Abstract
The human retina provides vision at light levels ranging from starlight to sunlight. Its supporting tissues regulate plasma-delivered lipophilic essentials for vision, including retinoids. The macula is an anatomic specialization for high-acuity and color vision that is also vulnerable to prevalent blinding diseases. The retina’s exquisite architecture comprises numerous cell types that are aligned horizontally, yielding structurally distinct cell, synaptic, and vascular layers that are visible in histology and in diagnostic clinical imaging. MALDI imaging mass spectrometry (IMS) is now capable of uniting low micrometer spatial resolution with high levels of chemical specificity. In this study, a multimodal imaging approach fortified with accurate multi-image registration was used to localize lipids in human retina tissue at laminar, cellular, and subcellular levels. Multimodal imaging results indicate differences in distributions and abundances of lipid species across and within single cell types. Of note are distinct localizations of signals within specific layers of the macula. For example, phosphatidylethanolamine and phosphatidylinositol lipids were localized to central RPE cells, whereas specific plasmalogen lipids were localized to cells of the perifoveal RPE and Henle fiber layer. Subcellular compartments of photoreceptors were distinguished by PE(20:0_22:5) in the outer nuclear layer, PE(18:0_22:6) in outer and inner segments, and cardiolipin CL(70:5) in the mitochondria-rich inner segments. Several lipids, differing by a single double bond, have markedly different distributions between the central fovea and the ganglion cell and inner nuclear layers. A lipid atlas, initiated in this study, can serve as a reference database for future examination of diseased tissues.
Keywords: age-related macular degeneration, macula, retinal pigment epithelium, photoreceptors, lipid atlas
Graphical Abstract
INTRODUCTION
The human retina is comprised of many specialized cell types including five neuronal classes: photoreceptors, bipolar cells, ganglion cells, horizontal cells, and amacrine cells1 that are collectively responsible for photon absorption, signal transduction, and signal transmission to the visual centers of the brain. In addition, the retina contains cells that support neurons including Müller (radial) glia, retina pigment epithelial (RPE) cells, and vascular endothelium. These cells are horizontally aligned in an exquisite architecture, giving rise to structurally distinct layers of cell bodies, synaptic endings, and vascular beds that are visible in histology and in diagnostic clinical imaging. The human retina is of particular interest because its unique anatomic specializations for high-acuity and color vision also render it vulnerable to prevalent blinding diseases.2 Although the cellular architecture of the human retina is well-characterized, its molecular composition is only partially understood. Complete knowledge of the molecular landscape of the human retina is essential for understanding normal biochemistry and physiology as well as for understanding cellular dysfunction and pathology in disease.
Research over decades has highlighted the importance of lipids for retinal function. The content of polyunsaturated long chain fatty acids varies with cell type, age, and ocular pathologies,3,4 and several monogenic inherited retinopathies directly impact lipid homeostasis and transfer.5,6 The characteristic extracellular deposits (drusen) of age-related macular degeneration (AMD) are dominated by lipoprotein-related lipids of intraocular and dietary origin.7–9 Retinoids (vitamin A derivatives) are essential for phototransduction and additionally feed into pathways resulting in amphiphilic fluorophores (bisretinoids) that are detectable in vivo and thus valuable for clinical autofluorescence imaging.10 Molecular analysis, however, is challenging because the neurons are 1/10 the size of those in the brain, and layers and spaces that are clinically relevant can be <10 μm thick.1,11
Matrix-assisted laser desorption ionization imaging mass spectrometry (MALDI IMS)12 provides the ability to map hundreds to thousands of molecular distributions in tissues at cellular resolution. With accurate coregistration to microscopy, these distributions can be correlated to very small histological features. MALDI IMS technology is particularly well-suited for examining tissues of the eye as demonstrated by published images of the retina,13–21 optic nerve,14,22–26 lens,27–31 and cornea.32 Cell layers of the retina distinguished by IMS have unique layer-specific lipid and metabolite signatures.13,14,16,20 With the development of multimodal imaging technologies incorporating data-rich IMS images with high spatial resolution methods like microscopy,33–36 cellular and subcellular localization of specific molecules provide a powerful tool to understand biochemistry in situ.
In this study, we used a recently developed method of high-accuracy registration33 to coregister high spatial resolution IMS images with histological images of the same tissue with single IMS pixel accuracy. This approach facilitated molecular characterization of aged healthy retinas to elucidate the molecular species in specific layers and cell types of central and peripheral human retina. Our work builds on prior IMS studies focusing on neurosensory retina14 or RPE only15,20 by enabling unbiased investigation of all chorioretinal layers in individual eyes. Our technique will aid evaluation of theories of AMD deposit biogenesis involving lipid trafficking among retinal neurons and glia7,8,37 and identification of signal sources for autofluorescence imaging.
METHODS
Tissue Acquisition and Characterization.
Whole eyes were obtained from deceased human donors via Advancing Sight Network (Birmingham AL; formerly the Alabama Eye Bank) as part of studies on age-related macular degeneration (AMD) that are approved by institutional review at University of Alabama at Birmingham (protocol no. N170213002), where tissues were collected. Donor criteria were ≥80 years of age, white, nondiabetic, and ≤6 h death-to-preservation. In this demographic, AMD is prevalent, and eyes were screened for AMD presence and staging using ex vivo multimodal imaging including optical coherence tomography (OCT), a widely used clinical diagnostic imaging technology. Eyes lacking detectable chorioretinal pathology were used for the studies described herein.
Tissue Handling and ex Vivo Imaging.
Methods were optimized for multimodal ex vivo clinical imaging of the ocular fundus,38 i.e., the retina, choroid, and sclera. To create a consistent opening with a smooth contour, globes were delivered intact by eye bank personnel and opened in-house as follows. To facilitate removing the cornea and a 2 mm wide scleral rim, a circular cut was scored with an 18 mm trephine (Stratis Healthcare, #6718L) and completed with a curved spring scissors (Roboz Cat No. RS-5681). To facilitate preservative penetration without disturbing the vitreous body and detaching the retina from RPE, the iris was slit. Globes with lens and iris in place were immersed in 0.1 M phosphate buffered 4% paraformaldehyde overnight. The iris and lens were removed before imaging.
For imaging with OCT and scanning laser ophthalmoscopy, globes were immersed in 4 °C phosphate buffer facing frontwards within a custom-built chamber with a 60-diopter lens.38 Spectral domain OCT (30° macula cube, 30 μm spacing, Automatic Real-time averaging ≥50), near-infrared reflectance, and autofluorescence (488 and 787 nm excitation wavelengths) images were captured with a Spectralis HRA&OCT and HRA2 (both Heidelberg Engineering). For color fundus photographs, globes were placed within a 30 cc quartz crucible (Fisher Scientific No. 08–074D). Images were taken at 3 magnifications (0.75×, 1.50×, 3.00×), and three lighting conditions (epi, flash, and dark field) with a Nikon D7200 camera mounted to an Olympus SZX9 Stereo microscope.
The goal of this study was to analyze macula and periphery together in continuous sections, since the two regions differ substantially by tissue mass, neuronal circuitry, and gene expression.39–41 With respect to embedding in carboxymethylcellulose (CMC) prior to cryosectioning, the posterior pole was trimmed to a 14 mm wide belt of retina, choroid, and sclera containing major landmarks (optic nerve head, fovea, and horizontal meridian of the visuotopic map) and extending anteriorly to pigmented tissue (ora serrata) at the edge of the ciliary body. To stabilize the globe and standardize dissection, posterior poles were placed in a custom-designed, chilled aluminum 3” × 4” × 1” billet with a 30 mm diameter hemispheric well and a set of parallel grooves 7 mm superior and inferior to the center of the well (Supplemental Figure 1). Globes were placed facing up in the well and the globe-billet combination oriented so the superior quadrant appeared at the left and the grooves were vertical, thus making the long edge of the tissue block parallel to the optic nerve head and fovea axis. To guide a knife cut, the globe was snipped vertically (10 mm) with scissors (Roboz No. 500216-G), guided by the parallel grooves. A tissue slicer (Thomas Sciences No. 6727C18) was placed in the two superior scissor cuts. With a guillotine motion, a superior cap was removed from the globe. While leaving the first blade in place to stabilize the globe, the inferior cap was removed by the same process at the second groove. The nasal periphery was removed with a slice 2 mm nasal to the optic nerve head.
