Abstract
BACKGROUND
Sporadic arteriovenous malformations of the brain, which are morphologically abnormal connections between arteries and veins in the brain vasculature, are a leading cause of hemorrhagic stroke in young adults and children. The genetic cause of this rare focal disorder is unknown.
METHODS
We analyzed tissue and blood samples from patients with arteriovenous malformations of the brain to detect somatic mutations. We performed exome DNA sequencing of tissue samples of arteriovenous malformations of the brain from 26 patients in the main study group and of paired blood samples from 17 of those patients. To confirm our findings, we performed droplet digital polymerase-chain-reaction (PCR) analysis of tissue samples from 39 patients in the main study group (21 with matching blood samples) and from 33 patients in an independent validation group. We interrogated the downstream signaling pathways, changes in gene expression, and cellular phenotype that were induced by activating KRAS mutations, which we had discovered in tissue samples.
RESULTS
We detected somatic activating KRAS mutations in tissue samples from 45 of the 72 patients and in none of the 21 paired blood samples. In endothelial cell– enriched cultures derived from arteriovenous malformations of the brain, we detected KRAS mutations and observed that expression of mutant KRAS (KRASG12V) in endothelial cells in vitro induced increased ERK (extracellular signal-regulated kinase) activity, increased expression of genes related to angiogenesis and Notch signaling, and enhanced migratory behavior. These processes were reversed by inhibition of MAPK (mitogen-activated protein kinase)–ERK signaling.
CONCLUSIONS
We identified activating KRAS mutations in the majority of tissue samples of arteriovenous malformations of the brain that we analyzed. We propose that these malformations develop as a result of KRAS-induced activation of the MAPK–ERK signaling pathway in brain endothelial cells. (Funded by the Swiss Cancer League and others.)
ARTERIOVENOUS MALFORMATIONS OF the brain are high-flow vascular malformations that occur in approximately 15 per 100,000 persons and cause hemorrhagic stroke in children.1–4 They are tortuous, morphologically abnormal vascular channels between arteries and veins that lack an intervening capillary network, allowing high-pressure arterial blood from feeding arteries to shunt directly into the venous outflow system. The underlying cause of sporadic arteriovenous malformations of the brain is unknown, but similar lesions are found in rare genetic syndromes, such as hereditary hemorrhagic telangiectasias (a group of disorders caused by inactivating germline mutations in regulators of the transforming growth factor β–SMAD pathway5–7), and in the capillary malformation–arteriovenous malformation syndrome (a RASopathy that is caused by inactivating mutations of RASA1 or EPHB48,9). This pattern indicates that a genetic cause may underlie the development of arteriovenous malformations of the brain, but most of these malformations occur as sporadic lesions in persons without a family history of the disease. We hypothesized that arteriovenous malformations of the brain arise from somatic mutations in the cranial vasculature.
METHODS
STUDY PATIENTS
Patients were eligible for inclusion in the study if they had sporadic, unifocal arteriovenous malformations of the brain with a defined nidus and evidence of arteriovenous shunting on digital subtraction angiography. In addition, patients had to have no family history of arteriovenous malformations and no documented history of genetic vascular disease. Clinical and surgical information was obtained from patients’ charts by the authorized study team. All diagnoses of arteriovenous malformations of the brain were confirmed by the study pathologists, radiologists, and surgeons (see Supplementary Appendix 1, available with the full text of this article at NEJM.org). All the patients, who were 18 years of age or older, provided written informed consent.
STUDY PROCEDURES, OUTCOMES, AND OVERSIGHT
The main study group comprised patients from Canada. Tissue samples of arteriovenous malformations of the brain and blood samples were obtained from these patients and deidentified, flash-frozen, and stored at Toronto Western Hospital, University Health Network, Toronto. Whole-exome sequencing was performed to detect somatic mutations. The results of DNA sequencing were confirmed with the use of droplet digital polymerase-chain-reaction (PCR) analysis. All the procedures performed with the use of samples obtained from patients were approved by the Institutional Research Ethics Review Board of the University Health Network. Details about the immunohistochemical analyses of the tissue samples are provided in Supplementary Appendix 1.
The independent validation group comprised patients from Finland. DNA samples were obtained from formalin-fixed, paraffin-embedded tissues of arteriovenous malformations of the brain from these patients. The samples were treated with a DNA glycosylase to reduce the risk of false positives on droplet digital PCR analysis, as described by Do and Dobrovic.10 Droplet digital PCR analysis was performed to detect KRAS mutations. All the procedures performed with the use of samples obtained from patients were approved by the Research Ethics Board at the University of Eastern Finland. Additional information regarding the tissue samples that were obtained from the patients is provided in Supplementary Appendix 1.
CELLULAR AND MOLECULAR STUDIES
Freshly resected tissue samples of arteriovenous malformations of the brain or control samples of normal tissue from temporal lobectomy specimens were obtained for the main study group from Toronto Western Hospital. Endothelial-cell cultures were established according to a modification of methods that had been described previously11 and were enriched and depleted with the use of anti-CD31 magnetic beads. Cell cultures were isolated from freshly resected tissue. Frozen tissues were used for exome sequencing and digital droplet PCR assays. The cell cultures were used for the digital droplet PCR assays and Western blotting. Details about the cellular and molecular studies to model mutant KRAS expression in cultured human cells are provided in Supplementary Appendix 1.
STATISTICAL ANALYSIS
Pairwise comparisons were made with the use of Student’s t-test. Comparison of three or more groups was performed with the use of a one-way analysis of variance with the Newman–Keuls post hoc test. A P value of 0.05 or less was considered to indicate statistical significance. The Q value is the adjusted P value that is found by means of an optimized false discovery rate approach, and the r value is the linear correlation coefficient.
