Abstract
Purpose
In vivo confocal microscopy (IVCM) is a noninvasive, reproducible, and inexpensive diagnostic tool for corneal diseases. However, widespread and effortless image acquisition in IVCM creates serious image analysis workloads on ophthalmologists, and neural networks could solve this problem quickly. We have produced a novel deep learning algorithm based on generative adversarial networks (GANs), and we compare its accuracy for automatic segmentation of subbasal nerves in IVCM images with a fully convolutional neural network (U-Net) based method.
Methods
We have collected IVCM images from 85 subjects. U-Net and GAN-based image segmentation methods were trained and tested under the supervision of three clinicians for the segmentation of corneal subbasal nerves. Nerve segmentation results for GAN and U-Net-based methods were compared with the clinicians by using Pearson's R correlation, Bland-Altman analysis, and receiver operating characteristics (ROC) statistics. Additionally, different noises were applied on IVCM images to evaluate the performances of the algorithms with noises of biomedical imaging.
Results
The GAN-based algorithm demonstrated similar correlation and Bland-Altman analysis results with U-Net. The GAN-based method showed significantly higher accuracy compared to U-Net in ROC curves. Additionally, the performance of the U-Net deteriorated significantly with different noises, especially in speckle noise, compared to GAN.
Conclusions
This study is the first application of GAN-based algorithms on IVCM images. The GAN-based algorithms demonstrated higher accuracy than U-Net for automatic corneal nerve segmentation in IVCM images, in patient-acquired images and noise applied images. This GAN-based segmentation method can be used as a facilitating diagnostic tool in ophthalmology clinics.
Translational Relevance
Generative adversarial networks are emerging deep learning models for medical image processing, which could be important clinical tools for rapid segmentation and analysis of corneal subbasal nerves in IVCM images.
Keywords: medical image analysis, convolutional neural networks, generative adversarial networks (GAN), image segmentation, cornea, in vivo confocal microscopy (IVCM)
Introduction
When Marvin L. Minsky designed the first confocal microscopy in 1957, he had no idea how his invention revolutionized his father's profession, ophthalmology.1 The three-dimensional nature of biological structures is always a major challenge in medical imaging and laser scanning confocal microscopy (LSCM), also known as in vivo confocal microscopy (IVCM). IVCM is one of the most successful imaging techniques for three-dimensional biological structures in clinical settings. After the late 1980s when the first LSCM was introduced, special attention has grown toward LSCM in developing new methods and applications, especially in medicine.2 After the clinical approval of the US Food and Drug Administration, IVCM has started to be used in ophthalmology clinics for diagnosis of corneal diseases.3 The main advantage of LSCM is its noninvasive, reproducible, and easily applicable nature, which utilizes light rather than a physical technique in sectioning the specimen.4 Because of this structural advantage, IVCM enables detailed assessment of sensory nerve anatomy in the dermis and cornea.5,6
The cornea is the most innervated tissue of the human body.7 Innervation of the cornea is made by subbasal corneal nerves, which penetrate from the periphery to the center of the cornea and create a plexus between Bowman's layer and the basal epithelium.8 They have significant roles in the homeostasis of the cornea, like temperature, tactile sensation, and nociception.9 Besides corneal diseases, various metabolic and neurological diseases affect corneal subbasal nerve plexus negatively.10 IVCM is an excellent diagnostic tool to observe corneal structure without any invasive procedure. Especially, morphological assessment of the subbasal nerve plexus of the cornea with IVCM guides clinicians about inflammatory and neurodegenerative processes which affect the cornea.8 IVCM imaging of the corneal subbasal nerve changes is essential for all corneal diseases, for instance, keratitis, keratoconus, corneal dystrophies, and dry eye disease.11 Moreover, IVCM imaging of corneal nerves helps to assess peripheral neuropathies caused by various metabolic conditions, such as type-1 diabetes or chemotherapy-induced peripheral neuropathies.12,13 The corneal subbasal nerve plexus is also an important indirect indicator of central neurodegeneration in multiple sclerosis (MS), Parkinson's disease, and various dementias.14–16
The ground-breaking properties of IVCM imaging also bring along its limitations. Widespread and frequent use of IVCM leads to a vast quantity of images to analyze, extensive time periods for manual segmentation and labeling of images, and increased subjective evaluation between ophthalmologists.17–20 Automated segmentation and analysis of IVCM images can easily solve these significant problems in the clinical decision making process.21–23 Even with novel image acquisition methods and image processing programs, inter-rater variability in manual image segmentation and analysis of IVCM images persist as a significant problem that can be solved with automated segmentation methods.24 However, conventional segmentation algorithms are not feasible for clinical applications because of the uneven illumination and noisy nature of IVCM images of the cornea and their high error rates compared to other clinical diagnostic tests.25
Deep learning-based methods have shown to be successful in various tasks in medical imaging analysis.26–28 A fully convolutional neural network (U-Net) architecture is one of the most commonly used deep learning architectures used for medical image segmentation.29–33 U-Net has also been shown to improve the performance of medical image segmentation by concatenating feature maps in the upsampling path with the corresponding cropped feature map in the downsampling path via skip connection.34 Convolutional neural networks (CNNs) are the backbones of the U-net architectures, and they have been shown to achieve performances close to human experts in the analysis of medical images. CNNs recently have become effective in various medical imaging tasks, including classification, segmentation, and registration and image reconstruction.27,35–41 Generative adversarial networks and their extensions have also provided solutions to many medical image analysis problems, such as image reconstruction, segmentation, detection, or classification.42–45 Furthermore, generative adversarial networks (GANs) have been shown to segment the images well and resolve the scarcity of the labeled data in the medical field with the help of generative models.