Tissues were embedded in 2.5% carboxymethyl cellulose (Sigma C9481). Carboxymethyl cellulose powder was mixed in deionized water, heated to 70 °C, stirred until dissolved (30 min to 1 h), and degassed before use. Tissue belts were placed into cryomolds (Peel-A-Way 22 × 30 mm molds, Polysciences No. 18646B) under visual guidance with a stereo microscope, oriented so the superior edge faced up (to be sectioned first). Molds were filled with 5 mL of cold carboxymethyl cellulose and frozen at −20 °C. For histopathologic evaluation, serial 10 μm cryosections were collected starting at the superior edge of the optic nerve head on prelabeled 1 × 3 in. glass slides coated with 10% poly-L-lysine (Sigma-Aldrich, St. Louis, MO) and maintained at 37° during sectioning to promote tissue adherence to the glass. At predefined intervals in the glass slide series, sections were captured on large, 45 × 45 mm in house, poly lysine coated indium–tin oxide (ITO) slides (Delta Technologies, Loveland, CO) for IMS analysis. A slide box was vacuum-packed with an oxygen-absorbing packet within a Bitran freezer bag to help prevent oxidation and deterioration of lipid signal and stored at −80 °C for transport to Vanderbilt. Signals in tissues stored in this manner were preserved for at least 12 weeks.
MALDI IMS Analysis.
The matrix 1,5-diaminonaphalene (DAN) (Sigma-Aldrich, St. Louis, MO) was applied to tissue sections using a sublimation device developed in-house for negative-ion mode analysis. MALDI IMS data were acquired with a 10–15 μm pixel size in full scan mode using a Bruker SolariX 9.4T FT ICR mass spectrometer (Bruker Daltonics, Billerica, MA) equipped with a modified source with a Gaussian profile Nd:YAG laser and optics to provide a laser spot diameter of 8 μm. Both red phosphorus42 and known lipid masses were used to calibrate the instrument prior to data acquisition. m/z 885.5499 was used for online calibration while generating peak picked sqlite files using ftmsProcessing software (Bruker Daltonics, Billerica, MA) postaquisition. Following data acquisition, an advanced image registration workflow33 was performed using both autofluorescence and bright-field microscopy images. IMS data were exported for accurate registration, and overlaid images were reconstructed using ImageJ Fiji freeware (NIH), while unregistered MALDI IMS images were generated using flexImaging (Bruker Daltonics, Billerica, MA). Data were acquired with 500 shots per pixel and a mass range of m/z 154–2000 using the 1 M data size resulting in a 0.5592 s transient, and mass resolution was ~60000 at m/z 699.499 and normalized to the root mean squared (RMS). The top 10 signals from each of the layers of the neural retina and RPE were extracted using flexImaging 5.0 ROI tool by selecting the ROI spectra and copying the CSV. This CSV data could then be opened in mMass43 via the copy from clipboard function in the file tab. Summed spectra from these regions then had multiple peaks labeled, and the compound search tool function was used to match negative-ion glycerophospholipids within 5 ppm. Summed spectra were then recalibrated based on known masses. Mass over charge values, intensity, and relative intensity values were extracted to Excel to generate tables.
Microscopy.
Pre-IMS autofluorescence (AF) images were acquired prior to matrix application on a Nikon Eclipse 90i (Nikon Instruments Inc., Melville, NY) with a 10× objective (numerical aperture 0.30, 0.92 pixel/μm) using filter cubes for DAPI (excitation 325–375 nm, emission 435–485 nm) and Texas Red (excitation 540–580 nm, emission 600–660 nm). To record the laser burn pattern and assess tissue morphology, post-IMS AF images were taken following MALDI IMS using only the DAPI filter and low-intensity transmitted light. Following post-IMS AF microscopy, the matrix was removed using a minimal amount of methanol until the slide was visibly clear of matrix. The tissue was stained with hematoxylin (Mayer’s, MHS32–1L, Sigma-Aldrich) and eosin (E6003, Sigma-Aldrich) (H&E), and bright-field images were acquired at 10× on the same microscope.
Registration.
Registration of the IMS data to microscopy was performed using an explicit IMS pixel-to-laser ablation mark registration method described previously,33 provided in the MSRC Registration Toolbox (https://github.com/nhpatterson/regToolboxMSRC), and outlined in Figure 1. Briefly, the post-IMS AF ablation mark microscopy image was used to link individual ablation marks to the theoretical pixel position image generated from the IMS metadata (x,y coordinates). Then, automated computational registration of H&E and pre-IMS AF images to the previously IMS registered post-IMS AF ablation mark image was performed using MSRC Registration Toolbox software. Finally, the three microscopy images (H&E, pre-IMS AF, and post-IMS AF) were registered directly to the IMS data via the ablation mark pattern. Spatial localization accuracy is performed by measuring the average distance between the center point of the ablation mark in the post-IMS microscopy with the center point of the IMS pixel after registration of the modalities. In this way, we align each IMS pixel to its corresponding ablation mark and calculate the global spatial error between this alignment.33
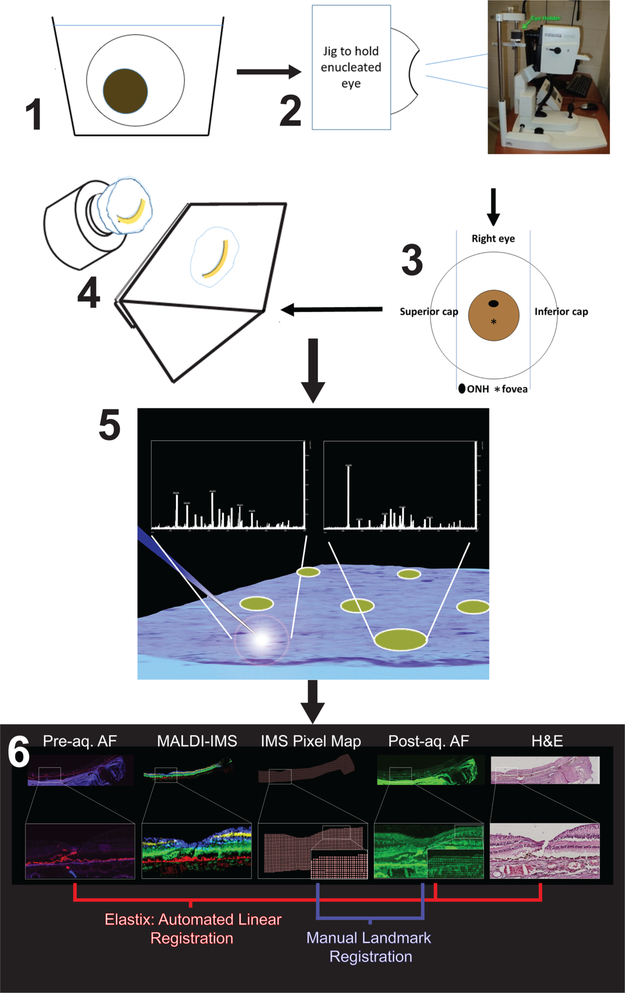
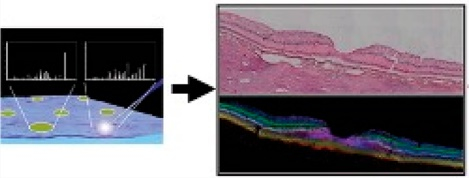
Figure 1.