RESULTS
PATIENTS
From January 2013 through October 2017, a total of 39 patients were included in the main study group and had samples of arteriovenous malformation stored in the neurosurgical tissue bank. Of those, 21 had matched blood samples and 6 had fresh arteriovenous-malformation tissue that was used to derive cell culture. In addition, fresh vessel samples that had been obtained from 3 patients undergoing temporal-lobe resection for epilepsy were included. Tissues from 2 patients with cavernous malformations and 1 with dural arteriovenous fistula as well as 2 samples of normal cortical vessels and 1 sample of gliosis tissue were included as controls. In the Finnish validation group, archived paraffin-embedded samples that had been obtained from 33 patients with arteriovenous malformations of the brain were included; 54 samples of arteries from the circle of Willis, 18 samples of cavernous malformations, and 2 samples of dural arteriovenous fistulas were also obtained for the control group.
DETECTION OF SOMATIC ACTIVATING KRAS MUTATIONS
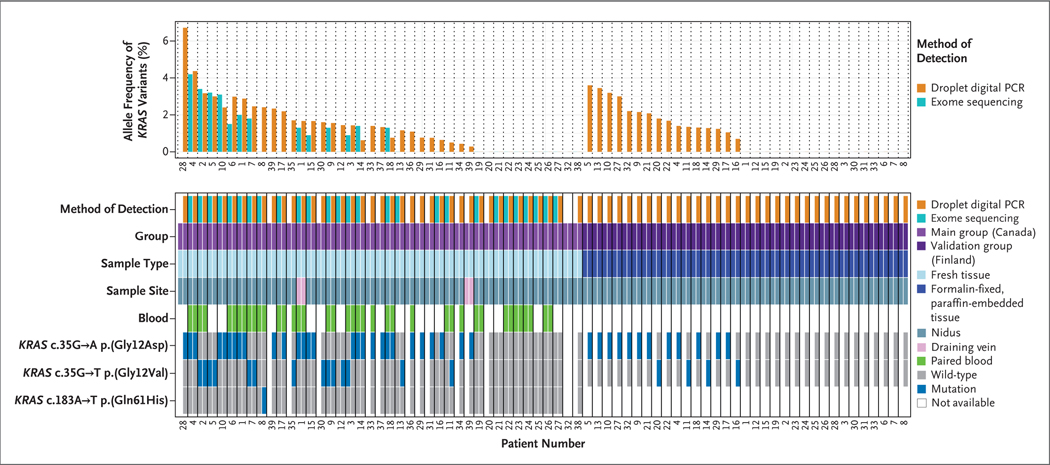
In the main study group, whole-exome sequencing was performed on tissue samples of arteriovenous malformations of the brain that were obtained from 26 patients. Using the default criteria of MuTect2 variant caller, we detected six somatic missense variants in three genes, including two variants of unknown clinical significance (one in PCSK5 and the other in TP53BP1) (see the Supplemental Methods Section in Supplementary Appendix 1). The other four identified variants mapped to the same genomic coordinate in KRAS (chr12:25398284), which corresponded to a c.35G→A (p.Gly12Asp) mutation (in Patients 1, 4, and 10) or a c.35G→T (p.Gly12Val) mutation (in Patient 2). These variants were present in 2.4 to 4.0% of the sequence reads per sample (mean sequence coverage, 339±64×). On analysis of the 17 paired blood samples, the exomes had good coverage for this site (mean sequence coverage, 121±41×); no sequence reads contained a variant (Fig. 1). On further analysis that was performed with the use of relaxed criteria (the presence of variants in >0.5% of sequence reads, with reads on both DNA strands; see the Supplemental Methods Section in Supplementary Appendix 1), KRAS mutations were present in tissue samples obtained from 12 of the 26 patients and in none of the 17 paired blood samples (Fig. 1). KRAS mutations were present in 0.9 to 4.1% of the sequence reads. Activating KRAS mutations were found at codon 12: the c.35G→T (p.Gly12Val) mutation was found in 4 patients and the c.35G→A (p.Gly12Asp) mutation in 8 patients, including 1 patient (Patient 1) in whom the nidus sample and the draining-vein sample contained the c.35G→A (p.Gly12Asp) mutation in 2.0% and 1.3% of the sequence reads, respectively. These mutations are known to confer constitutive activity on KRAS.12 (Details of the analyses are shown in Table S1 in Supplementary Appendix 2 and Tables S2 and S3 and Fig. S1 in Supplementary Appendix 1.)
Figure 1. Detection of KRAS Mutations in Samples Obtained from Patients.
The top chart shows the allele frequency of KRAS variants, determined by either the percentage of sequence reads that contained variants on whole-exome sequencing or the fractional abundance of variants on digital droplet polymerase-chain-reaction (PCR) analysis, in tissue samples of arteriovenous malformations of the brain. The samples are shown according to patient number in order of highest to lowest frequency, first in the main study group, which included 39 patients from Canada (29 [74%] with KRAS mutations), and then in the independent validation group, which included 33 patients from Finland (16 [48%] with KRAS mutations). The bottom chart shows details about the samples, including the sample type (fresh-frozen or formalin-fixed, paraffin-embedded tissue), sample site (nidus or draining vein), the presence or absence of a paired blood sample, and the specific activating mutation detected. No KRAS mutations were detected in paired blood samples. The CD31+ cells from Patient 32 in the main study group were positive for a c.35G→A KRAS mutation (data not shown in this figure), but there was insufficient tissue remaining for whole tissue analysis. Two patients from the main study group (Patients 1 and 39) had KRAS mutations in the draining-vein sample that matched those observed in the nidus sample.