In this study, we concentrate on the segmentation of the subbasal nerves in confocal microscopy images using deep learning methods. A corneal nerve segmentation network (CNS-Net) has previously been established with CNN for corneal subbasal nerve segmentation in IVCM.46 Supervised learning has been successfully applied to trace corneal nerves in IVCM using clinical data automatically.47,48 Deep convolutional neural network architectures for the automatic analysis of subbasal nerves have been used.49
The purpose of this study is to establish a generative adversarial network for automated corneal subbasal nerve fiber (CNF) segmentation and evaluation within IVCM. GANs are specific deep learning structures that have recently attracted extensive attention.50 GANs consist of two separate networks, which are generators and discriminators, and they have proved to be able to produce images which are similar to original ones. The original GANs use random noise data as input, whereas conditional GANs (CGANs) integrate input images as a conditioning variable. Thus, this allows the network to act as a picture-to-picture translator.51,52 We compare this GAN-based method with the U-net network, which is a widely applied network structure for image segmentation.34 Last, we have applied different types of noise on IVCM images to simulate realistic noise often encountered in the ophthalmology clinics and demonstrate performance changes in different neural network-based methods.
Methods
In Vivo Confocal Microscopy Image Acquisition
This study was approved by the local medical ethics committee and adhered to the tenets of the World Medical Association Declaration of Helsinki. After written informed consent was obtained from all subjects, a complete ophthalmological examination was performed and both corneas were photographed with an in vivo confocal microscopy (Heidelberg Retinal Tomography 3 Rostock Cornea Module, Heidelberg Engineering GmbH, Heidelberg, Germany).
In this study, the main purpose was the comparison of U-Net and GAN-based algorithms on automatic segmentation of corneal subbasal nerves in the IVCM images. Because of this, the healthy subjects and the patients with chronic ocular surface problems (peripheral neuropathy, meibomian gland dysfunction, etc.) were randomly included to simulate clinical settings. All subjects were surveyed regarding eye dryness, burning, aching, epiphora, and contact lens to remove the effects of any other factor on the subbasal nerve plexus. The subjects who did not meet with specified conditions were excluded from the study.
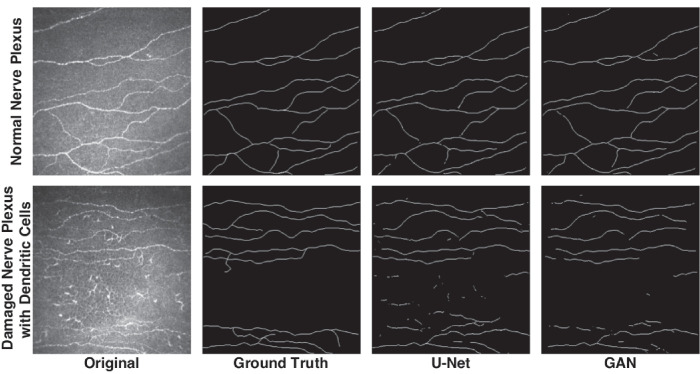
All subjects underwent corneal IVCM to analyze the total fiber length of the corneal subbasal nerve plexus (CNFL). Before the examination, two drops of local anesthetic (Oxybuprocaine hydrochloride 0.4%) and a drop of lubricant (Carbomer 2 mg/g liquid gel) were applied to both eyes. The IVCM images were taken in Heidelberg Retinal Tomography 3 Rostock Cornea Module. Each image size was 384 × 384 pixels, which covers 400 µm × 400 µm area on the cornea. The full thickness of the central cornea was scanned using the section mode. The duration of the examination was approximately 5 minutes for each eye. Three images per eye were selected, and a total of 510 images from 85 subjects were segmented by 2 blinded experienced graders (authors E.Y. and A.Y.T.). The segmented images were selected for deep learning methods by one blinded experienced IVCM expert (author A.S.) to validate corneal subbasal nerve segmentation. Graders made all image segmentation using ImageJ software (NIH, Bethesda, MD, USA; http://imagej.nih.gov/ij). NeuronJ, which is the most commonly used semi-automated nerve analysis plug-in of ImageJ, was used to quantify nerve fibers. All visible nerve fibers were traced in the image, and their total length was calculated as µm. After manual segmentation of IVCM images by three blinded graders, raw IVCM images and their nerve tracing masks were uploaded to the IVCM image database without any patient information (Fig. 1). Five hundred five (505) of 510 IVCM images had been uploaded with segmentation masks for deep learning. Five (5) of 510 IVCM images were excluded from the study due to several artifacts.
Figure 1.
Example IVCM images and masks for U-Net and GAN-based segmentation methods.
Design of Neural Networks for IVCM Image Analysis
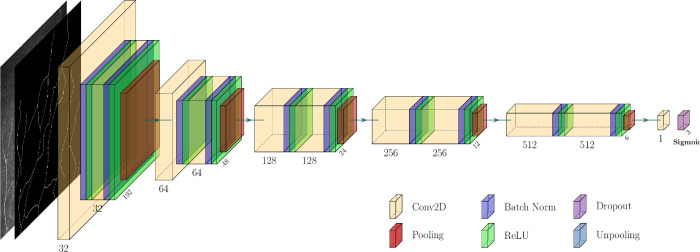
The downsampling and upsampling blocks consist of 2 convolutional layers with a kernel size of 3 × 3 and ReLU activations for U-Net (Fig. 2). The input IVCM image sizes were 384 × 384 in this study. At the end of the pipeline, the final convolutional layer was activated with the logistic (sigmoid) function. Two networks’ gradual improvement at each epoch was conducted at the end of the training process for GAN. The generator was trained to produce the real images (segmented neurite maps), and the discriminator was trained to differentiate the real ones (the ground-truth maps) from those generated by the generator. The generator was expected to become a predictor within an error rate when the IVCM image was given as input. The CGAN Network structure is shown in Figure 3.