Workflow of sample preparation and multimodal imaging. (1) The cornea is removed before fixation with 4% paraformaldehyde for 48 h at 4 °C. The iris and lens are removed before imaging. (2) The eye lacking cornea, iris, and lens mounted in jig to obtain OCT image. (3) The eye is placed in a dissection guide as described in the text to capture a belt with the optic nerve, macula, and temporal periphery. The belt is embedded in 2.6% CMC in a cryomold. The caps are discarded. (4) Cryosections at 12–14 μm throughout the entire eye are thaw-mounted on either glass or ITO slides. (5) ITO slides are imaged for autofluorescence (AF) before being coated with matrix via sublimation for acquisition of IMS data. (6) Highly accurate data registration is performed from IMS, postacquisition AF, pre AF, and H&E stained tissue.
LC–MS Methods.
Sectioned tissue was removed from polylysine-coated ITO glass slides with a clean razor blade and fine tipped forceps by first peeling the sclera away from the RPE and neural retina and discarding using forceps. The razor blade was then scraped across the surface of the glass to remove the remaining tissue. Tissue was placed in a 1 mL glass autosampler vial ensuring the tissue moved to the bottom. This process was repeated for six sections, pooling them into the same tube. A 200 μL portion of 2:1 chloroform/methanol was added, and the tube was sonicated for 3 min then vortex mixed for 1 min. The solution was washed twice with 20 μL of Milli-Q water mixing gently each time for 2 min and allowing the solution to separate before removing. The solution interface was then rinsed three times with pure “upper phase” (chloroform/methanol/water, 3:48:47, v/v/v) ensuring the lower phase is not disturbed while siphoning 20 μL of the upper phase each time and discarding to waste. The remaining lower phase was then dried under a stream of nitrogen and stored at −80 °C until analysis. Prior to analysis the dried material was reconstituted in 20 μL of methanol.
Data acquisition was performed using a Vanquish UHPLC (Thermo Fisher Scientific, San Jose, CA) interfaced to a Q Exactive HF high resolution Orbitrap mass spectrometer (Thermo Fisher Scientific, San Jose, CA). Chromatographic separation was accomplished with a reversed-phase Waters BEH C18 column, 2.1 mm × 150 mm, 1.7 μm (Waters, Milford, MA) at a flow rate of 300 μL/min. The mobile phases were 10 mM ammonium acetate in (A) water/acetonitrile (1:1) and (B) acetonitrile/2-propanol (1:1). Then 10 μL of sample extract was injected, and the elution gradient was as follows. The gradient was held at 20%B for the first minute, increased from 20%B to 100%B in 9 min where it was held for 30 s, and re-equilibrated at 20%B for 4.5 min. For all data acquired, spray voltage was set to 5 kV in negative mode with an S-lens RF level of 80 V, and capillary and probe temperatures of 320 and 200 °C, respectively. Sheath and auxiliary gases were set to 40 and 10 arbitrary units. Two different methods were used for collecting data in both untargeted and targeted modes:
Untargeted.
Mass spectra were collected over a scan range of m/z 200–1500 at a resolution of 45000. MS/MS spectra were collected using a top-7 data-dependent MS/MS method conducted at a resolution of 15 000, with an AGC target of 1e,5 a max inject time of 50 ms, and a normalized collision energy of 30.
Targeted.
Mass spectra were collected over a scan range of 500–1600 m/z, at a resolution of 30000. MS/MS data were collected using parallel reaction monitoring at a resolution of 15000 to isolate and fragment several ions of interest (see Supplemental Table 1). An isolation window of 2 m/z was used with a normalized collision energy of 30, AGC target of 2e,5 and a maximum injection time of 100 ms.
Tentative Molecular Assignments.
Assignments were made based solely on accurate mass measurements from IMS data when these had a <5 ppm error when fragmentation data were not obtained or were not interpretable.
RESULTS AND DISCUSSION
As the capabilities of MALDI IMS approach single-cell resolution, careful sample preparation and image registration between imaging modalities are required to maintain high quality tissue morphology and to assign exact molecular locations, respectively.
The preservation of native tissue morphology is paramount for obtaining high-quality MALDI IMS images. Thus, the workflow utilized for sample preparation and image registration was crucial for localizing molecules to layers of the neural retina. Figure 1 describes the workflow from eye preservation to multimodal imaging performed in this study. The fixation and embedding protocols preserve tissue morphology in a manner that allows a MALDI process without contamination. In an autofluorescence image acquired prior to matrix application, the RPE layer can be clearly visualized due to its content of autofluorescent lipofuscin and melanolipofuscin (long-lasting intracellular inclusion bodies).15,44–46 The explicit alignment of an IMS pixel to its corresponding laser burn pattern in images of autofluorescence and H&E-stained histology allows confident localization of signals to 0.5–2 μm accuracy, below the raster pitch of the IMS acquisition. Again, this capability is critical for imaging the precisely layered retina and single layer of RPE cells where inaccurate registration may lead to spurious tissue assignment of molecular signals. Utilizing morphology-rich microscopy images enables accurate registration to be achieved, providing high confidence when assigning laminar distributions of analytes. Notable features of our method are the use of paraformaldehyde-preserved tissues embedded in CMC for sectioning. This strategy preserves tissue morphology while allowing lipid IMS signals to be detected. Previous IMS studies of human retina and RPE included our own used fresh tissue.14,15,20 Moreover, one previous IMS study of human retina14 used a modified “optimal cutting temperature” compound that, in our experience, typically results in abundant polymer signals that suppress tissue analyte signals.
Central and Peripheral Retina Signals.
Figure 2A shows a schematic of major retinal cell types distributed across the layers. In this description, the directions “inner” and “outer” indicate toward the center and exterior of the eye, respectively. Rod and cone photoreceptors responsible for dim and bright light vision, respectively, are vertically compartmentalized and horizontally aligned at the outer surface of the neurosensory retina. These cells initiate phototransduction at the outer segments (OS). Mitochondria are numerous in the ellipsoid (outer) part of the inner segments. Photoreceptor cell bodies stacked in rows comprise the outer nuclear layer (ONL). In the outer plexiform layer (OPL), cones contact bipolar and horizontal cells (interneurons) at large synaptic terminals called pedicles, and rods, at spherules. These interneurons have cell bodies in the inner nuclear layer (INL), and in the inner plexiform layer (IPL), they contact processes of ganglion cells. Müller glia (support cells to neurons) span the retina from inner limiting to external limiting membrane and are intercalated among the neurons in every layer. External to the photoreceptors, the retinal pigment epithelium (RPE) forms a supporting layer that is a single cell thick. The retina has a dual circulation system, a retinal circulation within the neurosensory retina with a physiologic barrier like the blood–brain barrier, and the choroidal circulation, a vascular bed behind the RPE and part of the systemic circulation. Photoreceptors and RPE are served by the choroid, with photoreceptor terminals also supplied by a plexus of the retinal circulation.
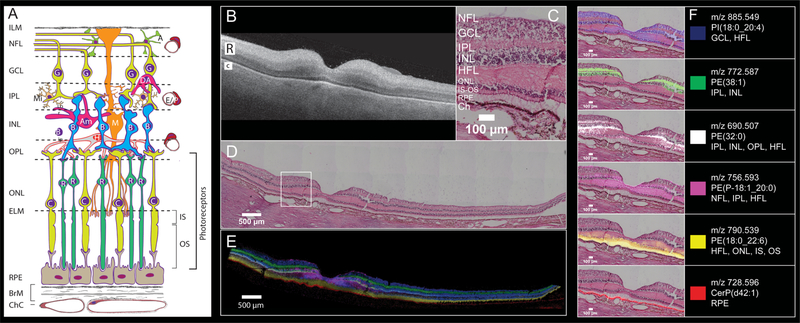
Figure 2.