CONFIRMATION OF KRAS MUTATIONS
To confirm the results of whole-exome sequencing and to detect additional KRAS variants that may have been missed because of low representation in the sample, we performed droplet digital PCR analysis of the tissue samples obtained from the 26 patients (primer sequences are shown in Table S4 in Supplementary Appendix 1). We detected 12 putative KRAS mutations that had been detected previously on whole-exome sequencing and 6 KRAS mutations in previously negative samples (Fig. 1). We also tested 13 additional tissue samples, 4 of which had matching blood samples, with the use of droplet digital PCR analysis only; we detected the c.35G→A (p.Gly12Asp) mutation in 9 of these samples and the c.35G→T (p.Gly12Val) mutation in 2 of them (Fig. 1). The other 2 samples were nonmutated. Altogether, droplet digital PCR analysis revealed KRAS mutations in tissue samples from 29 of the 39 patients: 19 samples had the c.35G→A (p.Gly12Asp) mutation, 9 had the c.35G→T (p.Gly12Val) mutation, and 1 had the c.183A→T (p.Gln61His) mutation. The fractional abundance of variants in the unpurified tissue samples ranged from 0.43 to 4.37% (Fig. 1), a finding consistent with the results of whole-exome sequencing (r = 0.72). The 21 paired blood samples were negative for KRAS mutations (fractional abundance, 0%; positive droplets, 0). Moreover, droplet digital PCR analysis of 3 samples from patients with vascular lesions of the brain that were not arteriovenous malformations (2 of cavernous malformations of the brain and 1 of a draining vein of a spinal dural arteriovenous fistula), 2 samples of normal cortical vessels, and 1 sample of gliosis tissue surrounding a cavernous malformation were negative for KRAS variants (fractional abundance, 0%; positive droplets, 0). (Details of the analyses are shown in Fig. S2 and in Tables S3 and S6 in Supplementary Appendix 1 and Table S5 in Supplementary Appendix 2.)
DETECTION OF KRAS MUTATIONS IN AN INDEPENDENT VALIDATION GROUP
The results in the main study group were validated in an independent validation group that involved 33 Finnish patients who had arteriovenous malformations of the brain. Genomic DNA was extracted from formalin-fixed, paraffin-embedded blocks of surgically resected tissue samples of arteriovenous malformations of the brain, and the DNA was analyzed with the use of droplet digital PCR analysis for the c.35G→A (p.Gly12Asp) and c.35G→T (p.Gly12Val) mutations in KRAS. To evaluate the level of sequence artifacts for these variants in DNA isolated from formalin-fixed, paraffin-embedded tissue,10 we analyzed formalin-fixed, paraffin-embedded tissue samples from normal intracranial arteries and from vascular lesions of the brain that were not arteriovenous malformations: 54 samples of cadaveric brain arteries from the circle of Willis, 18 samples of cavernous malformations, and 2 samples of dural arteriovenous fistulas. The false positives in these samples did not exceed a fractional abundance of 0.52% for the c.35G→A (p.Gly12Asp) mutation and 0.14% for the c.35G→T (p.Gly12Val) mutation (mean fractional abundance, 0.25±0.10%). Taking this into account, we selected a threshold for fractional abundance of 1.00% for the c.35G→A (p.Gly12Asp) mutation and 0.50% for the c.35G→T (p.Gly12Val) mutation, with a requirement of at least 5 positive droplets for either variant.
Droplet digital PCR analysis of 33 tissue samples showed that the fractional abundance of KRAS variants exceeded the thresholds in 16 samples (48%). In findings that were similar to the results in the main study group, the fractional abundance of KRAS variants was low in the independent validation group (0.70 to 3.60%) (Fig. 1).
In formalin-fixed, paraffin-embedded tissue samples, false positives on droplet digital PCR analysis for the c.35G→A (p.Gly12Asp) mutation can result from deamination of cytosine to uracil. To reduce the risk of false positives, all the tissue samples that had been positive for the c.35G→A (p.Gly12Asp) mutation in KRAS and some samples of arteries from the circle of Willis were treated with uracil DNA glycosylase. We observed a reduction in fractional abundance in the samples of arteries from the circle of Willis but no reduction in the samples of brain arteriovenous-malformation tissue that had been positive for the KRAS variant. In addition to paraffin-embedded tissue, fresh-frozen tissue samples of arteriovenous malformations of the brain were available in three patients in the independent validation group (Patients 2, 3, and 10). The droplet digital PCR results that were obtained from fresh-frozen tissue samples were similar to the results that were obtained with the use of paraffin-embedded tissue obtained from the same patients. (Details of the analyses are shown in Fig. S3 and in Tables S3 and S8 in Supplementary Appendix 1 and Table S7 in Supplementary Appendix 2.)
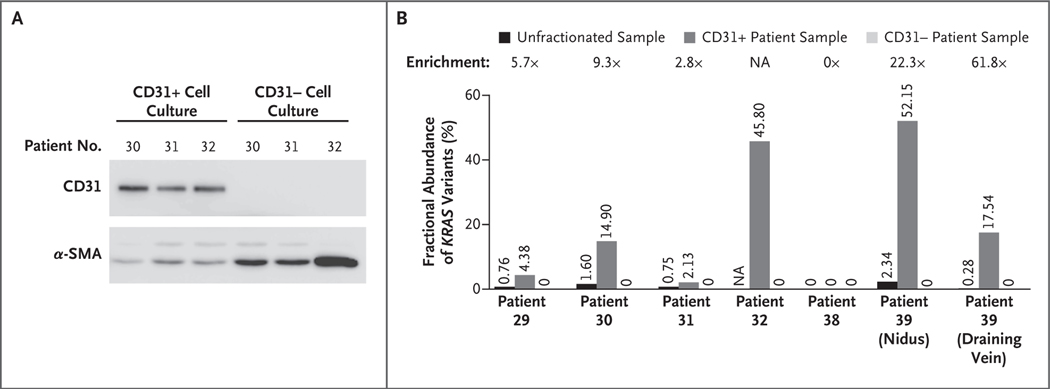
DETECTION OF KRAS MUTATIONS IN ENDOTHELIAL CELLS
To identify the types of cells that have KRAS mutations in arteriovenous malformations of the brain, we used magnetic-activated cell sorting to enrich and deplete endothelial cells from six fresh cell cultures derived from arteriovenous malformations of the brain (from Patients 29, 30, 31, 32, 38, and 39) and to enrich and deplete endothelial cells from three control cultures of normal vascular cells derived from cortical vessels (from three different patients undergoing temporal lobectomy for epilepsy). The endothelial-cell–enriched fractions of the cultures were, as expected, enriched with CD31+ cells (CD31 is an endothelial marker) (Fig. 2A); however, these fractions retained some cells that expressed alpha smooth-muscle actin (α-SMA). In contrast, the endothelial-cell–depleted fractions did not have cells that expressed CD31 and were highly enriched with α-SMA+ cells (Fig. 2A). Droplet digital PCR analysis showed that the CD31+ cultures derived from five of the six samples obtained from patients were positive for the c.35G→A (p.Gly12Asp) or c.35G→T (p.Gly12Val) mutation in KRAS. Those cultures also had a fractional abundance of variants that was 2.8 to 61.8 times as high as the level in whole tissue before fractionation, reaching 14.90% in Patient 30, 45.80% in Patient 32 (for whom whole unfractionated tissue was not available owing to the small amount of initial material), and 52.15% and 17.54% in the nidus and draining-vein samples, respectively, from Patient 39 (Fig. 2B).