Figure 2.
U-Net structure.
Figure 3.
General Conditional Generative Adversarial Network structure used in the study. Blue mask generated by the generator is concatenated with the input image and labeled as fake, while the green mask is obtained by experts and labelled as real.
A similar approach was previously applied to retinal fundus images to predict vessel maps, where the network structure was named V-GAN.53 Various types of a discriminator for generative adversarial networks, such as whole image (an ImageGAN), a single-pixel (a PixelGAN), or an NxN patch size (a PatchGAN), could be selected.51 The discriminator tries to evaluate the truthfulness of the generated region of the image in all these types. A patch size of 3 × 3 was evaluated in this study. A modified version of U-Net was constructed for the generator part of GAN.53 The generator and the discriminator (PatchGAN) network structures are presented in Figure 4 and Figure 5, respectively.
Figure 4.
Generator part of GAN structure.
Figure 5.
PatchGAN structure used for the discriminator part for GAN.
The Dataset and The Training Process
The dataset was constructed with 505 image pairs taken from 85 patients. Each image pair consisted of an input IVCM image and a corresponding neurite binary map image. The latter was created from .ndf files annotated by experts in the laboratory using the NeuronJ plugin of the ImageJ suite. The dataset was further divided into two subsets: training and testing. The distribution was randomly conducted, and the test group had 102 image pairs whereas the training group had 403. The selection in dividing the images into groups was based on individuals and not images to ensure that the network was tested on images not similar to the ones used in the training process. Training dataset input images were augmented with several image operations, including flipping horizontally, contrast, gamma, and brightness adjustments to increase the variance in the input space. Any augmentation function was randomly applied or not applied at each update step (i.e. with each batch in every epoch), with a probability of 0.5 from a uniform distribution.
The networks and training processes were utilized with the TensorFlow framework. The Adam optimizer function with an initial learning rate of 0.0002 and a beta value of 0.5 is run while the learning rate was decreased over time with a scheduler. In GAN training, both the generator and the discriminator were updated at each iteration step. The training was conducted on a single NVIDIA Tesla V100 GPU card (approximately 0.2 image per second) and loss curves for training and validation loss over epochs were generated to examine over-fitting issues54 (Supplementary Fig. S1).
In order to assess the robustness of the trained networks under noisy inputs, three separate noisy images were artificially generated and evaluated. These were (1) Gaussian distributed noise with the standard deviation of 0.1, (2) salt and pepper noise with the proportion value of 0.034, and (3) speckle noise with the standard deviation of 0.25. All noises were randomly distributed, and the parameters were chosen such that all of the noisy inputs had approximately the same peak signal-to-noise ratios (PSNRs) at 20.0 according to previous literature and international standards.55,56 The mean signal-to-noise ratio was calculated approximately as 12.0 (Supplementary Fig. S2).
Statistical Analysis
Intraclass correlation coefficient (ICC) was calculated from CNFL by an experienced third grader (author A.S.) to confirm inter-rater reliability between experts and to validate manual segmentation results of two graders (authors E.Y. and A.Y.T.). ICC between two graders had been found 0.97 (0.96–0.98) with a 95% confidence interval. Segmentation procedure repeated by experienced IVCM expert (author A.S.) in disputed images to increase the reliability of the manual segmentation.57
Corneal nerve fiber segmentation results of U-Net and GAN-based algorithms were compared with the manual segmentation results as the ground truth. Image to image comparison results of different algorithms analyzed with different statistical tests for all diagnostic parameters. Corneal subbasal nerve segmentation results of U-Net and GAN-based methods were statistically analyzed with Pearson's R correlation for correlation coefficient and Bland-Altman analysis for bias calculation.58 Accuracy, sensitivity, and specificity of U-Net and GAN-based methods were also calculated with receiver operating characteristics (ROC) from the confusion matrix. Two-sided p value < 0.05, which was also equal to 95% confidence interval and ±1.96 standard deviation (SD) interval, was considered statistically significant. For Pearson's R correlation, r > 0.80 was considered as a solid positive correlation.59 Statistical analysis results were indicated as (mean ± SD) or the coefficient of repeatability CR (−1.96 SD +1.96 SD), which is calculated according to the original article of Bland and Altman published in 1986.60
Results
U-Net and GAN-Based Segmentation Results
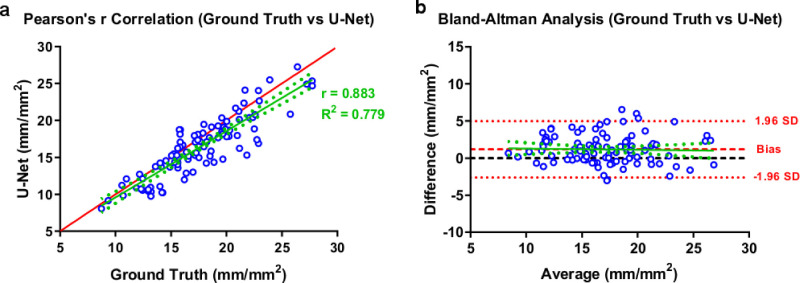
The U-Net based segmentation method demonstrated high levels of correlation (r = 0.883, R2 = 0.779) and low levels of bias (1.189 ± 1.929) compared to the segmentation results of the experts (Fig. 6). The coefficient of repeatability was also calculated as 4.425 (3.892–5.128). These results showed resemblance with U-Net based DeepNerve algorithm, which was used for corneal subbasal nerve segmentation of macaques.49
Figure 6.
Correlation (a) and Bland Altman (b) plots for U-Net structure.