Six MALDI IMS signals with distinct laminar localizations in human macula from an 83-year-old female donor. (A) Schematic diagram of retina, Bruch’s membrane, and choriocapillaris, indicating cell types in their characteristic layers. The outer nuclear layer (ONL) contains cell bodies of cones (C) and rods (R) and their inner fibers, all interleaved with Müller glia (M). Photoreceptor inner segments (IS) contain many mitochondria. Photoreceptor outer segments (OS) contain phototransduction-related proteins in membrane disks. The RPE apical processes are specialized for metabolic exchange with photoreceptors and retinoid processing. Layers: ILM, inner limiting membrane; NFL nerve fiber layer, GCL ganglion cell layer, IPL inner plexiform layer, INL inner nuclear layer, HFL Henle fiber layer, ELM, external limiting membrane; RPE retinal pigment epithelium, ChC, choriocapillaris; BrM, Bruch’s membrane; cell types, A, astrocyte; Am, amacrine; B, bipolar; E, endothelium; G, ganglion cell; H, horizontal cell; P, pericyte. (B) OCT B-scan of a donor eye macula prior to sample preparation, R and C indicating retina and choroid, respectively. The retina is at the top, the choroid (vasculature) is on the bottom, and the hyperreflective interface is the retinal pigment epithelium (RPE) layer. (C) Zoomed image of tissue section stained with H&E to indicate cell layers at the level shown in (A). NFL nerve fiber layer, GCL ganglion cell layer, IPL inner plexiform layer, INL inner nuclear layer, HFL Henle fiber layer, ONL outer nuclear layer, IS-OS photoreceptor inner and outer segments, RPE retinal pigment epithelium, Ch choroid. (D) Panoramic H&E stained tissue post-IMS data acquisition showing macula and extended stretch of temporal peripheral retina. (E) Overlay of MALDI IMS images of selected lipid signals localizing to distinct retinal layers and RPE as indicated. Colors defined in panel F. (F) Individual ion signals overlaid on top of H&E stained tissue image.
Figure 2B–E reveals retinal and choroidal structure in macular and peripheral regions of a normal retina of an 83-year-old donor. Figure 2B shows an optical coherence topography (OCT) B-scan of the preserved tissue before processing for IMS. Retinal layers are apparent despite loss of transparency after death. The two most reflective bands in the neurosensory retina (Figure 2B “R”) are the IPL and OPL, and the RPE is a reflective band along the top (inner) edge of the choroid (Figure 2B “C”). The 6 mm diameter macula is defined by neuronal content and includes the fovea (1 mm diameter with a 0.35 mm, all-cone, rod-free zone in the middle), the parafovea (an annulus of 0.5–1.5 mm radius from the foveal center), and the surrounding perifovea (1.5–3 mm radius). The term “central macula” in this report refers to fovea and parafovea together.
Parts C and D of Figure 2 shows a panoramic (D) and zoomed (C) view of post-IMS H&E stained tissue, indicating the cell and synaptic layers of human macula and temporal periphery (Figure 2D). The distributions of six different molecular species are shown as an overlay (Figure 2E) and as individual ion images overlaid on the registered H&E-stained tissue image (Figure 2F). These figures demonstrate that the observed lipid species are highly localized to distinct layers. The PI(18:0_20:4) (m/z 885.549, blue) signal localizes to the GCL and INL as well as in the HFL and ONL. A signal observed at m/z 772.587 (green) and tentatively assigned as PE(38:1) is observed in the IPL and INL spanning from the parafovea region to the far periphery. The signal observed at m/z 756.593 (pink) was identified as a phosphatidylethanolamine plasmalogen (PE-P-18:1_20:0) and is localized to the central fovea and in the HFL, IPL and NFL extending only to the parafovea, unlike the other signals. The distribution observed for m/z 756.593 bears a striking resemblance to the “bowtie” image of yellow xanthophyll carotenoid distribution, first described by Snodderly et al. using two-wavelength microdensitometry of sectioned monkey retina47–50 and recently replicated with resonance Raman microscopy of human tissue.51 Because strong evidence indicates that Müller glia in central macula are a major reservoir for xanthophylls as well as cones,52 it is possible that the m/z 756.593 bowtie localizes specifically to these cells.
A signal at m/z 690.507, tentatively identified as PE(32:0) (white), resides in the OPL, IPL, HFL, and INL, spanning the full breadth of the macula with lower abundance in the periphery. An intense signal at m/z 790.539 extending across the retina in the inner and outer segments (IS, OS) of photoreceptor (yellow) was identified as PE(18:0_22:6). This lipid is also observed in the HFL and ONL with lower abundance. This signal, previously observed in human photoreceptors by Zemski Berry et al.,14 is from a lipid that contains docosahexaenoic acid. This highly unsaturated fatty acid is abundant in neural tissue and especially in photoreceptors, where it is crucial for cellular function.53,54 External to the photoreceptors, a strong signal tentatively identified as a ceramide-1-phosphate (m/z 728.596, CerP(d42:1); red) is observed within the RPE layer across the entire retina (details below). Distributions of lipids in Figure 2 were replicated in the eye of an 81-year-old donor (Supplemental Figure 2). Lipid identification data can be found in Supplemental Table 1.
Signals Specific to Photoreceptors and RPE.
Rod photoreceptors are slender (2–3 μm) and can be very long (100 μm). They are the dominant photoreceptor in human retina (20:1 rod:cone ratio), being absent only in the coneonly foveal center. Signals unique to photoreceptor cells, a population necessarily dominated by rods in cross sections not including the fovea, are shown in Figure 3.
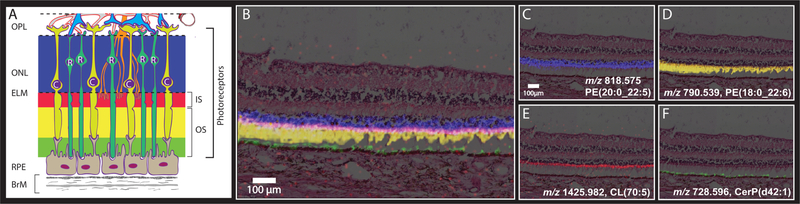
Figure 3.
MALDI IMS signals consistent with localization to photoreceptor and RPE compartments. (A) Schematic diagram of outer retina and Bruch’s membrane, excerpted from Figure 2A. Blue, pink, yellow, and green bands indicate layers formed by highly compartmentalized and vertically aligned photoreceptors and RPE cells in panels B and C. See Figure 2 for explanation of cellular and subcellular content of each layer. Layers: OPL, outer plexiform layer; ONL, outer nuclear layer; ELM, external limiting membrane; RPE, retinal pigment epithelium; BrM, Bruch’s membrane; R, Rod; C, cone photoreceptors. (B–F) Images and H&E stained tissue images overlaid in peripheral retina displaying signals from multiple lipid classes that localize to subcellular compartments of the photoreceptor cells. (B) Overlay showing four separate signals defined in panels C–F. (C) Localized to ONL. (D) Localized to photoreceptor inner and outer segments. (E) Localized to mitochondria-rich photoreceptor inner segments. (F) Localized to RPE apical processes.
Figure 3A shows part of the schematic from Figure 2A, focusing on photoreceptors and support cells. The RPE sends delicate processes in the apical direction to contact photoreceptor OS, near the RPE cell body for rods and 10–15 μm above the cell body for cones (because cone OS are short). The RPE sits on Bruch’s membrane, which serves as a flat vessel wall above the choriocapillaris endothelium. Figure 3A is color-coded to indicate photoreceptor and RPE compartments associated with IMS signals in Figure 3B (blue, red, yellow, green, for ONL, IS, OS, and RPE, respectively).
Figure 3B shows MALDI IMS images overlaid with an H&E image from this donor. The photoreceptors are attached to the RPE on the left of the image and detached from the RPE, a common artifact, on the right. A signal at m/z 818.575, identified as PE(20:0_22:6; blue), is observed with high abundance in the ONL, which contains photoreceptor cell bodies and processes of Müller glia. The m/z 818.575 signal was assigned to photoreceptors due to the lack of similar signal in other retinal layers where Müller glia are also present. Just external to this signal, a signal at m/z 1425.982 (red) can be seen in high abundance along a narrow band aligned with photoreceptor IS. This signal was identified as a cardiolipin CL(70:5). Cardiolipins are highly abundant in mitochondria, and the ellipsoid portion of photoreceptor IS contains many mitochondria providing energy for these metabolically active cells. This signal is also observed in inner retinal cells with abundant mitochondria. Below this layer are photoreceptor OS, where the docosahexaenoate-containing PE(18:0_22:6, Figure 2) is observed at m/z 790.539 (yellow). At the lowest part of the photoreceptor cells are OS that are interleaved with the apical processes of RPE cells. The signal observed at m/z 728.596 (green) is localized above and within the RPE, as discussed further below. Replicate data from an 81-year-old donor eye can be seen in Supplemental Figure 3.