Figure 2. Detection of KRAS Mutations in Endothelial-Cell–Enriched and Endothelial-Cell–Depleted Cultures.
Panel A shows the expression of CD31 and alpha smooth-muscle actin (α-SMA) in endothelial-cell–enriched (CD31+) and endothelial-cell–depleted (CD31−) fractions of cell cultures derived from tissue samples of arteriovenous malformations of the brain that were obtained from patients. The CD31+ fractions (isolated with anti-CD31 magnetic beads) were composed of CD31+ cells and some α-SMA+ cells, whereas the CD31− fractions were devoid of CD31+ cells but contained α-SMA+ cells. Panel B shows the fractional abundance of KRAS variants in CD31+ and CD31− cell cultures derived from tissue samples of arteriovenous malformations of the brain and in the whole tissue sample before fractionation. Enrichment is the factor change of the fractional abundance in the CD31+ cells as compared with the whole unfractionated sample. The samples are from patients in the main study group. For Patient 32, all available tissue was used for cell culture and no tissue was left for digital droplet polymerase-chain-reaction analysis. NA denotes not available.
In some samples, the fractional abundance of variants in the CD31+ cultures correlated with the percentage of CD31+ cells seen on immunostaining of low-passage (passage 1 to 2) CD31+ cultures: the nidus sample from Patient 32 (fractional abundance, 45.80% [i.e., >90% mutant KRAS cells]) had 87% CD31+ cells, and the draining-vein sample from Patient 39 (fractional abundance, 17.54% [i.e., >30% mutant KRAS cells]) had 30% CD31+ cells. These findings suggest that most endothelial cells derived from these samples were positive for KRAS mutations. In contrast, the CD31− cultures from the same samples were negative for KRAS mutations (Fig. 2B). Droplet digital PCR analysis of the control cell cultures did not reveal KRAS mutations in CD31+ or CD31− fractions. (Details of the analyses are shown in Figs. S2 and S4 and in Table S9 in Supplementary Appendix 1.)
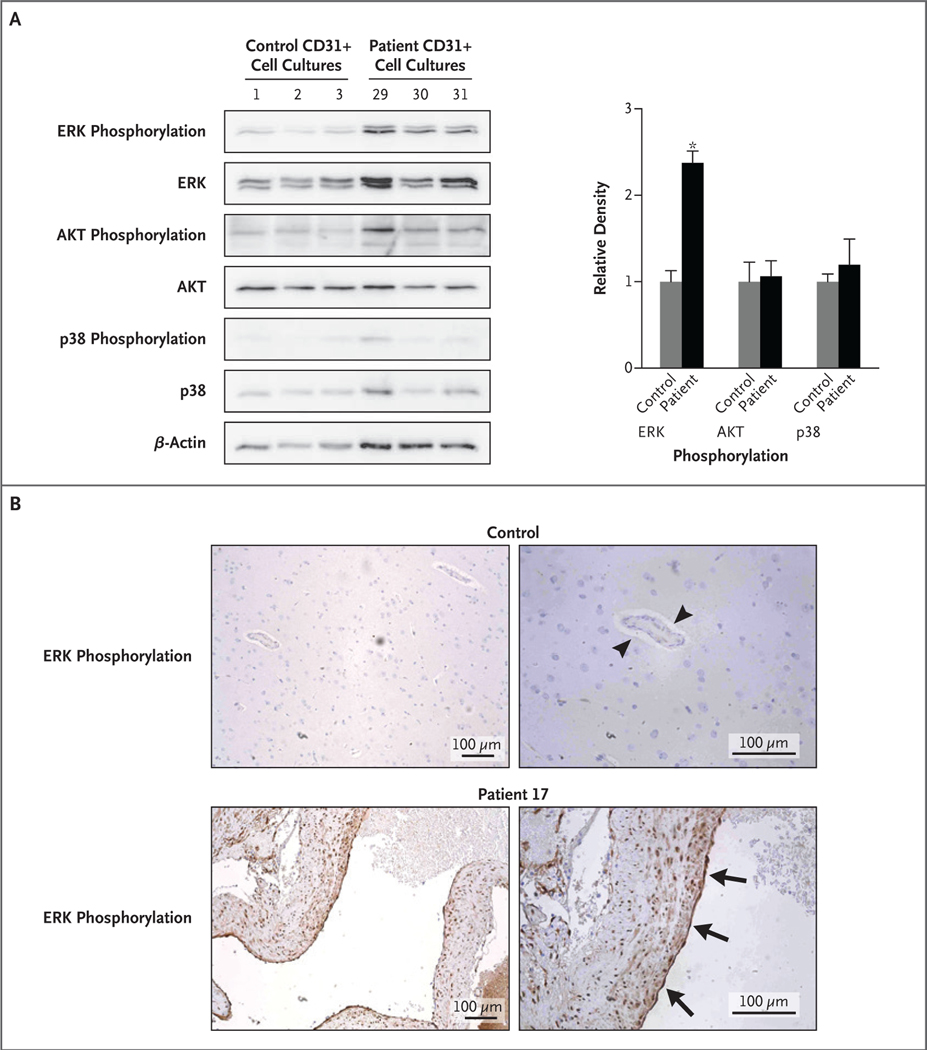
ERK1/2 ACTIVATION IN ENDOTHELIAL CELLS WITH KRAS MUTATIONS
KRAS is an effector molecule that lies downstream of receptor tyrosine kinases, and it activates diverse cellular signaling networks, such as the RAF–MEK (mitogen-activated protein kinase [MAPK] kinase)–ERK (extracellular signal-regulated kinase) (MAPK–ERK) signaling pathway and the PI3K (phosphoinositide 3 kinase)–AKT–mTOR (mechanistic target of rapamycin) pathway.12 We tested for phosphorylation of ERK1/2, AKT, and a noncanonical KRAS target, p38 MAPK,13 in endothelial cell–enriched cultures derived from arteriovenous malformations of the brain with KRAS mutations and in human umbilical-vein endothelial cells (HUVECs) that expressed one of two isoforms (proteins encoded by differently spliced messenger RNA) of mutant KRASG12V: KRAS4AG12V or KRAS4BG12V. The results for these cell cultures were compared with the results for primary endothelial cell–enriched cultures derived from normal brain vessels or HUVECs that received an empty expression construct. In both experiments, we observed increased levels of ERK1/2 phosphorylation in cells expressing mutant KRAS (Fig. 3A, and Fig. S5 in Supplementary Appendix 1) but did not observe increased levels of AKT or p38 phosphorylation. These findings suggest that mutant KRAS specifically activates the MAPK–ERK pathway in endothelial cells. Moreover, immunohistochemical staining of 25 tissue samples of arteriovenous malformations of the brain that were obtained from patients with arteriovenous malformations in the main study group and of 3 control samples of normal brain tissues was performed. There was strong staining for ERK phosphorylation in endothelial cells in the samples obtained from patients, regardless of the presence of a KRAS mutation, and little or no staining for ERK phosphorylation in the control samples (Fig. 3B).