The GAN algorithm also performed similarly to U-Net in correlation and Bland Altman analysis for subbasal nerve segmentation from IVCM images (Fig. 7). GAN-based segmentation of corneal subbasal nerves in IVCM images showed high correlation (r = 0.847, R2 = 0.717) and low levels of bias (3.279 ± 2.141) compared to the results of the experts’ segmentation. In Bland-Altman's analysis, the coefficient of repeatability was also calculated as 7.664 (6.741–8.882). It could be noticed that GAN displayed similar performance with U-Net based algorithm for segmentation of corneal subbasal nerves in IVCM images.
Figure 7.
Correlation (a) and Bland Altman (b) plots for GAN structure.
We also observed that GAN showed significantly higher accuracy for the segmentation of IVCM images when ROC curves were compared for both methods. Whereas the area under the curve for U-Net was 0.8934, the area under the curve for GAN was 0.9439 (Fig. 8). When IVCM images were compared one by one, U-Net and GAN-based algorithms masked similar areas in high-quality IVCM images. On the other hand, the U-Net based segmentation method showed a higher number of false-positive pixels for images with artifacts (e.g. uneven illumination and device-related noise) and distractor objects (e.g. dendritic cells, damaged nerves, and corneal foldings) compared to the GAN algorithm (see Fig. 1). Therefore, we had included three of the most common noise types in medical imaging to the IVCM images and then applied automatic segmentation with U-Net and GAN-based algorithms.
Figure 8.
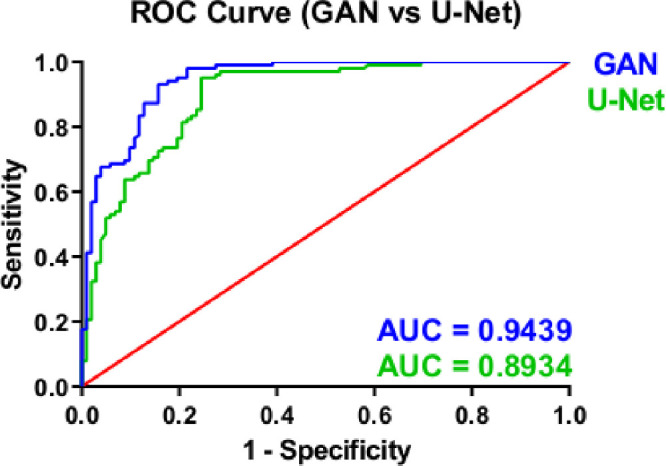
ROC curves for GAN and U-Net structures.
Noise Simulation Results
Three different types of noises were applied on IVCM images to simulate daily challenges in ophthalmology clinics for neural networks-based segmentation of IVCM images. Gaussian, salt and pepper (S&P), and speckle type noises with 12 signal-to-noise ratio (SNR), and 20 peak SNR levels were applied on IVCM images. The accuracy of the U-Net significantly decreased with speckle type noise (0.818 ± 0.029) compared to the original images (0.893 ± 0.022) and the images with other noises (0.883 ± 0.023 for Gaussian, 0.879 ± 0.024 for S&P). For the GAN-based method, Gaussian and S&P noises (0.915 ± 0.020 for Gaussian and 0.914 ± 0.020 for S&P) did not decrease the accuracy of corneal subbasal nerve segmentation significantly. However, speckle type noise (0.878 ± 0.024) significantly decreased the accuracy of the automatic segmentation of GAN compared to the results without noise (0.944 ± 0.015; Fig. 9).
Figure 9.
ROC curves for U-Net (a) and GAN (b) structures with different noises.
Discussion
In our study, we have compared U-Net and GAN-based corneal subbasal nerve segmentation methods in IVCM images. Although U-Net and GAN demonstrated equal performance in correlation and Bland Altman analyses, GAN showed significantly higher accuracy in ROC analysis. Additionally, we have simulated noise types, which clinicians often encounter during medical image acquisition, by applying Gaussian, S&P, and speckle type noises with equal SNR levels. Both U-Net and GAN-based segmentation methods were affected at most by speckle type noise. The decrease in U-Net based segmentation algorithm accuracy was significantly higher than the GAN-based segmentation algorithm in all types of noises.