The image of m/z 728.596 CerP(d42:1) highlights the challenges posed by the precisely vertical compartmentalization of the RPE, a critical layer in initiating vision and also central to AMD pathogenesis. The RPE performs demanding dual services to photoreceptors above and choriocapillaris below, as reflected in an internal gradient of organelles and molecules.55 For example, delicate apical processes contacting photoreceptors (Figures 2A and 3) and cell bodies share retinoid processing enzymes but differ in channels important for potassium currents.56,57 Figure 2 and Supplemental Figure 4 overlaying IMS with tissue autofluorescence show strong m/z 728.596 signal in RPE. Upon closer examination of Figure 2 this signal appears to extend above the RPE layer by up to 10 μm, depending on retinal position, with the greatest extensions observed at the fovea. Conversely, Figure 3 and Supplemental Figure 5 show signals possibly localized to only apical processes, i.e., not in the cell bodies, especially where apical processes are fortuitously standing upright. These discrepancies in m/z localizations are due to factors that impact alignment accuracy, e.g., OS attachment to RPE, angle of sectioning plane relative to the cell layer, and the different resolutions used (10 μm for Figures 2–5 vs 15 μm for Supplemental Figures 2 and 3). The differences between the results in the two figures will be resolved by processing more samples with intact retinas, at the higher resolution, in conjunction with specific markers for apical processes like ezrin.
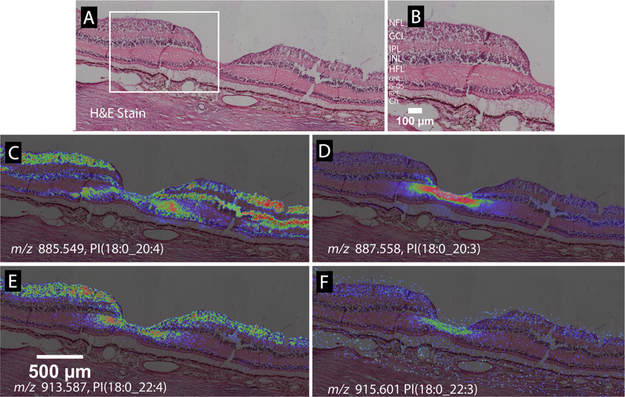
Figure 5.
Lipid species varying in only one double bond can exhibit different MALDI IMS distributions. (A) H&E stained tissue image of a normal macula, with the foveal pit in the center. (B) Zoomed view of panel A showing retinal layers. NFL, nerve fiber layer; GCL, ganglion cell layer; IPL, inner plexiform layer; INL, inner nuclear layer; HFL, Henle fiber layer; ONL, outer nuclear layer; PR/IS, photoreceptor inner segments; PR/OS, photoreceptor outer segments; RPE, retinal pigment epithelium; ChC, choriocapillaris. (C) m/z 885.5499, PI(18:0_20:4) distributes in the foveal center, GCL, INL, and HFL. (D) m/z 887.558, PI(18:0_20:3), with one double bond less than the species in panel C, localizes to the HFL of the foveal center and the inner INL. (E) m/z 913.587, PI(18:0_22:4) localizes to the foveal center, GCL, INL, and HFL, like the species in panel A, but with a lesser lateral extent of INL. (F) m/z 915.601, PI(18:0_22:3), with one double bond less than the species in panel E, localizes to the HFL of the foveal center.
Complementarity of Neurosensory Retina and RPE.
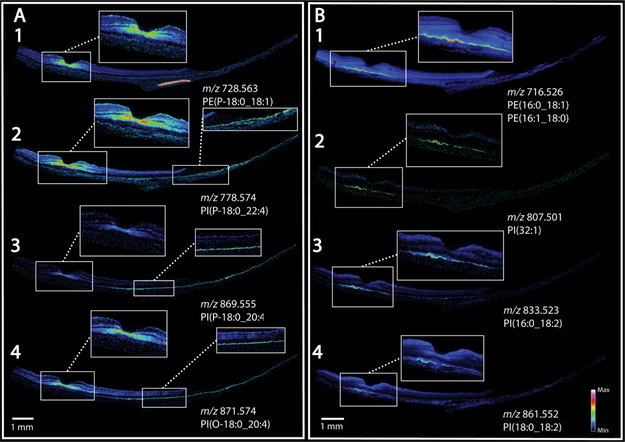
Figure 4 displays eight MALDI IMS images from lipid species that exhibit complementary localization patterns in the macula of neurosensory retina and underlying RPE. Figure 4A shows images generated from four plasmalogen phosphatidylglycerol lipid species. Panels A1–4 all exhibit high signal in the foveal center. Panels A1, 2, and 4 also have strong signal in the HFL, NFL, and IPL extending across the whole macula. Figure 4A3 displays a similar overall pattern with high signal in the peripheral retina but lower signal in the central macula. In all four panels, no signal is observed in RPE directly below the signal-rich area of neurosensory retina, yet signals are intense in peripheral RPE.
Figure 4.
MALDI IMS shows complementarity of signals in retina and RPE. (A) Signals localized to the foveal center and extending into the macular NFL, IPL, HFL also localize to the peripheral RPE. (B) Signals localized to RPE underlying the macula, not confined to the fovea.
Conversely, Figure 4B1–4 shows four signals that exhibit high abundance in the RPE in an area ~3 mm wide and centered under the fovea but not confined to the rod-free area, and exhibiting no signal in the periphery. The signal observed at m/z 716.526 in Figure 4B1 was attributed to two phosphatidylethanolamine lipids PE(16:0_18:1) and PE(16:1_18:0) that are exact isobars, as confirmed in LC–MS/MS experiments. The MALDI IMS image generated from three phosphatidylinositol lipids, one tentatively identified as PI(32:1) (Figure 4B2) and the other two identified as PI(16:0_18:2) and PI(18:0_18:2) (Figure 4B3,4) all display the same distribution but with lower relative intensities than that observed for the PE lipid in Figure 4B1. The positively identified PI lipids contain an 18:2 fatty acid side chain. The same distributions were also observed in an 81-year-old donor eye as seen in Supplemental Figure 6. The relevance of the apparent high abundance and specific localization of this particular fatty acid side chain has yet to be determined, although variations in abundance between midlife, aged normal, and AMD eyes was previously reported by Liu et al.3
Overall, Figure 4 clearly indicates not only differences in lipid composition between central and peripheral RPE but also a regional complementarity of neurosensory retina and RPE. Further, this arrangement is not confined to the rod-free fovea but rather involves the central macula, a region distinguished by high concentration of xanthophyll carotenoids.51 Regarding results in Figure 4A, plasmalogen lipids can act as free radical scavengers and can be associated with glial cells, which may explain a localization in the foveal HFL along with photoreceptors. The role of ambient light is often invoked to explain free radical generation58–62 in the retina. However, retinal illuminance in the human eye is nearly uniform for a wide portion of the visual field,63,64 and light distribution alone cannot explain the specific localizations observed in our studies. An alternative explanation for these localizations builds on the idea that in Müller glia in central macula have distinct properties, including xanthophyll sequestration and also, a separate retina-based retinoid regeneration cycle believed to serve cones.65 The latter pathways are distinct from the canonical RPE-based visual cycle.66,67
We speculate that retina-RPE complementarity in Figure 4 indicates that similar functions are performed by macular Müller cells and extramacular RPE. Further, we hypothesize that these functions include retinoid processing, due to a division of labor between Müller cells and RPE unique to macula, as a result of a retinoid cycle serving cones. The localized lipids may or may not be directly involved in retinoid processing per se. They may signify an overall distinct internal milieu in the cells harboring these pathways. Finally, our data suggest that the molecular inhomogeneity of human RPE first seen for bisretinoid A2E (low signal in macula, high in periphery)15,20 is part of a much larger pattern involving lipids in multiple essential functions in cells supporting neuronal function. The cellular and molecular basis for this retinal topography will be important to explore, because the layout of each species’ retina is evolved for efficient vision in its own habitat,68 under genetic control.