Figure 3. Detection of ERK Phosphorylation in Endothelial-Cell–Enriched Cell Cultures and Tissue Samples.
In Panel A, endothelial-cell–enriched (CD31+) cell cultures derived from tissue samples of arteriovenous malformations of the brain with KRAS mutations (from Patients 29, 30, and 31 in the main study group) show increased phosphorylation of ERK1/2 (extracellular signal-regulated kinase 1 or 2) but not of p38 or AKT, whereas three control CD31+ cell cultures derived from normal brain vasculature do not show increased phosphorylation of ERK1/2. Densitometry is shown on the right, normalized to beta-actin. The asterisk indicates a P value of 0.002. In Panel B, immunohistochemical staining of a tissue sample of an arteriovenous malformation of the brain with a KRAS mutation (from Patient 17 in the main study group) shows strong staining for ERK phosphorylation in endothelial cells lining the vascular lumen (arrows) and in vascular smooth-muscle cells in the vessel wall, whereas normal brain parenchymal vessels show little or no staining for ERK phosphorylation in endothelial cells (arrowheads).
PHENOTYPE OF ENDOTHELIAL CELLS WITH KRAS MUTATIONS
We performed RNA sequencing of HUVECs that expressed KRAS4AG12V, KRAS4BG12V, or control plasmid under growth factor–starved and serum-starved conditions. Assessment of categories of gene ontology that were altered in endothelial cells with KRASG12V expression revealed an enrichment of genes involved in the categories of angiogenesis or vascular development, proliferation, and migration. We also observed up-regulation of genes in the Notch pathway (e.g., DLL4, JAG1, JAG2, NOTCH1, HES1, and HEY2), which are involved in angiogenesis and arteriovenous specification14,15 and have been implicated in the pathogenesis of arteriovenous malformations.16 Arterial specification markers (e.g., NRP1 and EFNB2) and venous specification markers (e.g., EPHB4 and NR2F2) were unchanged. The expression of genes that are known to have a role in angiogenesis (e.g., VEGFA, VEGFC, DUSP5, and HLX) was also induced, as was the expression of genes that are characteristic of the endothelial-to-mesenchymal transition (e.g., SNAI1, SNAI2, ZEB1, and PCDH1), a process implicated in other vascular malformations.17 These results imply that active KRAS dysregulates angiogenesis and vascular remodeling in endothelial cells. (For details, see Fig. S6 in Supplementary Appendix 1.)
The angiogenic factors vascular endothelial growth factor (VEGF) and fibroblast growth factor (FGF) activate intracellular signaling in a RAS-dependent manner.18,19 Gene set enrichment analysis to compare the dysregulated genes in HUVECs that express KRASG12V with those in HUVECs that have been stimulated with VEGF20 showed a robust correlation (normalized enrichment score, 2.01; Q<0.0001 for false discovery rate) (Fig. S6D in Supplementary Appendix 1). This finding suggests that, in the absence of exogenous stimulation, HUVECs that express KRASG12V behave like angiogenic endothelial cells.
