooij, F.J., Johnson, A.D., Chen, M.H., Smith, A.V., Morris, A.P., Tanaka, T., Ferrucci, L. et al. (2014) Trans-ethnic meta-analysis of white blood cell phenotypes. Hum. Mol. Genet., 23, 6944–6960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tang, C.S., Zhang, H., Cheung, C.Y., Xu, M., Ho, J.C., Zhou, W., Cherny, S.S., Zhang, Y., Holmen, O., Au, K.W. et al. (2015) Exome-wide association analysis reveals novel coding sequence variants associated with lipid traits in Chinese. Nat. Commun., 6, 10206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sim, N.L., Kumar, P., Hu, J., Henikoff, S., Schneider, G. and Ng, P.C. (2012) SIFT web server: predicting effects of amino acid substitutions on proteins. Nucleic Acids Res., 40, W452–W457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Adzhubei, I.A., Schmidt, S., Peshkin, L., Ramensky, V.E., Gerasimova, A., Bork, P., Kondrashov, A.S. and Sunyaev, S.R. (2010) A method and server for predicting damaging missense mutations. Nat. Methods, 7, 248–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kircher, M., Witten, D.M., Jain, P., O'Roak, B.J., Cooper, G.M. and Shendure, J. (2014) A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet., 46, 310–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Pikman, Y., Lee, B.H., Mercher, T., McDowell, E., Ebert, B.L., Gozo, M., Cuker, A., Wernig, G., Moore, S., Galinsky, I. et al. (2006) MPLW515L is a novel somatic activating mutation in myelofibrosis with myeloid metaplasia. PLoS Med., 3, e270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Guglielmelli, P., Barosi, G., Specchia, G., Rambaldi, A., Lo, C.F., Antonioli, E., Pieri, L., Pancrazzi, A., Ponziani, V., Delaini, F. et al. (2009) Identification of patients with poorer survival in primary myelofibrosis based on the burden of JAK2V617F mutated allele. Blood, 114, 1477–1483. [DOI] [PubMed] [Google Scholar]
- 27. Eicher, J.D., Chami, N., Kacprowski, T., Nomura, A., Chen, M.H., Yanek, L.R., Tajuddin, S.M., Schick, U.M., Slater, A.J., Pankratz, N. et al. (2016) Platelet-related variants identified by Exomechip meta-analysis in 157,293 individuals. Am. J. Hum. Genet., 99, 40–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Auer, P.L., Johnsen, J.M., Johnson, A.D., Logsdon, B.A., Lange, L.A., Nalls, M.A., Zhang, G., Franceschini, N., Fox, K., Lange, E.M. et al. (2012) Imputation of exome sequence variants into population- based samples and blood-cell-trait-associated loci in African Americans: NHLBI GO Exome Sequencing Project. Am. J. Hum. Genet., 91, 794–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ma, W., Zhang, X., Wang, X., Zhang, Z., Yeh, C.H., Uyeji, J. and Albitar, M. (2011) MPL mutation profile in JAK2 mutation-negative patients with myeloproliferative disorders. Diagn. Mol. Pathol., 20, 34–39. [DOI] [PubMed] [Google Scholar]
- 30. Levine, R.L., Belisle, C., Wadleigh, M., Zahrieh, D., Lee, S., Chagnon, P., Gilliland, D.G. and Busque, L. (2006) X-inactivation-based clonality analysis and quantitative JAK2V617F assessment reveal a strong association between clonality and JAK2V617F in PV but not ET/MMM, and identifies a subset of JAK2V617F-negative ET and MMM patients with clonal hematopoiesis. Blood, 107, 4139–4141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Moliterno, A.R., Hankins, W.D. and Spivak, J.L. (1998) Impaired expression of the thrombopoietin receptor by platelets from patients with polycythemia vera. N. Engl. J. Med., 338, 572–580. [DOI] [PubMed] [Google Scholar]
- 32. Ballmaier, M., Germeshausen, M., Schulze, H., Cherkaoui, K., Lang, S., Gaudig, A., Krukemeier, S., Eilers, M., Strauss, G. and Welte, K. (2001) C-mpl mutations are the cause of congenital amegakaryocytic thrombocytopenia. Blood, 97, 139–146. [DOI] [PubMed] [Google Scholar]
- 33. Lek, M., Karczewski, K.J., Minikel, E.V., Samocha, K.E., Banks, E., Fennell, T., O'Donnell-Luria, A.H., Ware, J.S., Hill, A.J., Cummings, B.B. et al. (2016) Analysis of protein-coding genetic variation in 60,706 humans. Nature, 536, 285–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kawamata, N., Ogawa, S., Yamamoto, G., Lehmann, S., Levine, R.L., Pikman, Y., Nannya, Y., Sanada, M., Miller, C.W., Gilliland, D.G. et al. (2008) Genetic profiling of myeloproliferative disorders by single-nucleotide polymorphism oligonucleotide microarray. Exp. Hematol., 36, 1471–1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Liu, K., Martini, M., Rocca, B., Amos, C.I., Teofili, L., Giona, F., Ding, J., Komatsu, H., Larocca, L.M. and Skoda, R.C. (2009) Evidence for a founder effect of the MPL-S505N mutation in eight Italian pedigrees with hereditary thrombocythemia. Haematologica, 94, 1368–1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Tajuddin, S.M., Schick, U.M., Eicher, J.D., Chami, N., Giri, A., Brody, J.A., Hill, W.D., Kacprowski, T., Li, J., Lyytikainen, L.P. et al. (2016) Large-scale exome-wide association analysis identifies loci for white blood cell traits and pleiotropy with immune-mediated diseases. Am. J. Hum. Genet., 99, 22–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Chami, N., Chen, M.H., Slater, A.J., Eicher, J.D., Evangelou, E., Tajuddin, S.M., Love-Gregory, L., Kacprowski, T., Schick, U.M., Nomura, A. et al. (2016) Exome genotyping identifies pleiotropic variants associated with red blood cell traits. Am. J. Hum. Genet., 99, 8–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Mousas, A., Ntritsos, G., Chen, M.H., Song, C., Huffman, J.E., Tzoulaki, I., Elliott, P., Psaty, B.M., Auer, P.L., Johnson, A.D. et al. (2017) Rare coding variants pinpoint genes that control human hematological traits. PLoS Genet., 13, e1006925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Fan, F., Qi, L., Jia, J., Xu, X., Liu, Y., Yang, Y., Qin, X., Li, J., Li, H., Zhang, Y. et al. (2016) Noninvasive central systolic blood pressure is more strongly related to kidney function decline than peripheral systolic blood pressure in a Chinese community-based population. Hypertension, 67, 1166–1172. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