Spectral Specificity of High-Resolution IMS.
Figure 5 demonstrates markedly different localizations for molecules differing by a single double bond. Parts A and B, respectively, of Figure 5 show a panoramic and magnified image of an H&E stained macula with the fovea in the center. Figure 5C displays the distribution of PI(18:0_20:4) at m/z 885.549, localized to the foveal center, GCL, INL, and HFL. Figure 5D was generated from m/z 887.558, identified as PI(18:0_20:3) and differing by only one double bond from the lipid shown in Figure 5C, yet specifically localizing to the HFL in the fovea. Also localizing to these regions is another lipid pair varying in one double bond of a fatty acid chain, i.e., PI(18:0_22:4) and PI(18:0_22:3) shown in Figure 5E,F, respectively. Data shown in Figure 5 were replicated from an 81-year-old donor tissue as shown in Supplemental Figure 7. This observation of the same lipid with one extra double bond showing contrasting, and to some extent inverse, localization was recently observed in the human optic nerve.23 In this tissue, one species of sphingomyelin (SM(d36:2)) localized to axonal bundles while another (SM(d36:1)) was present in the surrounding glial cells and connective tissue.23
Registration of images from multimodal imaging experiments allows accurate assignment of IMS signals to retina layers, which, with knowledge of layer composition, can permit assignment to specific cell types and even subcellular compartments. The patterns shown in Figures 2–5 are only a subset of the many signals detected thus far. Supplemental Figures 8 and 9 display 100 images selected from the two donor tissues (83- and 81-year-old) indicating the diverse localizations of the many signals observed.
CONCLUSIONS
This multimodal imaging method provides a powerful tool to discover molecules in numerous cell types of the neural retina and supporting tissues. Use of ex vivo imaging with OCT allows identification of pathology before molecular analysis and quality control over post-mortem tissue destined for IMS. An atlas of lipid composition in normal tissue, such as described in this study, serves as a reference database for examination of diseased tissue. Data presented herein show striking topographies even within the same cell type. Of major significance is the regional molecular heterogeneity of human RPE, linked to the topography of overlying photoreceptors, an emerging concept for models of macular physiology and disease. Also significant is the potential of characterizing the composition of cells harboring xanthophyll macular pigment, an area of active investigation. Finally, our data will enable translation of specific m/z signals to the interpretation of clinical OCT, a widely used diagnostic technology.
Supplementary Material
ACKNOWLEDGMENTS
This project was supported by the National Institutes of Health P41 GM103391 (R.M.C.) and R01 EY027948 (C.A.C.). Support was also received by a Research to Prevent Blindness Catalyst Award for Innovation Research in Age-Related Macular Degeneration to K.L.S.
Footnotes
The authors declare no competing financial interest.
ASSOCIATED CONTENT
Supporting Information
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/jasms.0c00119.
Supplemental Figures 1–9 and Supplemental Table 1 (PDF)
Contributor Information
David M. G. Anderson, Department of Biochemistry and Mass Spectrometry Research Center, Vanderbilt University, Nashville, Tennessee 37240, United States
Jeffrey D. Messinger, Department of Ophthalmology and Visual Science, University of Alabama at Birmingham, Birmingham, AL 35294, United States
Nathan H. Patterson, Department of Biochemistry and Mass Spectrometry Research Center, Vanderbilt University, Nashville, Tennessee 37240, United States.
Emilio S. Rivera, Department of Biochemistry and Mass Spectrometry Research Center, Vanderbilt University, Nashville, Tennessee 37240, United States
Ankita Kotnala, Department of Biochemistry and Mass Spectrometry Research Center, Vanderbilt University, Nashville, Tennessee 37240, United States; Department of Ophthalmology and Visual Science, University of Alabama at Birmingham, Birmingham, AL 35294, United States.
Jeffrey M. Spraggins, Department of Biochemistry and Mass Spectrometry Research Center, Vanderbilt University, Nashville, Tennessee 37240, United States.
Richard M. Caprioli, Department of Biochemistry and Mass Spectrometry Research Center, Vanderbilt University, Nashville, Tennessee 37240, United States.
Christine A. Curcio, Department of Ophthalmology and Visual Science, University of Alabama at Birmingham, Birmingham, AL 35294, United States
Kevin L. Schey, Department of Biochemistry and Mass Spectrometry Research Center, Vanderbilt University, Nashville, Tennessee 37240, United States.
REFERENCES
- (1).Masland RH The Neuronal Organization of the Retina. Neuron 2012, 76 (2), 266–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Rossi EA; Roorda A The relationship between visual resolution and cone spacing in the human fovea. Nat. Neurosci. 2010, 13 (2), 156–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Liu A; Chang J; Lin Y; Shen Z; Bernstein PS Long-chain and very long-chain polyunsaturated fatty acids in ocular aging and age-related macular degeneration. J. Lipid Res. 2010, 51 (11), 3217–3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Agbaga M-P; Merriman DK; Brush RS; Lydic TA; Conley SM; Naash MI; Jackson S; Woods AS; Reid GE; Busik JV; Anderson RE Differential composition of DHA and very-long-chain PUFAs in rod and cone photoreceptors. J. Lipid Res. 2018, 59 (9), 1586–1596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Harkewicz R; Du H; Tong Z; Alkuraya H; Bedell M; Sun W; Wang X; Hsu YH; Esteve-Rudd J; Hughes G; Su Z; Zhang M; Lopes VS; Molday RS; Williams DS; Dennis EA; Zhang K Essential role of ELOVL4 protein in very long chain fatty acid synthesis and retinal function. J. Biol. Chem. 2012, 287 (14), 11469–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Sluch VM; Banks A; Li H; Crowley MA; Davis V; Xiang C; Yang J; Demirs JT; Vrouvlianis J; Leehy B; Hanks S; Hyman AM; Aranda J; Chang B; Bigelow CE; Rice DS ADIPOR1 is essential for vision and its RPE expression is lost in the Mfrp(rd6) mouse. Sci. Rep. 2018, 8 (1), 14339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Curcio CA Soft Drusen in Age-Related Macular Degeneration: Biology and Targeting Via the Oil Spill Strategies. Invest. Ophthalmol. Visual Sci. 2018, 59 (4), AMD160–AMD181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Curcio CA Antecedents of Soft Drusen, the Specific Deposits of Age-Related Macular Degeneration, in the Biology of Human Macula Why the Macula for Soft Drusen? Invest. Ophthalmol. Visual Sci. 2018, 59 (4), AMD182–AMD194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Bretillon L; Thuret G; Gregoire S; Acar N; Joffre C; Bron AM; Gain P; Creuzot-Garcher CP Lipid and fatty acid profile of the retina, retinal pigment epithelium/choroid, and the lacrimal gland, and associations with adipose tissue fatty acids in human subjects. Exp. Eye Res. 2008, 87 (6), 521–8. [DOI] [PubMed] [Google Scholar]