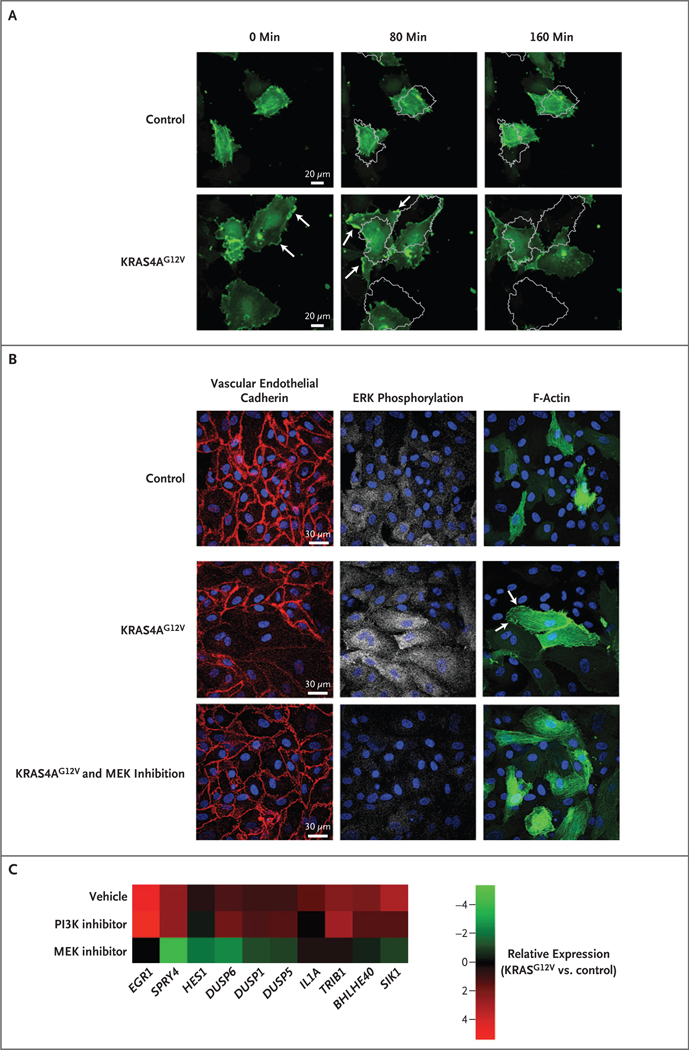
HUVECs with KRAS4AG12V expression had a highly elongated, mesenchymal-like phenotype (Fig. S7A in Supplementary Appendix 1). Time-lapse confocal imaging revealed that HUVECs that expressed KRAS4AG12V were more migratory than control cells on a scratch-wound assay performed in the absence of exogenous angiogenic factors and appeared to have more rapid rearrangements of actin during migration, as assessed by live imaging of actin dynamics with the use of LifeAct-GFP (Video 1, available at NEJM.org, and Fig. S7A and S7B in Supplementary Appendix 1). In contrast to control cells, HUVECs that expressed KRAS4AG12V were large and elongated and had rapid actin dynamics. F-actin was concentrated in lamellipodia on the periphery of the cells or moved in wavelike patterns through the cells. The cells were highly motile, even in the absence of a migratory signal (Fig. 4A and Video 2). Cell proliferation and apoptosis were unchanged in HUVECs that expressed KRAS4AG12V (Fig. S7C and S7D in Supplementary Appendix 1). A phenotype with similar morphologic features, migratory patterns, and actin dynamics was seen in human aortic cells that had been transfected with KRAS4AG12V (Video 3, and Fig. S8 in Supplementary Appendix 1).
Figure 4. Phenotype of Endothelial Cells Expressing Active KRAS.
Panel A shows time-lapse confocal images of human umbilical-vein endothelial cells (HUVECs) with KRAS4AG12V expression and control cells. In the absence of exogenous angiogenic factors or a migratory cue (e.g., scratch wound), KRAS4AG12V expression altered the actin dynamics (assessed by cotransfected LifeAct-GFP), including a reduction in the number of filopodia and an increase in lamellipodia formation (lamellipodia are indicated by arrows). Actin turnover was more rapid, and cells had increased motility; the images obtained at 80 minutes and 160 minutes show a gray outline of the position of the cells at 0 minutes. Panel B shows staining for vascular endothelial cadherin, ERK phosphorylation, and F-actin (assessed by cotransfected LifeAct-GFP) in HUVECs with KRAS4AG12V expression, HUVECs with KRAS4AG12V expression and MAPK–ERK inhibition with a MEK inhibitor for 16 hours, and control cells. KRAS4AG12V activated the MAPK–ERK pathway and promoted disassembly of adherens junctions. MAPK–ERK inhibition re-established the formation of adherens junctions and normalized actin localization (arrows) and cell shape. Panel C is a heat map showing the expression of vascular endothelial growth factor (VEGF)–regulated genes in HUVECs expressing KRAS4AG12V, treated with vehicle, MEK inhibitor, or phosphoinositide 3 kinase (PI3K) inhibitor for 6 hours in serum-starvation media. Data were compared with control cells treated with vehicle.
Since cells that expressed KRAS4AG12V did not form typical cobblestone monolayers (Fig. S7A in Supplementary Appendix 1), we assessed formation of adherens junctions. Expression of KRAS4AG12V led to disassembly of vascular endothelial cadherin junctions (Fig. 4B). The inhibition of MAPK–ERK signaling with the use of a MEK inhibitor suppressed ERK phosphorylation, restored localization of vascular endothelial cadherin to the junctions of endothelial cells, and appeared to reduce lamellipodia formation (Fig. 4B). In addition, inhibition of the MAPK–ERK pathway abrogated the VEGF-like gene signature that was induced by KRAS4AG12V, but inhibition of the PI3K pathway did not (Fig. 4C), a finding that reinforces the notion that the phenotype induced in endothelial cells by mutations that constitutively activate KRAS is specifically mediated by the MAPK–ERK pathway.
DISCUSSION
Somatic activating KRAS mutations were present in the majority of tissue samples of sporadic, nonfamilial arteriovenous malformations of the brain that we analyzed. They were accompanied by dysregulation of the MAPK–ERK pathway. This finding is consistent with the fact that other types of vascular malformation are caused by somatic mutations in genes in the PI3K or RAS–MAPK pathways. “Slow flow” vascular malformations are often associated with activating mutations in genes in the PI3K pathway,21,22 whereas “high flow” vascular anomalies, including sporadic arteriovenous malformations of the brain, tend to be associated with activating mutations in genes in the RAS–MAPK pathway.23–26
The finding of relatively low allele frequencies of KRAS variants in sporadic arteriovenous malformations of the brain (0.5 to 4%) is consistent with the cellular heterogeneity of arteriovenous malformations27 and the small fraction of endothelial cells therein and is consistent with the low frequencies of somatic mutations found in other genes in other types of vascular malformation (0.8 to 27%).22,23,25,26 The detection of KRAS mutations in endothelial cell–enriched fractions from primary cultures but not in endothelial cell–depleted fractions from the same cultures suggests that KRAS mutations are probably specific to endothelial cells and that dysregulation of the biology of endothelial cells is a key feature of the formation of arteriovenous malformations of the brain. Moreover, the allele frequencies of the KRAS variants correlated with the percentages of endothelial cells in enriched fractions, which suggests that these variants are present in most endothelial cells isolated from arteriovenous malformations of the brain, arise early in the development of the malformations, and are probably primary events in the pathogenesis of the malformations.