Our study is the first application of GAN-based algorithms on IVCM images, to the best of our knowledge. GAN-based algorithms have been studied to the segmentation of medical images from various imaging modalities, such as computed tomography,61 magnetic resonance,62 x-radiation,63 and ultrasound imaging.64 In ophthalmology, GANs are used for the segmentation of retinal vessels in fundus images.53,65 Traditional segmentation approaches, such as graph-cut methods, have utilized pixel-wise correspondence over the decades with some significant caveats, including artifacts and leakages. GANs have a potential to bring out the best of the approaches, where the discriminator unit is acting as a shape regulator.44 Although the effect of regularization is reported to be more eminent with compact shapes, unlike vessels or neurites, the discriminator's perception size might be set up from the whole image to a single pixel, as in the ImageGAN, PatchGAN, and PixelGAN examples. To regularize the network and overcome the collapses in the training process, Li and Shen66 proposed a method that combines CGAN with AC-GAN and introduces a classifier loss term to their structure in the cell segmentation task. Although a pre-processing step is conducted in a study compared to ours, the classification loss is shown to improve the resulting segmentations. Unannotated images and the annotated ones may also be fed into the segmentation workflow where the former ones will help the training, leading to a more robust discrimination process and more accurate generated segmentation maps.67 The main disadvantage of GAN-based algorithms is training separate networks together. During the training of the networks, GAN creates a zero-sum game to reach an equilibrium point between these networks, which is intuitively in contrast to the conventional algorithms, where the objective function is to minimize the loss of function.68 Therefore, several issues are observed in training of GANs, as experienced in this study as well, including oscillations, mode collapses, diminishing, or exploding gradients.69 In order to overcome and minimize these effects, a tedious fine-tuning approach must be followed. On the other hand, GANs have two significant advantages over U-Net or similar structures. Firstly, GANs let the generator networks produce near-realistic images. In medical imaging literature, this is efficiently utilized in image synthesis where the number of the images of a dataset is insufficient, as in many situations in medical image acquisition, to expand the datasets.70 Second, GANs are more robust to artifacts and digital image noise sources in general with the help of the discriminator enforcing the generator to produce better outputs (e.g. segmentation maps in this case).71
Segmentation of corneal subbasal nerves in IVCM images is still made manually by the experts in ophthalmology clinics. Existing automated corneal subbasal nerve analysis software still has lower accuracy compared to the experts in clinical applications.72 Neural networks-based algorithms are significantly superior to the other automated corneal subbasal nerve segmentation software, and preliminary studies hope about clinical applications.48,73 Different types of neural network-based methods have been used for corneal subbasal nerve segmentation, but most of the studies concentrated on U-Net based algorithms.46,49 Oakley et al. produced an auto-encoder and U-Net based neural network algorithm with high accuracy for corneal subbasal nerve segmentation in IVCM images of macaques.49 They have implemented pre-processing and post-processing algorithms to increase the accuracy of U-Net based segmentation methods. Wei et al. built a CNN-based structure for corneal subbasal nerve segmentation in IVCM images and tested it on patients acquired IVCM images in an ophthalmology clinic.46 Unfortunately, this study did not make inter-rater reliability measurement, subject separation, and correlation analysis to increase the reliability of the segmentation method. Our study compared the GAN-based corneal subbasal nerve segmentation algorithm with a similar CNN-based algorithm of Wei et al., and we showed significantly higher accuracy for corneal subbasal nerve segmentation compared to the CNN-based method. We applied a three-grader system and inter-rater reliability test to increase the reliability of manual segmentation of corneal subbasal nerves. We did not use any pre-processing or post-processing algorithms to show the actual capacity of the GAN-based segmentation algorithm. In addition to them, we made subject separation for test and training image sets and applied common types of medical imaging noises in the experiments.
In our study, the pre-processing methods for image analysis, such as background subtraction or edge detection algorithms, were not applied to the IVCM images to assess the actual capacities of U-Net and GAN-based algorithms. As with any deep learning-based technique, such a task requires numerous training images to generalize the underlying structures to be predicted. The more IVCM images collected and annotated by experts, the more robust and efficient the networks become. The model selection and adapting, hyperparameter optimization, and convergence of the networks are only some of the significant difficulties in most deep learning-based studies. Particularly, GANs are shown to be prone to some limitations in training due to several reasons, including nonconvergence issues where the model oscillates or never converges, vanishing or exploding gradients where the discriminator overwhelms over the generator, and mode collapses in which the generator does not improve.74 Even though the cohort size of the study was small compared to other studies, GANs showed significantly better performance than CNN based algorithms. Good performance of the GAN based algorithms in augmented and limited data sets has been shown in previous literature.75 Because of privacy concerns, access to biomedical images for deep learning applications decreased in recent years.76 We also applied image augmentation, and it helped the training process with small-sized data sets by increasing the variance within the input space.77 These augmentation operators were selected randomly whether to apply or not at each batch step of the training process.78 GANs with augmentation operators could be helpful for biomedical image analysis with a minimal number of images for various modalities.
GAN-based algorithms promise high hopes for biomedical imaging applications. In this first GAN study on IVCM images, we have focused on corneal subbasal nerve segmentation. However, applications of GAN-based algorithms in IVCM images could not be limited to the segmentation of the corneal nerve plexus. GAN-based algorithms also could be used in IVCM images of different layers of the cornea. GANs are currently used for image reconstruction, denoising, super-resolution, classification, object detection, and cross-modality image synthesis in various biomedical imaging modalities, and they also could be used in IVCM images for similar purposes.44 Because of their generator-discriminator reciprocal structure, they have significantly higher success rates than the other deep learning algorithms. Because of their structures, GANs could be precious diagnostic tools to discriminate lesions in specific conditions with a very limited number of images.79 In this study, we mainly focused on its capability of segmentation for corneal subbasal nerve plexus of the patients with ocular surface problems and healthy subjects. In further studies, we will investigate its capability of diagnostic classification for various corneal nerve-related lesions and diagnostic precision of various corneal pathologies in IVCM images.
Conclusions
The conjunction of Minsky's two important works, IVCM and artificial intelligence, is opening new horizons in medical imaging. Generative adversarial networks are still in their infancy, but their applications on various medical imaging modalities indicate great potentials. In our study, we compared GAN with U-Net, which is a commonly used deep learning method for image segmentation, for segmentation of IVCM images. Our study demonstrated the superiority of the GAN-based algorithm for accurate segmentation of corneal subbasal nerves compared to the U-Net based algorithm.
Supplementary Material
Acknowledgments
The authors thank the use of the services and facilities of the Koç University Research Center for Translational Medicine (KUTTAM), funded by the Presidency of Turkey, Presidency of Strategy and Budget.
Supported by the Scientific and Technological Research Council of Turkey (TÜBİTAK) under Grant 1180232.