- (10).Sparrow JR; Yoon KD; Wu Y; Yamamoto K Interpretations of fundus autofluorescence from studies of the bisretinoids of the retina. Invest. Ophthalmol. Visual Sci. 2010, 51 (9), 4351–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Curcio CA; Messinger JD; Sloan KR; McGwin G Jr; Medeiros NE; Spaide RF Subretinal drusenoid deposits in non-neovascular age-related macular degeneration: morphology, prevalence, topography, and biogenesis model. Retina 2013, 33, 265–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Caprioli RM; Farmer TB; Gile J Molecular imaging of biological samples: localization of peptides and proteins using MALDI-TOF MS. Anal. Chem. 1997, 69 (23), 4751–60. [DOI] [PubMed] [Google Scholar]
- (13).Anderson DM; Ablonczy Z; Koutalos Y; Spraggins J; Crouch RK; Caprioli RM; Schey KL High resolution MALDI imaging mass spectrometry of retinal tissue lipids. J. Am. Soc. Mass Spectrom. 2014, 25 (8), 1394–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Zemski Berry KA; Gordon WC; Murphy RC; Bazan NG Spatial organization of lipids in the human retina and optic nerve by MALDI imaging mass spectrometry. J. Lipid Res. 2014, 55 (3), 504–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Ablonczy Z; Higbee D; Anderson DM; Dahrouj M; Grey AC; Gutierrez D; Koutalos Y; Schey KL; Hanneken A; Crouch RK Lack of correlation between the spatial distribution of A2E and lipofuscin fluorescence in the human retinal pigment epithelium. Invest. Ophthalmol. Visual Sci. 2013, 54 (8), 5535–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Roy MC; Nakanishi H; Takahashi K; Nakanishi S; Kajihara S; Hayasaka T; Setou M; Ogawa K; Taguchi R; Naito T Salamander retina phospholipids and their localization by MALDI imaging mass spectrometry at cellular size resolution. J. Lipid Res. 2011, 52 (3), 463–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Garrett TJ; Dawson WW Lipid geographical analysis of the primate macula by imaging mass spectrometry. Methods Mol. Biol. 2009, 579, 247–60. [DOI] [PubMed] [Google Scholar]
- (18).Garrett TJ; Menger RF; Dawson WW; Yost RA Lipid analysis of flat-mounted eye tissue by imaging mass spectrometry with identification of contaminants in preservation. Anal. Bioanal. Chem. 2011, 401 (1), 103–13. [DOI] [PubMed] [Google Scholar]
- (19).Palmer AD; Griffiths R; Styles I; Claridge E; Calcagni A; Bunch J Sucrose cryo-protection facilitates imaging of whole eye sections by MALDI mass spectrometry. J. Mass Spectrom. 2012, 47 (2), 237–41. [DOI] [PubMed] [Google Scholar]
- (20).Anderson DMG; Ablonczy Z; Koutalos Y; Hanneken AM; Spraggins JM; Calcutt MW; Crouch RK; Caprioli RM; Schey KL Bis(monoacylglycero)phosphate lipids in the retinal pigment epithelium implicate lysosomal/endosomal dysfunction in a model of Stargardt disease and human retinas. Sci. Rep. 2017, 7 (1), 17352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Bowrey HE; Anderson DM; Pallitto P; Gutierrez DB; Fan J; Crouch RK; Schey KL; Ablonczy Z Imaging mass spectrometry of the visual system: Advancing the molecular understanding of retina degenerations. Proteomics: Clin. Appl. 2016, 10 (4), 391–402. [DOI] [PubMed] [Google Scholar]
- (22).Anderson DM; Mills D; Spraggins J; Lambert WS; Calkins DJ; Schey KL High-resolution matrix-assisted laser desorption ionization-imaging mass spectrometry of lipids in rodent optic nerve tissue. Mol. Vis 2013, 19, 581–92. [PMC free article] [PubMed] [Google Scholar]
- (23).Anderson DM; Spraggins JM; Rose KL; Schey KL High spatial resolution imaging mass spectrometry of human optic nerve lipids and proteins. J. Am. Soc. Mass Spectrom. 2015, 26 (6), 940–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Sato K; Saigusa D; Saito R; Fujioka A; Nakagawa Y; Nishiguchi KM; Kokubun T; Motoike IN; Maruyama K; Omodaka K; Shiga Y; Uruno A; Koshiba S; Yamamoto M; Nakazawa T Metabolomic changes in the mouse retina after optic nerve injury. Sci. Rep. 2018, 8 (1), 11930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Stark DT; Anderson DMG; Kwong JMK; Patterson NH; Schey KL; Caprioli RM; Caprioli J Optic Nerve Regeneration After Crush Remodels the Injury Site: Molecular Insights From Imaging Mass Spectrometry Optic Nerve Regeneration Imaging Mass Spectrometry. Invest. Ophthalmol. Visual Sci. 2018, 59 (1), 212–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Anderson DMG; Van de Plas R; Rose KL; Hill S; Schey KL; Solga AC; Gutmann DH; Caprioli RM 3-D imaging mass spectrometry of protein distributions in mouse Neurofibromatosis 1 (NF1)-associated optic glioma. J. Proteomics 2016, 149, 77–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Grey AC; Schey KL Age-related changes in the spatial distribution of human lens alpha-Crystallin products by MALDI imaging mass spectrometry. Invest. Ophthalmol. Visual Sci. 2009, 50 (9), 4319–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Stella DR; Floyd KA; Grey AC; Renfrow MB; Schey KL; Barnes S Tissue localization and solubilities of alphaA-Crystallin and its numerous C-terminal truncation products in pre- and postcataractous ICR/f rat lenses. Invest. Ophthalmol. Visual Sci. 2010, 51 (10), 5153–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Ellis SR; Wu C; Deeley JM; Zhu X; Truscott RJW; Panhuis M. i. h.; Cooks RG; Mitchell TW; Blanksby SJ Imaging of Human Lens Lipids by Desorption Electrospray Ionization Mass Spectrometry. J. Am. Soc. Mass Spectrom. 2010, 21 (12), 2095–2104. [DOI] [PubMed] [Google Scholar]
- (30).Pól J; Vidová V; Hyötyläinen T; Volný M; Novák P; Strohalm M; Kostiainen R; Havlíček V; Wiedmer SK; Holopainen JM Spatial distribution of glycerophospholipids in the ocular lens. PLoS One 2011, 6 (4), No. e19441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Le CH; Han J; Borchers CH Dithranol as a matrix for matrix assisted laser desorption/ionization imaging on a fourier transform ion cyclotron resonance mass spectrometer. J. Visualized Exp. 2013, No. 81, No. e50733–e50733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Chen Y; Jester JV; Anderson DM; Marchitti SA; Schey KL; Thompson DC; Vasiliou V Corneal haze phenotype in Aldh3a1-null mice: In vivo confocal microscopy and tissue imaging mass spectrometry. Chem.-Biol. Interact. 2017, 276, 9–14. [DOI] [PubMed] [Google Scholar]
- (33).Patterson NH; Tuck M; Van de Plas R; Caprioli RM Advanced Registration and Analysis of MALDI Imaging Mass Spectrometry Measurements through Autofluorescence Microscopy. Anal. Chem. 2018, 90 (21), 12395–12403. [DOI] [PubMed] [Google Scholar]
- (34).Svirkova A; Turyanskaya A; Perneczky L; Streli C; Marchetti-Deschmann M Multimodal imaging of undecalcified tissue sections by MALDI MS and μXRF. Analyst 2018, 143 (11), 2587–595. [DOI] [PubMed] [Google Scholar]
- (35).Van de Plas R; Yang J; Spraggins J; Caprioli RM Image fusion of mass spectrometry and microscopy: a multimodality paradigm for molecular tissue mapping. Nat. Methods 2015, 12 (4), 366–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Chughtai S; Chughtai K; Cillero-Pastor B; Kiss A; Agrawal P; MacAleese L; Heeren RMA A multimodal mass spectrometry imaging approach for the study of musculoskeletal tissues. Int. J. Mass Spectrom. 2012, 325–327, 150–160. [Google Scholar]
- (37).Spaide RF; Ooto S; Curcio CA Subretinal drusenoid deposits AKA pseudodrusen. Surv. Ophthalmol. 2018, 63 (6), 782–815. [DOI] [PubMed] [Google Scholar]