How might alterations in RAS signaling in endothelial cells induce arteriovenous malformations? The mutations that we have identified are known to drive strong and constitutive MAPK–ERK signaling28 and are important drivers of tumorigenesis.29 Arteriovenous malformations of the brain are not associated with cancer, which suggests a context-dependent role for KRAS mutations in the endothelium. The finding that somatic activating KRAS mutations in endometriosis do not cause cancer is consistent with this interpretation.30
Our initial in vitro exploration of endothelial-cell phenotypes after KRAS activation showed increased expression of angiogenic genes such as VEGFA/C and genes encoding proteins in the Notch signaling pathway. These transcriptional changes are accompanied by a cellular phenotype that includes enhanced migratory behavior and disassembly of adherens junctions. Previous studies have shown that increased VEGF and Notch signaling are relevant to the development and maintenance of arteriovenous malformations of the brain.31–37 Some of the in vitro phenotypes that we observed could be reverted to normal by means of inhibition of MAPK–ERK signaling. Our finding of increased MAPK–ERK signaling in endothelial cells from arteriovenous malformations of the brain without a KRAS variant suggests that activation of the MAPK–ERK pathway may be a defining feature of arteriovenous malformations of the brain. In the absence of available direct pharmacologic inhibitors of KRAS, small-molecule MEK inhibitors, which are used in clinical practice for treating cancers,38 represent candidates for testing in clinical trials to treat arteriovenous malformations of the brain.
Supplementary Material
Acknowledgments
Supported by research grants to Dr. Nikolaev from the Swiss Cancer League (LSCC 2939-02-2012 and KSF-3985-08-2016), Dinu Lipatti (2014), and Novartis (14B065). Ms. Khyzha was supported by a Canada Graduate Scholarship from the Natural Sciences and Research Council of Canada. Dr. DiStefano was supported by a Postdoctoral Fellowship from the Toronto General Hospital Research Institute. Dr. Suutarinen was supported by a research grant from the Petri Honkanen Foundation. Dr. Herman was supported by a grant from the National Institutes of Health (2T32HL007676). Dr. Wythe was supported by an American Heart Association Grant-in-Aid (16GRNT31330023). Dr. Antonarakis was supported by a grant from the European Research Council. Dr. Frösen was supported by research grants from the Finnish Medical Foundation and Kuopio University Hospital. Dr. Fish was supported by an operating grant from the Canadian Institutes of Health Research (CIHR) (MOP-119506) and a Team Project Award from the University of Toronto Medicine by Design initiative, which receives funding from the Canada First Research Excellence Fund and a Canada Foundation for Innovation equipment grant; he also received an Early Researcher Award from the Ontario Ministry of Research and Innovation and funding from the Canada Research Chair Program from the CIHR. Dr. Radovanovic was supported by the Timothy P. Susco Chair of Research Award from the Brain Aneurysm Foundation, the Toronto General and Western Hospital Foundation, and received seed support from the Department of Surgery and Division of Neurosurgery at the University Health Network.
We thank the staff of the Princess Margaret Genomics Centre and Bioinformatics Services (C. Virtanen and Z. Lu) for generating the RNA and DNA sequencing data used in this study; the staff of the Centre for Applied Genomics at the Toronto Hospital for Sick Children (T. Paton) for performing droplet digital polymerase-chain-reaction analyses; Zhiqi Chen from University Health Network for help with cell biology assays; and Melanie Peralta from the Pathology Research Program, University Health Network, Toronto, for technical help with immunohistochemical analyses.
Footnotes
Disclosure forms provided by the authors are available with the full text of this article at NEJM.org
REFERENCES
- 1.Brown RD Jr, Wiebers DO, Torner JC, O’Fallon WM. Incidence and prevalence of intracranial vascular malformations in Olmsted County, Minnesota, 1965 to 1992. Neurology 1996;46: 949–52. [DOI] [PubMed] [Google Scholar]
- 2.Berman MF, Sciacca RR, Pile-Spellman J, et al. The epidemiology of brain arteriovenous malformations. Neurosurgery 2000; 47:389–97. [DOI] [PubMed] [Google Scholar]
- 3.Al-Shahi R, Warlow C. A systematic review of the frequency and prognosis of arteriovenous malformations of the brain in adults. Brain 2001; 124: 1900–26. [DOI] [PubMed] [Google Scholar]
- 4.Al-Shahi R, Fang JS, Lewis SC, Warlow CP. Prevalence of adults with brain arteriovenous malformations: a community based study in Scotland using capturere-capture analysis. J Neurol Neurosurg Psychiatry 2002; 73:547–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McAllister KA, Grogg KM, Johnson DW, et al. Endoglin, a TGF-beta binding protein of endothelial cells, is the gene for hereditary haemorrhagic telangiectasia type 1. Nat Genet 1994; 8: 345–51. [DOI] [PubMed] [Google Scholar]
- 6.Johnson DW, Berg JN, Baldwin MA, et al. Mutations in the activin receptor-like kinase 1 gene in hereditary haemorrhagic telangiectasia type 2. Nat Genet 1996; 13:189–95. [DOI] [PubMed] [Google Scholar]
- 7.Gallione CJ, Repetto GM, Legius E, et al. A combined syndrome of juvenile polyposis and hereditary haemorrhagic telangiectasia associated with mutations in MADH4 (SMAD4). Lancet 2004; 363: 852–9. [DOI] [PubMed] [Google Scholar]
- 8.Revencu N, Boon LM, Mulliken JB, et al. Parkes Weber syndrome, vein of Galen aneurysmal malformation, and other fast-flow vascular anomalies are caused by RASA1 mutations. Hum Mutat 2008; 29: 959–65. [DOI] [PubMed] [Google Scholar]