Disclosure: E. Yıldız, None; A.T. Arslan, Techy Bilişim Ltd. (E); A. Yıldız Taş, None; A.F. Acer, None; S. Demir, Techy Bilişim Ltd. (E); A. Şahin, None; D. Erol Barkana, None
References
- 1. Winston PH, Marvin L.. Minsky (1927–2016). Nature. 2016; 530: 282–282. [DOI] [PubMed] [Google Scholar]
- 2. Davidovits P. Physics in biology and medicine. Waltham, MA: Academic Press; 2018. [Google Scholar]
- 3. Cavanagh HD, Petroll WM, Alizadeh H, He Y-G, McCulley JP, Jester JV.. Clinical and diagnostic use of in vivo confocal microscopy in patients with corneal disease. Ophthalmology. 1993; 100: 1444–1454. [DOI] [PubMed] [Google Scholar]
- 4. Paddock SW. Confocal microscopy: methods and protocols. New York, NY: Springer Science & Business Media; 1999. [Google Scholar]
- 5. González S, Tannous Z.. Real-time, in vivo confocal reflectance microscopy of basal cell carcinoma. J Am Acad Dermatol. 2002; 47: 869–874. [DOI] [PubMed] [Google Scholar]
- 6. Jalbert I, Stapleton F, Papas E, Sweeney D, Coroneo M.. In vivo confocal microscopy of the human cornea. Br J Ophthalmol. 2003; 87: 225–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Al-Aqaba MA, Dhillon VK, Mohammed I, Said DG, Dua HS.. Corneal nerves in health and disease. Prog Retin Eye Res. 2019; 73: 100762. [DOI] [PubMed] [Google Scholar]
- 8. Cruzat A, Qazi Y, Hamrah P.. In vivo confocal microscopy of corneal nerves in health and disease. The Ocular Surface. 2017; 15: 15–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Labetoulle M, Baudouin C, Calonge M, et al.. Role of corneal nerves in ocular surface homeostasis and disease. Acta Ophthalmol (Copenh). 2019; 97: 137–145. [DOI] [PubMed] [Google Scholar]
- 10. Shaheen BS, Bakir M, Jain S.. Corneal nerves in health and disease. Surv Ophthalmol. 2014; 59: 263–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cavalcanti BM, Cruzat A, Sahin A, Pavan-Langston D, Samayoa E, Hamrah P.. In vivo confocal microscopy detects bilateral changes of corneal immune cells and nerves in unilateral herpes zoster ophthalmicus. The Ocular Surface. 2018; 16: 101–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ferdousi M, Azmi S, Petropoulos IN, et al.. Corneal confocal microscopy detects small fibre neuropathy in patients with upper gastrointestinal cancer and nerve regeneration in chemotherapy induced peripheral neuropathy. PLoS One. 2015; 10: e0139394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Petropoulos IN, Ponirakis G, Khan A, et al.. Corneal confocal microscopy: ready for prime time. Clin Exp Optom. 2020; 103: 265–277. [DOI] [PubMed] [Google Scholar]
- 14. Arrigo A, Rania L, Calamuneri A, et al.. Early corneal innervation and trigeminal alterations in Parkinson disease: a pilot study. Cornea. 2018; 37: 448–454. [DOI] [PubMed] [Google Scholar]
- 15. Ponirakis G, Al Hamad H, Sankaranarayanan A, et al.. Association of corneal nerve fiber measures with cognitive function in dementia. Ann Clin Transl Neurol. 2019; 6: 689–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Testa V, De Santis N, Scotto R, et al.. Neuroaxonal degeneration in patients with multiple sclerosis: an optical coherence tomography and in vivo corneal confocal microscopy study. Cornea. 2020; 39: 1221–1226. [DOI] [PubMed] [Google Scholar]
- 17. Chen X, Graham J, Dabbah MA, Petropoulos IN, Tavakoli M, Malik RA.. An automatic tool for quantification of nerve fibers in corneal confocal microscopy images. IEEE Trans Biomed Eng. 2016; 64: 786–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Holmes TJ, Pellegrini M, Miller C, et al.. Automated software analysis of corneal micrographs for peripheral neuropathy. Invest Ophthalmol Vis Sci. 2010; 51: 4480–4491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kim J, Markoulli M.. Automatic analysis of corneal nerves imaged using in vivo confocal microscopy. Clin Exp Optom. 2018; 101: 147–161. [DOI] [PubMed] [Google Scholar]
- 20. Scarpa F, Grisan E, Ruggeri A.. Automatic recognition of corneal nerve structures in images from confocal microscopy. Invest Ophthalmol Vis Sci. 2008; 49: 4801–4807. [DOI] [PubMed] [Google Scholar]
- 21. Calvillo MP, McLaren JW, Hodge DO, Bourne WM.. Corneal reinnervation after LASIK: prospective 3-year longitudinal study. Invest Ophthalmol Vis Sci. 2004; 45: 3991–3996. [DOI] [PubMed] [Google Scholar]
- 22. Dabbah MA, Graham J, Petropoulos I, Tavakoli M, Malik RA.. Dual-model automatic detection of nerve-fibres in corneal confocal microscopy images. International Conference on Medical Image Computing and Computer-Assisted Intervention. New York, NY: Springer; 2010: 300–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Dabbah MA, Graham J, Petropoulos IN, Tavakoli M, Malik RA.. Automatic analysis of diabetic peripheral neuropathy using multi-scale quantitative morphology of nerve fibres in corneal confocal microscopy imaging. Med Image Anal. 2011; 15: 738–747. [DOI] [PubMed] [Google Scholar]
- 24. Patel DV, McGhee CN.. Quantitative analysis of in vivo confocal microscopy images: a review. Surv Ophthalmol. 2013; 58: 466–475. [DOI] [PubMed] [Google Scholar]