- (38).Pang CE; Messinger JD; Zanzottera EC; Freund KB; Curcio CA The Onion Sign in Neovascular Age-Related Macular Degeneration Represents Cholesterol Crystals. Ophthalmology 2015, 122 (11), 2316–2326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Li M; Jia C; Kazmierkiewicz KL; Bowman AS; Tian L; Liu Y; Gupta NA; Gudiseva HV; Yee SS; Kim M; Dentchev T; Kimble JA; Parker JS; Messinger JD; Hakonarson H; Curcio CA; Stambolian D Comprehensive analysis of gene expression in human retina and supporting tissues. Hum. Mol. Genet. 2014, 23 (15), 4001–4014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Quinn N; Csincsik L; Flynn E; Curcio C; Kiss S; Sadda S; Hogg R; Petö T; Lengyel I The clinical relevance of visualising the peripheral retina. Prog. Retinal Eye Res. 2019, 68, 83. [DOI] [PubMed] [Google Scholar]
- (41).Thoreson WB; Dacey DM Diverse Cell Types, Circuits, and Mechanisms for Color Vision in the Vertebrate Retina. Physiol. Rev. 2019, 99 (3), 1527–1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Sladkova K; Houska J; Havel J Laser desorption ionization of red phosphorus clusters and their use for mass calibration in time-of-flight mass spectrometry. Rapid Commun. Mass Spectrom. 2009, 23 (19), 3114–8. [DOI] [PubMed] [Google Scholar]
- (43).Strohalm M; Kavan D; Novak P; Volny M; Havlicek V mMass 3: a cross-platform software environment for precise analysis of mass spectrometric data. Anal. Chem. 2010, 82 (11), 4648–51. [DOI] [PubMed] [Google Scholar]
- (44).Feeney L Lipofuscin and melanin of human retinal pigment epithelium. Fluorescence, enzyme cytochemical, and ultrastructural studies. Invest Ophthalmol Vis Sci. 1978, 17 (7), 583–600. [PubMed] [Google Scholar]
- (45).Bermond K; Wobbe C; Tarau IS; Heintzmann R; Hillenkamp J; Curcio CA; Sloan KR; Ach T Autofluorescent Granules of the Human Retinal Pigment Epithelium: Phenotypes, Intracellular Distribution, and Age-Related Topography. Invest. Ophthalmol. Visual Sci. 2020, 61 (5), 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Ablonczy Z; Smith N; Anderson DM; Grey AC; Spraggins J; Koutalos Y; Schey KL; Crouch RK The utilization of fluorescence to identify the components of lipofuscin by imaging mass spectrometry. Proteomics 2014, 14 (7–8), 936–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Snodderly DM; Auran JD; Delori FC The macular pigment. II. Spatial distribution in primate retinas. Investigative Ophthalmology & Visual Science 1984, 25 (6), 674–685. [PubMed] [Google Scholar]
- (48).Powner MB; Gillies MC; Tretiach M; Scott A; Guymer RH; Hageman GS; Fruttiger M Perifoveal Müller Cell Depletion in a Case of Macular Telangiectasia Type 2. Ophthalmology 2010, 117 (12), 2407–2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Powner MB; Gillies MC; Zhu M; Vevis K; Hunyor AP; Fruttiger M Loss of Müller’s Cells and Photoreceptors in Macular Telangiectasia Type 2. Ophthalmology 2013, 120 (11), 2344–2352. [DOI] [PubMed] [Google Scholar]
- (50).Obana A; Sasano H; Okazaki S; Otsuki Y; Seto T; Gohto Y Evidence of Carotenoid in Surgically Removed Lamellar Hole-Associated Epiretinal Proliferation. Invest. Ophthalmol. Visual Sci. 2017, 58 (12), 5157–5163. [DOI] [PubMed] [Google Scholar]
- (51).Li B; George EW; Rognon GT; Gorusupudi A; Ranganathan A; Chang FY; Shi L; Frederick JM; Bernstein PS Imaging lutein and zeaxanthin in the human retina with confocal resonance Raman microscopy. Proc. Natl. Acad. Sci. U. S. A. 2020, 117, 12352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Pang CE; Maberley DA; Freund KB; White VA; Rasmussen S; To E; Matsubara JA LAMELLAR HOLEASSOCIATED EPIRETINAL PROLIFERATION: A Clinicopathologic Correlation. Retina 2016, 36 (7), 1408–1412. [DOI] [PubMed] [Google Scholar]
- (53).Bazan NG Cell survival matters: docosahexaenoic acid signaling, neuroprotection and photoreceptors. Trends Neurosci. 2006, 29 (5), 263–271. [DOI] [PubMed] [Google Scholar]
- (54).Bazan NG Neuroprotectin D1 (NPD1): A DHA-Derived Mediator that Protects Brain and Retina Against Cell Injury-Induced Oxidative Stress. Brain Pathol. 2005, 15 (2), 15–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Pollreisz A; Neschi M; Sloan KR; Pircher M; Mittermueller TJ; Dacey DM; Schmidt-Erfurth U; Curcio CA An atlas of human retinal pigment epithelium organelles significant for clinical imaging. Invest. Ophthalmol. Visual Sci. 2020, 61, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Zhang X; Hughes BA KCNQ and KCNE potassium channel subunit expression in bovine retinal pigment epithelium. Exp. Eye Res. 2013, 116, 424–432. [PMC free article] [PubMed] [Google Scholar]
- (57).Nawrot M; West K; Huang J; Possin DE; Bretscher A; Crabb JW; Saari JC Cellular Retinaldehyde-Binding Protein Interacts with ERM-Binding Phosphoprotein 50 in Retinal Pigment Epithelium. Invest. Ophthalmol. Visual Sci. 2004, 45 (2), 393–401. [DOI] [PubMed] [Google Scholar]
- (58).Messias MCF; Mecatti GC; Priolli DG; de Oliveira Carvalho P Plasmalogen lipids: functional mechanism and their involvement in gastrointestinal cancer. Lipids Health Dis. 2018, 17 (1), 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (59).Braverman NE; Moser AB Functions of plasmalogen lipids in health and disease. Biochim. Biophys. Acta, Mol. Basis Dis. 2012, 1822 (9), 1442–1452. [DOI] [PubMed] [Google Scholar]
- (60).Winkler BS; Boulton ME; Gottsch JD; Sternberg P Oxidative damage and age-related macular degeneration. Molecular vision 1999, 5, 32–32. [PMC free article] [PubMed] [Google Scholar]
- (61).Jarrett SG; Boulton ME Consequences of oxidative stress in age-related macular degeneration. Mol. Aspects Med. 2012, 33 (4), 399–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (62).Farooqui AA; Horrocks LA Book Review: Plasmalogens: Workhorse Lipids of Membranes in Normal and Injured Neurons and Glia. Neuroscientist 2001, 7 (3), 232–245. [DOI] [PubMed] [Google Scholar]
- (63).Kooijman AC Light distribution on the retina of a wide-angle theoretical eye. J. Opt. Soc. Am. 1983, 73 (11), 1544–1550. [DOI] [PubMed] [Google Scholar]
- (64).Weale RA Retinal illumination using a wide-angle model of the eye:comment. J. Opt. Soc. Am. A 1990, 7 (1), 170–171. [DOI] [PubMed] [Google Scholar]
- (65).Tang PH; Kono M; Koutalos Y; Ablonczy Z; Crouch RK New insights into retinoid metabolism and cycling within the retina. Prog. Retinal Eye Res. 2013, 32, 48–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (66).Goldman D Müller glial cell reprogramming and retina regeneration. Nat. Rev. Neurosci. 2014, 15 (7), 431–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (67).Kiser PD; Golczak M; Palczewski K Chemistry of the retinoid (visual) cycle. Chem. Rev. 2014, 114 (1), 194–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (68).Hughes A, The Topography of Vision in Mammals of Contrasting Life Style: Comparative Optics and Retinal Organisation. In The Visual System in Vertebrates; Crescitelli F, Dvorak CA, Eder DJ, Granda AM, Hamasaki D, Holmberg K, Hughes A, Locket NA, McFarland WN, Meyer DB, Muntz WRA, Munz FW, Olson EC, Reyer RW, Crescitelli F, Eds.; Springer: Berlin, 1977; pp 613–756. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.