- 9.Amyere M, Revencu N, Helaers R, et al. Germline loss-of-function mutations in EPHB4 cause a second form of capillary malformation-arteriovenous malformation (CM-AVM2) deregulating RAS-MAPK signaling. Circulation 2017; 136:1037–48. [DOI] [PubMed] [Google Scholar]
- 10.Do H, Dobrovic A. Dramatic reduction of sequence artefacts from DNA isolated from formalin-fixed cancer biopsies by treatment with uracil-DNA glycosylase. Oncotarget 2012;3: 546–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boscolo E, Pavesi G, Zampieri P, et al. Endothelial cells from human cerebral aneurysm and arteriovenous malformation release ET-1 in response to vessel rupture. Int J Mol Med 2006; 18: 813–9. [PubMed] [Google Scholar]
- 12.Simanshu DK, Nissley DV, McCormick F. RAS proteins and their regulators in human disease. Cell 2017; 170:17–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cuadrado A, Nebreda AR. Mechanisms and functions of p38 MAPK signalling. Biochem J 2010; 429:403–17. [DOI] [PubMed] [Google Scholar]
- 14.Benedito R, Hellström M. Notch as a hub for signaling in angiogenesis. Exp Cell Res 2013; 319:1281–8. [DOI] [PubMed] [Google Scholar]
- 15.Fish JE, Wythe JD. The molecular regulation of arteriovenous specification and maintenance. Dev Dyn 2015; 244: 391–409. [DOI] [PubMed] [Google Scholar]
- 16.ZhuGe Q, Zhong M, Zheng W, et al. Notch-1 signalling is activated in brain arteriovenous malformations in humans. Brain 2009; 132:3231–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maddaluno L, Rudini N, Cuttano R, et al. EndMT contributes to the onset and progression of cerebral cavernous malformations. Nature 2013; 498: 492–6. [DOI] [PubMed] [Google Scholar]
- 18.Kouhara H, Hadari YR, Spivak-Kroizman T, et al. A lipid-anchored Grb2-binding protein that links FGF-receptor activation to the Ras/MAPK signaling pathway. Cell 1997; 89:693–702. [DOI] [PubMed] [Google Scholar]
- 19.Meadows KN, Bryant P, Pumiglia K. Vascular endothelial growth factor induction of the angiogenic phenotype requires Ras activation. J Biol Chem 2001; 276: 49289–98. [DOI] [PubMed] [Google Scholar]
- 20.Zhang B, Day DS, Ho JW, et al. A dynamic H3K27ac signature identifies VEGFA-stimulated endothelial enhancers and requires EP300 activity. Genome Res 2013; 23:917–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Limaye N, Wouters V, Uebelhoer M, et al. Somatic mutations in angiopoietin receptor gene TEK cause solitary and multiple sporadic venous malformations. Nat Genet 2009; 41:118–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Limaye N, Kangas J, Mendola A, et al. Somatic activating PIK3CA mutations cause venous malformation. Am J Hum Genet 2015; 97: 914–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Couto JA, Vivero MP, Kozakewich HP, et al. A somatic MAP3K3 mutation is associated with verrucous venous malformation. Am J Hum Genet 2015; 96: 480–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Couto JA, Huang L, Vivero MP, et al. Endothelial cells from capillary malformations are enriched for somatic GNAQ mutations. Plast Reconstr Surg 2016; 137(1): 77e–82e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ayturk UM, Couto JA, Hann S, et al. Somatic activating mutations in GNAQ and GNA11 are associated with congenital hemangioma. Am J Hum Genet 2016; 98: 789–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Couto JA, Huang AY, Konczyk DJ, et al. Somatic MAP2K1 mutations are associated with extracranial arteriovenous malformation. Am J Hum Genet 2017; 100: 546–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Finby N, Begg CF. Arteriovenous malformation of the brain. N Y State J Med 1965;65: 554–8. [PubMed] [Google Scholar]
- 28.Cagnol S, Rivard N. Oncogenic KRAS and BRAF activation of the MEK/ERK signaling pathway promotes expression of dual-specificity phosphatase 4 (DUSP4/MKP2) resulting in nuclear ERK1/2 inhibition. Oncogene 2013; 32: 564–76. [DOI] [PubMed] [Google Scholar]
- 29.Park JT, Johnson N, Liu S, et al. Differential in vivo tumorigenicity of diverse KRAS mutations in vertebrate pancreas: a comprehensive survey. Oncogene 2015; 34:2801–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Anglesio MS, Papadopoulos N, Ayhan A, et al. Cancer-associated mutations in endometriosis without cancer. N Engl J Med 2017; 376: 1835–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Uranishi R, Baev NI, Ng PY, Kim JH, Awad IA. Expression of endothelial cell angiogenesis receptors in human cerebrovascular malformations. Neurosurgery 2001; 48:359–68. [DOI] [PubMed] [Google Scholar]
- 32.Hashimoto T, Lawton MT, Wen G, et al. Gene microarray analysis of human brain arteriovenous malformations. Neurosurgery 2004;54: 410–25. [DOI] [PubMed] [Google Scholar]
- 33.Murphy PA, Lam MT, Wu X, et al. Endothelial Notch4 signaling induces hallmarks of brain arteriovenous malformations in mice. Proc Natl Acad Sci U S A 2008;105: 10901–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Murphy PA, Lu G, Shiah S, Bollen AW, Wang RA. Endothelial Notch signaling is upregulated in human brain arteriovenous malformations and a mouse model of the disease. Lab Invest 2009; 89: 971–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Murphy PA, Kim TN, Lu G, Bollen AW, Schaffer CB, Wang RA. Notch4 normalization reduces blood vessel size in arteriovenous malformations. Sci Transl Med 2012;4: 117ra8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Murphy PA, Kim TN, Huang L, et al. Constitutively active Notch4 receptor elicits brain arteriovenous malformations through enlargement of capillary-like vessels. Proc Natl Acad Sci U S A 2014; 111: 18007–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li S, Wang R, Wang Y, et al. Receptors of the Notch signaling pathway are associated with hemorrhage of brain arteriovenous malformations. Mol Med Rep 2014; 9: 2233–8. [DOI] [PubMed] [Google Scholar]
- 38.Zhao Y, Adjei AA. The clinical development of MEK inhibitors. Nat Rev Clin Oncol 2014; 11:385–400. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.