- 25. Villani E, Baudouin C, Efron N, et al.. In vivo confocal microscopy of the ocular surface: from bench to bedside. Curr Eye Res. 2014; 39: 213–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chilamkurthy S, Ghosh R, Tanamala S, et al.. Deep learning algorithms for detection of critical findings in head CT scans: a retrospective study. Lancet North Am Ed. 2018; 392: 2388–2396. [DOI] [PubMed] [Google Scholar]
- 27. Le WT, Maleki F, Romero FP, Forghani R, Kadoury S.. Overview of machine learning: part 2: deep learning for medical image analysis. Neuroimaging Clin N Am. 2020; 30: 417–431. [DOI] [PubMed] [Google Scholar]
- 28. Tian Y, Fu S.. A descriptive framework for the field of deep learning applications in medical images. KBS. 2020; 210: 106445. [Google Scholar]
- 29. Du G, Cao X, Liang J, Chen X, Zhan Y.. Medical image segmentation based on u-net: A review. J Imaging Sci Technol. 2020; 64: 20508–20501. [Google Scholar]
- 30. Ibtehaz N, Rahman MS.. MultiResUNet: Rethinking the U-Net architecture for multimodal biomedical image segmentation. Neural Netw. 2020; 121: 74–87. [DOI] [PubMed] [Google Scholar]
- 31. Li C, Tan Y, Chen W, et al.. ANU-Net: Attention-based Nested U-Net to exploit full resolution features for medical image segmentation. Comput Graph. 2020; 90: 11–20. [Google Scholar]
- 32. Qin X, Wu C, Chang H, Lu H, Zhang X.. Match Feature U-Net: Dynamic Receptive Field Networks for Biomedical Image Segmentation. Symmetry. 2020; 12: 1230. [Google Scholar]
- 33. Yang Y, Feng C, Wang R.. Automatic segmentation model combining U-Net and level set method for medical images. Expert Syst Appl. 2020; 153: 113419. [Google Scholar]
- 34. Ronneberger O, Fischer P, Brox T.. U-Net: Convolutional Networks for Biomedical Image Segmentation. Cham, Switzerland: Springer International Publishing; 2015: 234–241. [Google Scholar]
- 35. Guedria S, De Palma N, Renard F, Vuillerme N. R2D2: A scalable deep learning toolkit for medical imaging segmentation. Software: Practice and Experience. 2020; 50: 1966–1985. [Google Scholar]
- 36. Han Y, Li J.. An Attention-oriented U-Net Model and Global Feature for Medical Image Segmentation. J Appl Sci Eng. 2020; 23: 731–738. [Google Scholar]
- 37. Lin D, Li Y, Nwe TL, Dong S, Oo ZM.. RefineU-Net: Improved U-Net with progressive global feedbacks and residual attention guided local refinement for medical image segmentation. Pattern Recognit Lett. 2020; 138: 267–275. [Google Scholar]
- 38. Feng S, Zhao H, Shi F, et al.. CPFNet: Context pyramid fusion network for medical image segmentation. IEEE Trans Med Imaging. 2020; 39: 3008–3018. [DOI] [PubMed] [Google Scholar]
- 39. Teng L, Li H, Karim S.. DMCNN: A deep multiscale convolutional neural network model for medical image segmentation. J Healthc Eng. 2019; 2019: 8597606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zhang Z, Wu C, Coleman S, Kerr D.. DENSE-INception U-net for medical image segmentation. Comput Methods Programs Biomed. 2020; 192: 105395. [DOI] [PubMed] [Google Scholar]
- 41. Chakravarty A, Sivaswamy J.. RACE-net: a recurrent neural network for biomedical image segmentation. IEEE J Biomed Health Inform. 2018; 23: 1151–1162. [DOI] [PubMed] [Google Scholar]
- 42. Kazeminia S, Baur C, Kuijper A, et al.. GANs for medical image analysis. Artif Intell Med. 2020; 109: 101938. [DOI] [PubMed] [Google Scholar]
- 43. Shi Z, Hu Q, Yue Y, Wang Z, AL-Othmani OMS, Li H.. Automatic Nodule Segmentation Method for CT Images Using Aggregation-U-Net Generative Adversarial Networks. Sensing and Imaging. 2020; 21: 1–16. [Google Scholar]
- 44. Yi X, Walia E, Babyn P.. Generative adversarial network in medical imaging: A review. Med Image Anal. 2019; 58: 101552. [DOI] [PubMed] [Google Scholar]
- 45. Nie D, Shen D.. Adversarial Confidence Learning for Medical Image Segmentation and Synthesis. Int J Comput Vision. 2020; 128: 2494–2513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wei S, Shi F, Wang Y, Chou Y, Li X.. A Deep Learning Model for Automated Sub-Basal Corneal Nerve Segmentation and Evaluation Using In Vivo Confocal Microscopy. Transl Vis Sci Technol. 2020; 9: 32–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Colonna A, Scarpa F, Ruggeri A.. Segmentation of Corneal Nerves Using a U-Net-Based Convolutional Neural Network. Cham, Switzerland: Springer International Publishing; 2018: 185–192. [Google Scholar]
- 48. Williams BM, Borroni D, Liu R, et al.. An artificial intelligence-based deep learning algorithm for the diagnosis of diabetic neuropathy using corneal confocal microscopy: a development and validation study. Diabetologia. 2020; 63: 419–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Oakley JD, Russakoff DB, McCarron ME, et al.. Deep learning-based analysis of macaque corneal sub-basal nerve fibers in confocal microscopy images. Eye Vis (Lond). 2020; 7: 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Goodfellow I, Pouget-Abadie J, Mirza M, et al.. Generative Adversarial Networks. Adv Neural Inf Process Syst. 2014; 3: 1–9. [Google Scholar]
- 51. Isola P, Zhu J-Y, Zhou T, Efros A.. Image-to-Image Translation with Conditional Adversarial Networks; 2017: 5967–5976.
- 52. Mirza M, Osindero S. Conditional Generative Adversarial Nets. 2014.
- 53. Son J, Park SJ, Jung K-H. Retinal Vessel Segmentation in Fundoscopic Images with Generative Adversarial Networks. 2017. [DOI] [PMC free article] [PubMed]
- 54. Keras Chollet F. Model Training APIs. 2015.
- 55. Keelan BW. ISO 12232 revision: Determination of chrominance noise weights for noise-based ISO calculation. Image Quality and System Performance II: International Society for Optics and Photonics. Proc SPIE. 2005; 5668: 139–147. [Google Scholar]
- 56. Thomos N, Boulgouris NV, Strintzis MG.. Optimized transmission of JPEG2000 streams over wireless channels. IEEE Trans. Image Process. 2005; 15: 54–67. [DOI] [PubMed] [Google Scholar]
- 57. Hallgren KA. Computing Inter-Rater Reliability for Observational Data: An Overview and Tutorial. Tutor Quant Methods Psychol. 2012; 8: 23–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Bunce C. Correlation, agreement, and Bland–Altman analysis: statistical analysis of method comparison studies. Am. J. Ophthalmol. 2009; 148: 4–6. [DOI] [PubMed] [Google Scholar]
- 59. Chan YH. Biostatistics 104: correlational analysis. Singapore Med J. 2003; 44: 614–619. [PubMed] [Google Scholar]
- 60. Bland JM, Altman D.. Statistical methods for assessing agreement between two methods of clinical measurement. Lancet North Am. Ed. 1986; 327: 307–310. [PubMed] [Google Scholar]
- 61. Dong X, Lei Y, Wang T, et al.. Automatic multiorgan segmentation in thorax CT images using U-net-GAN. Med Phys. 2019; 46: 2157–2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Hamghalam M, Wang T, Lei B.. High tissue contrast image synthesis via multistage attention-GAN: Application to segmenting brain MR scans. Neural Netw. 2020; 132: 43–52. [DOI] [PubMed] [Google Scholar]
- 63. Zhang Y, Miao S, Mansi T, Liao R.. Unsupervised X-ray image segmentation with task driven generative adversarial networks. Med. Image Anal. 2020; 62: 101664. [DOI] [PubMed] [Google Scholar]
- 64. Alsinan AZ, Patel VM, Hacihaliloglu I.. Bone shadow segmentation from ultrasound data for orthopedic surgery using GAN. Int J Comput Assist Radiol Surg. 2020; 15: 1477–1485. [DOI] [PubMed] [Google Scholar]
- 65. Park KB, Choi SH, Lee JY.. M-GAN: Retinal Blood Vessel Segmentation by Balancing Losses Through Stacked Deep Fully Convolutional Networks. IEEE Access. 2020; 8: 146308–146322. [Google Scholar]
- 66. Li Y, Shen L.. cC-GAN: A Robust Transfer-Learning Framework for HEp-2 Specimen Image Segmentation. IEEE Access. 2018; 6: 14048–14058. [Google Scholar]
- 67. Zhang Y, Yang L, Chen J, Fredericksen M, Hughes D, Chen D.. Deep adversarial networks for biomedical image segmentation utilizing unannotated images. Medical Image Computing and Computer Assisted Intervention. MICCAI. 2017: 408–416. [Google Scholar]
- 68. Goodfellow I, Pouget-Abadie J, Mirza M, et al.. Generative adversarial networks. Commun. ACM. 2020; 63: 139–144. [Google Scholar]
- 69. Gui J, Sun Z, Wen Y, Tao D, Ye J.. A review on generative adversarial networks: Algorithms, theory, and applications. arXiv. preprint arXiv:200106937 2020.
- 70. Mahapatra D, Bozorgtabar B, Garnavi R.. Image super-resolution using progressive generative adversarial networks for medical image analysis. Comput. Med. Imaging Graph. 2019; 71: 30–39. [DOI] [PubMed] [Google Scholar]
- 71. Chen Z, Zeng Z, Shen H, Zheng X, Dai P, Ouyang P. DN-GAN: Denoising generative adversarial networks for speckle noise reduction in optical coherence tomography images. Biomed. Signal Process. Control. 2020; 55: 101632. [Google Scholar]
- 72. Dehghani C, Pritchard N, Edwards K, Russell AW, Malik RA, Efron N.. Fully automated, semiautomated, and manual morphometric analysis of corneal subbasal nerve plexus in individuals with and without diabetes. Cornea. 2014; 33: 696–702. [DOI] [PubMed] [Google Scholar]
- 73. Zhang N, Francis S, Malik RA, Chen X.. A Spatially Constrained Deep Convolutional Neural Network for Nerve Fiber Segmentation in Corneal Confocal Microscopic Images Using Inaccurate Annotations. 2020 IEEE 17th International Symposium on Biomedical Imaging (ISBI); 2020: 456–460.
- 74. Salimans T, Goodfellow I, Zaremba W, Cheung V, Radford A, Chen X. Improved Techniques for Training GANs. 2016.
- 75. Karras T, Aittala M, Hellsten J, Laine S, Lehtinen J, Aila T. Training generative adversarial networks with limited data. arXiv. preprint arXiv:200606676 2020.
- 76. Prasser F, Spengler H, Bild R, Eicher J, Kuhn KA.. Privacy-enhancing ETL-processes for biomedical data. Int. J. Med. Inf. 2019; 126: 72–81. [DOI] [PubMed] [Google Scholar]
- 77. Perez L, Wang J.. The effectiveness of data augmentation in image classification using deep learning. arXiv. preprint arXiv:171204621 2017.
- 78. Taylor L, Nitschke G.. Improving deep learning using generic data augmentation. arXiv. preprint arXiv:170806020 2017.
- 79. Kearney V, Ziemer BP, Perry A, et al.. Attention-Aware Discrimination for MR-to-CT Image Translation Using Cycle-Consistent Generative Adversarial Networks. Radiol Artif Intell. 2020; 2: e190027. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.